Abstract
It is well-established that mitochondria are the powerhouses of the cell, producing adenosine triphosphate (ATP), the universal energy currency. However, the most significant strengths of the electron transport chain (ETC), its intricacy and efficiency, are also its greatest downfalls. A reliance on metal complexes (Fe-S clusters, hemes), lipid moities such as cardiolipin, and cofactors including alpha-lipoic acid and quinones render oxidative phosphorylation vulnerable to environmental toxins, intracellular reactive oxygen species (ROS) and fluctuations in diet. To that effect, it is of interest to note that temporal disruptions in ETC activity in most organisms are rarely fatal, and often a redundant number of failsafes are in place to permit continued ATP production when needed. Here, we highlight the metabolic reconfigurations discovered in organisms ranging from parasitic Entamoeba to bacteria such as pseudomonads and then complex eukaryotic systems that allow these species to adapt to and occasionally thrive in harsh environments. The overarching aim of this review is to demonstrate the plasticity of metabolic networks and recognize that in times of duress, life finds a way.
Keywords: mitochondrial dysfunction, energy, metabolism, metabolic reconfiguration, ATP
The electron transport chain (ETC) is a shooting gallery
Across all species, biological electron transport reactions are harnessed as a means to generate ATP and catalyze molecular transformations. While in eukaryotes, the specialized machinery of the ETC is housed in mitochondria, prokaryotic systems lack these organelles and the ETC is rather localized to the plasma membrane [1]. Arising early in evolution to fulfill energetic needs, the effectiveness of the ETC and oxidative phosphorylation has rendered these components remarkably well-conserved, with only minor variations across species [2]. The ETC consists of 4 complexes (I-IV) and ATP synthase, which rely on a reducing component in the form of electron carriers such as NADH and FADH2 [1]. These moieties are generated by the tricarboxylic acid (TCA) cycle, a series of enzymatic reactions beginning with citric acid, and likely one of the earliest metabolic components in cells. In conjunction with Embden–Meyerhof–Parnas (EMP) glycolysis, aerobic metabolism produces 30 ATP per molecule of glucose [3]. Unfortunately, the same enzymatic components underlying the intricacy and efficiency of oxidative phosphorylation are often its downfall. A reliance on metal cofactors, lipoic acid, cardiolipin and iron-sulfur (Fe-S) clusters which regularly succumb to nitro-oxidative stress renders TCA and ETC components susceptible to failure [4–8]. To that effect, the scope of this review is to demonstrate that, in several species, the malleability of metabolic networks provides the organism a means to meet their energetic needs when subjected to stressful environments.
While early estimates from in vitro studies suggested that the ETC produces 1–2% of the total superoxide cellular (O2•), it is now appreciated that this figure is markedly lower in vivo (~0.2%) [9,10]. Moreover, while reactive oxygen species (ROS) were once stigmatized as a causative factor of aging and disease, we now know that they play crucial roles in physiological processes, such as innate and adaptive immunity. For instance, knockdown of superoxide dismutase (SOD) in the roundworm Caenorhabditis elegans has been shown to have no effect on lifespan [11]. Additionally, knockdown of uncoupling protein 2 in mice leads to increased production of ROS and increased resistance against bacterial pathogens, likely via the role of ROS in T lymphocyte activation [12]. As such, there appears to be a “redox window”, wherein these species provide physiological benefits without inducing cellular dysfunction and death. However, numerous endogenous and exogenous factors can coalesce to tilt the balance of nitro-oxidative stress toward a pathological state. For example, alcohol consumption is known to trigger the activity of cytochrome p450, which in turn releases potentially damaging free radicals [13]. Ultraviolet light, ionizing radiation, inflammatory cytokines and environmental toxins such as heavy metals can also upset redox homeostasis [14–17]. Activation of inducible nitric oxide synthase (iNOS) in phagocytes to fend off bacterial invaders leads to the formation of nitric oxide (•NO) which rapidly reacts with O2• via radical-radical coupling to form peroxynitrite (ONOO−) [18]. If left unchecked, this inflammatory response can induce cellular dysfunction via the formation of nitrotyrosine-protein adducts [19].
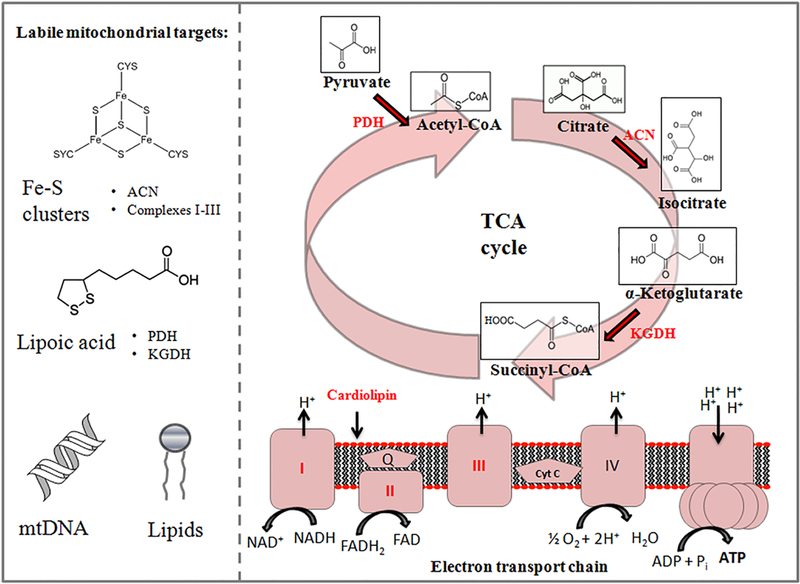
Whether in the plasma membrane or mitochondrion, ETC components and enzymes of the TCA cycle are vulnerable to oxidative stress due to their close proximity to sites of ROS production, such as complexes I and III [20]. While peroxynitrite has a longer half-life (10–20 ms) and can diffuse further, rendering it more selective in the nitrosylation of tyrosine residues, the short half-lives of O2• and hydroxyl radical (•OH) underlie their promiscuity, rapidly affecting surrounding proteins and lipids [21,22]. For instance, the activities of succinate dehydrogenase (SDH) and aconitase (ACN) rely on the presence of [4Fe-4S]2+ clusters which, in the presence of O2• and H2O2, can be converted to [3Fe-4S]1+ and subsequently into [2Fe-2S]2+ clusters which are ultimately degraded [23]. Lipoic acid residues covalently bound to the E2 subunit of alpha-ketoglutarate dehydrogenase (KGDH) and pyruvate dehydrogenase (PDH) are prime targets of ROS toxicity as well, leading to enzymatic inactivation [7]. Oxidative damage of cardiolipin, a key phospholipid for the stabilization of respiratory chain supercomplexes, also leads to marked reductions in ETC activity [7,8] (Figure 1). These examples do not include the mutations of mitochondrial DNA (mtDNA) in eukaryotes, which in turn can spawn a number of mitochondrial diseases. Indeed, a comprehensive review of both the physiological and pathological roles of reactive oxygen and nitrogen species is beyond the scope of this manuscript and has been covered extensively elsewhere. Here, we discuss two biological questions. How do organisms reprogram metabolic networks when aerobic respiration fails or under times of stress, as is inevitable, and how do they recover these components and resume business as usual?
Figure 1:
The tricarboxylic acid (TCA) cycle and electron transport chain are vulnerable to environmental stressors. Biomolecular components in red are prone to oxidative damage and degradation. ACN: aconitase; KGDH: α-ketoglutarate dehydrogenase; mtDNA: mitochondrial DNA; PDH: pyruvate dehydrogenase.
Inorganic pyrophosphate (PPi): An archaic energy carrier
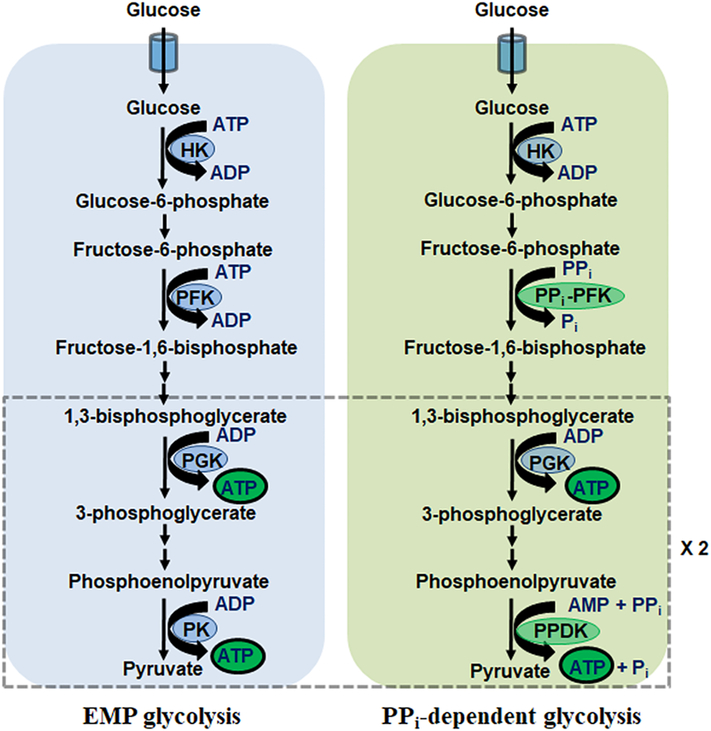
While ATP is often credited as the universal energy currency, it is far from the only high-energy compound on which organisms rely and is predated in evolution by PPi as well as polyphosphate chains of three or more [24]. Indeed, studies of the geochemical conditions that defined the early stages of life have demonstrated the potential for pyrophosphate to form spontaneously [25]. Cues that PPi can be used as an energy donor and as a substitute for ATP stem from the findings that several glycolytic enzymes, namely phosphofructokinase (PFK), pyruvate kinase (PK) and phosphoglycerate kinase (PGK) can incorporate PPi in their respective reactions [26]. More recently, a PPi-dependent kinase of the ribokinase family (TM0415) was identified in Entamoeba histolytica as well [27]. Remnants of pyrophosphate-dependent metabolism can be observed to this day in select bacteria but also parasitic anaerobic eukaryotes such as Giardia and Entamoeba spp [27–29]. For these organisms, the bioenergetic benefits of PPi-dependent glycolysis outweigh those of EMP glycolysis, producing 5 net ATP rather than the 2 yielded traditionally (Figure 2). This is possible owing to PPi-PFK and pyruvate phosphate dikinase (PPDK) variants which utilize the high-energy anhydride bond in PPi (ΔG = −19 kJ/mol) rather than ATP for cellular work. In E. histolytica, coupling of PPDK to the activity of the ubiquitous adenylate kinase (AK), which catalyzes the formation of ATP and AMP from 2 ADP, allows the regeneration of AMP for PPDK activity [30]. In the thermophilic microbe Caldicellulosiruptor saccharolyticus, PPDK plays a catalytic role when grown on glucose and xylose, and a membrane-bound H+-translocating pyrophosphatase allows the establishment of a proton motive force using the energy in PPi [31]. This catabolic role would indicate that in extreme environments, C. saccharolyticus has evolved a means to conserve energy. Pyrophosphate-dependent metabolism is not only restricted to microbial systems. In mammals, there is evidence that inorganic PPi can substitute for ATP in spermatozoa [32]. However, with occasional exceptions, PPi hydrolysis as an alternative to ATP is restricted to lower-level organisms. For this reason, PPi-dependent alternatives to glycolytic enzymes present themselves as potential targets for the development of novel antibiotics, as these would not be expected to affect resident flora and human cells [33] In plants, the glycolytic isoforms PPi-PFK and PPDK have been identified and their roles in stress responses characterized [32].
Figure 2:
Pyrophosphate-dependent glycolysis harnesses the high-energy bond of PPi for ATP production. EMP: Embden-Meyerhof-Parnas; HK: hexokinase; PFK: phosphofructokinase; PGK: phosphoglycerate kinase; PK: pyruvate kinase; PPDK: pyruvate phosphate dikinase.
Unlike mammalian systems, plants have comparatively low activity of cytosolic inorganic pyrophosphatase, allowing PPi to accumulate at concentrations of up to 0.5 mM [34]. While the majority of plants on earth are C3 plants (e.g., wheat, cotton) which use the Calvin cycle for photosynthesis, select species are C4 (e.g., maize, sugarcane) and use the Hatch-Slack pathway for carbon fixation. Although it is well-established that PPDK in C4 plants plays a key role in photosynthesis by regenerating the primary CO2 acceptor phosphoenolpyruvate (PEP) in chloroplasts, less is known about its role in the cytoplasm of C3 plants where it is a ubiquitous albeit low-abundance enzyme [35,36]. To that effect, numerous studies have pointed to PPDK of C3 plants as a component of the defense response to both abiotic (cold, drought, anoxia) and biotic (viral infection) stressors [37–43]. Accumulation of PPDK transcripts has been observed in the roots of rice seedlings (Oryza sativa ssp) during 60 hours of exposure to low-oxygen stress, suggesting that the bioenergetic advantage provided by PPi is favoured when ATP production via photosynthesis and oxidative phosphorylation is hindered [44]. Indeed, even in species sensitive to low oxygen such as Arabidopsis spp., O2 deficiency also triggers a metabolic reconfiguration seemingly aimed at ATP conservation by switching out ATP-dependent PFK for PPi-PFK [45]. Moreover, in anoxic rice seedlings, sucrose metabolism proceeds via sucrose synthase and UDP-glucose pyrophosphorylase, thus utilizing the high-energy bond of PPi rather than the two ATP required in the invertase/hexokinase (HK) pathway [45]. As such, plants readily employ PPi as an energetic compound to render themselves highly adaptable to environmental changes. Knowledge of bioenergetic transformations rendering plants more resistant to stress can be used to improve crop yield for human consumption via the introduction of transgenic species [46].
Harnessing PPi as an energy source in times of duress has also been observed in otherwise aerobic microbes, such as the metabolically malleable Pseudomonas fluorescens. Exposure to the •NO donor sodium nitroprusside (SNP) for 24 h at high doses (10 mM) leads to the inactivation of ETC complexes I, II and IV in the microbe [47]. However, rather than succumbing to the toxicity, the organism establishes the temporal formation of a metabolon to anaerobically catabolize citrate and generate ATP. Metabolons are multi-protein complexes previously observed to occur with TCA cycle enzymes [48]. Such an arrangement allows the efficient channeling of substrates into their desired end products without the intermediary metabolites equilibrating with the bulk cellular fluid. In the case of P. fluorescens, a phosphorylation event localized to PPDK allows for the formation of a metabolon consisting of ATP-independent citrate lyase (CL), phosphoenolpyruvate carboxylase (PEPC) and PPDK which effectively generates pyruvate and ATP without the ETC [47,48]. By decreasing the activity of inorganic pyrophosphatase, the organism further ensures itself a continued supply of this high-energy moiety, allowing for survival during nitro-oxidative stress. While P. fluorescens is not itself a pathogenic threat, it remains to be seen if similar adaptive responses are possible in other gram-negative organisms, such as the closely related Pseudomonas aeruginosa, for which there is growing resistance to commercially-available antibiotics.
The glyoxylate shunt is a TCA cycle variant built for stressful times
Present in protists, fungi, bacteria and plants, the glyoxylate shunt (GS) is a deviation of the TCA cycle wherein isocitrate is metabolized to glyoxylate and succinate via isocitrate lyase (ICL, encoded by the aceA gene) rather than α-ketoglutarate (αKG) using KGDH [49,50]. This metabolic pathway allows the organism to essentially bypass the decarboxylation steps of the Krebs cycle. With the help of malate synthase (MS, encoded by aceB or glcB), glyoxylate can then condense with acetyl-CoA to form malate which is shuttled to glucose production via gluconeogenesis [50]. Glucose can then be used for the biosynthesis of cell wall components or nucleotides. Ultimately, the glyoxylate shunt allows organisms to grow on fatty acids as their primary carbon source, particularly when sugars are unavailable [51]. For instance, in the hypoxic environment of macrophages, the pathogenic microbe Mycobacterium tuberculosis can assume a latent state for extended time periods thanks to the metabolic flexibility of the GS, using fatty acids as the building blocks for life [52]. Deletion of both ICL1 and ICL2 in M. tuberculosis leads to rapid elimination of the bacteria, the causative agent of tuberculosis, from the lungs of infected mice [53]. Indeed, the lack of GS enzymes in mammals has propelled interest in these proteins as targets for antibiotics, as evidenced by investigations into the mechanisms of 2-vinyl-D-isocitrate and 3-nitroproprionate [54].
More recently, the role of the GS in the response to harsh conditions such as oxidative stress, antibiotic stress, cold-/heat-shock has been elucidated. For instance, M. tuberculosis gains resistance to the antibiotics isoniazid, rifampicin and streptomycin by up-regulating the expression and activity of ICL [55]. As these antibiotics generate oxidative stress, the purported role of ICL is to bypass NADH-generating steps of the TCA cycle, thus reducing the flux of electrons funnelled to aerobic respiration and limiting further ROS generation. In P. aeruginosa, a common pathogen, expression of ICL is a virulence mechanism during infections, particularly in the cystic fibrosis lung [56]. As most of the cellular Fe (~94%) is allocated to respiratory chain components, the GS in P. aeruginosa is also activated in Fe-limiting conditions and when exposed to ROS, factors that disrupt ETC activity [57,58]. In the marine gammaproteobacterium, Photobacterium angustum S14, Fe withdrawal over a period of 24 h leads to a decrease in respiration and growth rates in both wild-type (WT) and ICL knockout mutants (ΔICL) [59].
However, these rates were ~30% lower in ΔICL organisms, and were restored when ICL was reintroduced [59]. In many organisms, GS activation by exogenous stress permits the rerouting of metabolism to generate compounds which fend off the stressful stimuli. For example, P. fluorescens can utilize ICL and acylating glyoxylate dehydrogenase to produce oxalate which can subsequently chelate and precipitate heavy metals in the extracellular environment, thus avoiding the noxious effects of metal toxicity [60]. Moreover, this organism can use the GS along with succinate semialdehyde dehydrogenase and α-ketoglutarate decarboxylase to pool αKG, a non-enzymatic scavenger of ROS [61]. In C. elegans as well as the yeast Saccharomyces cerevisiae, the glyoxylate shunt allows both organisms to convert fatty acids or acetate to trehalose, a disaccharide made up of two alpha-linked glucose moieties [62]. During harsh desiccation, trehalose acts as protein chaperone to prevent aggregation and also stabilize membranes against the effects of fast rehydration, essentially allowing the organisms to enter a long ametabolic state of anhydrobiosis [63].
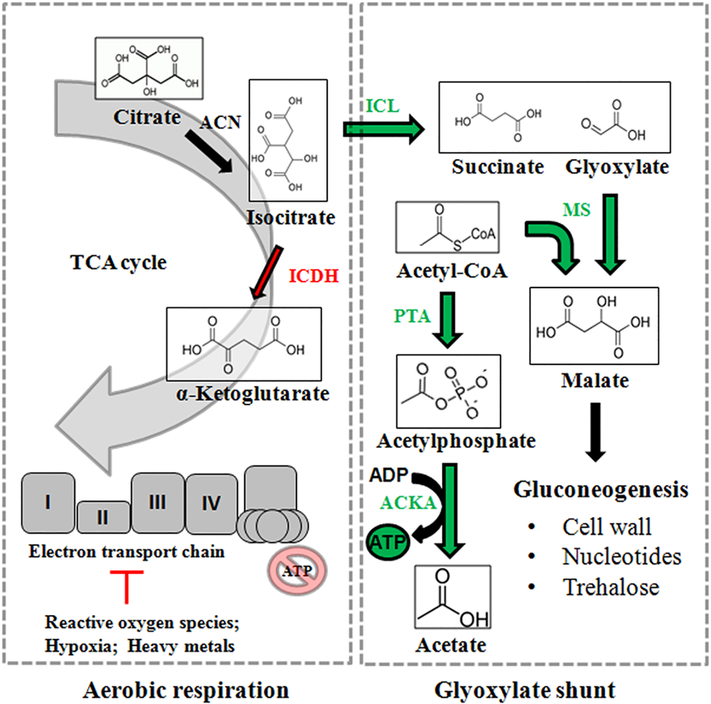
While the aforementioned organisms commonly employ the glyoxylate shunt for adaptation, use of the GS is particularly favored by species which thrive in extreme environments with limited nutrient availability. These include petroleum-polluted areas where temperatures, salinity, oxygen and levels of acidity vary greatly, wherein the GS permits select organisms to thrive on the degradation of long-chain alkanes [64]. Indeed, the growth of these extremophiles is of great interest in both the bioremediation and biotechnology industries. In some organisms, the GS allows them to bypass oxidative phosphorylation while still producing ATP via unconventional means. For instance, in Acinetobacter oleivorans DR1, numerous isoforms of alkane hydroxylases are upregulated to facilitate growth on the straight-chain alkane triacontane (C30) in comparison to the same bacteria grown on the TCA cycle intermediate succinate [65]. Moreover, TCA cycle enzymes show decreased expression during the assimilation of triacontane while ICL (aceA) becomes highly up-regulated, redirecting isocitrate towards the GS [65]. To produce energy in this metabolic reconfiguration, A. oleivorans DR1 elects to increase the expression of acetate kinase A (ackA) and phosphotransacetylase (pta), thus producing ATP via the conversion of acetyl-CoA to acetate and channelling carbon flux away from NADH production and the ETC [65,66] (Figure 3). In the ocean surface bacterium Dokdonia sp. MED134, light stimulation in the presence of alanine as a carbon source greatly upregulated (>300 fold) GS enzymes while proteorhodopsin produced ATP via light-driven proton pumping [67]. As such, it appears microorganisms with alternative ATP-producing machinery are readily capable of metabolizing isocitrate using the GS instead of the aerobic TCA cycle. A greater understanding of the pathways allowing extremophiles to thrive in nutrient-limited environments allows for the creation of genetically modified microorganisms which can safely remediate polluted environments (e.g., oil spills, radionuclides) [68].
Figure 3:
The glyoxylate shunt allows organisms to circumvent aerobic metabolism when faced with stress. Enzymatic reactions in red are down-regulated, while those in green see increased activity as part of the glyoxylate shunt. ACKA: acetate kinase A; ICDH: isocitrate dehydrogenase; ICL: isocitrate lyase; MS: malate synthase; PTA: phosphotransacetylase; TCA: tricarboxylic acid.
New insights into the bioenergetic roles of HIFs and HSPs
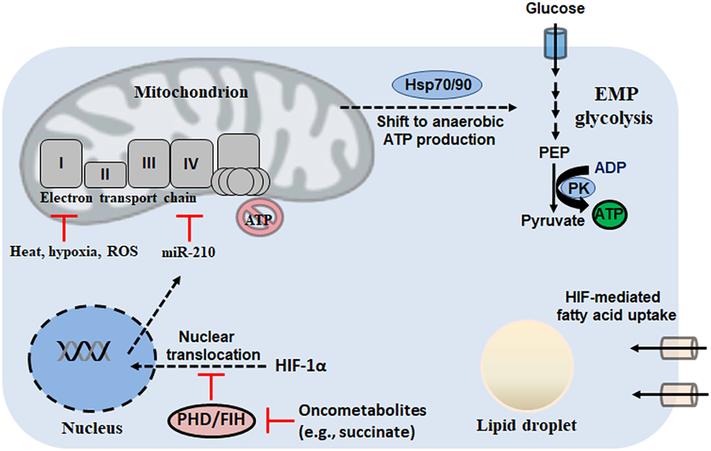
Higher mammals, including humans, lack the GS and PPi-dependent glycolytic variants seen in microbes and plants. As such, we are not privy to the metabolic reconfigurations and dynamic response to environmental stressors that have been elucidated in these organisms. To that effect, energy supporting biological functions is produced mostly via the mitochondrial ETC and partly by substrate-level phosphorylation (SLP) in the payoff phase of EMP glycolysis [69]. Coordination between the two during mitochondrial dysfunction and defects in aerobic respiration is mediated largely by hypoxia-inducible factors (HIF) as well as the complex interaction between nutrient and energy sensors including sirtuins, mammalian target of rapamycin complexes and adenosine monophosphate kinase (SIRT, mTOR and AMPK, respectively; reviewed extensively elsewhere) [70–73]. While HIFs primarily govern the response to hypoxia, heat shock proteins (HSPs) are a family of homologous chaperone proteins that regulate the response to environmental, chemical and physical stressors [74]. In response to reduced O2 availability, the transcriptional HIFs activate genes encoding glucose transport, glycolytic enzymes, lactate dehydrogenase and lactate export via MCT4 [75]. This reconfiguration facilitates ATP production via SLP while diverting carbon away from the TCA cycle. In normoxic conditions, two classes of oxygen-dependent dioxygenases hydroxylate HIF-α proteins. These consist of prolyl hydroxylase domain (PHD)-containing enzymes and the asparagine-targeting factor inhibiting HIF (FIH) enzyme [75]. Ultimately, proline hydroxylation of HIF-α leads to its ubiquitination by von-Hippel Lindau protein and subsequent proteasomal degradation. However, inhibition of PHDs leads to HIF stabilization and expression of hypoxic genes [75]. The central dogma of molecular biology, which dictates that molecular information flows from DNA to RNA and then protein, is challenged by the knowledge that small metabolites rising from environmental cues can also modulate cellular processes. Oncometabolites, moieties which are known to accumulate aberrantly in tumors and have pro-oncogenic roles, have also been shown to regulate hypoxia. For instance, the oncometabolite succinate was the first discovered oxygen-independent regulator of hypoxia with a role in the allosteric inhibition of PHDs [76]. It is now appreciated though that a number of metabolites, including pyruvate and lactate, can simulate a hypoxic environment (pseudohypoxia) and decrease PHD activity [77].
In addition to metabolites, other non-traditional regulators of cellular bioenergetics and adaptation to environmental cues have come to light. For instance, the role of microRNAs (miRNAs) in the control of genes responsible for mitochondrial dynamics and biogenesis has been elucidated. At 17–22 nucleotides of length, miRNAs are non-coding RNA molecules that can silence cognate mRNA and post-transcriptionally regulate gene expression [78]. Recently, it has been found that miRNAs can reinforce the adaptive response to hypoxia. For instance, miR-210 is potently induced by hypoxia and is dependent on HIF stability [79]. In the absence of O2, when the ETC can no longer produce ATP, miR-210 plays an indirect role in encouraging glycolytic SLP by directly targeting and down-regulating the expression of Fe-S cluster assembly proteins (ISCU 1/2), key components in the activity of ACN, SDH and complex I [80]. Moreover, miR-210 targets the complex IV assembly protein COX10, while also lowering cell energy requirements via its action on the cell cycle regulator E2F transcription factor 3 (E2F3) [80]. In the hypoxic tumour microenvironment, these strategies are exploited by cancer cells for continued proliferation. Cancer metastasis is enhanced via a clustering effect, whereby detached cells from the primary tumour site aggregate together to survive the loss of stromal interactions [81]. Labuschagne et al. recently described how detached cells upregulate cadherin expression to cluster together, inducing a hypoxic environment driving HIF-1α stabilization [82]. The latter induces mitophagy via BNIP3 and NIX, thus clearing damaged mitochondria and limiting ROS production to facilitate cell survival [83]. A by-product of this adaptation is a switch to glycolytic ATP production via SLP, a metabolic reconfiguration that can be targeted for therapy in detached, metastasizing cells. HIF-1α has also been shown to induce the uptake of fatty acids by increasing the expression of fatty acid binding proteins 3 and 7, leading to the accumulation of a significant lipid droplet (LD) during hypoxia [84]. Following reoxygenation and restored mitochondrial function, LDs are consumed for ATP production via β-oxidation, suggesting that HIFs facilitate energy production both during and after stressful stimuli.
Heat shock, as well as a number of stressors capable of inducing ETC dysfunction, triggers the up-regulation of HSPs to maintain cell survival and inhibit caspase activation and apoptosis. Indeed, in human volunteers exposed to extreme heat (~75°C) in a sauna, HSP expression was readily increased in peripheral blood mononuclear cells, while ATP production was shifted from oxidative phosphorylation to glycolytic SLP [85]. In cultured rat neurons, hyperthermia (43°C for 2 h) induces a progressive irreversible decline in O2 consumption after 30 min and mitochondrial dysfunction that progresses to cell death after 10 h, further suggesting heat stress negatively impacts oxidative phosphorylation [86]. Heat shock proteins are broadly characterized based on their size. For example, Hsp10 and Hsp60 are 10 and 60 kDa respectively and reside in the mitochondrion as chaperones for protein folding. Larger HSPs, however, such as Hsp70/90 (DnaK/HtpG in prokaryotes) contain C-terminal polypeptide-binding sites along with highly conserved N-terminal ATPase domains, linking them to cellular energy state [87]. Recent research has allowed for a better understanding of how HSPs regulate bioenergetics in response to environmental stress. Hsp70 induction in the myocardium undergoing ischemia allows the preservation of ATP levels [88]. In HeLa cells transfected with human Hsp70 cDNA, ATP levels are maintained by anaerobic glycolytic SLP while oxidative phosphorylation is attenuated, suggesting that challenging environments induce a shift away from mitochondrial ROS production [89]. In a model of hepatocellular carcinoma (HCC), Hsp90 has been shown to bind to the M2 isoform of PK (PKM2) and form a complex with the kinase GSK-3β which phosphorylates PKM2 and enhances its activity [90]. This metabolic reconfiguration results in potentiated glycolysis and proliferation while reducing apoptosis of HCC cells. Indeed, overexpression of both PKM2 and Hsp90 was a significant predictor of poor prognosis in HCC patents [90]. Collectively, studies on HIFs and HSPs have demonstrated that humans are adept at shifting ATP production away from mitochondria when faced with stress (Figure 4).
Figure 4:
In higher mammals, hypoxia-inducible factors (HIFs) and heat shock proteins (HSPs) facilitate the shift to anaerobic metabolism. EMP: Embden-Meyerhof-Parnas; FIH: asparagine-targeting factor inhibiting HIF; PEP: phosphoenolpyruvate; PHD: prolyl hydroxylase domain; PK: pyruvate kinase; ROS: reactive oxygen species.
Mitochondrial substrate-level phosphorylation: An overlooked role of succinate-CoA ligase
As described, the canonical role of the mitochondrion in eukaryotic systems is to house the TCA cycle and ETC, machinery that produces 9–11 ATP per turn of the cycle via oxidative phosphorylation. An intermediate step of the cycle is the production of succinate from succinyl-CoA, catalyzed by succinate-CoA ligase (SUCLG2) and producing GTP which is consumed in anabolic pathways (e.g., mitochondrial protein synthesis) [91]. To that effect, it has also been shown that variants in the β subunits of the enzyme exist which lead to a conformational change favouring ADP over GDP (SUCLA2) [91]. Moreover, differential tissue expression studies have demonstrated that the presence of the GDP-consuming isoform is favoured in anabolic tissues, whereas SUCLA2 is employed by catabolic organs such as the brain, heart and muscle [92,93]. In SUCLA2 +/− mice, brain mitochondrial function is perturbed, and in humans carrying SUCLA2 mutations, defective mitochondrial bioenergetics and mtDNA depletion are common pathological manifestations [93–97]. Theoretically, the presence of SUCLA2 allows for the production of 1 extra ATP per turn of the cycle, offering a bioenergetic advantage in these tissues. However, in some organisms, mitochondrial substrate-level phosphorylation (mSLP) is essential for growth. Trypanosoma brucei, the unicellular parasite responsible for human sleeping sickness, can readily cycle between the heterogeneous environments of the host bloodstream and the digestive track of the tsetse fly. As such, metabolic networks must be reconfigured to sufficiently generate ATP for cellular processes. Interestingly, in T. brucei, ablation of succinate dehydrogenase and the entry of electrons into the ETC does not cause a growth phenotype [98]. RNAi-induced knockdown of SUCL and mSLP does, however, decrease survival and suggests that anaerobic mitochondrial ATP production is necessary in the partly hypoxic conditions of the tsetse fly [98]. This metabolic reconfiguration can be capitalized upon by cells of the macrophage lineage, where the metabolite itaconate is produced as an antimicrobial compound. SUCLA2 can act on itaconate to produce itaconyl-CoA, decreasing the upstream supply of succinyl-CoA via a “CoA trap” and interfering with the ability of invading pathogens such as T. brucei to produce ATP anaerobically by mSLP [99].
Indeed, adaptation to hypoxia and mitochondrial damage appears to be a key purpose for SUCLA2 activity. It is known that mitochondrial F0-F1 ATP synthase is a reversible machine, consuming ATP when mitochondrial membrane potential (ΔΨm) is compromised. In a series of elegant studies, Chinopoulos et al. have demonstrated that during respiratory chain inhibition, mSLP via SUCLA2 provides ATP to F0-F1 ATP synthase to maintain ΔΨm without the need to import cytosolic ATP via adenine nucleotide translocase (ANT) [100]. As such, mSLP allows the maintenance of cytoplasmic energy stores to fend off stressful stimuli. In S. cerevisiae lacking a functional ATP synthase (Δfmc1), this respiratory growth defect can be rescued via the overexpression of the yeast oxodicarboxylate carrier Odc1p [101]. The latter allows the exchange of α-ketoadipate and αKG from the mitochondrial matrix and cytosol, respectively. Crucial to this adaptation appears to be the continual supply of αKG, as when mitochondrial function is impaired, genes leading to the production of this ketoacid come under the control of the retrograde-specific regulatory genes RTG1–3 [102]. While the main purpose of this metabolic re-engineering is the production of a precursor to glutamate for survival, Schwimmer et al. have demonstrated that mitochondria isolated from a Δfmc1/ODC1 yeast strain can produce ATP via mSLP using SUCLA2 when given αKG [103].
Perhaps our best understanding of the role SUCLA2 plays in bioenergetics under times of duress comes from cancer research. Glioblastoma multiforme (GBM) is the most common of adult brain cancers and carries the highest mortality rate. In GBM cells, mitochondria are abnormal in number, structure, cardiolipin composition and have defective oxidative phosphorylation suggesting severely compromised ability to produce ATP via aerobic respiration [104]. Indeed, estimates of ETC efficiency in cancer cells suggest that oxidative phosphorylation may operate at 50% capacity, however, this would drive F0-F1 ATP synthase in the reverse direction, consuming ATP to maintain ΔΨm and suggesting the ETC is not the primary contributor to energy production [105,106]. As in most tumour microenvironments, GBM cells have abundant access to glucose and glutamine, implicating that their survival is mediated by the Warburg effect (aerobic fermentation) and glutaminolysis respectively [107]. While SLP at the level of glycolytic PK could meet ATP requirements in cancer cells with defective oxidative phosphorylation, tumour cells often preferentially express the M2 isoform (PKM2) which has a low affinity for PEP [108]. As such, the role of glucose in select cancers may primarily be the production of key intermediates for tumour growth such as lipids and amino acids as well as ROS defense via shunting to the pentose phosphate pathway.
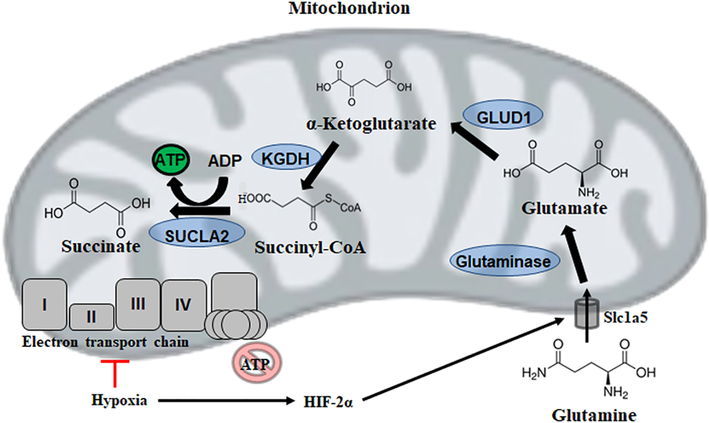
The high expression of the glutamine transporter Slc1a5 in GBM and other cancers would imply that this amino acid is essential for malignancy [109]. Indeed, it has recently been demonstrated in pancreatic cancer cells that HIF-2α induces the expression of a mitochondrial variant of Slc1a5 [110]. Moreover, glutaminase overexpression in cancer cells would favour the production of glutamate, which is then metabolized to αKG via glutamate dehydrogenase (GLUD1) or transamination. Evidence for the importance of this pathway in carcinogenesis stems from findings that tumour growth is significantly impaired when either enzyme is inhibited [111]. As in S. cerevisiae, this continual supply of αKG appears central to this metabolic re-engineering, providing a precursor to succinyl-CoA generated via KGDH. Production of ATP via SUCLA2 can then proceed, allowing tumour cells to, at least in part, meet energy requirements via mSLP despite mitochondrial abnormalities precluding oxidative phosphorylation (Figure 5). Indeed, in the invasive brain tumour model cell line VM-M3, both mycoplasma infection and hypoxia trigger rearrangements in the TCA cycle favouring the accumulation of succinate while also greatly increasing glutamine uptake, suggesting mSLP provides a fraction of ATP in the tumour microenvironment and permitting cancer cell growth [112].
Figure 5:
Mitochondrial substrate level phosphorylation via succinate-CoA ligase (SUCLA2) enables survival despite a defective electron transport chain. GLUD1: glutamate dehydrogenase 1; HIF: hypoxia-inducible factor; KGDH: α-ketoglutarate dehydrogenase.
Aside from energy production, it is also appreciated that glutamine is a key building block for cancer cells. Studies using 13C labelled carbon sources have demonstrated that, in a normoxic environment, labeling of acetyl-CoA in fatty acids from melanoma cells stems from glucose metabolism. However, during hypoxia, this labeling has its origins in glutamine metabolism [113]. The key to this reductive carboxylation is the reversible nature of select TCA cycle enzymes, such as isocitrate dehydrogenases 1 and 2 as well as aconitase. This allows the redirection of glutamine-derived αKG towards citrate which is metabolized via ATP-citrate lyase to acetyl-CoA units for lipogenesis [113]. Indeed, this reversed flux within the mitochondrion and TCA cycle enzymes is a common feature of hypoxia, as seen in diving mammals [114]. More recently, spatial-fluxomics has demonstrated that in HeLa cells with succinate dehydrogenase deficiency, citrate synthase activity can be reversed to produce oxaloacetate in mitochondria for pyrimidine synthesis when grown with glutamine [115]. For these reasons, therapeutic strategies aimed at limiting the bioavailability of glutamine for tumors, such as L-asparaginase treatment, have proven effective in select malignancies [116].
Phosphotransfer networks are the circuitry underlying rapid energy storage and access
In order to maintain energetic homeostasis, all biological systems require buffering networks for the storage and delivery of high-energy phosphate. While ATP itself is the universal energy currency, diffusion of nucleotides produced by F0-F1 ATP synthase is kinetically and thermodynamically inefficient, requiring significant concentration gradients. To that effect, in order to understand how ATP production via the ETC is linked to sites of ATP-consumption, the concept of ‘phosphowires’ was introduced. These consist of a network of enzymes including creatine kinase (CK), nucleoside-diphosphate kinase (NDPK) and the aforementioned AK which can transfer the high-energy phosphate to phosphocreatine (PCr) and alternative nucleotides, respectively [117]. Indeed, all three enzymes have been identified in the intermembrane space of the mitochondrion, providing further evidence for the existence of a phosphotransfer system [117]. The mean diffusion distance of creatine/PCr (37 and 57μm) is much higher than that of ADP/ATP (1.8 and 22 μm), allowing the shuttling of energy for functional work to far reaches of the cell, which is of particular benefit for energy-intensive tissues such as the heart and muscle in mammals [118]. While CK is found primarily in vertebrates; invertebrates such as insects, crustaceans and some unicellular organisms also have a buffering system for high-energy phosphate in the form of arginine kinase (ArgK) which stores ATP in the form of the phosphagen phosphoarginine.
Phosphagens are a family of high-energy storage compounds which can supply immediate energy when glycolytic SLP and the mitochondrial ETC are impaired or insufficient. Along with lombricine (earthworms), phosphoarginine and PCr comprise the best-characterized group of phosphagens, playing crucial roles as energy pools when organisms are faced with stressful stimuli. To delineate their importance, CK and ArgK buffering systems were installed in S. cerevisiae, which normally does not express these enzymatic networks [115]. The introduction of ArgK activity allowed for the creation of a 5 mM phosphoarginine pool in yeast, which were then subjected to transient pH reduction (pH of 2 for 1 h) and starvation to determine stress tolerance [119]. Versus WT yeast, those expressing ArgK had markedly improved resistance to both stressors, maintaining their ATP pool at the same pre-stress concentrations while WT yeast saw a 50% reduction [119]. In the versatile P. fluorescens, exposure to oxidative or nitrosative stresses incapacitates the ETC, favouring the production of ATP anaerobically via PPDK and pyruvate, water dikinase [120]. This metabolic reconfiguration allowing adaptation to ROS/RNS also sees the increased activity of NDPK and AK to permit the transfer of high-energy phosphate to other nucleosides and AMP production respectively, suggesting the presence of a buffering system and allowing continued anaerobic energy production [121]. Parasitic protozoans, such as the aforementioned T. brucei and Trypanosoma cruzi, the causative agent of Chagas disease, also rely heavily on phosphagen stores [122]. Indeed, ArgK undergoes a continuous increase in activity during the exponential phases of growth of T. cruzi [123]. Furthermore, ArgK overexpression in this parasite confers heightened resistance to pH and nutritional stress, providing a pool of energy when traditional glycolytic and TCA pathways are downregulated [123].
Owing to its role in human disease and metabolic disorders, PCr is the most-studied of all the phosphagens. Vertebrates express four distinct isoforms of CK: cytosolic CK in the muscle (CK-M) and brain (CK-B) and two mitochondrial enzymes located in muscle (sarcomeric mtCK) and ubiquitously (mtCK) [124]. Functional coupling and the reversibility of CK isoforms renders it possible for mtCKs to operate in one direction, forming high-energy PCr in mitochondria, while cytoplasmic isoforms generate ATP locally [124]. Heritable defects in creatine metabolism affecting its biosynthesis (AGAT: argine:glycine aminotransferase; GAMT: guanidinoacetate methyltransferase) and transport (SLC6A8) comprise a broad group of creatine deficiency syndromes characterized by mental retardation, seizures and speech disturbances [125]. Interestingly, murine GAMT KO studies have demonstrated that PCr is not necessary for the function of cardiomyocytes at rest [126]. Moreover, ineffective creatine biosynthesis had no effect on voluntary locomotor activity or exercise capacity until exhaustion. When mtCK and CK-M are knocked down, mice are incapable of maintaining ATP levels when submitted to an intense exercise protocol, despite no changes in baseline concentrations [127]. In humans, 45 obese volunteers were given dobutamine to increase cardiac work, but ATP delivery and CK kinetics were not increased to match the demand as compared to non-obese counterparts [128]. In breast cancer cells, human growth factor receptor 2 signaling-mediated phosphorylation of mtCK at Y153 increases its activity and buffers the cellular ATP pool in the tumour microenvironment [129]. Taken together, these findings imply that much of the bioenergetic importance of PCr is as an energy supply during stress, such as ischemia, hypoxia and ROS exposure.
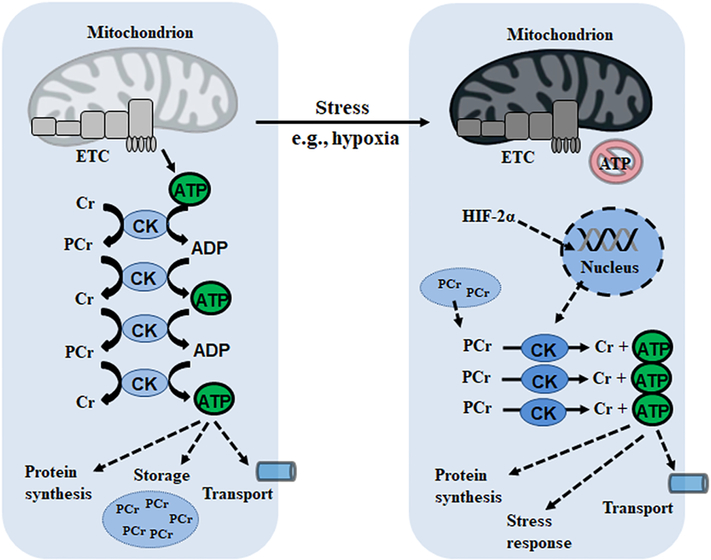
While creatine itself has been shown to have mild anti-oxidative properties, the tight coupling of ATP production to export by CK shuttling further lowers mitochondrial ROS production. Moreover, creatine is known to protect against mitochondrial toxins such as rotenone and paraquat as well as limit mtDNA damage [130]. The role of CK and PCr as a phosphagen in hypoxic environments has been well-delineated in intestinal epithelia, a barrier which must remain highly dynamic in order to accommodate nutrient and fluid transport [131]. The distance between mitochondrial and subcellular locations of ATP consumption in intestinal epithelial cells demands a CK/PCr phosphotransfer system, facilitating energy production coupled to myosin ATPase activity [130]. The latter mediates static tension and actin contractility which, if disrupted, affects barrier integrity and the assembly of apical junctions. As such, PCr is a source of rapid ATP generation allowing transepithelial transport. In inflammatory bowel disease (IBD), such as Crohn’s and ulcerative colitis, the mucosal barrier is subjected to intestinal inflammation and depletion of local oxygen which can progress to hypoxic lesions [132]. Using chromatin immunoprecipitation assays, Glover et al. demonstrated the association between HIF-2α and the promoter sequences of CK-B/CK-M, suggesting hypoxia triggers PCr consumption (Figure 6) [133]. Indeed, both mitochondrial and cytoplasmic CK activities were decreased in clinical IBD biopsies and reduced ATP levels have been observed in inflamed tissue, suggesting a key role for PCr as a phosphagen in this dynamic environment.
Figure 6:
Phosphotransfer systems permit the delivery and storage of high-energy phosphate in the form of phosphagens which are conducive to rapid ATP production during a stress response. Cr: creatine; ETC: electron transport chain; HIF: hypoxia-inducible factor; PCr: phosphocreatine.
Energy production in the human microbiome and disease relevance
Intestinal microbiota (also collectively referred to as the human microbiome) are bacterial populations that colonize the human gastrointestinal tract (GI) and have an integral role in normal gut physiology [137]. Thus, aberrations in the human microbiome (dysbiosis) are implicated in various disease states. An important metabolic function of these microbiota is fermentation of complex carbohydrates to short chain fatty acids, which are detectable in circulation [138]. This supports the concept that intestinal microbiota can communicate with other organs via metabolic byproducts, impacting physiologic function. Indeed, dysbiosis is correlated with GI diseases (e.g., irritable bowel syndrome (IBS)) and chronic diseases such as diabetes, cancer, asthma and potentially psychiatric conditions (e.g., autism spectrum disorder) [137, 139–141].
Metabolic substrates harnessed by gut microbiota include dietary carbohydrates, lipids, and amino acids [137]. Carbohydrate-derived short chain fatty acids (e.g. propionate, butyrate) are abundant in the intestine [142]. Propionate is found in the small and large intestine while butyrate is present in the cecum and colon. The former is a key energy source for epithelial cells and is transported to the liver where it regulates gluconeogenesis, reducing hepatic glucose and ultimately, adiposity [143]. Propionate is synthesized from five and six carbon sugars and arises via pyruvate and succinate intermediates; however, there are alternative pathways for propionate synthesis [137]. These pathways play an important role in fermentation of deoxy sugar moieties of host glycans and involve hydrolysis of propane-1,2-diol to propanol and eventually, propionyl-CoA [144]. Similarly, butyrate synthesis occurs via several pathways that utilize acetyl-CoA, glutarate, 4- aminobutyrate, or lysine as primary substrates [137]. In the microbiome of healthy human subjects, the primary pathway for butyrate synthesis is via acetyl-CoA and the secondary pathway is via lysine [145]. Interestingly, these pathways often occur together, and the acetyl-CoA pathways can utilize protein degradation byproducts allowing for processing of protein-rich diets [145]. Butyrate production itself potentially wields anti-cancer functions and is a key energy source for colonocytes, activating intestinal gluconeogenesis to regulate energy homeostasis [146].
Taken together, bacterial populations comprising the human microbiome are responsible for harnessing dietary molecules and activating a number of metabolic pathways. Importantly, alterations in substrate bioavailability impacts gene expression patterns and microbiota distribution due to competitive advantage of certain species [147]. Dysbiosis and altered metabolism underlies various disease states, and targeting the microbiome is an attractive potential therapeutic strategy [137]. To that end, regulating dietary substrates (e.g., FODMAP diet in IBS) or repopulating health gut bacteria via fecal microbiota transplant are potential interventions with broad clinical implications [148,149]. However, while microbiome research is currently rapidly advancing, there are many unanswered questions regarding elucidated metabolic pathways. Currently identified metabolites are likely only a small fraction of the total energy-producing biomolecules, necessitating further analysis of potential small molecules that can be harnessed for alternative energy-producing pathways in normal and human disease states [150]. Potentially, targeting substrate availability and activation of particular metabolic pathways in chronic diseases could be an attractive strategy, although further work is needed at this time.
Conclusion: Biotechnological and therapeutic implications
ATP is a key ingredient to sustain life across all organisms. However, the primary source of this moiety, oxidative phosphorylation, is prone to failure due to exogenous toxins, endogenous ROS, pathological inflammatory responses and environmental fluctuations. To counteract the frailty of the ETC, alternative metabolic networks have to rapidly and effectively be put in place to permit continued energy production under harsh conditions. Whether this includes capitalizing on the high-energy bond in pyrophosphate, rerouting carbon catabolism to the glyoxylate shunt or turning to phosphagen stores for energetic needs, biological systems have clearly evolved failsafes for survival in these precise scenarios (Table 1). However, a declining focus on exploratory basic science in favour of clinical research has limited the discovery of these novel metabolic pathways. This is an oversight, as an understanding of alternative ATP-producing machinery can have wide-ranging socioeconomic and healthcare benefits. Identification of the GS and PPi-dependent metabolism, owing to the absence of these processes in humans, has led to the identification of new antibiotic targets for which resistance has not been spread. In biorefineries, enhancing the ATP supply via metabolic engineering allows for the production of value-added products by microbial cell factories. Effective inhibition of SLP pathways in proliferating cancer cells with mitochondrial abnormalities can halt their progression. To that end, we conclude that a comprehensive understanding of the metabolic malleability allowing organisms to adapt to heterogeneous environments can provide cues for the design of novel therapeutics and sustainable manufacturing processes.
Table 1:
ATP-generating enzymes discovered across species.
| Enzyme name (EC number) | Reaction catalyzed | Organism(s) | References |
|---|---|---|---|
| Phosphoglycerate kinase (EC 2.7.2.3) | 1,3-bisphosphoglycerate + ADP ←→ glycerate 3-phosphate + ATP | All organisms | [26], [134] |
| Pyruvate kinase (EC 2.7.1.40) | Phosphoenolpyruvate + ADP → pyruvate + ATP | All organisms | [135], [136] |
| Pyruvate, water dikinase (EC 2.7.9.2) | Phosphoenolpyruvate + AMP + Pi ←→ pyruvate + H2O + ATP | Bacteria, Archaea, Some eukaryotes | [151] |
| Pyruvate phosphate dikinase (EC 2.7.9.1) | Phosphoenolpyurvate + AMP + PPi ←→ pyruvate + Pi + ATP | Bacteria, Archaea, Some eukaryotes (plants) | [35], [36] |
| Arginine kinase (EC 2.7.3.3) | Nω-phospho-L-arginine + ADP ←→ L-arginine + ATP | Bacteria, Eukaryotes (invertebrates) | [152], [153] |
| Creatine kinase (EC 2.7.3.2) | Phosphocreatine + ADP ←→ creatine + ATP | Higher mammals | [154] |
| Lombricine kinase (EC 2.7.3.5) | N-phospholombricine + ADP ←→ lombricine + ATP | Earthworms | [155] |
| Glycocyamine kinase (EC 2.7.3.1) | Phosphoguanidinoacetate + ADP ←→ guanidinoacetate + ATP | Sandworm | [156] |
| Hypotaurocyamine kinase (EC 2.7.3.6) | Nω-phosphohypotaurocyamine + ADP ←→ hypotaurocyamine + ATP | Sipunculan worm | [157] |
| Opheline kinase (EC 2.7.3.7) | N’-phosphoguanidinoethyl methyl phosphate +ADP ←→guanidinoethyl methyl phosphate + ATP | Ophelia neglecta | [158] |
| Taurocyamine kinase (EC 2.7.3.4) | N-phosphotaurocyamine + ADP ←→taurocyamine + ATP | Blood fluke | [159] |
| Succinate-CoA ligase (EC 6.2.1.5) | Succinyl-CoA + Pi + ADP ←→succinate + CoA + ATP | Bacteria, Archaea, Eukaryotes | [91] |
| ATP synthase (EC 7.1.2.2) | ADP + 4H+ (Side 1) + Pi ←→ ATP + H2O + 4H+ (Side 2) | Bacteria, Archaea, Eukaryotes | [161] |
| Butyrate kinase (EC 2.7.2.7) | Butyryl-phosphate + Pi + ADP ←→butyrate + ATP | Bacteria, Archaea, Eukaryotes | [137] |
Highlights.
The ETC is prone to temporal disruptions due to stressful environments
Organisms have engineered fail-safes to permit continued ATP production
Pyrophosphate and glycolytic variants are harnessed in times of duress
Mitochondrial substrate level phosphorylation is and overlooked energy source
Phosphagens and phosphotransfer networks are key to proper cellular function
Acknowledgements:
This work was supported by grants from the Canadian Institutes of Health Research (#123336), the Canada Foundation for Innovation Leader’s Opportunity Fund (#25407) and National Institutes of Health (R01GM133961).
Footnotes
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lynch M & Marinov GK. Membranes, energetics, and evolution across the prokaryote-eukaryote divide. eLife. 2017;6:e20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. The Evolution of Electron-Transport Chains. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26849. [Google Scholar]
- 3.Wolfe AJ. Glycolysis for the Microbiome Generation. Microbiol Spectr. 2015;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearce LL, Martinez-Bosch S, Manzano EL, et al. The Resistance of Electron Transport Chain Fe-S Clusters to Oxidative Damage during the Reaction of Peroxynitrite with Mitochondrial Complex II and Rat Heart Pericardium. Nitric Oxide. 2009;20(3):135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown GC & Borutaite V. Inhibition of mitochondrial respiratory complex I by nitric oxide, peroxynitrite and S-nitrosothiols. Biochim Biophys Acta. 2004;1658:44–49. [DOI] [PubMed] [Google Scholar]
- 6.Panov A, Dikalov S, Shalbuyeva N, et al. Rotenone model of Parkinson disease: multiple brain mitochondria dysfunctions after short term systemic rotenone intoxication. J Biol Chem. 2005;280:42026–42035. [DOI] [PubMed] [Google Scholar]
- 7.Auger C, Alhasawi A, Contavadoo M, et al. Dysfunctional mitochondrial bioenergetics and the pathogenesis of hepatic disorders. Front Cell Dev Biol. 2015;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paradies G, Petrosillo G, Pistolese M, et al. Reactive oxygen species affect mitochondrial electron transport complex I activity through oxidative cardiolipin damage. Gene. 2002;286:135–141. [DOI] [PubMed] [Google Scholar]
- 9.Boveris A & Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J. 1973;134(3):707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolfi-Donegan D, Braganza A, Shiva S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk JM & Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci USA. 2012;109(15):5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arsenijevic D, Onuma H, Pecqueur C, et al. Disruption of the uncoupling protein-2 gene in mice reveals a role in immunity and reactive oxygen species production. Nat Gen. 2000;26:435–439. [DOI] [PubMed] [Google Scholar]
- 13.Veith A & Moorthy B. Role of cytochrome P450S in the generation and metabolism of reactive oxygen species. Curr Opin Toxicol. 2018;7:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jager TL, Cockrell AE, Du Plessis SS. Ultraviolet Light Induced Generation of Reactive Oxygen Species. Adv Exp Med Biol. 2017;996:15–23. [DOI] [PubMed] [Google Scholar]
- 15.Azzam EI, Jay-Gerin J-P, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012;327(0):48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal D, Haque M, Sriramula S, et al. Role of Proinflammatory Cytokines and Redox Homeostasis in Exercise-Inducted Delayed Progression of Hypertension in Spontaneously Hypertensive Rats. Hypertension. 2009;54:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem. 2001;1(6): 529–539. [DOI] [PubMed] [Google Scholar]
- 18.Radi R Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. PNAS. 2018;115(23):5839–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doorn JA, Hurley TD, Petersen DR. Inhibition of human mitochondrial aldehyde dehydrogenase by 4-hydroxynon-2-enal and 4-oxonon-2-enal. Chem Res Toxicol. 2006;19:102–110. [DOI] [PubMed] [Google Scholar]
- 20.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial Reactive Oxygen Species (ROS) and ROS-induced ROS Release. Physiol Rev. 2014;94(3):909–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pacher P, Beckman JS, Liaudet L. Nitric Oxide and Peroxynitrite in Health and Disease. Physiol Rev. 2007;87(1):315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collin F Chemical Basis of Reactive Oxygen Species Reactivity and Involvement in Neurodegenerative Diseases. Int J Mol Sci. 2019;20(10):2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearce LL, Martinez-Bosch S, Manzano EL, et al. Resistance of Electron Transport Chain Fe-S Clusters to Oxidative Damage during the Reaction of Peroxynitrite with Mitochondrial Complex II and Rat Heart Pericardium. Nitric Oxide. 2009;20(3):135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller WEG, Schröder HC, Wang X. Inorganic Polyphosphates As Storage for and Generator of Metabolic Energy in the Extracellular Matrix. Chem Rev. 2019;24:12337–12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baykov AA, Malinen AM, Luoto HH, et al. Pyrophosphate-Fueled Na+ and H+ Transport in Prokaryotes. Microbiol Mol Biol Rev. 2013;77(2):267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abhrahamian M, Kagda M, Ah-Fong AMV, et al. Rethinking the evolution of eukaryotic metabolism: novel cellular partitioning of enzymes in stramenopiles links serine biosynthesis to glycolysis in mitochondria. BMC Evolutionary Biology. 2017;17(241). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata R, Fujihashi M, Sato T, et al. Identification of a pyrophosphate-dependent kinase and its donor selectivity determinants. Nat Commun. 2018;9(1765). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saavedra E, Encalada R, Vázquez C, et al. Control and regulation of the pyrophosphate-dependent glucose metabolism in Entamoeba histolytics. Mol Biochem Parasitol. 2019;229:75–87. [DOI] [PubMed] [Google Scholar]
- 29.Han J & Collins LJ. Reconstruction of Sugar Metabolic Pathways of Giardia lamblia. Int J Proteomics. 2012;2012:980829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varela-Gómez M, Moreno-Sánchez R, Pardo JP, et al. Kinetic Mechanism and Metabolic Role of Pyruvate Phosphate Dikinase from Entamoeba histolytica. JBC. 2004;279(52):54124–54130. [DOI] [PubMed] [Google Scholar]
- 31.van de Werken HJG, Verhaart MRA, VanFossen AL, et al. Hydrogenomics of the Extremely Thermophilic Bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol. 2008;74(21):6720–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi Y-J, Sutovsky M, Kennedy C, et al. Identification of the Inorganic Pyrophosphate Metabolizing, ATP Substituting Pathway in Mammalian Spermatozoa. PLoS One. 2012;7(4):e34524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C, Dunaway-Mariano D, Mariano PS. Design, synthesis, and evaluation of inhibitors of pyruvate phophate dikinase. J Org Chem. 2013;78:1910–1922. [DOI] [PubMed] [Google Scholar]
- 34.Atwell BJ, Greenway H, Colmer TD. Efficient use of energy in anoxia-tolerant plants with focus on germinating rice seedlings. New Phytol. 2015;206:36–56. [DOI] [PubMed] [Google Scholar]
- 35.Chastain CJ, Failing CJ, Manandhar L, et al. Functional evolution of C4 pyruvate, orthophosphate dikinase. J Exp Bot. 2011;62(9):3083–3091. [DOI] [PubMed] [Google Scholar]
- 36.Chastain CJ, Fries JP, Vogel JA, et al. Pyruvate, Orthophosphate Dikinase in Leaves and Chloroplasts of C3 Plants Undergoes Light-/Dark-Induced Reversible Phosphorylation. Plant Physiol. 2002;128(4):1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyskova V & Ryslava H. Unusual Properties and Functions of Plant Pyruvate, Orthophosphate Dikinase. Biochem Anal Biochem. 2016; 5(1). [Google Scholar]
- 38.Moons A, Valcke R, van Montagu M. Low-oxygen stress and water deficit induce cytosolic pyruvate orthophosphate dikinase (PPDK). Plant J. 1998;15:89–98. [DOI] [PubMed] [Google Scholar]
- 39.Wang DF, Portis AR, Moose SP, et al. Cool /c4 photosynthesis: Pyruvate Pi dikinase expression and activity corresponds to the exceptional cold tolerance of carbon assimilation in Miscanthus x giganteus. Pland Physiol. 2008;148:557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HM, Wang WJ, Wang HZ, et al. Effect of inland salt-alkaline stress on C4 enzymes, pigments, antioxidant enzymes, and photosynthesis in leaf, bark, and branch chlorenchyma of poplars. Photosynthetica. 2013;51:115–126. [Google Scholar]
- 41.Doubnerova V, Janoskova M, Synkova H, et al. Effect of Potato virus Y on activities of antioxidant and anaplerotic enzymes in transgenic Nicotiana tabacum L plants with the gene for P3 protein. Gen Appl Plant Physiol. 2007;33:123–140. [Google Scholar]
- 42.Hyskova DV, Miedzinska L, Dobra J, et al. Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J Plant Physiol. 2014;171:19–25. [DOI] [PubMed] [Google Scholar]
- 43.Ryslava H, Muller K, Semoradova S, et al. Photosynthesis and activity of phosphoenolpyruvate carboxylase in Nicotiana tabacum L. leaves infected by Potato virus A and Potato virus Y. Photosynthetica. 2003;41:357–363. [Google Scholar]
- 44.Moons A, Valcke R, Van Montagu M. Low-oxygen stress and water deficit induce cytosolic pyruvate orthophosphate dikinase (PPDK) expression in roots of rice, a C3 plant. Plant J. 1998;15:89–98. [DOI] [PubMed] [Google Scholar]
- 45.Mustroph A, Stock J, Hess N, et al. Characterization of the phosphofructokinase gene family in rice and its expression under oxygen deficiency stress. Front Plant Sci. 2013;4:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ku MSB, Ranade U, Hsu T-P, et al. Photosynthetic performance of transgenic rice plants overexpressing maize C4 photosynthesis enzymes. Studies in Plant Science. 2000;7:193–204. [Google Scholar]
- 47.Auger C, Lemire J, Cecchini D, et al. The Metabolic Reprogramming Evoked by Nitrosative Stress Triggers the Anaerobic Utilization of Citrate in Pseudomonas fluorescens. PLoS One. 2011;6(12):e28469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y & Fernie AR. Metabolons, Enzyme-Enzyme Assemblies that Mediate Substrate Channeling, and Their Roles in Plant Metabolism. Plant Communications. 2020;1:100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanni P, Giachetti E, Pinzauti G, et al. Comparative structure, function and regulation of isocitrate lyase, and important assimilatory enzymes. Comp Biochem Physiol B. 95:431–458. [DOI] [PubMed] [Google Scholar]
- 50.Ahn S, Jung J, Jang I-A, et al. Role of Glyoxylate Shunt in Oxidative Stress Response. J Biol Chem. 2016;291(22):11928–11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maloy SR, Bohlander M, Nuun WD. Elevated levels of glyoxylate shunt enzymes in Escherichia coli strains constitutive for fatty acid degradation. J Bacteriol. 1980;143:720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serafini A, Tan L, Horswell S, et al. Mycobacterium tuberculosis requires glyoxylate shunt and reverse methylcitrate cycle for lactate and pyruvate metabolism. Mol Microbiol. 2019;112(4):1284–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muñoz-Elias E & McKinney JD. M. tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2006;11(6):638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee Y-V, Wahab HA, Choong YS. Potential Inhibitors for Isocitrate Lyase of Mycobacterium tuberculosis and Non-M. tuberculosis: A summary. J Biomed Biotechnol. 2015;3:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nandakumar N, Nathan C, Rhee KY. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5(4306):1–10. [DOI] [PubMed] [Google Scholar]
- 56.Hagins JM, Locy R, Silo-Suh L. Isocitrate Lyase Supplies Precursors for Hydrogen Cyanide Production in a Cystic Fibrosis Isolate of Pseudomonas aeruginosa. J Bacteriol. 2009;191(20):6335–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tortell PD, Maldonado MT, Granger J, et al. Marine bacteria and biogeochemical cycling of iron in the oceans. FEMS Microbiol Ecol. 1999;29:1–11. [Google Scholar]
- 58.Ha S, Shin B, Park W. Lack of glyoxylate shunt dysregulates iron homeostasis in Pseudomonas aeruginosa. Microbiology. 2018;164:587–599. [DOI] [PubMed] [Google Scholar]
- 59.Koedooder C, Guéneuguès A, Van Geersdaële R, et al. The Role of the Glyoxylate Shunt in the Acclimation to Iron Limitation in Marine Heterotrophic Bacteria. Front Mar Sci. 2018;5(145). [Google Scholar]
- 60.Singh R, Lemire J, Mailloux RJ, et al. An ATP and Oxalate Generating Variant Tricarboxylic Acid Cycle Counters Aluminum Toxicity in Pseudomonas fluorescens. PLoS One. 2009;4(10):e7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mailloux RJ, Beriault R, Lemire J, et al. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS One. 2007;2:e690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erkut C, Gade VR, Laxman S, et al. The glyoxylate shunt is essential for desiccation tolerance in C. elegans and budding yeast. eLife. 2016;5:e13614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leprince O & Buitink J. Introduction to desiccation biology: from old borders to new frontiers. Planta. 2015;242:369–378. [DOI] [PubMed] [Google Scholar]
- 64.Park C & Park W. Survival and Energy Producing Strategies of Alkane Degraders Under Extreme Conditions and Their Biotechnological Potential. Front Microbiol. 2018;9:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park C, Shin B, Jung J, et al. Metabolic and stress responses of Acinetobacter oleivorans DR1 during long-chain alkane degradation. Microb Biotechnol. 2017;10(6):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park C & Park W. Survival and Energy Producing Strategies of Alkane Degraders Under Extreme Conditions and Their Biotechnological Potential. Front Microbiol. 2018;9:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palovaara J, Akram N, Baltar F, et al. Stimulation of growth by proteorhodopsin phototrophy involves regulation of central metabolic pathways in marine planktonic bacteria. Proc Natl Acad Sci USA. 2014;111(35):E3650–E3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prakash D, Gabani P, Chandel AK, et al. Bioremediation: a genuine technology to remediate radionuclides from the environment. Microb Biotechnol. 2013;6(4):349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernie AR, Zhang Y, Sweetlove LJ. Passing the Baton: Substrate Channelling in Respiratory Metabolism. Research. 2018;2018(9):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vachharajani VT, Liu T, Wang X, et al. Sirtuins Link Inflammation and Metabolism. J Immunol Res. 2016;2016:8167273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13(4):225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zoncu R, Efeyvan A, Sabatini DM. mTOR: from growth signal integrations to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canto C & Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beere HM. ‘The stress of dying’: the role of heat shock proteins in the regulation of apoptosis. J Cell Sci. 2004;117(13):2641–2651. [DOI] [PubMed] [Google Scholar]
- 75.Xie H & Simon MC. Oxygen availability and metabolic reprogramming in cancer. J Biol Chem. 2017;292(41):16825–16832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang M, Su H, Soga T, et al. Prolyl hydroxylase domain enzymes: important regulators of cancer metabolism. Hypoxia. 2014;2:127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iommarini L, Porcelli AM, Gasparre G, et al. Non-Canonical Mechanisms Regulating Hypoxia-Inducible Factor 1 Alpha in Cancer. Front Oncol. 2017;7:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fang Z, Du R, Edwards A, et al. The Sequence Structures of Human MicroRNA Molecules and Their Implications. PLoS One. 2013;8(1):e54215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivan M & Huang X. miR-210: Fine-Tuning the Hypoxic Response. Adv Exp Med Biol. 2014;772:205–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan SY, Zhang Y-Y, Hemann C, et al. MicroRNA-210 Controls Mitochondrial Metabolism during Hypoxia by Repressing the Iron-Sulfur Cluster Assembly Proteins ISCU1/2. Cell Metab. 2009;10(4):273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aceto N Bring along your friends: Homotypic and heterotypic circulating tumor cell clustering to accelerate metastasis. Biomed J. 2020;43(1):18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Labuschagne CF, Cheung EC, Blagih J, et al. Cell Clustering Promotes a Metabolic Switch that Supports Metastatic Colonization. Cell Metab. 2019;30(4):720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J & Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death & Differentiation. 2009;16:939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mylonis I, Simos G, Paraskeva E. Hypoxia-Inducible Factors and the Regulation of Lipid Metabolism. Cells. 2019;8(3):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bouchama A, Aziz MA, Al Mahri S, et al. A Model of Exposure to Extreme Environmental Heat Uncovers the Human Transcriptome to Heat Stress. Sci Rep. 2017;7(9429):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.White MG, Saleh O, Nonner D, et al. Mitochondrial dysfunction induced by heat stress in cultured rat CNS neurons. J Neurophysiol. 2012;108(8):2203–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smoch RG, Blackburn ME, Gierasch LM. Conserved, Disordered C Terminus of DnaK Enhances Cellular Survival upon Stress and DnaK in vitro Chaperone Activity. J Biol Chem. 2011;286(36):31821–31829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.George I, Geddis MS, Lill Z, et al. Myocardial Function Improved by Electromagnetic Field Induction of Stress Protein hsp70. J Cell Physiol. 2008;216(3):816–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Schumann U, Liu Y, et al. Heat shock protein 70 (Hsp70) inhibits oxidative phosphorylation and compensates ATP balance through enhanced glycolytic activity. J Appl Physiol. 1985;113(11):1669–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer. 2017;16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lambeth DO. Reconsideration of the significance of substrate-level phosphorylation in the citric acid cycle. BAMBED. 2006;34(1):21–29. [DOI] [PubMed] [Google Scholar]
- 92.Johnson J, Mehus J, Tews K, et al. Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J Biol Chem. 1998;273:27580–27586. [DOI] [PubMed] [Google Scholar]
- 93.Lambeth D, Tews K, Adkins S, et al. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J Biol Chem. 2004;279:36621–36624. [DOI] [PubMed] [Google Scholar]
- 94.Zhao Y, Tian J, Sui S, et al. Loss of succinyl-CoA synthase ADP-forming β subunit disrupts mtDNA stability and mitochondrial dynamics in neurons. Sci Rep. 2017;7:7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Hove JLK, Saenz MS, Thomas JA, et al. Succinyl-CoA ligase deficiency: a mitochondrial hepatoencephalomyopathy. Pediatr Res. 2010;68(2):159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller C, Wang L, Ostergaard E, et al. The interplay between SUCLA2, SUCLG2, and mitochondrial DNA depletion. Biochimica et biophysica acta. 2011;1812:625–629. [DOI] [PubMed] [Google Scholar]
- 97.Carrozzo R, Verrigni D, Rasmussen M, et al. Succinate-CoA ligase deficiency due to mutations in SUCLA2 and SUCLG1: phenotype and genotype correlations in 71 patients. Journal Inherit Metab Dis. 2016;39:243–252. [DOI] [PubMed] [Google Scholar]
- 98.Bochud-Allemann N & Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procylic Trypanosoma brucei. J Biol Chem. 2002;277(36):32849–32854. [DOI] [PubMed] [Google Scholar]
- 99.Tretter L, Patocs A, Chinopoulos C. Succinate, and intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. BBA-Bioenergetics. 2016;1857(8):1086–1101. [DOI] [PubMed] [Google Scholar]
- 100.Chinopoulus C, Gerencser AA, Mandi M, et al. Forward operation of adenine nucleotide translocase during F0F1-ATPase reversal: critical role of matrix substrate-level phosphorylation. FASEB J. 2010;24(7):2405–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Taffin de Tilques M, Tribouillard-Tanvier D, Tétaud E, et al. Overexpression of mitochondrial oxodicarboxylate carrier (ODC1) preserves oxidative phosphorylation in a yeast model of Barth syndrome. Dis Model Mech. 2017;10(4):439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liao X & Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. [DOI] [PubMed] [Google Scholar]
- 103.Schwimmer C, Lefebvre-Legendre L, Rak M, et al. Increasing mitochondrial substrate-level phosphorylation can rescue respiratory growth of an ATP synthase deficient yeast. J Biol Chem. 2005;280(35):30751–30759. [DOI] [PubMed] [Google Scholar]
- 104.Chinopoulos C & Seyfried TN. Mitochondrial Substrate-Level Phosphorylation as Energy Source for Glioblastoma: Review and Hypothesis. ASN Neuro. 2018;10:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chinopoulos C. Mitochondrial consumption of cytosolic ATP: Not so fast. FEBS Lett. 2011;585:1255–1259. [DOI] [PubMed] [Google Scholar]
- 106.Chinopoulos C. The “B Space” of mitochondrial phosphorylation. J Neurosci Res. 2011;89:1897–1904. [DOI] [PubMed] [Google Scholar]
- 107.Strickland M & Stoll EA. Metabolic Reprogramming in Glioma. Front Cell Dev Biol. 2017;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prakasam G, Iqbal MA, Bemezai RNK, et al. Posttranslational Modifications of Pyruvate Kinase M2: Tweaks that Benefit Cancer. Front Oncol. 2018;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bhutia YD & Ganapathy V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim Biophys Acta. 2016;1863(10):2531–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yoo HC, Park SJ, Nam M, et al. A Variant of SLC1A5 Is a Mitochondrial Glutamine Transporter for Metabolic Reprogramming in Cancer Cells. Cell Metab. 2020;31(2):267–283. [DOI] [PubMed] [Google Scholar]
- 111.Jin L, Alesi GN, Fan J, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27(2):257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flores RE, Brown AK, Taus L, et al. Mycoplasma infection and hypoxia initiate succinate accumulation and release in the VM-M3 cancer cells. Biochim Biphys Acta Bioenerg. 2018;1859(9):975–983. [DOI] [PubMed] [Google Scholar]
- 113.Filipp FV, Ratnikov B, De Ingeniis J, et al. Glutamine-fueled mitochondrial metabolism is decoupled from glycolysis in melanoma. Pigment Cell Melanoma Res. 2012;25(6):732–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hollinshead KER & Tennant DA. Mitochondrial metabolic remodeling in response to genetic and environmental perturbations. WIREs Mechanisms of Disease. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee WD, Mukha D, Aizenshtein E, Shlomi T. Spatial-fluxomics provides a subcellular-compartmentalized view of reductive glutamine metabolism in cancer cells. Nat Commun. 2019;10:1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lukey MJ, Wilson KF, Cerione RA. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med Chem. 2013;5(14):1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dzeja PP & Terzic A. Phosphotransfer networks and cellular energetics. J Exp Biol. 2003;206:2039–2047. [DOI] [PubMed] [Google Scholar]
- 118.Yoshizaki K, Watari H, Radda GK. Role of phosphocreatine in energy transport in skeletal muscle of bullfrog studied by 31P-NMR. Biochim Biophys Acta. 1990;1051(2):144–150. [DOI] [PubMed] [Google Scholar]
- 119.Canonaco F, Schlattner U, Pruett PS, et al. Functional Expression of Phosphagen Kinase Systems Confers Resistance to Transient Stresses in Saccharomyces cerevisiae by Buffering the ATP Pool. J Biol Chem. 277(35):31303–31309. [DOI] [PubMed] [Google Scholar]
- 120.Auger C, Lemire J, Cecchini D, et al. The Metabolic Reprogramming Evoked by Nitrosative Stress Triggers the Anaerobic Utilization of Citrate in Pseudomonas fluorescens. PLoS One. 6(12):e28469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Auger C & Appanna VD. A novel ATP-generating machinery to counter nitrosative stress is mediated by substrate-level phosphorylation. Biochem Biophys Acta. 2015;1850(1):43–50. [DOI] [PubMed] [Google Scholar]
- 122.Voncken F, Gao F, Wadforth C, et al. The Phosphoarginine Energy-Buffering System of Trypanosoma brucei Involves Multiple Arginine Kinase Isoforms with Different Subcellular Locations. PLoS One. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alonso GD, Pereira CA, Remedi MS, et al. Arginine kinase of the flagellate protozoa Trypanosoma cruzi Regulation of its expression and catalytic activity. FEBS Letters. 2001;498:22–25. [DOI] [PubMed] [Google Scholar]
- 124.Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochem Biophys Acta. 2006;1762(2):164–180. [DOI] [PubMed] [Google Scholar]
- 125.Sharer JD, Bodamer O, Longo N, et al. Laboratory diagnosis of creatine deficiency syndromes: a technical standard and guideline of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:256–263. [DOI] [PubMed] [Google Scholar]
- 126.Aksentijevic D, Zervou S, Eykyn TR et al. Age-Dependent Decline in Cardiac Function in Guanidinoacetate-N-Methyltransferase Knockout Mice. Front Physiol. 2019;10:1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gorselink M, Drost MR, Coumans WA, et al. Impaired muscular contractile performance and adenine nucleotide handling in creatine kinase-deficient mice. Am J Physiol Endocrinol Metab. 2001;281(3):E619–E625. [DOI] [PubMed] [Google Scholar]
- 128.Rayner JJ, Peterzan MA, Watson WD, et al. Myocardial Energetics in Obesity: Enhanced ATP Delivery Though Creatine Kinase With Blunted Stress Response. Circulation. 2020;141(14):1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ben-Sahra I & Puissant A. HER2 Signaling Hijacks the Creatine Shuttle to Fuel Breast Cancer Cell Growth. Cell Metab. 2018;28(6):805–807. [DOI] [PubMed] [Google Scholar]
- 130.Wallimann T, Tokarska-Schlattner M, Schlattner U. The creatine kinase system and pleiotropic effects of creatine. Amino Acids. 2011;20:1271–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kitzenberg D, Colgan SP, Glover LE. Creatine kinase in ischemic and inflammatory disorders. Clin Transl Med. 2016;5(31). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lanis JM, Kao DJ, Alexeev EE, et al. Tissue metabolism and the inflammatory bowel diseases. J Mol Med. 2017;95(9):905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Glover LE, Bowers BE, Saeedi B, et al. Control of creatine metabolism by HIF is an endogenous mechanism of barrier regulation in colitis. PNAS. 2013;110(49):19820–19825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chiarelli LR, Morera SM, Bianchi P, et al. Molecular Insights on Pathogenic Effects of Mutations Causing Phosphoglycerate Kinase Deficiency. PLoS One. 2017;7(2):e32065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fothergill-Gilmore LA & Michels PAM. Evolution of glycolysis. Prog Biophys Mol Biol. 1993;59(2):105–235. [DOI] [PubMed] [Google Scholar]
- 136.Zhong W, Cui L, Goh BC, et al. Allosteric pyruvate kinase-based “logic gate” synergistically senses energy and sugar levels in Mycobacterium tuberculosis. Nat Commun. 2017;8:1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Krishnan S, Alden N, Lee K. Pathways and Functions of Gut Microbiota Metabolism Impacting Host Physiology. Curr Opin Biotechnol. 2015;36:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 140.De Angelis M, Piccolo M, Vannini L, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder no otherwise specified. PLoS One. 2013;8:e76993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krajmalnik-Brown R, Lozupone C, Kang DW, Adams JB. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how complex community influences a complex disease. Microb Ecol Health Dis. 2015;26:26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol. 2011;45:S120–S127. [DOI] [PubMed] [Google Scholar]
- 143.De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. [DOI] [PubMed] [Google Scholar]
- 144.Reichardt N, Duncan SH, Young P, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Steliou K, Boosalis MS, Perrine SP, et al. Butyrate histone deacetylase inhibitors. Biores Open Access. 2012;1:192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rowland I, Gibson G, Heinken A, et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Halmos EP, Christophersen CT, Bird AR, et al. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93–100. [DOI] [PubMed] [Google Scholar]
- 149.Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota translplantation: in perspective. Therap Adv Gastronenterol. 2016;9(2):229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Feria-Gervasio D, Tottey W, Gaci N, et al. Three-stage continuous culture system with a self-generated anaerobia to study the regionalized metabolism of the human gut microbiota. J Microbiol Methods. 2014;96:111–118. [DOI] [PubMed] [Google Scholar]
- 151.Haferkamp P, Tjaden B, Shen L, et al. The Carbon Switch at the Level of Pyruvate and Phosphoenolpyruvate in Sulfolobus solfataricus P2. Front Microbiol. 2019; 10:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bragg J, Rajkovic A, Anderson C, et al. Identification and Characterization of a Putative Arginine Kinase Homolog from Myxococcus xanthus Required for Fruiting Body Formation and Cell Differentiation. J Bacteriol. 2012;194(10):2668–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Laino A, Lopez-Zavala AA, Garcia-Orozco KD, et al. Biochemical and structural characterization of a novel arginine kinase from the spider Polybetes pythagoricus. Peer J. 2017;5:e3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Borchel A, Verleih M, Rebl A, et al. Creatine metabolism differs between mammals and rainbow trout (Oncorhynchus mykiss). Springerplus. 2014;3:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang KY, Tull L, Cooper E, et al. Recombinant Protein Production of Earthworm Lumbrokinase for Potential Antithrombotic Application. Evid Based Complementary Altern Med. 2013(2–4):783971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Azzi A, Clark SA, Ellington WR, Chapman MS. The role of phosphagen specificity loops in arginine kinase. Prot Sci. 2009;13(3):575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Uda K, Iwai A, & Suzuki T. Hypotaurocyamine kinase evolved from a gene for arginine kinase. FEBS Letters. 2005;579(30):6756–6762. [DOI] [PubMed] [Google Scholar]
- 158.Van Thoai N, Di Jeso F, & Der Terrossian ED. A new ATP: guanidine phosphotransferase, opheline kinase. Biochim Biophys Acta. 1966;113(3):542–550. [PubMed] [Google Scholar]
- 159.Samoil V, Dagenais M, Ganapathy V, et al. Vesicle-based secretion in schistosomes: Analysis of protein and microRNA (miRNA) content of exosome-like vesicles derived from Schistosoma mansoni. Sci Rep. 2018;8:3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Koire AM & Cavalcanti ARO. Fusion of the subunits α and β of succinyl-CoA synthetase as a phylogenetic marker for Pezizomycotina fungi. Genet Mol Biol. 2011;34(4):669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Seelert H & Dencher NA. ATP synthase superassemblies in animals and plants: Two or more are better. Biochim Biophys Acta. 2011;1807(9):1185–1197. [DOI] [PubMed] [Google Scholar]