Abstract
The plasmid-encoded colistin resistance gene mcr-1 challenges the use of polymyxins and poses a threat to public health. Although IncI2-type plasmids are the most common vector for spreading the mcr-1 gene, the mechanisms by which these plasmids adapt to host bacteria and maintain resistance genes remain unclear. Herein, we investigated the regulatory mechanism for controlling the fitness cost of an IncI2 plasmid carrying mcr-1. A putative ProQ/FinO family protein encoded by the IncI2 plasmid, designated as PcnR (plasmid copy number repressor), balances the mcr-1 expression and bacteria fitness by repressing the plasmid copy number. It binds to the first stem-loop structure of the repR mRNA to repress RepA expression, which differs from any other previously reported plasmid replication control mechanism. Plasmid invasion experiments revealed that pcnR is essential for the persistence of the mcr-1-bearing IncI2 plasmid in the bacterial populations. Additionally, single-copy mcr-1 gene still exerted a fitness cost to host bacteria, and negatively affected the persistence of the IncI2 plasmid in competitive co-cultures. These findings demonstrate that maintaining mcr-1 plasmid at a single copy is essential for its persistence, and explain the significantly reduced prevalence of mcr-1 following the ban of colistin as a growth promoter in China.
INTRODUCTION
Horizontal gene transfer (HGT) refers to the transfer of genes between unrelated species, which increases genetic diversity and accelerates bacterial evolution (1,2). Conjugative plasmids are typical representatives of HGT and promote the spread of antibiotic resistance among pathogens, which has emerged as an important public health issue worldwide over the past decades (3–5). The acquired antibiotic-resistant plasmid can impose a fitness cost on the host bacteria, possibly because of the metabolic burden introduced by plasmid replication and plasmid-encoded gene expression (6,7). It, therefore, seems reasonable to assume that plasmids become lost over time in the absence of positive selection. However, laboratory studies have shown that epidemic antibiotic resistance plasmids can stably persist in host bacteria for long periods, even in the absence of antibiotics (8–10), indicating that these plasmids are well-adapted to the host bacteria following the amelioration of plasmid cost. However, the molecular mechanism through which antibiotic resistance plasmids have evolved to ameliorate the fitness cost is poorly understood. Several evolution studies have demonstrated that the reduction in plasmid cost can be attributed to compensatory mutations in the host genome or plasmid. For example, mutations in chromosomal genesgacA/gacS, which are involved in regulation of secondary metabolism, can reduce the fitness costs associated with the pQBR103 plasmid in Pseudomonas fluorescens, via decreased expression of both plasmid and chromosomal genes (11). Moreover, conjugative plasmids use specific regulators, such as the histone-like nucleoid structuring protein H-NS, to improve the plasmid fitness by controlling the energetically expensive conjugative process (12). Thus, the persistence of antibiotic resistance plasmids largely depends on the ability of host bacteria-plasmid co-adaption for minimizing the plasmid cost. Therefore, to understand the mechanisms of persistence for plasmid-mediated resistance, it is crucial to understand the molecular basis of the fitness of antibiotic resistance plasmids.
The emergence and spread of the plasmid-mediated colistin resistance genemcr-1 has created great challenges for public health (13,14). Epidemiological studies have revealed that mcr-1 has disseminated across in animals, humans, foods, and the environment in >50 countries and regions via plasmids (15). IncI2-type plasmids are described as the most epidemiological successful vectors for spreadingmcr-1 worldwide (16,17). In addition to mcr-1, IncI2 plasmids carry other clinically relevant resistance genes, such asblaCTX-M-55 (18) andblaKPC-3 (19). However, very little is known about the basic biology of IncI2 plasmids. Notably, the mechanisms underlying how IncI2 plasmids adapt to host bacteria and maintain resistance genes, particularly mcr-1, remain poorly understood. Our previous study showed that the first mcr-1-positive IncI2-type plasmid, pHNSHP45, conferred the greatest fitness advantage, rather than a cost, to the host bacteria among epidemic plasmids carrying mcr-1 (20). This indicates that the IncI2 plasmid contributes to the pervasive spread of mcr-1, in part due to the fitness advantage it bestowed on the host bacteria. pHNSHP45 also encodes a putative parA partitioning system and three putative toxin-antitoxin modules, relE/relB, hicA/hicB and hok/sok, for stable maintenance (13,21).
In this study, we investigated the regulatory mechanism controlling the fitness cost of the mcr-1-bearing IncI2 plasmids based on plasmid-encoded regulators. Our results reveal a novel plasmid-related adaptative mechanism that contributes to the persistence of IncI2 plasmids carrying mcr-1 in nature for long periods of time.
MATERIALS AND METHODS
Bacterial strains and bacterial growth
The bacterial strains used in this study are listed in Supplementary Table S1. All bacterial strains were cultured in LB broth in 37°C with shaking (250 rpm) or on LB agar plates at 37°C. Plasmid pHNSHP45 (GenBank accession number: KP347127) was introduced into Escherichia coli BW25113 by conjugation using E. coli C600/pHNSHP45 as the donor (13). The strains carrying pHNSHP45 and its derivatives (n = 3) were grown in LB broth overnight at 37°C, then diluted 1:200 to fresh medium and shaking (250 rpm) at 37°C for 16 h. The optical density at 600 nm (OD600) was measured every hour by Multiskan spectrum microplate spectrophotometer (Thermo Labsystems, Frankin, MA, USA).
Bacterial conjugation assays
The transfer frequency of pHNSHP45 and its derivatives were investigated by conjugation experiments using streptomycin-resistant E. coli C600 as recipients. Transconjugants were selected on LB agar plates supplemented with colistin and streptomycin. Transfer frequencies were calculated as the number of transconjugants per recipient. The data was assessed by two-tailed t-test.
Construction of plasmids and strains
Plasmids and primers used in this study are listed in Supplementary Table S2 and Supplementary Table S3. Complementation vectors were derived from pHSG575 and pBAD24M vectors. The pcnR and mcr-1 genes with their native promoter were amplified from pHNSHP45 using primers pro-pcnR-F/pro-pcnR-R and pro-mcr-1-F/pro-mcr-1-R, respectively, and cloned into pHSG575 to generate pHSG575-pcnR and pHSG575-mcr-1. The finO gene with native promoter was amplified from plasmid pHN7A8 (GenBank accession number: JN232517) using primers pro-finO-F/pro-finO-R and cloned into pHSG575 to generate pHSG575-finO. The ORF of pcnR was amplified from pHNSHP45 using primers pcnR-F/pcnR-R and cloned into pBAD24M to generate pBAD24M-pcnR. The ORF of pcnR was cloned into pET28b to generate an expression cassette in which the 6his-tag and thrombin recognition site were fused at the N-terminus of PcnR, and the resulting vector was named pET28b-pcnR-his. The antisense RNA (AS RNA) fragment with its promoter was amplified from pHNSHP45 using primers AS-RNA-F/AS-RNA-R and cloned into pUC19 to generate pUC19-AS RNA. The fragment containing the upstream region of repA (−497 to +45) was amplified from pHNSHP45 using primers repA-F/repA-R and cloned into the HindIII site of pHGR01 which contains a promoterless lacZ gene to produce PrepA-lacZ. Based on PrepA-lacZ, a stop codon was introduced into repR ORF by overlap PCR using primers repA-F/repR-stop-R and repR-stop-F/repA-R. Other mutations including ΔAS RNA, stem mutation, loop mutation, repR9 fusion and repR24 fusion were also based on PrepA-lacZ. Base mutation (CA-TT) was introduced into the –35 region of antisense RNA promoter by primers repA-F/CA-TT-R and CA-TT-F/repA-R to abolish the expression of antisense RNA (AS RNA), and this mutation was named ΔAS RNA. Base mutation (GAAA-CTCT) was introduced into repR gene using primers repA-F/GAAA-CTCT-R and GAAA-CTCT-F/repA-R to eliminate the stem structure of repR mRNA, which was named stem mutation (GAAA-CTCT). Loop mutation (GA-TT) was created in the same way as stem mutation (GAAA-CTCT) by primers repA-F/GA-TT-R and GA-TT-F/repA-R. The repR9 fusion and repR24 fusion were constructed by primers repA-F/repR9-R and primers repA-F/repR24-R and were cloned into the HindIII site of pHGR01 plasmid.
Deletion mutants of pHNSHP45 were constructed by λRed recombination system (22). The pHNSHP45 derivatives pHNSHP45ΔpcnR, pHNSHP45Δorf00050, pHNSHP45Δorf00039 using primers knock-pcnR-F/knock-pcnR-R, knock-orf50-F/ knock-orf50-R, knock-orf39-F/knock-orf39-R and pKD4 as templates, respectively. Deletion of mcr-1 was done on pHNSHP45 and pHNSHP45ΔpcnR using primers knock-mcr-1-F/knock-mcr-1-R. Deletion of gadW, gapdh, loiP, ack, hf, arnT, and tf were done using primers knock-gadW-F/knock-gadW-R, knock-gapdh-F/knock-gapdh-R, knock-loip-F/knock-loip-R, knock-ack-F/knock-ack-R, knock-hf-F/knock-hf-R, knock-arnT-F/knock-arnT-R and knock-tf-F/knock-tf-R, respectively.
Transposon library construction and transposon site identification
The transposon library was constructed by conjugation using a mariner-based transposon TnSC189. The donor strain SM10λpir/pSC189 (23) and recipient BW25113/pHNSHP45ΔpcnR were grown in 5 ml LB broth overnight at 37°C, respectively. The cell pellets of donor and recipient were collected by centrifugation and resuspend by 300 μl fresh LB broth. Then, donor and recipient cells were mixed in an appropriate ratio (1:2), dropped on LB agar plate and allowed to mate at 37°C for 3–6 h. The mating was then scraped up, suspended in fresh LB broth, and plated on LB agar plate containing colistin (2 μg/ml) and kanamycin (30 μg/ml) after appropriate dilution. After overnight culture at 37°C, the larger size colonies were pick up for bacteria growth assay.
Growth restored mutants were grown in 5 ml LB broth containing kanamycin (30 μg/ml) overnight at 37°C. Genomic DNA was amplified by thermal asymmetric interlaced PCR (TAIL-PCR) (24) to identify the transposon insertion sites. The special primers (SP) and arbitrary degenerate (AD) primers are listed in Supplementary Table S3. PCR procedures for TAIL-PCR refer to Tatjana Singer et al. (24). PCR products were purified for Sanger sequencing. The genome DNA of Tns1, Tns2, Tns3, Tns4 and Tns7 were chosen to perform whole-genome sequencing (WGS) (Illumina, USA) and Nanopore sequencing (Oxford Nanopore TechnologiesTM, UK). WGS reads and Nanopore reads were combined to produce a de novo hybrid assembly using Unicycler version 0.4.3 (25).
Electrophoretic mobility shift assay (EMSA)
Escherichia coli BL21(DE3) cells harbouring pET28b-pcnR-his were grown to OD600 of ∼0.5, then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) overnight. Cell pellets were resuspended in 20 ml of lysis buffer (150 mM NaCl, 5 mM imidazole, 50 mM potassium phosphate, pH 6.5) and lysed by sonication. The lysates were centrifuged at 12 000 g for 10 min at 4°C, and supernatant were collected. Purification of His-tag-PcnR was done by Ni-NTA affinity chromatography following the manufacturer's instructions (Qiagen, Germany). Briefly, the cleared lysate was mixed with 4 mL of the 50% Ni-NTA slurry in a column by shaking at 4°C for 2 h. The column was subsequently washed with washing buffer (150 mM NaCl, 10 mM imidazole, 50 mM potassium phosphate, pH 6.5) until OD280 was < 0.1. Then, His-tag-PcnR was eluted with elution buffer (150 mM NaCl, 250 mM imidazole, 50 mM potassium phosphate, pH 6.5). Fractions containing His-tag-PcnR were pooled and the buffer replaced with a solution of 150 mM NaCl, 50 mM Tris–HCl (pH 7.3) by ultrafiltration. Then, the purified His-tag-PcnR protein was digested by thrombin (0.3 U thrombin per 100 μg His-tag-PcnR) at 20°C for ∼12 h to cleave His-tag from PcnR. PcnR was purified from the digested product by cation exchange chromatography. The digested product was loaded onto a CM-Sephadex column and PcnR was eluted with a stepwise gradient of NaCl (0, 0.1, 0.2, 0.4, 0.6, 0.8 and 1M NaCl prepared in 50 mM Tris–HCl, pH7.3). The fractions containing PcnR were pooled and dialyzed against 50 mM Tris–HCl (pH7.3) with 150 mM NaCl, 10% glycerol. Protein was detected by 15% SDS-PAGE. Protein concentrations were determined by Bradford Protein Assay Kit (Takara, Japan), and bovine serum albumin as the standard.
The antisense RNA (AS RNA) were prepared by in vitro transcription reaction using bacteriophage T7 RNA polymerase (NEB, UK) with a DNA template –173 to –68 of repA. The RNA products were purified by RNA Clean Up Kit (Omega, USA) according to manufacturer's instructions. The first SL structure of repR mRNA (5′CUGAAGUGGAGUUUAAUUCCAUGAGAAGGAAAUAUCAACUCUUACAG3′) was synthesized by Sangon Biotech (Sangon Biotech, Inc., China). The EMSA experiments were performed using the Electrophoretic Mobility-Shift Assay (EMSA) kit according to the manufacturer's instructions (Thermo Fisher Scientific, USA). Briefly, the binding experiment was performed on ice in 10 μl reaction mixtures based on binding reaction buffer (150 mM KCl, 0.1 mM EDTA, 0.1 mM dithiothreitol, 10 mM Tris, pH7.4). An appropriate amount of RNA (50 ng) was mixed with different concentrations of PcnR protein and incubated for 30 min at 25°C. Then 2 μl of 6 × EMSA gel-loading solution was added and subjected to 6% nondenaturing PAGE (25 min at 220 V). Gels were stained by SYBR® Green and imaged using a Bio-Rad Molecular Imager Chemi Doc XRS+ Imaging System.
Rapid amplification of cDNA 5′ ends (5′RACE)
5′RACE was used to identify the transcription starts sites of repA and antisense RNA. The total RNA of strain BW25113/pHNSHP45 was extracted using HiPure Bacterial RNA Kit (Magen, China), and purified RNA samples were subjected to gDNA digestion using DNase On Column Kit (Magen, China) following the manufacturer's instructions. RNA integrity was checked on 1% agarose gels and quantified using NanoDrop (Thermo Scientific, USA). Reverse transcription was performed on 1 μg total RNA with special primers RepA-TSS or AS RNA-TSS using Goldenstar RT6 cDNA Synthesis Mix (TsingKe Biotech. China). The cDNA products were purified by MagPure cDNA Clean Up Kit (Magen, China). Purified cDNA was tailed by poly dC using terminal transferase (NEB, UK). Primer Race-TSS which contain poly dG and special primer RepA-TSS or AS RNA-TSS were used to perform PCR, and tailed cDNA as the template. Finally, the PCR products were cloned to pMD19T for Sanger sequencing.
Quantitative real-time PCR
The copy numbers of plasmids and mcr-1 gene were measured by qPCR. The repA and reference rpoB genes were cloned into pHSG575 to generate pHSG575-repA and pHSG575-rpoB. Plasmids pHSG575-repA, pHSG575-rpoB and pHSG575-mcr-1 were used as template DNA with primers qrepA-F/qrepA-R, qrpoB-F/qrpoB-R and qmcr-1-F/qmcr-1-R, respectively, to establish the corresponding standard curve for determining the copy numbers, respectively. The genomic DNA of BW25113/pHNSHP45, BW25113/pHNSHP45ΔpcnR, and transposon mutants was extracted using HiPure Bacterial DNA Kit (Magen, China), and were used as template with primers described above to perform qPCR. Copy numbers of plasmids and mcr-1 per cell could be calculated as repA/rpoB and mcr-1/rpoB ratios, respectively.
Expression of mcr-1 was detected by real-time reverse transcription PCR (RT-qPCR). Total RNA was prepared as described in 5′RACE assay. Reverse transcription was performed on 1 μg total RNA using Goldenstar RT6 cDNA Synthesis Mix (TsingKe Biotech. China). Primers qmcr-1-F/qmcr-1-R were used to perform qPCR, and GAPDH expression level was used as internal control using primers GAPDH-F/GAPDH-R. Relative expression values were obtained by the 2–△△Ct method (26).
Competition experiments in vitro and plasmid invasion assays
Competition experiments were used to measure the relative fitness of BW25113/pHNSHP45ΔpcnR::kan, BW25113/pHNSHP45Δorf00050::kan, BW25113/pHNSHP45Δorf00039::kan, BW25113/pHNSHP45ΔpcnRΔmcr-1::kan, BW25113/pHNSHP45ΔpcnR::kan-pHSG575-pcnR, BW25113/pHNSHP45Δmcr-1 and BW25113/pHNSHP45ΔpcnRΔmcr-1::kan-pHSG575-mcr-1. These strains were competed against BW25113/pHNSHP45. All competitions were carried out with three biological replicates. Overnight cultures were diluted 1:1000 in LB broth and mixed at 1:1 ratio. The mixture was then incubated for 24 h, and then the mixed population was 1:100 diluted into fresh LB broth. This procedure was repeated until the competition experiment had lasted for 72 h. The competition mixture at 0, 24, 48, 72 h were plated on LB agar containing colistin or kanamycin with properly dilution to count the colony number. The formula RF = ln(Nf, S1/Ni, S1)/ln (Nf, S2/Ni, S2) was used to calculate the relative fitness, where RF is the relative fitness of S1strain (compared to S2 strain), Ni, S1 and Nf, S1 are the densities of cells of S1 at the beginning and end of the competition, and Ni, S2 and Nf, S2 are the densities of cells of S2 at the beginning and end of the competition, respectively.
Plasmid invasion assays were carried out according to the method of previous study with some modifications (27). All invasion assays were carried out with three biological replicates. Briefly, overnight cultures of BW25113 were diluted 1:100 into 2ml LB broth and mixed with 1:100 000 dilutions of BW25113/pHNSHP45, BW25113/pHNSHP45ΔpcnR::kan, BW25113/pHNSHP45Δmcr-1::kan or BW25113/pHNSHP45ΔpcnRΔmcr-1::kan individually. Competitive co-culture experiments were done as similar steps described above. Cultures grew in 50 ml tubes containing 2 ml LB broth at 37°C, aerated by slow rolling (80 rpm), and diluted 100-fold into fresh LB broth every 24 for 120 h. Viable counts were determined at 24, 48, 72, 96, 120 h. For each time point, dilutions of the cultures were plated to antibiotic-free, kanamycin-containing and colistin-containing media. Cells containing pHNSHP45 grew on antibiotic-free plate and colistin-containing plates but were killed by kanamycin-containing media. Cells containing pHNSHP45ΔpcnR::kan grew on all plates. Cells containing pHNSHP45Δmcr-1::kan grew on antibiotic-free plate and kanamycin-containing media, but were killed by colistin-containing plate. Cells containing pHNSHP45ΔpcnRΔmcr-1::kan grew on kanamycin-containing plate but were killed by colistin-containing plate. For mixture containing BW25113, BW25113/pHNSHP45 and BW25113/pHNSHP45ΔpcnR::kan, colony counts from the three types of plates were used to calculate cell number by the following formula: BW25113 (plasmid-free) cells = antibiotic-free plate counts minus colistin-containing plate counts, BW25113/pHNSHP45 cells = colistin-containing plate counts minus kanamycin-containing plate counts, BW25113/pHNSHP45ΔpcnR::kan cells = kanamycin-containing plate counts. For mixture containing BW25113, BW25113/pHNSHP45 and BW25113/pHNSHP45ΔpcnRΔmcr-1::kan, colony counts from the three types of plates were used to calculate cell number by the following formula: BW25113 (plasmid-free) cells = antibiotic-free plate counts minus (colistin-containing plate counts plus kanamycin-containing plate counts), BW25113/pHNSHP45 cells = colistin-containing plate counts, and BW25113/ pHNSHP45ΔpcnRΔmcr-1::kan cells = kanamycin-containing plate counts. For mixture containing BW25113, BW25113/pHNSHP45 and BW25113/ pHNSHP45Δmcr-1::kan, colony counts from the three types of plates were used to calculate cell number by the following formula: BW25113 (plasmid-free) cells = antibiotic-free plate counts minus (colistin-containing plate counts plus kanamycin-containing plate counts), BW25113/pHNSHP45 cells = colistin-containing plate counts, and BW25113/ pHNSHP45Δmcr-1::kan cells = kanamycin-containing plate counts.
β-Galactosidase assay
β-Galactosidase assays were done according to Miller (28). Cells harbouring lacZ fusion vectors were cultured in LB broth containing kanamycin and/or chloramphenicol (ampicillin) overnight at 37°C. Overnight cultures were diluted 100-fold into LB broth containing antibiotics with or without 0.2% l-arabinose and shaking (250 rpm) at 37°C for 8 h. Cells were harvested and resuspended in lysis buffer (50 mM Tris–HCl pH 7.3, 1 mM DTT, 5% glycerol, 1 mM EDTA) and lysed by sonication. The lysates were centrifuged at 12 000 g for 10 min at 4°C, and supernatants were collected. Four microliters of the supernatant were taken for protein concentration determination using Bradford Protein Assay Kit (Takara, Japan). Fifty microliters of the supernatant were added directly to 0.95ml of Z buffer. Then 0.2 ml of 4 μg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) stock solution was added and incubated at room temperature. When color change was observed, the reactions were terminated by 500 μl of 1M Na2CO3. The optical density at 420 nm (OD420) was measured by Multiskan spectrum microplate spectrophotometer (Thermo Labsystems, Frankin, MA). The enzyme activity calculated by formula:
Miller units = OD420 × 1.7/(0.0045 × C × V × T), where C is the protein concentration of supernatant (μg/ml) and V is the volume of the added supernatant (ml), and T is the reaction time (min).
RESULTS
Identification of gene associated with the fitness of mcr-1-positive plasmid pHNSHP45
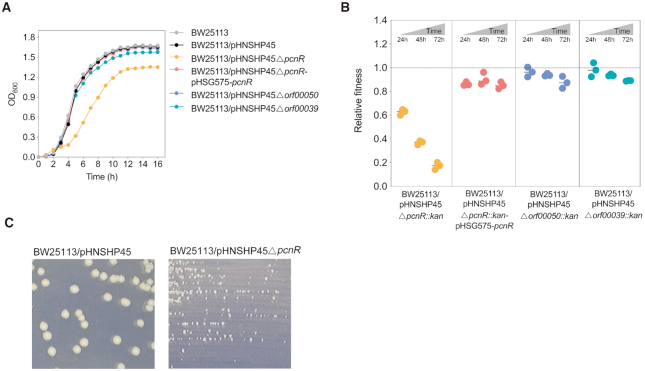
To understand the molecular basis of the fitness of mcr-1-positive IncI2 plasmid pHNSHP45 in E. coli, we analysed the structure of pHNSHP45 by comparative genomics. pHNSHP45 shares a large core set of genes between all members of the IncI2 group carrying mcr-1. Some conserved genes are involved in conjugative transfer and replication, but most have unknown functions (Supplementary Figure S1). Analysis of conserved genes of unknown function using Pfam database (29) revealed three putative transcriptional regulators (ORF00039, ORF00049 and ORF00050) (Supplementary Table S4). Previous studies showed that plasmid-encoded specific regulators, such as H-NS or PsiB, alleviate the plasmid cost by controlling the expression of certain genes carried by the plasmid or host bacteria (12,30). To determine whether these genes are involved in the regulation of pHNSHP45 fitness, we constructed in-frame deletions and determined the fitness cost of the resulting mutants by monitoring the growth curves of the mutants and performing competition experiments. Deletion of either orf00050, which codes for a putative CopG family protein, or orf00039, which codes for a putative CaiF/GrlA transcriptional regulator, had little effect on the growth and fitness of the host (Figure 1A and B). Interestingly, deletion of orf00049 (pcnR), which codes for a predicted ProQ/FinO family protein, delayed and reduced the growth of the host bacterium which produced smaller colonies (Figure 1A and C), and imposed a high fitness cost to the host (average relative fitness decreased from 0.626 to 0.173 within 72 h) (Figure 1B). Complementation of the ΔpcnR mutation by expressing the corresponding gene largely restored the growth and fitness of the host bacterium (Figure 1A and B), confirming that pcnR is essential for the fitness of pHNSHP45.
Figure 1.
Effects of pHNSHP45-encoded ProQ/FinO family protein PcnR on bacterial growth and fitness cost. (A) Effects of pHNSHP45 encoded putative regulators on bacterial growth. These data represent the mean of three independent experiments. (B) Relative fitness of E. coli BW25113 harbouring pHNSHP45Δorf00050::kan, pHNSHP45Δorf00039::kan, pHNSHP45ΔpcnR::kan, and its complementation strain pHNSHP45ΔpcnR::kan-pHSG575-pcnR in vitro competition. The reference strain is BW25113/pHNSHP45. Complementation assays were performed by expressing pcnR from its native promoter on pHSG575-pcnR. All competitions assays were carried out with three biological replicates and the relative fitness of each strain was detected at 24, 48 and 72 h. (C) The colonial morphology of E. coli BW25113/pHNSHP45 and BW25113/pHNSHP45ΔpcnR on LB agar plate.
Pfam analyses revealed that PcnR contains a FinO-like domain (Supplementary Table S4). The predicted structure of PcnR79–190 displays strong similarity to FinO35–152, even though the protein sequence is weakly related to FinO (Supplementary Figure S2A and S2B). FinO is an essential inhibitor of the F plasmid tra operon (31), and recent findings indicate that FinO acts as an RNA chaperone that binds with antisense RNA FinP and facilitates interactions between FinP and traJ mRNA (32–34). Previous studies have also demonstrated that deletion of finO leads to a 1000-fold increase in the conjugation frequency of the R1 plasmid (34), but a finO– strain has a larger adaptive cost than the finO+ strain (27), indicating that FinO may decrease the cost of plasmid to the host by repressing the expression of the tra operon.
Since the predicted structure of PcnR is similar to FinO, and deletion of pcnR increases the cost of pHNSHP45 to the host, we next examined whether PcnR decreases the conjugation frequency, as observed for FinO. As shown in Supplementary Figure S2C, deletion of pcnR resulted in an ∼8-fold increase in the plasmid transfer frequency (P < 0.0001), and complementation from pHSG575-pcnR restored the wild-type transfer frequency. However, the degree of influence of pcnR on the plasmid transfer frequency was much lower than that of FinO (35). Additionally, complementation of the ΔpcnR mutation with finO, which was cloned from an IncFII type plasmid pHN7A8 (36), did not restore the plasmid transfer frequency (P = 0.3118) or the wild-type growth rate (Supplementary Figure S2C and D), suggesting that PcnR and FinO may regulate different target genes.
Linking mcr-1 to the cost of pcnR-deficient plasmid
FinO-domain proteins, such as FinO of F-plasmid and RocC protein of Legionella pneumophila, generally serve as RNA chaperones to repress the translation of specific mRNAs (32,37). Therefore, since PcnR contains a FinO domain, it is possible that PcnR also serves as an RNA binding protein to regulate the fitness of pHNSHP45 by inhibiting expression of certain genes. To get a better understanding of the mechanisms governing the fitness cost of pHNSHP45ΔpcnR, a transposon library of E. coli BW25113 containing pHNSHP45ΔpcnR with random kanamycin-resistant mariner transposon insertions was screened on a plate containing kanamycin and colistin to generate growth restored mutants. Approximately 1000 colonies were chosen by visually screening large sized colonies. The growth curve of these colonies was determined, and seven colonies (Tns1 to Tns7) with growth rates close to that of the wild-type strain (BW25113/pHNSHP45) were chosen (Supplementary Figure S3A). To identify the transposon insertion sites, thermal asymmetric interlaced polymerase chain reaction (TAIL-PCR) (24) was performed on the growth-restored mutants (Tns1-Tns7); all insertion sites were chromosomal, including the arnT locus (Tns6) which codes for 4-amino-4-deoxy-l-arabinose lipid A transferase (Supplementary Table S5).
To investigate whether genes disrupted by transposons impacted the growth of E. coli BW25113/pHNSHP45ΔpcnR, individual deletions for each of the seven identified genes were constructed in this strain. Deletion of arnT in BW25113/pHNSHP45ΔpcnR was found to partially restore the wild-type growth rate, whereas deletion of other identified genes did not affect the growth of BW25113/pHNSHP45ΔpcnR (Supplementary Figure S3B).
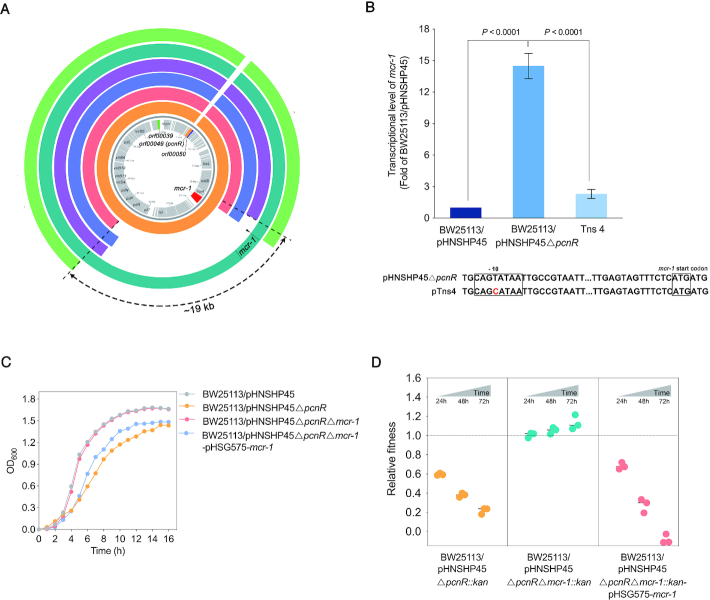
We then sought to determine whether any other mutations exist in growth-restored transposon mutants. To this end, Tns1, Tns2, Tns3, Tns4 and Tns7 were sequenced using HiSeq and Oxford Nanopore MinION to obtain the complete genome sequences of the mutants. Interestingly, while no other mutations were identified in the genome of these mutants besides genes inactivated by transposon insertion, a 15 to 19 kb region containing mcr-1 between nikB and pilP was found to be missing in the Tns1, Tns2, Tns3 and Tns7 mutants (Figure 2A). This finding indicates that deletion of fragments containing mcr-1 could compensate for the fitness cost caused by the ΔpcnR mutation. Furthermore, the mutant Tns4 contains a point mutation (T-C) in the –10 region of the mcr-1 promoter (Figure 2A and B). To determine whether this mutation affects the transcription of mcr-1, we measured the expression level of mcr-1 in BW25113/pHNSHP45ΔpcnR and the Tns4 mutant by RT-qPCR. As expected, transcription level of mcr-1 in Tns4 was significantly lower than that of BW25113/pHNSHP45ΔpcnR (P < 0.0001) (Figure 2B), indicating that the mutation in the promoter of mcr-1 improved the growth rate of the BW25113/pHNSHP45ΔpcnR strain. All these results suggested that the restored growth of transposon mutants might be relevant to the loss, or decreased expression of mcr-1. To verify the relationship between the ΔpcnR mutation and mcr-1, we constructed ΔpcnRΔmcr-1 mutants. As shown in Figure 2C and D, deletion of mcr-1 not only rescued the severe growth retardation of BW25113/pHNSHP45ΔpcnR, but also increased the relative fitness of the host. In addition, complementation of the ΔpcnRΔmcr-1 mutation by ectopic expression of mcr-1 from its native promoter led to severe growth retardation and a large fitness cost to the host (Figure 2C and D). Taken together, these results confirm that the high expression level of mcr-1 is responsible for the fitness burden of pHNSHP45ΔpcnR on BW25113.
Figure 2.
The fitness cost of pHNSHP45ΔpcnR is associated with mcr-1. (A) Comparison of the nucleotide sequence between pHNSHP45ΔpcnR and plasmids in transposon mutants. From inside to outside, coding sequences of pHNSHP45 appear on the innermost circle in grey. The second circle in orange represents the nucleotide sequence of pHNSHP45ΔpcnR. The following circles in red, blue, purple, cyan, and green represent plasmids pTns1, pTns2, pTns3, pTns4 and pTns7, respectively. The 15–19 kb regions containing mcr-1 between nikB and pilP genes are missing from pTns1, pTns2, pTns3 and pTns7 plasmids. A point mutation (shown by black triangle) was located in the -10 region of mcr-1 promoter on pTns4 plasmid. The circular BLAST Atlas was computed by the GView server (https://server.gview.ca/), and each plasmid was mapped against pHNSHP45 (GenBank accession number: KP347127). (B) Effect of point mutation (T-C) in the -10 region of promoter on the expression of mcr-1. Error bars represent the SD and P-values were calculated by One-way ANOVA test. (C) Deletion of mcr-1 restores the growth of BW25113/pHNSHP45ΔpcnR. Values represent the mean of three independent experiments. (D) Deletion of mcr-1 restore the relative fitness of BW25113/pHNSHP45ΔpcnR. E. coli BW25113 harbouring pHNSHP45ΔpcnR::kan, pHNSHP45ΔpcnRΔmcr-1::kan and pHNSHP45ΔpcnRΔmcr-1::kan-pHSG575-mcr-1 were competed with the reference strain BW25113/pHNSHP45 in vitro separately. All competitions assays were carried out with three biological replicates and the relative fitness of each strain was detected at 24, 48 and 72h.
PcnR balances the mcr-1 expression level and bacterial fitness by decreasing the plasmid copy number
We also measured the colistin minimum inhibitory concentrations (MICs) of transposon mutants, and found that the mutants (Tns1, Tns2, Tns3 and Tns7) lacking the 15- to 19-kb nikB-pilP region remained resistant to colistin (Table 1). We suspected that these transposon mutants might carry both a mcr-1-deficient plasmid and intact pHNSHP45 plasmid, causing the colistin resistance of these mutants. To verify this hypothesis, we performed PCR to detect the mcr-1 cassette. As expected, an intact mcr-1 cassette was observed in these mutants, suggesting that pHNSHP45 may exist as multiple copies in these mutants. As IncI2 is a group of low copy number plasmids, we hypothesized that the deletion of pcnR would increase the plasmid copy number, and that growth improvement of transposon mutants was caused by the deletion of mcr-1 from a subpopulation of plasmids.
Table 1.
Copy number of plasmids and mcr-1, and MICs of recombinants or mutants
| Strains | Plasmid copy number (repA) | mcr-1 copy number (mcr-1) | MIC (mg/l) of colistin or ceftazidime |
|---|---|---|---|
| BW25113 | - | - | 0.125 |
| BW25113/pHNSHP45 | 0.96 ± 0.19 | 1.03 ± 0.06 | 1 |
| BW25113/pHNSHP45ΔpcnR | 9.71 ± 0.74 | 10.91 ± 1.06 | 0.5 |
| BW25113/pHNSHP45ΔpcnR-pHSG575-pcnR | 1.40 ± 0.44 | 1.08 ± 0.07 | 1 |
| BW25113/pHN1122–1 | 0.85 ± 0.08 | - | 16a |
| BW25113/pHN1122–1ΔpcnR | 11.51 ± 1.94 | - | 128a |
| Tns1 | 8.52 ± 1.09 | 2.05 ± 0.24 | 0.5 |
| Tns2 | 9.12 ± 0.96 | 2.23 ± 0.20 | 1 |
| Tns3 | 11.17 ± 1.14 | 2.95 ± 0.82 | 0.5 |
| Tns4 | 9.95 ± 0.52 | 10.91 ± 1.06 | 1 |
| Tns7 | 9.59 ± 1.51 | 2.90 ± 0.08 | 0.5 |
aMICs of ceftazidime.
To verify this hypothesis, we detected the plasmid copy number by qPCR for all strains, including BW25113/pHNSHP45, BW25113/pHNSHP45ΔpcnR, BW25113/pHNSHP45ΔpcnR-pHS575-pcnR, and growth-restored transposon mutants (Table 1). Indeed, the copy numbers of pHNSHP45 and pHNSHP45ΔpcnR were ∼1 and ∼10, respectively, and trans-complementation of the ΔpcnR mutation restored the copy number of pHNSHP45ΔpcnR to a single copy (Table 1), strongly suggesting that pcnR negatively influences the plasmid copy number, hence the name plasmid copy number repressor (pcnR). In transposon mutants, the plasmid copy number was 8–12, whereas that of mcr-1 was 2–3 (except Tns4 which was 11) (Table 1). These results indicate that a high copy number of mcr-1 due to an increase in plasmid copy number imposed a fitness cost to the host. Hence the observed growth improvements of transposon mutants might be due to the lower mcr-1 copy number which were caused by the deletion of the fragment containing mcr-1 from most plasmid copies. In addition, the expression level of mcr-1 in BW25113/pHNSHP45ΔpcnR was ∼15-fold higher than that in BW25113/pHNSHP45 (P < 0.0001) (Figure 2B). These findings suggested that the high expression of mcr-1 caused by increased plasmid copy number led to a reduced growth rate of BW25113/pHNSHP45ΔpcnR.
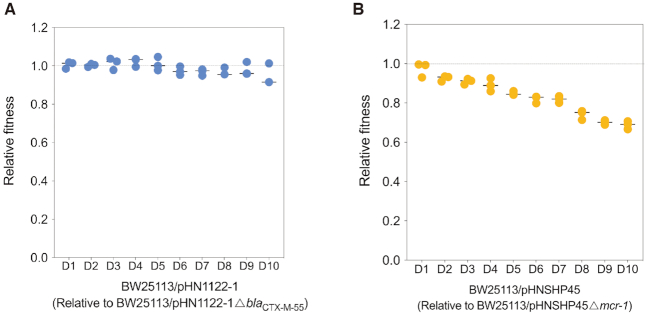
To investigate the effect of plasmid copy number on antimicrobial resistance, we determined the colistin MIC of BW25113/pHNSHP45 and BW25113/pHNSHP45ΔpcnR, and ceftazidime MIC of BW25113/pHN1122–1 as well as its derivate BW25113/pHN1122–1ΔpcnR. pHN1122–1 is another IncI2 plasmid carrying the blaCTX-M-55 gene and a pcnR gene (formerly named as yaeC) strictly identical to pcnR of pHNSHP45 (18). As shown in Table 1, the MIC of ceftazidime increased with increased plasmid copy number, whereas the colistin MIC was not positively related to the plasmid copy number. For example, colistin MIC of BW25113/pHNSHP45ΔpcnR was two-fold lower than that of the BW25113/pHNSHP45 strain. To elucidate the driving force for this difference, we compared the cost of mcr-1 and blaCTX-M-55 on E. coli BW25113. The mcr-1-bearing plasmid pHNSHP45 and blaCTX-M-55-bearing plasmid pHN1122–1 were competed with their corresponding resistant gene deficient mutants pHNSHP45Δmcr-1 and pHN1122–1ΔblaCTX-M-55, respectively. As shown in Figure 3A, the BW25113/pHN1122–1 strain showed no reduction (P = 0.0852) in fitness relative to BW25113/pHN1122–1ΔblaCTX-M-55 over 10 days. In contrast, BW25113/pHNSHP45 showed a significant reduction (P < 0.0001) in fitness relative to the BW25113/pHNSHP45Δmcr-1 in 10 days (Figure 3B). These results suggested that even a single copy of mcr-1 carried by its native plasmid imposed a cost to the host bacteria. Furthermore, in BW25113/pHNSHP45ΔpcnR, the mcr-1 copy number was ∼10-fold as high as that of its wild-type counterpart BW25113/pHNSHP45, thereby causing a higher cost to the host bacteria (Figure 2D). The increased cost caused by the high expression of mcr-1 may have resulted in the lower colistin MIC in BW25113/pHNSHP45ΔpcnR, implying that the high expression level of mcr-1 not only delays the growth of the host and imposes a high cost, but also negatively affects colistin resistance.
Figure 3.
The effect of antibiotic resistant genes (blaCTX-M-55 or mcr-1) on the fitness of E. coli BW25113/pHN1122-1(A) and BW25113/pHNSHP45 (B). E. coli BW25113/pHNSHP45 and BW25113/pHN1122-1 were competed with E. coli BW25113/pHNSHP45Δmcr-1 and BW25113/pHN1122–1ΔblaCTX-M-55, respectively. All competitions assays were carried out with three biological replicates and last for 10 days. The relative fitness of each strain was detected every day (from D1 to D10). P-values were calculated by one-way ANOVA test.
Consistent with the results of the previous studies that overexpression of mcr-1 impairs viability (38,39), and that mcr-1 is generally found in low-copy plasmids (40), our results suggest that maintaining the mcr-1 plasmid at a low copy number is essential for the fitness of the host bacteria. Furthermore, PcnR maintains a balance between the mcr-1 expression level and bacterial fitness by reducing the copy number of the IncI2 plasmid pHNSHP45 to ensure appropriate expression levels of mcr-1.
PcnR regulates RepA expression by binding to the repR mRNA
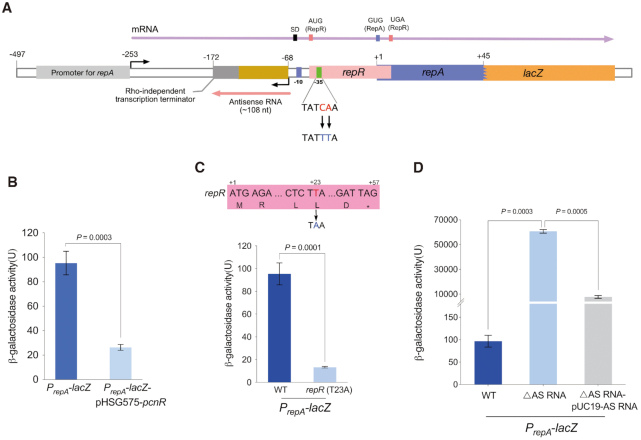
Since PcnR reduces the IncI2 plasmid copy number, we next determined whether it could repress the expression of repA. To this end, we fused the upstream fragment (−497 to +45) of repA (Figure 4A and Supplementary Figure S4) with a promoterless lacZ gene and monitored the β-galactosidase activity upon expression of pcnR by the pHSG575 plasmid. As expected, β-galactosidase activity decreased significantly upon expression of pcnR (P = 0.0003) (Figure 4B), confirming that pcnR directly represses the expression of repA.
Figure 4.
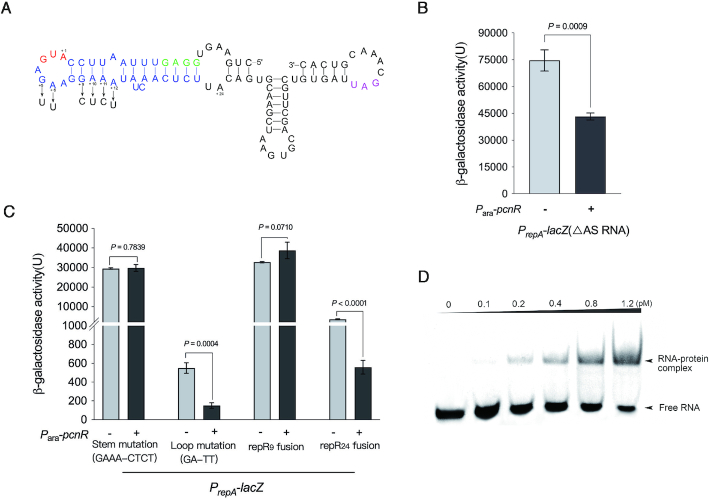
Regulation of RepA. (A) Schematic representation of the 5′untranslated region (UTR) of repA. The upstream nucleotide sequence (−497 to +45) of repA was fused with lacZ to monitor the expression of repA, and the resulting fusion was named PrepA-lacZ. The transcription of repA gene starts from the position -253, and the resulting polycistronic mRNA that contains ORFs encoding RepR and RepA is indicated by a purple arrow. The reading frame of repR overlaps the repA start codon (GUG). The transcription of antisense RNA (AS RNA) starts from the position -68, and a predicted rho-independent transcription terminator was located at the end of antisense RNA. The antisense RNA is indicated by red arrow. The -35 region of the promoter of antisense RNA (AS RNA) was mutated (CA-TT) to construct ΔAS RNA mutation. The rho-independent transcription terminator was predicted by ARNold: finding terminators web server (http://rssf.i2bc.paris-saclay.fr/toolbox/arnold/index.php). The upstream nucleotide sequence (−497 to +45) of repA is shown in Supplementary Figure S4. (B) Effect of pcnR on the expression of repA. Activity of PrepA was monitored from lacZ fusion (PrepA-lacZ) and pcnR is under control of its native promoter on pHSG575-pcnR plasmid. (C) The translation of RepA is coupled with RepR. Constructing T-A mutation at +23 position of repR to introduce a stop codon in repR open reading frame, and the resulting mutation was named repR (T23A). (D) Effect of antisense RNA (AS RNA) on the expression of RepA. Complementation assays were performed by expressing the AS RNA from its native promoter on pUC19-AS RNA. Error bars represent the SD and P-values were calculated by two-tailed t-tests.
To decipher the regulatory mechanism of pcnR to repA, we investigated the regulation of repA. Analysis of the region upstream the repA reading frame showed that the open reading frame of repR, coding for a 19-amino acid leader peptide, overlaps with the start codon of repA (Figure 4A and Supplementary Figure S4). In the IncFII type plasmid R1, the expression of repA is tightly coupled to a leader peptide and controlled by a small antisense RNA (41). We speculated that the replication control of pHNSHP45 may be similar to that of R1. To determine whether RepR is required for RepA synthesis, we introduced a stop codon at position +23 in repR (Figure 4C) and fused the lacZ gene to the repA reading frame. The effect of this mutation on repA expression was determined by measuring β-galactosidase activities. As shown in Figure 4C, expression of PrepA-lacZ was severely reduced by the introduced stop codon (P = 0.0001), suggesting that the translation of RepR is required for RepA synthesis.
In addition, we detected the existence of an antisense RNA (AS RNA) and the transcription start site (TSS) of repA by 5′ rapid amplification of cDNA ends (RACE). As shown in Figure 4A and Supplementary Figure S4, the AS RNA was transcribed from the non-coding strand of the DNA sequence that encodes the leader region of repA mRNA. This AS RNA is complementary to that the leader region of repA mRNA throughout its length, implying that the AS RNA may negatively regulate the expression of repA by binding at the leader region of repA mRNA. To verify this hypothesis, we abolished the AS RNA transcription by introducing mutations (CA-TT) in the –35 region of its promoter (Figure 4A and Supplementary Figure S4) and fused lacZ to the repA reading frame. As shown in Figure 4D, the β-galactosidase activity of PrepA-lacZ (ΔAS RNA) was dramatically increased compared with the wild-type (P = 0.0003). However, overexpression of AS RNA by pUC19 just partially complemented the ΔAS RNA mutation. Since these mutations (CA-TT) lie twelve nucleotides in the downstream of the start codon of repR (Supplementary Figure S4), we analysed the secondary structure of the repR mRNA by RNA fold software. Interestingly, as shown in Figure 5A, the RNA sequences near the start codon of RepR formed a SL structure, which may prevent the translation of RepR. The partial complementation of ΔAS RNA mutation may be attributable to an impairment of the stem structure of repR mRNA caused by the CA-UU mutations at position +16 and +17 (Supplementary Figure S5A). To investigate the effect of SL structure on the expression of repA in the ΔAS RNA strain, we introduced compensatory TA mutations (positions –7 and –4) (Supplementary Figure S5A) to restore the stem structure of the repR mRNA. As expected, the TA mutation decreased the expression of repA (∼ 32-fold). Meanwhile, overexpression of AS RNA in the ΔAS RNA (TA mutation) strain reduced the expression of repA to the wild-type level (Supplementary Figure S5B). These results indicate that the AS RNA inhibits the expression of repA.
Figure 5.
PcnR regulates the expression of RepA by binding to the repR mRNA. (A) Predicted RNA secondary structure of the repR mRNA. This structure was predicted by RNAfold web server. The Shine-Dalgarno (SD) sequence of repR is shown in green and the stem-loop structure containing the start codon of repR is highlighted in blue. The start codon of repR is shown in red, and A is numbered ‘+ 1’. The stop codon of repR is highlighted in purple. The mutational changes introduced for the present study are shown by arrows. The predicted secondary structure of the leader region of repA mRNA was shown in Supplementary Figure S8B. (B) The inhibition of PcnR on RepA expression is independent of antisense RNA. Activity of PrepA with ΔAS RNA mutation was monitored from the lacZ fusions, and expression of pcnR is controlled by Para with (+) or without (−) arabinose. Error bars represent the SD and P-values were calculated by two-tailed t-tests. (C) The inhibition of PcnR on RepA expression is dependent on the RNA secondary structure of repR mRNA. The stem structure of repR mRNA is predicted to be eliminated by GAAA-CTCT mutation and is denoted by stem mutation (GAAA-CTCT). The GA-TT mutation was located in the loop structure of repR mRNA and is denoted by loop mutation (GA-TT). The repR9 fusion and repR24 fusion represent the lacZ were fused with repR at + 9 and + 24 positions, respectively. Activity of PrepA with these mutations was monitored from the lacZ fusions. Expression of pcnR is controlled by Para with (+) or without (−) arabinose. Error bars represent the SD and P-values were calculated by two-tailed t-tests. (D) The first stem-loop structure of repR mRNA interacts with PcnR protein in vitro. 6  His-tagged PcnR was expressed in E. coli BL21(DE3) from pET28b-pcnR and purified by Ni-NTA affinity chromatography. Purified His-tag-PcnR was cut by thrombin to remove the His-tag and then purified PcnR was used for RNA-EMSA.
His-tagged PcnR was expressed in E. coli BL21(DE3) from pET28b-pcnR and purified by Ni-NTA affinity chromatography. Purified His-tag-PcnR was cut by thrombin to remove the His-tag and then purified PcnR was used for RNA-EMSA.
Considering that PcnR contains a FinO domain, we hypothesized that PcnR exerts an inhibitory effect on the expression of repA likely by binding to the AS RNA. His-tag-PcnR was expressed in E. coli BL21(DE3) from pET28b-pcnR, purified, and processed by thrombin to remove the His-tag and recover purified tag-free PcnR (Supplementary Figure S6) to use in an RNA-EMSA assay. However, PcnR failed to bind to the AS RNA (Supplementary Figure S7). To determine whether PcnR and the AS RNA synergistically inhibit the expression of repA, we introduced PcnR into BW25113/PrepA-lacZ (ΔAS RNA). In agreement with the RNA-EMSA results, PcnR also decreased the expression of repA in the absence of the AS RNA (P = 0.0009) (Figure 5B), indicating that the PcnR inhibition of RepA is independent of the AS RNA.
We, therefore, hypothesized that PcnR may stabilize the first SL structure embedded in the repR mRNA by binding to it. Hence, to determine whether this SL contributes to the inhibitory effect of PcnR on the expression of repA, a series of mutations were introduced into the SL, and lacZ was fused to the repA reading frame. Mutations (GAAA-CTCT) introduced at positions +9, +10, +11 and +12, were predicted to lie within the stem, while the mutations (GA-TT) at positions +5 and +6 in loop residues that were not expected to affect the stability of the SL structure (Figure 5A). Meanwhile, to investigate the inhibition of PcnR on the expression of repR in the absence or presence of the SL, lacZ was fused to positions +9 (repR9) and +24 (repR24), respectively.
Mutations at positions +9, +10, +11 and +12, predicted to disrupt the SL structure, nearly abolished the inhibitory effect of PcnR on the expression of repA (P = 0.7839) (Figure 5C), while also causing a large increase of repA expression relative to the wild-type construct. In contrast, mutations at positions +5 and +6 had little impact on the inhibitory effect of PcnR (P = 0.0004) (Figure 5C). This was expected, as these mutations within the loop region do not affect the stability of the SL structure. PcnR did not affect repR expression in the absence of the SL (RepR9 fusion) (P = 0.0710), whereas it repressed repR expression in the presence of the SL (RepR24 fusion) (P < 0.0001) (Figure 5C). These results indicate that the predicted SL structure likely plays a major role in the inhibitory effect exerted by PcnR on RepA.
To demonstrate the interaction between PcnR and the SL structure containing the RepR start codon, a small RNA (47 nt) containing this SL structure (Figure 5A) was used for an RNA EMSA assay. As shown in Figure 5D, incubation of the small RNA with the PcnR protein resulted in a robust shift of the RNA-protein complexes demonstrating that PcnR binds to this small RNA. Taken together, these results support the hypothesis that PcnR inhibits the expression of repA by binding at the first SL structure in the repR mRNA.
The ecological role of PcnR in the invasion of pHNSHP45 into bacterial populations
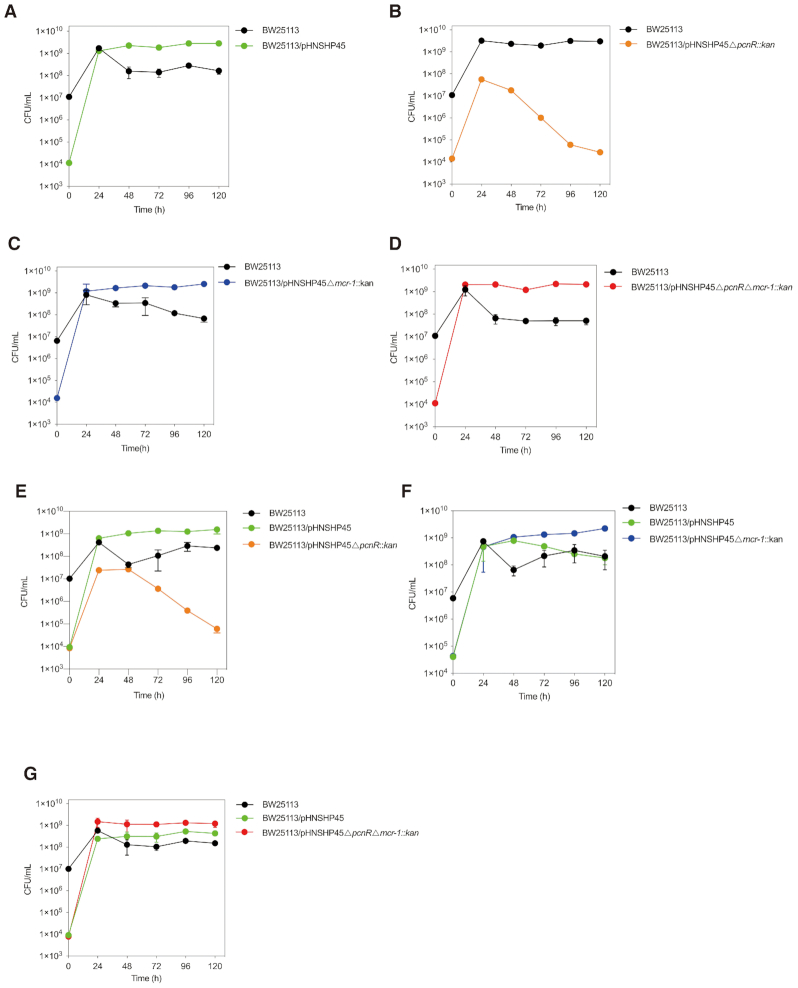
Although pHNSHP45ΔpcnR caused a high fitness cost to the host, its conjugative efficiency increased by ∼8-fold due to the increase in plasmid copy number compared with the wild-type plasmid (Supplementary Figure S2C). Plasmids persistence is usually attributed to low fitness cost and fast conjugation rate (42). To assess the role of pcnR in the persistence of the plasmid pHNSHP45, we examined the persistence of the pcnR+ plasmid pHNSHP45 and pcnR– plasmids pHNSHP45ΔpcnR::kan and pHNSHP45ΔpcnRΔmcr-1::kan by comparing their abilities to invade plasmid-free populations, individually and in competitive co-cultures. In addition, we investigated the effect of mcr-1 on the IncI2 plasmid invasiveness by comparing pHNSHP45 and pHNSHP45Δmcr-1::kan.
For each plasmid individually, the pcnR+ plasmid pHNSHP45 invaded and was present in the majority of cells after 24 h (Figure 6A), whereas the pcnR– plasmid pHNSHP45ΔpcnR::kan failed to invade the plasmid-free population and the number of BW25113 harbouring pHNSHP45ΔpcnR::kan plasmid decreased rapidly after 24 h (Figure 6B), indicating that plasmid horizontal transfer was not sufficient to offset the fitness cost. However, in the absence of mcr-1, the pcnR– plasmid could invade the majority of cells after 24 h (Figure 6D). The population dynamics in competitive co-cultures (Figure 6E) were similar with those of the individually co-cultures (Figure 6A and B). Remarkably, the pcnR–mcr-1– plasmid pHNSHP45ΔpcnRΔmcr-1::kan invaded more quickly and exhibited a long-term competitive advantage compared to pHNSHP45 (Figure 6D and G), which is consistent with the competitive data (Figure 2D). These observations suggest that pcnR is essential for the persistence and invasion of mcr-1-bearing IncI2 plasmids in bacterial populations. As shown in Figure 6A and C, both mcr-1+ plasmid pHNSHP45 and mcr-1– plasmid pHNSHP45Δmcr-1::kan invaded the majority of cells after 24 h. However, in the condition of competitive co-cultures (Figure 6F), the mcr-1– plasmid pHNSHP45Δmcr-1::kan showed a long-term competitive advantage over mcr-1+ plasmid pHNSHP45 after 48 h, suggesting that the presence of mcr-1 impairs the fitness of the host bacteria, thereby placing it at a disadvantage in competitive co-cultures.
Figure 6.
Bacterial population dynamics in co-cultures with plasmid-free and plasmid-containing E. coli BW25113. Plasmid-containing strains including E. coli BW25113/pHNSHP45, BW25113/pHNSHP45ΔpcnR::kan, BW25113/pHNSHP45Δmcr-1::kan,BW25113/pHNSHP45ΔpcnRΔmcr-1::kan were mixed with a 1000-fold excess of plasmid-free E. coli BW25113 at the beginning of the invasion assay. E. coli BW25113 is indicated by black dot, BW25113/pHNSHP45 is indicated by green dot, BW25113/pHNSHP45Δmcr-1::kan is indicated by blue dot, BW25113/pHNSHP45ΔpcnR::kan is indicated by orange dot, and BW25113/pHNSHP45ΔpcnRΔmcr-1::kan is indicated by red dot. (A) Co-cultures with E. coli BW25113 and BW25113/pHNSHP45. (B) Co-cultures with E. coli BW25113 and BW25113/pHNSHP45ΔpcnR::kan. (C) Co-cultures with E. coli BW25113 and BW25113/pHNSHP45Δmcr-1::kan. (D) Co-cultures with E. coli BW25113 and BW25113/pHNSHP45ΔpcnRΔmcr-1::kan. (E) Co-cultures with E. coli BW25113, BW25113/pHNSHP45, and BW25113/pHNSHP45ΔpcnR::kan. (F) Co-cultures with E. coli BW25113, BW25113/pHNSHP45 and BW25113/pHNSHP45Δmcr-1::kan. (G) Co-cultures with E. coli BW25113, BW25113/pHNSHP45, and BW25113/pHNSHP45ΔpcnRΔmcr-1::kan. Bars represent SD of three biological replicates.
DISCUSSION
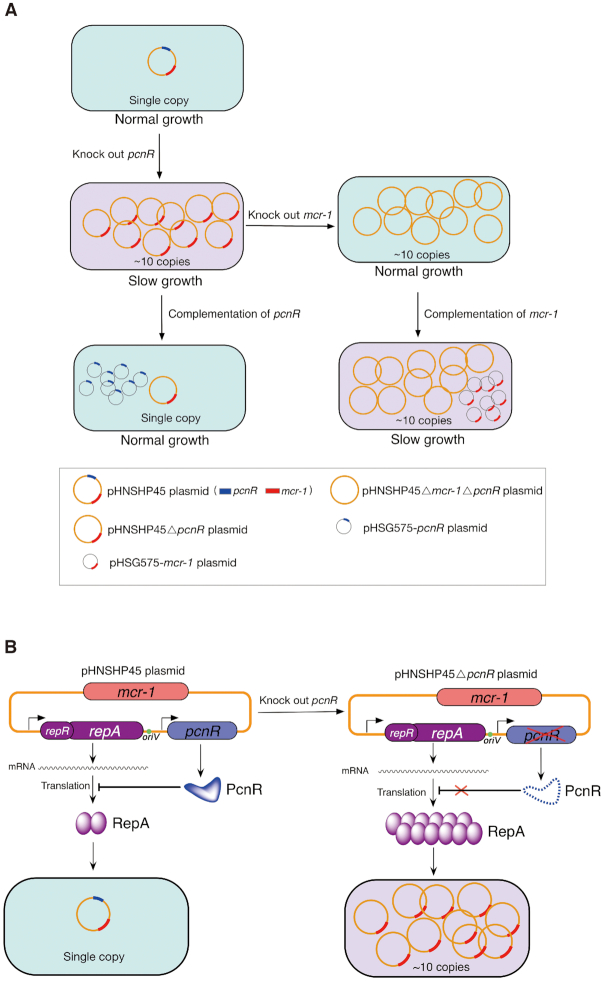
IncI2 is a group of low-copy number conjugative plasmids (43), which have recently garnered increasing attention as they carry mcr-1 (13). In this study, we investigated the role of IncI2 plasmid-encoded putative regulators in plasmid fitness to better understand the reason of evolutionarily successful spreading of mcr-1 by IncI2 plasmids. We found that pHNSHP45 lacking pcnR generated a large cost to the host that correlated with increasing plasmid copy numbers. This cost was fully compensated by the inactivation of mcr-1. Hence, the high cost was attributable to the high expression level of mcr-1 resulting from the increased plasmid copy number (Figure 7A). These results clearly demonstrate that plasmid-encoded ProQ/FinO family protein PcnR favours the fitness of the mcr-1-bearing IncI2 plasmids by controlling the plasmid copy number to ensure appropriate expression levels ofmcr-1. To the best of our knowledge, this is the first study dissecting an epidemic plasmid related adaptative mechanism for the colistin resistance gene.
Figure 7.
The pHNSHP45 plasmid-encoded PcnR protein favours the fitness of bacteria carrying mcr-1-bearing IncI2 plasmids. (A) PcnR enhances the fitness of E. coli BW25113 harbouring pHNSHP45 by repressing the plasmid copy number. pHNSHP45 plasmid is the first discovered mcr-1+-IncI2 plasmid. Deletion of pcnR increases plasmid copy number by 10-fold and largely impairs the growth of the host. Deletion of mcr-1 completely rescues the growth of pcnR– strain, although with high plasmid copy number. Complementation of mcr-1 in E. coli BW25113 harbouring pHNSHP45ΔpcnR confirmed that host bacteria are sensitive to multicopy mcr-1. Complementation of pcnR represses the copy number of pHNSHP45ΔpcnR. Summary: PcnR maintains a balance between the mcr-1 expression level and bacterial fitness by limiting the copy number of the IncI2 plasmid pHNSHP45 to ensure an appropriate expression level of mcr-1. (B) PcnR maintains the plasmid copy number at a single copy by inhibiting the translation of repA. Deletion of pcnR derepresses the expression of repA and leads to the plasmid copy number increase to ∼10 copies.
Although the acquisition of antimicrobial resistance by mutation or HGT is often associated with a fitness cost, bacterial evolution to reduce the cost of resistance contributes to the persistence of resistance in pathogens (6). High-level expression ofmcr-1 has previously been shown to influence bacterial fitness by impairing the growth rate and membrane structural integrity of the host bacteria (38,39). It is likely that maintaining a modest expression level of mcr-1 by a low copy number plasmid plays a key role in the persistence ofmcr-1 plasmids in bacterial populations. This idea was supported by our observation that a high copy number ofmcr-1 plasmids hindered the growth of host bacteria (Figure 7A), which was detrimental to the persistence of mcr-1 gene in bacterial populations. Remarkably, a high copy number ofmcr-1 plasmids negatively affects colistin resistance as a result of the toxic effect of excessive MCR-1 on the host bacteria, which explains whymcr-1 mediates a limited level of colistin resistance (MICs = 2–8 μg/ml) (13,44,45). These results suggest that maintainingmcr-1 at a low copy number is essential for its persistence. We also found that even a single copy mcr-1 imposed a high cost to its host bacterium, which negatively affected fitness and persistence of the mcr-1-bearing IncI2 plasmid in the bacterial population. In contrast, single copy blaCTX-M-55, as a negative control, has little effect on host bacterium fitness. These observations suggest that the role of PcnR in the persistence of IncI2 plasmids depends on the resistance gene carried by these plasmids and partly explain the significantly reduced prevalence of mcr-1 and IncI2 plasmids in E. coli following the ban of colistin as a growth promoter in China since April 30, 2017 (46–48).
In this study, we also investigated the regulation of expression of repA of IncI2 plasmids and found that some control components were similar to those in IncFII (R1) and IncI1 type plasmids. For example, the presence of a ∼108-nt AS RNA, which is transcribed constitutively from the complementary strand in the leader region of the repA mRNA, largely repressed the synthesis of RepA. The translation of RepA is coupled with the leader peptide RepR. The antisense RNA encoded by R1 plasmid binds to the leader region of the repA mRNA and regulates the expression of repA by inhibiting the synthesis of the leader peptide Tap (41). In IncI1 or IncB/O plasmid, the expression of the replication initiation gene repZ is also regulated by an antisense RNA which binds to the leader region of the repZ mRNA. This antisense RNA not only inhibits the translation of the leader peptide but also prevents the formation of a pseudoknot that is essential for the initiation of RepZ translation (49–51). While in the IncI2 plasmid pHNSHP45, plasmid-encoded antisense RNA also binds to the leader region of repA mRNA, but no pseudoknot similar to that in IncI1 plasmid could be found in this region (Supplementary Figure S8). Hence, we propose that the function of IncI2 plasmid-encoded antisense RNA may be different from that of IncI1 plasmid.
Besides these elements, we also identified a novel replication control component, PcnR, which is an RNA binding protein belonging to the ProQ/FinO family proteins. Deletion of pcnR leads to an approximate 10-fold increase in the copy number of IncI2 plasmid. Our results demonstrate that PcnR represses the synthesis of RepR by binding at the first SL structure of repR mRNA. We propose that the binding of PcnR to this SL structure could improve its stability and block the initiation of translation of the RepR leader peptide, thereby repressing the synthesis of RepA. Interestingly, IncI1 and IncB/O plasmids also encode putative ProQ/FinO family proteins (Supplementary Figure S9 and Supplementary Table S6). Besides, the RNA sequence containing the start codon of the leader peptide RepY is predicted to form a SL structure, although the structure is different from that of the repR mRNA (Supplementary Figure S8). Whether this SL structure and the putative ProQ/FinO family proteins are involved in regulating the expression of repZ in IncI1 or IncB/O plasmid remains to be demonstrated.
So far, the plasmid-encoded ProQ/FinO system has only been found to be involved in conjugation control of IncF plasmids (52). It stabilizes FinP sRNA and promotes the interaction with its traJ mRNA target (32–34). The chromosome-encoded RNA chaperone ProQ, which is structurally similar with FinO, promotes duplex formation between RaiZ sRNA and hupA mRNA to prevent the protein synthesis of HU-α (53). Hitherto, no role for a ProQ/FinO family protein in the regulation of plasmid replication has been found. Thus, PcnR is the first ProQ/FinO family protein identified to be involved in plasmid copy number control, which expands our knowledge of the functions of FinO-domain proteins.
To date, the ColE1 plasmid-encoded Rom protein is the only one RNA binding protein participating in plasmid replication control (52,54). Here, we identified PcnR as another plasmid-encoded RNA binding protein. However, unlike Rom, which negatively controls the plasmid copy number by stabilizing the sense RNA-antisense RNA kissing complex (55,56), PcnR inhibits the expression of repA by binding to the repR mRNA (Figure 7B), implying a novel plasmid replication control mechanism. Further studies are needed to explore the underlying regulatory mechanism of PcnR.
DATA AVAILABILITY
The authors declare that all data supporting the findings of this study are available within the article and the Supplementary data.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Xiaoxue Wang, South China Sea Institute of Oceanology, Chinese Academy of Sciences, for her helpful comments on this study.
Contributor Information
Jun Yang, College of Veterinary Medicine, National Risk Assessment Laboratory for Antimicrobial Resistant of Microorganisms in Animals, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs,Center for Emerging and Zoonotic Diseases, South China Agricultural University, Guangzhou, China; Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China.
Hai-Hong Wang, Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China; Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms, College of Life Sciences, South China Agricultural University, Guangzhou, China.
Yaoyao Lu, College of Veterinary Medicine, National Risk Assessment Laboratory for Antimicrobial Resistant of Microorganisms in Animals, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs,Center for Emerging and Zoonotic Diseases, South China Agricultural University, Guangzhou, China; Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China.
Ling-Xian Yi, College of Veterinary Medicine, National Risk Assessment Laboratory for Antimicrobial Resistant of Microorganisms in Animals, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs,Center for Emerging and Zoonotic Diseases, South China Agricultural University, Guangzhou, China; Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China.
Yinyue Deng, School of Pharmaceutical Sciences (Shenzhen), Sun Yat-sen University, Guangzhou 510275, China.
Luchao Lv, College of Veterinary Medicine, National Risk Assessment Laboratory for Antimicrobial Resistant of Microorganisms in Animals, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs,Center for Emerging and Zoonotic Diseases, South China Agricultural University, Guangzhou, China; Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China.
Vincent Burrus, Département de biologie, Université de Sherbrooke, Sherbrooke J1K 2R1, Québec, Canada.
Jian-Hua Liu, College of Veterinary Medicine, National Risk Assessment Laboratory for Antimicrobial Resistant of Microorganisms in Animals, Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, Key Laboratory of Zoonosis of Ministry of Agricultural and Rural Affairs,Center for Emerging and Zoonotic Diseases, South China Agricultural University, Guangzhou, China; Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31625026, 31830099, 31902322]; Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program [2019BT02N054], Guangdong Major Project of Basic and Applied Basic Research [2020B0301030007]; 111 Project [D20008]; Innovation Team Project of Guangdong University [2019KCXTD001]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gogarten J.P., Townsend J.P.. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005; 3:679–687. [DOI] [PubMed] [Google Scholar]
- 2. Jain R., Rivera M.C., Moore J.E., Lake J.A.. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 2003; 20:1598–1602. [DOI] [PubMed] [Google Scholar]
- 3. Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013; 303:298–304. [DOI] [PubMed] [Google Scholar]
- 4. Poirel L., Madec J.Y., Lupo A., Schink A.K., Kieffer N., Nordmann P., Schwarz S.. Antimicrobial resistance in Escherichia coli. Microbiol. Spectrum. 2018; 6:ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. San Millan A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018; 26:978–985. [DOI] [PubMed] [Google Scholar]
- 6. San Millan A., MacLean R.C.. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectrum. 2017; 5:MTBP-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. San Millan A., Toll-Riera M., Qi Q., MacLean R.C.. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 2015; 6:6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fischer E.A.J., Dierikx C.M., van Essen-Zandbergen A., van Roermund H.J.W., Mevius D.J., Stegeman A., Klinkenberg D.. The IncI1 plasmid carrying the blaCTX-M-1 gene persists in in vitro culture of a Escherichia coli strain from broilers. BMC Microbiol. 2014; 14:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enne V.I., Bennett P.M., Livermore D.M., Hall L.M.C.. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 2004; 53:958–963. [DOI] [PubMed] [Google Scholar]
- 10. Cottell J.L., Webber M.A., Piddock L.J.V.. Persistence of transferable extended-spectrum-β-lactamase resistance in the absence of antibiotic pressure. Antimicrob. Agents Chemother. 2012; 56:4703–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harrison E., Guymer D., Spiers A.J., Paterson S., Brockhurst M.A.. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015; 25:2034–2039. [DOI] [PubMed] [Google Scholar]
- 12. Dorman C.J. H-NS-like nucleoid-associated proteins, mobile genetic elements and horizontal gene transfer in bacteria. Plasmid. 2014; 75:1–11. [DOI] [PubMed] [Google Scholar]
- 13. Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X.et al.. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016; 16:161–168. [DOI] [PubMed] [Google Scholar]
- 14. Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z.et al.. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017; 2:16260. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., Liu J.H.. Monitoring colistin resistance in food animals, an urgent threat. Expert Rev. Anti Infect. Ther. 2018; 16:443–446. [DOI] [PubMed] [Google Scholar]
- 16. Quan J., Li X., Chen Y., Jiang Y., Zhou Z., Zhang H., Sun L., Ruan Z., Feng Y., Akova M.et al.. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet. Infect. Dis. 2017; 17:400–410. [DOI] [PubMed] [Google Scholar]
- 17. Wang R., van Dorp L., Shaw L.P., Bradley P., Wang Q., Wang X., Jin L., Zhang Q., Liu Y., Rieux A.et al.. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018; 9:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lv L., Partridge S.R., He L., Zeng Z., He D., Ye J., Liu J.-H.. Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob. Agents Chemother. 2013; 57:2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen L., Chavda K.D., Al Laham N., Melano R.G., Jacobs M.R., Bonomo R.A., Kreiswirth B.N.. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 2013; 57:5019–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu R., Yi L.-x., Yu L.-f., Wang J., Liu Y., Chen X., Lv L., Yang J., Liu J.-H.. Fitness advantage of mcr-1-bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 2018; 9:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meinersmann R.J. The biology of IncI2 plasmids shown by whole-plasmid multi-locus sequence typing. Plasmid. 2019; 106:102444. [DOI] [PubMed] [Google Scholar]
- 22. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiang S.L., Rubin E.J.. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002; 296:179–185. [DOI] [PubMed] [Google Scholar]
- 24. Singer T., Burke E.. High-throughput TAIL-PCR as a tool to identify DNA flanking insertions. Methods Mol. Biol. 2003; 236:241–272. [DOI] [PubMed] [Google Scholar]
- 25. Wick R.R., Judd L.M., Gorrie C.L., Holt K.E.. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017; 13:e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 27. Haft R.J.F., Mittler J.E., Traxler B.. Competition favours reduced cost of plasmids to host bacteria. ISME J. 2009; 3:761–769. [DOI] [PubMed] [Google Scholar]
- 28. Miller H.J. Assay of β-galactosidase. In: Experiments in Molecular Genetics. 1972; Cold Spring Harbor Laboratory; 352–355. [Google Scholar]
- 29. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A.et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2018; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bagdasarian M., Bailone A., Angulo J.F., Scholz P., Bagdasarian M., Devoret R.. PsiB, and anti-SOS protein, is transiently expressed by the F sex factor during its transmission to an Escherichia coli K-12 recipient. Mol. Microbiol. 1992; 6:885–893. [DOI] [PubMed] [Google Scholar]
- 31. Finnegan D Fau - Willetts N., Willetts N.. The nature of the transfer inhibitor of several F-like plasmids. Mol. Gen. Genet.: MGG. 1972; 119:57–66. [DOI] [PubMed] [Google Scholar]
- 32. Tim van Biesen T., Frost L.S.. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol. Microbiol. 1994; 14:427–436. [DOI] [PubMed] [Google Scholar]
- 33. Arthur D.C., Ghetu A.F., Gubbins M.J., Edwards R.A., Frost L.S., Glover J.N.. FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 2003; 22:6346–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arthur D.C., Edwards R.A., Tsutakawa S., Tainer J.A., Frost L.S., Glover J.N.. Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res. 2011; 39:4450–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dionisio F., Matic I., Radman M., Rodrigues O.R., Taddei F.. Plasmids spread very fast in heterogeneous bacterial communities. Genetics. 2002; 162:1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He L., Partridge S.R., Yang X., Hou J., Deng Y., Yao Q., Zeng Z., Chen Z., Liu J.-H.. Complete nucleotide sequence of pHN7A8, an F33:A-:B-type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J. Antimicrob. Chemother. 2012; 68:46–50. [DOI] [PubMed] [Google Scholar]
- 37. Attaiech L., Boughammoura A., Brochier-Armanet C., Allatif O., Peillard-Fiorente F., Edwards R.A., Omar A.R., MacMillan A.M., Glover M., Charpentier X.. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:8813–8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Q., Li M., Spiller O.B., Andrey D.O., Hinchliffe P., Li H., MacLean C., Niumsup P., Powell L., Pritchard M.et al.. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 2017; 8:2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y.Y., Zhu Y., Wickremasinghe H., Bergen P.J., Lu J., Zhu X.Q., Zhou Q.L., Azad M., Nang S.C., Han M.L.et al.. Metabolic perturbations caused by the over-expression of mcr-1 in Escherichia coli. Front. Microbiol. 2020; 11:588658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bontron S., Poirel L., Nordmann P.. Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J. Antimicrob. Chemother. 2016; 71:2318–2320. [DOI] [PubMed] [Google Scholar]
- 41. Blomberg P., Nordström K., Wagner E.G.. Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J. 1992; 11:2675–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hall J.P.J., Brockhurst M.A., Dytham C., Harrison E. The evolution of plasmid stability: are infectious transmission and compensatory evolution competing evolutionary trajectories. Plasmid. 2017; 91:90–95. [DOI] [PubMed] [Google Scholar]
- 43. Rozwandowicz M., Brouwer M.S.M., Fischer J., Wagenaar J.A., Gonzalez-Zorn B., Guerra B., Mevius D.J., Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018; 73:1121–1137. [DOI] [PubMed] [Google Scholar]
- 44. Wang Y., Tian G.B., Zhang R., Shen Y., Tyrrell J.M., Huang X., Zhou H., Lei L., Li H.Y., Doi Y.et al.. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet. Infect. Dis. 2017; 17:390–399. [DOI] [PubMed] [Google Scholar]
- 45. He Q.-W., Xu X.-H., Lan F.-J., Zhao Z.-C., Wu Z.-Y., Cao Y.-P., Li B.. Molecular characteristic of mcr-1 producing Escherichia coli in a Chinese university hospital. Ann. Clin. Microbiol. Antimicrob. 2017; 16:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu Y.Y., Zhou Q., He W., Lin Q., Yang J., Liu J.H.. mcr-1 and plasmid prevalence in Escherichia coli from livestock. Lancet Infect. Dis. 2020; 20:1126. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Q., Lv L., Huang X., Huang Y., Zhuang Z., Lu J., Liu E., Wan M., Xun H., Zhang Z.et al.. Rapid increase in carbapenemase-producing Enterobacteriaceae in retail meat driven by the spread of the blaNDM-5-carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob. Agents Chemother. 2019; 63:e00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Y., Xu C., Zhang R., Chen Y., Shen Y., Hu F., Liu D., Lu J., Guo Y., Xia X.et al.. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 2020; 20:1161–1171. [DOI] [PubMed] [Google Scholar]
- 49. Asano K., Mizobuchi K.. Copy number control of IncIα plasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J. 1998; 17:5201–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Asano K., Mizobuchi K.. Structural analysis of late intermediate complex formed between plasmid ColIb-P9 Inc RNA and its target RNA. How does a single antisense RNA repress translation of two genes at different rates. J. Biol. Chem. 2000; 275:1269–1274. [DOI] [PubMed] [Google Scholar]
- 51. Praszkier J., Pittard A.J.. Pseudoknot-dependent translational coupling in repBA genes of the IncB plasmid pMU720 involves reinitiation. J. Bacteriol. 2002; 184:5772–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olejniczak M., Storz G.. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers. Mol. Microbiol. 2017; 104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smirnov A., Wang C., Drewry L.L., Vogel J.. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 2017; 36:1029–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brantl S. Plasmid replication control by antisense RNAs. Microbiol. Spectr. 2014; 2:Plas-0001-2013. [DOI] [PubMed] [Google Scholar]
- 55. Tomizawa J, Som T.. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984; 38:871–878. [DOI] [PubMed] [Google Scholar]
- 56. Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986; 47:89–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and the Supplementary data.