Abstract
Quantifying severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in clinical samples is crucial for early diagnosis and timely medical treatment of coronavirus disease 2019. Here, we describe a digital warm-start CRISPR (dWS-CRISPR) assay for sensitive quantitative detection of SARS-CoV-2 in clinical samples. The dWS-CRISPR assay is initiated at above 50 °C and overcomes undesired premature target amplification at room temperature, enabling accurate and reliable digital quantification of SARS-CoV-2. By targeting SARS-CoV-2's nucleoprotein gene, the dWS-CRISPR assay is able to detect down to 5 copies/μl SARS-CoV-2 RNA in the chip. It is clinically validated by quantitatively determining 32 clinical swab samples and three clinical saliva samples. Moreover, it has been demonstrated to directly detect SARS-CoV-2 in heat-treated saliva samples without RNA extraction. Thus, the dWS-CRISPR method, as a sensitive and reliable CRISPR assay, facilitates accurate SARS-CoV-2 detection toward digitized quantification.
Keywords: SARS-CoV-2 detection, COVID-19, CRISPR-Cas12a, Digital warm-start CRISPR assay, Nucleic acid quantification
Graphical abstract
A digital warm-start CRISPR (dWS-CRISPR) assay for sensitive, quantitative detection of SARS-CoV-2 in clinical samples.

1. Introduction
Since December 2019 (Zhu et al., 2020), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as the causing agent of coronavirus disease 2019 (COVID-19) has spread worldwide, resulting in over one million deaths (WHO 2020). Sensitive and accurate quantification of SARS-CoV-2 plays a crucial role in early diagnosis and evaluating medical treatment of COVID-19. Presently, real-time fluorescence quantitative reverse transcription polymerase chain reaction (RT-qPCR) is the gold standard of SARS-CoV-2 detection (Feng et al., 2020; Ravi et al., 2020; Torrente-Rodríguez et al., 2020). However, RT-qPCR assays greatly depend on expensive real-time fluorescence PCR instrument and their quantitation accuracy is highly associated with well-designed TaqMan probes (Wang et al. 2020a, 2020d), not suitable for small clinics or community health settings. Alternatively, some isothermal nucleic acid amplification methods have been developed to rapidly detect SARS-CoV-2, such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) (Rabe and Cepko 2020; Yan et al., 2020; Yu et al., 2020), reverse transcription recombinase polymerase amplification (RT-RPA) (Behrmann et al., 2020; Xia and Chen 2020), reverse transcription recombinase-aided amplification (RT-RAA) (Wang et al., 2020b; Xue et al., 2020), and sensitive splint-based one-pot isothermal RNA detection (SENSR) (Woo et al., 2020). However, most of these isothermal amplification assays are either lack of the quantitative detection ability or subjected to undesired nonspecific amplification signals (or false positive signals).
As next-generation molecular diagnostics, nucleic acid detections based on clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) nucleases possess great prospects (Chertow 2018). In the CRISPR-Cas-based detections, the target-specific CRISPR RNAs (crRNAs) ensure high specificity and reliability of nucleic acid detection, and the Cas nucleases with collateral cleavage activities (e.g., Cas12a and Cas13a) produce amplified fluorescence signals (Abudayyeh et al., 2016; Chen et al., 2018; Li et al., 2018). Currently, CRISPR-Cas12a-based DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter) system and CRISPR-Cas13a-based SHERLOCK (Specific High-sensitivity Enzymatic Reporter UnLOCKing) system have been applied to detect SARS-CoV-2 (Broughton et al., 2020; Pang et al., 2020; Patchsung et al., 2020; Wang et al., 2020c). However, these assays are typically two-step approaches in which RT-RPA or RT-LAMP, as a separate target preamplification step, is indispensable, complicating the detection operation. To overcome this technical bottleneck, our lab and Zhang's lab developed all-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay (Ding et al., 2020) and SHERLOCK testing in one pot (STOP) assay (Joung et al., 2020) for SARS-CoV-2 detection, respectively. However, these one-pot CRISPR assays are not quantitative.
In nucleic acid quantification, digital nucleic acid detections (e.g., digital PCR) offer many advantages over conventional tube-based bulk assays, such as increased tolerance to inhibitory substances, higher sensitivity, and better detection precise (Gansen et al., 2012; Salipante and Jerome 2020). However, digital PCR assays rely on expensive and bulky PCR thermocyclers. Therefore, as alternatives to digital PCR, Yu's lab and Wang's lab reported digital CRISPR assays based on RPA/RT-RPA for SARS-CoV-2 detection, termed RADICA(Wu et al., 2020) and deCOViD (Park et al.), respectively. However, their quantitative accuracy is still a challenge by overestimating the initial target amount, since RPA/RT-RPA can be initiated at room temperature and cause undesired premature target amplification (Yeh et al., 2017). To minimize premature target amplification, the reaction mixture needs to be prepared on ice and loaded into chips within 1 min (Wu et al., 2020), which complicates detection procedure and remains a challenge in massive COVID-19 testing. Therefore, there is an urgent need for simple, reliable and accurate digital CRISPR assay for SARS-CoV-2 quantification.
Here, we present a digital warm-start CRISPR (dWS-CRISPR) assay for sensitive quantitative detection of SARS-CoV-2 in clinical COVID-19 samples. The dWS-CRISPR is established through partitioning the first warm-start CRISPR-Cas12a-based (WS-CRISPR) reaction into sub-nanoliter aliquots within a QuantStudio 3D digital chip. The WS-CRISPR reaction is a one-pot format combining low-temperature reverse transcription dual-priming isothermal amplification (RT-DAMP) and CRISPR-Cas12a-based detection. The dWS-CRISPR is efficiently initiated at 50–55 °C, thereby preventing premature target amplifications at room temperature and enabling accurate digital quantification of nucleic acids. Through targeting the SARS-CoV-2's nucleoprotein (N) gene, the dWS-CRISPR assay is developed to quantitatively determine 32 clinical swab samples and three clinical saliva samples. More importantly, the dWS-CRISPR assay is used to directly detect SARS-CoV-2 in heat-treated saliva samples without need for RNA extraction.
2. Material and methods
All the required materials, reagents, instrumentation and methods are provided in the Supplementary information.
3. Results and discussion
3.1. Overview of dWS-CRISPR assay
As shown in Fig. 1 a, the dWS-CRISPR reaction mixture is first prepared in a tube, containing the Cas12a-crRNA complex, six RT-DAMP primers (two outer primers of FO and RO, two inner primers of FI and RI, and two competition primers of FC and RC), single-stranded DNA fluorophore-quencher (ssDNA-FQ) reporter, SuperScript IV reverse transcriptase, Bst DNA polymerase, and SARS-CoV-2 RNA target. To achieve digital detection, the prepared reaction mixture is distributed into a QuantStudio 3D digital chip (Thermo Fisher Scientific). This chip is etched with 20,000 consistently-sized hexagon wells (the max width of 60 μm) in a silicon substrate and can partition the mixture into over ten thousand sub-nanoliter (about 0.7 nL) microreactions. After 90-min incubation at 52 °C, the microreactions with target RNA indicate strong green fluorescence (positive spots), whereas not in those without targets (negative spots). The target RNA can be quantified through testing and counting the number of positive spots of the chips.
Fig. 1.
Overview of dWS-CRISPR assay. (a) One-pot WS-CRISPR reaction mixture is first prepared in one tube. After distributed into QuantStudio 3D digital chip, over ten thousand sub-nanoliter (~0.7 nl) microreactions are isolated in microwells. When incubated at 52 °C, each microreaction with SARS-CoV-2 RNA target undertakes WS-CRISPR reaction and generates strong green fluorescence (positive spots), whereas not in those without target (negative spots). Scale bar is 300 μm. Through detecting and counting the positive microreactions (or spots), SARS-CoV-2 RNA can be quantified based on the proportion of positive spots. (b) Working principle of one-pot WS-CRISPR assay for SARS-CoV-2 detection. The WS-CRISPR reaction mixture contains Cas12a-crRNA complex, six DAMP primers (two outer primers of FO and RO, two inner primers of FI and RI, and two competition primers of FC and RC), ssDNA-FQ reporter, SuperScript IV reverse transcriptase, Bst DNA polymerase, and SARS-CoV-2 RNA in a one-pot format.
The dWS-CRISPR combines the firstly reported low-temperature RT-DAMP and CRISPR-Cas12a detection in a one-pot format. The RT-DAMP developed in our lab is a variant of RT-LAMP with a new primer design strategy (Ding et al., 2019). Each inner primer of RT-DAMP is formed with two target sites with a distance of below 40 nucleotides (nt) and a pair of competition primers is added to mediate pair-priming strand extension (Fig. 1b). Our previous study (Ding et al., 2019) has confirmed that the DAMP/RT-DAMP has not only improved the detection sensitivity, but also generated ultralow nonspecific signals. Given this, we chose RT-DAMP to develop our dWS-CRISPR assay.
In the dWS-CRISPR, six RT-DAMP primers recognize six distinct sites in target sequences to initiate self-priming and pair-priming (dual-priming) nucleic acid amplification, producing multiple amplicons with closed loop structures. Simultaneously, the Cas12a-cRNA complex specifically binds the complementary sites in the amplicons to activate the collateral cleavage activity, thereby indiscriminately cleaving surrounding ssDNA-FQ reporters to generate increased fluorescence. The ssDNA-FQ reporter is a 5-cytosine nucleotide single-stranded DNA (5′-CCCCC-3′) labeled with FAM (Fluorescein) at 5′ end and Iowa Black FQ quencher at 3′ end due to its higher affinity to Cas12a (Guanghui et al., 2020). Fluorescence is quenched via resonance energy transfer in intact ssDNA-FQ reporters, but can be recovered after the activated Cas12a cleaves the reporters.
3.2. Development of one-pot WS-CRISPR assay
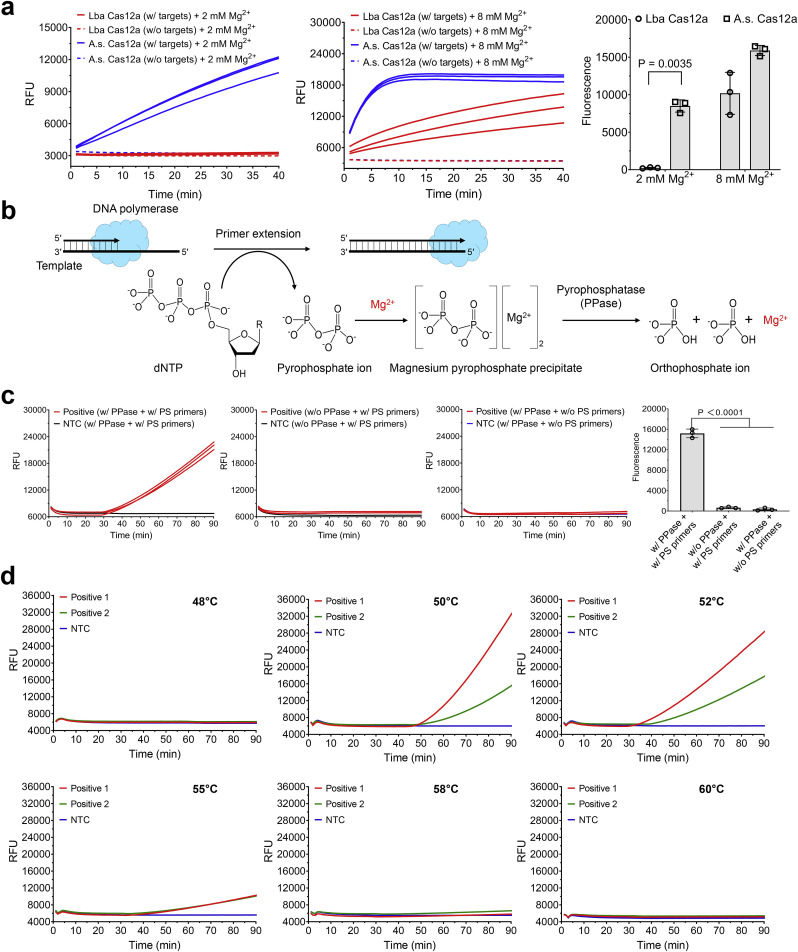
As of now, there still remains a challenge to directly couple LAMP or DAMP with CRISPR-Cas12a detection in a one-pot format due to the significant differences in their reaction buffer compositions and reaction temperature. One of the major concerns is the concentration of Mg2+. To lower Mg2+ concentration is beneficial to develop low temperature isothermal amplification (e.g., DAMP assay) due to the reduced melting points of the primers. However, the cleavage of Cas12a nucleases for both on-target and collateral activity is typically high-Mg2+-dependent (Fuchs et al., 2019; Jinek et al., 2012; Nguyen et al., 2020). To enable highly sensitive nucleic acid detection, two different Cas12a nucleases were evaluated and compared at various Mg2+ concentrations. As shown Fig. 2 a, when reducing the Mg2+ concentration from 8 mM to 2 mM, the Cas12a from Recombinant Acidaminococcus sp. BV3L6 (A.s. Cas12a) still has relatively high collateral cleavage activity at 2 mM Mg2+. Whereas, 2 mM Mg2+ completely inhibits the activity of Cas12a from Lachnospiraceae bacterium ND2006 (Lba Cas12a). Therefore, A.s. Cas12a is used to develop our one-pot WS-CRISPR assay for the rest of the experiments.
Fig. 2.
Optimization of one-pot WS-CRISPR assay. (a) Comparison of two different Cas12a nucleases for CRISPR-based fluorescence detection at different Mg2+ concentration. Lba Cas12a, EnGen Lba Cas12a from Lachnospiraceae bacterium ND2006 (New England Biolabs). A.s. Cas12a, Alt-R Cas12a (Cpf1) Ultra nuclease from Recombinant Acidaminococcus sp. BV3L6 (Integrated DNA Technologies). The used targets are 1 μM of synthetic SARS-CoV-2 N DNA fragments. Three replicates were run (n = 3). (b) The chemical reaction process of the generation and degradation of the magnesium pyrophosphate precipitate due to the existence of pyrophosphatase during primer extension or DNA polymerization. (c) Real-time fluorescence detection and endpoint fluorescence comparison of one-pot WS-CRISPR assay with PPase and/or PS primers at 52 °C. PS primers specifically denote two phosphorothioated inner primers of FI and RI and the rest of primers are non-phosphorothioated. “w/o PS primers” means reactions with the non-phosphorothioated inner primers. Positive, the reaction with 5 × 104 copies/μl SARS-CoV-2 RNA. Three replicates were run (n = 3). (d) Effect of reaction temperature on one-pot WS-CRISPR assay. Positive 1 and 2, the reactions with 3 × 106 and 5 × 104 copies/μl SARS-CoV-2 RNA, respectively. Three independent assays were conducted with the similar results. NTC, non-template control. Error bars represent the means ± standard deviation (s.d.) from replicates. The statistical significance was analyzed using unpaired two-tailed t-test.
In addition, during isothermal amplification, primer extension by DNA polymerase continuously consumes dNTPs and produces a large number of pyrophosphate ions that can chelate Mg2+ to form magnesium pyrophosphate precipitate as the reaction byproduct (Fig. 2b). The reduction of free Mg2+ in the solution further weakens the collateral cleavage activity of Cas12a nuclease. To this end, pyrophosphatase (PPase) is supplemented into the reaction system to degrade the magnesium pyrophosphate precipitate and release free Mg2+, maintaining a constant Mg2+ concentration. As shown in Fig. S4, the optimal concentration of PPase is 0.2 U/μl in our WS-CRISPR assay. Another major concern in developing one-pot CRISPR assay is the significant difference of the optimal reaction temperature between nucleic acid amplification and CRISPR-based detection. Most Cas12a nucleases have an optimal activity at 37 °C, but LAMP/DAMP powered by Bst DNA polymerase typically requires a relatively high temperature of 60–65 °C (Ding et al., 2019; Notomi et al., 2000). As reported by Ellington's lab (Cai et al., 2018), phosphorothioated inner primers are beneficial for developing low-temperature isothermal amplification. Therefore, through supplementing PPase and employing the phosphorothioated inner primers of FI and RI, one-pot WS-CRISPR assay was successfully developed (Fig. 2c).
To achieve the best performance, we optimized the one-pot WS-CRISPR reaction system in terms of the concentrations of Mg2+, Bst DNA polymerase, and SuperScript IV reverse transcriptase. As shown in Fig. S5–S7, the optimal concentrations are 0.2 U/μl PPase, 2 mM Mg2+, 24 U/μl Bst DNA polymerase, and 2 U/μl SuperScript IV. In addition, various DNA polymerases were investigated, including Bst DNA polymerase (large fragment), Bst 2.0 DNA polymerase, Bst 3.0 DNA polymerase, GspSSD 2.0 DNA polymerase, Bsm DNA polymerase (large fragment), IsoPol BST+ DNA polymerase, and IsoPol SD+ DNA polymerase. As shown in Fig. S8, the best DNA polymerase for our WS-CRISPR is Bst DNA polymerase (large fragment). Besides, we assessed the effect of reaction temperatures from 48 °C to 60 °C. Fig. 2d shows that this one-pot CRISPR reaction is typically initiated at 50–55 °C and has the optimal performance at 52 °C, providing a warm-start nucleic acid detection. Also, the one-pot WS-CRISPR reaction is only initiated when all the components of the RT-DAMP and CRISPR-Cas12a are mixed in one-pot (Fig. S9a). Unlike previous one-pot CRISPR assays coupled with RPA/RT-RPA (Ding et al., 2020; Park et al.; Wu et al., 2020), our WS-CRISPR assay provides a warm-start detection of nucleic acid and eliminates undesired premature target amplification at room temperature.
Next, we evaluated the WS-CRISPR's specificity and sensitivity. The specificity assay is evaluated by detecting SARS-CoV control, MERS-CoV, and Hs_RPP30 control. As shown in Fig. S9b, the WS-CRISPR assay has a high specificity to detect SARS-CoV-2. The sensitivity was investigated by detecting various concentrations of SARS-CoV-2 RNA. As shown in Fig. S9c, the WS-CRISPR assay is able to detection down to 500 copies/μl SARS-CoV-2 RNA in sample (equivalently 50 copies/μl RNA in the reaction) within 90 min. Furthermore, Fig. S9 demonstrates that the detection results of the WS-CRISPR assay can be visually read out based on the fluorescence imaging of reaction tubes under either LED blue light or UV light, enabling simple visual detection. Therefore, the developed one-pot WS-CRISPR assay has high sensitivity and specificity for SARS-CoV-2 detection in both real-time fluorescence monitoring and endpoint visual readout.
3.3. Optimization of dWS-CRISPR assay
The dWS-CRISPR assay is developed through partitioning the one-pot WS-CRISPR reaction mixture into sub-nanoliter microreactions in the QuantStudio 3D digital chips. As shown in Fig. 3 a, a typical workflow of dWS-CRISPR assay consists of RNA extraction from clinical samples, one-pot CRISPR reaction mixture preparation, distribution of the reaction mixture into the chip, and incubation at 52 °C. First, the dWS-CRISPR assays with various incubation times (e.g., 10, 30, 60, 90 and 120 min) were investigated. As shown in Fig. 3b and c , a 90-min incubation is enough for the dWS-CRISPR assay to reach the maximum percentage of positive spots.
Fig. 3.
The dWS-CRISPR assay for SARS-CoV-2 detection. (a) A typical workflow of dWS-CRISPR assay to detect SARS-CoV-2 in clinical samples. (b) Endpoint fluorescence micrographs of the QuantStudio digital chip for the SARS-CoV-2 detection with various incubation time (0, 10, 30, 60, 90 and 120 min) at 52 °C. In this dWS-CRISPR assay, 1 × 106 copies/μl SARS-CoV-2 RNA was loaded. (c) The percentage of positive spots comparison for the dWS-CRISPR assays with various incubation time at 52 °C.The number of positive spots was counted by setting the same threshold in the ImageJ software. Percentages of positive spots in each micrograph was calculated (n = 6). Error bars represent the means ± s.d. from replicates. The statistical significance was analyzed using unpaired two-tailed t-test. (d) Effect of various waiting time at room temperature on dWS-CRISPR assay and digital RT-AIOD-CRISPR assay during reaction solution preparation and distribution steps. After specific waiting time at room temperature, the chips were directly observed without incubation. Positive, the reaction with 5 × 105 copies/μl SARS-CoV-2 RNA. NTC, non-template control. Scale bars are 300 μm. Each micrograph is a representative of six distinct regions taken to cover about 2809 microreactions.
To avoid overestimating the initial amount of target nucleic acid by digital detection, it is crucial to prevent undesired premature target amplification during reagent preparation at room temperature. For example, some DNA polymerization reactions (e.g., RPA) can be initiated at room temperature (Yeh et al., 2017). To determine if there is any premature target amplification in our dWS-CRISPR assay, we set up various waiting times at room temperature during the reaction solution preparation and distribution steps (Fig. 3d). For comparison, we also assess the digital detection of our previously reported RT-AIOD-CRISPR assay, a one-pot RT-RPA-based CRISPR assay (Ding et al., 2020). As shown in Fig. 3d, positive spots can clearly be observed as short as 10 min-waiting time at room temperature in the digital RT-AIOD-CRISPR assay, confirming the premature target amplification occurs. This result is similar to that of the digital RADICA assay recently reported by Yu's lab (Wu et al., 2020). To minimize premature target amplification, they prepared reaction mixture on ice and quickly distributed it into the chips (within 1 min) (Wu et al., 2020). However, this operation complicates the assay's procedure and typically requires highly well-trained operators. On the contrary, no positive spots are observed in our dWS-CRISPR assay even after 720-min waiting time at room temperature (Fig. 3d). Thus, our dWS-CRISPR method provides a warm-start assay strategy and enables a simple, sensitive, and reliable quantification of SARS-CoV-2.
3.4. Analytical and clinical validation of dWS-CRISPR assay
The specificity assay of the dWS-CRISPR is first carried out by testing non-SARS-CoV-2 nucleic acids. As shown in Fig. 4a, positive spots are observed in the chip loaded with the SARS-CoV-2 positive control, whereas not for other non-target nucleic acids, such as SARS-CoV control, MERS-CoV, and Hs_RPP30 control, which is consistent to the results of the tube-based bulk reaction format (Fig. S9b). By testing various concentrations of SARS-CoV-2 RNA, the detection sensitivity is also investigated. As shown in Fig. 4b, dWS-CRISPR assay is able to detect down to 50 copies/μl SARS-CoV-2 RNA in the sample (equivalently 5 copies/μl RNA molecules in the chip), a 10-fold improvement compared with the bulk WS-CRISPR assay in the tube format (Fig. S9c). The improvement on sensitivity is probably contributed to the ultralocalized target and ssDNA-FQ reporter through sub-nanoliter microwells in the chip (Tian et al., 2020). In addition, Fig. 4c indicates that there is an excellent linear relationship (R2 = 0.99) between the concentration of targets (from 5 × 103 to 3 × 106 copies/μl) and the percentage of positive spots. In addition, according to the Poisson distribution (Gou et al., 2018; Whale et al., 2012), we obtained a slightly decreased linear relationship (R2 = 0.97) between lg(–ln(1 – f0)) and lg(Xdil), where f0 is the number of positive spots divided by the total number of reaction's microwells and Xdil refers to the dilution factor when using serially diluted targets (Fig. S10), which may attribute to the relatively low filling rate of microwells in the digital chip. In our dWS-CRISPR assay, we harnessed the commercially available QuantStudio digital chip that is specially designed for digital PCR/RT-PCR assay. The WS-CRISPR reaction system has a distinct wettability behavior on the digital chip due to the different reaction components. We will explore other digital chips (e.g., Clarity™ digital chip (Wu et al., 2020)) to test our dWS-CRISPR assay in the future.
Fig. 4.
Evaluation of dWS-CRISPR assay for SARS-CoV-2 detection. (a) Endpoint fluorescence micrographs of the QuantStudio digital chip for the specificity detection of the dWS-CRISPR assays. SARS-CoV-2 PC, SARS-CoV control, MERS-CoV control, and Hs_RPP30 PC were from Integrated DNA Technologies. (b) Endpoint fluorescence micrographs of the chip for the dWS-CRISPR assays testing various concentrations of SARS-CoV-2 RNA within 90-min incubation at 52 °C. (c) The linear relationship between percentage of positive spots (Y) and concentration of targets (X). The blue frame shows the enlarged view of low concentration range from 5 × 100 to 5 × 104 copies/μl. For each concentration's testing, total positive spots in all the six micrographs were used and three chips were taken to run three independent assays (n = 3). Error bars represent the means ± s.d. from replicates. (d) Heat map displaying the determined RNA concentration by RT-qPCR and dWS-CRISPR for detecting SARS-CoV-2 RNA extracted from 32 clinical swab samples (Swab S1–S32) and three saliva samples (Saliva S1–S3). The presented concentrations are the average values in three independent assays. Blank means SARS-CoV-2 negative. PC, SARS-CoV-2-positive control sample. NC, SARS-CoV-2-negative control sample. NTC, non-template control. Each micrograph is a representative of six distinct regions taken to cover about 2809 microreactions. Scale bars are 300 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To validate the clinical utility of the dWS-CRISPR assay, we detected SARS-CoV-2 RNA extracted from 32 clinical swab samples and three clinical saliva samples. For comparison, an in-home RT-qPCR assay using the U.S. CDC-approved SARS-CoV-2 N1 gene's primers and probes (provided by Integrated DNA Technologies) were set up as the parallel experiment. As shown in Fig. S11, 12 positive samples are consistently detected and identified by both dWS-CRISPR and RT-PCR assays, while all negative samples show negative signals. Therefore, in terms of qualitative analysis, the clinical testing result of dWS-CRISPR shows a 100% agreement with that of RT-qPCR method. On quantitative detection, Fig. 4d shows that the averaged viral loadings quantified by dWS-CRISPR range from 1.4 × 104 to 2.3 × 106 copies/μl, showing similar order of magnitude as those determined by RT-qPCR. However, Fig. 4d also indicates that dWS-CRISPR cannot quantify the Sample 12 and 19 with larger Cq values (Cq = 26.09 and 35.34 in Fig. S11) due to its limited quantitative range from 5 × 103 to 3 × 106 copies/μl (Fig. 4c). Despite it, the dWS-CRISPR assay is able to quantify the SARS-CoV-2 RNA extracted from both clinical swab and saliva samples, showing a comparable performance with conventional RT-qPCR method.
3.5. Direct SARS-CoV-2 detection in saliva samples by dWS-CRISPR assay
Recent research showed that saliva sampling is an attractive alternative to swab sampling in SARS-CoV-2 detection due to its simplicity, convenience and non-invasive nature (Wyllie et al., 2020). Especially, saliva samples can be self-collected by patients themselves, avoiding direct interaction between health care workers and patients. Given this, we investigated whether our dWS-CRISPR assay can directly be adapted to detect SARS-CoV-2 in crude saliva samples without RNA extraction step. As shown in Fig. 5 a, each saliva sample contains 90% (v/v) of human saliva obtained from healthy individual, 0%–10% (v/v) of spiked heat-inactivated SARS-CoV-2 from BEI Resources (Catalog # NR-52350), and 1× inactivation reagent developed by Rabe and Cepko (2020). After heated at 95 °C for 5 min, 1.5 μl of the saliva samples was directly added into the dWS-CRISPR reaction system. As shown in Fig. 5b, the dWS-CRISPR assay is successfully able to detect the SARS-CoV-2 spiked in the saliva samples without need for RNA extraction and purification, exhibiting high tolerance to potential inhibitors in saliva samples due to reaction partitioning. Thus, this interesting finding suggests that our dWS-CRISPR assay has the potential to directly detect SARS-CoV-2 from crude clinical saliva specimens through simple heating treatment, facilitating rapid and early molecular diagnostics of COVID-19 infection.
Fig. 5.
Direct detection of SARS-CoV-2 in crude saliva samples by dWS-CRISPR assay. (a) Workflow for direct SARS-CoV-2 testing in spiked saliva samples by dWS-CRISPR assay. (b) Endpoint fluorescence micrographs of the chip for direct detection of SARS-CoV-2 virus spiked in saliva samples. Saliva samples 1–5, the samples with 10%, 5%, 2.5%, 1%, and 0% of heat-inactivated SARS-CoV-2 virus. Each micrograph is a representative of six distinct regions taken to cover about 2809 microreactions. PC, SARS-CoV-2-positive control sample. NC, SARS-CoV-2-negative control sample. NTC, non-template control. Scale bars are 300 μm.
4. Conclusions
In this study, we propose a dWS-CRISPR assay for sensitive quantitative detection of SARS-CoV-2 in clinical samples. Compared to previous CRISPR-Cas-based assays, our WS-CRISPR/dWS-CRISPR assay offers several advantages (Table S2). First, the WS-CRISPR is the first assay combining CRISPR-Cas12a and Bst DNA polymerase-based isothermal amplifications (e.g., DAMP/RT-DAMP) in a one-pot format. Second, the WS-CRISPR is efficiently initiated at above 50 °C or so, addressing the challenge of premature target amplification in digital CRISPR detections coupled with RPA/RT-RPA. Third, dWS-CRISPR has high detection specificity and 10-fold higher sensitivity (down to 5 copies/μl SARS-CoV-2 RNA in the chip) than tube-based bulk assay format. Fourth, dWS-CRISPR shows high tolerance to inhibitors and can directly detect SARS-CoV-2 in crude saliva samples without RNA extraction. These advantages notwithstanding, new adaptive digital chips need to be further explored for the dWS-CRISPR toward absolute quantitative analysis in the future. Furthermore, the dWS-CRISPR assay can be integrated with smartphone-based portable detection platform to achieve onsite or point-of-care quantitative detection (Gou et al., 2018; Wei et al. 2013, 2014). Beyond COVID-19, we envision that the developed dWS-CRISPR assay will be extended to other biomedical applications, including cancer biomarker detection (e.g., liquid biopsy) (Geng et al., 2020; Yin et al., 2020), single-cell analysis(O'Hara et al., 2019), and antiretroviral therapy (e.g., HIV viral load testing) (Alteri et al., 2019; Rutsaert et al., 2018).
Author contribution
Xiong Ding: Conceptualization; Methodology; Investigation, Data curation, Data collection; Formal analysis; Validation; Writing – original draft, Writing – review & editing, Reviewing & Editing. All authors reviewed, discussed, and contributed to the final manuscript and approved it to be published. Kun Yin:, Data curation, Data collection; Formal analysis; Validation; Writing – review & editing, Writing –Reviewing & Editing. All authors reviewed, discussed, and contributed to the final manuscript and approved it to be published. Ziyue Li:, Data curation, Data collection; Formal analysis; Validation; Writing – review & editing, Writing –Reviewing & Editing. All authors reviewed, discussed, and contributed to the final manuscript and approved it to be published. Maroun M. Sfeir: Sample Collection; Resources; Validation; Writing – review & editing, Writing –Reviewing & Editing. All authors reviewed, discussed, and contributed to the final manuscript and approved it to be published. Changchun Liu: Conceptualization; Methodology; Funding acquisition; Project administration; Supervision; Writing – original draft, Writing – review & editing, Reviewing & Editing. All authors reviewed, discussed, and contributed to the final manuscript and approved it to be published.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported, in part, by the National Institutes of Health, United States [R01EB023607, R61AI154642, and R01CA214072].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113218.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., Koonin E.V., Zhang F. Science. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri C., Scutari R., Stingone C., Maffongelli G., Brugneti M., Falasca F., Martini S., Bertoli A., Turriziani O., Sarmati L., Coppola N., Andreoni M., Santoro M.M., Perno C.-F., Ceccherini-Silberstein F., Svicher V. J. Clin. Virol. 2019;117:61–67. doi: 10.1016/j.jcv.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Behrmann O., Bachmann I., Spiegel M., Schramm M., El Wahed A.A., Dobler G., Dame G., Hufert F.T. Clin. Chem. 2020;66(8):1047–1054. doi: 10.1093/clinchem/hvaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Jung C., Bhadra S., Ellington A.D. Anal. Chem. 2018;90(14):8290–8294. doi: 10.1021/acs.analchem.8b02062. [DOI] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow D.S. Science. 2018;360(6387):381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- Ding X., Xu Z., Yin K., Sfeir M., Liu C. Anal. Chem. 2019;91(20):12852–12858. doi: 10.1021/acs.analchem.9b02582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.-B., Cui M., Tao J., Tyrrell D.L., Zhang X.-E., Zhang H., Le X.C. Anal. Chem. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Fuchs R.T., Curcuru J., Mabuchi M., Yourik P., Robb G.B. 2019. bioRxiv; p. 600890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansen A., Herrick A.M., Dimov I.K., Lee L.P., Chiu D.T. Lab Chip. 2012;12(12):2247–2254. doi: 10.1039/c2lc21247a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Z., Li S., Zhu L., Cheng Z., Jin M., Liu B., Guo Y., Liu P. Anal. Chem. 2020;92(10):7240–7248. doi: 10.1021/acs.analchem.0c00818. [DOI] [PubMed] [Google Scholar]
- Gou T., Hu J., Wu W., Ding X., Zhou S., Fang W., Mu Y. Biosens. Bioelectron. 2018;120:144–152. doi: 10.1016/j.bios.2018.08.030. [DOI] [PubMed] [Google Scholar]
- Guanghui T., Gong J., Kan L., Zhang X., He Y., Pan J., Zhao L., Tian J., Lin S., Lu Z., Xue L., Wang C., Li Q. Research square. 2020. [DOI]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., Barretto R.P., Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.-L.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-Y., Cheng Q.-X., Liu J.-K., Nie X.-Q., Zhao G.-P., Wang J. Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.T., Smith B.M., Jain P.K. Nat. Commun. 2020;11(1):E4906. doi: 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28(12) doi: 10.1093/nar/28.12.e63. e63-e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara R., Tedone E., Ludlow A., Huang E., Arosio B., Mari D., Shay J.W. Genome Res. 2019;29(11):1878–1888. doi: 10.1101/gr.250480.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., Wu J., Xiao H., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H., Le X.C. Anal. Chem. 2020;92(24):16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Park, J.S., Hsieh, K., Chen, L., Kaushik, A., Trick, A.Y., Wang, T.H. Adv. Sci. 2003564. 10.1002/advs.202003564. [DOI] [PMC free article] [PubMed]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., et al. Nat. Biomed. Eng. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Rabe B.A., Cepko C. Proc. Natl. Acad. Sci. U. S. A. 2020;117(39):24450–24458. doi: 10.1073/pnas.2011221117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsaert S., Bosman K., Trypsteen W., Nijhuis M., Vandekerckhove L. Retrovirology. 2018;15(1):1–8. doi: 10.1186/s12977-018-0399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salipante S.J., Jerome K.R. Clin. Chem. 2020;66(1):117–123. doi: 10.1373/clinchem.2019.304048. [DOI] [PubMed] [Google Scholar]
- Tian T., Shu B., Jiang Y., Ye M., Liu L., Guo Z., Han Z., Wang Z., Zhou X. ACS Nano. 2021;15(1):1167–1178. doi: 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3(6):1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. J. Am. Med. Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Cai K., He X., Shen X., Wang J., Liu J., Xu J., Qiu F., Lei W., Cui L. Clin. Microbiol. Infect. 2020;26(8):1076–1081. doi: 10.1016/j.cmi.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z. Biosens. Bioelectron. 2020;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. J. Am. Med. Assoc. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Nagi R., Sadeghi K., Feng S., Yan E., Ki S.J., Caire R., Tseng D., Ozcan A. ACS Nano. 2014;8(2):1121–1129. doi: 10.1021/nn406571t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Qi H., Luo W., Tseng D., Ki S.J., Wan Z., Göröcs Z.n., Bentolila L.A., Wu T.-T., Sun R. ACS Nano. 2013;7(10):9147–9155. doi: 10.1021/nn4037706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whale A.S., Huggett J.F., Cowen S., Speirs V., Shaw J., Ellison S., Foy C.A., Scott D.J. Nucleic Acids Res. 2012;40(11) doi: 10.1093/nar/gks203. e82-e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO coronavirus disease (COVID-19) dashboard. 2020. https://covid19.who.int/
- Woo C.H., Jang S., Shin G., Jung G.Y., Lee J.W. Nat. Biomed. Eng. 2020:1–12. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Chan C., Lee Y.H., Springs S.L., Lu T.K., Yu H. medRxiv. 2020. [DOI]
- Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J. N. Engl. J. Med. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Chen X. Cell Discov. 2020;6(1):1–4. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G., Li S., Zhang W., Du B., Cui J., Yan C., Huang L., Chen L., Zhao L., Sun Y. Anal. Chem. 2020;92(14):9699–9705. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E.-C., Fu C.-C., Hu L., Thakur R., Feng J., Lee L.P. Sci. Adv. 2017;3(3) doi: 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Zou Z., Yin F., Liang H., Hu Z., Fang W., Lv S., Zhang T., Wang B., Mu Y. ACS Nano. 2020;14(8):10385–10393. doi: 10.1021/acsnano.0c04177. [DOI] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.