Abstract
The reed frog genus Hyperolius (Afrobatrachia, Hyperoliidae) is a speciose genus containing over 140 species of mostly small to medium-sized frogs distributed in sub-Saharan Africa. Its high level of colour polymorphism, together with in anurans relatively rare sexual dichromatism, make systematic studies more difficult. As a result, the knowledge of the diversity and taxonomy of this genus is still limited. Hyperolius robustus known only from a handful of localities in rain forests of the central Congo Basin is one of the least known species. Here, we have used molecular methods for the first time to study the phylogenetic position of this taxon, accompanied by an analysis of phenotype based on external (morphometric) and internal (osteological) morphological characters. Our phylogenetic results undoubtedly placed H. robustus out of Hyperolius into a common clade with sympatric Cryptothylax and West African Morerella. To prevent the uncovered paraphyly, we place H. robustus into a new genus, Congolius. The review of all available data suggests that the new genus is endemic to the central Congolian lowland rain forests. The analysis of phenotype underlined morphological similarity of the new genus to some Hyperolius species. This uniformity of body shape (including cranial shape) indicates that the two genera have either retained ancestral morphology or evolved through convergent evolution under similar ecological pressures in the African rain forests.
Subject terms: Herpetology, Phylogenetics, Speciation, Taxonomy
Introduction
African reed frogs, Hyperoliidae Laurent, 1943, are presently encompassing almost 230 species in 17 genera. These are split into two subfamilies, Hyperoliinae Laurent, 1943 with 12 genera, and the less speciose subfamily Kassininae Laurent, 1972 with 5 genera1,2 (Table 1). The phylogenetic relationships among the hyperoliid frogs are still not completely understood, and the systematics has undergone recent changes, e.g., revalidation of the subfamily Kassininae and reclassification of Acanthixalus to this subfamily1 or genus-level reorganizations in some species3. There is also a number of genera whose classification to the family is clear on the basis of morphological characters, but their exact phylogenetic position is still an unanswered question. In the Congo Basin, an important region for our study, it is the case of three monotypic genera. Callixalus pictus Laurent, 1950 and Chrysobatrachus cupreonitens Laurent, 1951 were described from high-altitude bamboo forests and grasslands of the Albertine Rift4,5, and Kassinula wittei Laurent, 1940 is known from wooded savanna uplands of south-eastern Democratic Republic of the Congo, northern Zambia, and recently from central Angola6,7. Some previous morphological studies provided rough ideas where these genera might be phylogenetically placed8,9, but the first two genera have never been studied by a molecular approach. All three genera are presently placed in Hyperoliinae1,2, neverthless, the hypothesized sister relationship of Callixalus and Acanthixalus8,9 keeps the question on the phylogenetic position of Callixalus open. Similar uncertainties apply to Chrysobatrachus and Kassinula. However, the latter has a characteristically different morphology, resembling either Kassina6,10, Paracassina8, or Afrixalus7,9. Newly, Kassinula was shown to be phylogenetically related to Afrixalus, possibly even embed within this genus7. A recent surprise in the hyperoliid systematics was the discovery of Morerella cyanophthalma Rödel, Assemian, Kouamé, Tohé & Perret, 2009, a monotypic genus from the coastal rain forests of Côte d’Ivoire in West Africa11. The study included phylogenetic analysis of three mitochondrial markers, which did place the genus in Hyperoliidae, but did not resolve its exact systematic position. Later studies, which comprised phylogenetic or phylogenomic analyses of nuclear DNA, uncovered affiliation of Morerella to Cryptothylax from Central African rain forests1,3.
Table 1.
Genera of the family Hyperoliidae, including their species richness and distribution2.
| Subfamily | Genus | Spp | Distribution |
|---|---|---|---|
| Hyperoliinae Laurent, 1943 | Afrixalus Laurent, 1944 | 35 | sub-Saharan Africa |
| Alexteroon Perret, 1988 | 3 | western Central Africa | |
| Arlequinus Perret, 1988 | 1 | western Central Africa | |
| Callixalus Laurent, 1950 | 1 | Albertine Rift | |
| Chrysobatrachus Laurent, 1951 | 1 | Albertine Rift | |
| Cryptothylax Laurent & Combaz, 1950 | 2 | Central Africa | |
| Heterixalus Laurent, 1944 | 11 | Madagascar | |
| Hyperolius Rapp, 1842 | 145 | sub-Saharan Africa | |
| Kassinula Laurent, 1940 | 1 | southern Central Africa | |
| Morerella Rödel, Kosuch, Grafe, Boistel & Veith, 2009 | 1 | West Africa | |
| Opisthothylax Perret, 1966 | 1 | western Central Africa | |
| Tachycnemis Fitzinger, 1843 | 1 | Seychelles Islands | |
| Kassininae Laurent, 1972 | Acanthixalus Laurent, 1944 | 2 | West and Central Africa |
| Kassina Girard, 1853 | 15 | sub-Saharan Africa | |
| Paracassina Peracca, 1907 | 2 | North-eastern Africa | |
| Phlyctimantis Laurent & Combaz, 1950 | 5 | West, Central and East Africa | |
| Semnodactylus Hoffman, 1939 | 1 | southern Africa |
More than a half of all hyperoliids belong to the genus Hyperolius distributed throughout sub-Saharan Africa2, 12. Hyperolius is a genus of mostly medium-sized and exclusively arboreal frogs with a great variation in colouration between and within species, often sexually dichromatic (otherwise rare in anurans), and with remarkably conservative morphology13. It is a genus inhabiting wide range of habitats from open savannas and bushland to closed-canopy rain forests13. One of the rain forest dwelling species is the little-known Hyperolius robustus Laurent, 1979 (Fig. 1 and Supplementary Fig. S1). It was described from a handful of localities in the central Congo Basin, Sankuru region of the present-day Democratic Republic of the Congo (DRC)14. Since the description, this species has only been reported by one author13,15, and only scarce biological data exist.
Figure 1.
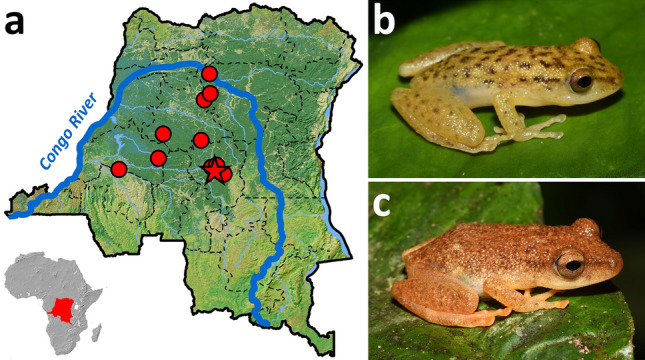
Hyperolius robustus—now in the new genus Congolius—and its distribution. (a) All known localities lie to the south of the wide arc of the Congo River (left bank), within the Democratic Republic of the Congo; star denotes type locality. Photographs of (b) adult male (IVB-H-CD18-143) and (c) adult female (NMP-P6V 76087/2) from the Kokolopori Bonobo Nature Reserve. Photos by V.G.
Hyperolius robustus is one of the larger species in the genus, with both sexes reaching up to 37–38 mm in body length13,15. Dark yellow to brown dorsum with varying intensity of dark spots in males differs from a reddish-brown dorsal colouration in females (Fig. 1b,c and Supplementary S2). Males bear large flat gular gland (disc, flap) of yellowish to white colour. The venter is coloured yellow in males and orange in females, especially limbs. Hyperolius robustus can be distinguished from its sympatric congeners by proportions of the head, especially relatively longer snout and closely-positioned nostrils. It resembles members of the H. concolor species group, specifically H. balfouri (Werner, 1908), H. cinnamomeoventris Bocage, 1866, or H. kivuensis Ahl, 1931 when compared to the species widespread across the Congo14. While Laurent14 suggested its probable relationship to the H. concolor group, he also noted that some of its morphological characters supported a possible close relationship to Cryptothylax, a genus superficially also resembling the H. concolor group, although reaching a larger adult size14. A later publication focusing more broadly on external morphology of Hyperolius gained an additional support for the similarity of H. robustus to H. balfouri and H. cinnamomeoventris16.
Recently, we collected new specimens of H. robustus at multiple sites in the central Congolian forests, and reviewed available museum material, including types (Supplementary Table S1, Appendices S1 and S2). This study aims to (1) clarify the phylogenetic and systematic position of Hyperolius robustus using a molecular multilocus approach, and (2) evaluate morphological variation based on morphometric and osteological approaches in a comparative framework.
Results
Molecular analyses
Conventional phylogenetics
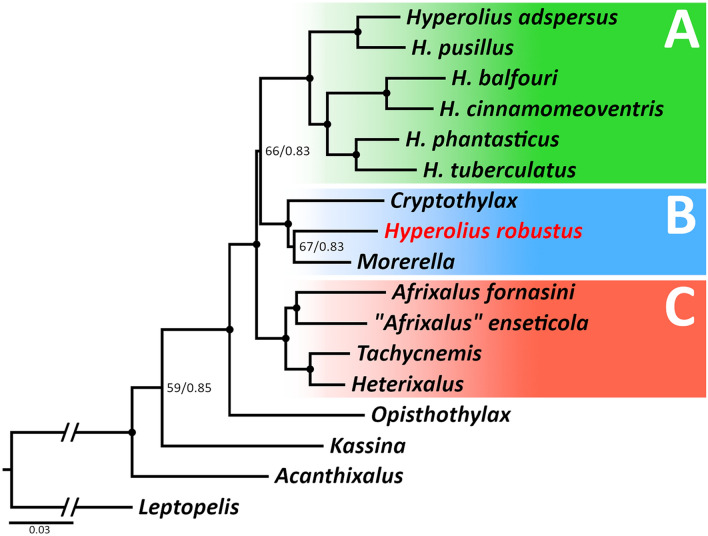
The phylogenetic analysis of the complete concatenated data set of four nuclear and one mitochondrial marker places H. robustus well outside the clade containing the genus Hyperolius (Fig. 2), rendering Hyperolius paraphyletic. The tree based on this complete concatenated data set has only minor difference in the topology compared to the tree based on the nuclear-only data set (Supplementary Fig. S3f). However, support values are lower in the complete data set. All phylogenetic inferences of individual markers or concatenated data put the taxa from the subfamily Hyperoliinae into a common clade, with variable support values depending on a molecular marker/data set. General topology within Hyperoliinae is also consistent across the markers/data sets (except of the mitochondrial tree, which has a lower resolution; Supplementary Fig. S3a). Opisthothylax is positioned as a basal genus in the subfamily Hyperoliinae and in the sister position to all other available Hyperoliinae genera forming three main clades (hereafter Clades A–C). Hyperolius robustus together with Cryptothylax and Morerella form the well-supported Clade B (received the full support in the complete and nuclear-only concatenated data sets), which is in the sister position to Clade A containing the remaining Hyperolius. The relationships within Clade B are uncertain but the sister relationship between H. robustus and Morerella received a full support in the nuclear-only data set. Clade A (Hyperolius sensu stricto) is a well-supported clade composed of three sub-clades. One subclade contains H. adspersus and H. pusillus (= H. nasutus complex15), and is in the sister position to the remaining two subclades. Another subclade (Clade 2 sensu Portik et al.1) is represented by H. balfouri and H. cf. cinnamomeoventris (an unresolved species complex of multiple distinct lineages17; hereafter as H. cinnamomeoventris) and is in the well-supported sister position to the third clade (Clade 1 sensu Portik et al.1), which includes H. phantasticus and H. tuberculatus (a similar case as the H. cinnamomeoventris species complex17; hereafter as H. tuberculatus). The third main clade, the well-supported Clade C contains Afrixalus from sub-Saharan Africa, Heterixalus from Madagascar, and Tachycnemis from Seychelles. Support values for the relationships within the subfamily Hyperoliinae (ingroup), among the three main Clades A–C, differ depending on a particular ML and BI analysis (Fig. 2 and Supplementary Fig. S3). However, all these analyses provide a strong support for the position of H. robustus outside of the genus Hyperolius. All nuclear markers, including the concatenated data set, place H. robustus into a common clade with Cryptothylax and Morerella, with three of the four nuclear markers (and the nuclear concatenated data set) supporting the sister relationship of H. robustus and Morerella with an intermediate to high support (only one marker infers the sister relationship between Cryptothylax and Morerella with an intermediate support). The phylogenetic analysis of the relatively short 16S mitochondrial marker places H. robustus in the sister position to Opisthothylax, but with a low support.
Figure 2.
Phylogenetic tree of hyperoliid frogs with a focus on the subfamily Hyperoliinae. The majority rule consensus tree inferred from Bayesian analyses of four nuclear and one mitochondrial marker (complete data set). Dots at nodes represent high supports (ML bootstrap ≥ 70/Bayesian posterior probability ≥ 0.95) while numbers show support values for nodes with a lower support. Sequence GenBank accession numbers are provided in Supplementary Table S1.
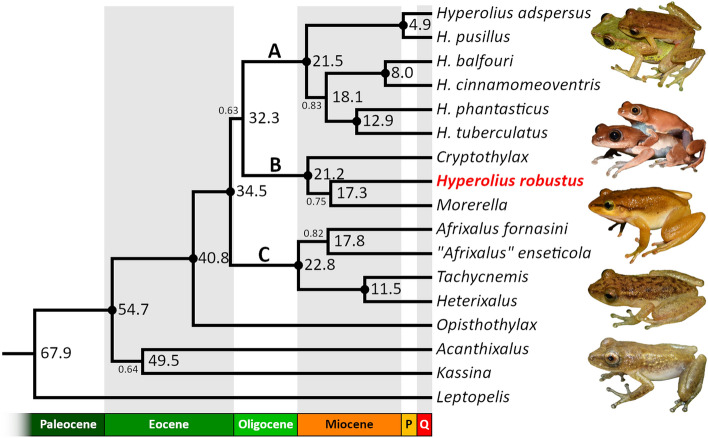
Dated species tree
The species-tree analysis yielded a tree with a similar topology as the conventional phylogenetic analyses (Fig. 3). Opisthothylax is placed with a high support in a basal position within the Hyperoliinae subfamily and its lineage diverged at approx. 40.8 Mya (± 5.5) in the middle Eocene. Similarly to the conventional phylogenetic analyses, all three main Clades A–C are well supported but their mutual relationships are not resolved. The diversification into the three main clades occurred during the Eocene–Oligocene transition (approx. 28–39 Mya, mean 33.4 Mya). Clade B received the full support, and—as in a majority of the conventional phylogenetic inferences—H. robustus is placed in the sister relationship to Morerella, albeit without a high support. The diversification within Clade B is dated in the early Miocene (21.2 ± 4.9 Mya), with the split between H. robustus and Morerella inferred at around 17.3 Mya (± 7.2). Clade A (the remaining Hyperolius) is also well supported, forming three subclades, but the sister relationship between the balfouri–cinnamomeoventris and phantasticus–tuberculatus subclades lacks a higher support (contrary to the conventional analyses). The initial radiation of the remaining studied Hyperolius began at around 21.5 Mya (± 3.7) in the early Miocene. The well-supported Clade C containing Afrixalus, Heterixalus, and Tachycnemis diversified approx. 22.8 Mya (± 5.1) in the early Miocene. The sister relationship of the two included Afrixalus species received only a weaker support, with the divergence dated to 17.8 Mya (± 8.6) in the Miocene. In fact, one of the Afrixalus species (A. enseticola) probably represents a distinct genus1. The youngest divergence between valid genera of Hyperoliinae was found between Heterixalus and Tachycnemis dated to11.5 Mya (± 5.8) in the late to middle Miocene.
Figure 3.
Time-calibrated species tree of the subfamily Hyperoliinae. Maximum clade credibility tree based on StarBEAST2 analysis of four nuclear markers with geological epochs as a time scale (abbreviation P stands for Pliocene and Q for Quaternary). Dots at nodes signify high supports (Bayesian posterior probability ≥ 0.95), median node ages are shown to the right from each node, lower support values are shown to the left of the relevant node. Leptopelis (Arthroleptidae) is outgroup. Photos on the right side from top to bottom (not to scale): Hyperolius cinnamomeoventris, H. phantasticus, Cryptothylax greshoffii (in a calling position; note the extensive gular gland and missing vocal sac), H. robustus (now Congolius robustus gen. et comb. n.), Morerella cyanophthalma. Photos by V.G. and T. F. Kpan (Morerella cyanophthalma).
Systematic account
Hereafter, we describe a new genus for H. robustus to remove the uncovered paraphyly of Hyperolius. To justify its genus level distinction, beside its specific morphological characters (see below), we refer to the deeper phylogenetic divergence than is found between Heterixalus from Madagascar and Tachycnemis from Seychelles, and a comparable divergence between Afrixalus (A. fornasini is type species of the genus) and a candidate, yet undescribed genus containing “Afrixalus” enseticola1.
Family Hyperoliidae Laurent, 1943.
Subfamily Hyperoliinae Laurent, 1943.
Congolius genus novum Nečas, Badjedjea & Gvoždík.
ZooBank registration: urn:lsid:zoobank.org:act:7C7D57F2-3D02-4456–9292-184A98A559E6.
Type species: Hyperolius robustus Laurent, 1979.
Suggested English name: Congo Frog.
Content: Congolius robustus (Laurent, 1979) combinatio nova (formerly Hyperolius robustus), Robust Congo Frog. Presently a monotypic genus.
Etymology
The generic name Congolius is of the masculine gender and it is a compound of the name Congo, referring to the region where the genus is endemic to, and the Greek word eleios = smooth in the Latinized form as the suffix –lius, which is used in the genus name Hyperolius to refer to its former affiliation of Congolius robustus to this genus.
Diagnosis and comparisons
The new genus Congolius can be distinguished from other genera within the family Hyperoliidae by following characters: pupil horizontal (vertical or rhomboidal in Acanthixalus, Afrixalus, Arlequinus, Cryptothylax, Heterixalus, Kassina, Kassinula, Tachycnemis, Opisthothylax, Paracassina, Phlyctimantis, and Semnodactylus; predominately horizontal but sometimes rhomboidal in Morerella, see photos in18); tympanum indistinct but visible in both sexes (indistinct in Acanthixalus, Callixalus, Chrysobatrachus, Heterixalus, Kassinula, Opisthothylax, and in most Hyperolius and Afrixalus); males with large gular gland with free lateral and posterior margins without free skin folds of dilatable skin beneath, Fig. 4c [gular gland present in males of all hyperoliid species; paired, laterally positioned in Acanthixalus; large without free margins obscuring whole gular region in Cryptothylax (Fig. 4e); positioned anteromedially and hidden under semi-transparent integument in Alexteroon; transverse in Arlequinus; rather indistinct transverse oval composed of two elements fused medially in Callixalus (Fig. 4k); distinct round composed of two elements fused medially in Chrysobatrachus (Fig. 4j); in a form of round median disc without extended dilatable skin beneath or around present in Morerella (Fig. 4d), Opisthothylax (Fig. 4m) and Tachycnemis; medioposteriorly positioned with free skin folds beneath the posterior margin in Afrixalus (Fig. 4l), Heterixalus, and most Hyperolius (Fig. 4f–i; exceptions exist in Hyperolius, e.g. H. zonatus Laurent, 1958 from West Africa;12); longitudinal oval or strap-shaped with free lateral margins in Kassina; round with free lateral margins in Paracassina, Phlyctimantis (Fig. 4n), and Semnodactylus]; dilatable vocal sac (pouch) present, when inflated forms a hemisphere extending to the pectoral region, Fig. 4a (extensively dilatable vocal sac absent in Arlequinus, Acanthixalus, Callixalus, Chrysobatrachus, Cryptothylax, and Morerella; see Fig. 4b for comparison with inflated vocal sac of Hyperolius balfouri); limbs without skin “fringes” (present in Alexteroon); dorsal skin slightly granular in both sexes (tuberculous in Acanthixalus); sexually dichromatic (colouration difference between sexes similar to Cryptothylax and Morerella, where females display more orange to reddish colouration than males, see Fig. 1b,c and Supplementary Fig. S2; sexually monochromatic genera: Acanthixalus, Afrixalus, Alexteroon, Arlequinus, Callixalus, Chrysobatrachus, some Heterixalus, some Hyperolius, Kassina, Kassinula, Opisthothylax, Paracassina, Phlyctimantis, and Semnodactylus).
Figure 4.
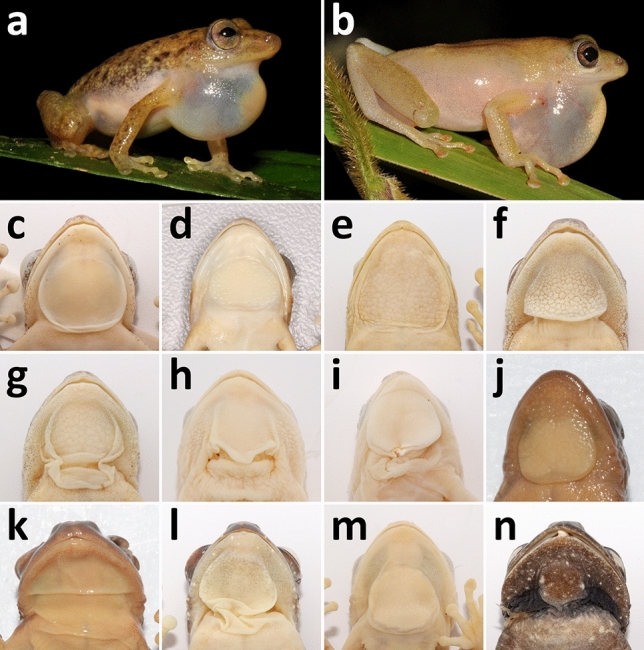
Gular region of selected hyperoliid taxa. (a) Congolius robustus gen. et comb. n. (IVB-H-CD18-255) with inflated vocal sac, (b) Hyperolius balfouri (IVB-H-CD15-062) with inflated vocal sac, (c) C. robustus gen. et comb. n. (ZMUC-R.771175), (d) Morerella cyanophthalma (ZMB 71588, paratype), (e) Cryptothylax greshoffii (NMP-P6V 76076/1), (f) H. balfouri (IVB-H-CD15-061), (g) H. cf. cinnamomeoventris (IVB-H-CD18-092), (h) H. phantasticus (IVB-H-CD18-203), (i) H. cf. adspersus (IVB-H-CD15-139), (j) Chrysobatrachus cupreonitens (MCZ A-64739, paratype), (k) Callixalus pictus (MCZ A-64707), (l) Afrixalus equatorialis (NMP-P6V 76073/1), (m) Opisthothylax immaculatus (NMP-P6V 76090/1), (n) Phlyctimantis verrucosus (NMP-P6V 76085/1). Photos by (a,b,j,k) V.G., (c,e–i,l–n) T.N. and (d) F. Tillack and M.-O. Rödel.
Osteologically, Congolius is characterized with sphenethmoid not dorsally exposed (dorsally visible in Cryptothylax, some Hyperolius, Kassina, Kassinula, Phlyctimantis, Semnodactylus, and Tachycnemis); ventroanterior portion of sphenethmoid unfused (fused in some Kassina, Paracassina, some Phlyctimantis, and Semnodactylus); praemaxilla and maxilla bear minute teeth; overlapping of praemaxilla and maxilla is minimal; nasals triangular, separated medially from each other and posteriorly from frontoparietals; canthal regions of nasals rounded; vomers lack teeth-bearing dentigerous processes, “vomerine teeth” (present in Cryptothylax, most Kassina, and Phlyctimantis); frontoparietals rectangular; quadratojugal anteriorly in contact with maxilla (not in contact with maxilla in Acanthixalus, some Afrixalus, Callixalus, Chrysobatrachus, some Heterixalus, some Hyperolius, some Kassina, and Kassinula; reduced or absent in some Afrixalus); columella present (reduced in Callixalus); neural arches non-imbricate (imbricate in Cryptothylax, Kassina, Paracassina, Phlyctimantis, and Semnodactylus); transverse processes of 8th vertebra angled perpendicularly to vertebral column (angled markedly forward in some Afrixalus, some Hyperolius, some Kassina, Kassinula, Opisthothylax, and Semnodactylus); relative length of vertebral column (ratio of length of vertebral column to transverse processes of 8th presacral vertebra) between values of 1.6 and 2.4 (longer vertebral column in Afrixalus, Callixalus, some Heterixalus, some Hyperolius, Kassina, Kassinula, Paracassina, Phlyctimantis, and Semnodactylus); medial margins of coracoids entire (centrally perforated in Kassinula, and Paracassina); omosternum greatly forked (unforked or less forked in Callixalus, some Hyperolius, and Paracassina); base of metasternum ossified (completely ossified in Morerella); carpal and tarsal bones not fused (Hyperoliidae synapomorphy); subarticular sesamoids absent (present in Kassina, Paracassina, some Phlyctimantis, and Semnodactylus); intercalary elements completely mineralized (cartilaginous centres in Afrixalus, Callixalus, Heterixalus, and Hyperolius); terminal phalanges long and peniform (long and claw-shaped in some Heterixalus, some Hyperolius, some Kassina, Kassinula, and Opisthothylax; short obtuse in some Kassina, Paracassina, and Phlyctimantis; bifurcated in Acanthixalus, some Kassina, and possibly Arlequinus based on external morphology, no osteological data for the latter). Osteological terminology follows Drewes8. See Supplementary Appendix S3 for literature resources.
Genetically, Congolius is differentiated, forms a clade with Cryptothylax and Morerella, and differs from other genera in the family Hyperoliidae by an uncorrected p-distance larger than 11.0% in the fragment of 16S rRNA (Supplementary Table S2).
Distribution
Congolius robustus is probably endemic to the central Congolian lowland forests, to the south of the wide arc of the Congo River (Fig. 1a). It has never been recorded from the right bank of the Congo River. For locality details, see Supplementary Appendix S2. It occupies occasionally flooded forests and dense farmbush along small streams. Congolius robustus has a nocturnal activity, and it can typically be found perching on vegetation 1.5–2 m high.
Nomenclatural note
This publication and the nomenclatural act are registered in ZooBank, the online Official Register of Zoological Nomenclature. The LSID (Life Science Identifier) of this publication from ZooBank is urn:lsid:zoobank.org:pub:F2045C29-D703-4FC8-B317-1BFD09815E15 and can be accessed at https://zoobank.org/.
Morphology
External morphology
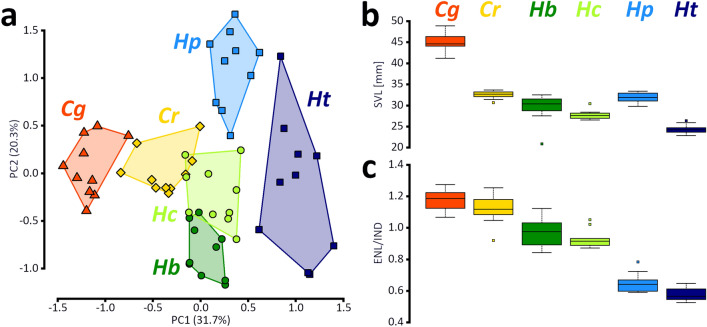
Body size of the six investigated taxa as average snout-vent length (SVL ± SD; in mm) in adult males is: Congolius robustus (32.5 ± 0.9), Cryptothylax greshoffii (45.2 ± 2.1), Hyperolius balfouri (29.4 ± 3.2), H. cinnamomeoventris (27.8 ± 1.0), H. phantasticus (32.0 ± 1.1), H. tuberculatus (24.4 ± 1.0) (Fig. 5b). Principal component analysis (PCA) was performed to evaluate body shape variation of the above-mentioned taxa. Factor loadings for the first three principal components (PCs) and most contributing variables are provided in Supplementary Table S3. The body shapes of the six taxa were found significantly different using MANOVA (F5,15 = 57.07, p = < 2.2e−16). The PC1 (31.7% of explained variation) can be interpreted as showing mostly the shape of head, especially snout. The PC2 (20.3%) was influenced mostly by the head width and variables characterizing the length of limbs. The PCA (Fig. 5a) placed Congolius along the PC1 axis between Cryptothylax and the four Hyperolius species in close proximity to H. cinnamomeoventris. The PC2 does not differentiate Congolius and Cryptothylax from representatives of Hyperolius, but differences among the species of the latter genus are apparent. The studied taxa are well separated from each other in the morphospace (PC1 x PC2), except of small overlaps of H. cinnamomeoventris with Congolius and H. balfouri putting the three taxa into a common cluster. The main distinguishing morphometric characters between Congolius and Hyperolius are the characters related to the shape of snout: eye-nostril distance (measured along body axis) is usually larger than internarial distance in Congolius (as well as Cryptothylax), and on average the same or smaller in the studied Hyperolius. The snout shape of the representatives of Clade 2 (H. balfouri, H. cinnamomeoventris) is more similar to Congolius than the snout shape of the representatives of Clade 1 (H. phantasticus, H. tuberculatus) (clades sensu Portik et al.1; Fig. 5c).
Figure 5.
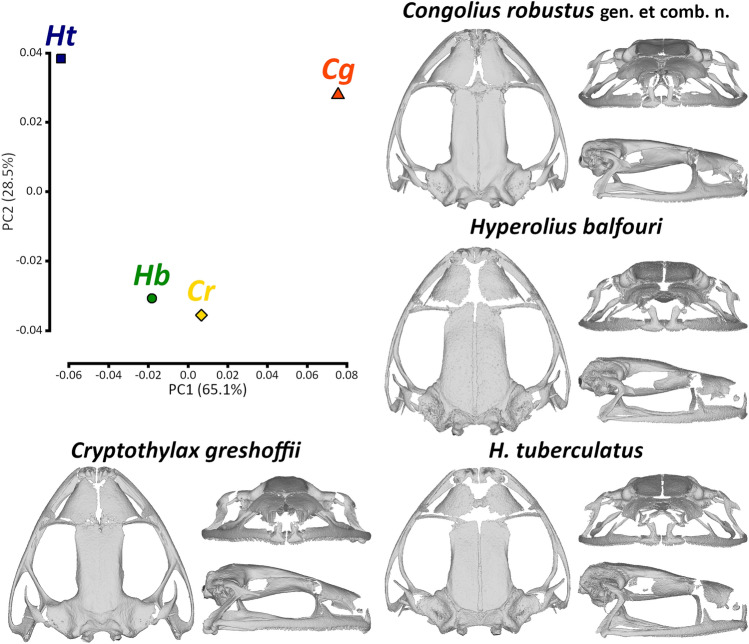
Morphometric analyses of external characters of Congolius gen. n. and relative (Cryptothylax) or similar (Hyperolius) genera from the lowland Congo Basin. Four Hyperolius species represent two phylogenetic clades (see Methods for details). (a) Principal component analysis of body shape, (b) variation of SVL as boxplots, (c) variation of the ENL/IND ratio as boxplots. Boxplots display average values (line), upper and lower quartiles (box), minimums and maximums without outliers (whiskers), and outliers (dots). Cg (Cryptothylax greshoffii), Cr (Congolius robustus gen. et comb. n.), Hb (Hyperolius balfouri), Hc (H. cinnamomeoventris), Hp (H. phantasticus), and Ht (H. tuberculatus).
Larval morphology is not presently known.
Osteology
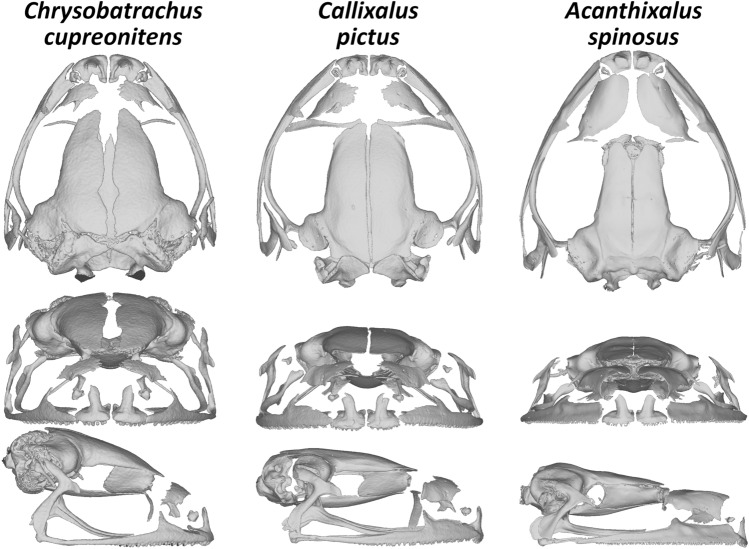
Skeletons with a special focus on skulls were studied in Congolius, its relative Cryptothylax, Hyperolius, two little-known genera Chrysobatrachus and Callixalus, and Acanthixalus. All are distributed in the Congo Basin, and thus of a systematic importance in respect to our erection of the new genus. Scans of skulls are shown in Figs. 6 and 7, for a female of C. robustus see Supplementary Figs. S4, S6. No differences were found in the osteological characters between male and female C. robustus. Our new osteological data were used in addition to the previously published8 for genus-level comparisons (see “Diagnosis and comparisons”). A special focus on the little-known genera distributed in the Congo uncovered that the skeletons of Acanthixalus, Chrysobatrachus and Callixalus exhibited number of characters clearly differentiating them from the new genus Congolius: bifurcated phalanges, and flat frontoparietals in Acanthixalus; moderately forked omosternum in Callixalus; reduced nasals in Callixalus and Chrysobatrachus; cartilaginous metasternum, and short and tall skull in Chrysobatrachus [8] (this study).
Figure 6.
Cranial morphology of Chrysobatrachus, Callixalus, and Acanthixalus. Reconstructions in dorsal, frontal, and lateral views are based on high-resolution X-ray microcomputed tomography. From left to right: Chrysobatrachus cupreonitens (CAS 145263, male), Callixalus pictus (CAS 145260, male), and Acanthixalus spinosus (CAS 153800, female). Skull views are scaled to a similar size among the three genera.
Figure 7.
Cranial morphology and shapes in a morphospace of Congolius gen. n. and relative (Cryptothylax) or similar (Hyperolius) genera from the lowland Congo Basin. The morphospace is expressed by a principal component analysis of geometric morphometric landmarks. Cg (Cryptothylax greshoffii, IVB-H-CD15-169, male), Cr (Congolius robustus gen. et comb. n., ZMUC-R.771175, male), Hb (Hyperolius balfouri, IVB-H-CD15-061, male), and Ht (H. tuberculatus, IVB-H-CD15-010, male). Reconstructions in dorsal, frontal, and lateral views are based on high-resolution X-ray microcomputed tomography. Skull views are scaled to a similar size among the four taxa. See Supplementary Fig. S4 for comparison of all scanned specimens.
Cranial data of Congolius and the remaining examined taxa, relative (Cryptothylax) or similar (Hyperolius) genera, were examined using the 3D geometric morphometrics. PCA generated two relevant PCs (Fig. 7), while the third contributed only 0.06% of the explained variation. PCA scores of the four analysed taxa are in Supplementary Table S4. The PC1 (65.1%) axis could be explained as a change of the cranial shape from rounded (negative PC1 values) to more triangular (positive PC1 values) from the dorsoventral view, including a shift of the mandibular joint to more posterior position or an elongation of frontoparietals and nasals (Fig. 7). Congolius lays in a close proximity to H. balfouri, while H. tuberculatus and Cryptothylax are positioned more distantly in different directions. The PC2 (28.5%) axis explains a change in the shape from a rather prolonged skull (negative PC2 values) to a wider skull (positive PC2 values). The examined taxa are clustered into two groups along the PC2 axis, Congolius and H. balfouri (negative PC2 scores), and Cryptothylax and H. tuberculatus (positive PC2 scores).
Discussion
The phylogenetic analyses confirmed our initial finding that “Hyperolius robustus”, now Congolius robustus, is phylogenetically placed outside of Hyperolius, as well as Laurent’s14 hypothesis of the possible relationship to Cryptothylax greshoffii. These two Central African taxa and Morerella cyanophthalma from West Africa form a common clade (Clade B), which is in the sister relationship to Hyperolius (Clade A, Figs. 2, 3). According to our results, the West African monotypic genus Morerella is the closest relative to Congolius diverging around 17 Mya (Figs. 2, 3). However, the sister relationship of the two genera is not always highly supported. For example, the analysis of the all-markers data set shows only a medium support for this relationship (Fig. 2). However, this could be caused by the implementation of the 16S mitochondrial marker, which is not optimal for deeper phylogenies due to saturated substitutions (see also Supplementary Fig. S3a). The phylogenetic analysis of the nuclear-markers data set gives to the Congolius–Morerella clade a high support (Supplementary Fig. S3f). A close relationship of Congolius and Morerella is further suggested by their similar superficial morphology as well as osteological characters. In particular, these two genera share a similar expression of sexual dichromatism (males brownish, females reddish), the general morphology of the gular region (lack of visible extended dilatable skin beneath or around gular gland), and the general osteological composition (the only main difference is in the degree of metasternum ossification).
The general topology of our phylogenetic reconstruction of the subfamily Hyperoliinae corresponds to the results of a recent phylogenomic study1. The only exception is “Afrixalus” enseticola, which is not in our results placed in the same clade with the genera Heterixalus and Tachycnemis1, but shares a clade with A. fornasini (type species of the genus). However, the divergence is relatively deep and the support only intermediate (Figs. 2, 3, Supplementary Fig. S3f.). The diversification into the three major clades in the subfamily Hyperoliinae (Clades A–C) took place during the Eocene–Oligocene transition (around 33 Mya). The Eocene–Oligocene boundary is associated with an extinction event on Earth19–21. This event is attributed to a rapid decrease in the global temperature caused mainly by the opening of the southern oceanic gateway, Earth orbital cycles, and a decrease of the atmospheric CO2 levels22–26. Most notably on the African continent, it was the accompanying aridification23,27. Our analyses lack a higher support in resolving the relationships among the three main clades, which suggests consecutive divergences in a short period of time. Such extinctions as at the Eocene–Oligocene transition might have opened previously occupied niches and could have led to radiations in vacant places in the ecosystem. The radiation within the clade B, which contains Congolius, is estimated to occur at around 21–17 Mya (Fig. 3). Similar ages were estimated for the radiations of Clades A and C occurring in the early Miocene (approx. 23–18 Mya). The Miocene is a geological epoch connected to yet another event of rapid global cooling at the Oligocene–Miocene transition (23 Mya)23,28. Although periods of the late Oligocene and the first half of Miocene were relatively warm (26 to 15 Mya)23, the mid-Miocene’s closure of the Tethys Ocean and continual northward drift of the African continent had major aridification effects23,28. Both events led to the reduction of the rainforest cover to refugia along coasts and major rivers by the late Miocene27. The rainforest withdrawal and fragmentation could have been a driver for the divergence in a common ancestor of Central African Congolius and West African Morerella occurring at approximately 17 Mya.
Representatives of the little-known genera Callixalus and Chrysobatrachus were not available for the molecular analyses. However, they were morphologically examined to compare with Congolius, including osteology (Fig. 6). Both genera, together with Acanthixalus from Kassininae, were found to have distinct morphological characters from the newly established genus (e.g., maxilla not in contact with quadratojugal). A possible close relationship of Acanthixalus and Callixalus was suggested earlier8,9 based on thorough morphological analyses. While a recent phylogenomic study placed Acanthixalus in Kassininae1, the monotypic genus Callixalus has conservatively remained in Hyperoliinae1,2, awaiting a focused study. Callixalus has not been recorded since its description in 1950 and hence, no recent genetic material is available. Our examination of Callixalus and Acanthixalus supports their similarity in a number of shared characters. However, we refrain from making systematic conclusions for these two genera, as our study was not designed to solve this question. A future research on the phylogenetic position of Callixalus, as well as Chrysobatrachus (rediscovered in 201129), Alexteroon (revision of the genus is in preparation30), Arlequinus, and Kassinula is needed to reveal a complete genus-level phylogenetic reconstruction of Hyperoliidae.
The distribution range of C. robustus seems to be confined to the central Congolian forests, south of the wide arc of the Congo River. Several recent surveys did not report any sightings of C. robustus on the right bank of the Congo River31–35. Therefore, the Congo River may represent a barrier that prevents C. robustus from reaching the northern parts of the Congo Basin. In contrast, the Congo River does not seem to be a dispersal barrier for some Hyperolius species17. The barrier for Congolius to the east could be either the Lualaba River (= upper part of the Congo River) or the Lomami River which flows in parallel to the west. Future herpetological field research in the Lomami National Park, which lies between these two rivers, could bring an answer to this question.
Morphologically, C. robustus is highly resembling some Hyperolius species, in particular from the H. concolor group14. The morphometric analyses of external characters and the body shape showed a great similarity between phylogenetically rather distant taxa (Fig. 5). The main distinguishing morphological character is the shape of snout, being longer than wide in Cryptothylax and Congolius compared to Hyperolius. Although some specimens of H. balfouri and H. cinnamomeoventris reach similar values of the ENL/IND ratio (Fig. 5c). Our results generally support the findings of Laurent14,16, who already pointed out on the similarity of Congolius and these two species from the H. concolor group. The analysis of the shape of skull yielded very similar results. It put Congolius more closely to phylogenetically more distant H. balfouri than to a close relative, Cryptothylax greshoffii.
There are two evolutionary processes that could be responsible for the origin of the morphological similarity between the newly erected Congolius and some Hyperolius species. First is the morphological stasis, which postulates tendency of some lineages to retain ancestral ecological niches, and thus, to reduce the morphological change during an evolutionary lineage’s existence36,37. This hypothesis would suggests that Congolius and some lineages of Hyperolius may have retained the same general body shape as possessed by their common ancestor, i.e. since the beginning of the Oligocene (approx. 32 Mya; our dating corresponds to a previously published study1). Second is the convergent evolution, which evolves when the same or similar selection pressures cause independent development of analogous characters in unrelated taxa38–40. Convergent evolution is a common characteristic feature in species-rich communities, like are typically present in rain forests. An independent parallel evolution of a similar body shape in Congolius and some Hyperolius could have been driven by similar ecological pressures across the African rain forests. However, further investigations into causalities of ecological factors and the phenotypic evolution of African reed frogs are needed to understand processes leading to the morphological uniformity of some unrelated hyperoliid taxa.
Conclusions
We present an evidence of the phylogenetic relationship of central Congolian “Hyperolius” robustus and sympatric Cryptothylax, as well as West African Morerella. We erect the new genus Congolius to prevent the uncovered paraphyly of Hyperolius, and place “H.” robustus to the new genus. Congolius morphologically greatly resembles some members of the genus Hyperolius, which might be attributable to either the morphological stasis or convergent evolution caused by similar ecological pressures in the African rain forests.
Material and methods
Sampling and molecular data
One museum15 and two recently collected “Hyperolius” robustus specimens from two locations (Kokolopori: ZMUC R.771176, IVB-H-CD18-255; Mombongo: NMP-P6V 76089; for details see Supplementary Table S1) were investigated genetically. Identifications were based on the comparison with the type material (Supplementary Fig. S1), the ZMUC material was already identified15. Preliminary phylogenetic analyses suggested that “H.” robustus might be placed out of Hyperolius. Taxon sampling was designed to focus on “H.” robustus and the subfamily Hyperoliinae (nucleotide sequences retrieved from GenBank; Supplementary Table S5), to include molecular data of all available genera (7/12), and representatives of main clades of Hyperolius and Afrixalus (sensu Portik et al.1). Two representatives of the hyperoliid subfamily Kassininae (Acanthixalus spinosus, Kassina senegalensis) and a representative of the sister family Arthroleptidae (Leptopelis notatus) were used as outgroups (retrieved from GenBank). Collection of osteological data focused on all systematically potentially important taxa from the Congo Basin, including those without molecular data (Callixalus, Chrysobatrachus). External morphometric examinations focused on Central African relative or morphologically similar taxa and selected “congeners” (Hyperolius) of “H.” robustus, see below for more details. Newly collected specimens were euthanized, fixed in 96% ethanol for a short period of time, and then transferred to 75% ethanol for preservation. Tissue samples for genetic analyses were extracted and fixed in 96% ethanol. Museum abbreviations follow the herpetological standards41 except for the abbreviation IVB (Institute of Vertebrate Biology of the Czech Academy of Sciences, Brno, Czech Republic) which is an institution registered with the Global Biodiversity Information Facility (http://grbio.org/cool/iskk-kcgq). IVB-H stands for a herpetological collection localised in the research facility Studenec.
DNA was extracted using a commercial DNA extraction kit following the manufacturer’s protocol (GeneAll Biotechnology). Single and two-step polymerase chain reactions (PCRs) were applied to amplify a mitochondrial fragment encoding 16S rRNA (hereinafter as 16S; 530 bp after trimming) and four nuclear markers: FIC Domain-containing gene (FICD; 529 bp), KIAA2013 gene (KIAA2013; 540 bp), Proopiomelanocortin gene (POMC; 595 bp) and Tyrosinase gene (Tyr; 532 bp). See Supplementary Table S6 for primers. PCR protocols followed original publications42–46 with the exception of 16S rRNA, where minor changes were applied47. The newly acquired sequences of all three “H.” robustus (now Congolius robustus) specimens were deposited in the online database GenBank (https://ncbi.nlm.nih.gov/genbank/). Sequences were checked by eye and assembled using Geneious Prime R11.0.348. MAFFT v7.450 plug-in for Geneious Prime was used to align all sequences49,50. See Supplementary Table S5 for GenBank accession numbers.
Phylogenetic inference and divergence dating
The three analysed samples of “H. robustus” from two distant locations exhibited only minor variations (16S: C/T on the 29th and A/G on the 183rd position, FICD: G/R on the 146th position, POMC: T/Y on the 293rd and C/M on the 313th positions, Tyr: A/G on the 91st position). To streamline interpretations of results, only one sample (IVB-H-CD18-255) was included in the final data sets. Maximum likelihood (ML) analyses and Bayesian inference (BI) were performed for each gene separately, concatenated nuclear markers, and concatenated all markers. ML analyses were performed in RAxML-NG v0.9.051 and BI using MrBayes v3.2.652, with substitution models and partition schemes selected in PartitionFinder 2 (following the Bayesian information criterion, BIC)53 (Supplementary Table S7). Nodal support values in ML analyses were received by the bootstrap method with the automatic bootstopping cut-off value 0.03, stopping at 200 replicates in all runs. All MrBayes analyses were run twice with four Markov chains Monte Carlo for 30 million generations with sampling every 3,000th generation. First 25% of produced trees were discarded as a “burn-in”. Clades supported with ML bootstrap values ≥ 70 and BI posterior probability values ≥ 0.95 were considered highly supported54, while bootstrap values ≥ 60 and posterior probability values ≥ 0.80 as intermediate support.
A dated coalescent-based species tree was inferred in StarBEAST2 v2.6.255 based on the four nuclear markers (FICD, KIAA2013, POMC, Tyr). A lognormal relaxed molecular clock and a normal distribution were used56. Calibration dates were taken as averages of recently published large phylogenomic studies of anuran amphibians, where calibrations were based on paleontological and geological records1, 57–59. Two calibration points were set: splits between Arthroleptidae and Hyperoliidae at ~ 64.5 (56.8–72.5) Mya, and Hyperoliinae and Kassininae at ~ 44.3 (36.8–51.8) Mya. The Yule tree model was set, and substitution models and partition schemes as suggested by PartitionFinder 2 (BIC) were applied (Supplementary Table S7). The analysis was run thrice for 100 million generations with sampling every 5,000th generation. Tracer 1.7.160 was used to inspect stationarity and ESS values for all parameters. The first 10% of samples from each analysis were discarded as “burn-in”, and the total of 54,000 remaining trees from all three analyses were combined in LogCombiner 1.7, and TreeAnnotator 2.6 (both available as part of the BEAST package61 at http://beast.bio.ed.ac.uk) was used to obtain a final species tree as a maximum clade credibility tree.
External morphology
Variation in external morphology, with a special interest to evaluate the similarity of the newly discovered genus and Hyperolius, was investigated on a total of 60 adult male specimens of six taxa (10 from each): Congolius robustus, Cryptothylax greshoffii, and four Hyperolius species. As the latter is a speciose genus, we included representatives of two main Hyperolius clades1. Two taxa were selected as representatives of Clade 21, the same taxa as identified earlier as the most similar to C. robustus14,16: H. balfouri and H. cinnamomeoventris from the H. concolor species group. Two other taxa were selected as representatives of Clade 11: H. tuberculatus from the H. viridiflavus group60 and H. phantasticus from the H. pardalis group62). Hyperolius cinnamomeoventris and H. phantasticus were also selected for their sympatric occurrence with C. robustus in the Kokolopori Bonobo Nature Reserve, Tshuapa, DRC. Representatives of the H. nasutus group, the third main Hyperolius clade1, were omitted for their characteristically different morphology (elongated slender body, sharp nose, translucent green colouration, sexes of same size and colouration)13,63. Cryptothylax greshoffii was selected as a sympatric relative of Congolius, and a representative of the sister clade of Hyperolius [1, 3] (this study). A list of all examined specimens is available in Supplementary Table S1.
Snout-vent length (SVL) and the following 15 measurements were taken with a digital calliper XTline P13430 150 mm to the nearest 0.1 mm (Supplementary Fig. S5): snout-urostyle length (SUL), from snout tip to posterior edge of urostyle; head width (HW), at greatest head width in close proximity to posterior edge of jaw; head length (HDL), from snout tip to posterior edge of jaw, measured along body axis; interorbital distance (IOD), shortest distance between upper eyelids; eye diameter (ED), between anterior and posterior corners of eye; eye-nostril length (ENL), from anterior corner of eye to centre of nostril, measured along body axis; internarial distance (IND), between centres of nostrils; snout length (SL), from anterior corner of eye to snout tip, measured along body axis; humerus length (HL), from body wall to outer edge of elbow; radioulna length (RL), from elbow to proximal edge of most proximal palmar tubercle; hand length (HaL), from most proximal palmar tubercle to tip of fourth manual digit; femur length (FL), from centre of vent to outer edge of knee; tibiofibula length (TL), from outer edge of knee to outer edge of tibiotarsal articulation; foot length (FoL), from most proximal edge of inner metatarsal tubercle to tip of fourth pedal digit; disc width (DW), at greatest width of adhesive disc of fourth pedal digit. These measurements follow recent taxonomic publications on Hyperolius64, and publications on general anuran morphometry65.
The measured values were subsequently statistically treated according to Mosimann66 to filter out the effect of body size, see also Gvoždík et al.67 or Dolinay et al.68. Principal component analysis (PCA) was performed in the programming language R v3.6.369 using R-package “vegan” v2.5–670 to explore the morphospace of the body shapes. Multivariate analysis of variance (MANOVA) was performed on the first three principal components to test body shape differentiation among the taxa, and descriptive statistics of body size and selected morphometric indices was calculated via boxplots69.
Osteology
A high-resolution X-ray microcomputed tomography (μCT) was used to compare osteological characters of Congolius robustus to other genera of Hyperoliinae present in the Congo Basin: Hyperolius (H. balfouri, H. cf. tuberculatus), Cryptothylax, Chrysobatrachus, and Callixalus (in order of initially presumed relationships or similarity8,9,14,16). Afrixalus and Kassinula were omitted for their clearly different morphology8,9. The μCT scan of Acanthixalus (Kassininae) was downloaded from MorphoSource (http://morphosource.org/Detail/MediaDetail/Show/media_id/13843) to allow a comparison to Callixalus, a little-known Congolian hyperoliid possibly related to this genus8,9. The X-ray computed tomography of the six Hyperoliinae taxa was conducted at the Institute of Theoretical and Applied Mechanics, Czech Academy of Sciences, Centre Telč, Czech Republic. A patented device TORATOM (European patent 2835631) was used together with an in-house developed software ToraBaC (developed in the Centre Telč, Institute of Theoretical and Applied Mechanics, Czech Academy of Sciences, Czech Republic; http://www.itam.cas.cz/Software/Torabac/index.html) for a flat field correction, defective pixel corrections, and a projection equalization. Scans were conducted with a resolution of 20 μm/px at 120 kV voltage and 83 μA target current with rotation step of 0.15° (2400 projections per full rotation, no averaging) and exposure time of 280 ms. Volume Graphics VGStudio Max 3.2 (Volume Graphics GmbH, Heidelberg, Germany) was used for reconstruction and visualization. Examination and terminology of osteological characters followed Drewes8. All acquired scans were deposited in MorphoSource (https://morphosource.org/, see Data availability for details) in a form of BMP image stacks. See Supplementary Table S1 for details on the osteological material.
To examine, if the similarity of the new genus and Hyperolius has its basis in osteological characters, we evaluated a variation in the cranial shape of Congolius gen. n., and relative (Cryptothylax) or similar (Hyperolius) genera from the lowland Congo Basin. Thirty-two morphometric landmarks were selected following Paluh et al.71 (Supplementary Fig. S6). 3D landmark coordinates were digitized using Stratovan Checkpoint v2018.06.18 (Stratovan Corporation, Davis, U.S.A.), and Procrustes and principal component analyses were performed in R using package “geomorph” v3.2.172 to evaluate the morphospace of the skull shape.
Graphics
Map shown in the Fig. 1a was produced using ArcGIS v10.6 (Esri Inc. 2017) with implementation of GlobCover 200973, and country boundaries downloaded from https://naturalearthdata.com. Images of skulls were produced in MeshLab v2020.0574. Photographs used were taken by authors if not stated otherwise. The final compositions and edits of all figures were done using GNU Image Manipulation Program v2.10.4 downloaded from https://gimp.org.
Ethics statement
Examined museum specimens were loaned from museums with permissions from the curators of the herpetological collections. Collection and handling of live animals was approved by the institutional review board of the Biodiversity Monitoring Centre, University of Kisangani, Democratic Republic of the Congo. All experiments were carried out in accordance with relevant guidelines and regulations. This study meets all the conditions of the ARRIVE guidelines.
Supplementary Information
Acknowledgements
We thank A. Lotana Lokasola (Kinshasa/Yetee) and the Kokolopori community for the invitation to conduct herpetological surveys of the Kokolopori Bonobo Nature Reserve. We are grateful to P. R. Møller and D. K. Johansson (Natural History Museum of Denmark, University of Copenhagen), L. A. Scheinberg (California Academy of Sciences, San Francisco), J. Rosado (Museum of Comparative Zoology, Harvard University, Cambridge), D. Meirte (Royal Museum for Central Africa, Tervuren), and J. Moravec (National Museum, Prague) for loaning or access to the material. We further thank D. Modrý (Brno) and M. Jirků (České Budějovice) for providing us additional comparative material, which was collected with permissions from Ministre de l’Education Nationale, de l’Alphabétisation, de l’Enseignement Supérieur et de la Recherche of the Central African Republic and the World Wildlife Fund, while the Primate Habituation Programme provided a logistical support. We would like to express our gratitude to A.-G. Zassi-Boulou and the directorate of IRSEN (Brazzaville) for field assistance and issuing permits, when a part of the comparative material was collected in the Republic of the Congo. J. Kielgast (Copenhagen) is acknowledged for providing a distribution record from the Salonga NP, T.F. Kpan (Abidjan), F. Tillack and M.-O. Rödel (Natural History Museum, Berlin) for providing photographs of Morerella cyanophthalma, O. Mikula (Brno) for providing access to the Stratovan Checkpoint software, D.C. Blackburn (Florida Museum of Natural History, Gainesville) for providing access to the μCT scan of Acanthixalus spinosus deposited in the MorphoSource online database.
Author contributions
T. N. conceived and designed the experiments, performed experiments, analysed the data, prepared figures and tables, authored and reviewed drafts of the paper, and approved the final draft. G. B. carried out fieldwork, authored and reviewed drafts of the paper, and approved the final draft. M. V. performed μCT experiments, authored and reviewed drafts of the paper, and approved the final draft. V. G. conceived and designed the experiments, carried out fieldwork, analysed the data, authored and reviewed drafts of the paper, and approved the final draft.
Funding
This work was funded by the Ministry of Culture of the Czech Republic (DKRVO 2019–2023/6.VII.c, National Museum, 00023272), institutional supports of IVB CAS (RVO: 68081766) and ITAM CAS (RVO: 68378297), Department of Botany and Zoology, Faculty of Science, Masaryk University, Brno, Czech Republic (MUNI/A/1098/2019), and G.B. was supported by the IFS grant No.: I-1-D-6074–1.
Data availability
The sequences obtained in this study are available in the GenBank online database (https://ncbi.nlm.nih.gov/genbank). IVB-H-CD18-255: MW626918, MW626921, MW626924, MW626927, MW626930; NMP-P6V 76089: MW626917, MW626920, MW626923, MW626926, MW626929; ZMUC R.771176: MW626916, MW626919, MW626922, MW626925, MW626928. The X-ray microcomputed tomography scans of the skulls obtained in this study are available in the MorphoSource online database (https://morphosource.org) in a form of BMP image stacks: Callixalus pictus CAS 145260: https://www.morphosource.org/biological_specimens/000S31537; Chrysobatrachus cupreonitens CAS 145263: https://www.morphosource.org/biological_specimens/000S31531; Congolius robustus ZMUC-R.771175 (male): https://www.morphosource.org/biological_specimens/000S31541; Congolius robustus ZMUC-R.771176 (female): https://www.morphosource.org/biological_specimens/000S31542; Cryptothylax greshoffii IVB-H-CD15-169: https://www.morphosource.org/biological_specimens/000S31540; Hyperolius balfouri IVB-H-CD15-061: https://www.morphosource.org/biological_specimens/000S31539; Hyperolius cf. tuberculatus IVB-H-CD15-010: https://www.morphosource.org/biological_specimens/000S31538. The μCT scan of Acanthixalus spinosus CAS 153800 was downloaded from https://www.morphosource.org/biological_specimens/000S10942, and used with the approval of the authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tadeáš Nečas, Email: tad.necas@gmail.com.
Václav Gvoždík, Email: vaclav.gvozdik@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-87495-2.
References
- 1.Portik DM, et al. Sexual dichromatism drives diversification within a major radiation of African amphibians. Syst. Biol. 2019;68:859–875. doi: 10.1093/sysbio/syz023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frost, D. R. Amphibian Species of the World: an Online Reference. Version 6.1https://amphibiansoftheworld.amnh.org/index.php; American Museum of Natural History, New York, USA. 10.5531/db.vz.0001 (2020).
- 3.Portik DM, Blackburn DC. The evolution of reproductive diversity in Afrobatrachia: A phylogenetic comparative analysis of an extensive radiation of African frogs. Evolution. 2016;70:2017–2032. doi: 10.1111/evo.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurent RF. Diagnoses préliminaires de treize batraciens nouveaux d’Afrique centrale. Rev. Zool. Bot. Afr. 1950;44:1–18. [Google Scholar]
- 5.Laurent RF. Deux reptiles et onze batraciens nouveaux d’Afrique centrale. Rev. Zool. Bot. Afr. 1951;44:360–381. [Google Scholar]
- 6.Laurent RF. Description d’un rhacophoride nouveau du Congo Belge (Batracien) Rev. Zool. Bot. Afr. 1940;33:313–316. [Google Scholar]
- 7.Conradie W, et al. New insights into the taxonomic status, distribution and natural history of De Witte’s Clicking Frog (Kassinula wittei Laurent, 1940) Afr. Zool. 2020;55:311–322. doi: 10.1080/15627020.2020.1821771. [DOI] [Google Scholar]
- 8.Drewes RC. A phylogenetic analysis of the Hyperoliidae (Anura): treefrogs of Africa, Madagascar, and Seychelles Islands. Occas. Pap. California Acad. Sci. 1984;139:1–70. [Google Scholar]
- 9.Channing A. A re-evaluation of the phylogeny of old World treefrogs. Afr. Zool. 1989;24:116–131. [Google Scholar]
- 10.Laurent RF, Combaz J. Sur l’attribution générique de certains batraciens appartenant à la sous-famille de Hyperoliinae. Rev. Zool. Bot. Afr. 1950;43:269–280. [Google Scholar]
- 11.Rödel M-O, et al. A new tree-frog genus and species from Ivory Coast, West Africa (Amphibia: Anura: Hyperoliidae) Zootaxa. 2009;2044:23–45. doi: 10.11646/zootaxa.2044.1.2. [DOI] [Google Scholar]
- 12.Channing, A. & Rödel, M.-O. Field Guide to the Frogs & Other Amphibians of Africa. (Struik Nature, 2019).
- 13.Schiøtz, A. Treefrogs of Africa (Edition Chimaira, 1999).
- 14.Laurent RF. Description de deux Hyperolius nouveaux du Sankuru (Zaire) Rev. Zool. Bot. Afr. 1979;93:779–791. [Google Scholar]
- 15.Schiøtz A. Notes on the genus Hyperolius (Anura, Hyperoliidae) in central République Démocratique du Congo. Alytes. 2006;24:40–60. [Google Scholar]
- 16.Laurent RF. Phennogrammes d’anoures bases sur la morphométrie. Monit. Zool. Ital. 1981;15:1–22. [Google Scholar]
- 17.Bell RC, et al. Idiosyncratic responses to climate-driven forest fragmentation and marine incursions in reed frogs from Central Africa and the Gulf of Guinea Islands. Mol. Ecol. 2017;26:5223–5244. doi: 10.1111/mec.14260. [DOI] [PubMed] [Google Scholar]
- 18.Soro N, Kouamé AM, Kouamé NG, Adepo-Gourène AB, Rödel M-O. Morerella cyanophthalma (Anura: Hyperoliidae) in south-eastern Ivory Coast: Additional data and implications for species’ conservation. Herpetol. Notes. 2019;12:1215–1223. [Google Scholar]
- 19.Rage J-C. The amphibians and reptiles at the Eocene-Oligocene transition in Western Europe: An outline of the faunal alterations. Dev. Palaeontol. Stratigr. 1986;9:309–310. doi: 10.1016/S0920-5446(08)70135-3. [DOI] [Google Scholar]
- 20.Prothero DR. The Late Eocene-Oligocene extinctions. Annu. Rev. Earth Planet. Sci. 1994;22:145–165. doi: 10.1146/annurev.ea.22.050194.001045. [DOI] [Google Scholar]
- 21.Pan AD, Jacobs BF, Dransfield J, Baker WJ. The fossil history of palms (Arecaceae) in Africa and new records from the Late Oligocene (28–27 Mya) of north-western Ethiopia. Bot. J. Linn. Soc. 2006;151:69–81. doi: 10.1111/j.1095-8339.2006.00523.x. [DOI] [Google Scholar]
- 22.Zachos JC, Quinn TM, Salamy KA. High-resolution (104 years) deep-sea foraminiferal stable isotope records of the Eocene-Oligocene climate transition. Paleoceanography. 1996;11:251–266. doi: 10.1029/96pa00571. [DOI] [Google Scholar]
- 23.Zachos JC, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 24.DeConto RM, Pollard D. Rapid Cenozoic glaciation of Antarctica induced by declining atmospheric CO2. Nature. 2003;421:245–249. doi: 10.1038/nature01290. [DOI] [PubMed] [Google Scholar]
- 25.Coxall HK, Wilson PA, Pälike H, Lear CH, Backman J. Rapid stepwise onset of Antarctic glaciation and deeper calcite compensation in the Pacific Ocean. Nature. 2005;433(7021):53–57. doi: 10.1038/nature03135. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, et al. Global cooling during the Eocene-Oligocene climate transition. Science. 2009;323(5918):1187–1190. doi: 10.1126/science.1166368. [DOI] [PubMed] [Google Scholar]
- 27.Plana V. Mechanisms and tempo of evolution in the African Guineo-Congolian rainforest. Philos. Trans. R. Soc. Lond. B. 2004;359:1585–1594. doi: 10.1098/rstb.2004.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zachos JC, Shackleton NJ, Revenaugh JS, Pälike H, Flower BP. Climate response to orbital forcing across the Oligocene-Miocene boundary. Science. 2001;292(5515):274–278. doi: 10.1126/science.1058288. [DOI] [PubMed] [Google Scholar]
- 29.Greenbaum E, Kusamba C. Conservation implications following the rediscovery of four frog species from the Itombwe Natural Reserve, Eastern Democratic Republic of the Congo. Herpetol. Rev. 2012;43:253–259. [Google Scholar]
- 30.Ernst R, Lautenschläger T, Branquima MF, Hölting M. At the edge of extinction: A first herpetological assessment of the proposed Serra do Pingano Rainforest National Park in Uíge Province, northern Angola. Zoosyst. Evol. 2020;96:237–262. doi: 10.3897/zse.96.51997. [DOI] [Google Scholar]
- 31.Jackson K, Blackburn DC. The amphibians and reptiles of Nouabale-Ndoki National Park, Republic of Congo (Brazzaville) Salamandra. 2007;43:149–164. [Google Scholar]
- 32.Jackson K, Zassi-Boulou A-G, Mavoungou L-B, Pangou S. Amphibians and reptiles of the Lac Télé Comunnity Reserve, Likouala Region, Republic of Congo (Brazzaville) Herpetol. Conserv. Biol. 2007;2:75–86. [Google Scholar]
- 33.Badjedjea G, et al. A preliminary survey of the amphibian fauna of Kisangani ecoregion, Democratic Republic of the Congo. J. Adv. Bot. Zool. 2015;3:V3I407. doi: 10.5281/zenodo.1000085. [DOI] [Google Scholar]
- 34.Badjedjea G, et al. Contribution to the knowledge of amphibians of Kponyo village (DR Congo) J. Adv. Bot. Zool. 2016;4:V4I104. doi: 10.5281/zenodo.898031. [DOI] [Google Scholar]
- 35.Masudi FM, et al. Preliminary data on amphibian diversity of the Okapi Wildlife Reserve (RFO) in Democratic Republic of the Congo. Am. J. Zool. 2019;2:38–43. doi: 10.11648/j.ajz.20190203.11. [DOI] [Google Scholar]
- 36.Fišer C, Robinson CT, Malard F. Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 2018;27:613–635. doi: 10.1111/mec.14486. [DOI] [PubMed] [Google Scholar]
- 37.Bickford D, et al. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- 39.Bravo GA, Remsen JV, Jr, Brumfield RT. Adaptive processes drive ecomorphological convergent evolution in antwrens (Thamnophilidae) Evolution. 2014;68:2757–2774. doi: 10.1111/evo.12506. [DOI] [PubMed] [Google Scholar]
- 40.Scheffer M, van Nes EH. Self-organized similarity, the evolutionary emergence of groups of similar species. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6230–6235. doi: 10.1073/pnas.0508024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabaj MH. Codes for natural history collections in ichthyology and herpetology. Copeia. 2020;108:593–669. doi: 10.1643/ASIHCODONS2020. [DOI] [Google Scholar]
- 42.Palumbi, S. et al. The Simple Fool’s Guide to PCR (University of Hawaii, 1991).
- 43.Bossuyt F, Milinkovitch MC. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6585–6590. doi: 10.1073/pnas.97.12.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith SA, Stephens PR, Wiens JJ. Replicate patterns of species richness, historical biogeography, and phylogeny in Holarctic treefrogs. Evolution. 2005;59:2433–2450. doi: 10.1111/j.0014-3820.2005.tb00953.x. [DOI] [PubMed] [Google Scholar]
- 45.Wiens JJ, Fetzner JW, Jr, Parkinson CL, Reeder TW. Hylid frog phylogeny and sampling strategies for speciose clades. Syst. Biol. 2005;54:778–807. doi: 10.1080/10635150500234625. [DOI] [PubMed] [Google Scholar]
- 46.Shen XX, Liang D, Feng YJ, Chen MY, Zhang P. A versatile and highly efficient toolkit including 102 nuclear markers for vertebrate phylogenomics, tested by resolving the higher level relationships of the Caudata. Mol. Biol. Evol. 2013;30:2235–2248. doi: 10.1093/molbev/mst122. [DOI] [PubMed] [Google Scholar]
- 47.Gvoždík V, et al. Evolutionary history of the Cameroon radiation of puddle frogs (Phrynobatrachidae: Phrynobatrachus), with descriptions of two critically endangered new species from the northern Cameroon Volcanic Line. PeerJ. 2020;8:e8393. doi: 10.7717/peerj.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronquist F, et al. MRBAYES 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 54.Huelsenbeck JP, Rannala B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst. Biol. 2004;53:904–913. doi: 10.1080/10635150490522629. [DOI] [PubMed] [Google Scholar]
- 55.Ogilvie HA, Bouckaert R, Drummond AJ. StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 2017;34:2101–2114. doi: 10.1093/molbev/msx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLOS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y-J, et al. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous-Paleogene boundary. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E5864–E5870. doi: 10.1073/pnas.1704632114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan Z-Y, et al. Natatanuran frogs used the Indian Plate to step-stone disperse and radiate across the Indian Ocean. Nat. Sci. Rev. 2018;6:10–14. doi: 10.1093/nsr/nwy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hime PM, et al. Phylogenomics reveals ancient gene tree discordance in the amphibian tree of life. Syst. Biol. 2020 doi: 10.1093/sysbio/syaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/sys032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouckaert R, et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dehling JM, Sinsch U. Partitioning of morphospace in larval and adult reed frogs (Anura: Hyperoliidae: Hyperolius) of the Central African Albertine Rift. Zool. Anz. 2019;280:65–77. doi: 10.1016/j.jcz.2019.04.003. [DOI] [Google Scholar]
- 63.Channing A, et al. Taxonomy of the super-cryptic Hyperolius nasutus group of long reed frogs of Africa (Anura: Hyperoliidae), with descriptions of six new species. Zootaxa. 2013;3620:301–350. doi: 10.11646/zootaxa.3620.3.1. [DOI] [PubMed] [Google Scholar]
- 64.Conradie W, et al. A new Reed Frog (Hyperoliidae: Hyperolius) from coastal northeastern Mozambique. Zootaxa. 2018;4379:177–198. doi: 10.11646/zootaxa.4379.2.2. [DOI] [PubMed] [Google Scholar]
- 65.Watters JL, Cummings ST, Flanagan RL, Siler CD. Review of morphometric measurements used in anuran species descriptions and recommendations for a standardized approach. Zootaxa. 2016;4072:477–495. doi: 10.11646/zootaxa.4072.4.6. [DOI] [PubMed] [Google Scholar]
- 66.Mosimann JE. Size allometry: Size and shape variables with characterizations of lognormal and generalized Gamma distributions. J. Am. Stat. Assoc. 1970;65:930–945. doi: 10.1080/01621459.1970.10481136. [DOI] [Google Scholar]
- 67.Gvoždík V, Moravec J, Kratochvíl L. Geographic morphological variation in parapatric Western Palearctic tree frogs, Hyla arborea and Hyla savignyi: Are related species similarly affected by climatic conditions? Biol. J. Linn. Soc. 2008;95:539–556. doi: 10.1111/j.1095-8312.2008.01056.x. [DOI] [Google Scholar]
- 68.Dolinay M, et al. Gene flow in phylogenomics: sequence capture resolves species limits and biogeography of Afromontane forest endemic frogs from the Cameroon Highlands. bioRxiv. 2020 doi: 10.1101/2020.10.09.332767. [DOI] [PubMed] [Google Scholar]
- 69.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, https://R-project.org, 2020).
- 70.Oksanen, J. et al. vegan: Community Ecology Package. R Package Version 2.5–6. https://CRAN.R-project.org/package=vegan (2019).
- 71.Paluh DJ, Stanley EL, Blackburn DC. Evolution of hyperossification expands skull diversity in frogs. Proc. Natl. Acad. Sci. U.S.A. 2020;117:8554–8562. doi: 10.1073/pnas.2000872117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams, D. C., Collyer, M. L. & Kaliontzopoulou, A. Software for Geometric Morphometric Analyses. R Package Version 3.2.1.https://cran.r-project.org/package=geomorph (2020).
- 73.Arino, O. et al. Global Land Cover Map for 2009 (European Space Agency & Université catholique de Louvain; 10.1594/PANGAEA.787668 (2012).
- 74.Cignoni et al. MeshLab: an open-source mesh processing tool. Sixth Eurographics Italian Chapter Conference 129–136 (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences obtained in this study are available in the GenBank online database (https://ncbi.nlm.nih.gov/genbank). IVB-H-CD18-255: MW626918, MW626921, MW626924, MW626927, MW626930; NMP-P6V 76089: MW626917, MW626920, MW626923, MW626926, MW626929; ZMUC R.771176: MW626916, MW626919, MW626922, MW626925, MW626928. The X-ray microcomputed tomography scans of the skulls obtained in this study are available in the MorphoSource online database (https://morphosource.org) in a form of BMP image stacks: Callixalus pictus CAS 145260: https://www.morphosource.org/biological_specimens/000S31537; Chrysobatrachus cupreonitens CAS 145263: https://www.morphosource.org/biological_specimens/000S31531; Congolius robustus ZMUC-R.771175 (male): https://www.morphosource.org/biological_specimens/000S31541; Congolius robustus ZMUC-R.771176 (female): https://www.morphosource.org/biological_specimens/000S31542; Cryptothylax greshoffii IVB-H-CD15-169: https://www.morphosource.org/biological_specimens/000S31540; Hyperolius balfouri IVB-H-CD15-061: https://www.morphosource.org/biological_specimens/000S31539; Hyperolius cf. tuberculatus IVB-H-CD15-010: https://www.morphosource.org/biological_specimens/000S31538. The μCT scan of Acanthixalus spinosus CAS 153800 was downloaded from https://www.morphosource.org/biological_specimens/000S10942, and used with the approval of the authors.