Abstract
A new convergent chemoenzymatic synthesis strategy, integrating enzymatic synthesis of heparan sulfate, sortase A ligation, copper(I)-catalyzed alkyne-azide cycloaddition, and solid phase peptide synthesis, has been established to efficiently synthesize a mimetic of heparan sulfate proteoglycan syndecan-1 glyco-polypeptide at a milligram scale. The mimic was able to bind with αvβ3 integrin faster and exhibit stronger inhibition of breast cancer cell migration compared to the glycan or the polypeptide alone. The novel approach could serve as a general approach for heparan sulfate proteoglycan mimetics synthesis.
Graphical Abstract
A new convergent chemoenzymatic synthesis strategy has been established to synthesize a mimetic of heparan sulfate proteoglycan syndecan-1 glyco-polypeptide.
Heparan sulfate proteoglycan (HSPG) consists of one or more heparan sulfate (HS) chains linked to serine residues in the core protein.1 Ubiquitous on mammalian cell surface and in the extracellular matrix, HSPGs are involved in a wide variety of important biological processes, including regulations of growth factors, cell adhesions and cell-cell communications.2–5 While heparan sulfate (HS) is generally considered to be the main determinant of HSPG activities, the core protein of HSPG can have significant impacts as well.6,7 However, due to the extreme heterogeneity of HS structures in nature, it is highly challenging to purify homogenous HSPGs from natural sources, presenting significant hurdles to decode the roles of HS and core protein in HSPG functions. Chemical synthesis of HS glycopeptide has been reported, which is a daunting task due to instabilities of the HS glycan under typical peptide synthesis conditions.8–10 Successful chemical synthesis requires sophisticated protective group design, tedious synthetic manipulation and route optimization, and meticulous synthetic operations. Herein, we report a new convergent strategy integrating chemical synthesis with enzymatic reactions to generate a well-defined glyco-polypeptide mimicking the complex structure of HSPG such as human syndecan-1. With significantly improved efficiency, milligram quantity of homogeneous HSPG mimetic could be obtained enabling the investigation of the roles of the glycan and the polypeptide in HSPG functions.
Syndecan-1 is a prototypical HSPG on mammalian cell surface, which can bind with integrins mediating cell adhesion, signaling, and migration. A 36 amino acid long polypeptide corresponding to residues 92–119 of human syndecan-1, termed synstatin (SSTN92–119), has been identified as the binding sites of integrins.11 To mimic the structural complexity of syndecan-1, we designed glyco-polypeptide analog 1, which contains a 48 amino acid residue polypeptide backbone including the full length SSTN sequence, as well as a HS glycan chain bearing the full structural features of HS encountered in nature, including iduronic acid, glucuronic acid, O-sulfation and N-sulfation.
To prepare the complex structure of HSPG mimetic 1, retrosynthetically, the target molecule is divided into glycopeptide module 2 and Gly5SSTN92–119 peptide 3 bearing a pentaglycine at its N-terminus (Scheme 1), which would be joined through a sortase A-mediated ligation. The glycopeptide 2 containing the ‘LPETG’ sorting sequence at the C-terminus could be assembled through the copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) of alkynyl peptide 4 and azido-oligosaccharide 5.
Scheme 1.
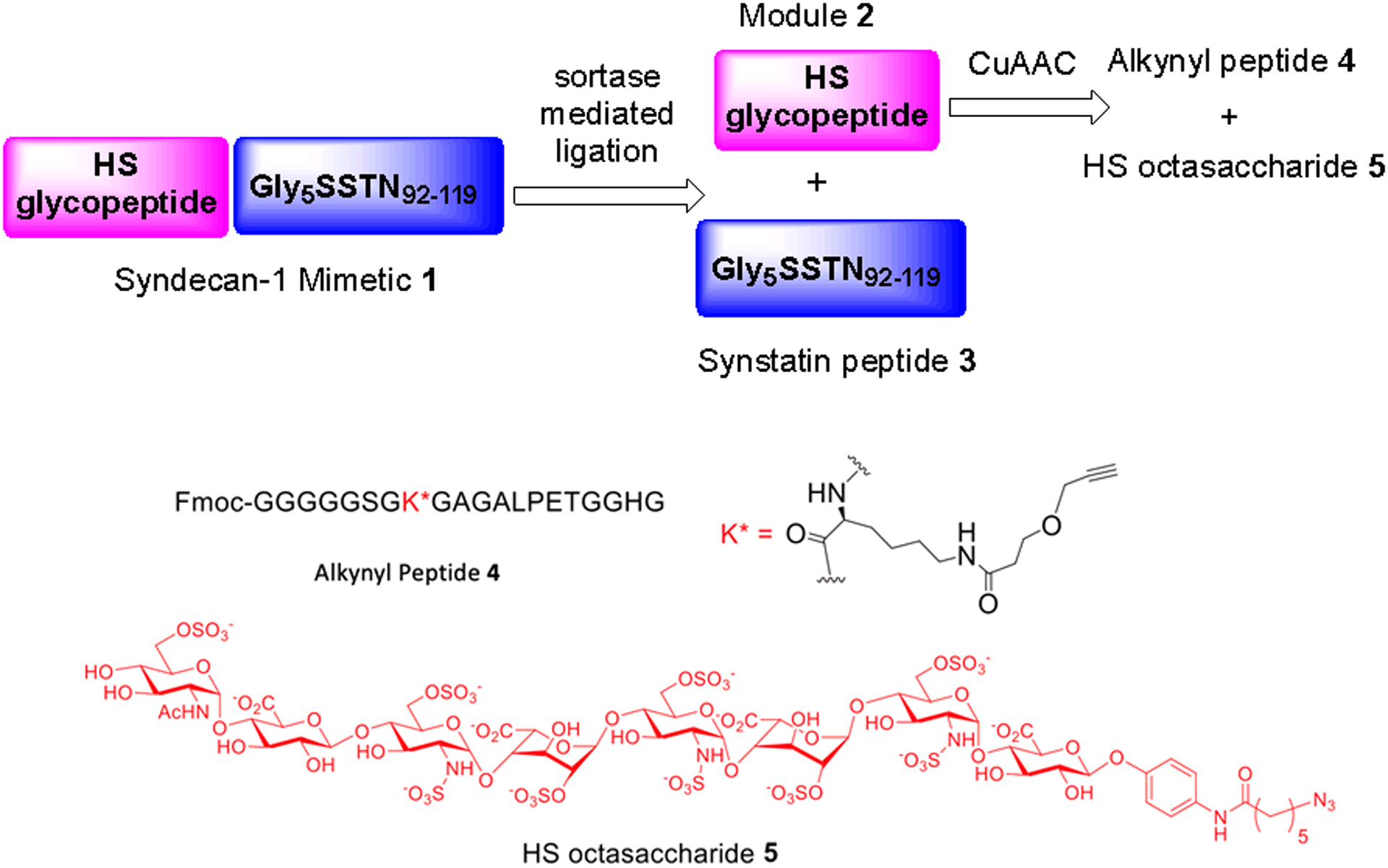
Retrosynthetic analysis of HSPG syndecan-1 mimetic 1.
The alkynyl peptide 4 was synthesized via microwave assisted solid phase supported peptide synthesis (SPPS) starting from Fmoc-glycine loaded resin 5 (Scheme 2A). As the peptide 4 is terminated with pentaglycine at its N-terminus, with the known synthetic difficulties of certain homooligopeptides,13,14 Fmoc-protected pentaglycine building block Fmoc-pentaglycine 6 was prepared separately via SPPS and purified with preparative HPLC assisted by the presence of the hydrophobic Fmoc moiety. 6 was then introduced to the N-terminus of the growing peptide chain on solid phase 5 (Scheme 2A). Subsequent acidic treatment (TFA/TIPS/H2O) cleaved the Fmoc-Gly5 terminated peptide 7 off the resin with all acid-labile protecting groups removed. After incubation of 7 with the propargyl alkyne NHS ester 8, the target peptide 4 was obtained with an 18% overall yield from 5. In a similar manner using microwave assisted SPPS, the 33-mer synstatin peptide Gly5SSTN92–119 3 with the N-terminus pentaglycine was prepared with an overall yield of 24% (Scheme S1).
Scheme 2.
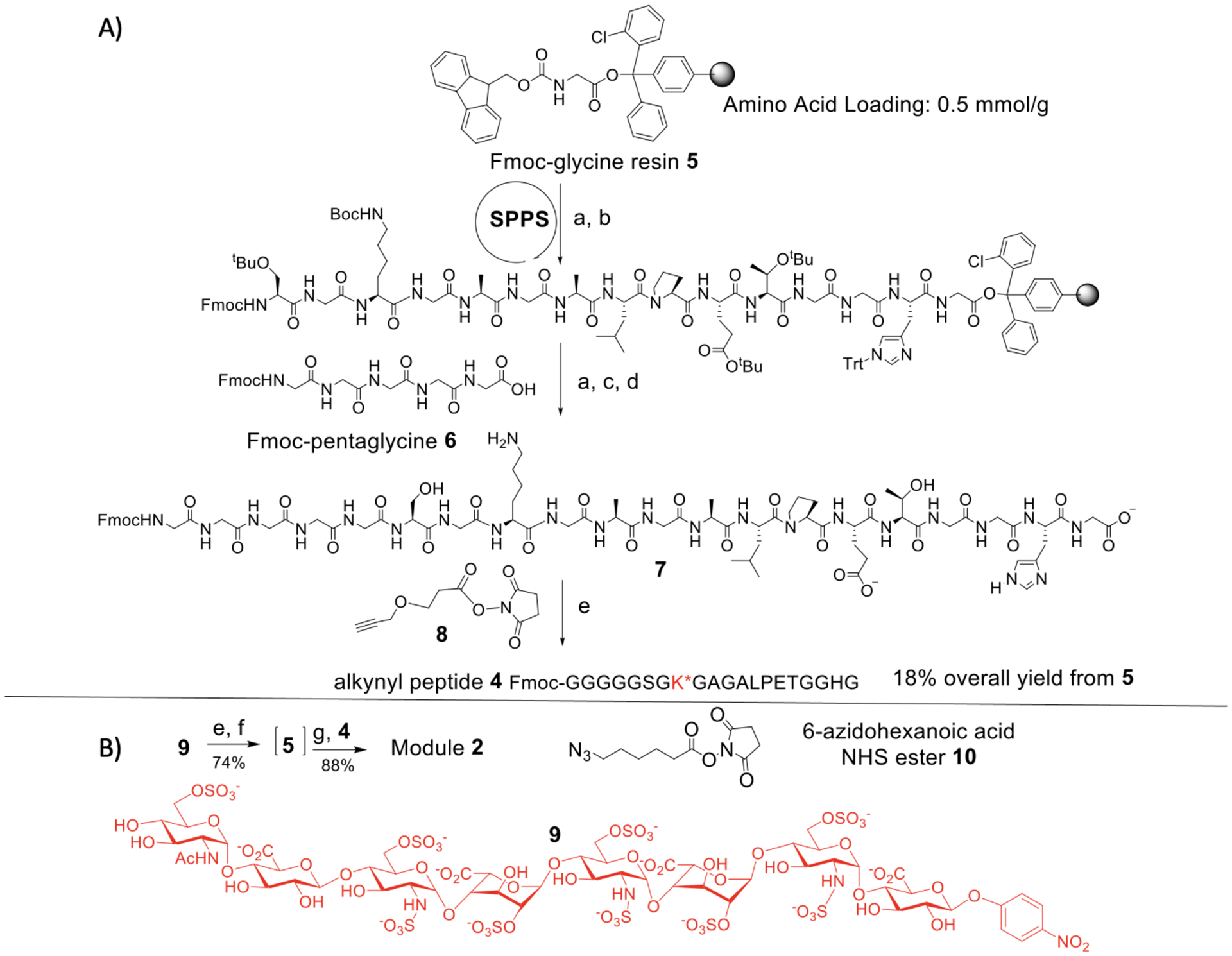
A) Microwave-assisted synthesis of alkyne-functionalized SorTag-containing peptide 4, and B) formation of glycopeptide mimetic 2 through the CuAAC. Reagents and conditions: (a) Fmoc- deprotection: 20% piperidine/DMF, 50 °C, 2 min, microwave; (b) Amino acid coupling: 5 eq. Fmoc-AA-OH, HBTU, HOBt, DIPEA, DMF, 50 °C, 10 min, microwave; (c) Oligopeptide coupling: 5 eq. Fmoc-pentaglycine 6, HATU, DIPEA, DMF, 50 °C, 10 min, microwave; (d) Resin cleavage: TFA/TIPS/H2O (95:2.5:2.5, v/v/v); (e) Propargyl alkyne NHS ester 8, aq. NaHCO3, 18% overall. (f) Pd/C, H2, H2O, 95%; (g) 6-azidohexanoic acid NHS ester 10, aq. NaHCO3, 78%. (h) CuSO4, tris-hydroxypropyltriazolylmethylamine, Na ascorbate, H2O, 88%.
To prepare the HSPG module 2, the p-nitrophenyl bearing heparin octasaccharide 9 was synthesized using HS biosynthetic enzymes,12 and its nitro moiety in the aglycon was reduced by catalytic hydrogenation (Scheme 2B). This was followed by the installation of the azide linker at the reducing end with 6-azidohexanoic acid NHS ester 10. The resulting azide functionalized HS octasaccharide 5 was mixed with the alkynyl peptide 4 (1:1 molar ratio) and subjected to copper catalyzed alkyne azide cycloaddition (CuAAC). The desired product module 2 was obtained in 65% overall yield from 9 following diethylaminoethyl cellulose (DEAE)-HPLC purification (Scheme 2B). The CuAAC condition is mild, which did not affect the structural integrity of the HS glycan or the glyco-polypeptide.
In order to extend the peptide backbone, the key ligation reaction was tested with sortase A (SrtA), a transpeptidase that cross-links the pilin subunits to assemble pili on the surface of gram-positive bacteria.15, 16 To achieve effective ligations, sortase A from Staphylococcus aureus (SrtAstaph) typically requires a LPXTG-containing peptide donor (X can be any natural amino acid) and an acceptor peptide having oligoglycine fragment at its N-terminus.17 SrtAstaph is able to irreversibly couple peptide fragments in the presence of nickel (II) sulfate if the donor peptide carries a Gly-His-Gly tripeptide at the C-terminus of the ‘LPETG’ sorting signal (SorTag), by forming Nickel-peptide complex with the histidine residue and therefore reducing the nucleophilicity of the cleaved peptide.16
The sortase mediated ligation was first explored using GAGALPETGGHG as the donor peptide and GGGGGLPAG as the acceptor peptide. Reaction conditions, including buffer strength, pH, temperature, reaction time and amount of NiSO4 added, were carefully optimized to minimize the undesired hydrolytic activities and improve the coupling efficiency. Incubation of SrtAstaph with the peptide donor at weakly acidic or neutral pH (pH 6.0 – 7.0) at 37 °C led to rapid hydrolysis of the donor. Increasing the pH of the reaction media to slightly basic (pH 8.0–8.5) and lowering the reaction temperature to 25 °C in the presence of 1.5 equivalent nickel (II) sulfate completely shut down the hydrolysis side reaction, while retaining a comparable rate of ligation reaction with the acceptor. In the presence of the donor substrate, a quantitative conversion of the substrate into the product was observed in 10 hours as monitored by LC-MS. When the optimized reaction condition was applied between glycopeptide module 2 and Gly5SSTN92–119 3 (Scheme 3), the desired ligation product 1 was obtained in 86% isolated yield on a milligram scale. 1H NMR and HPLC analysis confirmed product identity and purity. Glyco-polypeptide 1 has a Fmoc moiety at the N-terminus, which can be deprotected and potentially serve as a new acceptor for further peptide backbone extension via another sortase mediated ligation if necessary.
Scheme 3.
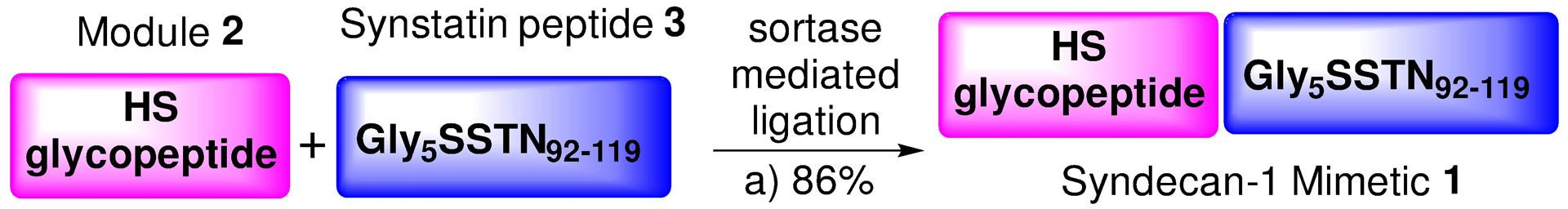
Sortase A-Mediated Ligation. Reagents and conditions: (a) SrtAstaph (5 mol%), 50 mM Tris-HCl buffer, 150 mM NaCl, 5 mM CaCl2, 0.5 mM mercaptoethanol, NiSO4 (1.5 eq. to 2), pH 8.5, 25°C, 4 hours, 86%.
With the glyco-polypeptide mimetic 1 in hand, we investigated its binding with integrin through biolayer interferometry (BLI). The Fmoc moiety of glyco-polypeptide mimic 1 was removed. The resulting free amine, Gly5SSTN92–119 3, and HS glycan 5 were biotinylated and immobilized onto streptavidin-coated sensors. Their bindings with soluble integrin αvβ3 were measured via BLI. While all three compounds were able to bind with integrin αvβ3, interestingly, little dissociation were observed in all cases under the conditions examined (Figure 2). Kinetic analysis indicated that the glyco-polypeptide mimetic 1 was able to more quickly engage integrin, with a kon rate more than 2-fold greater than the rates of glycan or synstatin peptide (Table 1).
Figure 2.
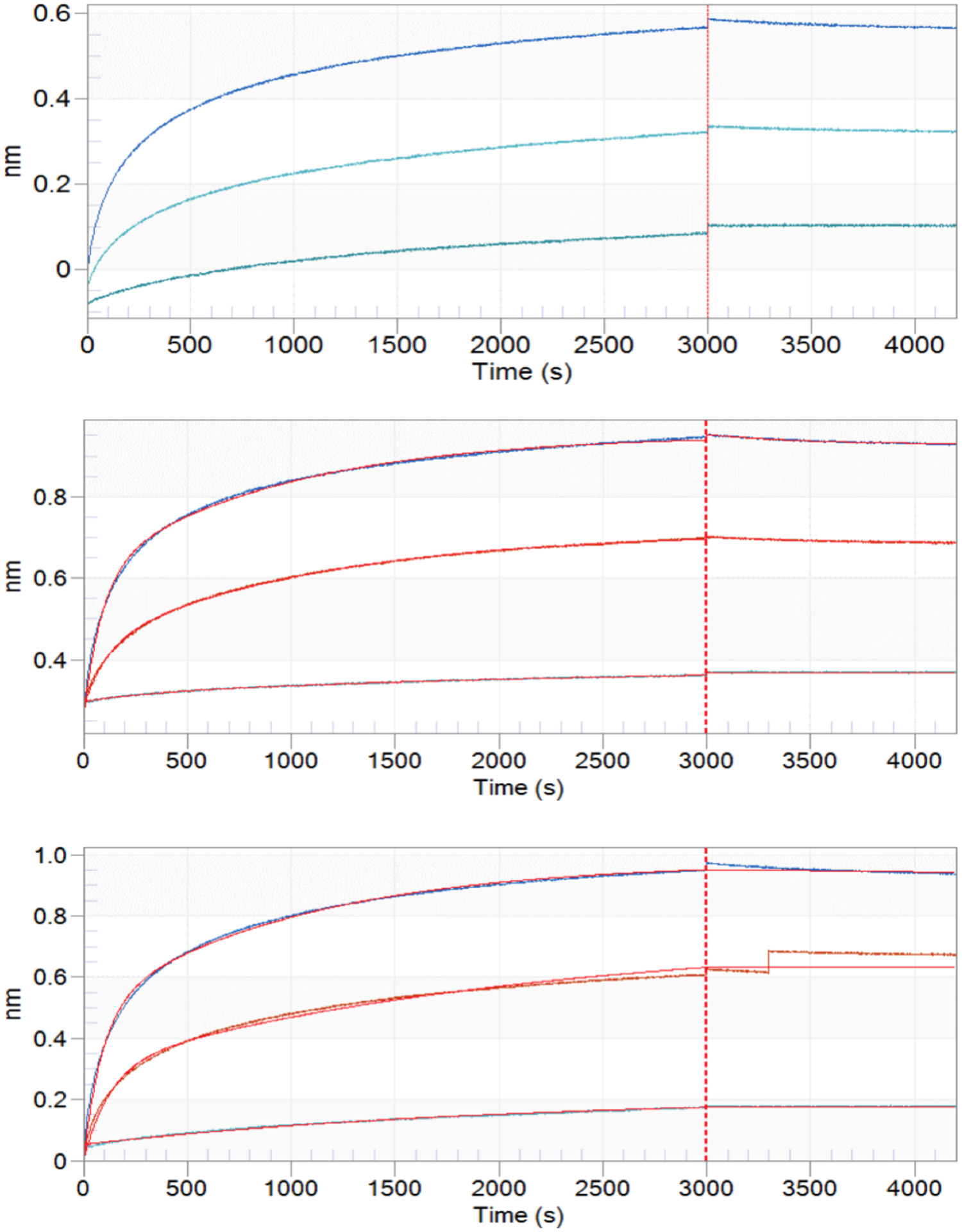
BLI sensorgrams of immobilized (a) HS 5, (b) Gly5SSTN92–119 3 and (c) glyco-polypeptide mimetic 1 binding with integrin αvβ3. Each set of binding curves was generated with integrin concentrations of 104.7 nM, 52.4 nM, and 13.1 nM, from top to bottom, respectively. Fitting curves were generated using 2:1 binding model from Octet Data Analysis 9.0.0.12.
Table 1.
The on-rates (kon) of 1, 3, and 5 with integrin αvβ3.
| Syndecan-1 Mimetic 1 | Gly5SSTN92–119 3 | HS glycan 5 | |
|---|---|---|---|
| kon (1/M) | 5.08 × 104 | 1.98 × 104 | 9.60 × 103 |
As the glyco-polypeptide mimetic 1 can bind with integrin strongly, we next measured its effect on cancer cells. MDA-MB-231 breast carcinoma cells activate the cell-surface integrin αvβ3 through the complex formation of syndecan-1, insulin-like growth factor-1 receptor and integrin to migrate.19 In addition to syndecan-1 mimetic 1 and Gly5SSTN92–119 3, heparin, which binds more tightly with integrin than heparan sulfate, was chosen to test the inhibitory effect on the migration of MDA-MB-231.3, 20 Over a 20-hour time span, heparin and Gly5SSTN92–119 peptide reached the maximal inhibitory effect at 6 μM (Figures 3a,3b). The maximal inhibition from heparin was ~18% reduction in relative migration (Figure 3b). The glycopeptide mimetic 1 at 6 μM achieved the strongest inhibition, >30% reduction in relative migration (Figure 3c).
Figure 3.
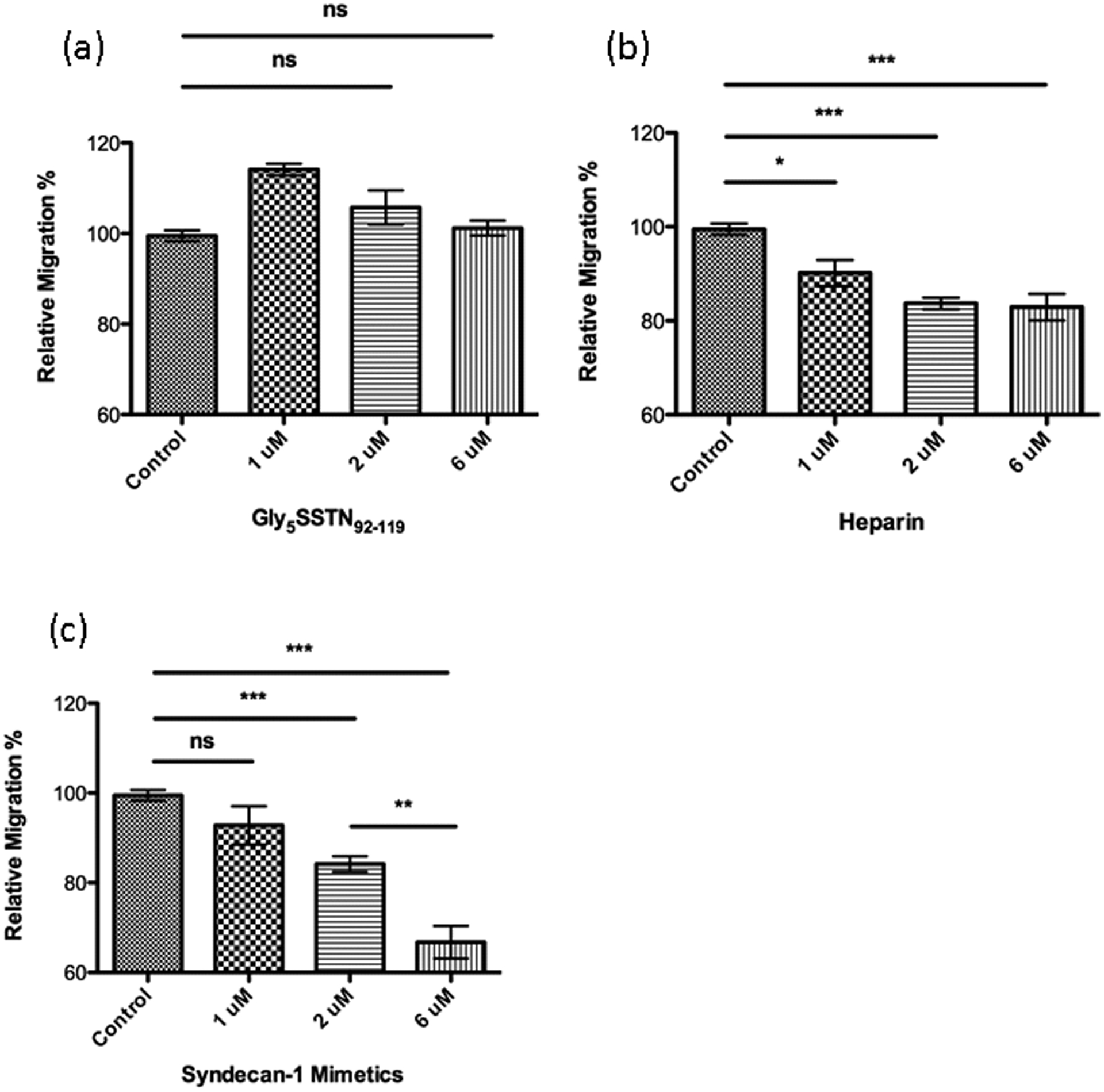
Wound-healing assay results of (a) Gly5SSTN92–119 3, (b) heparin, and (c) syndecan-1 mimetic 1. Each plot is displayed as mean ± S.D. of six biological replicates. The syndecan-1 mimetic 1 was able to more significantly reduce the migratory abilities of the tumor cells. T test was used for statistical analysis. *p<0.05, **p<0.01, ***p<0.001. The p values were determined through a two-tailed unpaired t-test using GraphPad Prism.
It is possible that the enhanced efficacy of the HSPG mimic 1 compared to glycan or peptide alone was due to the ability of the mimetic to simultaneously engage multiple binding sites on the integrin. To gain insights on the integrin αvβ3 binding process, in silico molecular docking simulations were performed, and potential integrin binding sites of Gly5SSTN92–119 peptide 3 and HS oligosaccharide 5 on the integrin were identified (Figures S2 and S3).21, 22 The syndecan-1 mimetics 1 was found to be large enough to bridge the synstatin and HS binding sites (Figure S4), which supports a potential synergy from SSTN92–119 and HS in integrin binding.
In conclusion, with the tremendous structural complexity of HSPG, access to HS glycopeptides with defined structures is highly challenging. Herein, we report an expedient approach to produce a HSPG syndecan-1 mimetic, which contains a 48 amino acid residue polypeptide backbone and the glycan chain with the full structural features of HS in nature including iduronic acid, glucuronic acid, 2-O, 6-O and 3-O sulfations, and N-sulfation. The deployment of CuAAC and sortase A-mediated ligation greatly shortens the synthetic routes and enhances the overall efficiency of the synthesis. The synthetic strategy is convergent, which can offer great potential flexibilities in varying the glyco-polypeptide structures with other peptide or glycan sequences.
The interaction of the glyco-polypeptide mimic 1 with integrin was investigated. Binding study showed that the glycopeptide was able to engage integrin αvβ3 faster than either HS glycan or synstatin peptide alone. Although, for all three ligands, dissociations are slow, the higher on-rate of HSPG mimetic suggested a potential cooperation of HS oligosaccharide and synstatin in integrin binding. Furthermore, the glycopeptide was more potent in inhibiting the migration of triple negative breast cancer cell MDA-MB-231, opening up the door to investigate the cellular functions of HSPG with structurally well-defined mimetics.
Supplementary Material
Figure 1.
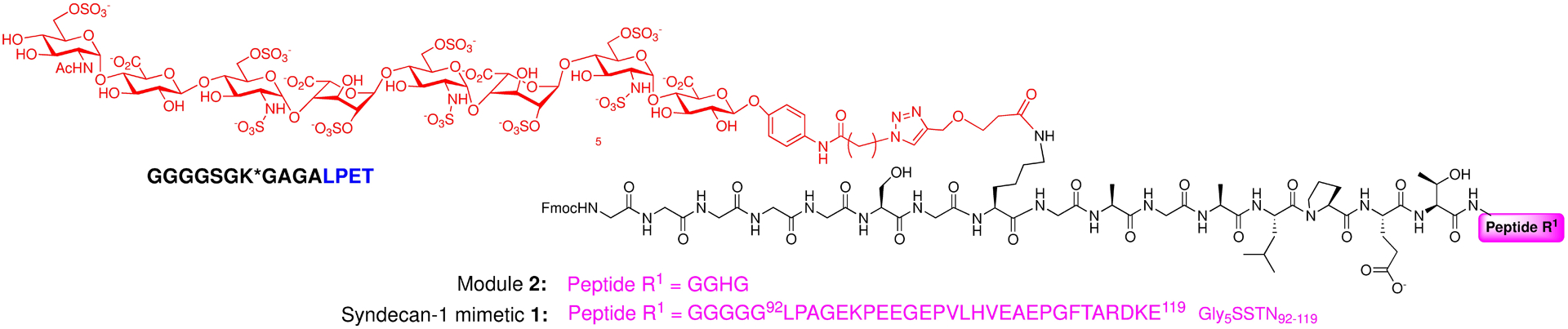
Structures of the HSPG syndecan-1 mimetics 1 and 2.
Acknowledgments
We are grateful to the National Institute of General Medical Sciences, National Institutes of Health (R01GM072667) and Michigan State University for financial support of our work.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Sugahara K and Kitagawa H, IUBMB Life, 2002, 54, 163. [DOI] [PubMed] [Google Scholar]
- 2.Sarrazin S, Lamanna WC and Esko JD, Cold Spring Harb. Perspect. Biol, 2011, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peysselon F and Ricard-Blum S, Matrix Biol., 2014, 35, 73. [DOI] [PubMed] [Google Scholar]
- 4.Ori A, Wilkinson MC and Fernig DG, J. Biol. Chem, 2011, 286, 19892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harburger DS and Calderwood DA, J. Cell Sci, 2009, 122, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkpatrick CA, Knox SM, Staatz WD, Fox B, Lercher DM and Selleck SB, Dev. Biol, 2006, 300, 570. [DOI] [PubMed] [Google Scholar]
- 7.Capurro MI, Xu P, Shi W, Li F, Jia A and Filmus J, Dev. Cell, 2008, 14, 700. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, Yoshida K, Yin Z, Dai H, Kavunja H, El-Dakdouki MH, Sungsuwan S, Dulaney SB and Huang X, Angew. Chem. Int. Ed, 2012, 51, 10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida K, Yang B, Yang W, Zhang Z, Zhang J and Huang X, Angew. Chem. Int. Ed, 2014, 53, 9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, Eken Y, Zhang J, Cole LE, Ramadan S, Xu Y, Zhang Z, Liu J, Wilson AK and Huang X, Chem. Sci, 2020, 11, 6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beauvais DM, Ell BJ, McWhorter AR and Rapraeger AC, J. Exp. Med, 2009, 206, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Cai C, Chandarajoti K, Hsieh P, Li L, Pham TQ, Sparkenbaugh EM, Sheng J, Key NS, Pawlinski R, Harris EN, Linhardt RJ and Liu J, Nat. Chem. Biol, 2014, 10, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams DJ, Atkins D, Cooper AI, Furzeland S, Trewin A and Young I, Biomacromolecules, 2008, 9, 2997. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Guan C, Tan X, Chen C, Guo Q and Li Y, Org. Biomol. Chem, 2015, 13, 1500. [DOI] [PubMed] [Google Scholar]
- 15.Hendrickx APA, Budzik JM, Oh S and Schneewind O, Nat. Rev, 2011, 9, 166. [DOI] [PubMed] [Google Scholar]
- 16.David Row R, Roark TJ, Philip MC, Perkins LL and Antos JM, Chem. Commun, 2015, 51, 12548. [DOI] [PubMed] [Google Scholar]
- 17.Mao H, Hart SA, Schink A and Pollok BA, J. Am. Chem. Soc, 2004, 126, 2670. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Bachelier R, Treilleux I, Pujuguet P, Peyruchaud O, Baron R, Cle P and Cle P, Cancer. Res, 2007, 67, 5821. [DOI] [PubMed] [Google Scholar]
- 19.Beauvais DM and Rapraeger AC, J. Cell Sci, 2010, 123, 3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faye C, Moreau C, Chautard E, Jetne R, Fukai N, Ruggiero F, Humphries MJ, Olsen BR and Ricard-Blum S, J. Biol. Chem, 2009, 284, 22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saladin A, Rey J, Thevenet P, Zacharias M, Moroy G and Tuffery P, Nucleic Acids Res., 2014, 42, W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mottarella SE, Beglov D, Beglova N, Nugent MA, Kozakov D and Vajda S, J. Chem. Inf. Model, 2014, 54, 2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


