Abstract
Background:
Cryptosporidium is a neglected zoonotic disease, but with the expansion of the human community into the animal environment, its incidence is increasing. Animals such as rats and pigs can act as intermediate hosts and transmit Cryptosporidium to humans due to their proximity. Transmission occurs due to the ability of Cryptosporidium to survive in any new host. The research aimed to identify and describe the transmission of Cryptosporidium from animals to humans.
Materials and Methods:
This research was a cross sectional study and samples were collected from 84 rats caught in residential areas, 205 pigs, and 438 humans in West Lombok. Fecal samples were examined using polymerase chain reaction (PCR) and sequencing to isolate the presence of Cryptosporidium, and identify the genetic similarity of the parasites found in rats and pigs with those that infect humans.
Results:
The PCR results found Cryptosporidium parvum in 4.76% (4/84) in rats; 6.34% 13/205) in pigs; and 0.91% (4/438) in humans. The sequencing results showed genetic kinship of C. parvum in rats, pigs, and humans. Based on sequence confirmation from Gene Banks and edited using ClustalW with MEGA X software, there are genetic similarities between Cryptosporidium isolates from West Lombok and C. suis isolates of cattle from Uganda and C. suis isolates of pigs from Slovakia.
Conclusion:
There are genetic similarities of Cryptosporidium in animals and humans, requiring that the Public Health programs in those contaminated areas must receive priority attention to prevent further transmission of these potentially fatal parasites.
Keywords: Zoonotic parasite, Cryptosporidium, Rats, Pigs and Humans
Introduction
Zoonotic diseases are increasing especially in developing countries and are becoming neglected diseases. Survey results from 1,407 pathogens in humans showed 58% of emerging infectious diseases and 75% of emerging infectious diseases are zoonotic diseases. An estimated 75 percent of new infectious diseases are zoonotic in origin, directly resulting from human and animal interactions (Woolhouse and Sequeria, 2005; Austin, 2021). Factors contributing to the increase of these diseases are as a result of an increase in population and human activities that have changed the forest environment to the human environment. Natural habitats that have changed their functions as agricultural land, plantations, and shelter, cause humans to live side by side with animals. Rats are reservoirs of infectious diseases because of their habitat and habit of looking for food in dirty places so that the diseases they carry can harm humans. Rat-based diseases began to increase by changes in habitat for animals and their closer proximity to the human environment (Thiermann, 2004; Woolhouse and Sequeria, 2005; Morand, 2015; Sun et al., 2018).
Rats can spread and transmit various infectious diseases to humans and other animals. Rats can carry 61 types of infectious diseases, including 20 types of viruses, 19 types of bacteria, and 22 types of parasites, including Cryptosporidium spp. The spread of parasitic zoonotic diseases from rats in the human environment needs to be carefully investigated for the source of parasitic zoonotic transmission. The proximity of rats to the human environment can be a risk factor for transmission of parasites from rats to humans and animals (Perec-Matysiak et al., 2015; Zahedi et al., 2016; Azzam KM, 2017; Krijger, 2020; ).
The presence of poorly organized pig farms and cattle farms has led to the rapid spread of zoonotic parasites. Pollution of soil, water and air around human housing by zoonotic parasites is a result of unsafe animal rearing. Research reports show that farming in a residential environment increases the incidence of diseases in animals and has the potential to spread zoonotic diseases (Mosites et al., 2016). Baqer et al. (2018) showed contamination by Cryptosporidium oocysts in a river adjacent to a cattle farm in Baghdad.
Cryptosporidium parvum is a zoonotic parasite that can cause gastrointestinal disorders with symptoms such as diarrhea. The incidence of diarrhea can increase morbidity and mortality rates especially in children and is a cause of death of four million lives in developing countries each year (Badry et al., 2014; Verkerke et al., 2014; Dupont, 2016; Yee et al., 2018). Prolonged infection results in dehydration and weight loss. This situation can be severe in children or people with low immunity. Pathological conditions that are caused by these parasitic infections include epithelium damage in the from of villious atrophy, mitochondrial changes and increased lysosomal activity in infected cells (Ridley, 2012; Bogitsh et al., 2013).
Cryptosporidium in humans is a recent case that often occurs in areas with poor hygiene, usually involving reports of contact between humans and rats causing the migration of Cryptosporidium to humans. Humans are often infected with C. parvum and C. hominis; cattle with C. parvum, C. bovis, C. ryanae, and C. andersoni; while sheep and goats are infected with C. parvum, C. ubiquitum, and C. xiaoi. Most of the species in these animals are also found in humans. So far, more than 20 species of Cryptosporidium have been identified in humans. Sequencing methods using the small subunit rRNA gene (SSU rRNA) showed the presence of C. parvum and C. muris in rats in China and detected C. parvum isolate 11dA15G1 identified using the gp60 gene which can infect humans (Zhao et al., 2015; Beser et al., 2020).
The research was conducted on the island of Lombok, where the pigs are often free to find their food around the house or are given leftovers. The poor pig farming practices result in the transmission of parasites between animals and humans. Zoonotic transmissions on the island of Lombok have never been reported. This research concentrated on the transmission of Cryptosporidium zoonotic diseases from animals to humans in West Lombok, Indonesia.
Materials and Methods
Study design.
The study design was a comparative cross-sectional research. The duration of the study was six months (January, 2019 to June, 2019).
Samples
The location of fecal sampling is based on the area that has pig farms. Residents who previously volunteered to fill out the informed consent were the sources of the research samples. Stool samples were collected from 84 rats caught and sacrificed from residential areas, 205 pigs, and 438 humans. Samples were taken in West Lombok, Indonesia at 191 locations. The freshness of the stool sample was maintained by the addition of a 5% potassium bichromate preservative.
Ethical Approval
This study was approved by the Medical and Health Research Ethics Committee of the Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada-Dr. Sardjito General Hospital, with approval number: KE/FK/1222/EC/2018, dated 21 November 2018.
Informed Consent
Informed consent was obtained from all individual participants included in this study.
Data collection
Interviews method and laboratory examinations of Cryptosporidium DNA found in humans, pigs, and mice by PCR and sequencing methods (Munshi, 2012) were employed to gather data for this study.
Laboratory Methods
DNA Extraction
Isolation of DNA was done using the procedures from QIAamp, Fast DNA Stool Mini Kit (Qiagen, German). Stool sample of 180-220 mg was inserted in a 2 ml tube with 1 ml InhibitEX Buffer, and vortexed until the sample was homogeneous. Samples were lysed using a mini Beadbeater for 5 minutes, and continued with the “freeze-Thaw” process, which was incubated at -80OC for 5 minutes and incubated 60OC in a water bath for 5 minutes (four times). To separate pellets, the sample was centrifuged for 1 minute at 10,000 rpm. 600 µL supernatant was then placed in a pipette, and 25 µL Protein K and 600 µL AL buffer was added, then vortexed for 15 seconds, and incubated 10 minutes at 70OC. Next, 600 µL of 96-100% ethanol was added to the lysate and vortexed. Lysate was then put in a spin column, and centrifuged at 10,000 rpm for 1 minute, then the filtrate was removed. Next, 500 µL AW1 buffer was added into the spin column with a new collection tube, centrifuged at 10,000 rpm for 1 minute, and the filtrate removed. Next, 500 µL Buffer AW 2 was added in a new collection tube, and centrifuged at 10,000 rpm for 3 minutes. Then, the spin column was removed from the collection tube then put in a new collection tube, and centrifuged at 10,000 rpm for 3 minutes. Next, the spin column was transferred to the 1.5 ml tube, and 100 - 200 µL buffer ATE was added to the spin column and incubated for 1 minute, then centrifuged at 10,000 rpm for 1 minute. Finally, the spin column was discarded, and the tube containing DNA was stored at -20OC.
PCR Amplification and Detection
Polymerase chain reaction (PCR) used the Bioline mix, with 1 µL DNA template, 7 µL ultrapure water and 10 µL master mix, with 1 µL primers. The primers used were: Cryptosporidium parvum gene 18S rRNA, F: 5’-TAAACGGTAGGGTATTGGCCT-3’; R: 5’-CAGACTTGCCCTCCAATTGATA-3 ’. The PCR conditions used were 35 cycles, with an initial activation temperature of 95OC for 5 minutes, denaturation temperature of 95OC for 30 seconds, annealing temperature of 59OC for 45 seconds, an extension temperature of 72OC for 3 minutes, and a final extension temperature of 72OC for 10 minutes. The final results were examined using electrophoresis at 2% agarose, for presence of Cryptosporidium parvum at 240bp (Zebardast et al., 2016).
Sequencing
Sequencing used the Applied Biosystem 3500 Genetic Analyzer 2500 tool with the Bigdye Terminator kit. DNA Cryptosporidium sequences from rats, pigs and human isolates were analyzed using the online BLAST program (NCBI), while sequences from Gene Banks that have genetic similarities with Cryptosporidium isolates from Lombok isolates were edited using ClustalW with MEGA X software. Phylogenetic trees were arranged based on neighbor-joining (Kumar et al., 2018).
Results
PCR results showed Cryptosporidium infection in rats 4.76% (4/84), pigs 6.34% (13/205) and humans 0.91% (4/438). Electrophoresis results on agarose 2% are shown in Figure 1.
Figure1.
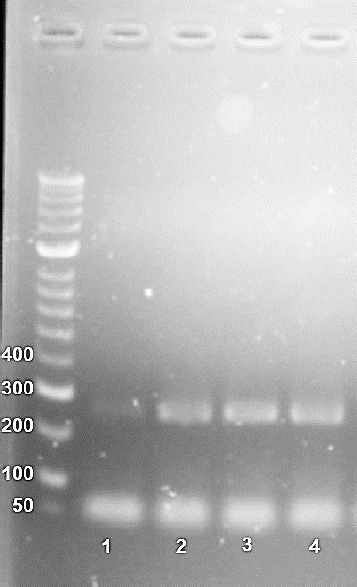
PCR results of Cryptosporidium parvum isolates from West Lombok, electrophoresis results in 2% agarose with FloroSafe stain, and HyperLadder Bioline, using 18S rRNA gene showed Cryptosporidium parvum 240 bp, No.1. Negative; No.2. Rats; No.3. Pig; and No.4. Human.
Cryptosporidium parvum from isolates of rats, pigs and humans in Lombok was spread in several districts, and the distribution is shown in Figure 2.
Figure 2.
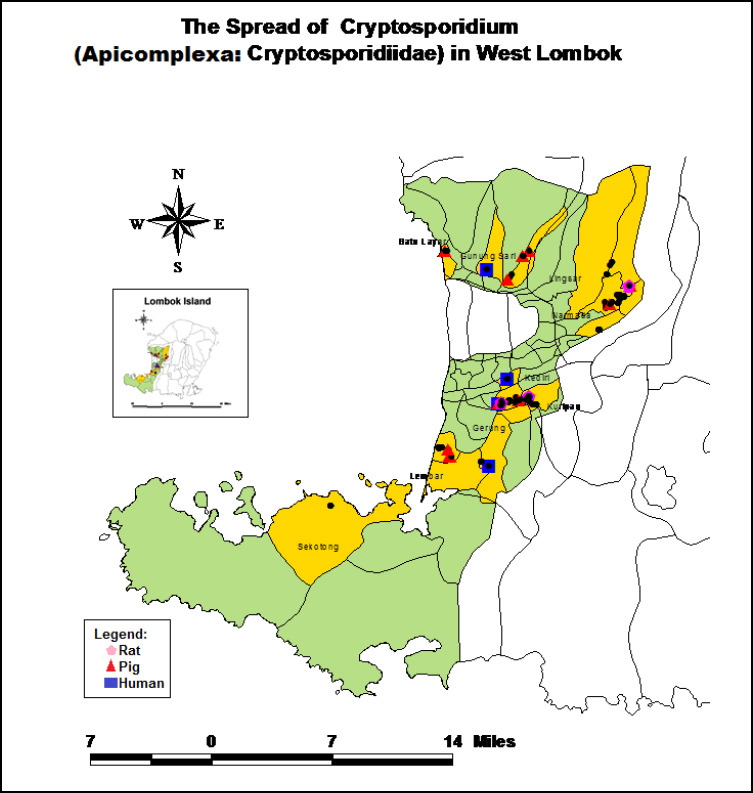
Cryptosporidium parvum from rats, pigs, and humans in West Lombok, C. parvum are found in the districts of Narmada, Kediri, and Gerung. C. parvum also was identified in Gunungsari and Lembar. The infection rate in rats was 4.76% (4/84), in pigs 6.34% (13/205) and in humans 0.91% (4/438).
Genetic similarity of Cryptosporidium parvum sequences of rat, pig and human isolates with DNA gene sequences from the NCBI Bank using the BLAST program online on the website (Http: //www.ncbi.nlm. nih.gov). DNA sequences of Cryptosporidium parvum isolates of rats, pigs, and humans based on the 18s rRNA gene are as follows:
Cryptosporidium in Rats
GTAGGGTATTGGCCTACCGTGGCAGTGACGGGTAACGGGGAATTAGGGTTCGATTCCGGAGAGGGAGCCTGAGAAACGGCTACCACATCTAAGGAAGGCAGCAGGCGCGCAAATTACCCAATGAAAACAGTTTCGAGGTAGTGACGAGAAATAACAATACAGGGCATTTTTTGCTCTGTAATTGGAATGATGGGAATGTAAAACCCTTTCCAGAGTATCAATTGGAGGGCAAGTC
Cryptosporidium in Pigs
GGTAGGGTATTGGCCTACCGTGGCAGTGACGGGTAACGGGGAATTAGGGTTCGATTCCGGAGAGGGAGCCTGAGAAACGGCTACCACATCTAAGGAAGGCAGCAGGCGCGCAAATTACCCAATGAAAACAGTTTCGAGGTAGTGACGAGAAATAACAATACAGGGCATTTTTTGCTCTGTAATGGAAATGATGGGTATAAGAAGGCCTTTCCAGAGTATCAATTGGAGGGCAAGTCTGGACTAG
Cryptosporidium in Humans
TTGGCCTACCGTGGCATTGACGGGTAACGGGGAATTAGGGTTTGATTCCGGAGAGGGAGCCTGAGAAACGGCTACCACATCTAAGGAAGGCAGCAGGCGCGCAAATTACGCAATATCAACGAAAGGGCGATAGGATCTAAACGGTCGGGTGTTAACCTTCTTAACAAGTATCAATTGGAGGGCAAGTCTGGAAC
BLAST of the DNA sequences of Cryptosporidium parvum isolates of rats, pigs, and human genes are aligned with the sequences of the Gene Bank shown in the Table 1.
Table 1.
Blast Cryptosporidium DNA sequences of gene Bank based on 18s rRNA.
| Description | Max. Score | Total Score | Query cover | E value | Per. Ident | Accession |
|---|---|---|---|---|---|---|
| C. parvum, obi24 | 303 | 303 | 100% | 8e-83 | 90.38% | FJ796279.1 |
| C. parvum, obi21 | 303 | 303 | 100% | 8e-83 | 90.38% | FJ796279.1 |
| C. parvum, 04IR(NG) | 303 | 303 | 100% | 8e-83 | 90.38% | AB441688.1 |
| C. hominis, 24937 | 195 | 249 | 75% | 2e-50 | 97.37% | MG952704.1 |
| C. hominis, 28-9 | 195 | 249 | 75% | 2e-50 | 97.37% | MK270514.1 |
| C. hominis, Human-IQ7 | 195 | 249 | 75% | 2e-50 | 97.37% | MK886605.1 |
| C. suis, B8 | 305 | 305 | 100% | 1e-85 | 90.38% | MG132078.1 |
| C. suis, HK107C | 267 | 267 | 97% | 6e-73 | 92.19% | MK301308.1 |
| C. suis, Swec705 | 305 | 305 | 100% | 7e-83 | 90.38% | MH187877.1 |
| C. suis | 305 | 305 | 100% | 7e-83 | 90.38% | KP704556.1 |
| C. suis, B14 | 305 | 305 | 100% | 7e-83 | 90.38% | KT223028.1 |
| C. suis, CWQ4 | 305 | 305 | 100% | 7e-83 | 90.38% | KJ790239.1 |
Phylogenetic Cryptosporidium parvum in rats, pigs, and humans in West Lombok, and DNA sequences from Gene Banks are used to construct phylogenetic trees. Phylogenetic tree is composed of the number Acc: FJ796279.1 C. parvum, Human, Japan; FJ796276.1 C. parvum, Human, Japan; AB441688.1 C. parvum, Human, Iran; MG952704.1 C. hominis, Macaca, Chinese; MK270514.1 C. hominis, Macaca, Chinese; MK886605.1 C. hominis, Human, Iraq; MH187877.1 C. suis, Homo sapiens, Estonia; MK301308.1 C. suis, cattle, Uganda; MG132078.1 C. suis Pig, china; KP704556.1 C. suis, Pig, Slovakia; KT223028.1 C. suis, Pig, Denmark; KJ790239.1C.suis, Pig, China. Genetic kinship analysis using sequence alignment is based on Gene Bank. Bootstrap tree consensus with the conclusion of 1000 replications and parasitic distance evolution was calculated based on the “Kimura 2” method. Phylogenetic analysis used “MEGA X” software (Kimura, 1980; Felsenstein, 1985; Saitou and Nei, 1987). Figure 3 shows the results.
Figure 3.
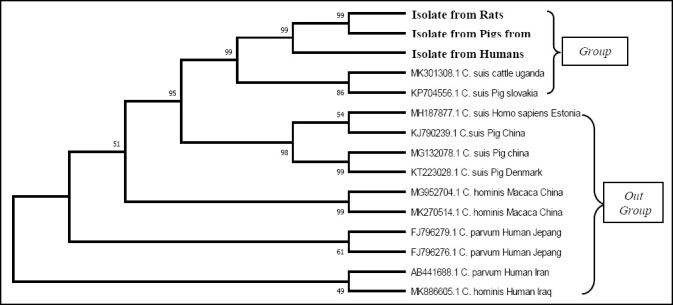
Phylogenetic tree of Cryptosporidium parvum isolates of rats, pigs and humans from the alignment with gene bank sequences based on the 18s rRNA gene. Cryptosporidium parvum isolates of Rats, pigs, and humans in West Lombok are one group with isolate MK301308.1 Cryptosporidium suis, Cattle, Uganda and isolate KP704556.1 Cryptosporidium suis, pigs, Slovakia. Cryptosporidium suis isolate MH187877.1 Homo sapiens, Estonia; KJ790239.1, MG132078.1 pig, Chinese; KT223028.1 pig, Denmark; Cryptosporidium hominis isolate MG952704.1, MK270514.1 Macaca, China; MK886605.1 Human, Iraq; Cryptosporidium parvum isolate MG952704.1, MK270514.1 Macaca China; MK886605.1 Human Iraq; Cryptosporidium parvum isolate FJ 796279.1 human, Japanese; and AB441688.1 Human, Iran is an outgroup.
Genetic kinship of Cryptosporidium isolates of rats, pigs and humans, confirmed by the pairwise distances calculation to the DNA sequence of the gene Bank (NCBI), with genetic distances are shown in Table 2.
Table 2.
Genetic distance of Cryptosporidium parvum DNA sequences of rat, pig and human isolates from DNA sequences from bank genes.
| No | Sample | Genetic distance | ||
|---|---|---|---|---|
| Rats | Pigs | Humans | ||
| 1 | Isolate from Rats | |||
| 2 | Isolate from Pigs | 0.0104 | ||
| 3 | Isolate from Humans | 0.2534 | 0.3166 | |
| 4 | FJ796279.1 C. parvum Human Japan | 0.1003 | 0.1462 | 0.3112 |
| 5 | FJ796276.1 C. parvum Human Japan | 0.1003 | 0.1462 | 0.3112 |
| 6 | AB441688.1 C. parvum Human Iran | 0.1003 | 0.1462 | 0.3112 |
| 7 | MG952704.1 C. hominis Macaca China | 0.1003 | 0.1462 | 0.3112 |
| 8 | MK270514.1 C. hominis Macaca China | 0.1003 | 0.1462 | 0.3112 |
| 9 | MK886605.1 C. hominis Human Iraq | 0.1003 | 0.1462 | 0.3112 |
| 10 | MH187877.1 C. suis Homo sapiens Estonia | 0.1056 | 0.1506 | 0.3030 |
| 11 | MK301308.1 C. suis cattle Uganda | 0.1056 | 0.1377 | 0.2710 |
| 12 | MG132078.1 C. suis Pig China | 0.1056 | 0.1506 | 0.3030 |
| 13 | KP704556.1 C. suis Pig Slovakia | 0.1056 | 0.1506 | 0.3030 |
| 14 | KT223028.1 C. suis Pig Denmark | 0.1056 | 0.1506 | 0.3030 |
| 15 | KJ790239.1 C. suis Pig China | 0.1056 | 0.1506 | 0.3030 |
The DNA sequence of Cryptosporidium parvum isolates of rats, pigs, and humans has a tight genetic range, which is: 0.0104-03112 from the Gene Bank sequence. The sequences from Gene Bank are: C. parvum human isolates from Japan, Iran; C. hominis Macaca isolate from China, human from Iraq, Estonian Homo sapiens; and C. suis pig isolates from China, Slovakia, Denmark.
Discussion
Cryptosporidium identified in rats, pigs and humans in West Lombok was 4.76% (4/84), 6.34% (13/205) and 0.9% (4/438), respectively, by PCR at 240bp. Cryptosporidium was also identified in other countries using PCR analysis. El-Bakri et al. (2018) identified twenty-six individuals (19.4%) who were positive for Cryptosporidium among asymptomatic healthy expatriate workers in Sharjah, United Arab Emirates. Meanwhile, Elmatrawy et al. (2017) identified Cryptosporidium spp. in children with diarrhea in Egypt 6% (9/150); while Bodager et al. (2015), identified Cryptosporidium with gene signatures from rat 2.08% (1/48), pig 1.65% (3/17) and human 0.83% from (1/120) human samples in Ranomafana National Park, Madagascar.
The similarity of DNA sequences of Cryptosporidium parvum isolates of rats, pigs and humans are shown by the Mega X program. These results identify genetic similarities of C. parvum that infect rats, pigs, and humans. Genetic similarity is related to the emergence of C. parvum zoonotic infections from rats and pigs to humans. Molecular analysis of the Cryptosporidium parvum uses the 18S rRNA gene because it is a reference gene that is often used as an internal control in the analysis of gene expression. Reference genes are genes whose expression is stable, not induced by certain treatments, abundant in all tissues, and follow the stages of the eukaryotic development. The 18S rRNA gene encodes an 18S ribosomal RNA gene, as a constituent of the eukaryotic small subunit ribosome in the process of recognition and hybridization of mRNA that is translated in the ribosome (Thellin et al., 1999; Dresios et al., 2006) The 18S rRNA gene is also used to detect Cryptosporidium from porcupine isolates in the UK, which was detected in 8% (9/111). The 18S rRNA gene also showed good results for detecting Cryptosporidium in pigs in Australia with the Next Generation Sequencing (NGS) method (Paparini et al., 2015; Sangster et al., 2016).
Cryptosporidium parvum isolates of rats, pigs and humans have a genetic similarity to DNA sequences from NCBI (the max score. 303, total score 303, query cover 97%, E-value 53-73 and per. ident 92.71%). The taxonomic results of the gene C. parvum have the highest scores than other species. Phylogenetic tree kinship analysis used the neighbor-joining method with bootstrap 1000x. The genetic relationship between C. parvum isolates of rats and pigs in West Lombok is monophyletic and C. parvum isolates of humans in West Lombok are synapomorphic with isolates of rats and pigs. Pairwise distance calculation between Cryptosporidium isolates of rats, pigs, and humans in West Lombok was 0.000-0.3170, indicating there is a close genetic relationship between C. parvum in rats, pigs, and humans in West Lombok.
C. parvum identified in humans in West Lombok comes from rats and pigs. C. parvum found in West Lombok is a zoonotic parasite, and there has been a transmission of Cryptosporidium infection from rats and pigs to humans. The presence of C. parvum in rats and pigs is a new source of transmission from C. parvum in West Lombok. The results of research on zoonotic Cryptosporidium were also reported by Deng et al. (2020), while C. parvum was identified in 8.6% (27/314) from the phylogenetic analysis of red squirrel pets sold in Sichuan, China. The presence of C. parvum in red squirrels is suspected as the source of transmission of C. parvum to humans and causes diarrhea. Phylogenetic analysis of C. parvum isolates of rats, pigs and humans are located in a genetic kinship group with isolates KP704556.1 C. suis pig, Slovakia and MK301308.1 C. suis, cattle, Uganda (Gordon, 2003).
Pairwise distance calculation analysis showed the genetic relationship between Cryptosporidium rats, pigs, and humans in West Lombok with Cryptosporidium Gene Bank isolates in Slovakia with genetic distance 0.1060-0.3030 and pigs in Uganda with genetic distance 0.1060-0.2710. Pairwise distance calculation analysis is used to determine the transition substitution and transversions through the many different nucleotides per base pair. Species with genetic distance that are getting closer have a strong genetic relationship (Dharmayanti, 2018).
Cryptosporidium parvum infection in West Lombok may be derived from rats and pigs, while the results of alignment with Gene Bank isolates showed a genetic relationship with C. parvum isolates of rats and pigs. These results concur with the study by Zou et al. (2017), identifying Cryptosporidium in pig farms in China using the 18S rRNA gene finding Cryptosporidium infection of 8% - 23%, which can act as a zoonotic source to humans. Utsi et al. (2016) found that a cryptosporidiosis outbreak in America was caused by visitors infected with Cryptosporidium, after returning from a petting farm.
Volunteers who have provided stools samples in this study had a house close to the pig farm. The pigs’ fecal litter was strewn in the yard and sometimes thrown into the garden or the river. The source of pigs fodder comes from recycling food scraps from residents.
The presence of rats around cages and houses, and the activity of rats moving from pigpens to poeple’s homes can carry and transmit diseases from rats to humans or to other animals. C. parvum identified in rats, pigs, and humans in West Lombok shows that there is a connection between Cryptosporidium transmission from rats, pigs, and humans. C. parvum which infects rats, pigs and humans is a zoonotic intestinal parasite, and this parasite may contribute to the incidence of diarrhea in West Lombok. The same case has occurred in Korea, where C. parvum was identified in 0.37% (32/8,571) of hospital diarrhea samples (Ma et al., 2019). Cryptosporidium zoonosis has also occurred in Madagascar and infects rats, pigs and humans around the national park (Bodager et al., 2015).
Cryptosporidium parvum infection in West Lombok can originate from rats and pigs. Zoonoses occur due to risk factors for environmental hygiene, raising pigs, and the presence of rats. This is in accordance with the opinion of Innes et al. (2020), that the presence of pigs, cows, horses and other wild animals such as rats around residential areas presents a high risk of polluting the environment by animal feces containing Crytptosporidium oocytes. Contamination of the environment by animal feces facilitates transmission of Cryptosporidium parvum to humans.
Feng et al. (2018) identified transmission between Cryptosporidium from goats to cattle that can even infect humans. Most Cryptosporidium species and genotypes have a host specificity so that one to four of the Cryptosporidium species can be found in one host (Widmer et al., 2020). The ability of the parasite to adapt within the new host and geographical pressure causes the formation of unique subtypes and phenotypic properties, especially those found in humans (C. parvum and C. hominis) (Feng et al., 2018). The intensity of transmission, genetic diversity, and genetic recombination form the genetic tree structure of Cryptosporidium. Molecular research on Cryptosporidium spp. can help increase our understanding of various patterns of transmission of Cryptosporidium to new hosts (Thompson, 2013).
Conclusions
There are genetic similarities of Cryptosporidium spp. that infect rats, pigs and humans in the West Lombok Regency, West Nusa Tenggara Province. This allows the transmission of the parasitic zoonoses Cryptosporidium which infect rats, pigs and humans in the West Lombok Regency, West Nusa Tenggara Province based on parasitic genetic kinship. The results of this study require that Public Health programs in contaminated areas receive priority attention to prevent further transmission of this potentially fatal parasite. More research is needed to see what risk factors contribute to zoonoses.
Abbreviations:
- (SSU rRNA)
Small Subunit rRNA gene
- (DNA)
Deoxyribonucleic Acid
- (RNA)
Ribonucleic Acid
- (PCR)
Polymerase chain reaction
- (BLAST)
Basic Local Alignment Search Tool
- (NCBI)
National Center for Biotechnology Information
- (MEGA X)
Molecular Evolutionary Genetics Analysis X
- (NGS)
Next Generation Sequencing
Acknowledgment
We extend our thanks to Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, Indonesia; Department of Medical Laboratory Technology, Politeknik Kesehatan Mataram, Mataram, Indonesia.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Austin K.F. Degradation and disease:ecologically unequal exchanges cultivate emerging pandemics. World Development. 2021;137:105163. doi: 10.1016/j.worlddev.2020.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzam K.M, El-Abd N.M, El-Hady A, Eman A. Survey of Endoparasites of Different Rodent Species in Egypt. Egyptian Journal of Biological Pest Control. 2016;26:4. [Google Scholar]
- 3.Badry A.H.H, Jameel A.Y, Mero W.M.S. Pathogenic microorganisms associated with diarrhea in infants and children in Duhok province, Kurdistan region/Iraq. Journal of University of Zakho. 2014;2(2):266–275. [Google Scholar]
- 4.El Bakri A, Mogane L, Ezzedine S, Potgieter N, Bessong P, Odeh R.A, Samie A. Prevalence of CRYPTOSPORIDIUM SPP among asymptomatic healthy expatriate workers in Sharjah, United Arab Emirates. African journal of infectious diseases. 2018;12(2):7–13. doi: 10.21010/ajid.v12i2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baqer N.N, Hammood A.H, Hassan K.F, Hassan E.S.A. Detection of water-borne parasites in drinking water of Baghdad, Iraq. African journal of infectious diseases. 2018;12(2):1–6. doi: 10.21010/ajid.v12i2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beser J, Bujila I, Wittesjö B, Lebbad M. From mice to men:Three cases of human infection with Cryptosporidium ditrichi. Infection, Genetics and Evolution. 2020;78:104120. doi: 10.1016/j.meegid.2019.104120. [DOI] [PubMed] [Google Scholar]
- 7.Bodager J.R, Parsons M.B, Wright P.C, Rasambainarivo F, Roellig D, Xiao L, Gillespie T.R. Complex epidemiology and zoonotic potential for Cryptosporidium suis in rural Madagascar. Veterinary parasitology. 2015;207(1-2):140–143. doi: 10.1016/j.vetpar.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Bogitsh B.J, Carter C.E, Oeltmann T.N, et al. Human parasitology. Academic Press 2013 [Google Scholar]
- 9.Deng L, Chai Y, Luo R, Yang L, Yao J, Zhong Z, Wang W, Xiang L, Fu H, Liu H. Occurrence and genetic characteristics of Cryptosporidium spp and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Scientific Reports. 2020;10(1):1–10. doi: 10.1038/s41598-020-57896-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dharmayanti N.L.P. Molecular Phylogenetic:Organism Taxonomy Method Based on Evolution History 2018 [Google Scholar]
- 11.Dresios J, Chappell S.A, Zhou W, Mauro V.P. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nature structural and molecular biology. 2006;13(1):30. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 12.Dupont H.L. Persistent diarrhea a clinical review. JAMA - Journal of the American Medical Association. 2016;315(24):2712–2723. doi: 10.1001/jama.2016.7833. [DOI] [PubMed] [Google Scholar]
- 13.Elmatrawy O.M, Hassan M.A, Morsy S.M, Rubio J.M, El-Badry A.A. Molecular diagnosis of Cryptosporidium spp Versus microscopy in diarrheic patients in cairo. Journal of the Egyptian Society of Parasitology. 2017;47(2):303–308. [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies:an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Ryan U.M, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends in parasitology. 2018 doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Gordon M. Methods of Phylogenetic Analysis:New Improvements on Old Methods. Biochem 218 final project 2003 [Google Scholar]
- 17.Innes E.A, Chalmers R.M, Wells B, Pawlowic M.C. A One Health Approach to Tackle Cryptosporidiosis. Trends in Parasitology. 2020 doi: 10.1016/j.pt.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of molecular evolution. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.Krijger I.M. Rodent-borne health risks in farming systems 2020 [Google Scholar]
- 20.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D.-W, Lee M.-R, Hong S.-H, Cho S.-H, Lee S.-E. Molecular Prevalence and Genotypes of Cryptosporidium parvum and Giardia duodenalis in Patients with Acute Diarrhea in Korea, 2013–2016. The Korean journal of parasitology. 2019;57(5):531. doi: 10.3347/kjp.2019.57.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morand S. Global parasite and Rattus rodent invasions:The consequences for rodent-borne diseases. Integrative Zoology. 2015;10:267–281. doi: 10.1111/1749-4877.12143. [DOI] [PubMed] [Google Scholar]
- 23.Mosites E, Thumbi S.M, Otiang E, McElwain T.F, Njenga M.K, Rabinowitz P.M, Rowhani-Rahbar A, Neuhouser M.L, May S, Palmer G.H. Relations between Household Livestock Ownership, Livestock Disease, and Young Child Growth–3. The Journal of nutrition. 2016;146(5):1118–1124. doi: 10.3945/jn.115.225961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munshi A. DNA sequencing–methods and applications. InTech Rijeka 2012 [Google Scholar]
- 25.Paparini A, Gofton A, Yang R, White N, Bunce M, Ryan U.M. Comparison of Sanger and next generation sequencing performance for genotyping Cryptosporidium isolates at the 18S rRNA and actin loci. Experimental parasitology. 2015;151:21–27. doi: 10.1016/j.exppara.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Perec-Matysiak A, Bunkowska-Gawlik K, Zalesny G, Hildebrand J. Small rodents as reservoirs of Cryptosporidium spp and Giardia spp. in south-western Poland. Annals of Agricultural and Environmental Medicine. 2015;22(1) doi: 10.5604/12321966.1141359. [DOI] [PubMed] [Google Scholar]
- 27.Ridley J.W. Parasitology for medical and clinical laboratory professionals. Cengage Learning 2012 [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method:a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Sangster L, Blake D.P, Robinson G, Hopkins T.C, Sa R.C.C, Cunningham A.A, Chalmers R.M, Lawson B. Detection and molecular characterisation of Cryptosporidium parvum in British European hedgehogs (Erinaceus europaeus) Veterinary parasitology. 2016;217:39–44. doi: 10.1016/j.vetpar.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Sun P, Wronski T, Bariyanga J.D, Apio A. Gastro-intestinal parasite infections of Ankole cattle in an unhealthy landscape:An assessment of ecological predictors. Veterinary Parasitology. 2018;252(January):107–116. doi: 10.1016/j.vetpar.2018.01.023. https://doi.org/10.1016/j.vetpar.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 31.Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E. Housekeeping genes as internal standards:use and limits. Journal of biotechnology. 1999;75(2-3):291–295. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 32.Thiermann A. Emerging diseases and implications for global trade. 2004;23(2):701–708. doi: 10.20506/rst.23.2.1509. [DOI] [PubMed] [Google Scholar]
- 33.Thompson R.C.A. Parasite zoonoses and wildlife:one health spillover and human activity. International journal for parasitology. 2013;43(12-13):1079–1088. doi: 10.1016/j.ijpara.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utsi L, Smith S.J, Chalmers R.M, Padfield S. Cryptosporidiosis outbreak in visitors of a UK industry-compliant petting farm caused by a rare Cryptosporidium parvum subtype:a case-control study. Epidemiology and Infection. 2016;144(5):1000–1009. doi: 10.1017/S0950268815002319. [DOI] [PubMed] [Google Scholar]
- 35.Verkerke H.P, Sobuz S.U, Petri W.A. Molecular diagnosis of infectious diarrhea:Focus on enteric protozoa. Expert Review of Molecular Diagnostics. 2014;14(8):935–946. doi: 10.1586/14737159.2014.951035. http://dx.doi.org/10.1586/14737159.2014.951035. [DOI] [PubMed] [Google Scholar]
- 36.Widmer G, Köster P.C, Carmena D. Cryptosporidium hominis infections in non-human animal species:revisiting the concept of host specificity. International journal for parasitology. 2020;50(4):253–262. doi: 10.1016/j.ijpara.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Woolhouse and Sequeria. (2005) Host Range and Emerging and Reemerging Pathogens. 11(12) doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yee E.U, Kuo E, Goldsmith J.D. Pathologic Features of Infectious Gastritis. Advances in Anatomic Pathology. 2018;00(00):1–16. doi: 10.1097/PAP.0000000000000187. [DOI] [PubMed] [Google Scholar]
- 39.Zahedi A, Paparini A, Jian F, Robertson I, Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife:Critical insights into better drinking water management. International Journal for Parasitology:Parasites and Wildlife. 2016;5(1):88–109. doi: 10.1016/j.ijppaw.2015.12.001. http://dx.doi.org/10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zebardast N, Yeganeh F, Gharavi M.J, Abadi A, Seyyed Tabaei S.J, Haghighi A. Simultaneous detection and differentiation of Entamoeba histolytica, E dispar, E. moshkovskii, Giardia lamblia and Cryptosporidium spp. in human fecal samples using multiplex PCR and qPCR-MCA. Acta Tropica. 2016;162:233–238. doi: 10.1016/j.actatropica.2016.07.004. http://dx.doi.org/10.1016/j.actatropica.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Z, Wang R, Zhao W, Qi M, Zhao J, Zhang L, Li J, Liu A. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology. 2015;142(6):800–806. doi: 10.1017/S0031182014001929. [DOI] [PubMed] [Google Scholar]
- 42.Zou Y, Ma J.-G, Yue D.-M, Zheng W.-B, Zhang X.-X, Zhao Q, Zhu X.-Q. Prevalence and risk factors of Cryptosporidium infection in farmed pigs in Zhejiang, Guangdong, and Yunnan provinces, China. Tropical animal health and production. 2017;49(3):653–657. doi: 10.1007/s11250-017-1230-y. [DOI] [PubMed] [Google Scholar]


