Abstract
Coordinated changes in gene expression allow for the single fertilized oocyte to develop into a complex, multi-cellular organism. These changes in expression are controlled by transcription factors that gain access to discrete cis-regulatory elements in the genome, allowing them to activate gene expression. While nucleosomes present barriers to transcription-factor occupancy, pioneer transcription factors have unique properties that allow them to bind DNA in the context of nucleosomes, define cis-regulatory elements and facilitate the subsequent binding of additional factors that determine gene expression. In this capacity, pioneer factors act at the top of gene regulatory networks to control developmental transitions. Nonetheless, developmental context also influences pioneer-factor binding and activity. Here we discuss the interplay between pioneer factors and development, both their role in driving developmental transitions and the influence that cellular environment has on pioneer-factor binding and activity.
During development, a single DNA genome is differentially interpreted to give rise to all of the distinct cell types of the organism. This process is mediated by sequence-specific, DNA-binding transcription factors that occupy discrete cis-regulatory regions and drive gene expression. However, the histone proteins that package the genome into chromatin limit the ability of many transcription factors to bind the underlying DNA (Luger et al., 1997; Zhu et al., 2018). A specialized class of transcription factors, termed pioneer factors, are uniquely capable of binding to histone-wrapped DNA, establishing accessible chromatin domains and facilitating the binding of additional transcription factors (reviewed in Zaret, 2020). These distinctive properties enable pioneer factors to define the cis-regulatory regions that are subsequently bound by additional transcription factors to drive gene expression. In this capacity, pioneer factors act at the top of gene-regulatory networks to affect transitions in cell fate. The unique features of pioneer factors contribute to their ability to drive reprogramming events both in culture and during development, as well as to their role in developmental diseases and cancer when mutated or misexpressed.
Pioneer factors are able to target DNA on the surface of nucleosomes, allowing them to bind regions of chromatin that are inaccessible to other transcription factors. Despite this shared central feature, pioneer factors utilize a multitude of different protein domains for this purpose (Fernandez Garcia et al., 2019). These domains enable pioneer factors to recognize specific DNA motifs even within the context of nucleosomes and can also allow for non-specific affinity for nucleosomes. Indeed, nonspecific nucleosome interactions may be essential for the function of some pioneer factors (Lerner et al., 2020). Systematic studies of interactions between transcription factors and nucleosomes identified diverse modes of binding, including some factors that bind to the nucleosomes dyad access and others that bind to the edge or across the DNA gyres (Zhu et al., 2018). Together these biochemical studies reflect the fact that nucleosome binding by pioneer factors is not achieved by a single mechanism (Zaret, 2020). Pioneer factors similarly use a diversity of mechanisms to drive chromatin accessibility, including partially unwrapping DNA from the nucleosome, inhibiting inter-nucleosome interactions, evicting histones and recruiting ATP-dependent chromatin-modifying enzymes (Zaret, 2020). While pioneer factors engage silent regions of chromatin using a variety of methods, this intrinsic biochemical property is a necessary feature that determines their ability to drive cell fate and cellular reprogramming.
Despite the specialized properties of pioneer factors, they do not occupy all the sequences in vivo that they can bind in vitro, and binding is often cell-type specific. Indeed, studies of pioneer factors at multiple stages of development have begun to elucidate barriers to pioneer-factor binding and function, and it has become evident that pioneer factors often cooperate with additional factors to drive changes in gene expression. To understand the cell-type specific differences in pioneer-factor binding and function, it has been important to complement biochemical experiments with functional studies of these factors over development. We have therefore focused this review on pioneer factors during development and refer you to a number of recent comprehensive reviews on the molecular mechanisms by which pioneer factors engage chromatin (Iwafuchi-Doi, 2019; Mayran and Drouin, 2018; Zaret, 2020). We discuss a subset of the major developmental transitions regulated by pioneer factors, provide examples of the role of developmental context in influencing pioneer-factor binding and activity, define specific barriers to pioneer-factor function and provide examples of how misexpression of pioneer factors can lead to disease.
Pioneer factors in developmental transitions
Dramatic changes in the transcriptome are required to drive developmental transitions. The ability of pioneer factors to access their binding sites even within the context of a nucleosome makes them uniquely capable of broadly restructuring chromatin accessibility, shifting the transcriptional profile of the cell and, in so doing, driving changes in cell fate. The original identification of pioneer factors was based on their role in liver development. Studies of the liver-specific alb1 enhancer identified FoxA and Gata4 occupancy in the embryo, well before gene expression was activated, suggesting that early these factors primed the gene for later expression (Gualdi et al., 1996). FoxA was shown to bind nucleosomes and create nuclease accessible regions in nucleosome arrays (Cirillo et al., 2002) and along with Gata4 is required for hepatic induction from endoderm (Lee et al., 2005). Since this original identification, numerous additional developmental transitions have been found to be regulated by transcription factors with pioneering activity.
Pioneer transcription factors are required for reprogramming of specified cell types to pluripotency both in culture and during early embryonic development (Figure 1). Over development, differentiated cells rarely return to a less differentiated state. Nonetheless, in culture, a cocktail of transcription factors (Oct4 (Pou5f4), Sox2, Klf4, and c-Myc; OSKM) reprograms mouse or human differentiated fibroblasts to induced pluripotent stem cells (iPSC) (Takahashi and Yamanaka, 2006). This reprogramming necessitates both the silencing of the fibroblast regulatory network and the activation of genes that drive pluripotency. Oct4, Sox2, and Klf4 (OSK) are required for silencing somatic enhancers as well as activation of pluripotency enhancers (Chronis et al., 2017). They function as pioneer factors, binding to closed chromatin and facilitating c-Myc binding (Soufi et al., 2012). The ability of OSK to access closed chromatin during reprogramming reflects their ability to bind nucleosomes in vitro, which is achieved by recognition of partial DNA motifs exposed on nucleosomes (Soufi et al., 2015). While each of these factors have essential features of pioneer factors, they do not function entirely independently and cooperative interactions amongst these factors are required to stabilize interactions with the genome. Thus, multiple pioneer factors function together to drive reprogramming in culture.
Figure 1: Pioneer factors drive developmental transitions.
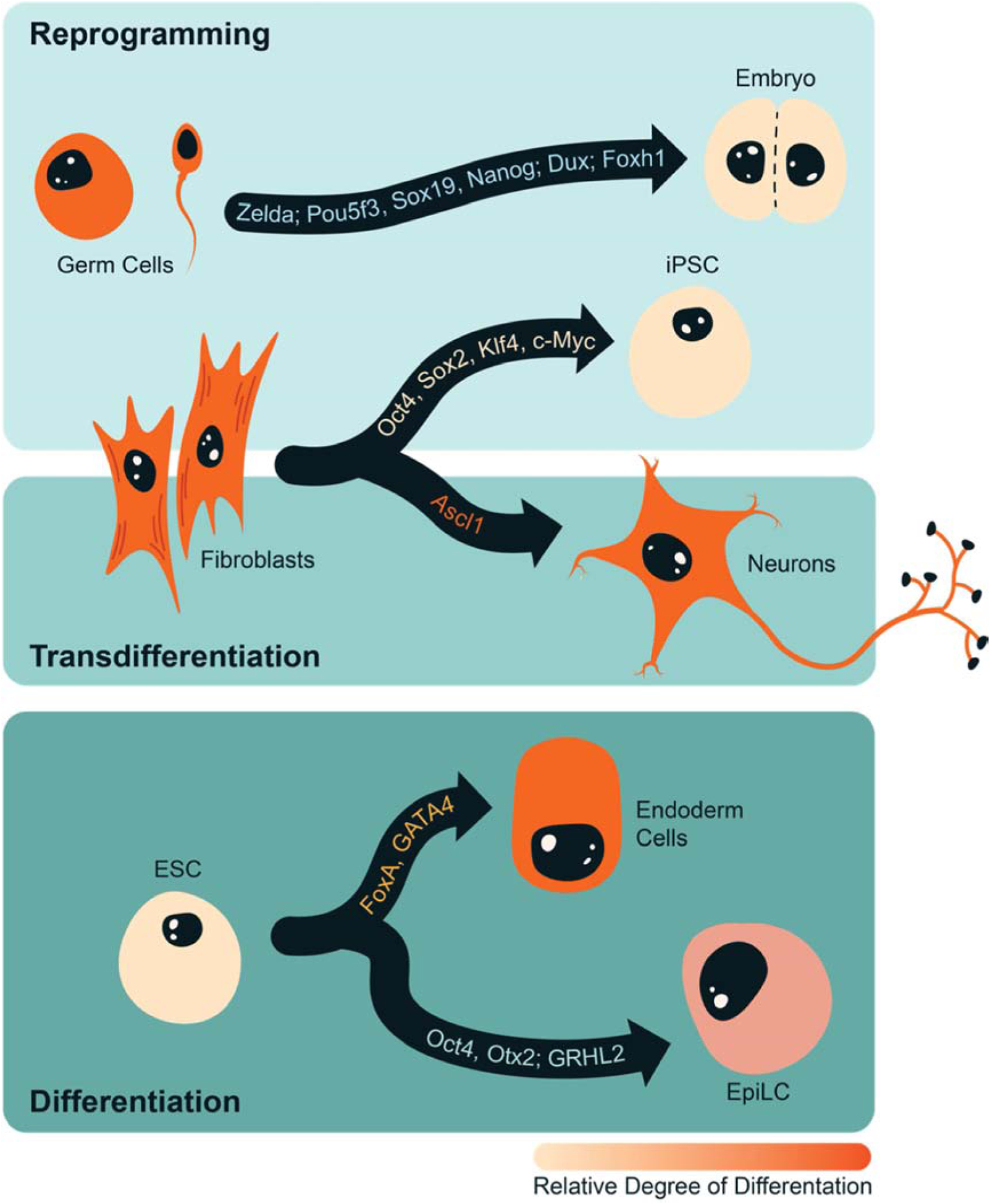
Pioneer factors reprogram specialized cell types (germ cells, fibroblasts) to pluripotency (iPSC, early embryonic cells). Expression of other pioneer factors can transdifferentiate fibroblasts to additional cell types, such as neurons and can also drive differentiation from naïve (ESC) to more specialized cell types (endoderm, EpiLC). A subset of pioneer factors involved in each conversion are listed along the arrows, which indicate the direction of the conversion events. The color of the cell represents the relative degree of differentiation with dark orange being the most differentiated.
Pioneer factor-mediated reprogramming in culture is a slow and inefficient process that takes weeks with only a small subset of cells converting to pluripotency. By contrast, a similar, but much more rapid and efficient process takes place immediately following fertilization as the specialized germ cells are reprogrammed to the totipotent cells of the early embryo. This conserved developmental transition is initiated by maternally deposited mRNAs and proteins, and the zygotic genome is largely transcriptionally silent. The gradual activation of transcription from the zygotic genome is coordinated with the degradation of the maternal products during this maternal-to-zygotic transition (MZT). Pioneer factors drive the reprogramming of the zygotic genome during these initial steps of development. In Drosophila, Zelda is essential for activating transcription from the zygotic genome and therefore development beyond the MZT (Harrison et al., 2011; Liang et al., 2008). Zelda is a maternally encoded pioneer factor necessary for maintaining or establishing chromatin accessibility at the cis-regulatory elements that drive the initial wave of zygotic gene expression (McDaniel et al., 2019; Schulz et al., 2015; Sun et al., 2015). Indeed, Zelda-mediated chromatin accessibility facilitates the binding of additional transcription factors necessary for patterning the early embryo (Xu et al., 2014; Yamada et al., 2019; Yáñez-Cuna et al., 2012). In zebrafish and frogs, orthologs of the reprogramming factors Sox2 and Oct4 are instrumental in activating the zygotic genome (Gentsch et al., 2019; Lee et al., 2013; Leichsenring et al., 2013). In zebrafish, Pou5f3 (Oct4), Sox19b and Nanog activate expression of the zygotic genome, and loss of all three factors leads to decreased chromatin accessibility at developmental enhancers (Miao et al., 2020; Pálfy et al., 2020; Veil et al., 2019). In Xenopus, Pou5f3 (Oct4) and Sox3 similarly remodel condensed chromatin at regulatory elements allowing the zygotic genome to be activated (Gentsch et al., 2019). Zygotic genome activation in Xenopus also relies on the additional pioneer factor Foxh1, which primes enhancers for activation and is required for the subsequent binding of additional transcription factors (Charney et al., 2017; Paraiso et al., 2019). In mammals, it is likely that multiple activators function together to drive expression from the zygotic genome. In both mice and humans, members of the DUX family of transcription factors activate hundreds of genes expressed during zygotic genome activation, similar to the role of pioneer factors in the early development of other species (Hendrickson et al., 2017; Iaco et al., 2017; Whiddon et al., 2017). In human myoblasts, DUX4 functions as a pioneer factor, opening regions of chromatin and driving transcription (Choi et al., 2016). Furthermore, Dux expression in mouse embryonic stem cells (mESCs) drives changes in chromatin accessibility and gene expression that are similar to the totipotent, two-cell (2C) embryo (Hendrickson et al., 2017). Thus, in vertebrates and invertebrates pioneer factors drive the cellular reprogramming that takes place during the initial stages of development.
Beyond reprogramming to pluripotency, pioneer factors mediate the transcriptional changes that control additional developmental transitions and function to define tissue-specific fates (Figure 1). As an organism develops, cells gradually become more differentiated, and this cell-type specification is driven by proteins with pioneering activity. These developmental transitions can be reflected in culture, such as the shift from the naïve pluripotency of embryonic stem cells (ESC) to the primed pluripotency of epiblast-like cells (EpiLCs), which is instructed by pioneer factors, including Oct4, Sox2 and Nanog (Buecker et al., 2014). The pioneer factor Grainy head like-2 (GRHL2) also functions during this transition to define novel enhancers and maintain gene expression as cells exit naïve pluripotency (Chen et al., 2018). The paradigmatic pioneer factor, FoxA1, is instructive in hepatic differentiation and functions by opening chromatin at tissue-specific enhancers (Cirillo et al., 2002; Gualdi et al., 1996). Demonstrating that the pioneering activity of FoxA factors is required for development, a protein domain in the Forkhead box family member FoxA2 important for interactions with nucleosomes is essential for embryonic development and FoxA2-mediated gene expression (Iwafuchi et al., 2020). In pituitary development, Pax7 expression drives intermediate lobe identity fate by both pioneering at a subset of enhancers and more rapidly activating another subset of enhancers (Budry et al., 2012; Mayran et al., 2018). Pioneer factors are similarly required for the conversions in cell fate required for direct-lineage reprogramming (Morris, 2016). For example, Ascl1 gains access to fibroblast chromatin and is necessary for the transdifferentiation of fibroblasts to neurons (Wapinski et al., 2017). While this is by no means an exhaustive list of pioneer factors nor their role in development, these examples highlight the important role that pioneer factors play in defining cell-type specific enhancers and directing the transcriptional changes necessary to drive developmental transitions.
Pioneer factor occupancy and activity is developmentally regulated
Despite the unique ability of pioneer factors to bind silenced regions of the genome, in cells pioneer factors bind only a subset of their defined sequence-recognition motifs and these binding sites differ depending on cell type. Genome-wide mapping of OSK occupancy and gene expression dynamics throughout reprogramming revealed distinct binding during early and late stages of reprogramming (Chronis et al., 2017). Widespread reorganization of Oct4 genomic occupancy is also evident as cells transition from naïve to primed pluripotency (Buecker et al., 2014). Similarly, the pioneer factor FOXA2 exhibits cell-type specific binding when assayed across several different human cell lines, including liver carcinoma (Hep2), lung carcinoma(A549), and ESC-derived endoderm (dEN). Even upon ectopic expression, FOXA2 does not bind the same set of sites or motifs in all cell types, and DNA sequence is insufficient to direct FOXA2 binding as it only binds 6–14% of its recognition motifs in any given cell type (Donaghey et al., 2018). Mouse FoxA2 also demonstrates cell-type specific binding during in vitro differentiation from ESC to definitive endoderm (Cernilogar et al., 2019). A majority (>65%) of FOXA1 binding sites differ between those identified in the breast cancer MCF7 cell line and those in the prostate cancer LncaP cell lines, and in MCF7 cells only 3.7% of FOXA1 recognition motifs are bound (Lupien et al., 2008). These cell-type specific differences in pioneer factor occupancy occur largely at enhancers (Lupien et al., 2008; Mayran et al., 2018). Chromatin accessibility at enhancers is more correlated with tissue-specific gene expression than accessibility of promoters, which are often accessible even in tissues in which the gene they regulate is not expressed (Reddington et al., 2020). Together, these data indicate that pioneer-factor binding is context dependent and that they function preferentially to regulate chromatin accessibility at enhancers to drive cell-type specific patterns of gene expression.
While the majority of pioneer factors show tissue-specific genomic occupancy, in Drosophila Grainy head binds to the majority of the same loci throughout embryonic development and in larval tissues (Jacobs et al., 2018; Nevil et al., 2017). Similarly, throughout the reprogramming events in the early Drosophila embryo Zelda remains bound to same regions of the genome. Furthermore, Zelda binding at this time in development is distinctive as compared to other pioneer factors in that it is driven largely by DNA sequence with 64% of the canonical recognition motifs occupied at the earliest time point (Harrison et al., 2011). Thus, while there are some factors that show distinctive binding properties, overall it is clear that developmental context influences pioneer factor genomic occupancy and that specific chromatin features act as barriers even to pioneer-factor binding.
Developmental context not only affects pioneer-factor binding, but also influences pioneer-factor activity in establishing accessible chromatin. One defining feature of pioneer factors is their ability to determine regions of accessible chromatin. However, even in an individual cell type not every pioneer factor-bound locus requires the pioneer factor for accessibility. In the early Drosophila embryo, Zelda is only required for chromatin accessibility at a subset of the sites it occupies (Schulz et al., 2015; Sun et al., 2015). Similarly, while induction of either FOXA2 or the melanotrope-specifying pioneer factor Pax7 result in many sites that gain accessibility, there are many more binding sites that remain inaccessible and therefore resistant to pioneer function (Donaghey et al., 2018; Mayran et al., 2018). This locus-specific requirement of a pioneer factor indicates that additional features within the cell can influence pioneer-factor activity. The necessity of a pioneer factor for defining accessible cis-regulatory regions may also vary depending on developmental context. While Grainy head occupies largely the same genomic sites in tissues assayed through days of development, Grainy head is only necessary for defining regions of open chromatin in the late-stage Drosophila embryo and the larvae. Earlier in development Grainy head is dispensable for chromatin-accessible regions (Jacobs et al., 2018; Nevil et al., 2020). Together these examples highlight that both pioneer-factor binding and activity are regulated by cell-specific features that vary with developmental stage. These pioneer-factor extrinsic features include chromatin structure and cofactor expression (Figure 2).
Figure 2: Chromatin and cofactor expression modulate pioneer-factor occupancy and activity.
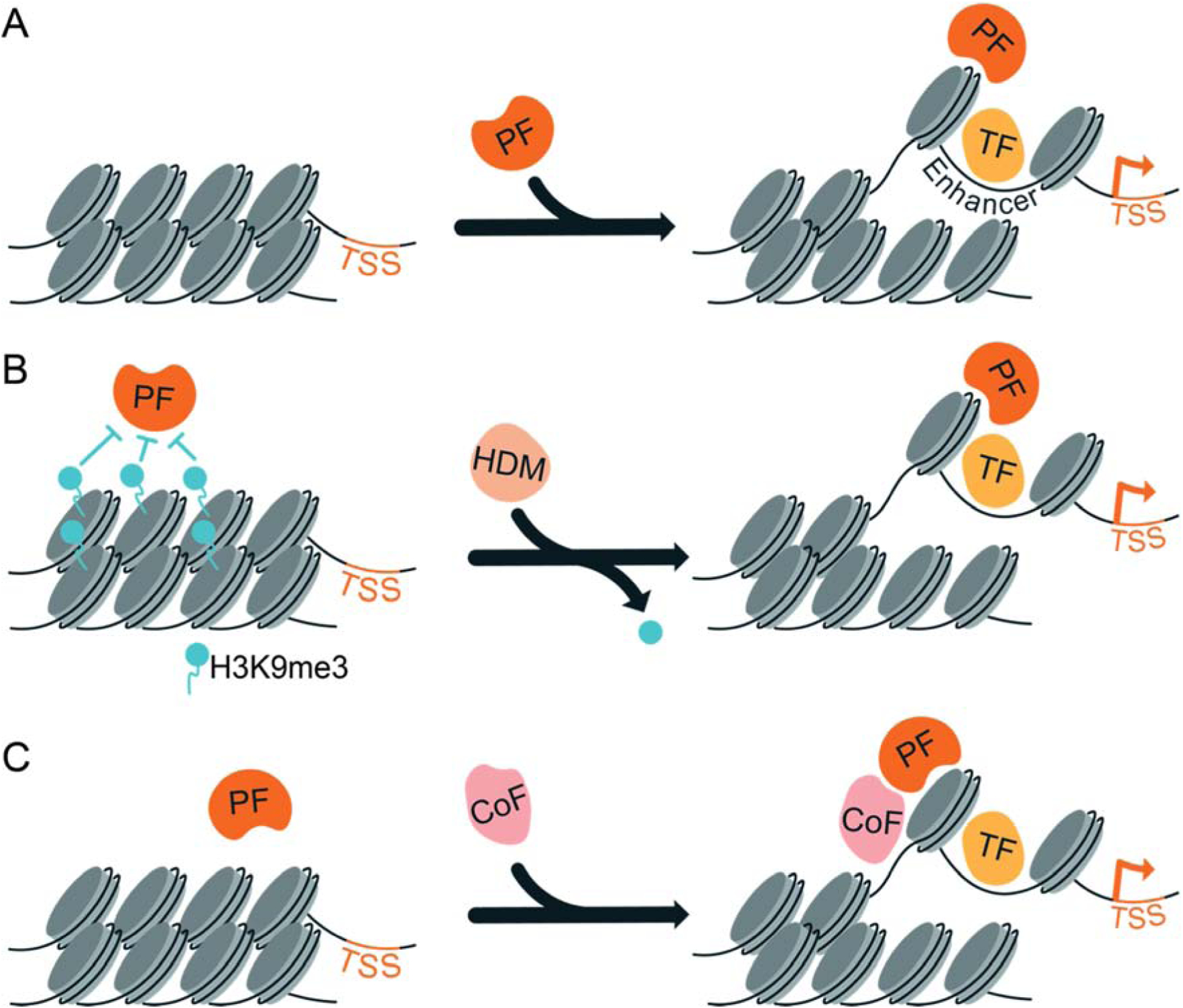
A. Pioneer factors (PF) bind unmarked, silent chromatin and establish regions of accessible chromatin at enhancers to allow for the subsequent binding of lineage-specific transcription factors (TF) that drive gene expression. B. Specific chromatin modifications (such as histone methylation) act as a barrier to pioneer-factor binding. Expression of enzymes that remove these marks, such as histone demethylases (HDM), enable pioneer factors to overcome this barrier, drive accessibility at enhancers and promote gene expression. C. The expression of cofactors (CoF) can stabilize pioneer-factor binding and promote genomic occupancy. TSS, transcription start site.
Chromatin influences pioneer-factor occupancy and activity
Pioneer factors are capable of binding silenced chromatin and defining accessible cis-regulatory regions. However, the fact that in cells pioneer factors occupy only a subset of their binding motifs demonstrates that DNA sequence alone is insufficient to direct binding and there must be specific cellular features that influence pioneer-factor binding. One such feature is the chromatin structure, including histone variants and post-translational modifications to both the DNA and histones that wrap the DNA (Figure 2B). Specific post-translational modifications on histones, such as methylation of histone H3 on lysine 9 (H3K9me3), act as barriers to pioneer-factor binding. For example, heterochromatic regions containing H3K9me3 are refractory to OSK binding (Soufi et al., 2012). These limitations to pioneer-factor binding likely influence reprogramming efficiency as knockdown of the enzyme that deposits H3K9me3 both facilitates OSK binding and increases reprogramming efficiency (Onder et al., 2012; Soufi et al., 2012). Additional chromatin barriers to reprogramming exist and limit pioneer-factor binding. A chemical screen demonstrated that inhibiting proteins that influence H3K27 and H3K79 methylation can increase reprogramming efficiency, enhance OCT-factor occupancy of regulatory regions for pluripotency genes and even allow additional transcription factors to substitute for OCT4 in this process (Kim et al., 2020). Thus, multiple histone modifications act as epigenetic impediments to OCT4 binding and, in so doing, limit efficient reprogramming. FOXA1 binding is similarly preferentially excluded from regions enriched for H3K9 methylation (Lupien et al., 2008). DNA methylation is another chromatin feature associated with repressive chromatin, but unlike H3K9 methylation does not act as a barrier to chromatin binding by either Pax7 or FOXA2 (Donaghey et al., 2018; Mayran et al., 2018).
It is less clear whether there are chromatin marks that actively promote pioneer-factor binding. Methylation of H3K4 is part of a lineage-specific epigenetic signature that is correlated with cell-type specific recruitment of FOXA1 to chromatin, suggesting this histone modification may promote FOXA1 occupancy (Lupien et al., 2008; Wang et al., 2015). Regions bound by FoxA2 during in vitro endoderm differentiation are similarly somewhat enriched for marks of active chromatin in ESC (Cernilogar et al., 2019). Most pioneer factors, like FOXA2, GATA4, and OSK, predominantly bind to regions devoid of both activating and repressive modifications (Donaghey et al., 2018; Soufi et al., 2012). The neurogenic reprogramming pioneer factor Ascl1 preferentially localizes to chromatin with H3K4me1, H3K27ac, and H3K9me3, and the enrichment for this trivalent set of histone modifications helps to determine cell-type specific occupancy (Wapinski et al., 2017). Together these data suggest that pioneer factors are generally excluded from binding to actively silenced, heterochromatic regions, but factor-specific preferences in chromatin binding exist.
In addition to only binding a subset of their recognition motifs in vivo, once bound pioneer factors do not drive accessibility at all occupied genomic regions. For example, upon Pax7 induction there are a subset of loci that are bound by Pax7, but resistant to Pax7-mediated chromatin opening. These regions are enriched for markers of definitive heterochromatin and for binding of the insulator CTCF (Mayran et al., 2018). Therefore, in addition to shaping DNA binding, pre-existing chromatin state can influence the ability of a pioneer factor to open chromatin once bound.
Chromatin structure changes dramatically during development, facilitating the necessary widespread changes in gene expression. While pioneer factors are major drivers of these chromatin changes, evidence supports a role for repressive chromatin structure in limiting both pioneer-factor binding and activity. Understanding the barrier that pre-existing chromatin structure plays in pioneer-factor mediated reprogramming will have important implications in both development and disease.
Cell-type specific pioneer factors and lineage-specific transcription factors cooperate to define developmental enhancers
Another major cell-type specific feature that regulates pioneer-factor occupancy and activity is the set of additional transcription factors expressed in any given cell type. Based on the canonical definition of pioneer factors, lineage-specific transcription factors bind to the cis-regulatory regions made accessible by the pioneer factor and activate gene expression (Figure 2A). Thus, the complement of lineage-specific factors expressed in any cell type will determine the pioneer-factor mediated gene expression profile. However, these lineage-specific factors can also enable pioneer-factor binding to novel genomic loci, stabilize transient binding and facilitate chromatin accessibility at pioneer-factor bound regions.
In many developmental contexts, multiple factors with pioneering function are required to drive changes in cell fate. Notably, reprogramming of both mouse and human cells in culture requires a cocktail of proteins with pioneer-factor activity (OSK) (Figure 3A) (Chronis et al., 2017; Soufi et al., 2012). Highlighting the importance of interactions amongst these pioneer factors in driving genome occupancy, analysis of Klf4-binding sites both in the presence and absence of Oct4 and Sox2 revealed Klf4 was dependent on these factors for genomic occupancy at a large number of loci during reprogramming. Similarly, analysis of Oct4 and Sox2 binding in the absence of other factors further demonstrated that while each factor has some individual activity, cooperative binding of OSK is necessary for genomic targeting during reprogramming (Chronis et al., 2017). Furthermore, while each factor is required for chromatin accessibility at a subset of loci, collaboration amongst all three factors is essential for defining enhancers of pluripotency genes (Chronis et al., 2017; Li et al., 2017). The reprogramming of the early embryonic genome in zebrafish similarly requires the cooperative function of multiple factors (Nanog, Pou5f3 (Oct4) and Sox19b), which function synergistically to establish chromatin accessibility and activate transcription (Gao et al., 2020; Lee et al., 2013; Miao et al., 2020; Pálfy et al., 2020; Veil et al., 2019). At some regions these factors co-bind to elicit open regions of chromatin, while at others individual factors function independently. It is likely that genomic reprogramming in the early mammalian embryo similarly requires multiple pioneering factors. While DUX drives a gene expression program similar to that of the totipotent two-cell embryo, it is not absolutely required for mouse development, suggesting additional factors can function in its absence to activate the zygotic genome (Chen and Zhang, 2019; De Iaco et al., 2020). These examples demonstrate the requirement for multiple pioneer factors in reprogramming the specified genome and establishing pluripotency both in culture and in the early embryo.
Figure 3: Cooperation amongst pioneer factors during development.
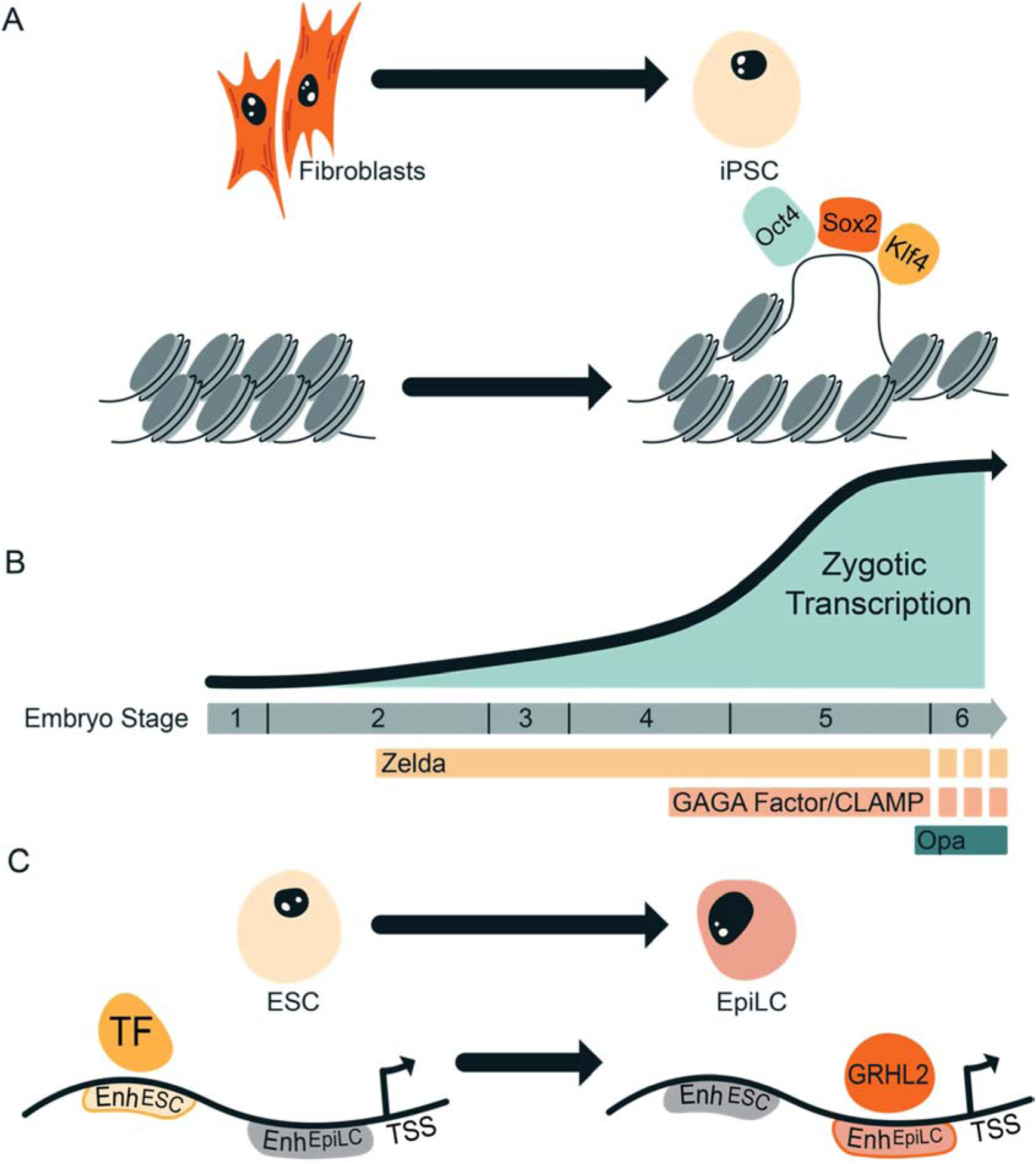
A. Simultaneous binding of multiple pioneer factors may be necessary for the formation of accessible chromatin at cis-regulatory elements. For example, during reprogramming of fibroblasts to iPSCs, Oct4, Sox2, and Klf4 are required to function together to define a subset of enhancers. B. Multiple pioneer factors (Zelda, GAGA factor, CLAMP, and Odd-paired (Opa)) function sequentially to regulate early embryonic development in Drosophila. C. Sequential action of pioneer factors can define cell-type specific enhancers that maintain gene expression during differentiation. As ESCs exit naïve pluripotency and become primed EpiLCs, GRHL2 defines novel enhancers to maintain expression of a gene regulatory network necessary for defining EpiLCs.
Pioneer factors can be redirected to new genomic loci by cell-type specific proteins. Pioneer factors essential for driving pluripotency and reprogramming, including Oct4, Sox2 and Nanog, are also essential for the transition from naïve to primed pluripotency in culture. As naïve ESCs transition to primed epiblast-like cells (EpiLC), Oct4 binding is widely reorganized at distal enhancers. The Oct4 motifs underlying bound regions in both cell-types are similar, suggesting features beyond DNA-sequence recognition are driving these widespread changes in pioneer-factor binding. Motif searches suggest that interactions with additional factors shape the changes in binding. Recognition motifs for factors known to function with Oct4 in pluripotency, including Esrrb and Klf4, were enriched near strong Oct4-binding sites in ESC (Buecker et al., 2014). Indeed, Esrrb expression can facilitate binding of OSK to pluripotency enhancers during reprogramming, supporting a role for this ESC-expressed transcription factor in guiding pioneer-factor binding (Chronis et al., 2017). Binding motifs for other factors, Otx2 and Zic2/3, were associated with Oct4 binding in EpiLC. Upon overexpression in ESC, Otx2 can access a subset of its EpiLC-binding sites and globally redirect Oct4 binding, providing a mechanism for the differential roles of Oct4 in regulating naïve and primed pluripotency (Buecker et al., 2014). These examples highlight how the cohort of cofactors expressed in a given cell type along with the sequences they bind can influence pioneer-factor occupancy and, in so doing, redefine the cellular transcriptional landscape.
While in some cases cofactors may redirect pioneer-factor binding in a tissue-specific manner, cofactors can also serve to stabilize weak interactions with the genome (Figure 2C). The pioneer factor FOXA2 demonstrates cell-type specific binding. Loci that are strongly enriched for binding in derived definitive endoderm cells showed a low signal upon FOXA2 induction in fibroblasts, suggesting additional factors might stabilize binding in a cell-type specific manner. Indeed, simultaneous expression of GATA4 could increase FOXA2 occupancy at a subset of these regions, showing that co-expression could stabilize the weak FOXA2 binding. Nonetheless, this GATA4-dependent increase in FOXA2 binding did not result in increased chromatin accessibility (Donaghey et al., 2018). Therefore, additional factors must be responsible for working with FOXA2 to drive accessibility at these regions in definitive endoderm. When nucleosomal versus direct DNA-binding by FOXA2 was assayed during endoderm differentiation in culture, no clear differences in chromatin configuration could explain the cell-type specific binding. Instead, regions in which pioneer transcription factors engaged with nucleosomes were enriched for binding by additional transcription factors and highlights the coordination of multiple factors in accessing chromatin during endoderm differentiation (Meers et al., 2019). In cultured cells, FOXA-responsive enhancers show intrinsic cell-type specificity, suggesting that both the underlying DNA sequence and the factors that it recruits function together to drive tissue-specific gene expression patterns (Sérandour et al., 2011). In ESC, binding of the pioneer factor Sox2 at a subset of sites (~10%) depends on the DNA-binding activity of PARP-1. These PARP-1 dependent sites are preferentially enriched for regions that are occupied by nucleosomes, show low chromatin accessibility and have suboptimal Sox motifs. Thus, at a subset of loci Sox2 requires a cofactor for nucleosome binding, and in vitro data support a model in which PARP-1 facilitates Sox2 binding to motifs that are positioned suboptimally on the nucleosome (Liu and Kraus, 2017). Together these studies provide examples of how transcription factors stabilize pioneer-factor binding at a subset of loci in a cell-type specific manner and function together to define novel cis-regulatory elements during development.
In addition to affecting genomic occupancy, cofactors can influence the ability of pioneer factors to drive chromatin accessibility subsequent to DNA binding. When overexpressed in ESCs, FoxA2 is preferentially recruited to regions primed with specific histone modifications, but chromatin accessibility at these loci is only established when additional lineage-specific factors are expressed (Cernilogar et al., 2019). Similarly, during in vitro hepatic differentiation, FOXA factors prime enhancers for activation by lineage-specific transcription factors, like PDX1 (Wang et al., 2015). In Drosophila, Grainy head binding in the early embryo is not required for chromatin accessibility, and the gene expression program regulated by Grainy head differs depending on developmental stage (Nevil et al., 2020). Therefore, Grainy head activity is regulated by tissue-specific properties that function after Grainy head binding. Thus, in some tissues and at a subset of loci pioneer-factor occupancy alone is unable to establish chromatin accessibility, but instead requires the activity of additional factors.
Sequential function of pioneer factors during differentiation
While reprogramming both in culture and in the early embryo requires the simultaneous action of multiple pioneer factors, the sequential action of multiple pioneer factors can also be necessary for dramatic developmental changes in cellular identity. The earliest stages of embryonic reprogramming in Drosophila are primarily driven by the pioneer factor Zelda. However, recent evidence has demonstrated essential roles for additional pioneering factors after Zelda as the zygotic genome becomes broadly activated. Odd-paired, GAGA factor, and CLAMP are required for defining chromatin accessibility at cis-regulatory elements important in patterning the embryo independently of Zelda (Gaskill et al., 2020; Koromila et al., 2020; Soluri et al., 2020). While Zelda, GAGA factor and CLAMP are ubiquitously expressed, maternally encoded proteins, Odd-paired is zygotically expressed in a tissue-specific pattern and is essential in defining the gene expression patterns necessary for embryonic segmentation (Bhat et al., 1996; Duan et al., 2020; Liang et al., 2008; Soluri et al., 2020). Thus, embryonic development requires the sequential activity of pioneer factors to define the cis-regulatory regions that pattern the embryo (Figure 3B). Activation of the zygotic genome in Xenopus similarly requires a hand-off between maternally encoded and zygotically expressed pioneer factors. Maternally provided Foxh1 binds to enhancers that are subsequently bound by FoxA, which is expressed zygotically at gastrulation (Charney et al., 2017). These examples highlight how the sequential activity of pioneering factors can drive embryonic patterning during differentiation.
The sequential activity of pioneer factors in defining cis-regulatory regions is also evident in cell culture systems that recapitulate differentiation from the stem cell fate. Similar to the binding of maternal Foxh1 in Xenopus embryos, FoxD3 is already bound to the FoxA1-responsive alb1 enhancer in mouse ESC and functions to maintain unmethylated DNA at this locus. In this case, FoxD3 may function as a “placeholder” to prevent the establishment of repressive chromatin at this enhancer and facilitate subsequent FoxA1 binding (Xu et al., 2009). Enhancers are defined by the function of FoxD3 in ESC and later activated upon differentiation. By contrast, during the exit from naïve pluripotency GRHL2 defines a distinct set of enhancers that maintain an epithelial gene expression program (Figure 3C). The GRHL2-target genes are expressed in both naïve ESC and in formative EpiLC following in vitro differentiation, but rely on switching from a set of enhancers that drive expression in ESC to a set of GRHL2-defined enhancers for expression in EpiLC (Chen et al., 2018). Thus, the pioneer factor GRHL2 is part of a larger set of transcription factors that drive the rewiring of enhancers during the transition from ESC to EpiLC, allowing target gene expression to remain constant despite the changes in enhancer usage. It is evident from these examples that the sequential action of enhancer-defining pioneer factors refines the gene expression network as cells differentiate.
Conservation of pioneer factors and the developmental transitions they control
Because of the fundamental importance of pioneer factors in shaping gene expression during development, it is perhaps unsurprising that both pioneer factors and their importance in specific developmental transitions are conserved. The pioneering features of FoxA were first defined based on the role of FoxA in hepatic development in mice (Gualdi et al., 1996). In Caenorhabditis elegans, PHA-4, the FoxA homolog, is essential for defining foregut identity and functions as a pioneer factor to facilitate chromatin decompaction (Fakhouri et al., 2010; Hsu et al., 2015). Like FoxA, the pioneer factor Grainy head is widely conserved in metazoans (Traylor-Knowles et al., 2010). Indeed, Grainy head orthologs in worms, flies, and mammals all bind to the same consensus motif and are essential in defining epithelial cell fates (reviewed in Wang and Samakovlis, 2012). Grainy head protein family members drive regions of chromatin accessibility in Drosophila larval tissues and in human tissue culture cells (Chen et al., 2018; Jacobs et al., 2018). Through their conserved functions these pioneer factors act at the top of gene regulatory networks to drive cell-type specification.
During conserved developmental transitions, the essential role of pioneering factors may be shared despite the fact that the proteins themselves may not be conserved across species. Perhaps the clearest example of the shared requirement for pioneer-factor function is during the rapid and efficient reprogramming that occurs during the initial stages of embryogenesis. In all species studied to date, pioneer factors are required to regulate the activation of the zygotic genome (Schulz and Harrison, 2019). In Drosophila, the pioneer factor Zelda primes the genome for activation (Liang et al., 2008; Schulz et al., 2015; Sun et al., 2015). Zelda is a zinc-finger transcription factor that is not found outside of insects and crustaceans (Ribeiro et al., 2017). Nonetheless, in other organisms the role of pioneer factors in genome activation is conserved, even if the specific proteins are not. In zebrafish, the pioneer factors Nanog, Pou5f3 (Oct4) and Sox19b function together to activate the zygotic genome (Lee et al., 2013; Leichsenring et al., 2013; Miao et al., 2020; Pálfy et al., 2020; Veil et al., 2019). In other species, Pou- and Sox-domain containing proteins may also be involved in determining chromatin accessibility during this conserved transition. In human embryos OCT4 has been implicated in regulating zygotic genome accessibility and transcription (Gao et al., 2018). Pou5f3 and Sox3 also function to open and mark regions of chromatin important for germ layer formation in the early Xenopus embryo (Gentsch et al., 2019). Despite these similarities, additional species-specific pioneering proteins have been identified. In frogs, Otx1, Vegt, and Foxh1 function together at enhancers to control zygotic genome activation (Paraiso et al., 2019). In mice and human, DUX family transcription factors activate a subset of genes during the earliest stages of development. However, the viability of DUX4 knockout mice makes it clear additional factors can drive this essential developmental transition (Chen and Zhang, 2019; De Iaco et al., 2020). Studies of the MZT in multiple species have highlighted the conserved role of pioneer factors, if not the factors themselves, in driving this essential, conserved developmental transition.
Pioneer factors in disease
Because pioneer factors function as master regulators of cell fate, they are particularly prone to driving disease when mutated or misexpressed. Notably, expression of multiple different individual pioneer factors has been linked to the toxic cell proliferation that characterizes cancers, and expression levels are correlated with poor prognoses (Dobersch et al., 2019; Jozwik and Carroll, 2012). For example, studies of cancer stem cells have demonstrated that these cells share features with other stem cell populations and express the core-reprogramming pioneer factors OCT4, NANOG, and SOX2. The gene networks regulated by these core pluripotency transcription factors are overexpressed in relatively undifferentiated tumors and are associated with poor clinical outcomes (Ben-Porath et al., 2008). Indeed, inducible expression of these reprogramming factors in mice leads to tumor formation, suggesting a causal link between pioneer factor-mediated reprogramming and tumor initiation (Ohnishi et al., 2014). Correlations have also been found between the expression of these transcription factors in tumor cells, stem cell-like characteristics, and resistance to anti-tumor therapies (reviewed in Dobersch, Rubio and Barreto, 2019).
Misexpression of other early embryonic reprogramming factors in later stages of development can also result in detrimental gene expression profiles. Because pioneer factors are uniquely capable of accessing the genome, these proteins are able to bind and reprogram the expression profile of the cell. For example, Facioscapulohumeral muscular dystrophy (FSHD) is caused by misexpression of the mammalian pioneer factor DUX4 in muscle cells (reviewed in Campbell et al., 2018). The expression of this pioneer factor in tissues in which it is not normally expressed results in the activation of genes expressed in the preimplantation embryo. The activation of this novel gene expression program results in apoptosis and wasting of the muscles in which DUX4 is expressed (Geng et al., 2012; Whiddon et al., 2017; Young et al., 2013). Emphasizing the contribution of the cellular environment to the disease outcome of pioneer-factor misexpression, increased DUX4 expression is also linked to cancers, where it promotes escape from immune surveillance (Chew et al., 2019).
Pioneer factors also function in disease through their ability to redirect cofactor binding. FOXA1 regulates the genome occupancy of hormone receptors in a large number of cells during normal development (Hurtado et al., 2011; Paakinaho et al., 2019; Swinstead et al., 2016). The biological significance of these interactions is highlighted by the frequent mutation of FOXA1 in prostate and breast cancers that depend on these hormone receptors. Gain of function mutations demonstrate that FOXA1 functions as an oncogene, and the nature of the mutations suggests pioneering activity is essential for the oncogenic properties of the mutated proteins (Adams et al., 2019; Parolia et al., 2019). Expression of FOXA1 in an immortalized prostate cell line can reprogram androgen-receptor binding to reflect the binding profile in prostate cancer (Pomerantz et al., 2015). Similarly, in estrogen receptor positive breast cancer cells, FOXA1 overexpression drives genome-wide enhancer reprogramming (Fu et al., 2019). Despite the identified role of FOXA1 in directing estrogen-receptor binding, FOXA1 also functions independently to regulate gene expression in therapy-resistant, breast-cancer cell lines (Cocce et al., 2019). These estrogen-receptor independent loci are co-occupied by the additional pioneer factor GRHL2, suggesting that two pioneer factors may collaborate to establish novel enhancers, which drive expression of pathways leading to therapy resistance (Cocce et al., 2019).
In addition to the evidence in numerous epithelial cancers that support an oncogenic role for GRHL2, GRHL2 has also been implicated as a tumor suppressor. This tumor-suppressive function is based on the ability of GRHL2 to promote epithelial cell fate, which suppresses the cellular migration and invasion necessary for metastasis (reviewed in Reese, Harrison and Alarid, 2019). These conflicting roles highlight the importance of studying pioneer-factor function within the endogenous cellular context and the role of cell intrinsic properties, including chromatin state and cofactor expression, in regulating GRHL2 activity. Together, the complex roles of pioneer factors in human disease reflect both the unique capacity of these factors to reprogram the cellular transcriptome and the effect of developmental context on this activity.
Conclusions
The complex network of gene expression that controls metazoan development is determined by the ability of transcription factors to access cis-regulatory elements and drive transcription. The ability of pioneer transcription factors to define these regulatory regions endows them with the capacity to restructure the gene expression profile of a cell and, in so doing, result in dramatic changes in cell fate. The unique features of pioneer factors enable them to reprogram cell fate, whereas misexpression or mutation can lead to disease. However, cell-specific features, such as chromatin structure and co-factor expression, influence pioneer-factor binding and activity. Indeed, in many cases pioneer factor-defined cis-regulatory modules must subsequently be bound by lineage-specific transcription factors for transcription to initiate. Biochemical studies have begun to elucidate the molecular mechanism allowing pioneer factors to bind nucleosomes and scan the genome for specific binding sites. These have been complemented with cell-culture studies that have reinforced the importance of pioneer factors in defining cis-regulatory modules. Nonetheless, future studies defining the barriers to pioneer-factor function and the complex interplay between pioneer factors and cofactors will be essential for understanding how the genome is differentially interpreted during development and how failures in this process can lead to disease.
It is clear that while pioneer factors share certain unique functions, pioneer factors use different features to both gain access to chromatin and to increase accessibility at binding sites. In the case of FoxA, chromatin binding alone may be able to drive accessibility by evicting the linker histone H1 (Iwafuchi-Doi et al., 2016). However, other factors bind chromatin using different strategies and interact with chromatin remodeling enzymes to open the chromatin (Iwafuchi-Doi, 2019; Soufi et al., 2015). In other cases, the mechanisms by which the pioneer factor increases chromatin accessibility are less clear. It is possible that some pioneer factors may form subnuclear hubs that increase the localization of other transcription factors, which may result in changes to the local chromatin structure. For example, Zelda is localized to transient hubs in the nucleus that potentiate binding by the transcription factors Bicoid and Dorsal (Dufourt et al., 2018; Mir et al., 2018; Yamada et al., 2019). It will be important to determine if these differences in the mechanisms of pioneer factor-mediated chromatin accessibility are cell-type dependent.
While studies of pioneer factors during in vitro differentiation have provided important insights into their function during development, more studies are needed of pioneer factors within an organism to understand both how developmental context influences pioneer-factor activity and to identify new pioneering factors essential for determining distinct cellular fates. Given the conservation of both the factors themselves and the transitions they regulate, model organisms provide a powerful platform for interrogating pioneer-factor function during development. The advances in high-resolution live imaging coupled with low-cell and single-cell genomic technologies lay the groundwork for a better understanding of the role of pioneer factors both in organisms and in cellular reprogramming in culture.
Acknowledgments
We thank Marissa Gaskill, Tyler Gibson, Andrew Mehle and members of the Harrison lab for helpful discussions. We apologize to those whose research we did not discuss or cite due to space limitations. Research in the Harrison lab is supported by an ACS Research Scholars Grant, R01 GM111694, R01 NS111647 and R35 GM136298 from the National Institutes of Health (NIH) and a Vallee Scholar Award (M.M.H). A.J.M was supported by an NSF predoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, Cho H, DiLoreto R, Chhangawala S, Liu Y, et al. (2019). FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571, 408–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, and Weinberg RA (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, and Schedl P (1996). The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development 122, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L’Honoré A, Vallette S, Brue T, Figarella-Branger D, Meij B, and Drouin J (2012). The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 26, 2299–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, and Wysocka J (2014). Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 14, 838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AE, Belleville AE, Resnick R, Shadle SC, and Tapscott SJ (2018). Facioscapulohumeral dystrophy: activating an early embryonic transcriptional program in human skeletal muscle. Hum. Mol. Genet 27, R153–R162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar FM, Hasenöder S, Wang Z, Scheibner K, Burtscher I, Sterr M, Smialowski P, Groh S, Evenroed IM, Gilfillan GD, et al. (2019). Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res. 47, 9069–9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney RM, Forouzmand E, Cho JS, Cheung J, Paraiso KD, Yasuoka Y, Takahashi S, Taira M, Blitz IL, Xie X, et al. (2017). Foxh1 Occupies cis-Regulatory Modules Prior to Dynamic Transcription Factor Interactions Controlling the Mesendoderm Gene Program. Dev. Cell 40, 595–607.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, and Zhang Y (2019). Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet 51, 947–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AF, Liu AJ, Krishnakumar R, Freimer JW, DeVeale B, and Blelloch R (2018). GRHL2-Dependent Enhancer Switching Maintains a Pluripotent Stem Cell Transcriptional Subnetwork after Exit from Naive Pluripotency. Cell Stem Cell 23, 226–238.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew GL, Campbell AE, De Neef E, Sutliff NA, Shadle SC, Tapscott SJ, and Bradley RK (2019). DUX4 Suppresses MHC Class I to Promote Cancer Immune Evasion and Resistance to Checkpoint Blockade. Dev. Cell 50, 658–671.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, and Kyba M (2016). DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44, 5161–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, and Plath K (2017). Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell 168, 442–459.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, and Zaret KS (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289. [DOI] [PubMed] [Google Scholar]
- Cocce KJ, Jasper JS, Desautels TK, Everett L, Wardell S, Westerling T, Baldi R, Wright TM, Tavares K, Yllanes A, et al. (2019). The Lineage Determining Factor GRHL2 Collaborates with FOXA1 to Establish a Targetable Pathway in Endocrine Therapy-Resistant Breast Cancer. Cell Rep. 29, 889–903.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobersch S, Rubio K, and Barreto G (2019). Pioneer Factors and Architectural Proteins Mediating Embryonic Expression Signatures in Cancer. Trends Mol. Med 25, 287–302. [DOI] [PubMed] [Google Scholar]
- Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, et al. (2018). Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet 50, 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JE, Rieder LE, Huang A, Jordan WT, McKenney M, Watters S, Fawzi NL, and Larschan EN (2020). CLAMP and Zelda function together as pioneer transcription factors to promote Drosophila zygotic genome activation. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourt J, Trullo A, Hunter J, Fernandez C, Lazaro J, Dejean M, Morales L, Nait-Amer S, Schulz KN, Harrison MM, et al. (2018). Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat. Commun 9, 5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri THI, Stevenson J, Chisholm AD, and Mango SE (2010). Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Garcia M, Moore CD, Schulz KN, Alberto O, Donague G, Harrison MM, Zhu H, and Zaret KS (2019). Structural Features of Transcription Factors Associating with Nucleosome Binding. Mol. Cell 75, 921–932.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Pereira R, De Angelis C, Veeraraghavan J, Nanda S, Qin L, Cataldo ML, Sethunath V, Mehravaran S, Gutierrez C, et al. (2019). FOXA1 upregulation promotes enhancer and transcriptional reprogramming in endocrine-resistant breast cancer. Proc. Natl. Acad. Sci. U. S. A 116, 26823–26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. (2018). Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 173, 248–259.e15. [DOI] [PubMed] [Google Scholar]
- Gao M, Veil M, Rosenblatt M, Gebhard A, Hass H, Buryanova L, Yampolsky LY, Grüning B, Timmer J, and Onichtchouk D (2020). Pluripotency factors select gene expression repertoire at Zygotic Genome Activation. BioRxiv 2020.02.16.949362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskill M, Gibson T, Larson E, and Harrison M (2020). The pioneer factor GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. BioRxiv 2020.07.15.204248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, vanderMaarel SM, Ruzzo WL, Gentleman RC, Tawil R, et al. (2012). DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Dev. Cell 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Owens NDL, and Smith JC (2019). The Spatiotemporal Control of Zygotic Genome Activation. IScience 16, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, and Zaret KS (1996). Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes Dev. 10, 1670–1682. [DOI] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, and Eisen MB (2011). Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Obstet. Gynecol. Surv 72, 483–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HT, Chen HM, Yang Z, Wang J, Lee NK, Burger A, Zaret K, Liu T, Levine E, and Mango SE (2015). Erratum: Recruitment of RNA polymerase II by the pioneer transcription factor PHA4 (Science (2015) 19 (1372–1376)). Science (80-.). 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, and Carroll JS (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet 43, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D (2017). A family of double-homeodomain transcription factors regulates zygotic genome activation in placental mammals. Nat. Genet 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Verp S, Offner S, Grun D, and Trono D (2020). DUX is a non-essential synchronizer of zygotic genome activation. Dev. 147, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M (2019). The mechanistic basis for chromatin regulation by pioneer transcription factors. Wiley Interdiscip. Rev. Syst. Biol. Med 11, e1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, and Zaret KS (2016). The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol. Cell 62, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi M, Cuesta I, Donahue G, Takenaka N, Osipovich AB, Magnuson MA, Roder H, Seeholzer SH, Santisteban P, and Zaret KS (2020). Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat. Genet 52, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J, Atkins M, Davie K, Imrichova H, Romanelli L, Christiaens V, Hulselmans G, Potier D, Wouters J, Taskiran II, et al. (2018). The transcription factor Grainy head primes epithelial enhancers for spatiotemporal activation by displacing nucleosomes. Nat. Genet 50, 1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwik KM, and Carroll JS (2012). Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer 12, 381–385. [DOI] [PubMed] [Google Scholar]
- Kim KP, Choi J, Yoon J, Bruder JM, Shin B, Kim J, Arauzo-Bravo MJ, Han D, Wu G, Han DW, et al. (2020). Permissive epigenomes endow reprogramming competence to transcriptional regulators. Nat. Chem. Biol [DOI] [PubMed] [Google Scholar]
- Koromila T, Gao F, Iwasaki Y, He P, Pachter L, Peter Gergen J, and Stathopoulos A (2020). Odd-paired is a pioneer-like factor that coordinates with zelda to control gene expression in embryos. Elife 9, 1–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, and Kaestner KH (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944–947. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, and Giraldez AJ (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mos̈sner R, Driever W, and Onichtchouk D (2013). Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science (80-.). 341, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Lerner J, Gomez-Garcia PA, McCarthy RL, Liu Z, Lakadamyali M, and Zaret KS (2020). Two-Parameter Mobility Assessments Discriminate Diverse Regulatory Factor Behaviors in Chromatin. J. Clean. Prod 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, Qin Y, Guo L, He J, Yu S, et al. (2017). Chromatin Accessibility Dynamics during iPSC Reprogramming. Cell Stem Cell 21, 819–833.e6. [DOI] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, and Rushlow C (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Kraus WL (2017). Catalytic-Independent Functions of PARP-1 Determine Sox2 Pioneer Activity at Intractable Genomic Loci. Mol. Cell 65, 589–603.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, and Richmond TJ (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, and Brown M (2008). FoxA1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell 132, 958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, and Drouin J (2018). Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem 293, 13795–13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, Khetchoumian K, Hariri F, Pastinen T, Gauthier Y, Balsalobre A, and Drouin J (2018). Pioneer factor Pax7 deploys a stable enhancer repertoire for specification of cell fate. Nat. Genet 50, 259–269. [DOI] [PubMed] [Google Scholar]
- McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, and Harrison MM (2019). Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation. Mol. Cell 74, 185–195.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Janssens DH, and Henikoff S (2019). Pioneer Factor-Nucleosome Binding Events during Differentiation Are Motif Encoded. Mol. Cell 75, 562–575.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Tang Y, Bonneau AR, Chan SH, Kojima ML, Pownall ME, Vejnar CE, and Giraldez AJ (2020). Synergistic activity of Nanog, Pou5f3, and Sox19b establishes chromatin accessibility and developmental competence in a context-dependent manner. BioRxiv 2020.09.01.278796. [Google Scholar]
- Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, and Eisen MB (2018). Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. Elife 7, e40497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA (2016). Direct lineage reprogramming via pioneer factors; a detour through developmental gene regulatory networks. Dev. 143, 2696–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevil M, Bondra ER, Schulz KN, Kaplan T, and Harrison MM (2017). Stable binding of the conserved transcription factor grainy head to its target genes throughout Drosophila melanogaster development. Genetics 205, 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevil M, Gibson TJ, Bartolutti C, Iyengar A, and Harrison MM (2020). Establishment of chromatin accessibility by the conserved transcription factor Grainy head is developmentally regulated. Dev. 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. (2014). Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156, 663–677. [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakinaho V, Swinstead EE, Presman DM, Grøntved L, and Hager GL (2019). Meta-analysis of Chromatin Programming by Steroid Receptors. Cell Rep. 28, 3523–3534.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálfy M, Schulze G, Valen E, and Vastenhouw NL (2020). Chromatin accessibility established by Pou5f3, Sox19b and Nanog primes genes for activity during zebrafish genome activation. PLoS Genet. 16, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KD, Blitz IL, Coley M, Cheung J, Sudou N, Taira M, and Cho KWY (2019). Endodermal Maternal Transcription Factors Establish Super-Enhancers during Zygotic Genome Activation. Cell Rep. 27, 2962–2977.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, Wang X, Vats P, Cao X, Pitchiaya S, et al. (2019). Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, Cejas P, Vazquez F, Cook J, Shivdasani RA, et al. (2015). The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet 47, 1346–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddington JP, Garfield DA, Sigalova OM, Karabacak Calviello A, Marco-Ferreres R, Girardot C, Viales RR, Degner JF, Ohler U, and Furlong EEM (2020). Lineage-Resolved Enhancer and Promoter Usage during a Time Course of Embryogenesis. Dev. Cell [DOI] [PubMed] [Google Scholar]
- Reese RM, Harrison MM, and Alarid ET (2019). Grainyhead-like Protein 2: The Emerging Role in Hormone-Dependent Cancers and Epigenetics. Endocrinology 160, 1275–1288. [DOI] [PubMed] [Google Scholar]
- Ribeiro L, Tobias-Santos V, Santos D, Antunes F, Feltran G, de Souza Menezes J, Aravind L, Venancio TM, and Nunes da Fonseca R (2017). Evolution and multiple roles of the Pancrustacea specific transcription factor zelda in insects. PLoS Genet. 13, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, and Harrison MM (2019). Mechanisms regulating zygotic genome activation. Nat. Rev. Genet 20, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, and Harrison MM (2015). Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Métivier R, et al. (2011). Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 21, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soluri IV, Zumerling LM, Parra OAP, Clark EG, and Blythe SA (2020). Zygotic pioneer factor activity of odd-paired/zic is necessary for late function of the drosophila segmentation network. Elife 9, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, and Zaret KS (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, and Zaret KS (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nien CY, Chen K, Liu HY, Johnston J, Zeitlinger J, and Rushlow C (2015). Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinstead EE, Miranda TB, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis LD, et al. (2016). Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 165, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Traylor-Knowles N, Hansen U, Dubuc TQ, Martindale MQ, Kaufman L, and Finnerty JR (2010). The evolutionary diversification of LSF and Grainyhead transcription factors preceded the radiation of basal animal lineages. BMC Evol. Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veil M, Yampolsky LY, Grüning B, and Onichtchouk D (2019). Pou5f3, SoxB1, and Nanog remodel chromatin on high nucleosome affinity regions at zygotic genome activation. Genome Res. 29, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, and Samakovlis C (2012). Grainy Head and Its Target Genes in Epithelial Morphogenesis and Wound Healing. Curr. Top. Dev. Biol 98, 35–63. [DOI] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. (2015). Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16, 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Lee QY, Chen AC, Li R, Corces MR, Ang CE, Treutlein B, Xiang C, Baubet V, Suchy FP, et al. (2017). Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons. Cell Rep. 20, 3236–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watts JA, Pope SD, Gadue P, Kamps M, Plath K, Zaret KS, and Smale ST (2009). Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 23, 2824–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Chen H, Ling J, Yu D, Struffi P, and Small S (2014). Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 28, 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Whitney PH, Huang SK, Eck EC, Garcia HG, and Rushlow CA (2019). The Drosophila Pioneer Factor Zelda Modulates the Nuclear Microenvironment of a Dorsal Target Enhancer to Potentiate Transcriptional Output. Curr. Biol 29, 1387–1393.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, and Stark A (2012). Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 22, 2018–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, and Tapscott SJ (2013). DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis. PLoS Genet. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS (2020). Pioneer Transcription Factors Initiating Gene Network Changes. Annu. Rev. Genet 54, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al. (2018). The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]


