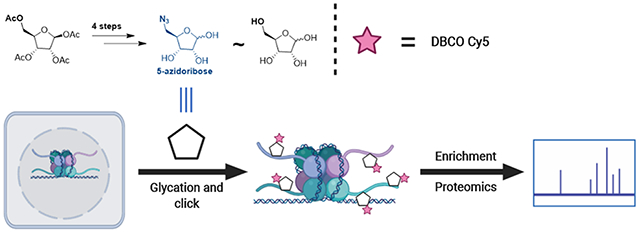
Abstract
Reactive cellular metabolites can modify macromolecules and form adducts known as nonenzymatic covalent modifications (NECMs). The dissection of the mechanisms, regulation, and consequences of NECMs, such as glycation, has been challenging due to the complex and often ambiguous nature of the adducts formed. Specific chemical tools are required to directly track the formation of these modifications on key targets in order to uncover their underlying physiological importance. Here, we present the novel chemoenzymatic synthesis of an active azido-modified ribose analog, 5-azidoribose (5-AR), as well as the synthesis of an inactive control derivative, 1-azidoribose (1-AR), and their application toward understanding protein ribose-glycation in vitro and in cellulo. With these new probes we found that, similar to methylglyoxal (MGO) glycation, ribose glycation specifically accumulates on histones. In addition to fluorescent labeling, we demonstrate the utility of the probe in enriching modified targets, which were identified by label-free quantitative proteomics and high-resolution MS/MS workflows. Finally, we establish that the known oncoprotein and hexose deglycase, fructosamine 3-kinase (FN3K), recognizes and facilitates the removal of 5-AR glycation adducts in live cells, supporting the dynamic regulation of ribose glycation as well as validating the probe as a new platform to monitor FN3K activity. Altogether, we demonstrate this probe’s utilities to uncover ribose-glycation and deglycation events as well as track FN3K activity toward establishing its potential as a new cancer vulnerability.
Graphical Abstract
INTRODUCTION
Metabolites, either endogenously generated or assimilated from the microenvironment, induce rapid cellular responses ranging from the upregulation of signaling cascades to longterm adjustments in the transcriptional program.1 These instigate cellular changes by feeding into other pathways, acting as cofactors of enzymes, or directly reacting with endogenous macromolecules such as lipids, nucleic acids, and proteins. The resulting appendages, known as nonenzymatic covalent modifications (NECMs), have been identified in numerous proteins and demonstrated to compromise their structure, function, and stability.2
Glycation, which is one of the more prevalent NECMs, is characterized by the condensation of the aldehyde form of monosaccharides (such as glucose or fructose) or glycolytic byproducts (such as methylglyoxal, MGO) with nucleophilic amino acid side chains (primarily lysines and arginines).3–5 The initial Maillard glycation adducts can undergo several iterations of subsequent oxidations, rearrangements, and crosslinking reactions to ultimately form a plethora of species collectively referred to as advanced glycation end products (AGEs).6 These AGEs are particularly prevalent in diseases such as diabetes and cancer and are strongly linked to the physiological consequences of these pathologies.7,8
One formidable challenge in characterizing the heterogeneous AGE mixture is in determining the identity of the initial reactive carbonyl species. This is principally due to a combination of two factors: the sheer variety of adducts rearranged from the original sugar adduct, which may be in the thousands, and the substantial overlap in structures and molecular weights of the modifications that arise from distinct electrophilic reactants.9,10 Despite this, significant work has been published regarding the characterization of relevant AGEs, their cellular repercussions, and the adapted cellular regulatory mechanisms in place.11,12 Commercial antibodies against several glycation adducts on proteins as well as high-resolution mass spectrometry workflows have been used to characterize, for example, MGO glycation.13–15 Yet, there remains a scarcity of modification-specific antibodies and a bottleneck in mass spectrometry-based approaches that has hindered progress in uncovering the specific mechanisms, roles, and regulation of protein glycation in vivo. Alternatively, chemical biology approaches have been developed using analogs of the desired sugars with appended enrichment moieties and have been applied in the study of a broad range of glycobiology processes ex vivo and in vivo.16,17 These analogs simultaneously possess the temporal and pharmacokinetic benefits of small molecules while maintaining high specificity.17 For example, bioorthogonal handle-containing sugar mimics have been synthesized and applied in studying glycopolymer mechanisms of action.18
Using similar tools, we and others have shown that while many proteins are potential substrates for glycation, the key regulatory histone proteins are prime targets due to their very long half-lives and accessible lysine and arginine-rich tails.13,19–21 Moreover, histone proteins undergo extensive post-translational modifications (PTMs), including acetylation and methylation, primarily on lysines and arginines, which were shown to regulate cellular transcription and are often disrupted in disease.22,23 We now have a deeper understanding of these enzymatically installed histone PTMs and their assorted functions; however, very little is known about the roles that NECMs play in similar cellular processes. Indeed, we found that in rapidly dividing and metabolically active cells, such as cancer cells, the adducts formed on histones compete with PTMs and change the cellular epigenetic landscape. In addition, we found they alter local chromatin architecture by initially disrupting salt-bridges and ultimately forming irreversible cross-links between positively charged histones and the negatively charged phosphodiester backbone of DNA.13
While ribose glycation has been linked to extreme physiological pathologies such as protein aggregation in alpha synucleinopathy, the effect it has on finely tuned processes such as chromatin regulation has never been investigated.24,25 This is again largely due to the complexity and ambiguity of the adducts and a lack of specific chemical and biological tools that will allow their high-resolution analysis. We hypothesized that like MGO, ribose glycation adducts accumulate on histones in cancer cells where ribose is required for rapid nucleic acid synthesis and is thus found at higher concentrations.26,27 In support, in vitro histone glycation has been reported to be 20–30 times faster with ribose than other reducing sugars such as glucose.24,28
To evaluate histone ribose glycation in cells, we report here the chemoenzymatic synthesis of an active azidoribose probe as well as the synthesis of its glycation-inactive counterpart. The probes are used to demonstrate the specific reactivity of ribose toward histones in their recombinant form, isolated nuclei as well as live cells. In addition to labeling, we establish the enrichment capabilities of the azidoribose probe and identify a variety of key nuclear proteins, in addition to histones, that are modified by the pentose sugar. Finally, we reveal that in its physiological context, a known cytosolic ribose, glucose, and fructose deglycase, fructosamine 3-kinase (FN3K) recognizes and removes azidoribose glycation adducts.
RESULTS AND DISCUSSION
Probe Design and Synthesis.
In cells, reducing sugars such as ribose 5-phosphate and ribose are either endogenously present as intermediates within the pentose phosphate pathway or actively imported via the Glut1 transporter (Scheme S1).27,29 Like all reducing sugars, ribose, a metabolite involved in several crucial pathways, exists in both “open” aldopentose and “closed” cyclic hemiacetal forms. Although a reactive carbonyl is exposed in the “open” linear conformation, ribose predominantly exists in the hemiacetal pyranose and furanose states.30 However, the unstable puckered conformation of the furanose ring increases the exposure of the electrophilic aldehyde, subsequently facilitating ribose’s reactivity in the Maillard reaction.31,32 Once the respective condensation and Schiff base formation reactions have occurred, a subsequent rearrangement produces a stabilized ketoamine adduct (RiboLys). This adduct is important as it represents the first stabilized glycation intermediate that retains the identity of the initial reacting sugar and begins the cascade toward a myriad of AGEs (Scheme 1).32
Scheme 1.
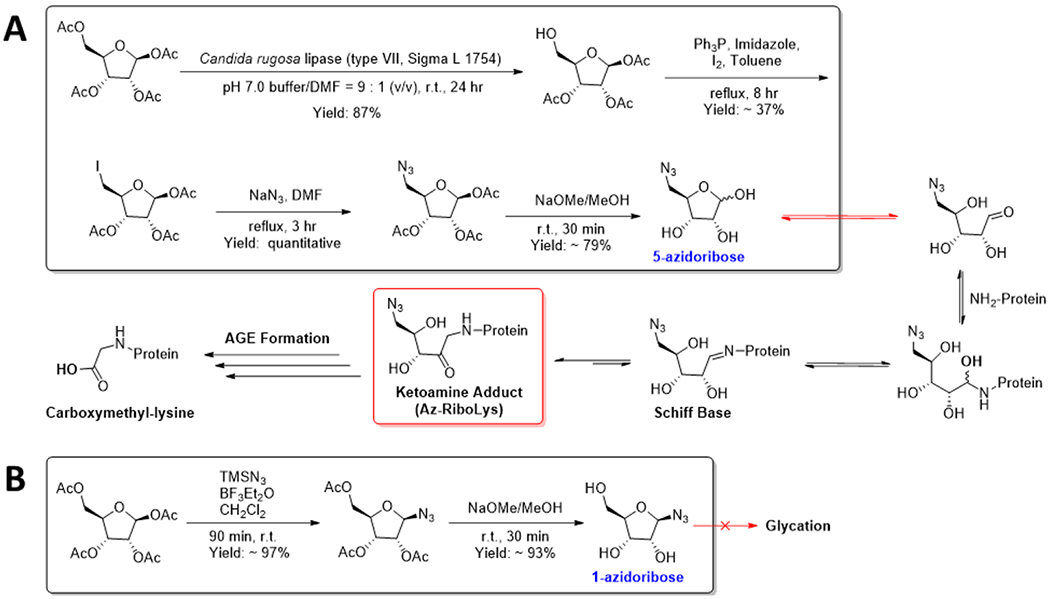
Synthesis of (A) 5-Azidoribose (5-AR) and (B) 1-Azidoribose (1-AR) Probes to Track Ribose Glycation
The identification, characterization, and exploration of the effects of such modifications are extremely challenging without a specific antibody or custom mass-spectrometry workflow. Rather, chemical biology approaches involving the synthesis of close derivatives of the native metabolites have been used to great effect to label, track, and enrich modifications.33–37 Employing a similar approach, we aimed to generate a click handle-containing ribose that would not disrupt its capacity to react nor be lost after the reaction. Thus, we chose to append an azido moiety onto the C-5 of ribose to retain its linear-ring isomerization and glycation activity (5-azidoribose, 5-AR). After the completion of glycation and rearrangement, the relatively stable ribulosamine (Az-RiboLys) will be the predominant adduct present and can be subsequently labeled via the azide handle. In parallel, we generated a control probe where we introduced an azide onto C-1 (1-azidoribose, 1-AR). This inactive probe possesses similar chemical characteristics, i.e., it can partake in a click reaction but cannot readily isomerize into the active aldopentose form which ablates its glycation activity. The azido-placement in each molecule additionally reduces the likelihood of either molecule being incorporated into endogenous nucleic acid biosynthesis pathways while maintaining high resemblance to ribose through their furanose rings and C-2 and C-3 hydroxyls. This is because both free hydroxyl C-1 and C-5 positions are required for the addition of a nucleobase and phosphate, respectively. Importantly, 5-AR is highly unlikely to be utilized in ADP-ribosylation, a transfer reaction in which NAD+ is cleaved leading to the covalent attachment of ADP to a nucleophilic residue.38 Although the C-1 hydroxyl in 5-AR may be a substrate for phosphorylation and subsequent conversion into azido-nicotinate d-ribonucleotide (Az-NaMN), an analog of a precursor in the biosynthesis of NAD+, the lack of a C-5 hydroxyl prevents activation by phosphorylation and subsequent NAD+ formation. Concurrently, 1-AR lacks a C-1 hydroxyl, preventing it from being phosphorylated and subsequently converted into NaMN. Ablation of their ability to partake in enzymatic processes potentially increases the probes’ cellular half-lives and reduces background.39,40
For the synthesis of 5-AR, the C-5 acetyl of commercial β-d-ribofuranose1,2,3,5-tetraacetate was selectively deprotected using the hydrolase activity of C. rugosa, an engineered lipase.41 The resulting free hydroxyl was converted via an Appel reaction into an iodide, which was subsequently transformed into an azide using sodium azide in dimethyl-formamide. A final global deprotection yielded the desired active 5-AR probe.42 Conversely, the C-1 acetyl of the same commercial β-d-ribofuranose 1,2,3,5-tetraacetate was converted into an azide using trimethylsilyl azide in the presence of the Lewis acid, boron trifluoride etherate.43 Subsequent global deprotection yielded the 1-AR probe. Both probes as well as all the intermediates were structurally characterized by 1HNMR, 13CNMR, and high-resolution mass spectrometry (HRMS), and the incorporation of the azides was supported through the presence of FT-IR peaks corresponding to azide-stretches. The de novo chemoenzymatic as well as the 1-AR syntheses were performed on small and large scales with high efficiency to produce the final products with the indicated yields (Scheme 1).
Azidoribose Glycates Recombinant Histones.
Due to their long-lived nature, exposed and lysine-rich tails, and key roles in chromatin physiology, histones are uniquely at risk of chronic NECM damage and have been shown to be prime glycation targets in cells with potentially devastating cellular consequences.13,20,44 To address the potential reactivity between histones and ribose, we first sought to evaluate the chemical composition of the adducts generated from this reaction. We incubated ribose, 5-AR, and 1-AR with free lysine at equimolar concentrations at 37 °C for 16 h and subseqeuently analyzed the products by mass-spectrometry (MS). As shown in Figures S1 and S2, ribose and 5-AR, but not 1-AR, generated the predicted RiboLys and Az-RiboLys ketoamine adducts, respectively. We next evaluated the reactivity of the ribose probes toward histones in vitro by incubating 5 mM of each of the probes, d-ribose, or phosphate-buffered saline (PBS) with each of the four canonical histones for 8 h at 37 °C. We have previously shown that these acute high exposure conditions sufficiently mimic chronic low-exposure histone glycation reactivity in cells.13 We then utilized copper-free click chemistry to fluorescently label the modifications by reacting our glycated histones with a strained alkyne (dibenzocyclooctyne, DBCO) conjugated to a Cyanine-5 (Cy5) fluorophore (Figure 1A).45 After separating the reaction mixture via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferring the proteins onto a PVDF membrane, we visualized the modified proteins using the Cy5 fluorescent signal. The results presented in Figure 1B, demonstrate the detection of strong labeled bands for each of the four histones in the 5-AR reactions but practically no background in the 1-AR, d-ribose, or PBS controls. Notably, we chose the 8-h time point after performing a time course reaction on H3, which showed a diminishing Cy5 signal after prolonged incubations (>24 h) (Figure S3). We next analyzed the same reactions side-by-side to determine the relative reactivity of 5-AR with each of the histone proteins. Interestingly, while all four histones are glycated by 5-AR, there is a noticeable difference in intensity among them, with H4 being the most reactive (Figure 1C and Figure S4). While reasons for the preference remain debatable, this result is in alignment with previous results that we and others have reported regarding the reactivity between histones and MGO.13,14 Finally, we confirmed that our probe indeed glycates histones via the same mechanism as unmodified d-ribose by performing competition experiments between them in vitro using both pre- (Figure S5) and coincubation (Figure S6) conditions. Collectively, our experiments demonstrate the sensitivity and utility of 5-AR in visualizing histone ribose glycation at its early stages after acute exposure.25,32
Figure 1.
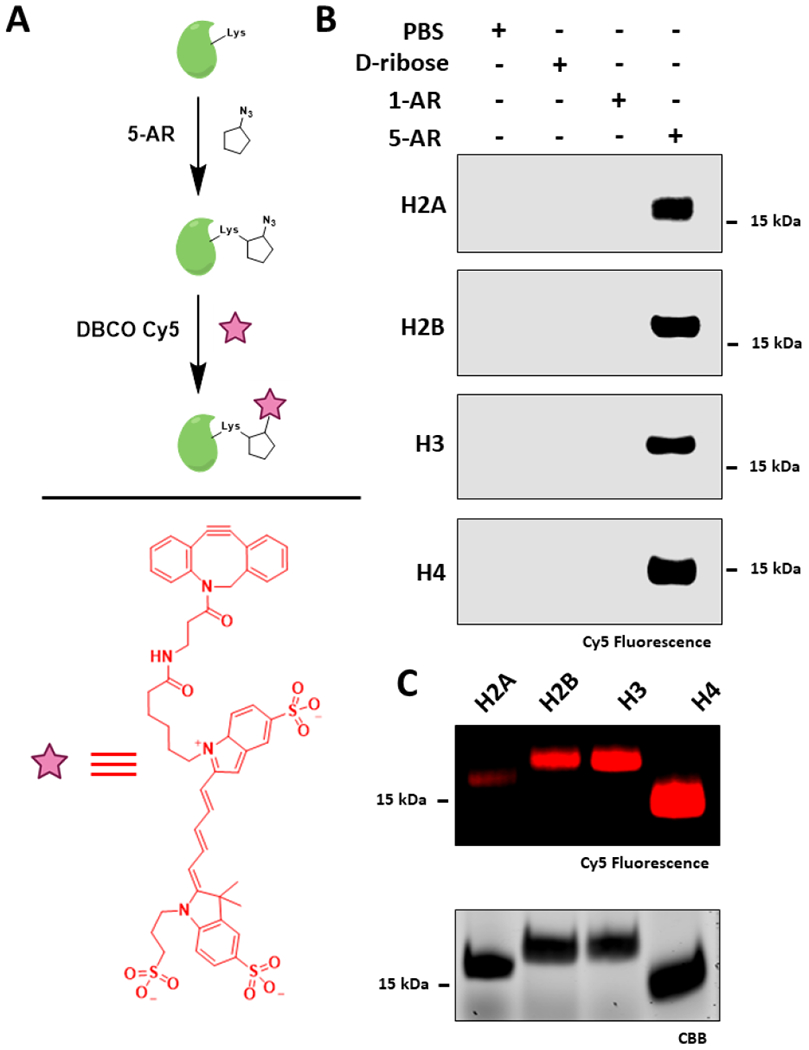
5-AR glycates histones in vitro. (A) Schematic depicting incubation of histones with probes followed by click labeling with DBCO Cy5. (B) Incubation of recombinant histones with PBS, 5 mM of d-ribose, 1-AR, or 5-AR for 8 h followed by click-labeling and in-blot fluorescence analysis. (C) Histones glycated with 5 mM of 5-AR, labeled with DBCO Cy5, analyzed side-by-side by in-blot fluorescence and Coomassie Brilliant Blue (CBB) for loading control.
Azidoribose Glycation Accumulates on Histones in Isolated Nuclei and Live Cells.
After validating that our probe reacts with purified histones in vitro, we turned to investigate this glycation event in a more physiological context. We thus evaluated our probes’ abilities to glycate proteins in isolated nuclei, which have been established as accurate and reasonable systems to study nuclear processes.46 Initially, we isolated nuclei from 293T cells through hypotonic lysis and incubated them with PBS or 5 mM of d-ribose, 1-AR, or 5-AR. After washing away unreacted sugar, we fluorescently tagged the resuspended nuclei with DBCO Cy5, lysed them through sonication, and separated the lysate by SDS-PAGE. Once more, through in-blot fluorescence, we observe specific labeling by the active probe over its control counterpart with histones as the major targets (Figure 2A).
Figure 2.
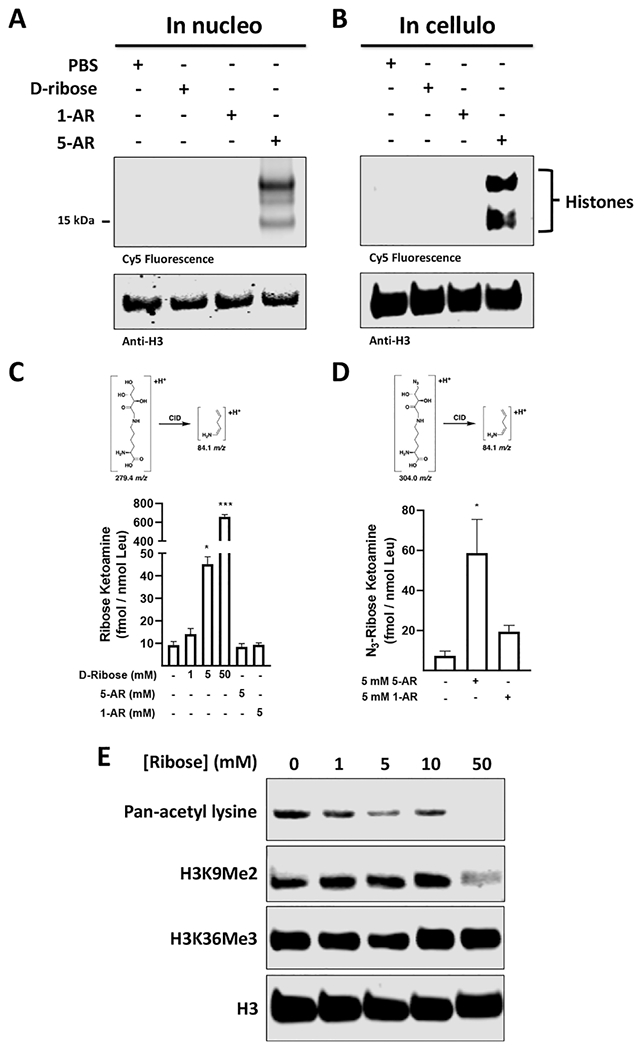
Ribose permeates cells and nuclei to glycate histones. (A) Glycation of isolated 293T nuclei with PBS or 5 mM of d-ribose, 1-AR, or 5-AR for 8 h, followed by DBCO Cy5 labeling and in-blot fluorescence visualization and western blot analysis. (B) Glycation of live 293T cells with PBS or 5 mM of d-ribose, 1-AR, or 5-AR, followed by hypotonic isolation of nuclei, DBCO Cy5 click labeling, in-blot fluorescence and western blot analysis. (C) Quantification of RiboLys on chromatin isolations from 293T cells incubated with 5-AR, 1-AR, or increasing concentrations of d-ribose. (D) Quantification of Az-RiboLys on chromatin isolations from 293T cells incubated with 5-AR or 1-AR. The MRM transitions used for quantification are shown. (E) Glycation of live 293T cells with increasing amounts of d-ribose followed by high-salt histone extraction, and western blot post-translational modification (PTM) analysis with the indicated antibodies.
Next, we evaluated whether our active probe crosses the plasma membrane, is internalized, and reacts with histones in live cells. We incubated cultured 293T cells with media containing PBS, or 5 mM of d-ribose, 1-AR, or 5-AR for 8 h. We then washed away unreacted sugar, harvested the cells, and fluorescently tagged the nuclei. After washing away excess fluorophore, we lysed the nuclei through sonication and separated the nuclear lysate via SDS-PAGE. As with the in nucleo experiment, histones were shown to be the primary targets (Figure 2B). We verified that azidoribose modifies histones in cells via the same mechanism as d-ribose, by demonstrating dose-dependent competition when coincubating 5-AR (5 mM) with increasing concentrations of d-ribose (0, 1, 5, and 10 mM) in cultured 293Ts (Figure S7).
To validate that ribose and 5-AR form lysine ketoamine adducts on chromatin in live cells, we next incubated live 293T cells with increasing concentrations of d-ribose or 5 mM of either 1-AR or 5-AR for 8 h at 37 °C. Chromatin was isolated from the harvested cells, digested to individual amino acids, and subjected to multiple reaction monitoring (MRM)-MS analysis.47 Importantly, our results identify basal accumulation of RiboLys and a dose-dependent increase with additional ribose exposure (Figure 2C). Similar analyses demonstrate that the addition of 5-AR, but not 1-AR, to cells induces the significant accumulation of Az-RiboLys on chromatin (Figure 2D). Interestingly, 5-AR exposure did not reduce the basal RiboLys signal, likely due to an excess of available cellular target sites. Notably, the low level 5-AR background in the absence of probe stems from noise in the MRM signal in cells.
Ribose glycation Affects the Epigenetic Landscape.
Given that histone NECMs such as MGO-glycation were shown to crosstalk with key histone PTM marks and ribose also glycates histones, we reasoned that ribose glycation might also affect the epigenetic landscape either by directly competing for the same lysine residues or by affecting binding of neighboring writers.13,44 To this end, we cultured 293T cells in media infused with increasing concentrations of d-ribose for 48 h. Thereafter, we isolated the histones, separated them via SDS-PAGE, and analyzed the levels of several key representative PTMs by western blotting. Notably, the signals corresponding to general acetyl lysine (pan-KAc) and histone H3 lysine-9 dimethyl (H3K9Me2) marks decrease in the presence of 5- and 50-mM ribose, respectively, whereas the signal corresponding to the H3 lysine 36 trimethyl (H3K36Me3) is unchanged (Figure 2E and Figure S8). These marks were selected as they are associated with and regulate several chromatin states by different mechanisms. Namely, histone acetylation “relaxes” chromatin by directly affecting the electrostatic interactions between the histone octamer and DNA.48,49 Conversely, histone methylation modulates transcription and chromatin landscaping via effector proteins that specifically recognize the methylation mark.22,23 The fact that the levels of H3K36 trimethylation are unaffected by ribose glycation suggests a degree of complexity and selectivity in the crosstalk and not a simple competition for all sites.50
Enrichment of 5-AR Adducts Used to Determine Nuclear Ribose Glycation Targets.
While histones are key substrates, glycation has been shown to impact cellular function through the modification of other proteins, such as transcription factors.51,52 We thus sought to utilize the enrichment functionality of 5-AR, in conjunction with proteomic workflows, to map all the potential chromatin substrates of ribose glycation (Figure 3A). First, we validated the probe’s capacity to be used for enrichment. Following nuclear isolation, incubation with 5 mM of 5-AR or 1-AR and subsequent DBCO Cy5 click-labeling, we incubated the labeled nuclear lysate with Dynabeads bound to anti-Cy5 antibody. After unbound material was washed away, the beads were boiled and analyzed by in-blot fluorescence as described above. The fluorescent signal as well as subsequent immunoblotting for histone H3 supported the successful selective enrichment of 5-AR-modified proteins (Figure 3B). It is noteworthy that higher migrating bands that appear in the 1-AR control are removed through stringent washing during the enrichment step, suggesting that they are not covalently labeled after the click reaction and are thus designated as background. Additionally, although the signals corresponding to the histones are distinctive, they are seemingly underrepresented when compared to those of the remainder of the nuclear proteome. This is partly due to the fact that while they are the prime targets, histones only make up one family of proteins in the nucleus and are likely outnumbered by the remainder of the glycated nuclear proteome. Moreover, the low turnover time associated with histones becomes more effective at promoting the accumulation of glycation adducts over chronic and not acute exposures.13 The appearance of these specific higher molecular weight signals visualized in the fluorescence channel support the notion of additional nuclear glycation events and emphasize the need for their characterization.
Figure 3.
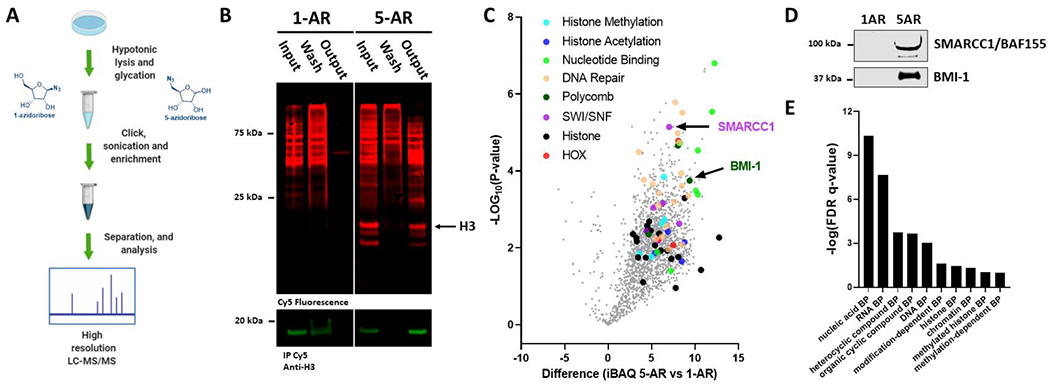
Azidoribose probes can be used to selectively enrich ribose glycation adducts from 293T cells for further analyses. (A) Schematic depicting treatment of isolated nuclei with 5-AR and 1-AR, DBCO Cy5 click labeling, enrichment, digestion, and label-free quantitative proteomics (LFQ) analysis. (B) In-blot fluorescence and western blot showing background labeling and selective enrichment of glycated proteins from AR-treated isolated nuclei. (C) Volcano plot depicting differences in iBAQ values from 5-AR- over 1-AR-enriched sample groups against negative logarithmic (base 10) p-values of the t-test performed from multiple replicates. All identified proteins represented in gray, validated proteins BMI-1 and SMARCC1 labeled, and select protein groups highlighted in separate colors. (D) Western blot of representative output fractions validating the labeling and enrichment of two chromatin remodeling proteomic hits. (E) Binding protein (BP) gene ontology analysis of gene list from set of protein target dataset (p < 0.05 and unique peptides >2).
In order to identify all the enriched proteins, triplicate experiments were analyzed by label-free quantitative (LFQ) proteomics, which was chosen due to the inherent statistical power of the obtained results.53 Enriched glycation targets were eluted from the Dynabead support, trypsinized, separated by chromatography, and quantified by high-resolution mass spectrometry. The resulting logarithmic ratios of protein intensities in the 5-AR- over the control 1-AR-enriched group were calculated and are represented in a volcano plot against the negative logarithmic p-values of the t-test performed from the replicates (Figure 3C). These data reveal that in addition to the core histones, several other protein targets, whose functions and misregulation are implicated in physiological pathways and disease, are specifically and significantly enriched. Importantly, very few proteins were detected in the control 1-AR enrichment, which supports both the probe design as well as the use of DBCO Cy5 (full proteomics data available as Dataset S1 and Dataset S2). Among the positive hits, we observed several notable classes of important nuclear proteins such as chromatin remodelers, nucleotide binders, and DNA repair proteins. These underly important nuclear pathways highlighted by key epigenetic landscape-establishing enzymes such as histone acetyltransferase (KAT7), histone deacetylase (HDAC2), and histone N-methyltransferases (EHMT1, NSD2, EZH2, KMT2A, and NSD3). The entire glycated nuclear proteome is presented as a volcano plot in Figure 3C with specific proteins depicted as different colors and listed in Dataset S3. To validate our proteomic analyses, two of the identified targets, SMARCC1/BAF155 and BM1-1, were selected and analyzed by immunoblotting with their respective antibodies, showing significant enrichment after 5-AR treatment and pulldown (Figure 3D). Interestingly, both proteins are key determinants of chromatin structure and function, which might serve as another mechanism for epigenetic landscape editing following glycation.13,14 SMARCC1/BAF155 are subunits of the SWI/SNF chromatin remodeling complex and BMI-1 is a member of the polycomb repressor complex 1 whose activity ultimately results in chromatin folding, compaction, and transcription attenuation.54,55 These findings are particularly important as, to the best of our knowledge, no NECMs have ever been identified on either of these two proteins, and this could potentially indicate novel physiologically important events.51,56 After validating the proteomic analyses, we determined the underlying pathways of the enriched glycated proteins (Gene list S1) by utilizing a Gene Ontology enRIchment anaLysis and visuaLizAtion tool (GOrilla).57 The results of the analysis, presented in Figure 3E, display a significant presence ofnuclear regulatory systems including chromatin remodeling-and DNA-binding proteins.
Collectively, these findings support the enrichment utility of the 5-AR probe in combination with DBCO Cy5 fluorescent labeling and the resulting specific enrichment of histones. The proteomic analyses placed histones among prime glycation targets, however, unlike in the fluorescence analyses, histones were not among the most enriched proteins in the proteomic datasets. We believe this is primarily due to sample processing bias. Since histones are highly enriched in lysines and arginines, upon trypsin treatment, most of the fragments formed are too short to be captured by a reverse phase column and thus detected by the mass spectrometry.58 In order to target specific tail modifications, specialized sample processing techniques that involve the chemical treatment of lysines prior to trypsin digestion, must be used during sample preparation.59 However, such treatments are incompatible with the general proteome as the peptides generated will be too long for subsequent mass spectrometry analyses. It is thus notable that despite these technical constraints surrounding the detection of core histones, our data indicates a significant representation of all the core histones. In this case, we speculate that of the histones that were enriched via N-terminal glycation, upon trypsin digestion, only their globular (less lysine/arginine-rich) domains were detectable. Finally, the presence of proteins from several important regulatory systems in the enriched set supports the motivation to further investigate the effects of ribose glycation.
Ribose Glycation is a Reversible Mark.
While glycation is a nonenzymatic modification, cellular enzymatic mechanisms have evolved to regulate glycation levels. The functions of these enzymes range from scavengers (Glo1/2), deglycases (DJ-1), or rewriters (peptidyl arginine deiminase, type IV, or PAD4).11,15,44 Another known regulatory enzyme is the kinase FN3K which was shown to catalyze the phosphorylation of C-3 hydroxyls of hexosamines, resulting in spontaneous degradation yielding a free amine and a reactive 3-deoxy-hexosone species.60
Recently, the glucose deglycation activity of FN3K has been linked to the oncogenic activity of NRF2 in vivo.51 We thus sought to investigate whether early Az-RiboLys modifications are substrates of FN3K in its physiological context. We overexpressed FN3K in 293T cells for 24 h, before adding 5 mM of 5-AR for an additional 12 h. After washing off the excess sugar, cells were harvested and fractionated to nuclei and cytosolic fractions. Both fractions were fluorescently labeled with DBCO Cy5, separated by SDS-PAGE, and visualized by Cy5 fluorescence or western blot analyses, with anti-actin and anti-H3, to verify overexpression. Our analysis, presented in Figure 4A, shows decreased glycation signals in FN3K-overexpression samples in the cytosolic fractions but not the nuclear histones. This finding is consistent with the fact that FN3K is known to localize to mitochondria and cytosolic cellular fractions, where it is active.51 We also observe, via their fluorescence intensities, that while the majority of cytosolic glycation decreases with FN3K overexpression, some proteins seem to be more glycated. We speculate that this may be due to secondary protein expression effects associated with FN3K overexpression. Future experiments analyzing the total glycated proteome and the changes in its composition following FN3K overexpression and knockout could uncover the potential mechanisms behind these observations. Collectively, these data validate the 5-AR probe as a viable method for tracking FN3K deglycation activity. And finally, while FN3K is not a histone ribose deglycase, this in cellulo data encourages the idea that other similar regulatory proteins may exist (Figure 4B).
Figure 4.
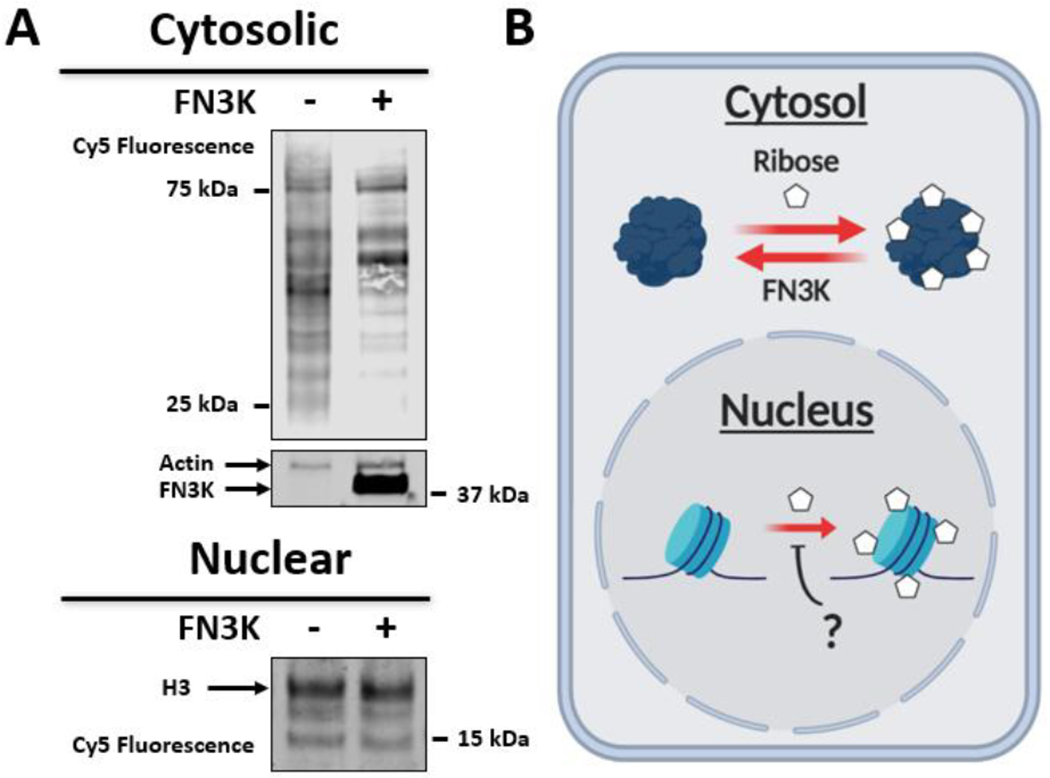
FN3K deglycates 5-AR-glycated cytosolic proteins in live cells. (A) Cytosolic and nuclear fractions of FN3K-transfected and wild-type 293T cells after 5-AR glycation and DBCO Cy5 labeling. (B) Model depicting cytosolic but not nuclear ribose deglycation activity of FN3K.
CONCLUSIONS
Here, we have designed and synthesized a pair of ribose analog probes to track ribose glycation in cells. The active 5-AR probe contains an azide instead of a hydroxyl at the C-5 position for visualization and enrichment utilities and the inactive 1-AR probe has an azide attached at the C-1 position, abating its capacity to isomerize but retaining its ability to partake in any click chemistry. Via bioorthogonal copper-free click chemistry, we validate the utility of the probes to visualize histone glycation in vitro and in cellulo and find that histones are selectively labeled. We determine by mass-spectrometry analysis that, like ribose, the azidoribose probe can form a ketoamine adduct on lysine side-chains in live cells. Additionally, we demonstrate that ribose-glycation competes with some enzymatic writers but not others for histone-tail lysine side chains, exposing an underlying degree of selectivity. Thereafter, we utilized the probe pair to enrich nuclear ribose-glycated proteins, which we identified via label-free quantitative proteomics. Using the proteomics data and subsequent immunoblot validation, we uncover that several proteins involved in vital transcription regulatory pathways, including chromatin remodeling and DNA repair, are glycated by ribose in live cells. Finally, we show that the kinase FN3K deglycates 5-AR-modified cytosolic proteins but not nuclear proteins. This work collectively is a testament to the utility of the developed probes to monitor and isolate ribose glycation adducts, through a stabilized ketoamine intermediate, as well as to assess endogenous regulatory repair proteins in cellulo.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Søren Heissel, Ph.D., Dr. Hanan Alwaseem, Ph.D., and Dr. Henrik Molina, Ph.D. of the Rockefeller Proteomics Resource Center for their invaluable assistance in LFQ data collection and analysis and Dr. George Sukenick and Rong Wang of the MSK Analytical Core Facility for expert NMR and mass spectral support. Work in the David lab is supported by the Josie Robertson Foundation, the Pershing Square Sohn Cancer Research Alliance, the NIH (CCSG core grant P30 CA008748, MSK SPORE P50 CA192937, and R21 DA044767), the Parker Institute for Cancer Immunotherapy (PICI), and the Anna Fuller Trust. In addition, the David lab is supported by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center. Elements of figures were prepared with BioRender.
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at:
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.0c01325. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE repository with the dataset identifier PXD019204.61
Complete experimental procedures and characterization data for all new compounds (PDF)
Raw proteomics dataset S1 (XLSX)
Raw proteomics dataset S2 (XLSX)
Contributor Information
Igor Maksimovic, Tri-Institutional PhD Program in Chemical Biology, New York, New York 10065, United States; Chemical Biology Program, Memorial Sloan Kettering Cancer Center, New York, New York 10065, United States.
Qingfei Zheng, Chemical Biology Program, Memorial Sloan Kettering Cancer Center, New York, New York 10065, United States.
Marissa N. Trujillo, Department of Pharmaocology and Toxicology, College of Pharmacy, University of Arizona, Tucson, Arizona 85721, United States
James J. Galligan, Department of Pharmaocology and Toxicology, College of Pharmacy, University of Arizona, Tucson, Arizona 85721, United States.
Yael David, Tri-Institutional PhD Program in Chemical Biology, New York, New York 10065, United States; Chemical Biology Program, Memorial Sloan Kettering Cancer Center, New York, New York 10065, United States; Department of Pharmacology and Department of Physiology, Biophysics and Systems Biology, Weill Cornell Medicine, New York, New York 10065, United States.
REFERENCES
- (1).Schvartzman JM; Thompson CB; Finley LWS Metabolic regulation of chromatin modifications and gene expression. J. Cell Biol 2018, 217 (7), 2247–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cloos PA; Christgau S Non-enzymatic covalent modifications of proteins: mechanisms, physiological consequences and clinical applications. Matrix Biol. 2002, 21 (1), 39–52. [DOI] [PubMed] [Google Scholar]
- (3).Harmel R; Fiedler D Features and regulation of non-enzymatic post-translational modifications. Nat. Chem. Biol. 2018, 14 (3), 244–252. [DOI] [PubMed] [Google Scholar]
- (4).Hellwig M; Henle T Baking, ageing, diabetes: a short history of the Maillard reaction. Angew. Chem., Int. Ed 2014, 53 (39), 10316–29. [DOI] [PubMed] [Google Scholar]
- (5).Wang T; Kartika R; Spiegel DA Exploring post-translational arginine modification using chemically synthesized methylglyoxal hydroimidazolones. J. Am. Chem. Soc. 2012, 134 (21), 8958–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Huebschmann AG; Regensteiner JG; Vlassara H; Reusch JE Diabetes and advanced glycoxidation end products. Diabetes Care 2006, 29 (6), 1420–32. [DOI] [PubMed] [Google Scholar]
- (7).Ang SH; Thevarajah M; Alias Y; Khor SM Current aspects in hemoglobin A1c detection: a review. Clin. Chim. Acta 2015, 439, 202–11. [DOI] [PubMed] [Google Scholar]
- (8).Sims GP; Rowe DC; Rietdijk ST; Herbst R; Coyle AJ HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 2010, 28, 367–88. [DOI] [PubMed] [Google Scholar]
- (9).Hemmler D; Roullier-Gall C; Marshall JW; Rychlik M; Taylor AJ; Schmitt-Kopplin P Insights into the Chemistry of Non-Enzymatic Browning Reactions in Different Ribose-Amino Acid Model Systems. Sci. Rep. 2018, 8 (1), 16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Vallejo-Cordoba B; Gonzalez-Cordova AF CE: a useful analytical tool for the characterization of Maillard reaction products in foods. Electrophoresis 2007, 28 (22), 4063–71. [DOI] [PubMed] [Google Scholar]
- (11).Richarme G; Mihoub M; Dairou J; Bui LC; Leger T; Lamouri A Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem. 2015, 290 (3), 1885–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Suji G; Sivakami S Glucose, glycation and aging. Biogerontology 2004, 5 (6), 365–73. [DOI] [PubMed] [Google Scholar]
- (13).Zheng Q; Omans ND; Leicher R; Osunsade A; Agustinus AS; Finkin-Groner E; D’Ambrosio H; Liu B; Chandarlapaty S; Liu S; David Y Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun. 2019, 10 (1), 1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Galligan JJ; Wepy JA; Streeter MD; Kingsley PJ; Mitchener MM; Wauchope OR; Beavers WN; Rose KL; Wang T; Spiegel DA; Marnett LJ Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (37), 9228–9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nokin MJ; Durieux F; Peixoto P; Chiavarina B; Peulen O; Blomme A; Turtoi A; Costanza B; Smargiasso N; Baiwir D; Scheijen JL; Schalkwijk CG; Leenders J; De Tullio P; Bianchi E; Thiry M; Uchida K; Spiegel DA; Cochrane JR; Hutton CA; De Pauw E; Delvenne P; Belpomme D; Castronovo V; Bellahcene A Methylglyoxal, a glycolysis side-product, induces Hsp90 glycation and YAP-mediated tumor growth and metastasis. eLife 2016, 5, 19375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Prescher JA; Bertozzi CR Chemistry in living systems. Nat. Chem. Biol. 2005, 1 (1), 13–21. [DOI] [PubMed] [Google Scholar]
- (17).Sletten EM; Bertozzi CR Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem., Int. Ed. 2009, 48 (38), 6974–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bertozzi CR; Kiessling LL Chemical glycobiology. Science 2001, 291 (5512), 2357–64. [DOI] [PubMed] [Google Scholar]
- (19).Zheng Q; Maksimovic I; Upad A; Guber D; David Y Synthesis of an Alkynyl Methylglyoxal Probe to Investigate Non-enzymatic Histone Glycation. J. Org. Chem. 2020, 85, 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Galligan JJ; Marnett LJ Histone Adduction and Its Functional Impact on Epigenetics. Chem. Res. Toxicol. 2017, 30 (1), 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Commerford SL; Carsten AL; Cronkite EP Histone turnover within nonproliferating cells. Proc. Natl. Acad. Sci. U. S. A. 1982, 79 (4), 1163–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bartholomew B Regulating the chromatin landscape: structural and mechanistic perspectives. Annu. Rev. Biochem. 2014, 83, 671–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jenuwein T; Allis CD Translating the histone code. Science 2001, 293 (5532), 1074–80. [DOI] [PubMed] [Google Scholar]
- (24).Han C; Lu Y; Wei Y; Liu Y; He R D-ribose induces cellular protein glycation and impairs mouse spatial cognition. PLoS One 2011, 6 (9), No. e24623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wei Y; Han CS; Zhou J; Liu Y; Chen L; He RQ D-ribose in glycation and protein aggregation. Biochim. Biophys. Acta, Gen. Subj. 2012, 1820 (4), 488–94. [DOI] [PubMed] [Google Scholar]
- (26).Lee DY; Chang GD Methylglyoxal in cells elicits a negative feedback loop entailing transglutaminase 2 and glyoxalase 1. Redox Biol. 2014, 2, 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Patra KC; Hay N The pentose phosphate pathway and cancer. Trends Biochem. Sci. 2014, 39 (8), 347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Talasz H; Wasserer S; Puschendorf B Nonenzymatic glycation of histones in vitro and in vivo. J. Cell. Biochem. 2002, 85 (1), 24–34. [PubMed] [Google Scholar]
- (29).Zheng L; Nagar M; Maurais AJ; Slade DJ; Parelkar SS; Coonrod SA; Weerapana E; Thompson PR Calcium Regulates the Nuclear Localization of Protein Arginine Deiminase 2. Biochemistry 2019, 58 (27), 3042–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Quesada-Moreno MM; Azofra LM; Aviles-Moreno JR; Alkorta I; Elguero J; Lopez-Gonzalez JJ Conformational preference and chiroptical response of carbohydrates D-ribose and 2-deoxy-D-ribose in aqueous and solid phases. J. Phys. Chem. B 2013, 117 (47), 14599–614. [DOI] [PubMed] [Google Scholar]
- (31).Harvey SC; Prabhakaran M Ribose puckering: structure, dynamics, energetics, and the pseudorotation cycle. J. Am. Chem. Soc. 1986, 108 (20), 6128–6136. [Google Scholar]
- (32).Wei Y; Chen L; Chen J; Ge L; He RQ Rapid glycation with D-ribose induces globular amyloid-like aggregations of BSA with high cytotoxicity to SH-SY5Y cells. BMC Cell Biol. 2009, 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Vocadlo DJ; Hang HC; Kim EJ; Hanover JA; Bertozzi CR A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (16), 9116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kho Y; Kim SC; Jiang C; Barma D; Kwon SW; Cheng J; Jaunbergs J; Weinbaum C; Tamanoi F; Falck J; Zhao Y A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (34), 12479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sprung R; Nandi A; Chen Y; Kim SC; Barma D; Falck JR; Zhao Y Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 2005, 4 (3), 950–7. [DOI] [PubMed] [Google Scholar]
- (36).Kostiuk MA; Corvi MM; Keller BO; Plummer G; Prescher JA; Hangauer MJ; Bertozzi CR; Rajaiah G; Falck JR; Berthiaume LG Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J. 2008, 22 (3), 721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Martin DD; Vilas GL; Prescher JA; Rajaiah G; Falck JR; Bertozzi CR; Berthiaume LG Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 2008, 22 (3), 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liu C; Yu X ADP-ribosyltransferases and poly ADP-ribosylation. Curr. Protein Pept. Sci. 2015, 16 (6), 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Novogrodsky A; Hurwitz J The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid. I. Phosphorylation at 5’-hydroxyl termini. J. Biol. Chem. 1966, 241 (12), 2923–32. [PubMed] [Google Scholar]
- (40).Jones ME Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 1980, 49, 253–79. [DOI] [PubMed] [Google Scholar]
- (41).Chien TC; Chern JW A convenient preparation of 1,2,3-tri-O-acetyl-beta-D-ribofuranose by enzymatic regioselective 5-O-deacetylation of the peracetylated ribofuranose. Carbohydr. Res. 2004, 339 (6), 1215–7. [DOI] [PubMed] [Google Scholar]
- (42).Ebner M; Arnold S Glucose isomerase catalysed isomerisation reactions of (2 R,3 R)-configured aldofuranoses into the corresponding open-chain 2-ketoses. Carbohydr. Res. 1997, 305 (3–4), 331–336. [Google Scholar]
- (43).El Akri K; Bougrin K; Balzarini J; Faraj A; Benhida R Efficient synthesis and in vitro cytostatic activity of 4-substituted triazolyl-nucleosides. Bioorg. Med. Chem. Lett. 2007, 17 (23), 6656–9. [DOI] [PubMed] [Google Scholar]
- (44).Zheng Q; Adewola O; David Y Protein Arginine Deiminase 4 Antagonizes Methylglyoxal-Induced Histone Glycation. bioRxiv 2019, 826818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (43), 16793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Dignam JD; Lebovitz RM; Roeder RG Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983, 11 (5), 1475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Galligan JJ; Kingsley PJ; Wauchope OR; Mitchener MM; Camarillo JM; Wepy JA; Harris PS; Fritz KS; Marnett LJ Quantitative Analysis and Discovery of Lysine and Arginine Modifications. Anal. Chem. 2017, 89 (2), 1299–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ren Q; Gorovsky MA Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell 2001, 7 (6), 1329–35. [DOI] [PubMed] [Google Scholar]
- (49).Carrozza MJ; Utley RT; Workman JL; Cote J The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003, 19 (6), 321–9. [DOI] [PubMed] [Google Scholar]
- (50).Kolasinska-Zwierz P; Down T; Latorre I; Liu T; Liu XS; Ahringer J Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009, 41 (3), 376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Sanghvi VR; Leibold J; Mina M; Mohan P; Berishaj M; Li Z; Miele MM; Lailler N; Zhao C; de Stanchina E; Viale A; Akkari L; Lowe SW; Ciriello G; Hendrickson RC; Wendel HG The Oncogenic Action of NRF2 Depends on De-glycation by Fructosamine-3-Kinase. Cell 2019, 178 (4), 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Bollong MJ; Lee G; Coukos JS; Yun H; Zambaldo C; Chang JW; Chin EN; Ahmad I; Chatterjee AK; Lairson LL; Schultz PG; Moellering RE A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature 2018, 562 (7728), 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Cox J; Hein MY; Luber CA; Paron I; Nagaraj N; Mann M Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13 (9), 2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Pasini D; Bracken AP; Helin K Polycomb group proteins in cell cycle progression and cancer. Cell Cycle 2004, 3 (4), 396–400. [PubMed] [Google Scholar]
- (55).Smith-Roe SL; Nakamura J; Holley D; Chastain PD 2nd; Rosson GB; Simpson DA; Ridpath JR; Kaufman DG; Kaufmann WK; Bultman SJ SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget 2015, 6 (2), 732–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Research Watch. Deglycation of NRF2 by FN3K Promotes Oncogenesis and Drug Resistance. Cancer Discovery 2019, 9 (10), OF8. [Google Scholar]
- (57).Eden E; Navon R; Steinfeld I; Lipson D; Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 2009, 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Karch KR; Denizio JE; Black BE; Garcia BA Identification and interrogation of combinatorial histone modifications. Front. Genet. 2013, 4, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Huang H; Lin S; Garcia BA; Zhao Y Quantitative Proteomic Analysis of Histone Modifications. Chem. Rev. 2015, 115 (6), 2376–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Szwergold BS; Howell S; Beisswenger PJ Human fructosamine-3-kinase: purification, sequencing, substrate specificity, and evidence of activity in vivo. Diabetes 2001, 50 (9), 2139–47. [DOI] [PubMed] [Google Scholar]
- (61).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


