Abstract
Metagenomic studies using next-generation sequencing technologies have revealed rich human intestinal microbiome, which likely influence host immunity and health conditions including cancer. Evidence indicates a biological link between altered microbiome and cancers in the digestive system. Escherichia coli and Bacteroides fragilis have been found to be enriched in colorectal mucosal tissues from patients with familial adenomatous polyposis that is caused by germline APC mutations. In addition, recent studies have found enrichment of certain oral bacteria, viruses, and fungi in tumor tissue and fecal specimens from patients with gastrointestinal cancer. An integrative approach is required to elucidate the role of microorganisms in the pathogenic process of gastrointestinal cancers, which develop through the accumulation of somatic genetic and epigenetic alterations in neoplastic cells, influenced by host genetic variations, immunity, microbiome, and environmental exposures. The transdisciplinary field of molecular pathological epidemiology (MPE) offers research frameworks to link germline genetics and environmental factors (including diet, lifestyle, and pharmacological factors) to pathologic phenotypes. The integration of microbiology into the MPE model (microbiology-MPE) can contribute to better understanding of the interactive role of environment, tumor cells, immune cells, and microbiome in various diseases. We review major clinical and experimental studies on the microbiome, and describe emerging evidence from the microbiology-MPE research in gastrointestinal cancers. Together with basic experimental research, this new research paradigm can help us develop new prevention and treatment strategies for gastrointestinal cancers through targeting of the microbiome.
Keywords: big data, bioinformatics, inflammation, gene-by-environment interaction, omics, population health science
Introduction
Carcinomas that arise in the digestive system, including upper and lower gastrointestinal tract, liver, gallbladder, extrahepatic bile duct, and pancreas, are collectively leading causes of death worldwide (Cortes et al. 2020). The human intestine contains more than 100 trillion microorganisms, which can influence the immune system and health conditions including cancer (Tilg et al. 2020). Emerging longitudinal studies from the Integrative Human Microbiome Project have demonstrated associations of changes in the human microbiome with preterm birth (Fettweis et al. 2019), inflammatory bowel diseases (Lloyd-Price et al. 2019), and prediabetes (Zhou et al. 2019). Accumulating evidence suggests that gastrointestinal cancers develop through the accumulation of somatic mutations and epigenetic alterations in tumor cells with complex influences of host genetic variations, microbiome, immunity, and environmental exposures (Fig. 1). Hence, it has been a challenge to elucidate the role of microbes in the pathogenic process of human gastrointestinal cancers.
Fig. 1.
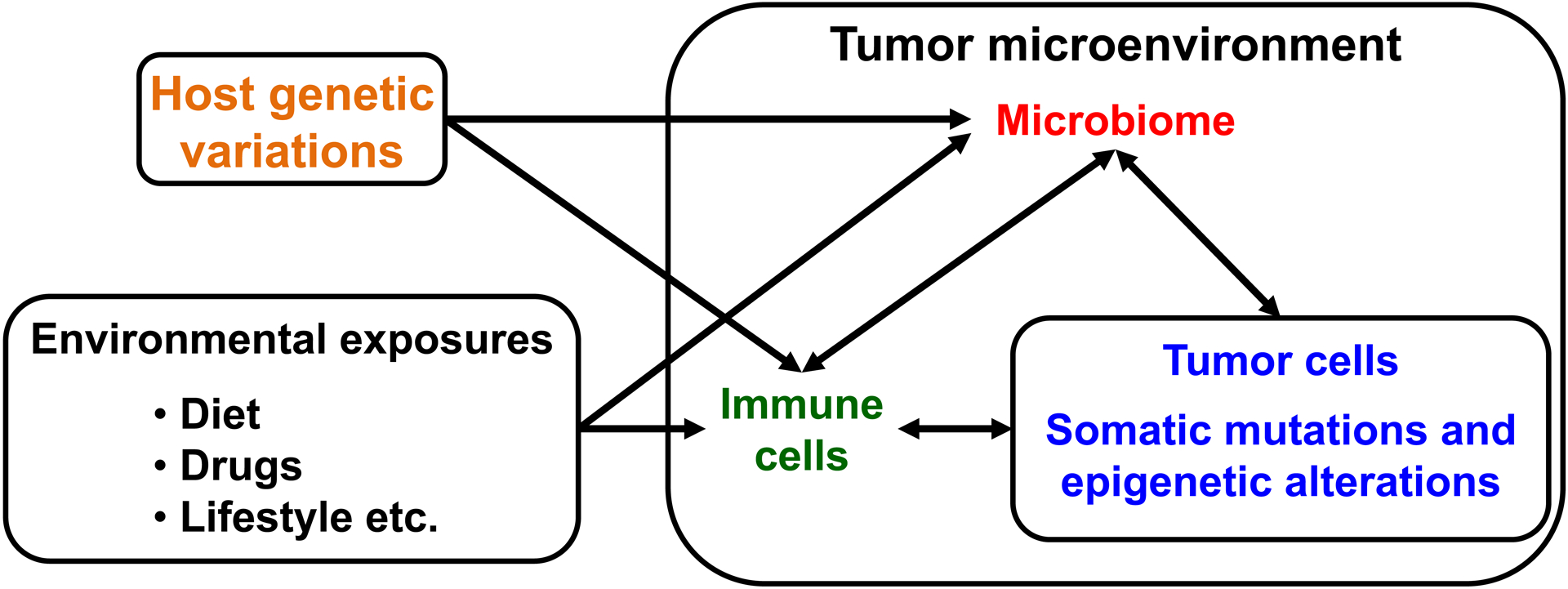
Influences of host genetic variations, microbiome, immunity, and environmental exposures on tumor genetic and epigenetic alterations. Gastrointestinal cancers develop through the accumulation of somatic mutations and epigenetic alterations in tumor cells with complex influences of microbiome and immunity in the tumor microenvironment, host genetic variations, and environmental exposures.
The integration of molecular pathology and epidemiology has generated the transdisciplinary field of ‘molecular pathological epidemiology (MPE)’ (Ogino and Stampfer 2010), which aims to link germline genetics and modifiable factors (including environment, diet, lifestyle, and pharmacological factors) to pathologic features, most commonly tumor characteristics (Ogino et al. 2011; Ogino et al. 2019; Ogino et al. 2018). The concept of MPE as a distinct field has been widespread (Carr et al. 2018; Gunter et al. 2019; Hughes et al. 2017; Rescigno et al. 2017; Waluga et al. 2018; Wang et al. 2020). Ogino et al. have shown basic approaches of MPE research in our previous review (Ogino et al. 2011; Ogino et al. 2016; Ogino et al. 2018). Although an interventional study is a gold standard, to date no interventional MPE studies have been published. Hence, better approach of MPE research is a prospective cohort study, which can reduce potential bias related to case-case and case-control designs. In a prospective cohort study, disease incidence analyses of MPE can compute risk estimates of environmental exposures, including diet, nutrition, and lifestyle, for the incidence of disease with specific subtypes according to phenotypic characteristics (e.g. KRAS mutation), and disease consequence (e.g. patient survival) analyses of MPE can examine prognostic associations of environmental exposures according to phenotypic characteristics of diseases. Utilizing colorectal cancer cases in two U.S. nationwide prospective cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study), MPE studies have shown that cigarette smoking is associated with an increased risk, especially of microsatellite instability (MSI)-high, CpG island methylator phenotype (CIMP)-high, and BRAF-mutated colorectal cancers (Limsui et al. 2010; Nishihara et al. 2013a), and that high levels of MSI and CIMP are common features of colorectal cancers arising within 5 years after colonoscopy (Nishihara et al. 2013b). In addition, MPE studies have shown that regular aspirin use after diagnosis is associated with longer survival, especially in patients with PTGS2-overexpressing colorectal cancer (Chan et al. 2009), PIK3CA-mutated colorectal cancers (Liao et al. 2012), and CD274-low colorectal cancers (Hamada et al. 2017). These MPE studies demonstrate that the MPE approach can contribute to precision cancer medicine and prevention. In addition to cancer research, MPE approach can be applied to non-neoplastic diseases such as cardiovascular diseases, obesity, diabetes mellitus, drug toxicity, and immunity-related and infectious diseases (Ogino et al. 2016).
Genome-wide association studies (GWAS) suggest interactions of host genetic variations with diet, lifestyle, and other environmental exposures in the development of gastrointestinal cancers (Table 1). There are few gene-environment interaction studies considering microorganisms or immunity in gastrointestinal cancers to date. A gene-environment interaction study has shown a statistically significant interaction of the rs2294008 or rs2976392 single nucleotide polymorphism (SNP) at the PSCA gene with Helicobacter pylori infection in risk for gastric cancer (Nan et al. 2015). In a study of hepatocellular carcinoma (HCC), the rs7574865 SNP at the STAT4 gene and the rs9275319 SNP at the HLA-DQA1 gene showed a statistically significant interaction with hepatitis B virus infection in risk for HCC (Nan et al. 2013). A study using the integration of immunology and MPE into GWAS has revealed that the rs11676348 SNP was associated with colorectal cancer exhibiting Crohn’s-like lymphoid reaction or high-level of MSI (Khalili et al. 2015).
Table 1.
Major studies of interactions between host genetic variations and environmental exposures in gastrointestinal cancers
| Environmental exposure | Gene (References) |
|---|---|
| Esophageal adenocarcinoma | |
| Smoking |
ADH1B and ALDH2 (Tanaka et al. 2010) RNF144A (Dong et al. 2018) |
| Recurrent gastroesophageal reflux disease symptoms | RND3 (Dong et al. 2018). |
| Gastric adenocarcinoma | |
| Helicobacter pylori infection | PSCA (Cai et al. 2017) |
| Alcohol drinking | SLC52A3 (Cai et al. 2017) |
| Colorectal carcinoma | |
| Processed meat or total red meat | NAT2 (Wang et al. 2015a) |
| Vegetables | EIF3H (Hutter et al. 2012) |
| Smoking | SMAD7 and TGFBR1 (Zhong et al. 2013) |
| Alcohol drinking |
MFSD14B (Gong et al. 2016) DUSP10 (Song et al. 2018) |
| Aspirin and/or anti-inflammatory drugs use | MGST1, IL16 (Nan et al. 2015; Nan et al. 2013). |
| Use of estrogen plus progesterone therapy | CYP24A1 (Garcia-Albeniz et al. 2016) |
| Hepatocellular carcinoma | |
| Hepatitis B virus infection | STAT4, HLA-DQA1 (Jiang et al. 2013) |
| Pancreatic cancer | |
| Smoking |
XRCC3 (Duell et al. 2008) EPHX1 (Jang et al. 2012) |
| Obesity |
IGF1 (Nakao et al. 2011) FTO and ADIPOQ (Tang et al. 2011) |
| Alcohol drinking | IGF2R, IRS1 (Dong et al. 2012) |
| Diabetes mellitus | PTGS1 (Tang et al. 2014) |
The relationship between the complex gut microbiome, tumor cells, and immune cells in humans cannot be completely recapitulated in any in vivo or in vitro model. Hence, the integration of microbiology into the MPE model (microbiology-MPE) can contribute to better understanding of the complex interactions of environment, tumor cells, immune cells, and microbiome during the development and progression of gastrointestinal cancers (Chen et al. 2019a; Hamada et al. 2019; Luo et al. 2019). Herein, we review major clinical and experimental studies on the microbiome and gastrointestinal cancers, and describe emerging evidence from microbiology-MPE studies.
Gut microbiome and gastrointestinal tract cancer
Esophageal cancer
Esophageal carcinoma largely consists of two histological types, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC is a common type of esophageal cancer worldwide and predominates in certain high-risk areas, such as China and Japan (Stewart et al. 2014). In contrast, EAC is a predominant type in European and North American countries (Ajayi et al. 2018; Baba et al. 2017). Cigarette smoking, alcohol consumption, caustic injury, poor oral health, and poor nutritional status are major risk factors for ESCC; whereas the risk factors of EAC include advanced age, male sex, obesity, gastro-esophageal reflex disease, cigarette smoking, and diet low in vegetables and fruit (Smyth et al. 2017). Genetic polymorphisms of the GSTM1, the ALDH2 and ADH1B (alcoholic metabolic enzymes), or the MTHFR (folate metabolic enzyme) have been associated with an increased risk of esophageal carcinoma (Tian et al. 2019a). Human studies suggest associations of the microbiome with ESCC and EAC (Table 2).
Table 2.
Specific microorganism or dysbiosis of microbiome in esophageal and gastric cancers
| Specific microorganism or dysbiosis of the microbiome | Findings (References) |
|---|---|
| Esophageal cancer | |
| Oral microbiome |
|
| Fusobacterium nucleatum |
|
| Escherichia coli |
|
| Campylobacter concisus |
|
| Gastric cancer | |
| Helicobacter pylori |
|
| Gastric mucosal microbiome |
|
| Tongue-coating microbiome |
|
Low microbial diversity measured by the number of detectable bacterial genera sample in the oral cavity has been associated with the presence of ESCC (Chen et al. 2015; Yu et al. 2014). Peters et al. have performed 16S rRNA gene sequencing in prediagnostic mouthwash specimens, and found that a high amount of Tannerella forsythia in the oral cavity may be associated with higher risk of EAC, and that Porphyromonas gingivalis may be associated with higher risk of ESCC (Peters et al. 2017). Yamamura et al. have demonstrated that a high amount of Fusobacterium nucleatum, which is common species in the oral microbiota, in tumor tissue is associated with shorter patient survival following resection of esophageal cancer, including both ESCC and EAC, and that the amount of Fusobacterium nucleatum correlates with tumor expression of chemokine CCL20, which has been shown to promote the accumulation of regulatory T cells (Yamamura et al. 2016). Epidemiological studies have demonstrated an association of indicators of poor oral health, such as high numbers of lost teeth, with an increased risk of ESCC (Chen et al. 2017). These data suggest that dysbiosis in the oral microbiome may play a role in esophageal carcinogenesis.
Other studies have shown that Escherichia coli and Campylobacter concisus, which have been shown to potentiate carcinogenesis through specific toxins that can induce DNA damage (He et al. 2019; Wilson et al. 2019), are enriched in Barrett’s esophagus (Blackett et al. 2013; Zaidi et al. 2016).
Gastric cancer
Gastric cancer remains one of the leading causes of cancer-related mortality worldwide and the most prevalent cancer in Eastern Asia (Ajani et al. 2017). The worldwide incidence of gastric cancer has declined rapidly over the recent few decades due to a decline in Helicobacter pylori infection rates. Gastric cancer can be classified according to tumor location as cardia (the upper part of the stomach) and non-cardia (the mid and distal stomach). The incidence of gastric cardia cancer and EAC has increased. Chronic infection with Helicobacter pylori, gram-negative species that colonizes gastric epithelium, has been associated with an increased risk of gastric cancer, and Helicobacter pylori is categorized as a class I carcinogen by the World Health Organization (Ajani et al. 2017). Risk factors for cancers arising from cardia and non-cardia regions of the stomach may be different. Common risk factors for both cardia and non-cardia gastric cancer include advanced age, male sex, cigarette smoking, radiation, and family history (Ajani et al. 2017). Factors associated cardia gastric cancer include obesity and gastro-esophageal reflex disease; whereas the risk factors of non-cardia gastric cancer include Helicobacter pylori infection, low socioeconomic status, low consumption of fruits and vegetables and high intake of salty and smoked food (Karimi et al. 2014). Genetic polymorphisms of the APEX1, CASP8, DNMT1, ERCC5, GSTT1, IL1B, IL1RN, IL10, IL17F, MDM2, PPARG, TLR4, or TNF have been associated with higher risk for and gastric cancer (El-Omar et al. 2000; El-Omar et al. 2003; Tian et al. 2019b). A germline CDH1 mutation is associated with familial diffuse gastric cancer (Guilford et al. 1998). In addition to the infection with Helicobacter pylori, human metagenomic studies suggest a potential link between the gastric microbiome and gastric cancer (Table 2).
A Chinese pilot study showed that low microbial diversity in gastric nontumor tissues was associated with advanced tumor grade in patients with gastric cardia cancer, and that patients with metastatic gastric cardia cancer had lower relative abundance of Lactobacillales in gastric nontumor tissues compared to gastric cardia cancer patients without metastasis (Yu et al. 2017). Metagenomic analyses of gastric mucosal microbiota have shown a gradual decrease in microbial diversity from gastritis to intestinal metaplasia to gastric cancer (Aviles-Jimenez et al. 2014), and that lower microbial diversity, lower amount of Helicobacter, and higher amounts of certain members of the oral microbiota, including Parvimonas micra, Peptostreptococcus stomatis, and Fusobacterium nucleatum, were observed in both cardia and non-cardia gastric cancer tissues, compared with nontumor tissues (Coker et al. 2018; Ferreira et al. 2018). Dysbiosis in the tongue coating microbiome was observed in patients with gastric cancer (Wu et al. 2018). These findings suggest that oral microbes may play a role in the development of gastric cancer.
Colorectal cancer
Colorectal cancers are a heterogeneous group of diseases that result from the accumulation of genomic and epigenomic alterations, and tumor-host interactions, which is influenced by environmental exposures including, diet, nutrition, and lifestyle, the microbiome, and host immunity (Tilg et al. 2018; Wong and Yu 2019). Modifiable risk factors for colorectal cancer include cigarette smoking, alcohol consumption, overweight and obesity, physical inactivity, high consumption of red and processed meat, and low consumption of dietary fiber, whole grains, and other healthful nutrients (Islami et al. 2018). Genome-wide association studies of colorectal caner have reported that genetic polymorphisms associated with higher risk for colorectal cancer are located either inside or near protein-coding genes that include ATOH1, APOBEC1, BMPR1B, BMP5, CDKN2A, CYP17A1, EIF3H, FKBP5, MED13L, PDLIM5, PTGER4, PTPN1, RTEL1, RPS21, SMARCAD1, SPSB2, TERT, or TFEB (Schmit et al. 2019; Zeng et al. 2016). Germline mutations in the APC gene or the DNA mismatch repair genes (MSH2, MLH1, PMS1, PMS2, and MSH6) have been associated with familial adenomatous polyposis or hereditary nonpolyposis colorectal cancer. Germline mutations of the MUTYH, STK11, SMAD4, BMPR1A, or PTEN have been associated with other familial polyposis syndromes (Kuipers et al. 2015). Human metagenomic studies demonstrate associations of the microbiome in mucosal tissue and fecal specimens with colorectal neoplasms (Table 3).
Table 3.
Specific microorganism or dysbiosis of microbiome in colorectal cancer
| Specific microorganism or dysbiosis of the microbiome | Findings (References) |
|---|---|
| Colorectal cancer | |
| Oral microbes |
|
| Viral microbiome |
|
| Fungal microbiome |
|
| Fusobacterium nucleatum |
|
| Campylobacter species |
|
| Escherichia coli |
|
| Bacteroides fragilis |
|
| Enterococcus faecalis |
|
| Streptococcus gallolyticcus |
|
Colorectal mucosal microbiome has been significantly different from the fecal microbiome in patients with colorectal cancer (Chen et al. 2012; Flemer et al. 2017). Metagenomic analyses of the microbiome in human colorectal tumor tissues have demonstrated that the tumor microbiome changes across time periods from adenoma to adenocarcinoma, and that some members of the oral microbiota, including Fusobacterium, Gemella, Peptostreptococcus, Parvimonas, and Leptotrichia, were enriched in colorectal adenocarcinoma tissues (Nakatsu et al. 2015). Metagenomic analyses of the fecal microbiome in patients with colorectal neoplasms by Yachida et al. identified that Atopobium parvulum and Actinomyces odontolyticus were significantly increased in patients with adenomas and early stage adenocarcinoma, and that Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum, and Peptostreptococcus anaerobius were enriched in patients with metastatic colorectal adenocarcinoma (Yachida et al. 2019). Meta-analyses of human metagenomic studies of the fecal microbiome have revealed that oral microbes, including Fusobacterium nucleatum, Parvimonas micra, and Peptostreptococcus stomatis, were enriched in patients with colorectal carcinoma (Thomas et al. 2019; Wirbel et al. 2019). Experimental evidence indicates that Peptostreptococcus anaerobius can activate the NFKB signaling pathway in colorectal cancer cell lines and inhibit T-cell-mediated immune responses against colorectal tumors through the recruitment of myeloid-derived suppressor cells and tumor-associated macrophages into the tumor microenvironment in the ApcMin/+ mouse model (Long et al. 2019). Epidemiologic studies have demonstrated an association of periodontal disease with colorectal cancer risk (Michaud et al. 2018; Momen-Heravi et al. 2017). These findings demonstrate potential roles of oral and intestinal microbes in the progression of colorectal neoplasm.
In addition to altered composition of the bacteria in colorectal mucosal tissue and fecal specimens, emerging evidence suggests that dysbiosis of the viral microbiome (virome) or the fungal microbiome (mycobiome) is associated with colorectal cancer (Coker et al. 2019; Nakatsu et al. 2018). Nakatsu et al. found that some viral taxa, such as Orthobunyavirus, Inovirus, or Tunalikevirus, were enriched in fecal specimens from patients with colorectal cancer, and that dysbiosis of intestinal virome was associated with worse clinical outcomes in colorectal cancer (Nakatsu et al. 2018). Analyses by Coker et al. found that altered composition of the intestinal mycobiome was associated with colorectal cancer, and that fecal specimens from patients with colorectal cancer had higher amounts of Malasseziomycetes and lower amounts of Saccharomycetes and Pneumocystidomycetes compared to cancer-free individuals (Coker et al. 2019).
Accumulating evidence suggests enrichment of Fusobacterium nucleatum in human colorectal adenomas and carcinomas compared with adjacent normal tissue (Mima et al. 2017). Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity (Komiya et al. 2019). In addition, higher amount of tissue Fusobacterium nucleatum has been associated with advanced disease stage (Castellarin et al. 2012; Flanagan et al. 2014; Kostic et al. 2012), a lower density of T cells in colorectal carcinoma tissue (Mima et al. 2015), and worse patient survival (Mima et al. 2016b). Fusobacterium nucleatum has been detected not only in primary tumors, but also in metastatic lymph nodes and liver metastases (Bullman et al. 2017; Yu et al. 2016). In addition to Fusobacterium nucleatum, several fusobacterial species, including Fusobacterium gonidiaformans, Fusobacterium periodonticum, and Fusobacterium varium have been enriched in colorectal cancer from southern Chinese populations (Yeoh et al. 2020). Experimental evidence implies potential roles of Fusobacterium nucleatum in the development and progression of colorectal cancer. Fusobacterium nucleatum may inhibit T-cell-mediated immune responses against colorectal tumors through the recruitment of myeloid-derived suppressor cells into the tumor microenvironment in the ApcMin/+ mouse model (Kostic et al. 2013). The Fap2 protein of Fusobacterium nucleatum has been shown to interact with T cell immunoglobulin and ITIM domain (TIGIT) receptor, and inhibit activities of NK cells and T cells (Gur et al. 2015). Fusobacterium nucleatum expresses the virulence factor FadA on the bacterial cell surface, which has been shown to bind to CDH1, activate the WNT signaling pathway in colorectal carcinoma cells, and promote colorectal tumor growth (Rubinstein et al. 2013). The Fap2 protein has been shown to mediate attachment of Fusobacterium nucleatum to colorectal cancers that express host Gal-GalNAc, and be transmitted hematogenously to colorectal carcinoma tissue (Abed et al. 2016). Fusobacterium nucleatum has been shown to activate the NFKB signaling pathway and up-regulate MIR21 expression in colorectal cancer cell lines, and promote the development of intestinal tumors in ApcMin/+ mouse model (Yang et al. 2017).
Cytolethal distending toxin (CDT) is a well-characterized genotoxin (Ge et al. 2007; Graillot et al. 2016; Guidi et al. 2013; Nesic et al. 2004). Enrichment of Campylobacter species has been observed in colorectal cancer tissue and fecal specimens from patients with colorectal cancer (Warren et al. 2013; Wu et al. 2013). In a mouse model, Campylobacter jejuni can potentiate the development of intestinal tumors through the genotoxic action of the CDT (He et al. 2019). Colibactin is encoded by the polyketide synthase (pks) island present in Escherichia coli from phylogroup B2, and has been found to induce DNA damage (Cuevas-Ramos et al. 2010; Nougayrede et al. 2006; Wilson et al. 2019), and promote colon carcinogenesis in Il10−/− mice (Arthur et al. 2014; Arthur et al. 2012). Pleguezuelos-Manzano et al. have demonstrated that pks-positive Escherichia coli could induce mutations characterized by a specific signature in human intestinal organoids and promote carcinogenesis (Pleguezuelos-Manzano et al. 2020). Clinical studies with a limited sample size suggest that the amount of Escherichia coli is higher in colorectal carcinoma tissue than in adjacent normal tissue, and that higher amount of Escherichia coli may be associated with advanced disease stage (Bonnet et al. 2014; Kohoutova et al. 2014).
Enterotoxigenic Bacteroides fragilis expresses the virulence factor Bacteroides fragilis toxin, which has been shown to activate the WNT, NFKB, and STAT3 signaling pathways in colonic epithelial cells, and to potentiate the development of intestinal tumors in ApcMin/+ mice (Chung et al. 2018; Wu et al. 2003; Wu et al. 2004; Wu et al. 2007). Accumulating evidence indicates that T helper 17 (TH17) cells, which produce IL17 and IL22, can promote tumor development and progression in the gastrointestinal tract (Chae et al. 2010; Gaffen et al. 2014; Grivennikov et al. 2012). Enterotoxigenic Bacteroides fragilis induces TH17 cells, which activate the STAT3 signaling pathway in tumor cells in the ApcMin/+ mouse model of colon cancer (Wang et al. 2009; Wu et al. 2009). Some human studies suggest that enterotoxigenic Bacteroides fragilis is detected significantly more often in colon mucosa tissue or fecal specimens of colorectal cancer cases than cancer-free individuals, and that higher amount of enterotoxigenic Bacteroides fragilis is associated with advanced disease stage (Boleij et al. 2015; Toprak et al. 2006; Wei et al. 2016).
Familial adenomatous polyposis is caused by germline APC mutations (Groden et al. 1991; Nishisho et al. 1991). Dejea et al. have identified that pks-positive Escherichia coli and enterotoxigenic Bacteroides fragilis were more commonly found in colorectal tissues from patients with familial adenomatous polyposis (68% and 60%, respectively), compared to those from healthy individuals (22% and 30%, respectively). In mouse models, co-colonization with these two microbes can potentiate intestinal carcinogenesis through increased DNA damage in colonic epithelium and IL17 induction in the colon (Dejea et al. 2018). These findings suggest a potential link between the microbiome and germline genetics in colorectal carcinogenesis.
Enterococcus faecalis has been shown to produce extracellular superoxide that induces DNA damage and genomic instability in colonic epithelial cells (Huycke et al. 2002; Wang et al. 2008; Wang and Huycke 2007), and activates macrophages to produce 4-hydroxy-2-nonenal, which promotes colon carcinogenesis in Il10−/− mice (Wang et al. 2013; Wang et al. 2012). One human study showed that Enterococcus faecalis was detected more often in fecal specimens of colorectal cancer cases than controls (Balamurugan et al. 2008). Studies demonstrated that the amount of Streptococcus gallolyticcus in human colorectal carcinomas was higher than in control tissue, and that the amount of Streptococcus gallolyticcus was correlated with PTGS2 (cyclooxygenase-2) expression level in colorectal cancer (Abdulamir et al. 2010; Boleij and Tjalsma 2013; Gupta et al. 2010).
Gut microbiome and hepatobiliary-pancreatic cancer
Hepatocellular carcinoma (HCC)
HCC is the most common primary liver malignancy, and the major risk factors for HCC include hepatitis B and C, alcohol consumption, non-alcoholic fatty liver disease, and liver cirrhosis (Llovet et al. 2016). Numerous polymorphisms in the genes, which are associated with oxidative stress (GSTM1 and GSTT1 genes) and detoxifying (CAT gene) systems, iron metabolism (HFE gene), inflammation (TNF, IL1B, TGFB1, and NFKB1 genes), DNA repair mechanisms (MTHFR gene) cell cycle regulation (MDM2 and TP53 genes), growth factors (EGF gene), or immune response (CD24 gene), have been reported as risk factors of HCC (Nahon and Zucman-Rossi 2012). Clinical studies suggest associations of microbes and microbial dysbiosis with the development of HCC (Table 4).
Table 4.
Specific microorganism or dysbiosis of microbiome in hepatobiliary-pancreatic cancers
| Specific microorganism or dysbiosis of the microbiome | Findings (References) |
|---|---|
| Hepatocellular carcinoma | |
| Fecal microbiome |
|
| Helicobacter species |
|
| Escherichia coli |
|
| Clostridium species |
|
| Biliary tract cancer | |
| Fecal microbiome |
|
| Bile microbiome |
|
| Helicobacter species |
|
| Biliary mucosal microbiome |
|
| Salmonella typhi |
|
| Pancreatic cancer | |
| Pancreatic tissue microbiome |
|
| Fungal microbiome |
|
| Oral microbiome |
|
| Fusobacterium |
|
Metagenomic analyses of the fecal microbiome in patients with liver cirrhosis or HCC revealed that Gemmiger, Parabacteroides, and Paraprevotella were enriched in patients with HCC, compared with those with liver cirrhosis (Ren et al. 2019). Helicobacter pylori and other Helicobacter species have been detected human HCC tissue specimens (Huang et al. 2004; Kruttgen et al. 2012; Rocha et al. 2005). Experimental studies have shown that Helicobacter hepaticus may colonize the hepatic bile canaliculi and the large intestine of mice, and potentiate liver tumor development by increasing tumor cell proliferation, damaging DNA, activating the WNT and NFKB signaling pathways in tumor cells, and suppressing the innate immunity to recognize and eliminate tumor cells in a mouse model of aflatoxin- and hepatitis C virus-induced HCC (Fox et al. 2010; Ward et al. 1994). Escherichia coli has been shown to induce double-strand DNA breaks and promote colon carcinogenesis in Il10−/− mice (Arthur et al. 2014; Arthur et al. 2012). The increased amount of Escherichia coli in the fecal specimens is associated with the presence of HCC in patients with liver cirrhosis, suggesting that intestinal overgrowth of Escherichia coli may contribute to the development of HCC (Grat et al. 2016).
Evidence suggests an association of diabetes, obesity, non-alcoholic fatty liver disease, and non-alcoholic steatohepatitis with the development HCC (Anstee et al. 2019). Experimental studies using mouse models of obesity-induced HCC have shown that microbial dysbiosis correlates with high levels of bile acids, including deoxycholic acid, in the liver, which can potentiate tumor development by up-regulating the expressions of inflammation-related genes such as IL6 and TNF (Xie et al. 2016a; Xie et al. 2016b). Yoshimoto et al. have shown that deoxycholic acid produced by gut microbiota can potentiate tumor development by provoking the senescence-associated secretory phenotype in hepatic stellate cells and up-regulation of IL6 in a mouse model of obesity-induced HCC (Yoshimoto et al. 2013). Among various microbial species, Clostridium species were enriched in these mouse models of obesity-induced HCC (Niwa et al. 2015; Xie et al. 2016a; Xie et al. 2016b; Yoshimoto et al. 2013). Ma and colleagues demonstrate that in multiple mouse models, Clostridium species can inhibit the accumulation of hepatic natural killer T (NKT) cells, and suppress antitumor immune response against both primary and secondary liver tumors (Ma et al. 2018). Colonization with a commensal Clostridium species, which are gram-positive bacteria and involved in the conversion of primary to secondary bile acids, decreased hepatic NKT cells and increased liver tumor metastases. Analysis of Clostridium species in human HCC tissue specimens would be required for clinical application.
Biliary tract cancer
Cholangiocarcinomas are cancers of the intrahepatic or extrahepatic bile ducts (Banales et al. 2016). Primary sclerosing cholangitis, biliary infections with Opisthorchis viverrini and Clonorchis sinensis, biliary malformations such as Caroli’s disease and choledochal cysts, hepatolithiasis, recurrent bacterial cholangitis, carcinogens such as thorotrast and dioxins, and hepatitis C and liver cirrhosis are major risk factors for cholangiocarcinomas (Ray 2015). Genetic polymorphisms in CXCR2 and the drug metabolizing enzyme genes (CYP1A2, NAT1, NAT2, GSTM1, GSTT1, or MTHFR) have been associated with biliary tract cancers (Hsing et al. 2008; Kukongviriyapan 2012). Genetic polymorphisms of the IL10 and VEGFA genes are associated with high risk for gallbladder cancer (Hsing et al. 2008). Although there are a few experimental studies on microbes in relation to biliary tract cancers, clinical studies have shown associations of microbes and microbial dysbiosis with the development of biliary tract cancer (Table 4).
A metagenomic study of the fecal microbiome in patients with intrahepatic cholangiocarcinoma has revealed that amounts of four genera (Lactobacillus, Actinomyces, Peptostreptococcaceae, and Alloscardovia) were increased in fecal specimens from patients with intrahepatic cholangiocarcinoma, compared with those from healthy individuals (Jia et al. 2019). In preliminary studies of the bile microbiome, dysbiosis in the bile microbiome was associated with biliary mucosal dysplasia or cholangiocarcinoma (Chen et al. 2019b; Pereira et al. 2017).
Epidemiologic studies have demonstrated associations of Helicobacter species, including Helicobacter pylori, Helicobacter bilis, and Helicobacter hepatics, with an increased risk of cholangiocarcinoma (Bulajic et al. 2002; Fukuda et al. 2002; Murphy et al. 2014; Segura-Lopez et al. 2015; Shimoyama et al. 2010; Zhou et al. 2013). Experimental evidence suggests that Helicobacter bilis can activate the NFKB signaling pathway and increase the production of VEGFA, which leads to enhancement of angiogenesis in human cholangiocarcinoma cell lines (Takayama et al. 2010). Clinical studies using metagenomic analyses have shown that Bifidobacteriaceae, Enterobacteriaceae and Enterococcaceae are enriched in tumor tissue specimens of cholangiocarcinoma (Chng et al. 2016), and that amounts of Methylophilaceae, Fusobacterium, Prevotella, Actinomyces, Novosphingobium and Helicobacer pylori were increased in cholangiocarcinoma tissue specimens compared with nontumor tissue specimens (Aviles-Jimenez et al. 2016).
Epidemiologic studies have shown that chronic Salmonella typhi infection is associated with an increased risk of gallbladder cancer (Nagaraja and Eslick 2014). Experimental evidence suggests that Salmonella typhi can induce malignant transformation in the ApcMin/+ mouse model, murine gallbladder organoids, and fibroblasts that exhibit TP53 inactivation and MYC amplification through activation of the AKT and MAPK signaling pathways (Scanu et al. 2015).
Pancreatic cancer
Pancreatic cancer is associated with an extremely poor prognosis: the 5-year survival rate is 6–10% and approximately 367,000 new cases were diagnosed worldwide in 2015 (Kleeff et al. 2016). Although 10–20% of patients with pancreatic cancer have surgically resectable disease at the time of presentation, only 15–20% of those survive for 5 years or more. Cigarette smoking, a high intake of fat or meat, and diabetes mellitus are major risk factors for pancreatic cancer (Kleeff et al. 2016). Genetic variations as well as environmental exposures have been associated with the development of pancreatic cancer. Genetic polymorphisms associated with an increased risk for pancreatic cancer have been located either inside or near protein-coding genes that include ABO, BCAR1, DAB2, DPP6, FOXQ1, HNF1B, HNF4G, GRP, NOC2L, NR5A2, ETAA1, SUGCT, PDX1, TERT, TFF1, and TP63 (Klein et al. 2018; Wolpin et al. 2014; Wu et al. 2011). Major germline mutations associated with an increased risk of pancreatic cancer includes mutations in the STK11, CDKN2A, or BRCA2. The pancreas is anatomically connected to the gastrointestinal tract via the pancreatic duct and communicates with the liver via the common bile duct (Thomas and Jobin 2020). An increasing body of evidence suggests possible roles of microbes in the development of pancreatic tumors (Table 4).
Pushalkar et al. have found that Pseudomonas and Elizabethkingia were enriched in tumor tissue and fecal specimens from patients with pancreatic cancer, and that intestinal microbes can migrate from the gut to the pancreas and inhibit T-cell-mediated immune responses against pancreatic tumors through the recruitment of myeloid-derived suppressor cells into the tumor microenvironment in a mouse model (Pushalkar et al. 2018). Metagenomic analyses of human pancreatic cancer microbiome by Riquelme et al. have found that high amount of Pseudoxanthomonas, Streptomyces, or Saccharopolyspora in pancreatic cancer tissue specimens was associated with high density of CD8+ T-cells in tumor tissues and better overall survival (Riquelme et al. 2019). Analyses of fungal microbiome (mycobiome) in pancreatic cancer tissue specimens by Aykut et al. have revealed that Malassezia was enriched in human pancreatic cancer tissue specimens, and that Malassezia can potentiate the development of pancreatic tumors in a mouse model (Aykut et al. 2019).
Epidemiologic studies have shown positive associations of periodontitis with an increased risk of development of pancreatic cancer (Hujoel et al. 2003; Michaud et al. 2007). Human studies using metagenomic analyses suggest that high amounts of pathogenic periodontal microbes, such as Neisseria elongata and Porphyromonas gingivalis, in saliva are associated with an increased risk of development of pancreatic cancer (Farrell et al. 2012). Metagenomic analyses of the oral microbiome in patients with pancreatic cancer from a prospective cohort study have revealed that dysbiosis in the oral microbiome was associated with the incidence of pancreatic cancer, and that the presence of Porphyromonas gingivalis or Aggregatibacter actinomycetemcomitans in the oral cavity was associated with higher incidence of pancreatic cancer (Fan et al. 2018). Gaiser et al. have found enrichment of oral microbes, including Fusobacterium nucleatum and Granulicatella adiacens, in tumor tissue specimens of intraductal papillary mucinous neoplasms with high-grade dysplasia, which have been precursors to invasive pancreatic cancer (Gaiser et al. 2019). Mitsuhashi et al. have shown that a high amount of Fusobacterium species in tumor tissue is associated with worse prognosis in patients with pancreatic cancer (Mitsuhashi et al. 2015).
Emerging findings of the microbiology-MPE research
The concept and study designs of the microbiology-MPE research have been figured and discussed in our previous review (Hamada et al. 2019). Using this approach, we can examine associations of germline genetic variations and environmental exposures, including lifestyle factors, dietary patterns, medications, with specific cancer subtypes according to microbial profile, which are not detectable in conventional epidemiology and microbiology research. If the microbial data before cancer diagnosis are available in prospective studies, we can link microbial profile and the incidence of specific cancer subtypes classified by tumor molecular characteristics (e.g. somatic mutations and epigenetic alterations in tumor cells) or the tumor microenvironment (e.g. antitumor immunity). In patient survival analysis, the microbiology-MPE approach enables us to examine prognostic associations of the environmental exposures according to specific cancer subtypes classified by microbial profile. In addition, we can examine an association of microbial profile with patient survival according to specific cancer subtypes classified by molecular characteristics.
The envirome (or the exposome), which broadly includes dietary and lifestyle factors, have been implicated in the development of colorectal tumors. Smoking, adiposity (body fatness), alcohol drinks, and red and processed meat have been associated with an increased risk of colorectal cancer, whereas regular aspirin use, physical activity, plasma vitamin D level, and high intakes of dietary fiber, whole grains, calcium, and marine omega-3 fatty acid may decrease risk of colorectal cancer (Song et al. 2020). The microbiology-MPE studies have shown that a so-called prudent diet that is rich in whole grains and fiber was associated with a lower risk of colorectal carcinoma with detectable levels of Fusobacterium nucleatum but not with a lower risk of carcinoma without Fusobacterium nucleatum (Fig. 2A) (Mehta et al. 2017), and that an inflammatory dietary pattern (rich in red and processed meat, refined grains, and sugar) has been associated with a higher risk of Fusobacterium nucleatum-positive proximal colon carcinoma, but not with a risk of Fusobacterium nucleatum-negative proximal colon carcinoma (Fig. 2B) (Liu et al. 2018b). These findings support a potential role for the gut microbes in mediating the effect of diet on colorectal carcinogenesis. Although mechanistic studies have been a major part of microbiology research on carcinogenesis, insights from microbiology-MPE research would serve as particularly valuable evidence for the microbial etiologies and pathogenesis of human neoplasms.
Fig. 2.
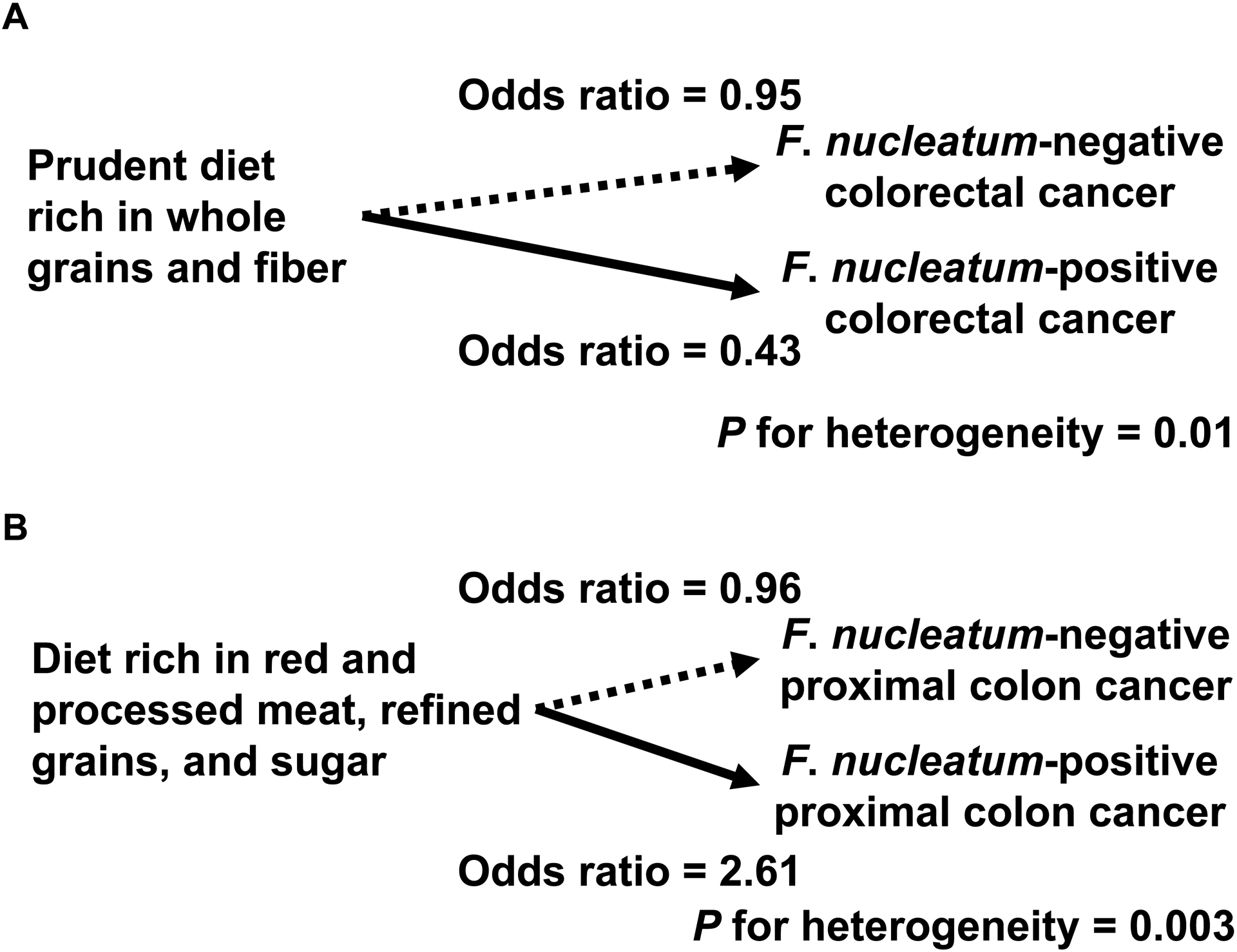
Illustration of the microbiome-MPE approach using tumor microbial status. Fusobacterium nucleatum can inhibit antitumor immune response and potentiate colonic neoplasia development in animal models. Using data on colorectal cancer cases and tumor microbial profile in two U.S. nationwide prospective cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study), the microbiome-MPE studies have revealed that a so-called prudent diet that is rich in whole grains and fiber was associated with a lower risk of colorectal carcinoma with detectable levels of Fusobacterium nucleatum but not with a lower risk of carcinoma without Fusobacterium nucleatum (A), and that an inflammatory dietary pattern (rich in red and processed meat, refined grains, and sugar) was associated with a higher risk of Fusobacterium nucleatum-positive colorectal carcinoma, but not with a risk of Fusobacterium nucleatum-negative carcinoma (B).
The proportions of colorectal cancers with specific molecular features such as high-level MSI, high-level CIMP, and BRAF and PIK3CA mutations have been shown to gradually increase along the bowel subsites from rectum to ascending colon (Yamauchi et al. 2012) and these findings have been replicated in other datasets (Phipps et al. 2012; Phipps et al. 2015; Rosty et al. 2013). These findings led to the colorectal continuum concept that most likely reflected the influence of the gut microbiome on local tissue microenvironment and carcinogenesis. Studies have demonstrated that a high amount of Fusobacterium nucleatum in carcinoma tissue is associated with high-level MSI (Ito et al. 2015; Mima et al. 2015; Tahara et al. 2014), autophagy status (Haruki et al. 2019), lower density of T cells in tumor tissue (Mima et al. 2015), and worse patient survival (de Carvalho et al. 2019; Kunzmann et al. 2019; Mima et al. 2016b), and that the proportion of colorectal cancers containing high amounts of Fusobacterium nucleatum increased gradually along the bowel subsites from rectum to cecum (Mima et al. 2016a). In addition, our further analyses revealed that Fusobacterium nucleatum in tumor tissue was associated with lower-level tumor-infiltrating lymphocytes (TIL) in MSI-high colorectal carcinoma, while it was associated with high-level TIL in non-MSI-high carcinoma; these findings might reflect divergent effects of the bacteria on the tumor-immune microenvironment according to the amount of tumor neoantigens (Hamada et al. 2018). MSI-high colorectal cancers are genetically characterized by a hypermutator phenotype associated with a high number of neoantigens (Giannakis et al. 2016). These data would inform future mechanistic studies to examine the interplay of Fusobacterium nucleatum and tumor characteristics in colorectal carcinogenesis.
Future perspectives, challenges, and conclusions
Considering that oral health, diet, lifestyle, pharmacological factors (including antibiotics), probiotics, and prebiotics can influence the composition of intestinal microbiota (Biedermann et al. 2013; O’Keefe et al. 2015; Zitvogel et al. 2015), future investigations need to examine potential influences of those modifiable factors on the gut microflora and tumorigenic processes. Although to date, no clinical trials demonstrate the efficacy of modulating the microbiome in the development or progression of gastrointestinal cancers, some clinical studies have shown the effect of microbiome modulation in inflammatory bowel diseases and toxicity of cancer chemotherapy. Fecal microbiota transplantation (FMT) has been effective treatment for recurrent or refractory Clostridium difficile infection (Costello et al. 2015; van Nood et al. 2013). Ulcerative colitis is a chronic inflammatory bowel disease characterized by colonic mucosal inflammation, and is associated with an increased risk of colorectal cancer (Castano-Milla et al. 2014). FMT has been shown to induce clinical remission and endoscopic improvement in active ulcerative colitis (Moayyedi et al. 2015; Paramsothy et al. 2017; Rossen et al. 2015). Probiotic refers to bacteria or a combination of live bacteria that confer a health benefit to hosts when consumed in adequate amounts (Suez et al. 2019). In 15 patients with colorectal cancer, administration of probiotics may reduce amounts of Fusobacterium and Peptostreptococcus (Hibberd et al. 2017). Patients with colorectal cancer who received 5-FU and Lactobacillus rhamnosus were less like to have diarrhea (Osterlund et al. 2007).
Escherichia coli and Bacteroides fragilis have been enriched in colorectal mucosal tissues from patients with familial adenomatous polyposis that is caused by germline APC mutations, suggesting a potential link between the microbiome and germline genetics in carcinogenesis. Further investigations need to examine potential combined influences of germline mutations and the microbiome on carcinogenesis in hereditary neoplasms. The integrative approach of microbiology-MPE can be a novel tool that potentially expands our knowledge of the etiologies and pathogenesis of cancers not only in the digestive system but also in other body sites. Future microbiology-MPE analyses can reveal additional links of host genetic variations and modifiable lifestyle factors with specific subtypes of neoplasms, which will contribute to the development of precision prevention and treatment strategies. With the advances in next-generation sequencing technologies, large population-based research on genomics, metagenomics, and other omics (epigenomics, transcriptomics, proteomics, and metabolomics) has become reality, but necessitates our efforts to new transdisciplinary frameworks of our science and research enterprise, including interdisciplinary education system (Ogino et al. 2012). Successful microbiology-MPE studies may be among research examples that can inform such efforts.
Although clinical studies have linked specific microbes and microbial dysbiosis to gastrointestinal cancers, there are considerable study-to-study differences in reported specific microbes and microbial dysbiosis, which may be due to limitations including small sample sizes, undefined tissue sampling sites, procedures for biospecimen collection, processing, and storage, methods for microbiome analysis, and limited data on clinical features and tumor molecular features. Challenges exist in the microbiology-MPE research, as previously discussed (Hamada et al. 2019; Ogino et al. 2011). Sample size is generally limited based on biospecimen availability, which can also lead to selection bias. Hence MPE investigators should make efforts to maximize the number of cases available for research. Statistical analysis methods to test hypothesis on etiological heterogeneity between disease subtypes (Lu et al. 2018; Wang et al. 2015b; Wang et al. 2016) and to address missing data (Liu et al. 2018a; Nevo et al. 2018) in MPE research have been validated in various study designs. Standardized procedures for collecting, processing, and storing biospecimens, and methods of microbiome analyses are required for collaborative projects on microbiology-MPE. In addition, transdisciplinary education and transdisciplinary research teams are also important to perform the MPE research due to the nature of microbiology-MPE. We have started training programs of molecular pathology, epidemiology, microbiology, and immunology for physicians and researchers (e.g. Lectures and/or hands-on training of epidemiology and biostatistics in departments of pathology).
In conclusion, accumulating evidence supports the influential role of the microbiome in cancers of the digestive system and likely neoplasms of other organs and tissues. The microbiology-MPE research can provide a novel methodological framework to integrate data on host genetic variations and modifiable factors into analyses of the microbiome and tumor characteristics, to generate novel insights into tumor-immune-microbiome interactions. This new type of research effort can inform cancer prevention and treatment strategies targeting the microbiome.
Funding:
This work was supported by U.S. National Institutes of Health (NIH) grants [R35 CA197735 to S.O.; R01 CA151993 to S.O.; R21 CA230873 to S.O.; R01 CA248857 to S.O.], and Cancer Research UK Grand Challenge Award (OPTIMISTICC, C10674/A27140 to S.O.). K.M. is supported by grants from JSPS KAKENHI (grant number 17H05094 and 20K17658). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Note added in Proofs:
The MPE approach combined with microbiome research is expected to reveal new risk factors for early-onset colorectal cancer that has been increasing worldwide for uncertain reasons (Akimoto et al. in press), and there is an urgent need for research.
Competing interests: All authors declare that they have no competing financial interest.
Use of standardized official symbols: We use HUGO (Human Genome Organization)-approved official symbols (or root symbols) for genes and gene products; all of which are described at www.genenames.org. The official gene symbols are italicized to differentiate from non-italicized gene product symbols.
References
- Abdulamir AS, Hafidh RR, Bakar FA (2010) Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer 9: 249. doi: 10.1186/1476-4598-9-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G (2016) Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 20: 215–25. doi: 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto N, Ugai T, Zhong R, Hamada T, Fujiyoshi K, Giannakis M, Wu K, Cao Y, Ng K, Ogino S (in press) Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol. [DOI] [PMC free article] [PubMed]
- Ajani JA, Lee J, Sano T, Janjigian YY, Fan D, Song S (2017) Gastric adenocarcinoma. Nat Rev Dis Primers 3: 17036. doi: 10.1038/nrdp.2017.36 [DOI] [PubMed] [Google Scholar]
- Ajayi TA, Cantrell S, Spann A, Garman KS (2018) Barrett’s esophagus and esophageal cancer: Links to microbes and the microbiome. PLoS Pathog 14: e1007384. doi: 10.1371/journal.ppat.1007384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M (2019) From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol 16: 411–428. doi: 10.1038/s41575-019-0145-7 [DOI] [PubMed] [Google Scholar]
- Arthur JC, Gharaibeh RZ, Muhlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, Fodor AA, Jobin C (2014) Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun 5: 4724. doi: 10.1038/ncomms5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338: 120–3. doi: 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviles-Jimenez F, Guitron A, Segura-Lopez F, Mendez-Tenorio A, Iwai S, Hernandez-Guerrero A, Torres J (2016) Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect 22: 178 e11–22. doi: 10.1016/j.cmi.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J (2014) Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep 4: 4202. doi: 10.1038/srep04202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, Guo Y, Saxena A, Vardhan M, Diskin B, Wang W, Leinwand J, Kurz E, Kochen Rossi JA, Hundeyin M, Zambrinis C, Li X, Saxena D, Miller G (2019) The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574: 264–267. doi: 10.1038/s41586-019-1608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Iwatsuki M, Yoshida N, Watanabe M, Baba H (2017) Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann Gastroenterol Surg 1: 99–104. doi: 10.1002/ags3.12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS (2008) Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol 23: 1298–303. doi: 10.1111/j.1440-1746.2008.05490.x [DOI] [PubMed] [Google Scholar]
- Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D (2016) Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 13: 261–80. doi: 10.1038/nrgastro.2016.51 [DOI] [PubMed] [Google Scholar]
- Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E, Rehman A, Ott SJ, Steurer-Stey C, Frei A, Frei P, Scharl M, Loessner MJ, Vavricka SR, Fried M, Schreiber S, Schuppler M, Rogler G (2013) Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 8: e59260. doi: 10.1371/journal.pone.0059260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, Macfarlane GT, Dillon JF (2013) Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett’s and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther 37: 1084–92. doi: 10.1111/apt.12317 [DOI] [PubMed] [Google Scholar]
- Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, Ellis B, Carroll KC, Albesiano E, Wick EC, Platz EA, Pardoll DM, Sears CL (2015) The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 60: 208–15. doi: 10.1093/cid/ciu787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A, Tjalsma H (2013) The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect Dis 13: 719–24. doi: 10.1016/S1473-3099(13)70107-5 [DOI] [PubMed] [Google Scholar]
- Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Dechelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A (2014) Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 20: 859–67. doi: 10.1158/1078-0432.CCR-13-1343 [DOI] [PubMed] [Google Scholar]
- Bulajic M, Maisonneuve P, Schneider-Brachert W, Muller P, Reischl U, Stimec B, Lehn N, Lowenfels AB, Lohr M (2002) Helicobacter pylori and the risk of benign and malignant biliary tract disease. Cancer 95: 1946–53. doi: 10.1002/cncr.10893 [DOI] [PubMed] [Google Scholar]
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon YCS, Fasani R, Aguirre AJ, Ng K, Elez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M (2017) Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358: 1443–1448. doi: 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Dai S, Chen W, Xia C, Lu L, Dai S, Qi J, Wang M, Wang M, Zhou L, Lei F, Zuo T, Zeng H, Zhao X (2017) Environmental factors, seven GWAS-identified susceptibility loci, and risk of gastric cancer and its precursors in a Chinese population. Cancer Med 6: 708–720. doi: 10.1002/cam4.1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PR, Alwers E, Bienert S, Weberpals J, Kloor M, Brenner H, Hoffmeister M (2018) Lifestyle factors and risk of sporadic colorectal cancer by microsatellite instability status: a systematic review and meta-analyses. Ann Oncol 29: 825–834. doi: 10.1093/annonc/mdy059 [DOI] [PubMed] [Google Scholar]
- Castano-Milla C, Chaparro M, Gisbert JP (2014) Systematic review with meta-analysis: the declining risk of colorectal cancer in ulcerative colitis. Aliment Pharmacol Ther 39: 645–59. doi: 10.1111/apt.12651 [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22: 299–306. doi: 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL (2010) Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci U S A 107: 5540–4. doi: 10.1073/pnas.0912675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, Ogino S, Fuchs CS (2009) Aspirin use and survival after diagnosis of colorectal cancer. JAMA 302: 649–58. doi: 10.1001/jama.2009.1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Du G, Guo J, Zhang Y (2019a) Bugs, drugs, and cancer: can the microbiome be a potential therapeutic target for cancer management? Drug Discov Today 24: 1000–1009. doi: 10.1016/j.drudis.2019.02.009 [DOI] [PubMed] [Google Scholar]
- Chen B, Fu SW, Lu L, Zhao H (2019b) A Preliminary Study of Biliary Microbiota in Patients with Bile Duct Stones or Distal Cholangiocarcinoma. Biomed Res Int 2019: 1092563. doi: 10.1155/2019/1092563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Liu F, Ling Z, Tong X, Xiang C (2012) Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7: e39743. doi: 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Winckler B, Lu M, Cheng H, Yuan Z, Yang Y, Jin L, Ye W (2015) Oral Microbiota and Risk for Esophageal Squamous Cell Carcinoma in a High-Risk Area of China. PLoS One 10: e0143603. doi: 10.1371/journal.pone.0143603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yuan Z, Lu M, Zhang Y, Jin L, Ye W (2017) Poor oral health is associated with an increased risk of esophageal squamous cell carcinoma - a population-based case-control study in China. Int J Cancer 140: 626–635. doi: 10.1002/ijc.30484 [DOI] [PubMed] [Google Scholar]
- Chng KR, Chan SH, Ng AH, Li C, Jusakul A, Bertrand D, Wilm A, Choo SP, Tan DM, Lim KH, Soetinko R, Ong CK, Duda DG, Dima S, Popescu I, Wongkham C, Feng Z, Yeoh KG, Teh BT, Yongvanit P, Wongkham S, Bhudhisawasdi V, Khuntikeo N, Tan P, Pairojkul C, Ngeow J, Nagarajan N (2016) Tissue Microbiome Profiling Identifies an Enrichment of Specific Enteric Bacteria in Opisthorchis viverrini Associated Cholangiocarcinoma. EBioMedicine 8: 195–202. doi: 10.1016/j.ebiom.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F (2018) Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 23: 203–214 e5. doi: 10.1016/j.chom.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J (2018) Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67: 1024–1032. doi: 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J (2019) Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68: 654–662. doi: 10.1136/gutjnl-2018-317178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J, Perez-Garcia JM, Llombart-Cussac A, Curigliano G, El Saghir NS, Cardoso F, Barrios CH, Wagle S, Roman J, Harbeck N, Eniu A, Kaufman PA, Tabernero J, Garcia-Estevez L, Schmid P, Arribas J (2020) Enhancing global access to cancer medicines. CA Cancer J Clin 70: 105–124. doi: 10.3322/caac.21597 [DOI] [PubMed] [Google Scholar]
- Costello SP, Chung A, Andrews JM, Fraser RJ (2015) Fecal Microbiota Transplant for Clostridium difficile Colitis-Induced Toxic Megacolon. Am J Gastroenterol 110: 775–7. doi: 10.1038/ajg.2015.70 [DOI] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP (2010) Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 107: 11537–42. doi: 10.1073/pnas.1001261107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho AC, de Mattos Pereira L, Datorre JG, Dos Santos W, Berardinelli GN, Matsushita MM, Oliveira MA, Duraes RO, Guimaraes DP, Reis RM (2019) Microbiota Profile and Impact of Fusobacterium nucleatum in Colorectal Cancer Patients of Barretos Cancer Hospital. Front Oncol 9: 813. doi: 10.3389/fonc.2019.00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wick EC, Wang H, Wu S, Pardoll DM, Housseau F, Sears CL (2018) Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359: 592–597. doi: 10.1126/science.aah3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Levine DM, Buas MF, Zhang R, Onstad L, Fitzgerald RC, Stomach, Oesophageal Cancer Study C, Corley DA, Shaheen NJ, Lagergren J, Hardie LJ, Reid BJ, Iyer PG, Risch HA, Caldas C, Caldas I, Pharoah PD, Liu G, Gammon MD, Chow WH, Bernstein L, Bird NC, Ye W, Wu AH, Anderson LA, MacGregor S, Whiteman DC, Vaughan TL, Thrift AP (2018) Interactions Between Genetic Variants and Environmental Factors Affect Risk of Esophageal Adenocarcinoma and Barrett’s Esophagus. Clin Gastroenterol Hepatol 16: 1598–1606 e4. doi: 10.1016/j.cgh.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Li Y, Tang H, Chang P, Hess KR, Abbruzzese JL, Li D (2012) Insulin-like growth factor axis gene polymorphisms modify risk of pancreatic cancer. Cancer Epidemiol 36: 206–11. doi: 10.1016/j.canep.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Duell EJ, Bracci PM, Moore JH, Burk RD, Kelsey KT, Holly EA (2008) Detecting pathway-based gene-gene and gene-environment interactions in pancreatic cancer. Cancer Epidemiol Biomarkers Prev 17: 1470–9. doi: 10.1158/1055-9965.EPI-07-2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, Lanyon G, Martin M, Fraumeni JF Jr., Rabkin CS (2000) Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404: 398–402. doi: 10.1038/35006081 [DOI] [PubMed] [Google Scholar]
- El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, Stanford JL, Mayne ST, Goedert J, Blot WJ, Fraumeni JF Jr., Chow WH (2003) Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124: 1193–201. doi: 10.1016/s0016-5085(03)00157-4 [DOI] [PubMed] [Google Scholar]
- Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, Ravel J, Hayes RB, Ahn J (2018) Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67: 120–127. doi: 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61: 582–8. doi: 10.1136/gutjnl-2011-300784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C (2018) Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 67: 226–236. doi: 10.1136/gutjnl-2017-314205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, Huang B, Arodz TJ, Edupuganti L, Glascock AL, Xu J, Jimenez NR, Vivadelli SC, Fong SS, Sheth NU, Jean S, Lee V, Bokhari YA, Lara AM, Mistry SD, Duckworth RA 3rd, Bradley SP, Koparde VN, Orenda XV, Milton SH, Rozycki SK, Matveyev AV, Wright ML, Huzurbazar SV, Jackson EM, Smirnova E, Korlach J, Tsai YC, Dickinson MR, Brooks JL, Drake JI, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Wijesooriya NR, Hendricks-Munoz KD, Jefferson KK, Strauss JF 3rd, Buck GA (2019) The vaginal microbiome and preterm birth. Nat Med 25: 1012–1021. doi: 10.1038/s41591-019-0450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ (2014) Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33: 1381–90. doi: 10.1007/s10096-014-2081-3 [DOI] [PubMed] [Google Scholar]
- Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, O’Riordain M, Shanahan F, O’Toole PW (2017) Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 66: 633–643. doi: 10.1136/gutjnl-2015-309595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Feng Y, Theve EJ, Raczynski AR, Fiala JL, Doernte AL, Williams M, McFaline JL, Essigmann JM, Schauer DB, Tannenbaum SR, Dedon PC, Weinman SA, Lemon SM, Fry RC, Rogers AB (2010) Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut 59: 88–97. doi: 10.1136/gut.2009.183749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Kuroki T, Tajima Y, Tsuneoka N, Kitajima T, Matsuzaki S, Furui J, Kanematsu T (2002) Comparative analysis of Helicobacter DNAs and biliary pathology in patients with and without hepatobiliary cancer. Carcinogenesis 23: 1927–31. [DOI] [PubMed] [Google Scholar]
- Gaffen SL, Jain R, Garg AV, Cua DJ (2014) The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol 14: 585–600. doi: 10.1038/nri3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiser RA, Halimi A, Alkharaan H, Lu L, Davanian H, Healy K, Hugerth LW, Ateeb Z, Valente R, Fernandez Moro C, Del Chiaro M, Sallberg Chen M (2019) Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 68: 2186–2194. doi: 10.1136/gutjnl-2018-317458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Albeniz X, Rudolph A, Hutter C, White E, Lin Y, Rosse SA, Figueiredo JC, Harrison TA, Jiao S, Brenner H, Casey G, Hudson TJ, Thornquist M, Le Marchand L, Potter J, Slattery ML, Zanke B, Baron JA, Caan BJ, Chanock SJ, Berndt SI, Stelling D, Fuchs CS, Hoffmeister M, Butterbach K, Du M, James Gauderman W, Gunter MJ, Lemire M, Ogino S, Lin J, Hayes RB, Haile RW, Schoen RE, Warnick GS, Jenkins MA, Thibodeau SN, Schumacher FR, Lindor NM, Kolonel LN, Hopper JL, Gong J, Seminara D, Pflugeisen BM, Ulrich CM, Qu C, Duggan D, Cotterchio M, Campbell PT, Carlson CS, Newcomb PA, Giovannucci E, Hsu L, Chan AT, Peters U, Chang-Claude J (2016) CYP24A1 variant modifies the association between use of oestrogen plus progestogen therapy and colorectal cancer risk. Br J Cancer 114: 221–9. doi: 10.1038/bjc.2015.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Rogers AB, Feng Y, Lee A, Xu S, Taylor NS, Fox JG (2007) Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell Microbiol 9: 2070–80. doi: 10.1111/j.1462-5822.2007.00939.x [DOI] [PubMed] [Google Scholar]
- Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, Sukawa Y, Stewart C, Rosenberg M, Mima K, Inamura K, Nosho K, Nowak JA, Lawrence MS, Giovannucci EL, Chan AT, Ng K, Meyerhardt JA, Van Allen EM, Getz G, Gabriel SB, Lander ES, Wu CJ, Fuchs CS, Ogino S, Garraway LA (2016) Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 15: 857–865. doi: 10.1016/j.celrep.2016.03.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Hutter CM, Newcomb PA, Ulrich CM, Bien SA, Campbell PT, Baron JA, Berndt SI, Bezieau S, Brenner H, Casey G, Chan AT, Chang-Claude J, Du M, Duggan D, Figueiredo JC, Gallinger S, Giovannucci EL, Haile RW, Harrison TA, Hayes RB, Hoffmeister M, Hopper JL, Hudson TJ, Jeon J, Jenkins MA, Kocarnik J, Kury S, Le Marchand L, Lin Y, Lindor NM, Nishihara R, Ogino S, Potter JD, Rudolph A, Schoen RE, Schrotz-King P, Seminara D, Slattery ML, Thibodeau SN, Thornquist M, Toth R, Wallace R, White E, Jiao S, Lemire M, Hsu L, Peters U, Ccfr, Gecco (2016) Genome-Wide Interaction Analyses between Genetic Variants and Alcohol Consumption and Smoking for Risk of Colorectal Cancer. PLoS Genet 12: e1006296. doi: 10.1371/journal.pgen.1006296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graillot V, Dormoy I, Dupuy J, Shay JW, Huc L, Mirey G, Vignard J (2016) Genotoxicity of Cytolethal Distending Toxin (CDT) on Isogenic Human Colorectal Cell Lines: Potential Promoting Effects for Colorectal Carcinogenesis. Front Cell Infect Microbiol 6: 34. doi: 10.3389/fcimb.2016.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grat M, Wronka KM, Krasnodebski M, Masior L, Lewandowski Z, Kosinska I, Grat K, Stypulkowski J, Rejowski S, Wasilewicz M, Galecka M, Szachta P, Krawczyk M (2016) Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant Proc 48: 1687–91. doi: 10.1016/j.transproceed.2016.01.077 [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, Datz C, Feng Y, Fearon ER, Oukka M, Tessarollo L, Coppola V, Yarovinsky F, Cheroutre H, Eckmann L, Trinchieri G, Karin M (2012) Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491: 254–8. doi: 10.1038/nature11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. (1991) Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66: 589–600. doi: 10.1016/0092-8674(81)90021-0 [DOI] [PubMed] [Google Scholar]
- Guidi R, Guerra L, Levi L, Stenerlow B, Fox JG, Josenhans C, Masucci MG, Frisan T (2013) Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell Microbiol 15: 98–113. doi: 10.1111/cmi.12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE (1998) E-cadherin germline mutations in familial gastric cancer. Nature 392: 402–5. doi: 10.1038/32918 [DOI] [PubMed] [Google Scholar]
- Gunter MJ, Alhomoud S, Arnold M, Brenner H, Burn J, Casey G, Chan AT, Cross AJ, Giovannucci E, Hoover R, Houlston R, Jenkins M, Laurent-Puig P, Peters U, Ransohoff D, Riboli E, Sinha R, Stadler ZK, Brennan P, Chanock SJ (2019) Meeting report from the joint IARC-NCI international cancer seminar series: a focus on colorectal cancer. Ann Oncol 30: 510–519. doi: 10.1093/annonc/mdz044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Madani R, Mukhtar H (2010) Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis 12: 164–71. doi: 10.1111/j.1463-1318.2009.01814.x [DOI] [PubMed] [Google Scholar]
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklic K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O (2015) Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42: 344–55. doi: 10.1016/j.immuni.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Cao Y, Qian ZR, Masugi Y, Nowak JA, Yang J, Song M, Mima K, Kosumi K, Liu L, Shi Y, da Silva A, Gu M, Li W, Keum N, Zhang X, Wu K, Meyerhardt JA, Giovannucci EL, Giannakis M, Rodig SJ, Freeman GJ, Nevo D, Wang M, Chan AT, Fuchs CS, Nishihara R, Ogino S (2017) Aspirin Use and Colorectal Cancer Survival According to Tumor CD274 (Programmed Cell Death 1 Ligand 1) Expression Status. J Clin Oncol 35: 1836–1844. doi: 10.1200/JCO.2016.70.7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Nowak JA, Milner DA Jr., Song M, Ogino S (2019) Integration of microbiology, molecular pathology, and epidemiology: a new paradigm to explore the pathogenesis of microbiome-driven neoplasms. J Pathol 247: 615–628. doi: 10.1002/path.5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, Zhang X, Mima K, Bullman S, Sukawa Y, Nowak JA, Kosumi K, Masugi Y, Twombly TS, Cao Y, Song M, Liu L, da Silva A, Shi Y, Gu M, Li W, Koh H, Nosho K, Inamura K, Keum N, Wu K, Meyerhardt JA, Kostic AD, Huttenhower C, Garrett WS, Meyerson M, Giovannucci EL, Chan AT, Fuchs CS, Nishihara R, Giannakis M, Ogino S (2018) Fusobacterium nucleatum in Colorectal Cancer Relates to Immune Response Differentially by Tumor Microsatellite Instability Status. Cancer Immunol Res 6: 1327–1336. doi: 10.1158/2326-6066.CIR-18-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki K, Kosumi K, Hamada T, Twombly TS, Vayrynen JP, Kim SA, Masugi Y, Qian ZR, Mima K, Baba Y, da Silva A, Borowsky J, Arima K, Fujiyoshi K, Lau MC, Li P, Guo C, Chen Y, Song M, Nowak JA, Nishihara R, Yanaga K, Zhang X, Wu K, Bullman S, Garrett WS, Huttenhower C, Meyerhardt JA, Giannakis M, Chan AT, Fuchs CS, Ogino S (2019) Association of autophagy status with amount of Fusobacterium nucleatum in colorectal cancer. J Pathol. doi: 10.1002/path.5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, Pons B, Mirey G, Vignard J, Hendrixson DR, Jobin C (2019) Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut 68: 289–300. doi: 10.1136/gutjnl-2018-317200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgard L, Wettergren Y (2017) Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol 4: e000145. doi: 10.1136/bmjgast-2017-000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing AW, Sakoda LC, Rashid A, Andreotti G, Chen J, Wang BS, Shen MC, Chen BE, Rosenberg PS, Zhang M, Niwa S, Chu L, Welch R, Yeager M, Fraumeni JF Jr., Gao YT, Chanock SJ (2008) Variants in inflammation genes and the risk of biliary tract cancers and stones: a population-based study in China. Cancer Res 68: 6442–52. doi: 10.1158/0008-5472.CAN-08-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fan XG, Wang ZM, Zhou JH, Tian XF, Li N (2004) Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J Clin Pathol 57: 1273–7. doi: 10.1136/jcp.2004.018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes LAE, Simons C, van den Brandt PA, van Engeland M, Weijenberg MP (2017) Lifestyle, Diet, and Colorectal Cancer Risk According to (Epi)genetic Instability: Current Evidence and Future Directions of Molecular Pathological Epidemiology. Curr Colorectal Cancer Rep 13: 455–469. doi: 10.1007/s11888-017-0395-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujoel PP, Drangsholt M, Spiekerman C, Weiss NS (2003) An exploration of the periodontitis-cancer association. Ann Epidemiol 13: 312–6. [DOI] [PubMed] [Google Scholar]
- Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, Nan H, Lemire M, Rangrej J, Figueiredo JC, Jiao S, Harrison TA, Liu Y, Chen LS, Stelling DL, Warnick GS, Hoffmeister M, Kury S, Fuchs CS, Giovannucci E, Hazra A, Kraft P, Hunter DJ, Gallinger S, Zanke BW, Brenner H, Frank B, Ma J, Ulrich CM, White E, Newcomb PA, Kooperberg C, LaCroix AZ, Prentice RL, Jackson RD, Schoen RE, Chanock SJ, Berndt SI, Hayes RB, Caan BJ, Potter JD, Hsu L, Bezieau S, Chan AT, Hudson TJ, Peters U (2012) Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res 72: 2036–44. doi: 10.1158/0008-5472.CAN-11-4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke MM, Abrams V, Moore DR (2002) Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23: 529–36. [DOI] [PubMed] [Google Scholar]
- Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, McCullough ML, Patel AV, Ma J, Soerjomataram I, Flanders WD, Brawley OW, Gapstur SM, Jemal A (2018) Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin 68: 31–54. doi: 10.3322/caac.21440 [DOI] [PubMed] [Google Scholar]
- Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, Yoshii S, Takenouchi T, Hasegawa T, Okita K, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y (2015) Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer 137: 1258–68. doi: 10.1002/ijc.29488 [DOI] [PubMed] [Google Scholar]
- Jang JH, Cotterchio M, Borgida A, Gallinger S, Cleary SP (2012) Genetic variants in carcinogen-metabolizing enzymes, cigarette smoking and pancreatic cancer risk. Carcinogenesis 33: 818–27. doi: 10.1093/carcin/bgs028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Lu S, Zeng Z, Liu Q, Dong Z, Chen Y, Zhu Z, Hong Z, Zhang T, Du G, Xiang J, Wu D, Bai W, Yang B, Li Y, Huang J, Li H, Safadi R, Lu Y (2019) Characterization of Gut Microbiota, Bile Acid Metabolism, and Cytokines in Intrahepatic Cholangiocarcinoma. Hepatology. doi: 10.1002/hep.30852 [DOI] [PubMed] [Google Scholar]
- Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP, Wang Z, Jiang W, Chen TY, Gao Y, Sun LD, Long JR, Huang HX, Wang D, Yu H, Zhang P, Tang LS, Peng B, Cai H, Liu TT, Zhou P, Liu F, Lin X, Tao S, Wan B, Sai-Yin HX, Qin LX, Yin J, Liu L, Wu C, Pei Y, Zhou YF, Zhai Y, Lu PX, Tan A, Zuo XB, Fan J, Chang J, Gu X, Wang NJ, Li Y, Liu YK, Zhai K, Zhang H, Hu Z, Liu J, Yi Q, Xiang Y, Shi R, Ding Q, Zheng W, Shu XO, Mo Z, Shugart YY, Zhang XJ, Zhou G, Shen H, Zheng SL, Xu J, Yu L (2013) Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet 45: 72–5. doi: 10.1038/ng.2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F (2014) Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 23: 700–13. doi: 10.1158/1055-9965.EPI-13-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili H, Gong J, Brenner H, Austin TR, Hutter CM, Baba Y, Baron JA, Berndt SI, Bezieau S, Caan B, Campbell PT, Chang-Claude J, Chanock SJ, Chen C, Hsu L, Jiao S, Conti DV, Duggan D, Fuchs CS, Gala M, Gallinger S, Haile RW, Harrison TA, Hayes R, Hazra A, Henderson B, Haiman C, Hoffmeister M, Hopper JL, Jenkins MA, Kolonel LN, Kury S, LaCroix A, Marchand LL, Lemire M, Lindor NM, Ma J, Manson JE, Morikawa T, Nan H, Ng K, Newcomb PA, Nishihara R, Potter JD, Qu C, Schoen RE, Schumacher FR, Seminara D, Taverna D, Thibodeau S, Wactawski-Wende J, White E, Wu K, Zanke BW, Casey G, Hudson TJ, Kraft P, Peters U, Slattery ML, Ogino S, Chan AT, Gecco, Ccfr (2015) Identification of a common variant with potential pleiotropic effect on risk of inflammatory bowel disease and colorectal cancer. Carcinogenesis 36: 999–1007. doi: 10.1093/carcin/bgv086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH, Neoptolemos JP (2016) Pancreatic cancer. Nat Rev Dis Primers 2: 16022. doi: 10.1038/nrdp.2016.22 [DOI] [PubMed] [Google Scholar]
- Klein AP, Wolpin BM, Risch HA, Stolzenberg-Solomon RZ, Mocci E, Zhang M, Canzian F, Childs EJ, Hoskins JW, Jermusyk A, Zhong J, Chen F, Albanes D, Andreotti G, Arslan AA, Babic A, Bamlet WR, Beane-Freeman L, Berndt SI, Blackford A, Borges M, Borgida A, Bracci PM, Brais L, Brennan P, Brenner H, Bueno-de-Mesquita B, Buring J, Campa D, Capurso G, Cavestro GM, Chaffee KG, Chung CC, Cleary S, Cotterchio M, Dijk F, Duell EJ, Foretova L, Fuchs C, Funel N, Gallinger S, JM MG, Gazouli M, Giles GG, Giovannucci E, Goggins M, Goodman GE, Goodman PJ, Hackert T, Haiman C, Hartge P, Hasan M, Hegyi P, Helzlsouer KJ, Herman J, Holcatova I, Holly EA, Hoover R, Hung RJ, Jacobs EJ, Jamroziak K, Janout V, Kaaks R, Khaw KT, Klein EA, Kogevinas M, Kooperberg C, Kulke MH, Kupcinskas J, Kurtz RJ, Laheru D, Landi S, Lawlor RT, Lee IM, LeMarchand L, Lu L, Malats N, Mambrini A, Mannisto S, Milne RL, Mohelnikova-Duchonova B, Neale RE, Neoptolemos JP, Oberg AL, Olson SH, Orlow I, Pasquali C, Patel AV, Peters U, Pezzilli R, Porta M, Real FX, Rothman N, Scelo G, Sesso HD, Severi G, Shu XO, Silverman D, Smith JP, Soucek P, et al. (2018) Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun 9: 556. doi: 10.1038/s41467-018-02942-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohoutova D, Smajs D, Moravkova P, Cyrany J, Moravkova M, Forstlova M, Cihak M, Rejchrt S, Bures J (2014) Escherichia coli strains of phylogenetic group B2 and D and bacteriocin production are associated with advanced colorectal neoplasia. BMC Infect Dis 14: 733. doi: 10.1186/s12879-014-0733-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, Uchiyama S, Matsumoto M, Nakajima A (2019) Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut 68: 1335–1337. doi: 10.1136/gutjnl-2018-316661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS (2013) Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14: 207–15. doi: 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22: 292–8. doi: 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruttgen A, Horz HP, Weber-Heynemann J, Vucur M, Trautwein C, Haase G, Luedde T, Roderburg C (2012) Study on the association of Helicobacter species with viral hepatitis-induced hepatocellular carcinoma. Gut Microbes 3: 228–33. doi: 10.4161/gmic.19922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T (2015) Colorectal cancer. Nat Rev Dis Primers 1: 15065. doi: 10.1038/nrdp.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukongviriyapan V (2012) Genetic polymorphism of drug metabolizing enzymes in association with risk of bile duct cancer. Asian Pac J Cancer Prev 13 Suppl: 7–15. [PubMed] [Google Scholar]
- Kunzmann AT, Proenca MA, Jordao HW, Jiraskova K, Schneiderova M, Levy M, Liska V, Buchler T, Vodickova L, Vymetalkova V, Silva AE, Vodicka P, Hughes DJ (2019) Fusobacterium nucleatum tumor DNA levels are associated with survival in colorectal cancer patients. Eur J Clin Microbiol Infect Dis 38: 1891–1899. doi: 10.1007/s10096-019-03649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, Sun R, Nosho K, Meyerhardt JA, Giovannucci E, Fuchs CS, Chan AT, Ogino S (2012) Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med 367: 1596–606. doi: 10.1056/NEJMoa1207756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsui D, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, Laird PW, Lynch CF, Anderson KE, French AJ, Haile RW, Harnack LJ, Potter JD, Slager SL, Smyrk TC, Thibodeau SN, Cerhan JR, Limburg PJ (2010) Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst 102: 1012–22. doi: 10.1093/jnci/djq201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Nevo D, Nishihara R, Cao Y, Song M, Twombly TS, Chan AT, Giovannucci EL, VanderWeele TJ, Wang M, Ogino S (2018a) Utility of inverse probability weighting in molecular pathological epidemiology. Eur J Epidemiol 33: 381–392. doi: 10.1007/s10654-017-0346-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tabung FK, Zhang X, Nowak JA, Qian ZR, Hamada T, Nevo D, Bullman S, Mima K, Kosumi K, da Silva A, Song M, Cao Y, Twombly TS, Shi Y, Liu H, Gu M, Koh H, Li W, Du C, Chen Y, Li C, Li W, Mehta RS, Wu K, Wang M, Kostic AD, Giannakis M, Garrett WS, Hutthenhower C, Chan AT, Fuchs CS, Nishihara R, Ogino S, Giovannucci EL (2018b) Diets That Promote Colon Inflammation Associate With Risk of Colorectal Carcinomas That Contain Fusobacterium nucleatum. Clin Gastroenterol Hepatol 16: 1622–1631 e3. doi: 10.1016/j.cgh.2018.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z (2019) Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 40: 336–348. doi: 10.1016/j.ebiom.2018.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G (2016) Hepatocellular carcinoma. Nat Rev Dis Primers 2: 16018. doi: 10.1038/nrdp.2016.18 [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vazquez-Baeza Y, White RA 3rd, Investigators I, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C (2019) Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569: 655–662. doi: 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, Yu J (2019) Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol 4: 2319–2330. doi: 10.1038/s41564-019-0541-3 [DOI] [PubMed] [Google Scholar]
- Lu W, Wang X, Zhan X, Gazdar A (2018) Meta-analysis approaches to combine multiple gene set enrichment studies. Stat Med 37: 659–672. doi: 10.1002/sim.7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Zhang Y, Xv C, Ji J, Lou G, Guo X, Chen M, Zhang Y, Wei H, Guo M, Huang R, Yu S (2019) Fusobacterium nucleatum, the communication with colorectal cancer. Biomed Pharmacother 116: 108988. doi: 10.1016/j.biopha.2019.108988 [DOI] [PubMed] [Google Scholar]
- Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan W, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF (2018) Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360. doi: 10.1126/science.aan5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, Nowak JA, Kosumi K, Hamada T, Masugi Y, Bullman S, Drew DA, Kostic AD, Fung TT, Garrett WS, Huttenhower C, Wu K, Meyerhardt JA, Zhang X, Willett WC, Giovannucci EL, Fuchs CS, Chan AT, Ogino S (2017) Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol 3: 921–927. doi: 10.1001/jamaoncol.2016.6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud DS, Joshipura K, Giovannucci E, Fuchs CS (2007) A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J Natl Cancer Inst 99: 171–5. doi: 10.1093/jnci/djk021 [DOI] [PubMed] [Google Scholar]
- Michaud DS, Lu J, Peacock-Villada AY, Barber JR, Joshu CE, Prizment AE, Beck JD, Offenbacher S, Platz EA (2018) Periodontal Disease Assessed Using Clinical Dental Measurements and Cancer Risk in the ARIC Study. J Natl Cancer Inst 110: 843–854. doi: 10.1093/jnci/djx278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, Shi Y, Song M, da Silva A, Gu M, Li W, Hamada T, Kosumi K, Hanyuda A, Liu L, Kostic AD, Giannakis M, Bullman S, Brennan CA, Milner DA, Baba H, Garraway LA, Meyerhardt JA, Garrett WS, Huttenhower C, Meyerson M, Giovannucci EL, Fuchs CS, Nishihara R, Ogino S (2016a) Fusobacterium nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin Transl Gastroenterol 7: e200. doi: 10.1038/ctg.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S (2016b) Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65: 1973–1980. doi: 10.1136/gutjnl-2015-310101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Ogino S, Nakagawa S, Sawayama H, Kinoshita K, Krashima R, Ishimoto T, Imai K, Iwatsuki M, Hashimoto D, Baba Y, Sakamoto Y, Yamashita YI, Yoshida N, Chikamoto A, Ishiko T, Baba H (2017) The role of intestinal bacteria in the development and progression of gastrointestinal tract neoplasms. Surg Oncol 26: 368–376. doi: 10.1016/j.suronc.2017.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, Kostic AD, Giannakis M, Watanabe H, Bullman S, Milner DA, Harris CC, Giovannucci E, Garraway LA, Freeman GJ, Dranoff G, Chan AT, Garrett WS, Huttenhower C, Fuchs CS, Ogino S (2015) Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol 1: 653–61. doi: 10.1001/jamaoncol.2015.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y (2015) Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 6: 7209–20. doi: 10.18632/oncotarget.3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, Lee CH (2015) Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 149: 102–109 e6. doi: 10.1053/j.gastro.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Momen-Heravi F, Babic A, Tworoger SS, Zhang L, Wu K, Smith-Warner SA, Ogino S, Chan AT, Meyerhardt J, Giovannucci E, Fuchs C, Cho E, Michaud DS, Stampfer MJ, Yu YH, Kim D, Zhang X (2017) Periodontal disease, tooth loss and colorectal cancer risk: Results from the Nurses’ Health Study. Int J Cancer 140: 646–652. doi: 10.1002/ijc.30486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G, Michel A, Taylor PR, Albanes D, Weinstein SJ, Virtamo J, Parisi D, Snyder K, Butt J, McGlynn KA, Koshiol J, Pawlita M, Lai GY, Abnet CC, Dawsey SM, Freedman ND (2014) Association of seropositivity to Helicobacter species and biliary tract cancer in the ATBC study. Hepatology 60: 1963–71. doi: 10.1002/hep.27193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja V, Eslick GD (2014) Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther 39: 745–50. doi: 10.1111/apt.12655 [DOI] [PubMed] [Google Scholar]
- Nahon P, Zucman-Rossi J (2012) Single nucleotide polymorphisms and risk of hepatocellular carcinoma in cirrhosis. J Hepatol 57: 663–74. doi: 10.1016/j.jhep.2012.02.035 [DOI] [PubMed] [Google Scholar]
- Nakao M, Hosono S, Ito H, Watanabe M, Mizuno N, Yatabe Y, Yamao K, Ueda R, Tajima K, Tanaka H, Matsuo K (2011) Interaction between IGF-1 polymorphisms and overweight for the risk of pancreatic cancer in Japanese. Int J Mol Epidemiol Genet 2: 354–66. [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ (2015) Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 6: 8727. doi: 10.1038/ncomms9727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Lam TY, Yu AC, Wang X, Chen Z, Wong MC, Ng SC, Chan MTV, Chan PKS, Chan FKL, Sung JJ, Yu J (2018) Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 155: 529–541 e5. doi: 10.1053/j.gastro.2018.04.018 [DOI] [PubMed] [Google Scholar]
- Nan H, Hutter CM, Lin Y, Jacobs EJ, Ulrich CM, White E, Baron JA, Berndt SI, Brenner H, Butterbach K, Caan BJ, Campbell PT, Carlson CS, Casey G, Chang-Claude J, Chanock SJ, Cotterchio M, Duggan D, Figueiredo JC, Fuchs CS, Giovannucci EL, Gong J, Haile RW, Harrison TA, Hayes RB, Hoffmeister M, Hopper JL, Hudson TJ, Jenkins MA, Jiao S, Lindor NM, Lemire M, Le Marchand L, Newcomb PA, Ogino S, Pflugeisen BM, Potter JD, Qu C, Rosse SA, Rudolph A, Schoen RE, Schumacher FR, Seminara D, Slattery ML, Thibodeau SN, Thomas F, Thornquist M, Warnick GS, Zanke BW, Gauderman WJ, Peters U, Hsu L, Chan AT, Ccfr, Gecco (2015) Association of aspirin and NSAID use with risk of colorectal cancer according to genetic variants. JAMA 313: 1133–42. doi: 10.1001/jama.2015.1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H, Morikawa T, Suuriniemi M, Imamura Y, Werner L, Kuchiba A, Yamauchi M, Hunter DJ, Kraft P, Giovannucci EL, Fuchs CS, Ogino S, Freedman ML, Chan AT (2013) Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J Natl Cancer Inst 105: 1852–61. doi: 10.1093/jnci/djt331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Hsu Y, Stebbins CE (2004) Assembly and function of a bacterial genotoxin. Nature 429: 429–33. doi: 10.1038/nature02532 [DOI] [PubMed] [Google Scholar]
- Nevo D, Nishihara R, Ogino S, Wang M (2018) The competing risks Cox model with auxiliary case covariates under weaker missing-at-random cause of failure. Lifetime Data Anal 24: 425–442. doi: 10.1007/s10985-017-9401-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara R, Morikawa T, Kuchiba A, Lochhead P, Yamauchi M, Liao X, Imamura Y, Nosho K, Shima K, Kawachi I, Qian ZR, Fuchs CS, Chan AT, Giovannucci E, Ogino S (2013a) A prospective study of duration of smoking cessation and colorectal cancer risk by epigenetics-related tumor classification. Am J Epidemiol 178: 84–100. doi: 10.1093/aje/kws431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT (2013b) Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med 369: 1095–105. doi: 10.1056/NEJMoa1301969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P (1991) Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 253: 665–9. doi: 10.1126/science.1651563 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Ishikawa K, Ishigami M, Honda T, Achiwa K, Izumoto T, Maekawa R, Hosokawa K, Iida A, Seino Y, Hamada Y, Goto H, Oiso Y, Arima H, Tsunekawa S (2015) Effect of hyperglycemia on hepatocellular carcinoma development in diabetes. Biochem Biophys Res Commun 463: 344–50. doi: 10.1016/j.bbrc.2015.05.066 [DOI] [PubMed] [Google Scholar]
- Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E (2006) Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313: 848–51. doi: 10.1126/science.1127059 [DOI] [PubMed] [Google Scholar]
- O’Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PG, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG (2015) Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 6: 6342. doi: 10.1038/ncomms7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Chan AT, Fuchs CS, Giovannucci E (2011) Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut 60: 397–411. doi: 10.1136/gut.2010.217182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, King EE, Beck AH, Sherman ME, Milner DA, Giovannucci E (2012) Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am J Epidemiol 176: 659–67. doi: 10.1093/aje/kws226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nishihara R, VanderWeele TJ, Wang M, Nishi A, Lochhead P, Qian ZR, Zhang X, Wu K, Nan H, Yoshida K, Milner DA Jr., Chan AT, Field AE, Camargo CA Jr, Williams MA, o EL (2016) Review Article: The Role of Molecular Pathological Epidemiology in the Study of Neoplastic and Non-neoplastic Diseases in the Era of Precision Medicine. Epidemiology 27: 602–11. doi: 10.1097/EDE.0000000000000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nowak JA, Hamada T, Milner DA Jr., Nishihara R (2019) Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol 14: 83–103. doi: 10.1146/annurev-pathmechdis-012418-012818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nowak JA, Hamada T, Phipps AI, Peters U, Milner DA Jr., Giovannucci EL, Nishihara R, Giannakis M, Garrett WS, Song M (2018) Integrative analysis of exogenous, endogenous, tumour and immune factors for precision medicine. Gut 67: 1168–1180. doi: 10.1136/gutjnl-2017-315537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Stampfer M (2010) Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst 102: 365–7. doi: 10.1093/jnci/djq031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, Kouri M, Elomaa I, Joensuu H (2007) Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br J Cancer 97: 1028–34. doi: 10.1038/sj.bjc.6603990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, Xuan W, Lin E, Mitchell HM, Borody TJ (2017) Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389: 1218–1228. doi: 10.1016/S0140-6736(17)30182-4 [DOI] [PubMed] [Google Scholar]
- Pereira P, Aho V, Arola J, Boyd S, Jokelainen K, Paulin L, Auvinen P, Farkkila M (2017) Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS One 12: e0182924. doi: 10.1371/journal.pone.0182924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, Jacobs EJ, Gapstur SM, Hayes RB, Ahn J (2017) Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res 77: 6777–6787. doi: 10.1158/0008-5472.CAN-17-1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps AI, Buchanan DD, Makar KW, Burnett-Hartman AN, Coghill AE, Passarelli MN, Baron JA, Ahnen DJ, Win AK, Potter JD, Newcomb PA (2012) BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev 21: 1792–8. doi: 10.1158/1055-9965.EPI-12-0674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, Sinicrope FA, Rosty C, Buchanan DD, Potter JD, Newcomb PA (2015) Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 148: 77–87 e2. doi: 10.1053/j.gastro.2014.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C, Puschhof J, Huber AR, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, van der Linden R, van Elst S, Genomics England Research C, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H (2020) Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. doi: 10.1038/s41586-020-2080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, Mohan N, Aykut B, Usyk M, Torres LE, Werba G, Zhang K, Guo Y, Li Q, Akkad N, Lall S, Wadowski B, Gutierrez J, Kochen Rossi JA, Herzog JW, Diskin B, Torres-Hernandez A, Leinwand J, Wang W, Taunk PS, Savadkar S, Janal M, Saxena A, Li X, Cohen D, Sartor RB, Saxena D, Miller G (2018) The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 8: 403–416. doi: 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K (2015) Biliary tract: Therapeutic strategies for cholangiocarcinoma-wishing on WNT inhibitors? Nat Rev Gastroenterol Hepatol 12: 187. doi: 10.1038/nrgastro.2015.39 [DOI] [PubMed] [Google Scholar]
- Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Chen X, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S (2019) Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68: 1014–1023. doi: 10.1136/gutjnl-2017-315084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescigno T, Micolucci L, Tecce MF, Capasso A (2017) Bioactive Nutrients and Nutrigenomics in Age-Related Diseases. Molecules 22. doi: 10.3390/molecules22010105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, Quesada P, Sahin I, Chandra V, San Lucas A, Scheet P, Xu H, Hanash SM, Feng L, Burks JK, Do KA, Peterson CB, Nejman D, Tzeng CD, Kim MP, Sears CL, Ajami N, Petrosino J, Wood LD, Maitra A, Straussman R, Katz M, White JR, Jenq R, Wargo J, McAllister F (2019) Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 178: 795–806 e12. doi: 10.1016/j.cell.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha M, Avenaud P, Menard A, Le Bail B, Balabaud C, Bioulac-Sage P, de Magalhaes Queiroz DM, Megraud F (2005) Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut 54: 396–401. doi: 10.1136/gut.2004.042168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen NG, Fuentes S, van der Spek MJ, Tijssen JG, Hartman JH, Duflou A, Lowenberg M, van den Brink GR, Mathus-Vliegen EM, de Vos WM, Zoetendal EG, D’Haens GR, Ponsioen CY (2015) Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients With Ulcerative Colitis. Gastroenterology 149: 110–118 e4. doi: 10.1053/j.gastro.2015.03.045 [DOI] [PubMed] [Google Scholar]
- Rosty C, Young JP, Walsh MD, Clendenning M, Sanderson K, Walters RJ, Parry S, Jenkins MA, Win AK, Southey MC, Hopper JL, Giles GG, Williamson EJ, English DR, Buchanan DD (2013) PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One 8: e65479. doi: 10.1371/journal.pone.0065479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW (2013) Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14: 195–206. doi: 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J (2015) Salmonella Manipulation of Host Signaling Pathways Provokes Cellular Transformation Associated with Gallbladder Carcinoma. Cell Host Microbe 17: 763–74. doi: 10.1016/j.chom.2015.05.002 [DOI] [PubMed] [Google Scholar]
- Schmit SL, Edlund CK, Schumacher FR, Gong J, Harrison TA, Huyghe JR, Qu C, Melas M, Van Den Berg DJ, Wang H, Tring S, Plummer SJ, Albanes D, Alonso MH, Amos CI, Anton K, Aragaki AK, Arndt V, Barry EL, Berndt SI, Bezieau S, Bien S, Bloomer A, Boehm J, Boutron-Ruault MC, Brenner H, Brezina S, Buchanan DD, Butterbach K, Caan BJ, Campbell PT, Carlson CS, Castelao JE, Chan AT, Chang-Claude J, Chanock SJ, Cheng I, Cheng YW, Chin LS, Church JM, Church T, Coetzee GA, Cotterchio M, Cruz Correa M, Curtis KR, Duggan D, Easton DF, English D, Feskens EJM, Fischer R, FitzGerald LM, Fortini BK, Fritsche LG, Fuchs CS, Gago-Dominguez M, Gala M, Gallinger SJ, Gauderman WJ, Giles GG, Giovannucci EL, Gogarten SM, Gonzalez-Villalpando C, Gonzalez-Villalpando EM, Grady WM, Greenson JK, Gsur A, Gunter M, Haiman CA, Hampe J, Harlid S, Harju JF, Hayes RB, Hofer P, Hoffmeister M, Hopper JL, Huang SC, Huerta JM, Hudson TJ, Hunter DJ, Idos GE, Iwasaki M, Jackson RD, Jacobs EJ, Jee SH, Jenkins MA, Jia WH, Jiao S, Joshi AD, Kolonel LN, Kono S, Kooperberg C, Krogh V, Kuehn T, Kury S, LaCroix A, Laurie CA, Lejbkowicz F, Lemire M, Lenz HJ, Levine D, et al. (2019) Novel Common Genetic Susceptibility Loci for Colorectal Cancer. J Natl Cancer Inst 111: 146–157. doi: 10.1093/jnci/djy099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Lopez FK, Aviles-Jimenez F, Guitron-Cantu A, Valdez-Salazar HA, Leon-Carballo S, Guerrero-Perez L, Fox JG, Torres J (2015) Infection with Helicobacter bilis but not Helicobacter hepaticus was Associated with Extrahepatic Cholangiocarcinoma. Helicobacter 20: 223–30. doi: 10.1111/hel.12195 [DOI] [PubMed] [Google Scholar]
- Shimoyama T, Takahashi R, Abe D, Mizuki I, Endo T, Fukuda S (2010) Serological analysis of Helicobacter hepaticus infection in patients with biliary and pancreatic diseases. J Gastroenterol Hepatol 25 Suppl 1: S86–9. doi: 10.1111/j.1440-1746.2010.06224.x [DOI] [PubMed] [Google Scholar]
- Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, Cunningham D (2017) Oesophageal cancer. Nat Rev Dis Primers 3: 17048. doi: 10.1038/nrdp.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Chan AT, Sun J (2020) Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 158: 322–340. doi: 10.1053/j.gastro.2019.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Shin A, Oh JH, Kim J (2018) Effects of interactions between common genetic variants and alcohol consumption on colorectal cancer risk. Oncotarget 9: 6391–6401. doi: 10.18632/oncotarget.23997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart BW, Wild C, International Agency for Research on Cancer, World Health Organization (2014) World cancer report 2014. International Agency for Research on Cancer WHO Press, Lyon, France Geneva, Switzerland [Google Scholar]
- Suez J, Zmora N, Segal E, Elinav E (2019) The pros, cons, and many unknowns of probiotics. Nat Med 25: 716–729. doi: 10.1038/s41591-019-0439-x [DOI] [PubMed] [Google Scholar]
- Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estecio MR, Issa JP (2014) Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 74: 1311–8. doi: 10.1158/0008-5472.CAN-13-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Takahashi H, Matsuo Y, Okada Y, Takeyama H (2010) Effect of Helicobacter bilis infection on human bile duct cancer cells. Dig Dis Sci 55: 1905–10. doi: 10.1007/s10620-009-0946-6 [DOI] [PubMed] [Google Scholar]
- Tanaka F, Yamamoto K, Suzuki S, Inoue H, Tsurumaru M, Kajiyama Y, Kato H, Igaki H, Furuta K, Fujita H, Tanaka T, Tanaka Y, Kawashima Y, Natsugoe S, Setoyama T, Tokudome S, Mimori K, Haraguchi N, Ishii H, Mori M (2010) Strong interaction between the effects of alcohol consumption and smoking on oesophageal squamous cell carcinoma among individuals with ADH1B and/or ALDH2 risk alleles. Gut 59: 1457–64. doi: 10.1136/gut.2009.205724 [DOI] [PubMed] [Google Scholar]
- Tang H, Dong X, Hassan M, Abbruzzese JL, Li D (2011) Body mass index and obesity- and diabetes-associated genotypes and risk for pancreatic cancer. Cancer Epidemiol Biomarkers Prev 20: 779–92. doi: 10.1158/1055-9965.EPI-10-0845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Wei P, Duell EJ, Risch HA, Olson SH, Bueno-de-Mesquita HB, Gallinger S, Holly EA, Petersen GM, Bracci PM, McWilliams RR, Jenab M, Riboli E, Tjonneland A, Boutron-Ruault MC, Kaaks R, Trichopoulos D, Panico S, Sund M, Peeters PH, Khaw KT, Amos CI, Li D (2014) Genes-environment interactions in obesity- and diabetes-associated pancreatic cancer: a GWAS data analysis. Cancer Epidemiol Biomarkers Prev 23: 98–106. doi: 10.1158/1055-9965.EPI-13-0437-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, Gandini S, Serrano D, Tarallo S, Francavilla A, Gallo G, Trompetto M, Ferrero G, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Wirbel J, Schrotz-King P, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G, Cordero F, Dias-Neto E, Setubal JC, Tett A, Pardini B, Rescigno M, Waldron L, Naccarati A, Segata N (2019) Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 25: 667–678. doi: 10.1038/s41591-019-0405-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RM, Jobin C (2020) Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol 17: 53–64. doi: 10.1038/s41575-019-0242-7 [DOI] [PubMed] [Google Scholar]
- Tian J, Liu C, Liu G, Zuo C, Chen H (2019a) Cumulative evidence for association between genetic polymorphisms and esophageal cancer susceptibility: A review with evidence from meta-analysis and genome-wide association studies. Cancer Med 8: 1289–1305. doi: 10.1002/cam4.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Liu G, Zuo C, Liu C, He W, Chen H (2019b) Genetic polymorphisms and gastric cancer risk: a comprehensive review synopsis from meta-analysis and genome-wide association studies. Cancer Biol Med 16: 361–389. doi: 10.20892/j.issn.2095-3941.2018.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H, Adolph TE, Gerner RR, Moschen AR (2018) The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 33: 954–964. doi: 10.1016/j.ccell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Tilg H, Zmora N, Adolph TE, Elinav E (2020) The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol 20: 40–54. doi: 10.1038/s41577-019-0198-4 [DOI] [PubMed] [Google Scholar]
- Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G (2006) A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect 12: 782–6. doi: 10.1111/j.1469-0691.2006.01494.x [DOI] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ (2013) Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368: 407–15. doi: 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- Waluga M, Zorniak M, Fichna J, Kukla M, Hartleb M (2018) Pharmacological and dietary factors in prevention of colorectal cancer. J Physiol Pharmacol 69. doi: 10.26402/jpp.2018.3.02 [DOI] [PubMed] [Google Scholar]
- Wang H, Iwasaki M, Haiman CA, Kono S, Wilkens LR, Keku TO, Berndt SI, Tsugane S, Le Marchand L (2015a) Interaction between Red Meat Intake and NAT2 Genotype in Increasing the Risk of Colorectal Cancer in Japanese and African Americans. PLoS One 10: e0144955. doi: 10.1371/journal.pone.0144955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H (2009) IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med 206: 1457–64. doi: 10.1084/jem.20090207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Kuchiba A, Ogino S (2015b) A Meta-Regression Method for Studying Etiological Heterogeneity Across Disease Subtypes Classified by Multiple Biomarkers. Am J Epidemiol 182: 263–70. doi: 10.1093/aje/kwv040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Spiegelman D, Kuchiba A, Lochhead P, Kim S, Chan AT, Poole EM, Tamimi R, Tworoger SS, Giovannucci E, Rosner B, Ogino S (2016) Statistical methods for studying disease subtype heterogeneity. Stat Med 35: 782–800. doi: 10.1002/sim.6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ST, Cui WQ, Pan D, Jiang M, Chang B, Sang LX (2020) Tea polyphenols and their chemopreventive and therapeutic effects on colorectal cancer. World J Gastroenterol 26: 562–597. doi: 10.3748/wjg.v26.i6.562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM (2008) Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res 68: 9909–17. doi: 10.1158/0008-5472.CAN-08-1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen TD, Yang Y, Moore DR, Huycke MM (2013) Cyclooxygenase-2 generates the endogenous mutagen trans-4-hydroxy-2-nonenal in Enterococcus faecalis-infected macrophages. Cancer Prev Res (Phila) 6: 206–16. doi: 10.1158/1940-6207.CAPR-12-0350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huycke MM (2007) Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology 132: 551–61. doi: 10.1053/j.gastro.2006.11.040 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang Y, Moore DR, Nimmo SL, Lightfoot SA, Huycke MM (2012) 4-hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology 142: 543–551 e7. doi: 10.1053/j.gastro.2011.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Fox JG, Anver MR, Haines DC, George CV, Collins MJ Jr., Gorelick PL, Nagashima K, Gonda MA, Gilden RV, et al. (1994) Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst 86: 1222–7. [DOI] [PubMed] [Google Scholar]
- Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA (2013) Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 1: 16. doi: 10.1186/2049-2618-1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Cao S, Liu S, Yao Z, Sun T, Li Y, Li J, Zhang D, Zhou Y (2016) Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients’ survival? A pilot study on relevant mechanism. Oncotarget. doi: 10.18632/oncotarget.10064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carra A, Brennan CA, Chun E, Ngo L, Samson LD, Engelward BP, Garrett WS, Balbo S, Balskus EP (2019) The human gut bacterial genotoxin colibactin alkylates DNA. Science 363. doi: 10.1126/science.aar7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, Sunagawa S, Coelho LP, Schrotz-King P, Vogtmann E, Habermann N, Nimeus E, Thomas AM, Manghi P, Gandini S, Serrano D, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Waldron L, Naccarati A, Segata N, Sinha R, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G (2019) Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 25: 679–689. doi: 10.1038/s41591-019-0406-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, Beane-Freeman L, Bracci PM, Buring J, Canzian F, Duell EJ, Gallinger S, Giles GG, Goodman GE, Goodman PJ, Jacobs EJ, Kamineni A, Klein AP, Kolonel LN, Kulke MH, Li D, Malats N, Olson SH, Risch HA, Sesso HD, Visvanathan K, White E, Zheng W, Abnet CC, Albanes D, Andreotti G, Austin MA, Barfield R, Basso D, Berndt SI, Boutron-Ruault MC, Brotzman M, Buchler MW, Bueno-de-Mesquita HB, Bugert P, Burdette L, Campa D, Caporaso NE, Capurso G, Chung C, Cotterchio M, Costello E, Elena J, Funel N, Gaziano JM, Giese NA, Giovannucci EL, Goggins M, Gorman MJ, Gross M, Haiman CA, Hassan M, Helzlsouer KJ, Henderson BE, Holly EA, Hu N, Hunter DJ, Innocenti F, Jenab M, Kaaks R, Key TJ, Khaw KT, Klein EA, Kogevinas M, Krogh V, Kupcinskas J, Kurtz RC, LaCroix A, Landi MT, Landi S, Le Marchand L, Mambrini A, Mannisto S, Milne RL, Nakamura Y, Oberg AL, Owzar K, Patel AV, Peeters PH, Peters U, Pezzilli R, Piepoli A, Porta M, Real FX, Riboli E, Rothman N, Scarpa A, Shu XO, Silverman DT, Soucek P, Sund M, Talar-Wojnarowska R, Taylor PR, Theodoropoulos GE, et al. (2014) Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 46: 994–1000. doi: 10.1038/ng.3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Yu J (2019) Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol 16: 690–704. doi: 10.1038/s41575-019-0209-8 [DOI] [PubMed] [Google Scholar]
- Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, Yang X, Cao G, Hu Z, Zhou Y, Zuo C, Wang C, Zhang X, Zhou Y, Yu X, Dai W, Li Z, Shen H, Liu L, Chen Y, Zhang S, Wang X, Zhai K, Chang J, Liu Y, Sun M, Cao W, Gao J, Ma Y, Zheng X, Cheung ST, Jia Y, Xu J, Tan W, Zhao P, Wu T, Wang C, Lin D (2011) Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet 44: 62–6. doi: 10.1038/ng.1020 [DOI] [PubMed] [Google Scholar]
- Wu J, Xu S, Xiang C, Cao Q, Li Q, Huang J, Shi L, Zhang J, Zhan Z (2018) Tongue Coating Microbiota Community and Risk Effect on Gastric Cancer. J Cancer 9: 4039–4048. doi: 10.7150/jca.25280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, Luan C, Liu Y, Wang B, Xiang C, Wang Y, Zhao F, Gao GF, Wang S, Li L, Zhang H, Zhu B (2013) Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 66: 462–70. doi: 10.1007/s00248-013-0245-9 [DOI] [PubMed] [Google Scholar]
- Wu S, Morin PJ, Maouyo D, Sears CL (2003) Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 124: 392–400. doi: 10.1053/gast.2003.50047 [DOI] [PubMed] [Google Scholar]
- Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL (2004) Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappaB pathway. Infect Immun 72: 5832–9. doi: 10.1128/IAI.72.10.5832-5839.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL (2009) A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15: 1016–22. doi: 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Zhang M, Franco A, Sears CL (2007) Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci 120: 1944–52. doi: 10.1242/jcs.03455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Wang X, Huang F, Zhao A, Chen W, Yan J, Zhang Y, Lei S, Ge K, Zheng X, Liu J, Su M, Liu P, Jia W (2016a) Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int J Cancer 139: 1764–75. doi: 10.1002/ijc.30219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Wang X, Liu P, Wei R, Chen W, Rajani C, Hernandez BY, Alegado R, Dong B, Li D, Jia W (2016b) Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 7: 19355–66. doi: 10.18632/oncotarget.8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T (2019) Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 25: 968–976. doi: 10.1038/s41591-019-0458-7 [DOI] [PubMed] [Google Scholar]
- Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H (2016) Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated with Prognosis. Clin Cancer Res 22: 5574–5581. doi: 10.1158/1078-0432.CCR-16-1786 [DOI] [PubMed] [Google Scholar]
- Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, Chan AT, Giovannucci E, Fuchs C, Ogino S (2012) Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut 61: 847–54. doi: 10.1136/gutjnl-2011-300865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer MP, Wang P, Cai S, Goel A, Qin H, Ma Y (2017) Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 152: 851–866 e24. doi: 10.1053/j.gastro.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh YK, Chen Z, Wong MCS, Hui M, Yu J, Ng SC, Sung JJY, Chan FKL, Chan PKS (2020) Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut. doi: 10.1136/gutjnl-2019-319635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N (2013) Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499: 97–101. doi: 10.1038/nature12347 [DOI] [PubMed] [Google Scholar]
- Yu G, Gail MH, Shi J, Klepac-Ceraj V, Paster BJ, Dye BA, Wang GQ, Wei WQ, Fan JH, Qiao YL, Dawsey SM, Freedman ND, Abnet CC (2014) Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev 23: 735–41. doi: 10.1158/1055-9965.EPI-13-0855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Hu N, Wang L, Wang C, Han XY, Humphry M, Ravel J, Abnet CC, Taylor PR, Goldstein AM (2017) Gastric microbiota features associated with cancer risk factors and clinical outcomes: A pilot study in gastric cardia cancer patients from Shanxi, China. Int J Cancer 141: 45–51. doi: 10.1002/ijc.30700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y (2016) Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer 139: 1318–26. doi: 10.1002/ijc.30168 [DOI] [PubMed] [Google Scholar]
- Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN, Matsui D, Boyd NH, Gazarik KE, Heit MI, Nistico L, Kasi PM, Spirk TL, Byers B, Lloyd EJ, Landreneau RJ, Jobe BA (2016) Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer 16: 52. doi: 10.1186/s12885-016-2093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Matsuda K, Jia WH, Chang J, Kweon SS, Xiang YB, Shin A, Jee SH, Kim DH, Zhang B, Cai Q, Guo X, Long J, Wang N, Courtney R, Pan ZZ, Wu C, Takahashi A, Shin MH, Matsuo K, Matsuda F, Gao YT, Oh JH, Kim S, Jung KJ, Ahn YO, Ren Z, Li HL, Wu J, Shi J, Wen W, Yang G, Li B, Ji BT, Genetics, Epidemiology of Colorectal Cancer C, Brenner H, Schoen RE, Kury S, Colorectal Transdisciplinary S, Gruber SB, Schumacher FR, Stenzel SL, Colon Cancer Family R, Casey G, Hopper JL, Jenkins MA, Kim HR, Jeong JY, Park JW, Tajima K, Cho SH, Kubo M, Shu XO, Lin D, Zeng YX, Zheng W (2016) Identification of Susceptibility Loci and Genes for Colorectal Cancer Risk. Gastroenterology 150: 1633–1645. doi: 10.1053/j.gastro.2016.02.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Liu L, Zou L, Sheng W, Zhu B, Xiang H, Chen W, Chen J, Rui R, Zheng X, Yin J, Duan S, Yang B, Sun J, Lou J, Liu L, Xie D, Xu Y, Nie S, Miao X (2013) Genetic variations in the TGFbeta signaling pathway, smoking and risk of colorectal cancer in a Chinese population. Carcinogenesis 34: 936–42. doi: 10.1093/carcin/bgs395 [DOI] [PubMed] [Google Scholar]
- Zhou D, Wang JD, Weng MZ, Zhang Y, Wang XF, Gong W, Quan ZW (2013) Infections of Helicobacter spp. in the biliary system are associated with biliary tract cancer: a meta-analysis. Eur J Gastroenterol Hepatol 25: 447–54. doi: 10.1097/MEG.0b013e32835c0362 [DOI] [PubMed] [Google Scholar]
- Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, Zhang MJ, Rao V, Avina M, Mishra T, Johnson J, Lee-McMullen B, Chen S, Metwally AA, Tran TDB, Nguyen H, Zhou X, Albright B, Hong BY, Petersen L, Bautista E, Hanson B, Chen L, Spakowicz D, Bahmani A, Salins D, Leopold B, Ashland M, Dagan-Rosenfeld O, Rego S, Limcaoco P, Colbert E, Allister C, Perelman D, Craig C, Wei E, Chaib H, Hornburg D, Dunn J, Liang L, Rose SMS, Kukurba K, Piening B, Rost H, Tse D, McLaughlin T, Sodergren E, Weinstock GM, Snyder M (2019) Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 569: 663–671. doi: 10.1038/s41586-019-1236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, Galluzzi L, Viaud S, Vetizou M, Daillere R, Merad M, Kroemer G (2015) Cancer and the gut microbiota: an unexpected link. Sci Transl Med 7: 271ps1. doi: 10.1126/scitranslmed.3010473 [DOI] [PMC free article] [PubMed] [Google Scholar]


