Abstract
In order to identify the appropriate times for growth and development, organisms must sense and process information about the availability of nutrients, energy status, and environmental cues. For sessile eukaryotes like plants, integrating such information can be critical in life or death decisions. For nearly 30 years, the conserved phosphatidylinositol 3-kinase-related protein kinases (PIKKs) target of rapamycin (TOR) has been established as a central hub for integrating external and internal metabolic cues. Despite the functional conservation across eukaryotes, the TOR complex has evolved specific functional and mechanistic features in plants. Here we present recent findings on the plant TOR complex that highlight conserved and unique nature of this critical growth regulator and its role in multiple aspects of plant life.
Keywords: cellular nutrition and homeostasis, energy sensing, symbiotic relationships, circadian cycle, autophagy, phytohormones, photosynthesis
Should I stay or should I grow now?
All living organisms need to be able to identify when nutrient availability and environmental cues are conducive to growth and development. In eukaryotes, the conserved TOR kinase serves as a key regulator of growth by receiving inputs from numerous metabolic processes and environmental cues. When conditions are favorable for growth, TOR is active and promotes anabolic processes to drive growth while repressing catabolic processes, but when nutrients are limited or environmental stresses are present, TOR is inactivated and catabolic processes are promoted [1, 2].
The yeast Saccharomyces cerevisiae has two TOR genes: TOR1 and TOR2. Either TOR1 or TOR2 can form the rapamycin-sensitive TOR complex 1 (TORC1), while only TOR2 can form the rapamycin-insensitive TORC2 [3]. In mammalian systems, there is only one TOR gene, but the protein exists in two complexes that are highly similar to yeast TORC1 and TORC2 [4]. These two signaling complexes differ in core components and the substrates and cellular processes they affect. Aside from TOR, the core components of both mammalian and yeast TORC1 include RAPTOR/KOG1 (see Glossary) and LST8, while TORC2 includes SIN1/AVO1, RICTOR/AVO3 and LST8 [5] (Figure 1). While all plant systems examined to date have homologs of the TORC1 components RAPTOR and LST8, SIN1 and RICTOR appear to be absent in the plant lineage (Figure 2), indicating that plants may not form a conserved TORC2 [6].
Figure 1.
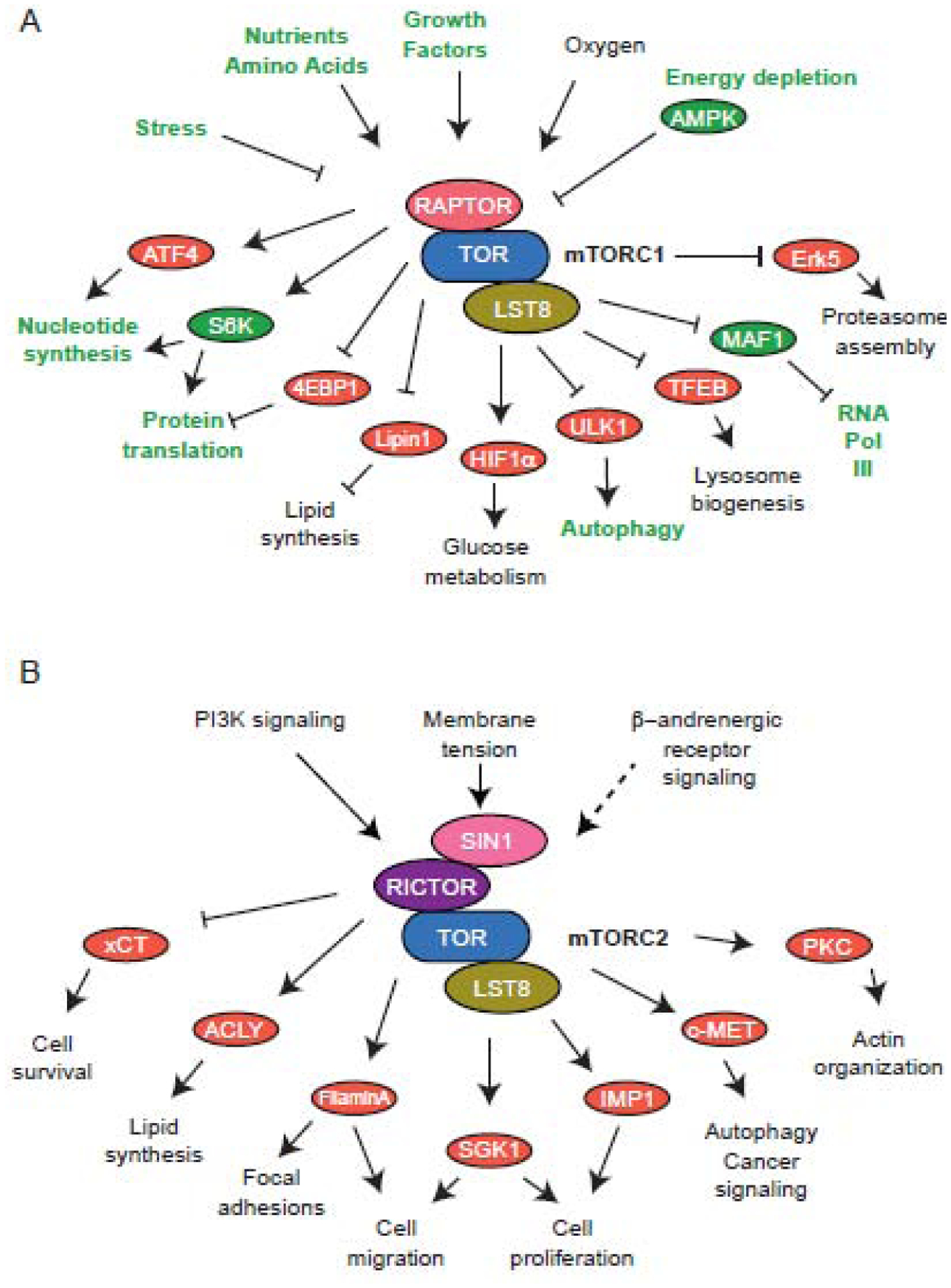
Overview of mammalian TOR complex signaling. A) Upstream activators/repressors and downstream effectors of mTORC1 regulate numerous metabolic and catabolic processes. Activators, effectors, and processes in green are conserved TOR–regulated processes in plants. B) Signaling through mTORC2 modulates diverse cellular functions.
Figure 2.
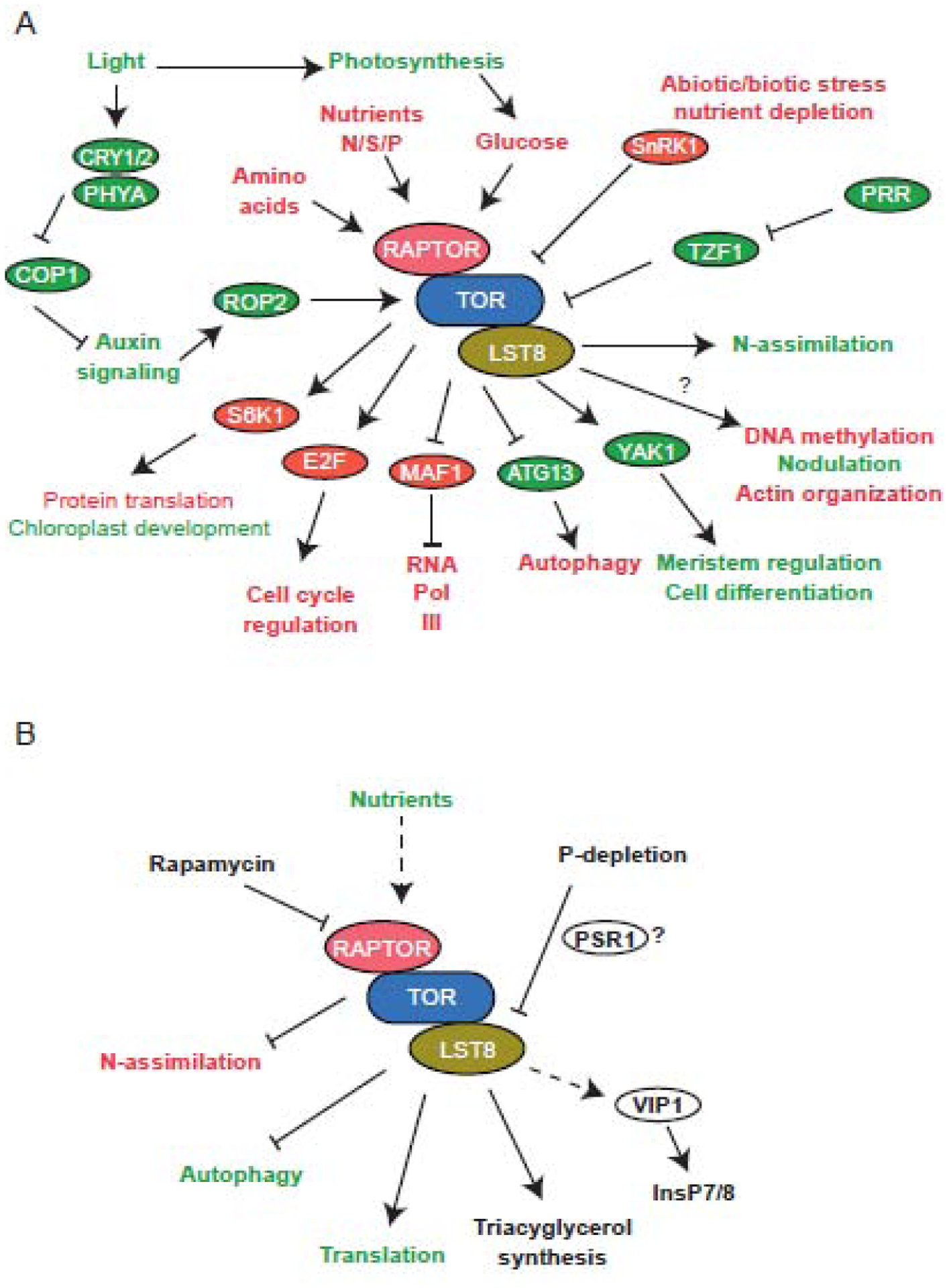
Summary of plant TOR signaling in A. thaliana and C. reinhardtii. A) Inputs and mechanisms for the activation/inhibition of TOR signaling and the downstream processes regulated by TOR in A. thaliana. Activators/inhibitors, effectors, and cellular processes in red are conserved with mammalian TOR signaling. Those in green are plant specific. B) Inputs and mechanisms for the activation/inhibition of TOR signaling and the downstream processes regulated by TOR in C. reinhardtii. Activators/inhibitors, effectors, and cellular processes in green are conserved with the Arabidopsis TOR signaling. N-assimiliation in C. reinhardtii is highlighted in red as TOR seems to have the opposite regulation on this process compared to Arabidopsis.
TORC1 is the better understood TOR signaling complex and is involved in the regulation of a host of cellular and developmental processes conserved between animals and plants such as growth and proliferation, protein translation, cell cycle, embryogenesis, stress responses, regulation of stem cells, and biosynthesis of nucleotides, lipids, and amino acids (Figure 1A) [1,7,8]. Some metazoan-specific processes controlled by TORC1 include lysosome biogenesis, neural development, and oncogenesis, while plant-specific examples include cell wall assembly, activation of meristems, growth of roots and leaves, and flowering [9,10]. TORC2 in mammalian cells regulates processes like cell survival, cell migration, focal adhesions, and metabolism of glucose and lipids (Figure 1B) [11,12]. In yeast, TORC2 regulates processes such as turgor pressure, membrane tension and homeostasis, actin cytoskeleton remodeling, and the pentose phosphate pathway [13,14]. While several TORC2-controlled processes are specific to metazoans, other processes that occur in yeast are relevant for plant growth and development, including cell wall integrity sensing, actin cytoskeleton organization, plasma membrane tension and homeostasis, and turgor pressure [11,13,14]. The absence of the canonical TORC2 components, namely RICTOR and SIN1, in plants poses that either certain processes are controlled by other regulators, or that plant TOR is able to regulate the same processes via TORC1 or perhaps through a novel protein complex.
Our knowledge of TOR signaling in plants lags behind metazoans and yeast. While there are many similarities, plants lack homologs of some of the genes involved signaling up or downstream of TOR. Plants also perform unique processes and, being sessile organisms, face different environmental challenges than animals. Here we present a broad overview of recent developments in plant TOR signaling, focusing on findings in the model alga and plant Chlamydomonas reinhardtii and Arabidopsis thaliana, respectively, and present some pressing questions to be addressed as we move onward in the study of TOR signaling in plants.
A TOR umbrella for the signaling of key nutrients
Carbon (C), nitrogen (N), sulfur (S), and phosphorus (P) are essential nutrients for building key macromolecules. Little is known about how plants sense levels of C, N, S, and P, but evidence supports an involvement of TOR. C levels are sensed through the levels of photosynthates, such as glucose. The mechanisms by which glucose activates plant TOR signaling are not yet understood, but it has been established that the glucose-TOR signaling activates genes involved the biosynthesis of nucleotides, amino acids, proteins, and lipids while repressing genes involved in the catabolism of these products (Figure 2A) (for an in-depth review, see [15]).
Rapamycin-mediated inhibition of TOR in C. reinhardtii leads to an increase in N assimilation with a subsequent increase in amino acid production (Figure 2B) [16], while silencing of TOR or the TOR effector Type 2A phosphatase-associated protein of 46 kDa (TAP46) genes in Arabidopsis results in down-regulation of genes involved in N-assimilation and increased expression of genes involved in N-recycling [17,18], indicating that TOR signaling does play a role in regulating N uptake and incorporation but that role may differ between Arabidopsis and C. reinhardtii. Plant TOR signaling has also been implicated in sensing available N, but the exact mechanisms remain unclear [19]. Known N-sensing mechanisms, such as the general control non-derepressible 2 (GCN2) pathway, sensing of glutamate by PII plastidic proteins and the family of glutamate receptor-like (GLR) proteins, are potential candidates to serve as an upstream N-sensor for TOR signaling [19]. GCN2, a eukaryotic initiation factor 2α (eIF2α) kinase, has been implicated as a sensor for C and N precursors for cysteine biosynthesis [20].
Depletion of S precursors for cysteine biosynthesis through the mutation of sulfite reductase sir1–1 inhibits TOR activity through the down-regulation of glucose metabolism, resulting in decreases in protein translation, meristematic activity, and elevated autophagy [20]. This same group found that combining the sir1–1 mutant with cad2–1, a cadmium-sensitive mutant that exhibits reduced glutathione synthesis, ameliorates some of the growth reduction observed in sir1–1 plants and increases ribosome abundance when TOR is activated, indicating that TOR regulates the allocation of cysteine between glutathione and protein synthesis [21]. Such a mechanism of detecting cysteine precursors may enable plants to finely tune responses to limitations of specific nutrients.
In plants, whether TOR is also involved in P signaling and in the well-known role of P in organ growth [22] is yet to be established. In Arabidopsis, the inositol pentakisphosphate 2-kinase (AtIPK1) gene plays an important role in the regulation of phosphate homeostasis, as partial loss-of-function mutants have reduced levels of inositol hexakisphosphate (InsP6) and altered expression of genes involved in Pi uptake, allocation and starvation response genes [23] In C. reinhardtii, phosphate starvation regulator protein 1 (PSR1) is a key transcription factor in P depletion conditions in C. reinhardtii [24]. In response to P depletion LST8 levels decrease, which results in lower TOR activity and reduced phosphorylation of the ribosomal protein S6 (RPS6), an established indirect downstream target of TOR signaling. On the other hand, in psr1 mutants LST8 levels do not decrease in P deplete conditions and RPS6 phosphorylation is actually increased [25]. Phosphate starvation response 1 (PHR1) in Arabidopsis belongs to the same family of transcription factors as PSR1 in C. reinhardtii [26]. Although it is not as responsive to Pi starvation as PSR1, further investigation to determine if PHR1 is involved in TOR signaling could be worthwhile. Mutation of VIP1, an inositol hexakisphosphate kinase in C. reinhardtii, displays increased sensitivity to rapamycin, and reduced levels of the signaling molecules inositol phosphates InsP7 and InsP8 [27]. These results imply that PSR1 is involved in TOR signaling for P availability, possibly through InsP signaling, at least in algae; however, the mechanisms that connect PSR1 to changes in TOR are yet unknown (Figure 2B).
In all, the available evidence to date hints to a functional connection between nutrient sensing and TOR activity, but the underlying mechanisms are yet to be determined. Some of such mechanisms may be direct or indirect (i.e., signaling for one nutrient may share or impact signaling components for a different nutrient), underscoring that, as key nutrients’ signaling and monitoring proteins are identified or studied in the context of TOR signaling, it will be critical to establish how and if they relay information directly to TOR.
Amino acid signaling and homeostasis controlled by TOR
Similar to mammals and yeast, amino acids (AAs) are one of the many inputs that stimulate TOR activity to promote growth in plants (Figure 2A) [8]. However, plants lack the mammalian and yeast RAG GTPases and Ragulator complex responsible for AA sensing [6,10]. Thus, the mechanisms for AA sensing in plants are unknown, but several recent studies have started to elaborate on the effects of particular AAs on TOR signaling. For example, sufficiently high levels of proline and alanine promote the activation of TOR. This in turn downregulates mitochondrial pathways that consume AAs for respiration and upregulates protein synthesis [28]. Loss-of-function mutations of isopropylmatae synthase 1 (IPMS1), a leucine biosynthetic gene, lead to increased accumulation of branched-chain AAs (BCAAs), most notably valine [29,30]. This results in activation of TOR leading to a variety defects in plant growth and development, such as an increase in the number of meristematic cells that are smaller and defects in chloroplast development compared to wild type [29,30]. Additional work is needed to determine if levels of specific AAs or AA biosynthetic precursors serve as identifiers of nutrient levels and how this information is incorporated into TOR signaling. Interestingly, increased TOR activity due to high levels of BCAAs results in a higher degree of actin bundling, which could be repressed by inducible RNA interference (RNAi) of TOR (Figure 2A) [29]. Furthermore, BCAA treatment was still able to induce actin bundling in raptor1b mutants, suggesting this likely occurs in a TORC1 independent manner [29]. Indeed, because actin organization is dependent on TORC2 in yeast and metazoans, these findings imply that, while the canonical TORC2 components are absent in plants, TOR is able to regulate the actin cytoskeleton in a unique manner in plants [29].
TOR influences symbiotic relationships
Plants have the ability to form symbiotic relationships with soil dwelling bacteria. Because these relationships involve nutrient exchange between plants and bacteria as well as organ development and growth, perhaps a role of TOR signaling in the establishment of symbiotic relationships is not entirely surprising. Indeed, TOR expression increases during nodulation of bean (Phaseolus vulgaris) and decreased TOR activity impairs infection thread development and alters nodule development [31]. Consistently, by producing auxin, the plant growth-promoting rhizobacterium Azospirillum brasilense increases TOR expression in the root meristem, stimulating root meristem cell division and lateral root formation [32]. One interesting process to look at in the future is the role of TOR in sensing nitrogen availability in relation to nodule development, which is inhibited when nitrogen availability is non-limiting [33].
Plant meristem regulation, cell differentiation, and an unexpected connection with the circadian clock
The apical meristems are key sources of stem cells, which can elongate to lengthen the shoot or root, or differentiate for organogenesis. TOR has a critical role in the proper maintenance of both root and shoot apical meristems, but the requirements for TOR activation in each are different. Glucose signaling is sufficient for activation of TOR in the root meristem; however, both glucose and light are required for TOR activation in the shoot meristem and for the transition to photoautotropic growth (Figure 2A) [34]. A picture of the components of TOR signaling in plants has been recently emerging. Treatment with the TOR inhibitor AZD8055 treatment reduces root meristem cell number and induces early cell differentiation. Loss-of-function mutants of YAK1, a downstream effector of TOR, are resistant to AZD8055, while YAK1 overexpression confers hypersensitivity to AZD8055 [35]. When TOR activity is inhibited, YAK1 activates siamese-related (SMR) cyclin dependent kinase inhibitors to induce cell differentiation (Figure 2A) [35]. Interestingly, TOR also plays a role in cell dedifferentiation. For instance, TOR phosphorylation and stabilization of the transcription factor E2Fa is critical for sugar-induced callus formation from differentiated tissue through the activation of S-phase genes necessary for cell cycle progression (Figure 2A) [36].
Links between meristem regulation through TOR signaling and the circadian clock have been recently uncovered. Members of the circadian-signaling Pseudo Response Regulators (PRR) family inhibit Tandem Zinc Finger 1 (TZF1), which binds to and affects the stability of TOR mRNA to represses TOR signaling-mediated root cell proliferation (Figure 2A) [37]. Furthermore, nicotinamide blocks glucose-TOR signaling effects on root meristem activation, root growth, and circadian period adjustment by inhibiting ATP production [38]. Similarly, chemical inhibition of TOR or knockdown of TOR transcripts lengthens the circadian period, similar to what is observed during chemical inhibition of the electron transport chain [39]. These findings support that TOR signaling may use ATP sensing as an input to modulate the circadian period and plant growth.
The emergent role of TOR in plant autophagy
The process of autophagy enables organisms to recycle cellular components and in plants is up-regulated in conditions of nutrient starvation and other growth-limiting stresses [40]. TOR signaling inhibits autophagy through pathways that are now emerging in plants. In Arabidopsis, the sucrose non-fermenting-related protein kinase 1 (SnRK1) acts upstream of TOR to repress TOR activity and promote autophagy (Figure 2A) [41]. This is likely executed via the SnRK1 subunit KIN10. Indeed, overexpression of KIN10 leads to elevated autophagy levels in stress conditions, while kin10 mutant plants fail to activate autophagy under similar conditions [41]. Conversely, overexpression of TOR inhibits activation of autophagy under starvation, salt, and osmotic stresses, but not oxidative or endoplasmic reticulum stress [42]. Similar results were observed when exogenous auxin was applied to activate TOR signaling [42].
Insights into the TOR-mediated inhibitory mechanisms of autophagy are provided by an analysis of ATG13, one of the members of the largely conserved autophagy-related proteins (ATG) that are necessary for various steps in autophagy. Similar to other RAPTOR-interacting proteins, such as ribosomal protein S6 kinase β−1 (S6K1), and protein phosphatase 2A/B (PP2A/B), ATG13 contains a conserved five AA motif, which was found to be required for interaction with RAPTOR and subsequent phosphorylation by TOR [43]. Similar to yeast, hyper-phosphorylated forms of ATG13 are found in nutrient-rich conditions, and in nutrient limiting conditions the ATG1/13 complex is degraded through autophagy [44,45]. TOR-phosphorylation of ATG13 in Arabidopsis appears to have a similar inhibitory effect on autophagy as supported by the evidence that the deletion of this motif from ATG13 in Arabidopsis leads to elevated levels of autophagy (Figure 2A) [43]. In C. reinhardtii, TOR inhibition by rapamycin treatment leads to elevated levels of ATG8, similar to the effects of oxidative stress, ER stress, and C/N limiting conditions, and is indicative of the activation of autophagy [46]. These results illustrate a feedback loop between TOR signaling and autophagy to increase the recycling of key resources in times of stress through modulation of the activity of proteins directly involved in autophagy.
Photosynthesis: a multilayered involvement of TOR
Photosynthesis is one of the most important processes in plants, producing sugars from carbon dioxide, water, and sunlight. TOR appears to be involved in several steps preceding photoautotrophy and maintenance of photosynthesis. As previously discussed, both glucose and light-induced auxin biosynthesis are required for TOR activation in the shoot meristem to facilitate conversion to photoautotropic growth in Arabidopsis [34]. Mechanistically, constitutive photomorphogenesis 1 (COP1) inhibits TOR activity during skotomorphogenic development of etiolated seedlings. Light perception by phytochrome A and cryptochrome inactivate COP1 to promote the auxin–mediated activation of TOR and the downstream phosphorylation of RPS6, enhancing protein translation in de-etiolating seedlings to promote cotyledon opening and the transition to photoautotrophic growth (Figure 2A) [47].
TOR signaling also induces expression of genes involved in chloroplasts and photosynthesis, while TOR inhibitors have been shown to reduce chloroplast number and size and thus impair photoautotrophic growth [48,49]. In rice, suppression of s6k1 and/or raptor2 yields pale yellow-green leaves and altered thylakoid membrane structure, and further investigation has demonstrated that S6K1 signaling is vital for galactolipid biosynthesis for the thylakoid membrane and fatty acid homeostasis (Figure 2A) [50].
Glucose production in leaves promotes TOR activity and thus TOR activity is higher in mature source leaves while developing sink leaves that still do not produce sufficient glucose leaves have lower TOR activity [51]. Higher TOR activity facilitates loading glucose into the phloem through plasmodesmata, channels between cells that allow the exchange of cytoplasmic contents, allowing for the transport of sugars from source to sink leaves [51].
An examination in C. reinhardtii of proteins that exhibited changes in oxidation during TOR inhibition revealed a reduction in photosynthetic electron flow and identified 20 proteins involved in photosynthesis with altered oxidation status [52]. The reduction in electron flow during inhibition of TOR was also observed in another study that also found that TOR inhibition caused altered chloroplast morphology, increased non-photochemical quenching and damage to photosystem II reaction centers [53]. In this same study it was observed that inhibition of TOR also led to an increase in fragmentation of mitochondria and increased mitochondrial respiration, suggesting that TOR balances chloroplast and mitochondria functions in algae [53].
While research has demonstrated that TOR signaling is important for the initiation of photoautotropic growth and the photosynthetic machinery, what is not known is if any other signals besides glucose allow TOR to sense and regulate photosynthetic processes once established.
TOR signaling and phytohormones
Phytohormones are small signaling molecules that are involved in developmental processes and responses to external stimuli [54]. In relation to TOR activity, these can be separated into two groups based on whether they promote or inhibit growth (Figure 3). When TOR activity is inhibited, the genes involved in the signaling pathways of phytohormones that promote growth (auxin, gibberellic acid, brassinosteroids, and cytokinins) are repressed, while those that inhibit growth (abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), and ethylene) are up-regulated [48]. Some of these changes in gene expression may result from changes in DNA methylation, as a new study has shown that inhibition of TOR results in decreased methylation of the genome and many of the differentially methylated genes are involved in plant hormone signaling and metabolic processes [55]. An involvement of Arabidopsis MAF1, a conserved stress-responsive Pol III repressor that is regulated by TOR [56], is also plausible but yet untested.
Figure 3.
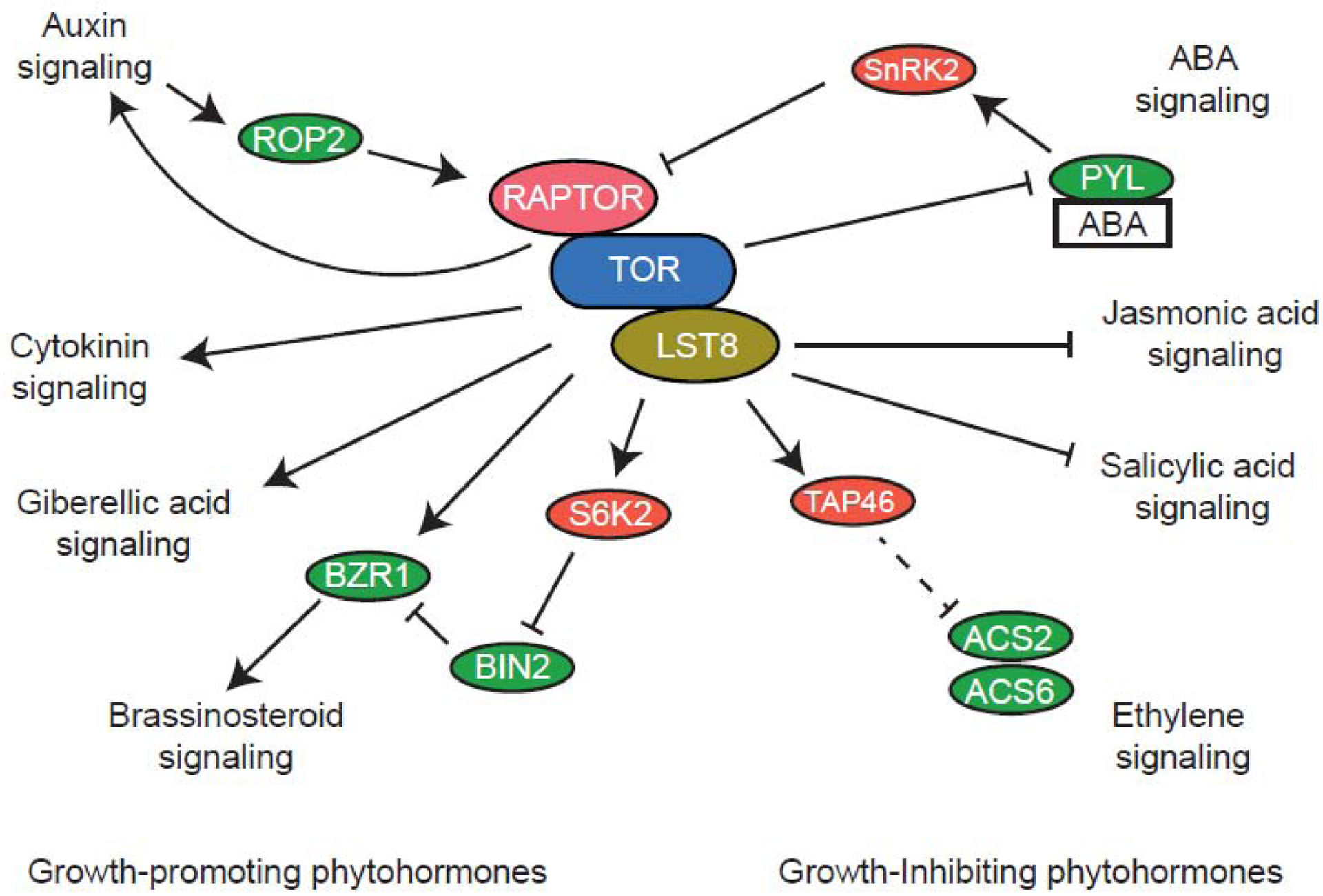
Overview of interaction between TOR-signaling and phytohormones in A. thaliana. Proteins in red are conserved regulators or effectors of TOR signaling in the mammalian system.
Here we report some of the recent discoveries about the interaction between phytohormone and TOR signaling.
Growth-promoting phytohormones
Auxin and its analogs are crucial for plant growth by regulating cell division in the shoot and root meristems, elongation of maturing cells, cell differentiation, and growth phenomena, such as gravitropism and phototropism [57]. The plant responses to auxin treatments are attenuated in the presence of TOR inhibitors, causing shorter primary roots, a reduced number of lateral roots, and a lack of gravitropism compared to plants treated with auxin alone [58]. When activated by exogenous auxin, the small GTPase Rho of Plants (ROP2) binds and activates TOR and, through the S6K1, promotes translation re-initiation at upstream open reading frames of genes involved in the regulation of stems cells and cell differentiation (Figure 3) [59,60]. Hence, ROP2 may be a component of the auxin-TOR integrating machinery for the control of cell differentiation and growth.
Brassinosteroids contribute to the regulation many cellular processes, including cell division and elongation, stress responses, and photomorphogenesis [61]. Chemical or genetic inhibition of TOR activity reduces the levels of brassinazole resistant 1 (BZR1), a brassinosteroid signaling transcription factor that promotes growth and the expression of brassinosteroid-responsive genes [62]. BZR1 is destabilized via phosphorylation by brassinosteroid insensitive 2 (BIN2), which was found to be a target of TOR through the ribosomal protein S6 kinase β−2 (S6K2), indicating that TOR-S6K2 signaling promotes the accumulation of BZR1 to facilitate this transition from heterotrophic to photoautotrophic growth (Figure 3) [49,63].
These findings uncover the existence of positive regulators of growth that respond to TOR. Future work may identify the growth-cease factors connected with TOR signaling that supersede the positive role of endogenous auxin and brassinosteroids to halt growth during development.
Growth-inhibiting phytohormones
ABA is a key regulator of plant responses to abiotic stresses, such as drought, and is also important for seed development and germination [64]. In the absence of stress, TOR signaling phosphorylates ABA receptors of the pyrabactin-like (PYL) protein family to inhibit ABA signaling (Figure 3) [65]. Conversely, in response to stress, ABA levels rise; ABA binds to PYL and inhibits PP2C proteins, allowing for the release of SnRK2 from the complex; SnRK2 then phosphorylates RAPTOR to inhibit TOR activity and growth (Figure 3) [65]. Inhibition of TOR or mutations in LST8–1 or RAPTOR lead to decreased ABA levels due to lowered expression of ABA biosynthetic genes, as well as an increased sensitivity to ABA in seed germination and plant growth [66]. Mutations in YAK1 suppress hypersensitivity to ABA in lst8–1 mutants [67]. In mammalian and yeast cells, TAP42 interacting protein of 41 kDa (TIP41) was shown to be a downstream effector of TOR interacting with PP2A, which facilitates multiple ABA-regulated processes in plants. Consistently, tip41 mutant plants are smaller and hypersensitive to both TOR inhibitors and ABA [68,69]. Together, these discoveries highlight a regulatory loop between ABA and TOR signaling but also that multiple factors may contribute to the antagonistic interactions between TOR and ABA signaling. These are likely needed to tune plants’ ability to control growth in response to the type and entity of environmental stresses.
JA plays a role in numerous plant growth and developmental processes, but may be best known for the role it plays in plant responses to abiotic and biotic stresses, such as wounding and challenge by pathogens [70]. Recent developments have started to uncover the antagonistic relationship between JA and TOR signaling. Compared to wild type, raptor1b mutants exhibit constitutively reduced JA levels in leaves. Nonetheless, upon wounding, these mutants can produce JA. Furthermore, treatment with the TOR inhibitor AZD8055 induces high levels of JA in both WT and raptor1b mutants [71]. Similarly, AZD8055 combined with methyljasmonate inhibits plant growth, indicating crosstalk between TOR and JA signaling [71,72]. In support of this, similar to the antagonistic relationship found between TOR activity and ABA response, plants with increased TOR activity were found to be more vulnerable to pathogens while TOR repression yields smaller, but more resistant plants [73]. Further investigation may uncover the mechanism for how JA can inhibit TOR signaling to shift from growth to defense, perhaps through a regulatory loop similar to that found for ABA.
Ethylene is a key phytohormone in the processes of ripening, leaf senescence and abscission, as well as plant growth and development and stress responses [74]. Similar to ABA and JA, inhibition of TOR increases expression of genes involved in senescence, ethylene signaling and biosynthesis [68]. Furthermore, ethylene-insensitive mutants display a reduced sensitivity to AZD8055 and ethylene signaling reduces the inhibition of hypocotyl growth in AZD8055-treated seedlings [75]. TAP46 interacts with the ethylene biosynthesis pathway proteins 1-aminocyclopropane-1-carboxylate synthase 2 (ACS2) and ACS6, which accumulate when TOR is inhibited, suggesting that TOR regulates ethylene signaling through modulation of ethylene production [68]. These findings underscore an antagonistic growth regulation by ethylene and TOR. It is possible that ethylene signaling may be also involved in inhibiting TOR signaling during senescence and perhaps future research can uncover the underlying mechanisms.
Concluding remarks
Compared to the noticeable progress in the study of TOR signaling mechanisms in mammalian and yeast systems, the knowledge of plant and algal TOR has many more gaps (see Outstanding Questions). Notably, while addition of individual AA has been shown to stimulate TOR activity, the mechanisms by which plants sense AA is yet unknown. Similarly, identifying the sensing mechanisms for essential nutrients and how these impact TOR signaling will be crucial for our understanding of how the processing of multiple inputs factors into plant growth. This knowledge may allow for a more in-depth examination of plant symbiosis with rhizobium and mycorrhizae. Understanding the crosstalk between phytohormone and TOR signaling pathways will allow for identifying key growth-promoting and growth-cease factors that direct plant growth and development, as well as stress responses. While plants lack the RICTOR and SIN1 components to form TORC2, many of those functions regulated by TORC2 in yeast such as membrane tension, turgor pressure, cell wall integrity, and actin organization are crucial for plants as well. While there is evidence that TOR activity impacts the actin cytoskeleton, it remains to be seen whether these other processes are regulated by TOR, and if so, is it done through TORC1 signaling or some novel, plant-specific mechanism. Lastly, while the role of TOR has been established in chloroplast development and the inhibition of TOR has been shown to impact photosynthesis, it seems that the status of such a crucial process would likely be sensed by TOR through more than just the production of sugars. Whether TOR activity is responsive to processes such as photodamage or photinhibition, or is involved in chloroplast repositioning during changes in light intensity, needs to be investigated.
Outstanding Questions.
How do plants monitor the availability of nutrients and how is that information relayed to TOR?
How are amino acid levels sensed in plants and how is that information relayed to TOR?
Do plants still utilize TOR to regulate cellular processes traditionally mediated by TORC2?
How do phytohormone signaling and TOR signaling pathways interact with each other?
Does a mechanism exist for TOR to monitor the photosynthesic apparatus?
Several mechanisms and functions attributed to TOR seem to have plant-specific features. While this makes it difficult to translate knowledge of TOR signaling in non-plant species to plants, it is also exciting. This is because we may be able to identify new pathways in plant TOR, which may eventually reveal convergent features with non-plant TOR signaling. For example, the involvement of plant TOR in symbiosis may reveal foundational mechanisms underpinning the establishment and maintenance of the microbiome in mammals. Above all, the availability of viable mutants in plant TOR signaling and the possibility to generate whole higher-order mutants of TOR with disrupted signaling pathways will be invaluable to query mechanisms for conserved processes with metazoans (e.g., cell growth and proliferation, organ elongation).
Highlights.
Plant and mammalian TOR signaling pathways share many conserved methods of activation, inhibition, and effectors, and regulate some overlapping cellular processes but some of the mammalian TOR core components are missing in plants.
TOR signaling in plants has evolved to regulate multiple plant-specific processes, some using known TOR effectors from mammalian TOR signaling.
Crosstalk between TOR and phytohormone signaling enables plants to modulate the growth response to adapt to a wide variety of biotic and abiotic stressors.
Acknowledgements
This work was funded primarily by National Science Foundation (MCB1727362), the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (award number DE-FG02-91ER20021), and AgBioResearch (MICL02598) to FB.
Glossary
- Cryptochrome
photoreceptor proteins that are sensitive to blue light
- Galactolipids
the dominant class of lipids present in plastid membranes; these are essential for the function of photosynthesis
- Gravitropism
differential growth in response to gravity
- Infection thread
an internal tubule formed within the root hair as rhizobia begin to infiltrate the plant during nodulation
- LST8
Lethal with SEC13 protein 8; a WD-40 repeat containing protein found in both mTORC1 and mTORC2 and serves as a scaffold for protein interaction
- Nodulation
the process of forming nitrogen-fixing root nodules containing symbiotic bacteria (rhizobia) in members of the legume family
- PII proteins
prokaryotic signal transduction proteins involved in nitrogen metabolism that are also found in plant plastids
- Phototropism
differential growth in response to light
- Phytochrome
photoreceptor proteins that respond to red and far-red light
- Pyrabactin
a synthetic sulfonamide that mimics ABA
- RAPTOR
regulatory-associated protein of mTOR; a component of mTORC1 that binds to substrates of TOR such as S6K
- RICTOR
rapamycin-insensitive companion of mTOR; a protein necessary for mTORC2 assembly
- SIN1
SAPK-interacting protein 1; a protein necessary for mTORC2 assembly
- Skotomorphogenesic development
the development of a seedling growing in the dark, characterized by a rapidly elongating hypocotyl and tight apical hook
- TAP42
type 2A associated phosphatase-associated protein 42; involved in the negative regulation of TOR signaling
- Thylakoid membranes
a system of interconnected membranes within the chloroplast that carry out the light reactions of photosynthesis
- YAK1
yet another kinase 1; a member of the dual specificity tyrosine phosphorylated kinase family
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dobrenel T et al. (2016). TOR Signaling and Nutrient Sensing. Annu Rev Plant Biol. 67(1): 261–285. [DOI] [PubMed] [Google Scholar]
- 2.Saxton RA and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 168(6): 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loewith R et al. (2002). Two TOR Complexes, Only One of which Is Rapamycin Sensitive, Have Distinct Roles in Cell Growth Control. Mol Cell 10(3): 457–468. [DOI] [PubMed] [Google Scholar]
- 4.Russell RC et al. (2011). An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development. 138(16): 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wullschleger S et al. (2006). TOR Signaling in Growth and Metabolism. Cell. 124(3): 471–484. [DOI] [PubMed] [Google Scholar]
- 6.Tatebe H and Shiozaki K (2017). Evolutionary Conservation of the Components in the TOR Signaling Pathways. Biomolecules. 7(4): 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Sahra I and Manning BD (2017). mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 45: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González A and Hall MN (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36(4): 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leiber RM et al. (2010). The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22(6): 1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L et al. (2018). TOR signaling in plants: conservation and innovation. Development. 145(13): dev160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie J et al. (2018). Who does TORC2 talk to? Biochem J. 475(10): 1721–1738. [DOI] [PubMed] [Google Scholar]
- 12.Luo Y et al. (2018). Weighing In on mTOR Complex 2 Signaling: The Expanding Role in Cell Metabolism. Oxid med cell longev. 7838647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaubitz C et al. (2016). TORC2 Structure and Function. Trends Biochem Sci. 41(6): 532–545. [DOI] [PubMed] [Google Scholar]
- 14.Riggi M et al. (2020). The flipside of the TOR coin - TORC2 and plasma membrane homeostasis at a glance. J Cell Sci. 133(9). [DOI] [PubMed] [Google Scholar]
- 15.Wu Y et al. (2019). Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J Exp Bot. 70(8): 2227–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mubeen U et al. (2018). Target of Rapamycin Inhibition in Chlamydomonas reinhardtii Triggers de Novo Amino Acid Synthesis by Enhancing Nitrogen Assimilation. Plant Cell. 30(10): 2240–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldana C et al. (2013). Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J 73(6): 897–909. [DOI] [PubMed] [Google Scholar]
- 18.Ahn CS et al. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23(1): 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gent L and Forde BG (2017). How do plants sense their nitrogen status? J Exp Bot. 68(10): 2531–2539. [DOI] [PubMed] [Google Scholar]
- 20.Dong Y et al. (2017). Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun. 8(1):1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Speiser A,M et al. (2018). Sulfur Partitioning between Glutathione and Protein Synthesis Determines Plant Growth. Plant Physiol. 177(3): 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motte H et al. (2019). Molecular and Environmental Regulation of Root Development. Annu Rev Plant Biol. 70(1): 465–488. [DOI] [PubMed] [Google Scholar]
- 23.Kuo HF et al. (2014). Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J 80(3): 503–515. [DOI] [PubMed] [Google Scholar]
- 24.Bajhaiya AK et al. (2016). PSR1 Is a Global Transcriptional Regulator of Phosphorus Deficiency Responses and Carbon Storage Metabolism in Chlamydomonas reinhardtii. Plant Physiol. 170(3): 1216–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couso I et al. (2020). Phosphorus Availability Regulates TORC1 Signaling via LST8 in Chlamydomonas. Plant Cell 32(1): 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubio V et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(16): 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couso I et al. (2016). Synergism between Inositol Polyphosphates and TOR Kinase Signaling in Nutrient Sensing, Growth Control, and Lipid Metabolism in Chlamydomonas. Plant Cell. 28(9): 2026–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Leary BM et al. (2020). Metabolite Regulatory Interactions Control Plant Respiratory Metabolism via Target of Rapamycin (TOR) Kinase Activation. Plant Cell. 32(3): 666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao P et al. (2019). Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis. Elife. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaufelberger M et al. (2019). Mutations in the Arabidopsis ROL17/isopropylmalate synthase 1 locus alter amino acid content, modify the TOR network, and suppress the root hair cell development mutant lrx1. J Exp Bot. 70(8): 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanjareddy K et al. (2016). A Legume TOR Protein Kinase Regulates Rhizobium Symbiosis and Is Essential for Infection and Nodule Development. Plant Physiol. 172(3): 2002–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Méndez-Gómez M et al. (2020). TARGET OF RAPAMYCIN signaling plays a role in Arabidopsis growth promotion by Azospirillum brasilense Sp245. Plant Sci. 293: 110416. [DOI] [PubMed] [Google Scholar]
- 33.Mortier V et al. (2012). Never too many? How legumes control nodule numbers. Plant Cell Environ. 35(2): 245–258. [DOI] [PubMed] [Google Scholar]
- 34.Li X et al. (2017). Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA. 114(10): 2765–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrada A et al. (2019). A TOR-YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis. Development. 146(3). [DOI] [PubMed] [Google Scholar]
- 36.Lee K and Seo PJ (2017). Arabidopsis TOR signaling is essential for sugar-regulated callus formation. J Integr Plant Biol. 59(10): 742–746. [DOI] [PubMed] [Google Scholar]
- 37.Li B et al. (2019). PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis. Nucleic Acids Res. 47(10): 5001–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N et al. (2019). Metabolite-mediated TOR signaling regulates the circadian clock in Arabidopsis. Proc Natl Acad Sci USA. 116(51): 25395–25397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y et al. (2020). Attenuated TOR signaling lengthens circadian period in Arabidopsis. Plant Signal Behav. 15(2): 1710935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P et al. (2018). New advances in autophagy in plants: Regulation, selectivity and function. Semin Cell Dev Biol. 80: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soto-Burgos J and Bassham DC (2017). SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS One. 12(8): e0182591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pu Y, et al. (2017). TOR-Dependent and -Independent Pathways Regulate Autophagy in Arabidopsis thaliana. Front Plant Sci. 8: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Son O et al. (2018). Involvement of TOR signaling motif in the regulation of plant autophagy. Biochem Biophys Res Commun. 501(3): 643–647. [DOI] [PubMed] [Google Scholar]
- 44.Puente C et al. (2016). Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy. J Biol Chem 291(11): 6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suttangkakul A et al. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23(10): 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pérez-Pérez ME et al. (2010). Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152(4): 1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen GH et al. (2018). TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc Natl Acad Sci USA 115(50): 12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong P et al. (2015). Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front Plant Sci. 6: 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong F et al. (2017). Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 213(1): 233–249. [DOI] [PubMed] [Google Scholar]
- 50.Sun L et al. (2016). Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochim Biophys Acta. 1861(7): 639–649. [DOI] [PubMed] [Google Scholar]
- 51.Brunkard JO et al. (2020). TOR dynamically regulates plant cell-cell transport. Proc Natl Acad Sci USA. 117(9): 5049–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ford MM et al. (2019). Inhibition of TOR in Chlamydomonas reinhardtii Leads to Rapid Cysteine Oxidation Reflecting Sustained Physiological Changes. Cells 8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Upadhyaya S and Rao BJ (2019). Reciprocal regulation of photosynthesis and mitochondrial respiration by TOR kinase in Chlamydomonas reinhardtii. Plant Direct 3(11): e00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubio V et al. (2009). Plant hormones and nutrient signaling. Plant Mol Biol. 69(4): 361–373. [DOI] [PubMed] [Google Scholar]
- 55.Zhu T et al. (2020). Target of Rapamycin Regulates Genome Methylation Reprogramming to Control Plant Growth in Arabidopsis. Front in Genet. 11(186). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahn CS, et al. (2019). Characterization of Maf1 in Arabidopsis: function under stress conditions and regulation by the TOR signaling pathway. Planta. 249(2): 527–542. [DOI] [PubMed] [Google Scholar]
- 57.Enders TA and Strader LC (2015). Auxin activity: Past, present, and future. Am J Bot. 102(2): 180–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng K et al. (2016). Target of Rapamycin Is a Key Player for Auxin Signaling Transduction in Arabidopsis. Front Plant Sci. 7(291). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schepetilnikov M et al. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32(8): 1087–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schepetilnikov M et al. (2017). GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 36(7): 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nolan TM et al. (2020). Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell. 32(2): 295–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z et al. (2016). TOR Signaling Promotes Accumulation of BZR1 to Balance Growth with Carbon Availability in Arabidopsis. Curr Biol. 26(14): 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He JX et al. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 99(15): 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng LM et al. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol Sin. 35(5): 567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang P et al. (2018). Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol Cell. 69(1): 100–112 e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kravchenko A et al. (2015). Mutations in the Arabidopsis Lst8 and Raptor genes encoding partners of the TOR complex, or inhibition of TOR activity decrease abscisic acid (ABA) synthesis. Biochem Biophys Res Commun. 467(4): 992–997. [DOI] [PubMed] [Google Scholar]
- 67.Forzani C et al. (2019). Mutations of the AtYAK1 Kinase Suppress TOR Deficiency in Arabidopsis. Cell Rep 27(12): 3696–3708. [DOI] [PubMed] [Google Scholar]
- 68.Punzo P et al. (2018). TIP41 network analysis and mutant phenotypes predict interactions between the TOR and ABA pathways. Plant Signal Behav. 13(12): e1537698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Punzo P et al. (2018). The PP2A-interactor TIP41 modulates ABA responses in Arabidopsis thaliana. Plant J. 94(6): 991–1009. [DOI] [PubMed] [Google Scholar]
- 70.Ruan J et al. (2019). Jasmonic Acid Signaling Pathway in Plants. Int J Mol Sci. 20(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salem MA and Giavalisco P (2019). Mutation in the Arabidopsis regulatory-associated protein TOR 1B (RAPTOR1B) leads to decreased jasmonates levels in leaf tissue. Plant Signal Behav. 14(10): e1649567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song Y et al. (2017). The crosstalk between Target of Rapamycin (TOR) and Jasmonic Acid (JA) signaling existing in Arabidopsis and cotton. Sci Rep. 7: 45830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Vleesschauwer D et al. (2018). Target of rapamycin signaling orchestrates growth-defense trade-offs in plants. New Phytol. 217(1): 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dubois M et al. (2018). The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci. 23(4): 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuo F et al. (2020). Target of Rapamycin (TOR) Negatively Regulates Ethylene Signals in Arabidopsis. Int J Mol Sci. 21(8). [DOI] [PMC free article] [PubMed] [Google Scholar]


