Abstract
In multifocal intrahepatic cholangiocarcinoma (IHC), intrahepatic metastases (IM) represent a contraindication to surgical resection, whereas satellite nodules (SN) do not. However, no consensus criteria exist to distinguish IM from SN. The purpose of this study was to determine genetic alterations and clonal relationships in surgically resected multifocal IHC. Next-generation sequencing of 34 spatially separated IHC tumors was performed using a targeted panel of 201 cancer-associated genes. Proposed definitions in the literature were applied of SN located in the same liver segment and ≤2 cm from the primary tumor; and IM located in a different liver segment and/or >2 cm from the primary tumor. Somatic point mutations concordant across tumors from individual patients included BAP1, SMARCA4 and IDH1. Small insertions and deletions (indels) present at the same genome positions among all tumors from individuals included indels in DNA repair genes, CHEK1, ERCC5, ATR and MSH6. Copy number alterations were also similar between all tumors in each patient. In this cohort of multifocal IHC, genomic profiles were concordant across all tumors in each patient, suggesting a common progenitor cell origin, regardless of the location of tumors in the liver. The decision to perform surgery should not be based upon a perceived distinction between IM and SN.
Multiple intrahepatic tumors in intrahepatic cholangiocarcinoma are classified as satellite nodules or metastases, based on tumor location in the liver. In this study, genomic profiles were concordant across all tumors in individual patients, indicative of monoclonal metastatic seeding.
Introduction
Intrahepatic cholangiocarcinoma (IHC) is the second most common primary liver cancer, following hepatocellular carcinoma, and has a rising incidence globally (1). Surgery remains the only potentially curative treatment, with 5-year overall survival rates between 20 and 35% (2). However, two-thirds of patients suffer postoperative disease recurrence, most commonly in the remnant liver. Approximately 30% of patients undergo resection of multiple anatomically separate tumors and have significantly worse survival compared with patients undergoing resection of solitary tumors (1,3). In multifocal IHC, individual tumors are thought to represent disseminated metastases or regional extensions of the primary tumor. Peritumoral satellite nodules (SN) are considered potentially resectable, whereas intrahepatic metastases (IM) represent a contraindication to surgical resection (4,5). However, radiologic or pathologic criteria that distinguish between regional SN and disseminated IM are lacking.
Previous reports have proposed classifying SN and IM based on tumor location and distribution in the liver (6–9). To our knowledge, no prior study has examined molecular differences between SN and IM. To address this gap, we aimed to determine whether IM and SN are genomically distinct entities to inform clinical decision-making. Here, we report that somatic point mutations, copy number alterations (CNAs) and small insertions and deletions (indels) are concordant across all tumors in individual patients, indicative of a shared origin from the same initiating cell. These findings suggest that multiple intrahepatic tumors in IHC represent metastases, regardless of their location in the liver.
Materials and methods
Patient samples
Thirty-four tumor samples and nine matched control samples of adjacent non-tumoral liver were collected from nine patients who underwent resection of multifocal IHC at the University of Texas MD Anderson Cancer Center from 2010 to 2017. Hematoxylin and eosin stained slides were reviewed by a gastrointestinal pathologist to determine the proportion of malignant cells relative to necrosis, fibrosis and inflammatory infiltrate. Five µm thick sections were prepared from formalin-fixed, paraffin-embedded tissue. Slides were deparaffinized, and tissue was scraped and placed in Eppendorf tubes. Genomic DNA was extracted from all samples as described previously (10).
Tumor number and size were recorded from pathology reports. The collection and analysis of patient samples were approved by the Institutional Review Board.
Intrahepatic metastases versus satellite nodules
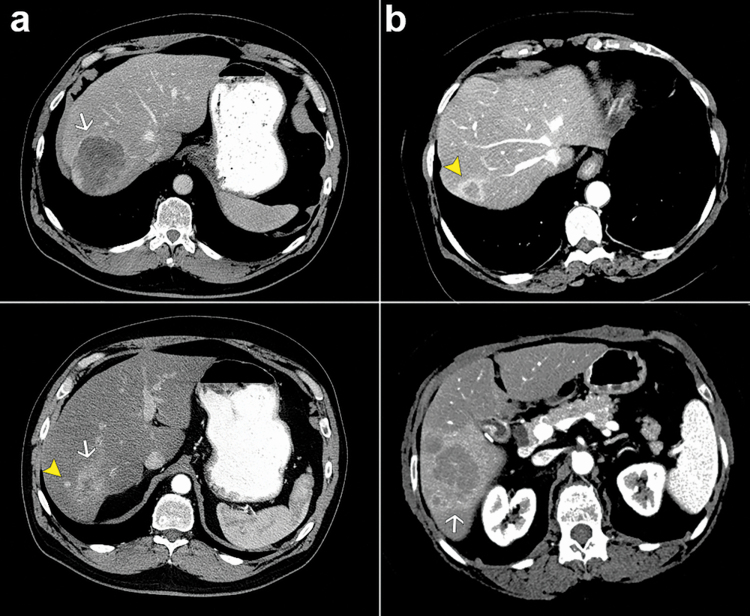
Preoperative radiologic imaging and surgical pathology reports were used to classify SN and IM, based on a proposed definition in the literature (6,11). SN were defined as smaller lesions located in the same liver segment, ≤2 cm from the larger primary tumor (Figure 1). IM were defined as lesions in a different liver segment and/or >2 cm from the primary tumor.
Figure 1.
Representative preoperative computed tomography images showing main mass (white arrows) and smaller tumors (yellow arrowheads). (a) SN, patient 1. (b) IM, patient 9.
Targeted DNA sequencing
T200 is a deep targeted sequencing platform that comprises 4874 exons encoding 938 607 bases, with median coverage of 300 reads in 201 cancer-related genes (10). This platform was chosen because it has been optimized for formalin-fixed, paraffin-embedded specimens and reliably detects low-frequency mutations and CNAs with a false discovery rate of 1%. The 201 genes were selected for their biological relevance in cancer based on mutational data in the Catalogue of Somatic Mutations in Cancer and The Cancer Genome Atlas.
Sequence alignment and variant calling
Sequencing data were mapped to the human reference sequence GRCh37 (hg19) using the Burrows-Wheeler Aligner on default settings. Aligned reads were further processed following the Genome Analysis Toolkit Best Practices of duplicate removal, indels realignment and base recalibration. Platypus was used to call SNVs and indels. The MuTect algorithm was used to identify somatic mutations by comparing tumors with matched non-tumoral liver samples. Indels were identified using Pindel (12). In addition to build-in filters, the following filtering criteria were applied: (i) variant allele frequency in tumor DNA >5%; (ii) sequence depth >10; (iii) presence of variant on both strands; (iv) variants rare in the population, with allele frequency <0.01 in the Exome Aggregation Consortium Data and (v) removal of variants in positions listed in the dbSNP 138 database. Substitutions and indels were annotated using Annotate Variation software (ANNOVAR) based on the University of Santa Cruz (USCS) Known Genes (http://genome.ucsc.edu).
Jaccard Index
Similarities in SNVs among samples from individual patients and between samples in different patients were calculated with the Jaccard Index using the R package set (https://cran.rproject.org/web/packages/sets). All SNVs in 201 cancer-related genes were selected to estimate similarities.
Copy number alterations
Tumor and adjacent non-tumoral DNA was used to obtain tumor-specific, somatic CNAs from T200 data using an in-house R package. The log2 ratios of tumor versus normal reads were calculated for each tumor region after adjusting for total mapped reads in that tumor region, then segmented by circular binary segmentation algorithm. The Genomic Identification of Significant Targets in Cancer (GISTIC) 2.0 algorithm was applied to the segmental copy number profiles to identify significant amplifications and deletions (13). The standard q value cutoff of 0.25 was used to define significant regions of CNAs. For two-dimensional visualization of the data, t-stochastic neighbor embedding (t-SNE) was applied using the R package, Rtsne (14).
Results
Patient characteristics
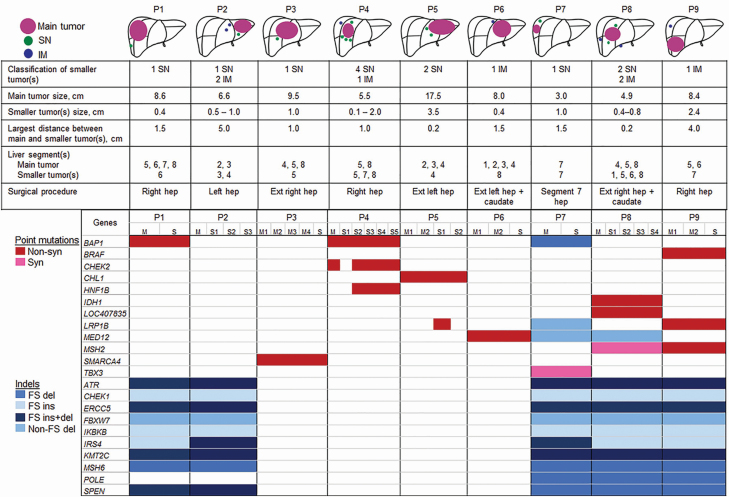
A total of 34 tumor samples from 9 patients were analyzed using T200 targeted sequencing (Figure 2). In some patients, multiple samples were obtained from the main tumor and/or smaller tumor (S). All patients underwent hepatectomy with curative intent. Six patients received perioperative chemotherapy. After median follow-up of 30 months (range, 10–76 months), seven patients (78%) suffered disease recurrence and five patients (56%) died of disease. The first site of disease recurrence involved the liver in six of seven patients.
Figure 2.
Schematic representations, clinicopathologic factors and mutation heat map of nine patients undergoing resection of multifocal IHC. Mutated genes comprise rows, and columns represent patient samples. The presence and type of mutations are indicated by different colors, as shown, and the absence of mutation indicated by white. del, deletion; ext, extended; FS, frameshift; hep, hepatectomy; indel, small insertion and deletion; ins, insertion; M, main tumor; P, patient; S, smaller tumor(s); syn, synonymous.
Smaller tumors (S) were located in the same segment as the dominant mass in four patients and different segments in five patients (Figure 2). Median distance between the primary and smaller tumors was 1.5 cm (range, 0.2–5 cm). One patient (patient 8) had bilateral tumors in both the right and left liver.
Exonic somatic point mutations and indels
Eight of the nine patients had exonic somatic point mutations in cancer-associated genes shared between the primary and smaller tumors (Figure 2). Non-synonymous mutations identified in all samples from individual patients were BAP1 from patients 1 and 4, SMARCA4 from patient 3, CHL1 from patient 5, MED12 from patient 6, IDH1 and LOC407835 from patient 8 and BRAF, MSH2 and LRP1B from patient 9. Synonymous mutations common for all samples in individual patients were TBX3 in patient 7 and MSH2 in patient 8.
Five of the nine patients had exonic indels at the same genome positions identified in all samples (Figure 2). Shared indels in the five patients were ATR, CHEK1, ERCC5, FBXW7, IKBKB, IRS4, KMT2C, MSH6 and SPEN. Three patients had indels in POLE concordant across all samples.
The Jaccard Index, which measures similarity and diversity of sample sets, showed that mutational profiles were similar among multiple samples from the same individual and heterogeneous between patients (Figure 3a).
Figure 3.
Concordance of single-nucleotide variants and CNAs across tumors from individual patients. (a) Jaccard similarity plot based on single-nucleotide variants showing that tumors within the same patient are similar, whereas tumors between patients are distinct. (b) Data from CNAs from all samples in each patient are plotted with t-SNE, a graph layout algorithm that gives each data point a location in a multidimensional map.
Copy number alterations
To investigate the clonal relationship between lesions, somatic CNA analysis was performed. Most CNAs were present in all samples sequenced from the same patient (Figure 4). Dimension reduction using t-distributed stochastic neighboring embedding (t-SNE) showed CNA profiles of samples from individual patients clustered together, indicating clonal similarity of multiple tumors from the same patient (Figure 3b).
Figure 4.
Heat map of CNAs for multiple samples in each patient with multifocal IHC. Red and blue indicate copy number amplifications and deletions, respectively. Chromosome boundaries and clinical data are annotated. M, main tumor; OS, overall survival; P, patient; PFS, progression-free survival; Preop Ctx, preoperative chemotherapy; S, smaller tumor.
Gene-level copy number status was determined via the GISTIC2.0 algorithm (Figure 5). Recurrent CNAs included amplifications in chromosomes 5p15.33 (patients 1, 2 and 8) and 11q13.2 (patients 2, 5 and 6). Recurrent deletions included chromosomes 9p21.3 (patients 4, 5 and 6) and 18q23 (patients 3, 4 and 7).
Figure 5.
GISTIC2.0 analysis of CNAs for multiple samples in each patient. Regions of amplifications or deletions are designated with their chromosomal cytogenetic band.
Discussion
In patients with multifocal IHC, a distinction is made between potentially resectable SN and unresectable IM. However, criteria that distinguish between SN and IM are lacking. In this study, molecular alterations were concordant across all intrahepatic tumors in individual patients. These data suggest that multiple tumors in IHC are derived from the same initiating cell and represent monoclonal metastatic seeding, regardless of tumor location or distribution in the liver. Our findings challenge the notion that SN and IM are different by providing preliminary evidence that all multifocal disease represents metastatic dissemination. Thus, the decision to perform surgery in multifocal IHC should not be based upon a perceived biologic distinction between IM and SN.
Clinically actionable mutations in driver genes such as IDH1 and FGFR2 have been identified in up to 40% of patients with IHC (15,16). In this study, somatic point mutation analysis revealed that eight of the nine patients’ tumors harbored driver mutations concordant across all samples in each patient. Shared non-synonymous mutations included BRAF and epigenetic regulators, SMARCA4, MED12, IDH1 and BAP1. Analogous to the results for somatic point mutations, CNA profiles were concordant between tumors in each patient. Deletions in chromosome 9p21.3, a locus harboring CDKN2A and CDKN2B tumor suppressor genes, were identified in all samples from three of the nine patients. Recurrent CDKN2A/B deletions have been reported in 8–18% of patients with IHC and are associated with a poor prognosis (17–20). Three patients had amplifications in 11q13.2, which includes CCND1, FGF family members (FGF3, 4 and 19) and ORAOV1. Sia et al. reported amplification of 11q13.2 in IHC tumors characterized by activation of RAS and mitogen-activated protein kinase signaling pathways, which was associated with worse survival (18).
An unexpected finding of this study was concordant mutations in DNA repair genes in five of the nine patients. Indels present at the same genome positions in all samples from five patients included CHEK1, ERCC5, ATR and MSH6. In addition, three patients had shared indels in POLE and one patient had a non-synonymous point mutation in MSH2. Alterations in DNA repair genes have previously been identified in 12.5% of patients with IHC (17). This has important therapeutic implications, since tumors harboring mutations affecting DNA damage response and genomic instability can potentially be targeted with agents such as PARP inhibitors (21). Furthermore, frameshift indels, shared among all samples in five of our patients, have been shown to trigger neoantigenic peptides highly distinct from self, leading to increased tumor immunogenicity and higher likelihood of response to immunotherapy (22). Clinical trials investigating immune checkpoint inhibitors in cholangiocarcinoma are ongoing (21).
Limitations of this study include small sample size and targeted sequencing of 201 genes, which underestimates the total burden of molecular alterations. However, the main finding of similar genomic profiles of samples from individual patients was consistent for all patients in the study. The sample size was insufficient to evaluate intratumoral heterogeneity of IHC. Another major limitation is the lack of analysis of FGFR2 fusions or rearrangements, which are reportedly present in 10–20% of patients with IHC and important targets for therapy (23–25). Furthermore, epigenomic and non-coding regulatory factors, which may contribute to differences between SN and IM, were not examined. This study was limited to patients who had resected, synchronous tumors and molecular alterations in patients with more advanced, unresectable or metachronous IHC were not analyzed. Future studies with more comprehensive molecular analysis and larger cohorts that include patients with unresectable and metachronous disease will be important to further elucidate the clinical impact of molecular alterations in multifocal IHC.
In conclusion, in this study of multifocal IHC, SNVs, indels and CNAs were concordant across spatially separated tumors within an individual and divergent between tumors from different individuals. These findings are clinically important because they challenge the current practice of differentiating between potentially resectable SN and unresectable IM. Future investigations are needed for adjuvant therapy strategies to eradicate intrahepatic micrometastases in multifocal IHC.
Funding
This work was supported by the University of Texas MD Anderson Cancer Center Research Start-up Funds and the National Institutes of Health/National Cancer Institute (P30-CA016672 and R01CA237327).
Conflict of Interest Statement: None declared.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Glossary
Abbreviations
- CNA
copy number alteration
- IHC
intrahepatic cholangiocarcinoma
- IM
intrahepatic metastases
- SN
satellite nodules
References
- 1. Bridgewater, J., et al. (2014) Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol., 60, 1268–1289. [DOI] [PubMed] [Google Scholar]
- 2. Moeini, A., et al. (2016) Molecular pathogenesis and targeted therapies for intrahepatic cholangiocarcinoma. Clin. Cancer Res., 22, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Igami, T., et al. (2011) Staging of peripheral-type intrahepatic cholangiocarcinoma: appraisal of the new TNM classification and its modifications. World J. Surg., 35, 2501–2509. [DOI] [PubMed] [Google Scholar]
- 4. Weber, S.M., et al. (2015) Intrahepatic cholangiocarcinoma: expert consensus statement. HPB (Oxford), 17, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blumgart, L.H., et al. (2007) Surgery of the Liver, Biliary Tract, and Pancreas. Saunders Elsevier, Philadelphia, PA. [Google Scholar]
- 6. Baheti, A.D., et al. (2014) Correlation of CT patterns of primary intrahepatic cholangiocarcinoma at the time of presentation with the metastatic spread and clinical outcomes: retrospective study of 92 patients. Abdom. Imaging, 39, 1193–1201. [DOI] [PubMed] [Google Scholar]
- 7. Conci, S., et al. (2018) Patterns of distribution of hepatic nodules (single, satellites or multifocal) in intrahepatic cholangiocarcinoma: prognostic impact after surgery. Ann. Surg. Oncol., 25, 3719–3727. [DOI] [PubMed] [Google Scholar]
- 8. Aherne, E.A., et al. (2018) Intrahepatic cholangiocarcinoma: can imaging phenotypes predict survival and tumor genetics? Abdom. Radiol. (NY), 43, 2665–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Addeo, P., et al. (2019) Prognostic impact of tumor multinodularity in intrahepatic cholangiocarcinoma. J. Gastrointest. Surg., 23, 1801–1809. [DOI] [PubMed] [Google Scholar]
- 10. Chen, K., et al. (2015) Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin. Chem., 61, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tabrizian, P., et al. (2015) Outcomes following resection of intrahepatic cholangiocarcinoma. HPB (Oxford), 17, 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye, K., et al. (2009) Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics, 25, 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mermel, C.H., et al. (2011) GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol., 12, R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Der Maaten, L., et al. (2008) Visualizing data using t-SNE. J. Mach. Learn. Res., 9, 2579–2605. [Google Scholar]
- 15. Chun, Y.S., et al. (2017) Systemic and adjuvant therapies for intrahepatic cholangiocarcinoma. Cancer Control, 24, 1073274817729241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valle, J.W., et al. (2017) New horizons for precision medicine in biliary tract cancers. Cancer Discov., 7, 943–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Javle, M., et al. (2016) Biliary cancer: utility of next-generation sequencing for clinical management. Cancer, 122, 3838–3847. [DOI] [PubMed] [Google Scholar]
- 18. Sia, D., et al. (2013) Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene, 32, 4861–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farshidfar, F., et al. ; Cancer Genome Atlas Network. (2017) Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep., 19, 2878–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lowery, M.A., et al. (2018) Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin. Cancer Res., 24, 4154–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bogenberger, J.M., et al. (2018) Emerging role of precision medicine in biliary tract cancers. NPJ Precis. Oncol., 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Turajlic, S., et al. (2017) Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol., 18, 1009–1021. [DOI] [PubMed] [Google Scholar]
- 23. Goyal, L., et al. (2017) Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov., 7, 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Javle, M., et al. (2018) Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J. Clin. Oncol., 36, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abou-Alfa, G.K., et al. (2020) Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol., 21, 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.