Abstract
Proteins that can bring together separate DNA sites, either on the same or on different DNA molecules, are critical for a variety of DNA-based processes. However, there are no general and technically simple assays to detect proteins capable of DNA looping in vivo nor to quantitate their in vivo looping efficiency. Here, we develop a quantitative in vivo assay for DNA-looping proteins in Escherichia coli that requires only basic DNA cloning techniques and a LacZ assay. The assay is based on loop assistance, where two binding sites for the candidate looping protein are inserted internally to a pair of operators for the E. coli LacI repressor. DNA looping between the sites shortens the effective distance between the lac operators, increasing LacI looping and strengthening its repression of a lacZ reporter gene. Analysis based on a general model for loop assistance enables quantitation of the strength of looping conferred by the protein and its binding sites. We use this ‘loopometer’ assay to measure DNA looping for a variety of bacterial and phage proteins.
INTRODUCTION
Proteins that bind to specific DNA sites and are able to interact with each other to bring together separate DNA molecules or to form loops within the same DNA molecule are critical in essential DNA processes, such as transcription, replication, recombination and DNA organization (1–3). The primary role of the interaction can be to affect the function of the proteins at one or both sites. DNA looping provides cooperative binding over large distances, increasing site occupancy by the proteins (1,2,4), and can be used to target catalytic activities in transcriptional control, such as in activation of σ54 promoters (5) or eukaryotic enhancers (6,7). Bridging interactions between replicases bound to separate plasmids can inhibit replication initiation to provide plasmid copy number control (8,9). Alternatively, the primary role of the interaction can be the juxtaposition of the two DNA segments. Promoters can be repressed by being enclosed in small DNA loops (10,11), and the large chromatin loops formed by the interaction between insulator elements partition eukaryotic chromosomes into topological domains that are important for enhancer–promoter specificity (12). In many cases, the interaction combines both roles. Site-specific recombinases must juxtapose their DNA sites for correct recombination and the interactions in the synapsed complex also activate the recombinase catalytic steps (13). The long-range interaction between the OL- and OR-binding sites for the phage λ CI repressor increases CI binding at these sites, improving repression of the lytic promoters (14,15), while the DNA loop formed also juxtaposes a distal UP element near to the PRM promoter where it can activate lysogenic transcription (16,17).
Despite the importance of DNA looping and bridging, a general method for identifying proteins capable of bringing DNA together in vivo is not currently available. For most proteins, an in vivo looping capability is first implicated when the function of a protein at one binding site is affected by a distal binding site. For example, in the first described example of DNA looping by a transcription factor, looping by AraC was suspected because deletion of a binding site 270 bp upstream of the araBAD promoter affected AraC regulation of the promoter (18). To further support DNA looping, additional tests, usually specific to each system, are needed to rule out an independent function of the distal site. Helical phasing experiments, where the functional interaction between the sites is sensitive to insertion of nonintegral DNA turns in the intervening DNA, are often used to confirm DNA looping in vivo (18–21). However, helical phasing sensitivity is not present for long DNA loops (>500 bp), or for trans interactions, and may be minimized by protein flexibility. An alternative route to identifying DNA-looping proteins in vivo is provided by ligation-based proximity assays such as 3C (22). These techniques have revealed vast numbers of DNA loops in eukaryotic and bacterial genomes (23,24). However, additional approaches are needed to identify the proteins responsible for these loops. Thus, confirmation of the DNA looping or bridging activity of a specific protein often requires in vitro techniques such as DNA footprinting (25), electrophoretic mobility shift (26), enhancement of ligation (9) or single-molecule methods such as electron or atomic force microscopy (14,27) and tethered particle motion (28). However, these in vitro approaches need purified active protein and often require specialized expertise and equipment.
In addition, an important limitation of all these approaches is that they do not reveal the in vivo strength of the protein-mediated DNA looping, that is the fraction of time that the sites spend in direct contact in cells. Extraction of looping frequencies from functional in vivo assays has been possible only for highly characterized regulatory proteins, such as the Lac and CI repressors (29). Ligation-mediated proximity assays and techniques such as Dam-C can provide relative but not absolute frequencies of close proximity (30). Absolute proximity measures can be obtained by microscopic imaging of intact cells (31,32), but none of these proximity approaches is able to confirm direct protein-mediated contact. In vitro studies can quantitate looping; however, these measurements are made under conditions that may not reflect in vivo looping. Knowing how often the loops form in vivo provides critical information about the interplay between DNA looping and function, while identification of DNA-looping proteins capable of directing strong DNA looping will also aid in more effective manipulation of DNA looping in cells.
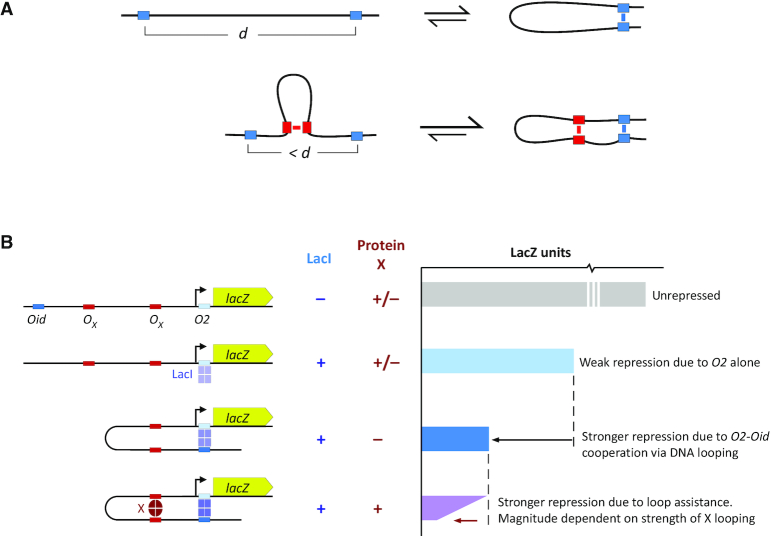
Here, we report the development of an in vivo assay for DNA-looping proteins in Escherichia coli that does not depend on detailed knowledge of the protein’s function and requires only basic DNA cloning techniques and a LacZ assay. Furthermore, the assay provides a quantitative measurement of the strength of looping exerted by the protein and its binding sites. The assay is based on loop assistance, where the formation of one DNA loop assists the formation of another DNA loop (Figure 1A). When a pair of sites for one DNA-looping protein is nested within a pair of sites for another DNA-looping protein, DNA looping by the internal protein assists looping by the external protein by bringing its binding sites closer together (33–36). In the loopometer, the external loop is formed by two operators for the E. coli Lac repressor (LacI) that repress the expression of a lacZ reporter gene (Figure 1B). The proximal operator directly represses the promoter and the distal operator cooperates by DNA looping to increase LacI occupation of the proximal operator. This allows LacI looping to be detected and measured by the increase in repression in the presence of the distal operator (33,34). To assay DNA looping by a candidate protein, a pair of binding sites is inserted internal to the LacI loop and the protein is expressed. DNA looping by the candidate protein improves LacI looping by shortening the effective distance between the lac operators, detected by increased LacI repression of the reporter (Figure 1B). Analysis of the measurements using a general model for loop assistance (35) enables the looping strength of the protein and its binding sites to be estimated. We validate the assay using the λ CI protein and test a variety of bacterial and phage proteins for DNA-looping activity.
Figure 1.
Assaying DNA looping by loop assistance. (A) The DNA loop formed between an internal pair of sites (red) assists the formation of the loop between the external sites (blue) by reducing the effective DNA distance, d, between them. (B) DNA looping by a candidate protein (X) and its DNA-binding sites (OX) is detected and measured in the loopometer by its enhancement of loop-dependent LacI repression of a promoter for a lacZ reporter. In the absence of the strong upstream lacOid operator, a LacI tetramer binds poorly to the weak lacO2 operator, giving weak repression of the promoter. In the presence of Oid, occupation of O2 is increased due to cooperative DNA looping, and repression is increased. DNA looping by the candidate protein shortens the distance between Oid and O2, increasing LacI looping and further improving repression.
MATERIALS AND METHODS
General strains and media
All reporter strains are derivatives of E4643, itself a derivative of BW30270 MG1655 rph+ (CGSC7925) with the lacIZYA (MG1655:360,527–366,797) region removed (29). Plasmid replication of pLOM2–500 and pCYMR, which are derived from the CRIM plasmids of Haldimann and Wanner (37), is dependent on the R6Kγ π replicase protein Pir. We thus maintain these plasmids in the Pir-expressing (pir+) strain E4644 = EC100D (Epicentre) = F–mcrA Δ(mrr-hsdRMS-mcrBC) φ80d(lacZΔM15) ΔlacX74 recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ–rpsL StrRnupG pir+(DHFR).
LB (1% Bacto-tryptone, 1% NaCl, and 0.5% yeast extract, pH 7.0) is used for routine growth of strains. M9MM-glyc = 1 × M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.01 mM (NH4)2Fe(SO4)2·6H2O, and 0.4% glycerol [10 × M9 salts = 67.8 g of NaH2PO4, 30.0 g of KH2PO4, 10 g NH4Cl and 5 g NaCl/l H2O] were used for LacZ assays. Antibiotics used were (μg/ml for selection of integrated copy/plasmid copies): Ap, ampicillin (-/100); Cm, chloramphenicol (20/30); Km, kanamycin (20/50); Sp, spectinomycin (20/50); Tc, tetracycline (3/20). A 100 mM stock of cumic acid (CA; Sigma-Aldrich Cat. 268402) was prepared in ethanol.
Constructing loopometer strains
Cloning protein binding sites into pLOM2-500
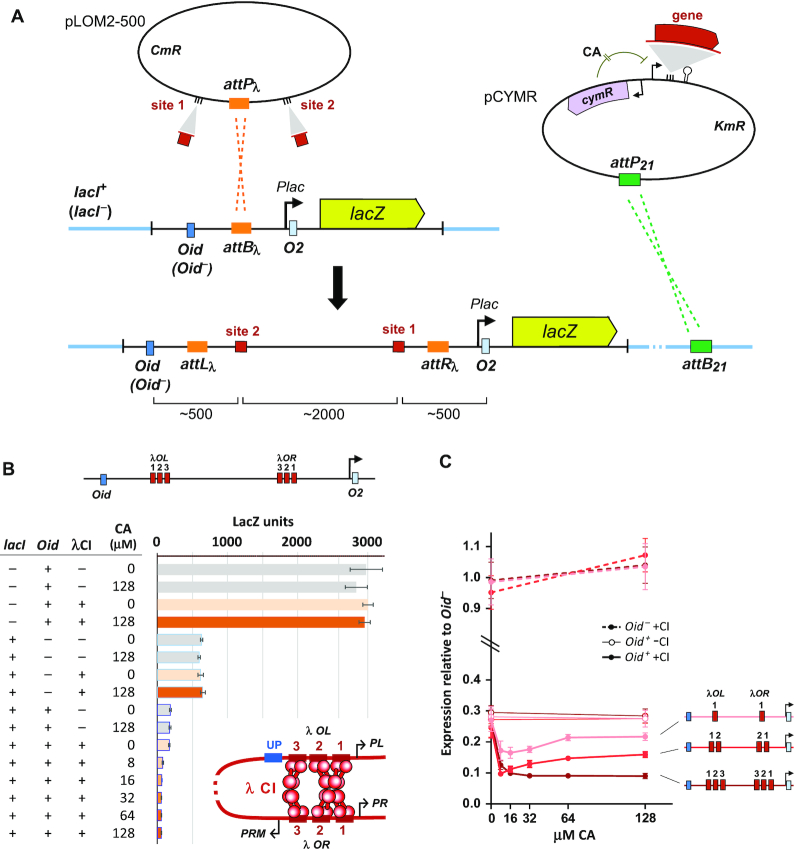
The restriction sites flanking sites 1 and 2 in pLOM2–500 (Figure 2A and Supplementary Figure S1A) allow various strategies for cloning candidate binding sites at these positions. We use Gibson isothermal assembly (38), either by 2-fragment assemblies with a single DNA fragment comprising both inserts and attPλ (PCR-generated or purchased commercially) or by 4-fragment assemblies of two polymerase chain reaction (PCR)-generated insert fragments, an attPλ fragment and digested and isolated plasmid backbone. pLOM2–500 plasmids were maintained in E4644 (LB + Cm, 30 μg/ml). Supplementary Figure S2B shows the detailed structure of pLOM2–500 and the reporter after its integration into the loopometer landing pad. Primers #2238 (TGGCGACGCTCATGTATGTG) and #2239 (CTCTTACGTGCCGGAAGT) were used for sequencing the site 1–site 2 region.
Figure 2.
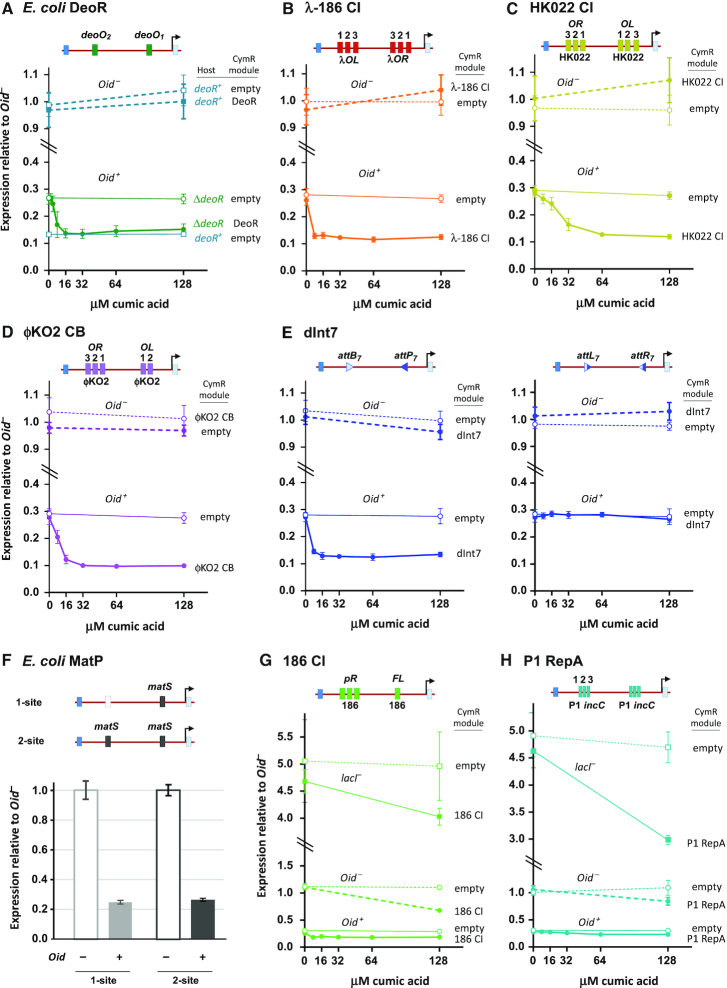
The loopometer system. (A) The basic procedure for the construction of the looping reporter strains. Protein-binding sites are cloned into the pLOM2–500 plasmid and the resulting plasmid is inserted by λ integrase into a reporter landing pad in the Escherichia coli chromosome, causing the two sites to be nested inside a pair of lac operators, Oid and O2, which control the PlacUV5 promoter for a lacZ.O2– reporter gene. LacI is supplied by a PlacI.lacI+ gene inserted elsewhere in the chromosome. The pLOM2–500 plasmid is also integrated into control strains lacking lacI+ or Oid. The gene for the candidate protein is cloned into the pCYMR expression vector, under the control of the CymR repressor that can be inactivated by cumic acid (CA). This expression vector or the empty control is inserted by φ21 integrase-mediated recombination into the chromosome of the three strains carrying pLOM2–500. (B) LacZ assay results for λ CI and λ OL123-OR123. DNA looping is indicated by a LacI-dependent, Oid-dependent, candidate protein-dependent decrease in units. Error bars are 95% confidence limits (Student's t), n= 6. The insert shows DNA looping by a CI octamer and tetramer at the native OL and OR sites. (C) Graphic display of results for λ CI and OL123-OR123, OL12-OR12, OL1-OR1 reporters (lacI+ only). LacZ units within each set are normalized to the average of the four Oid– values (±CI, ±CA).
Cloning protein genes into pCYMR
The pCYMR plasmids (Figure 2A and Supplementary Figure S3A) contain a multiple cloning site (SpeI.ApaL1.XmaI.PvuII.AflII.AccIII.NarI) located downstream of the CymR-binding site (cymO), followed by transcription terminators. We generally used 2-fragment Gibson assembly to insert PCR-generated fragments of protein genes into the PvuII site. Sequences for translation of the protein message need to be included in the insert. pCYMR plasmids were maintained in E4644 (LB + Km, 50 μg/ml). Inserted sequences were confirmed using primers #2037 (CATGATCACCATAGATCCTTTCTCC) and #787 (ACCGGTTAATTAACGGCACCACCGAA). Three variants of pCYMR were made with altered activity of the expression promoter: pCYMR.1 (standard vector), pCYMR.4 (higher expression) and pCYMR.6 (lower expression; Supplementary Figure S3). The detailed structure of the pCYMR expression plasmids and the cloning junctions for the genes inserted are given in Supplementary Figures S3 and S4.
Integrating pLOM-500
The pLOM-500 reporter plasmid containing the protein-binding sites is separately integrated into three strains: AH6112 (lacI–Oid+), AH6113 (lacI+Oid–) and AH6114 (lacI+Oid+). These strains differ in whether Oid is present at the distal site in the loopometer landing pad (Oid+) or absent (Oid–; Figure 2, sequences in Supplementary Figure S2D), and whether the pIT3-SH.lacI-rev plasmid (lacI+; Supplementary Figure S2E) or the empty vector pIT3-SH (lacI–) is integrated at attPHK022. Strains AH6112, AH6113 and AH6114 also contain the pINTts λ Int helper plasmid (37), which express λ Int at temperatures above 30°C and carry a temperature-sensitive pSC101 repA101 gene needed for its maintenance. For transformation and integration, these cells are grown at 30°C in LB + Ap, 100 μg/ml to OD600 ∼0.2–0.4, transferred to 39°C for 20 min, before being made competent and transformed with pLOM-500 vectors by the TSS method (39), with selection for integrants on LB + Cm, 20 μg/ml plates at 37°C. Purified colonies are tested for correct pLOM-500 integration by PCR (40) with a mix of primers #462 (ATGACAGAGGCAGGGAGTGG), #1729 (TTGTGCTTCTCTGGAGTGCG), #1685 (GTATCCCCACTCACTTAGTC) and #1686 (CCAGTGAATCCGTAATCATGG), which give 351, 841, 470 and 727 bp attP, attB, attL and attR fragments, respectively.
Integrating pCYMR
The three resulting strains are then transformed with the pAH121 φ21 Int helper plasmid (37), with outgrowth in LB at 30°C and selection on LB + Ap, 100 μg/ml plates at 30°C. Integration competent cells of these strains are then made and transformed (as above) with empty pCYMR or the pCYMR plasmid containing the candidate protein gene, with selection on LB + Km, 20 μg/ml plates at 37°C. Purified colonies are tested for correct integration by PCR (40) with a mix of primers #465 (GGGAATTAATTCTTGAAGACG), #467 (ACTTAACGGCTGACATGG), #871 (ATCGCCTGTATGAACCTG) and #2404 (GCAGCCTAACAAAAAACAACG), which give 1226, 420, 621 and 1025 bp attP, attB, attL and attR fragments, respectively. Alternative methods for expressing the candidate protein may be used, as long as they do not require selection for chloramphenicol (the reporter module) or spectinomycin (the LacI expression module), and are not under LacI control.
LacZ assays
The standard loopometer assay involves LacZ (β-galactosidase) assay of six strains, all with the same protein-binding sites inserted at loopometer sites 1 and 2. The strains have three different combinations of lacI and Oid: lacI–Oid+, lacI+Oid– and lacI+Oid+, and carry either the integrated empty pCYMR vector or the pCYMR vector expressing the relevant protein.
Strains were grown and kinetic LacZ assays were performed in 96-well flat-bottomed microtiter plates. Six fresh colonies of each strain on LB + Cm, 20 μg/ml plates (except for ID1292 and ID1293, which were selected with Km20) were picked with a 1000 μl micropipette tip and resuspended in 40 μl M9MM + Cm20. About 5 μl of each resuspension was added to 95 μl M9MM+ Cm, 20 μg/ml + CA in a microtiter plate, sealed and incubated with shaking at 37°C overnight. These cultures were then diluted 5 μl into 95 μl fresh medium and incubated for ∼3 h. The five control strains were grown in 0 and 128 μM CA, while the lacI+ Oid+ (pCYMR-X) strain was grown with 0, 8, 16, 32, 64 and 128 μM CA (see Figure 2B).
Cultures were grown to OD600 0.3–0.8, measured using a Labsystems Multiskan Ascent plate reader with a 620 nm filter. The OD620 values were converted to OD600 (1 cm path length) values using an empirically derived relationship. For the assay, 5 μl of culture was added to 195 μl warmed assay buffer in a fresh microtiter plate, consisting of 88 μl TZ8 buffer, 60 μl 4 mg/ml o-nitrophenyl-β-D galactoside (Sigma-Aldrich Cat. N1127) in TZ8, 2 μl 10 mg/ml chicken egg white lysozyme (Sigma-Aldrich Cat. L6876, 40,000 units/mg) in TZ8, 4 μl 20 mg/ml polymyxin B (Sigma-Aldrich Cat. P-4932) in H2O, 6 μl H2O and 35 μl M9MM. TZ8 buffer is 100 mM Tris-HCl, pH 8.0, 1 mM MgSO4, 10 mM KCl. The plate was incubated at 28°C in the plate reader, with OD414 readings taken every 2 min for 1 h. Enzyme activity was determined as the slope of the line of best fit of OD414 versus time (readings with OD414 > 2.5 were ignored). LacZ units were calculated as 200 000 × (OD414/min)/(OD600 x 5 μl). The improved linearity compared with our previous assay (41) is examined in Supplementary Figure S5.
Quantitation of looping
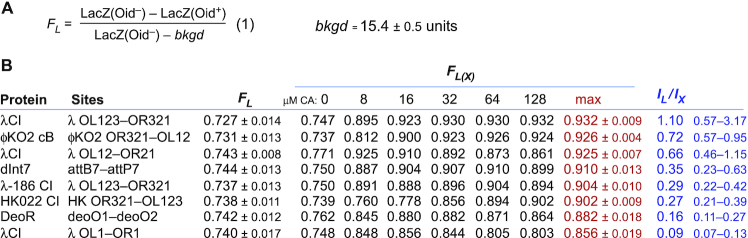
FL(X), the fractional LacI looping in the presence of the candidate protein, is obtained from the LacZ activities of the lacI+Oid+ and lacI+Oid– reporters with the expression of the protein, combined with the background LacZ activity of strain ID1285, using Equation (1) (Figure 4A).
Figure 4.
Calculating fractional looping. (A) Equation (1) for calculation of fractional LacI looping from reporter LacZ units is from Hao et al. (34). Background units were measured with the lacI+Oid+ Plac– reporter strain ID1285 (n = 16). (B) FL values were calculated from paired Oid+ and Oid– assays in the absence of the candidate protein (n = 12; except n = 10 for λCI:OL1-OR1; 0 and 128 μM CA values pooled). FL(X) values for LacI looping in the presence of the candidate protein at each CA concentration were calculated the same way (n = 6, except n = 5 for λCI:OL1-OR1). Values are ±95% confidence limits (Student's t). The 95% confidence intervals for the IL/IX values were determined numerically from 1000 calculations of IL/IX in which the values for FL(X), p, q and r were individually varied according to their errors.
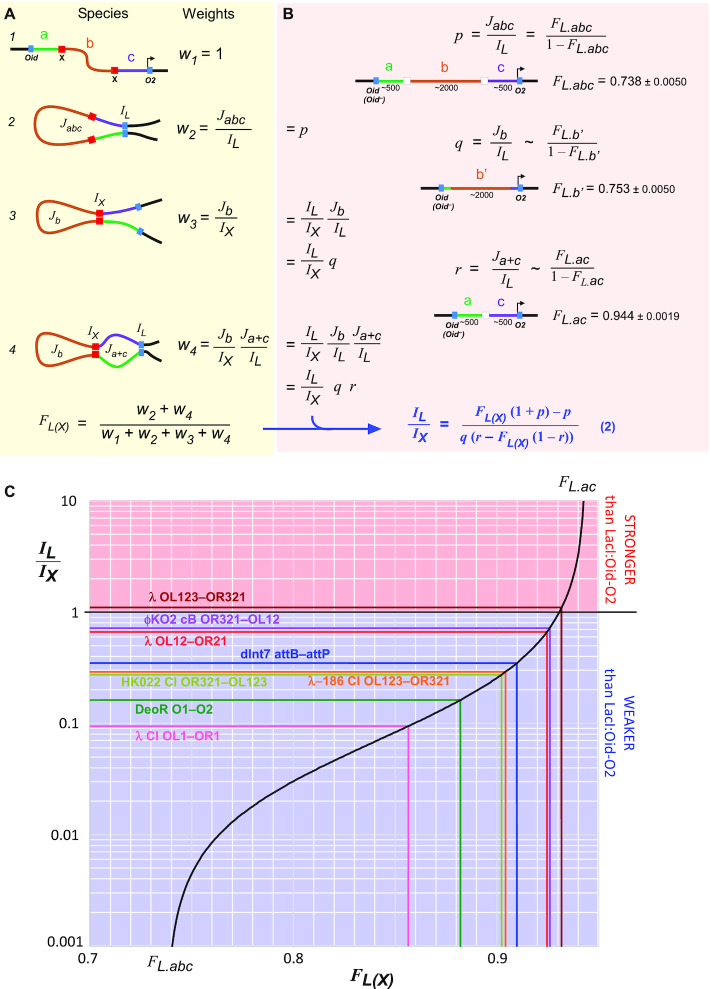
Estimation of IL/IX from this FL(X) value uses Equation (2) (Figure 5B), which requires additional FL measurements to estimate the weights p, q and r. The derivation of Equation (2) is based on our statistical-mechanical model for loop assistance (Figure 5; 35). The weight p is obtained from the FL measurement of LacI looping of the lacI+Oid+ and lacI+Oid– versions of the loopometer in the presence of empty pCYMR. The weight q is estimated from assays of the lacI+Oid+ and lacI+Oid– strains ID1290 and ID1291, in which Oid and Plac.O2 are separated by a b’ sequence similar to that between sites 1 and 2 (b, Figure 5B). The weight r is estimated from assays of the lacI+Oid+ and lacI+Oid– strains ID1292 and ID1293, in which Oid and Plac.O2 are separated by a concatenation of the 500 bp Oid-site 1 and site 2-Plac.O2 a and c arms separated by a 45 bp λ OR21 segment (Figure 5B).
Figure 5.
Estimating looping strength relative to LacI/Oid-O2. (A) Our statistical–mechanical model for two nested DNA loops applied to the case of the loopometer reporter (see text). (B) Derivation of Equation (2), which relates IL/IX (the looping strength of the candidate protein and sites relative to LacI:Oid-O2) to loopometer measurements of FL and FL(X) and the statistical weights p, q and r for looping of particular DNA segments by LacI alone (see text). FL.abc is the mean of the eight FL values in Figure 4 (n = 8), but values for individual reporters could also be used. FL.b’ and FL.ac values (n = 32 and 28) were obtained from strains ID1290 and ID1291 or ID1292 and ID1293, respectively. Values are ± 95% confidence limits (Student's t). (C) The plot of Equation (2), using the p, q and r values of (B), showing the measured FL(X) values and derived IL/IX values of the proteins/sites tested.
Availability
Plasmids pAS0007 ( = pLOM2–500 with φKO2 operators: Addgene # 164868), pID1072 ( = pCYMR-1 with φKO2 cB: Addgene # 164869) and plasmid pAH121 (Addgene #164901), as well as strains AH6112–4, ID1285, and ID1290–3 will be deposited in Addgene.
Details of system construction
pLOM plasmids
Plasmid pLOM1–500 is an earlier version of pLOM2–500 that does not carry flanking restriction sites on both sides of sites 1 and 2, and was used for reporters with some binding sites (Supplementary Figure S1B). The sequences of protein binding sites cloned into these vectors are listed in Supplementary Figure S1.
pCYMR plasmids
The CymR repressed promoter in these vectors was derived from the T5 promoter.cymO sequence of pNEW (42), which includes an unwanted overlapping lac operator and also displayed leaky CymR repression. We randomized the sequences between the -35 and -10 hexamers (including the first -10 basepair) to remove this operator and alter promoter activity to generate the pCYMR-1, pCYMR-4 and pCYMR-6 variants (Supplementary Figure S3).
Loopometer recipient strains
The reporter landing pad present in strains AH6112, AH6113 and AH6114 was constructed in two steps by recombineering (Supplementary Figure S2A) using pSIM6 (43). First, a PCR fragment containing a kanamycin resistance gene (KmR) and a distal portion of the lacZ gene was used to replace attBλ in the E4643 chromosome to give strain AH6101 (Supplementary Figure S2A). Second, after reintroduction of pSIM6, a PCR fragment carrying Oid+ (or Oid–), attBλ, PlacUV5.lacO2 and the proximal portion of the lacZ(O2–) gene (Supplementary Figure S2A) was used to replace KmR, with screening for lacZ+ on LB + X-gal (20 μg/ml) plates. These two strains were transformed with the pAH69 HK022 Int helper plasmid (37) and pIT3-SH.lacI-rev (Supplementary Figure S2E), or the empty vector pIT3-SH were then integrated at attPHK022. The resulting strains were transformed with the pINTts λ integrase helper plasmid (37) to give AH6112 (lacI–Oid+), AH6113 (lacI+Oid–) and AH6114 (lacI+Oid+).
ΔdeoR strains
Strains equivalent to AH6112 (lacI–Oid+) and AH6114 (lacI+Oid+) but carrying a deletion of the deoR gene were made by recombineering, first replacing sequences between MG1655:CATCAACTTAATGCG 881996 and 882732 ATAATCCCTCTGAA with a KmR cassette flanked by FRT sites, followed by FLP-mediated removal of the KmR gene using the pE.FLP plasmid (40), resulting in an in-frame deletion of almost the entire deoR gene.
The Plac– strain for measuring reporter background
A DNA segment spanning the leftward FRT site, chloramphenicol resistance gene (CmR) and PlacUV5 was obtained from AH6114 carrying an integrated pLOM2–500 vector (Supplementary Figure S2B) and cloned into a plasmid, where mutations were introduced into the –35 and –10 sequences of PlacUV5 (Supplementary Figure S2D). A PCR fragment (Supplementary Figure S2C) from this Plac–plasmid was used for recombineering into an AH6114 (pLOM2–500) strain in which the sequences between the FRT sites (Supplementary Figure S2B) had been removed by treatment with pE.FLP (40), allowing selection for the CmR gene in the insert. The resulting Plac–strain, ID1285 is lacI+Oid+, with matS null sequences (Supplementary Figure S1) at sites 1 and 2.
Strains for calibration of LacI looping
The DNA sequences for construction of b’ and ac strains (Supplementary Figure S6) were assembled in plasmids, which were then used to generate PCR fragments (Supplementary Figure S6) for recombineering into the chromosome of AH6101 (Supplementary Figure S2A), with selection for CmR (b’) or screening for lacZ+ (ac). Successive integration of pIT3-SH.lacI-rev and empty pCYMR gave b’ strains ID1290/ID1291 (Oid+/Oid–) and ac strains ID1292/ID1293 (Oid+/Oid–). The b’ segment shares 1870 bp with the loopometer b segment, with divergent sequences at each end near Oid and Plac.O2. The ac strain carries a 45 bp λ OR21 insert between the fused sites 1 and 2.
Extended reporters were made by insertion of plasmid pID1302 into the attB3 site of the b’ reporters ID1290/ID1291, and full-length lacI+Oid+reporters (abc) carrying the matS null site (Supplementary Figure S1) at both sites 1 and 2. Plasmid pID1302 (Supplementary Figure S6B) carries the attP3 site, the pir-dependent R6Kγ origin, a tetracycline resistance gene (TcR) and ∼6 kb of spacer sequence derived from the E. coli ftsK and rne genes. The int3 gene and the Int3 attachment sites are from the collection of Yang et al. (44). Integration of pID1302 (Supplementary Figure S6B) was mediated by expression of Int3 from the helper plasmid pAH6046, which was derived from pINTts (37) by replacing λ int with the int3 gene.
Sequences of strains and plasmids are available on request.
RESULTS AND DISCUSSION
Procedure for testing DNA looping by a candidate protein
The standard assay uses a looping reporter and a separate module for the controlled expression of the protein, both of which are integrated into the bacterial chromosome (Figure 2A).
The looping reporters are constructed by insertion of two potential binding sites for the candidate protein at sites 1 and 2 on either side of the bacteriophage λ attP attachment site (attPλ) in plasmid pLOM2–500, using standard cloning techniques (‘Materials and Methods’ section). The resulting plasmid is then integrated into the chromosome of three different E. coli strains by recombination into an attBλ site on a specially constructed ‘landing pad’ (Figure 2A). Integration is catalyzed by λ integrase expressed from a separate helper plasmid (37). Integration results in the two binding sites being located ∼2 kb apart, separated by the inactive plasmid replication origin and the chloramphenicol resistance gene, with the proximal binding site located ∼500 bp upstream of a lacO2 operator controlling a promoter (PlacUV5) for a lacZ reporter gene. Integration into strain AH6114 produces the intact loop reporter, which has a second lac operator, Oid, located ∼500 bp upstream of the distal binding site, and a lacI gene elsewhere on the chromosome. The AH6112 and AH6113 strains lack either lacI or Oid, respectively, and serve as controls.
For expression of the candidate looping protein, we routinely use a chemically inducible expression system based on the CymR repressor from Pseudomonas putida F1 (42), which is inactivated by added CA. This allows testing of a range of protein concentrations, which may be important if high protein levels give submaximal looping, as seen for LacI (29), or are toxic. The gene for the candidate looping protein is inserted into the pCYMR plasmid, and the resulting plasmid is transformed into the three reporter strains previously transformed with the phage φ21 integrase helper plasmid (37), for catalyzing integration at the φ21 attachment site on the bacterial chromosome (attB21) (Figure 2A). Our standard pCYMR.1 vector gives a low level of uninduced ‘leak’, with strong induction by CA (Supplementary Figure S3B). Nevertheless, we generally also integrate an empty pCYMR.1 plasmid into the reporter strains to provide control strains with no candidate protein.
Once the strains have been constructed, LacZ assays are done to test whether CA-induced expression of the candidate protein gives an Oid-dependent increase in LacI repression of the lacZ gene (Figure 1B). We use a microtiter plate-based version of the basic Miller LacZ assay (‘Materials and Methods’ section); however, standard Miller assays are adequate.
Validation of the assay with λ CI
We tested the assay with the λ CI repressor (45). Binding of CI dimers to individual operators via the N-terminal domains, and further association of these dimers to tetramers and octamers mediated by the C-terminal domain (46,47), is able to produce a variety of DNA loops. The natural cooperative binding of two CI dimers to pairs of adjacent operators at its OR- and OL-binding sites is due to mini-loops, with the operators, which are spaced ∼2 DNA turns apart, being ‘looped’ by a CI tetramer (17,48). DNA looping has also been seen between single operators spaced 5 or 6 DNA turns apart (25). Longer DNA loops—beyond 2 kb in vitro and in vivo—have been observed for the interaction of two tetramer-bound sites, resulting in four operators linked by a CI octamer (14,17,28). The natural three-operator groupings at OL and OR are capable of even stronger looping (28,35), presumably because the two operators not bound by the octamer are bridged by an additional CI tetramer (49) (Figure 2B). We have shown that the full OL and OR sequences can form a 20 kb loop with ∼16% efficiency in vivo (35).
Figure 2B shows the results of CA-controlled expression of λ CI protein with λ OR and OL inserted at sites 1 and 2 in the reporter. In the lacI– background, the reporter is unrepressed, giving ∼3000 LacZ units, and this activity is unaffected by the CI expression module, with or without full induction by CA (128 μM). In the lacI+ background but in the absence of the upstream Oid operator, the reporter is repressed ∼5-fold due to weak binding of LacI to the proximal O2 site. Again, this activity is unaffected by CI expression. These controls check that any binding of the candidate protein to its sites does not directly affect the promoter or affect LacI repression in the absence of the upstream Oid. In the presence of Oid and the absence of CI, LacI repression of the reporter increases ∼4-fold. This loop-dependent repression effect is due to the strong Oid site binding a LacI tetramer and thus fixing the free DNA-binding domain of the tetramer at the end of a 3 kb tether attached to O2. At this distance, the effective concentration of the LacI DNA-binding domain seen by O2 is substantially higher than the concentration of the LacI DNA-binding domains of free LacI tetramers, resulting in frequent DNA looping and increased occupation of O2.
In the presence of CI, repression in the lacI+Oid+ reporter is increased even further (Figure 2B). Given the results of the control strains, this increased repression can be interpreted as DNA looping by CI between OL and OR that, at least some of the time, shortens the distance between Oid and O2 to increase their relative concentration and increase LacI looping (Figure 1).
We generally test a range of induction levels for the candidate protein, as DNA looping generally has a concentration optimum (29,34). Too low protein concentration gives insufficient occupation of the DNA sites; too high concentration causes loop blockage, where the sites are occupied by higher order multimers that are unable to interact further (e.g. LacI tetramers at both lac operators).
To test whether the assay could display differences in the strength of looping, we compared λ CI looping of the native 3-operator OL and OR sites with looping by 2-operator or single operator sites. The equivalent assay to Figure 2B was done for reporters with these sites, and a subset of the results for all three combinations is plotted in Figure 2C. These plots are normalized to the average units obtained for each Oid– reporter (± CI, 0/128 μM CA) to account for day-to-day variation in the LacZ assay.
As expected, the single operator OL1-OR1 reporter displayed weaker looping than the multioperator combinations, with CI expression giving less assistance to LacI repression at all induction levels. We note that this is the first time that looping has been observed for single CI operators spaced more than a few DNA turns apart. Looping by the 2-operator OL12-OR12 combination was also weaker than the 3-operator pair at most CI induction levels.
Interestingly, the 1- and 2-operator combinations gave maximal looping at low CI and less looping as CI levels increased. Formally, this can be explained as being due to the formation of higher order looping-incompetent CI multimers at each site. However, we were surprised to see this effect for λ CI, as it implies that CI octamers are forming independently at 1- or 2-operator sites. This effect was not seen for the 3-operator OL and OR sites, presumably because loop blockage is counterbalanced by loop-promoting bridging between the third operators (49).
Having validated the assay, we then used it to test other known or suspected DNA-looping proteins.
DeoR
The E. coli DeoR protein utilizes DNA looping in its repression of an operon for catabolism of nucleosides. DeoR binds to single operators (O1 and O2) spaced ∼600 bp apart at each of a pair of tandem promoters, with a third site (OE) located ∼300 bp further upstream. Repression at either promoter is improved in the presence of the other sites and is detectable when O1 is placed ∼5 kb away from O2 (50). DNA loops between all sites have been observed by electron microscopy (51).
We inserted the deoO1 and O2 operators into sites 1 and 2 of the loopometer reporter, the deoR gene into the pCYMR-1 expression module and tested loop assistance in strains in which the endogenous deoR gene was deleted (‘Materials and Methods’ section). A clear Oid-dependent increase in LacI repression was seen with increasing expression of DeoR (Figure 3A). A similar degree of looping was seen for the reporter exposed to the endogenous level of DeoR protein in a deoR+ strain (Figure 3A).
Figure 3.
Testing candidate proteins and sites for DNA looping. LacZ units produced by loopometer reporters in lacI+Oid+ and lacI+Oid– strains, normalized to the average of the Oid– values. Protein expression was induced with CA from chromosomally integrated pCYMR vectors or an empty vector control. Measurements in lacI–Oid+ strains showed no substantial effects of candidate proteins in the absence of LacI and are omitted to save space, except for (G) and (H). Error bars are 95% confidence limits (Student's t), n = 6 unless otherwise stated. Sequences of inserts at loopometer sites 1 and 2 are given in Supplementary Figure S1; protein sequences inserted in pCYMR are given in Supplementary Figure S4. (A) DNA looping between deoO1 and deoO2 operators by endogenous or exogenous E. coli DeoR. (B) DNA looping between full λ OL123 and OR123 sites by a hybrid protein with the λ CI DNA-binding N-terminal domain and the phage 186 CI C-terminal association domain. (C) DNA looping between phage HK022 OL and OR by HK022 CI. (D) DNA looping between phage φKO2 OL and OR by φKO2 CB. (E) DNA looping by integrase dInt7 (catalytically inactivated) between its attB and attP sites (left), but a lack of looping between its attL and attR sites (right). (F) A lack of looping between matS-binding sites by endogenous E. coli MatP is indicated by the absence of decreased LacZ units compared to a control reporter with only a single matS site. (G and H) Inconclusive looping assays for phage 186 CI looping between its pR sites and regulatory FL site, and for phage P1 RepA looping between P1 triple-iteron sites. In both cases, the expressed proteins have confounding lacI-independent and Oid-independent effects.
Hybrid λ-186 CI repressor
We have previously shown that a hybrid repressor in which the λ CI DNA-binding N-terminal domain (NTD) is fused to the phage 186 CI oligomerization C-terminal domain (CTD) is able to regulate the λ PR and PRM promoters at OR in a way that responds to the presence of OL located 3.8 kb away (52). The intact 186 CI protein is a nonlambdoid repressor that uses distal binding sites for regulation in vivo (53) and forms DNA loops in vitro (27). The 186 CI CTD forms a wheel-like 14-mer structure that should present attached NTDs on its rim for DNA binding (52). It is thus very likely that this hybrid protein can form DNA loops; however, this has not been independently confirmed.
When we expressed the λ-186 CI hybrid from integrated pCYMR-1 (‘Materials and Methods’ section) in the full λ OL-OR loopometer reporter, we saw an increase in Oid-dependent LacI repression indicative of DNA looping (Figure 3B). Looping was weaker than we saw with intact λ CI (Figure 2C).
Lambdoid phage repressors: HK022 CI and φKO2 CB
φHK022 is a lambdoid phage with a similar basic genomic arrangement to λ, including divergent lytic operons controlled by OL and OR elements separated by the immunity region, including the cI gene (54). The natural distance between OL and OR is ∼700 bp in HK022, compared to 2.3 kb in λ. The HK022 CI repressor shows homology with other lambdoid repressors and is known to bind cooperatively to adjacent operators within OR and OL (54). DNA looping by HK022 CI seems possible given these similarities to λ, and is further suggested by loop-like DNase I sensitivities induced by CI in the 53 bp between OL2 and OL3, and by the location of a CI operator 300 bp downstream of OR that is involved in immunity (54,55). However, DNA looping has not been tested.
The entire HK022 OR and OL regions (with mutations to inactivate the pL and pR promoters; Supplementary Figure S1A) were inserted at the loopometer sites 1 and 2 (Supplementary Figure S2). Expression of HK022 CI produced an Oid-dependent increase in LacI repression of the reporter, clearly showing DNA looping (Figure 3C).
Klebsiella oxytoca phage φKO2 is another lambdoid phage, closely related to N15. These ‘telomeric’ phages are unusual in forming nonintegrated linear prophages and are classed as lambdoid based on their operon structures and sequence similarities (56). The φKO2 immunity repressor CB shows homology with lambdoid repressors and represses lytic transcription from OL and OR regions located ∼630 bp apart, either side of the cB gene (57). The similarities to λ led Hammerl et al. (57) to speculate that φKO2 CB protein might form a DNA loop between OL and OR. However, this has not been experimentally validated.
We inserted φKO2 OL and OR (with mutations to inactivate promoters; Supplementary Figure S1A) into the loopometer and expressed the φKO2 CB protein. Increased LacI repression seen in the presence of CB induction confirmed OL-OR looping by CB (Figure 3D).
Thus, all three of the lambdoid phage repressors tested—λ, HK022 and φKO2—display DNA looping between the operators that control early lytic transcription. The genomic arrangement of divergent lytic promoters separated by a short immunity region containing the repressor gene is a module common to many lambdoid phages, suggesting that long-range repressor looping is widespread. In λ, CI looping increases repression of the lytic promoters (14,15), and also provides complex control of the immunity promoter (17). Looping-dependent cooperative repression of the lytic promoters seems likely to be a shared feature; however, differences in the operator arrangements in HK022 and φKO2 suggest that these and other lambdoid phages may use looping differently in control of the immunity promoter.
dInt7—an inactivated serine integrase
Site-specific recombination between sites either on the same or different DNA molecules must involve the formation of a protein bridge between the two DNA sites. However, the strength of this DNA bridging capability is not clear, since the catalytic steps of recombination may be sufficiently fast that only a transient DNA contact is required.
Serine integrases are a large class of site-specific recombinases that, in the absence of a recombination directionality factor (RDF), catalyze a unidirectional reaction between their attachment sites—attP + attB → attL + attR—without the need of cofactors such as IHF (58). The bridging complex is an Int tetramer with the two DNA sites each bound to an Int dimer.
To test the DNA-looping capacity of serine integrases, we used Int7, one of a set of 34 large serine integrases isolated and characterized by Yang et al. (44). We made an inactive variant—dInt7—by mutating the catalytic serine (Ser10) to alanine, and inserted this into the pCYMR-1 expression module. The cognate attB7 and attP7 sites were inserted into sites 1 and 2 of the loopometer reporter, in an inverted orientation. We saw a clear Oid-dependent increase in LacI repression when dInt7 was expressed (Figure 3E), indicative of substantial DNA looping between attB7 and attP7.
The inability of the serine integrases to recombine attL and attR is thought to be due to poor synapsis between Int-bound attL and attR sites. Electrophoretic mobility shift assays with φC31 and BxB1 integrases found synaptic complexes only between attP and attB (59,60). A model stimulated by structural studies of the LI integrase bound to attP DNA suggests that upon completion of the DNA strand cleavage and rejoining reactions between attP and attB, new cis interactions form between the Int monomers bound at attL and attR that prevent Int tetramerization (61).
To test the expected lack of synapsis between the attL7 and attR7 sites, we used transient expression of active Int7 to recombine attP7 and attB7 in the loopometer reporter, generating reporters with an inverted 2 kb internal segment flanked by attL7 and attR7 sites. Expression of dInt7 did not result in detectable DNA looping in these reporters (Figure 3E). This result provides in vivo confirmation of an inability of the integrase alone to bring attL and attR together, at least for Int7. However, the assay does not by itself show whether the looping defect is due to a lack of Int binding to attL or attR or whether Int is bound but cannot loop.
In general, it is not possible to be certain from a negative loopometer result that a protein and sites are incapable of looping, because the assay does not provide independent confirmation that the protein is expressed, is active and is binding its DNA sites. In the case of dInt7, we at least know that the expressed protein is active by its looping of attP7-attB7, but while other serine integrases are known to bind to attL and attR sites in vitro (59,60), it is possible that this binding is not occurring for dInt7 in vivo.
A negative result for MatP:matS
Another example of a negative result is provided by our examination of looping by MatP. The E. coli MatP protein binds to 13 bp sites termed matS that are clustered around the replication terminus of the chromosome. Some 23 matS sites are distributed over an ∼800 kb region corresponding to the Ter macrodomain, with MatP and matS sites functioning to structure and insulate this domain (62). X-ray crystallography of MatP–matS complexes showed a MatP tetramer forming a bridge between two separate matS DNAs, with DNA looping also observed in vitro by electron and atomic force microscopy (63), leading to the idea that the Ter domain might involve a network of MatP–matS looping. However, more recent Hi-C measurements did not reveal strong contacts between matS sites. This and other results indicate that MatP exerts its effects without DNA looping (64).
To test MatP/matS looping, we constructed loopometer reporters carrying a matS sequence at sites 1 and 2. To avoid effects on the cells due to the elimination of the endogenous MatP protein (62), we made control reporters without matS at site 1. We found that LacI looping was not enhanced by the presence of the second matS site, indicating a lack of looping (Figure 3F).
This result supports the idea that MatP does not loop matS sites in vivo. However, our assay does not confirm the binding of MatP to our matS sites, and it is possible that the DNA sequence context or the cellular location somehow reduces binding at our sites relative to the natural sites at Ter. The conflict between the in vitro evidence for looping and the in vivo results may be due to a variety of factors. The in vivo concentration of MatP may be far from optimal for looping; either too low to give significant occupancy of matS sites (note that ChIP assays (62) can be positive even when occupancy is low) or so high that MatP tetramers bind independently to each matS site. Bound MatP dimers may also interact with other proteins that block dimer–dimer contacts in vivo.
Inconclusive results: 186 CI and P1 RepA
In two cases, testing of DNA looping in the loopometer gave inconclusive results because the expression of the candidate protein affected LacZ units in the control strains lacking either LacI or Oid.
In attempting to test DNA looping by intact phage 186 CI repressor between its FL and pR regulatory sites (53), we found that while expression of 186 CI reduced LacZ units in the lacI+Oid+ strain, it also reduced LacZ units in the lacI+Oid– and lacI–Oid+ control strains. This effect was also seen (Figure 3G) when low levels of expression of 186 CI were used to avoid potential cellular effects by the use of the pCYMR-4 variant, which uses a weaker expression promoter (Supplementary Figure S3). Expression of 186 CI also inhibited LacZ activity in reporters that contain non-186 sequences at sites 1 and 2, suggesting that 186 CI is somehow affecting LacI expression or its repression of Plac. The cause of this effect is currently unresolved and prevents conclusions about 186 CI DNA looping from the assay.
We also attempted to test looping by the phage P1 RepA DNA replicase protein, which has been proposed to interact when bound to iteron sequences located on different DNA molecules in a ‘handcuffing’ interaction that is thought to help control the copy number of the plasmid P1 prophage (8). However, we found that the expression of P1 RepA also affected LacZ activity of the control strains (Figure 3H).
These results show that the loopometer is not a foolproof assay for DNA looping and highlight the importance of using the control reporter strains.
It has been pointed out to us that these control strains would not reveal the effects of an expressed protein on general factors that could specifically affect DNA looping, such as levels of DNA supercoiling (65) or nucleoid-associated proteins (21,66). In cases where such effects are suspected, we recommend testing the effect of the expressed protein on Oid+ and Oid– reporters that do not contain binding sites for the protein, in order to detect any confounding effects on LacI looping.
Model-based quantitation of looping strength
A simple display of whether or not a candidate protein and its sites interact to loop DNA provides useful information about the mechanism of action of the protein. The fractional reduction in LacZ units in the lacI+Oid+ reporter relative to the lacI+Oid– reporter (Figure 3) allows a comparison of the looping effect of the different proteins, sites and concentrations, but does not provide quantitation of the relative strengths of looping. Quantitation of looping strength can provide more information about the effect of concentrations or site variants, the likely importance of looping in the protein’s activity and allows better comparison of DNA looping by different proteins.
To quantitate looping strength, the first step is to calculate the fractional LacI looping FL, that is, the fraction of time that Oid and O2 are looped by LacI. This is obtained using Equation (1) (Figure 4A; (34)), which takes into account background LacZ units that result from low level lacZ expression from sources other than Plac. This background can be measured using strain ID1285, which carries a mutated Plac promoter (‘Materials and Methods’ section), giving ∼15 LacZ units with our assay. The fractional decrease in units seen with the lacI+Oid+ reporter relative to the lacI+Oid– reporter gives a linear read-out of the fractional looping; when there is no looping, the Oid+ and Oid– units are the same, giving FL = 0; if looping were 100%, Plac would be fully repressed and the Oid+ units would equal the background, giving FL = 1.
Figure 4B shows the FL values calculated from the data of Figures 2 and 3. In the absence of the test proteins, the lacI+Oid– and lacI+Oid+ reporters give FL values (for LacI alone) of ∼0.74. That is, at the LacI concentration in our reporters, the 3 kb loop between Oid and O2 is formed ∼74% of the time. FL(X), the fraction of LacI looping in the presence of the various internal looping proteins, is increased over these FL values (Figure 4B). Slight increases in looping are seen with uninduced expression (0 μM CA) due to some leak in CymR repression. The maximal values of FL(X) allow the looping strengths of the test proteins and sites to be ranked. In this set, the strongest looping was exhibited by λCI/OL123-OR123 and the weakest looping by λCI/OL1-OR1, with the other proteins and sites giving intermediate looping strengths. We note that this comparison between different proteins is based on the assumption that differences in the structure of the bridge formed by the different proteins between sites 1 and 2 do not substantially affect LacI looping, that is, that the measured differences in LacI looping are due solely to differences in the frequency of formation of the internal loop. The large size of the external loop, 500 + 500 bp, gives us some confidence in this assumption.
The FL and FL(X) values can be used to move beyond a simple ranking and to obtain estimates of the relative looping strengths by estimating the strength of looping relative to LacI.
The approach uses a simple model for loop assistance (Figure 5A; 35). The model specifies four species due to the looped or nonlooped state of each pair of sites. Each of these species can be assigned a statistical weight or relative propensity (w1 to w4). The propensity to form a DNA loop is a balance between the energetic cost of bringing together two DNA sites (primarily entropic at these long distances), and the energetic benefit provided by the interaction of the sites due to the protein–DNA and protein–protein interactions involved. The cost is inversely related to the effective relative concentration of the two DNA sites, J, which for sites in cis is a function of the distance between them along the DNA (29,35,36). We represent the benefit by the factor I, which is effectively a loop dissociation constant, being inversely related to the benefit and having units of concentration (35). I quantitates the ‘looping strength’ of a protein and its DNA sites, with lower I indicating higher looping strength. I is determined in a complex way by the specific DNA:protein and protein:protein binding constants, the protein’s concentration and the various looping and nonlooping complexes formed at the two DNA sites. Each of the weights for the single-looped species is given by the ratio of the J factor for the DNA loop and the I factor for the protein-mediated pairing (Figure 5A). The weight for the double-looped species is the weight for the internal loop (w3) multiplied by a weight representing the closure by LacI of the small loop comprising the a and c arms bridged by the candidate protein (Figure 5A; 35). The fractional LacI looping in the presence of the candidate protein, FL(X), is then the sum of the weights for the LacI looped species divided by the sum of all weights (Figure 5A).
Using the loopometer data and two additional measurements, it is possible with this model to obtain an estimate of the ratio of the I value for the candidate protein (IX) relative to that of LacI between Oid and O2 (IL), IL/IX. The weights w2 to w4 can be expressed in terms of IL/IX and three weights for LacI looping of the various DNA segments: JL.abc/IL, JL.b/IL and JL.a+c/IL, designated p, q and r for brevity (Figure 5B). Substituting these terms into the equation for FL(X) allows IL/IX to be obtained from FL(X) if p, q and r are known (Figure 5B, Equation 2). Estimates of these three weights can be obtained because the weight for a single DNA loop is related to F for that loop: J/I = F/(1–F) (35). The weight p can be obtained from the FL measurement of LacI looping of the loopometer in the absence of the internal protein (Figure 5B). To estimate q, we constructed and assayed Oid+ and Oid– versions of reporters with Oid and O2 separated by a 2 kb b-like DNA segment (Figure 5B; strains ID1290 lacI+Oid+ and ID1291 lacI+Oid–; ‘Materials and Methods’ section ). This b’ segment contains most of the internal loopometer b segment, but with different sequences near Oid and Plac.O2. Measurement of LacI looping in these reporters measures FL.b’, giving an estimate for q = Jb/IL. We note that equal looping of these b’ and b sequences by LacI has not been demonstrated. To estimate r, we constructed and assayed reporters in which the internal b segment was removed, leaving Oid and O2 separated by a 1 kb ac segment comprised of the joined 500 bp a and c arms (strains ID1292 lacI+Oid+ and ID1293 lacI+Oid–), allowing measurement of FL.ac. Assuming that the looping of the DNA-joined ac segment is similar to the looping of the a+c segments when they are brought together by the b-looping bridge formed by the candidate protein, we can use FL.ac as a proxy for FL.a+c. We note that we saw some divergence from this assumption in a previous study, suggesting that the internal b loop or the protein bridge may affect the looping of the a and c arms (35). Thus, the FL and FL(X) measurements from the loopometer, combined with assays of four pre-made strains allows estimation of IL/IX for the candidate protein and its sites.
The IL/IX values calculated in this way for the tested looping proteins and their sites are given in Figure 4B. Note that higher IL/IX values indicate higher looping strength of the candidate protein relative to LacI/Oid-O2 (as I is inversely related to the interaction strength). The relationship between IL/IX and FL(X) is plotted in Figure 5C, allowing a simple read-out of IL/IX from the loopometer measurement of FL(X). The various errors in the measurements result in substantial uncertainties in these IL/IX values, particularly for the stronger looping proteins. For λCI looping between OL123-OR123, we obtained IL/IX = 1.1, with a 95% confidence interval of 0.57–3.17 (Figure 4B). This range spans the IL/IX value of 2.7 obtained previously by our more direct comparison of λCI/OL123-OR123 and LacI/Oid-O2 looping (35). The steepness of the IL/IX versus FL(X) plot at high FL(X) values indicates that the loopometer is not well suited for distinguishing between very strong DNA-looping proteins. However, the curve shows that the loopometer should be capable of detecting and measuring very weak looping, down to looping ∼100-fold weaker than LacI/Oid-O2.
The IL/IX values do not provide absolute measurements of looping strength for individual proteins, but the internal LacI standard allows the looping strengths of the different proteins to be quantitatively compared. For example, the data indicate that maximal looping by λCI between OL12 and OR12 is some 2-fold weaker than for the full 3-operator sites, with single operator OL1-OR1 looping a further 5-fold weaker. Importantly, the use of these IL/IX values should assist the comparison of looping strengths of proteins that may be assayed under different conditions, aiding comparisons between assays done in different labs.
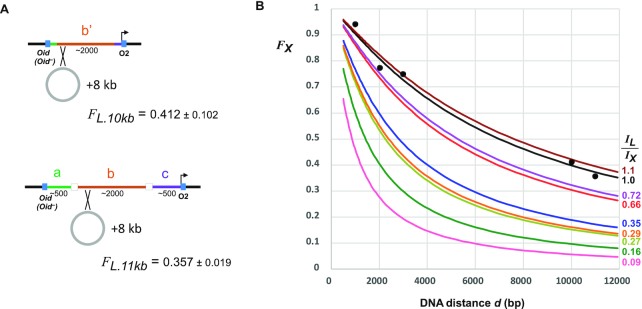
Ideally, the measurement of IL/IX in the loopometer would allow estimation of the fractional looping of the protein and its sites at any DNA distance. The fractional looping for a single loop is given by the simple relation: F = J/(J + I), and since we previously estimated IL = 83 nM (35), we should be able to obtain IX from the IL/IX value. We also determined a power law relationship between J and DNA distance d (bp): J = 1.04 × 106 × d1.15 nM by measuring LacI looping over distances ranging from 300 to 50 000 bp in the E. coli chromosome (summarized in (36)). Together, this should allow the F versus d relationship to be obtained for any IL/IX value. However, these previous measurements were made with an older version of our LacZ assay, which we now know gives an underestimate of LacI looping (Supplementary Figure S5). To recalibrate the relationship between LacI looping and DNA distance, we constructed loopometer reporters in which the Oid-O2 distance was increased by 8 kb by the integration of a plasmid into an integrase attachment site that had been included in our original constructs (Figure 6A; ‘Materials and Methods’ section). By assaying Oid+ and Oid– versions of these reporters, we thus obtained FL values for ∼10 and 11 kb spacings. Combining these with the FL measurements for ∼1, 2 and 3 kb spacings (Figure 5B), we found a reasonable match between these five values and the predictions of the F versus d relationship (Figure 6B; IL/IX = 1 curve), if either IL was about 2-fold lower, or the power law prefactor was about 2-fold higher, than our previously measured values. This recalibration allows the fractional looping of the proteins and their sites at any DNA distance on the E. coli chromosome to be estimated from the IL/IX values (Figure 5). For example, the loopometer measurement of IL/IX = 0.16 for DeoR/deoO1-O2 predicts ∼72% looping between these sites at their natural 600 bp spacing (65–82% based on the 95% confidence interval for IL/IX) and 20% looping at a 5 kb separation. The predicted looping between λ OL123 and OR123 at their natural 2.3 kb spacing is ∼79%, in agreement with an independent measurement using high-resolution live-cell microscopy ((31); though uncertainty in IL/IX gives a large range for predicted OL123-OR123 looping: 48–92%). Note that prediction of looping frequencies for spacings below 500 bp is unreliable due to the onset of helical phasing sensitivity at these distances.
Figure 6.
Estimating fractional looping for different DNA distances. (A) Reporters with increased Oid-Plac.O2 spacings were made by integrating an 8 kb plasmid into the abc and b’ segments of reporter strains to make strains ID1318/ID1319 (abc: Oid–/Oid+) and strains ID1320/ID1321 (b’: Oid–/Oid+). FL values (± 95% confidence limits, Student's t, n = 8) were measured as in Figure 4. (B) Plot of predicted fractional looping by the candidate protein, FX, versus DNA distance, d (bp), for different values of IL/IX, using the relationships F = J/(J+I) (35) and J = 1.04 × 106 × d1.15 nM (36) and assuming IL = 38.3 nM. Points are FL looping values for LacI/Oid-O2 from Figure 5 and (A) and are reasonably fitted by the FX curve for IL/IX = 1.
CONCLUSIONS
The loopometer represents a simple assay that can provide strong evidence for DNA looping in vivo. At a minimum, it requires only the construction of two plasmids—one for the binding sites and one to express the candidate protein—followed by sequential integration of these (and the empty expression plasmid) to make six bacterial strains for the LacZ assay. Quantitation of the looping strength relative to LacI/Oid-O2 is enabled by further assays of five pre-made strains, for measuring background and for LacI looping calibration.
Aside from the detection and measurement of DNA looping, the assay permits examination of the effect of protein concentration on looping, which may be an important factor in regulation. Additional constructions and assays can provide comparisons of looping between different sites, including testing the effect of binding site mutations. Similarly, the effects of protein mutations on looping can readily be examined, though the assay does not by itself distinguish between effects on protein–protein versus protein–DNA interactions.
The in vivo fluorescence microscopy-based assay of DNA looping by Hensel et al. (31) is the only approach that is currently able to provide similar information to the loopometer. In this study, the OL- and OR-binding sites for λ CI were placed adjacent to sites bound by fluorescently tagged proteins, and the distances between these sites were measured in live cells in the presence of CI. While this assay could readily be adapted to other proteins, it requires specialized equipment and expertise and is likely to be limited to strong DNA-looping proteins, as only these can efficiently loop DNA over the long distances required to resolve looped and unlooped states microscopically.
The loopometer assay currently has a number of limitations, some of which should be able to be overcome by further development: (i) The assay can fail if there are confounding effects of the expressed protein on looping-independent expression of lacZ. This could potentially be overcome by the use of a different reporter or by replacing LacI with a different looping protein. (ii) Comparison of looping strength between different proteins relies on the untested assumption that the precise structure of the bridge formed by the different proteins between sites 1 and 2 does not substantially affect LacI’s ability to loop the two 500 bp DNA arms. We suspect that DNA arms of different lengths could be used to test and validate this assumption. (iii) The assay only tests homotypic interactions, that is looping by a single protein. However, the assay could be easily modified to detect heterotypic interactions by introducing a second protein expression system. (iv) The assay can only test proteins that have specific binding sites and can be expressed in the active form in E. coli, which may limit testing of eukaryotic proteins. By itself, the assay does not provide evidence for the activity of the protein unless looping is observed. The use of fusions to bacterial DNA-binding domains (e.g. the λ CI NTD) may extend the range of testable proteins, since the ability to fold an active DNA-binding domain would not be required. We note that such fusions could provide an alternative bacterial one- or two-hybrid assay, allowing detection of interacting domains by DNA looping. (v) A bacterial assay may not be appropriate for eukaryotic looping proteins because of the lack of necessary accessory factors (e.g. nucleosomes). However, the principle of detecting DNA-looping proteins by loop assistance should also work in eukaryotic cells, as studies have shown that DNA-looping interactions can increase gene expression by bringing an enhancer closer to the promoter (67–69). Thus, insertion of a DNA sequence flanked by binding sites for a candidate DNA-looping protein between a promoter and enhancer, combined with an inducible protein expression system, could be used to generate a qualitative eukaryotic loopometer.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kim Sneppen, Sandeep Krishna and members of the Shearwin lab for discussions.
Notes
Present address: Adrienne E. Sullivan, Quantitative Cell Biology Laboratory, The Francis Crick Institute, London, UK.
Contributor Information
Nan Hao, Department of Molecular and Biomedical Science, University of Adelaide, Adelaide, SA 5005, Australia; CSIRO Synthetic Biology Future Science Platform, Canberra, ACT 2601, Australia.
Adrienne E Sullivan, Department of Molecular and Biomedical Science, University of Adelaide, Adelaide, SA 5005, Australia.
Keith E Shearwin, Department of Molecular and Biomedical Science, University of Adelaide, Adelaide, SA 5005, Australia.
Ian B Dodd, Department of Molecular and Biomedical Science, University of Adelaide, Adelaide, SA 5005, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [GNT1100651]; Australian Research Council [DP160101450]; CSIRO (to N.H., in part via Synthetic Biology Future Science Platform. Funding for open access charge: University of Adelaide [75125608].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schleif R. DNA looping. Annu. Rev. Biochem. 1992; 61:199–223. [DOI] [PubMed] [Google Scholar]
- 2. Matthews K.S. DNA looping. Microbiol. Rev. 1992; 56:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cournac A., Plumbridge J.. DNA looping in prokaryotes: experimental and theoretical approaches. J. Bacteriol. 2013; 195:1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dröge P., Müller-Hill B.. High local protein concentrations at promoters: strategies in prokaryotic and eukaryotic cells. BioEssays. 2001; 23:179–183. [DOI] [PubMed] [Google Scholar]
- 5. Bush M., Dixon R.. The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol. Mol. Biol. Rev. 2012; 76:497–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Long H.K., Prescott S.L., Wysocka J.. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016; 167:1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schoenfelder S., Fraser P.. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 2019; 20:437–455. [DOI] [PubMed] [Google Scholar]
- 8. Das N., Chattoraj D.K.. Origin pairing (‘handcuffing’) and unpairing in the control of P1 plasmid replication. Mol. Microbiol. 2004; 54:836–849. [DOI] [PubMed] [Google Scholar]
- 9. Schumacher M.A., Tonthat N.K., Kwong S.M., Chinnam N.B., Liu M.A., Skurray R.A., Firth N.. Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Becker N.A., Greiner A.M., Peters J.P., Maher L.J.. Bacterial promoter repression by DNA looping without protein–protein binding competition. Nucleic Acids Res. 2014; 42:5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choy H.E., Park S.W., Parrack P., Adhya S.. Transcription regulation by inflexibility of promoter DNA in a looped complex. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chetverina D., Aoki T., Erokhin M., Georgiev P., Schedl P.. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays. 2014; 36:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grindley N.D.F., Whiteson K.L., Rice P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006; 75:567–605. [DOI] [PubMed] [Google Scholar]
- 14. Révet B., Von Wilcken-Bergmann B., Bessert H., Barker A., Müller-Hill B.. Four dimers of λ repressor bound to two suitably spaced pairs of λ operators form octamers and DNA loops over large distances. Curr. Biol. 1999; 9:151–154. [DOI] [PubMed] [Google Scholar]
- 15. Svenningsen S.L., Costantino N., Court D.L., Adhya S.. On the role of Cro in lambda prophage induction. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:4465–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Michalowski C.B., Little J.W.. Role of cis-acting sites in stimulation of the phage λ Prm promoter by CI-mediated looping. J. Bacteriol. 2013; 195:3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui L., Murchland I., Shearwin K.E., Dodd I.B.. Enhancer-like long-range transcriptional activation by CI-mediated DNA looping. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:2922–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunn T.M., Hahn S., Ogden S., Schleif R.F.. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:5017–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikopoulou C., Panagopoulos G., Sianidis G., Psarra E., Ford E., Thanos D. The transcription factor ThPOK orchestrates stochastic interchromosomal interactions required for IFNB1 virus-inducible gene expression. Mol. Cell. 2018; 71:352–361. [DOI] [PubMed] [Google Scholar]
- 20. Müller J., Oehler S., Müller-Hill B.. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996; 257:21–29. [DOI] [PubMed] [Google Scholar]
- 21. Becker N.A., Kahn J.D., Maher L.J.. Bacterial repression loops require enhanced DNA flexibility. J. Mol. Biol. 2005; 349:716–730. [DOI] [PubMed] [Google Scholar]
- 22. McCord R.P., Kaplan N., Giorgetti L.. Chromosome conformation capture and beyond: toward an integrative view of chromosome structure and function. Mol. Cell. 2020; 77:688–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le T.B.K., Imakaev M. V., Mirny L.A., Laub M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science (80-.). 2013; 342:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hochschild A., Ptashne M.. Cooperative binding of λ repressors to sites separated by integral turns of the DNA helix. Cell. 1986; 44:681–687. [DOI] [PubMed] [Google Scholar]
- 26. Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B.. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987; 6:1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H., Dodd I.B., Dunlap D.D., Shearwin K.E., Finzi L.. Single molecule analysis of DNA wrapping and looping by a circular 14mer wheel of the bacteriophage 186 CI repressor. Nucleic Acids Res. 2013; 41:5746–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zurla C., Manzo C., Dunlap D., Lewis D.E.A., Adhya S., Finzi L. Direct demonstration and quantification of long-range DNA looping by the λ bacteriophage repressor. Nucleic Acids Res. 2009; 37:2789–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Priest D.G., Cui L., Kumar S., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of the DNA tethering effect in long-range DNA looping in vivo and in vitro using the Lac and λ repressors. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Redolfi J., Zhan Y., Valdes-Quezada C., Kryzhanovska M., Guerreiro I., Iesmantavicius V., Pollex T., Grand R.S., Mulugeta E., Kind J.et al.. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nat. Struct. Mol. Biol. 2019; 26:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hensel Z., Weng X., Lagda A.C., Xiao J.. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E. coli cells. PLoS Biol. 2013; 11:e1001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J., Hsu A., Hua Y., Wang G., Cheng L., Ochiai H., Yamamoto T., Pertsinidis A.. Single-gene imaging links genome topology, promoter–enhancer communication and transcription control. Nat. Struct. Mol. Biol. 2020; 27:1032–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Priest D.G., Kumar S., Yan Y., Dunlap D.D., Dodd I.B., Shearwin K.E.. Quantitation of interactions between two DNA loops demonstrates loop domain insulation in E. coli cells. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E4449–E4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hao N., Shearwin K.E., Dodd I.B.. Programmable DNA looping using engineered bivalent dCas9 complexes. Nat. Commun. 2017; 8:1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hao N., Sneppen K., Shearwin K.E., Dodd I.B.. Efficient chromosomal-scale DNA looping in Escherichia coli using multiple DNA-looping elements. Nucleic Acids Res. 2017; 45:5074–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hao N., Shearwin K.E., Dodd I.B.. Positive and negative control of enhancer–promoter interactions by other DNA loops generates specificity and tunability. Cell Rep. 2019; 26:2419–2433. [DOI] [PubMed] [Google Scholar]
- 37. Haldimann A., Wanner B.L.. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure–function studies of bacteria. J. Bacteriol. 2001; 183:6384–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 39. Chung C.T., Niemela S.L., Miller R.H.. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. St-Pierre F., Cui L., Priest D.G., Endy D., Dodd I.B., Shearwin K.E.. One-step cloning and chromosomal integration of DNA. ACS Synth. Biol. 2013; 2:537–541. [DOI] [PubMed] [Google Scholar]
- 41. Hao N., Krishna S., Ahlgren-Berg A., Cutts E.E., Shearwin K.E., Dodd I.B.. Road rules for traffic on DNA—systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res. 2014; 42:8861–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choi Y.J., Morel L., François T. Le, Bourque D., Lucie B., Groleau D., Massie B., Miguez C.B. Novel, versatile, and tightly regulated expression system for Escherichia coli strains. Appl. Environ. Microbiol. 2010; 76:5058–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Datta S., Costantino N., Court D.L.. A set of recombineering plasmids for Gram-negative bacteria. Gene. 2006; 379:109–115. [DOI] [PubMed] [Google Scholar]
- 44. Yang L., Nielsen A.A.K., Fernandez-Rodriguez J., McClune C.J., Laub M.T., Lu T.K., Voigt C.A. Permanent genetic memory with >1-byte capacity. Nat. Methods. 2014; 11:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ptashne M. A Genetic Switch: Phage Lambda Revisited. 2004; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 46. Bell C.E., Lewis M.. Crystal structure of the λ repressor C-terminal domain octamer. J. Mol. Biol. 2001; 314:1127–1136. [DOI] [PubMed] [Google Scholar]
- 47. Rusinova E., Ross J.B.A., Laue T.M., Sowers L.C., Senear D.F. Linkage between operator binding and dimer to octamer self-assembly of bacteriophage γ cI repressor. Biochemistry. 1997; 36:12994–13003. [DOI] [PubMed] [Google Scholar]
- 48. Stayrook S., Jaru-Ampornpan P., Ni J., Hochschild A., Lewis M.. Crystal structure of the λ repressor and a model for pairwise cooperative operator binding. Nature. 2008; 452:1022–1025. [DOI] [PubMed] [Google Scholar]
- 49. Dodd I.B., Shearwin K.E., Perkins A.J., Burr T., Hochschild A., Egan J.B.. Cooperativity in long-range gene regulation by the λ CI repressor. Genes Dev. 2004; 18:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dandanell G., Valentin-Hansen P., Erik Løve Larsen J., Hammer K.. Long-range cooperativity between gene regulatory sequences in a prokaryote. Nature. 1987; 325:823–826. [DOI] [PubMed] [Google Scholar]
- 51. Amouyal M., Mortensen L., Buc H., Hammer K.. Single and double loop formation when deoR repressor binds to its natural operator sites. Cell. 1989; 58:545–551. [DOI] [PubMed] [Google Scholar]
- 52. Pinkett H.W., Shearwin K.E., Stayrook S., Dodd I.B., Burr T., Hochschild A., Egan J.B., Lewis M.. The structural basis of cooperative regulation at an alternate genetic switch. Mol. Cell. 2006; 21:605–615. [DOI] [PubMed] [Google Scholar]
- 53. Dodd I.B., Egan J.B.. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol. Microbiol. 2002; 45:697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carlson N.G., Little J.W.. Highly cooperative DNA binding by the coliphage HK022 repressor. J. Mol. Biol. 1993; 230:1108–1130. [DOI] [PubMed] [Google Scholar]
- 55. Carlson N.G., Little J.W.. A novel antivirulence element in the temperate bacteriophage HK022. J. Bacteriol. 1993; 175:7541–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ravin N. V. N15: the linear phage-plasmid. Plasmid. 2011; 65:102–109. [DOI] [PubMed] [Google Scholar]
- 57. Hammerl J.A., Jäckel C., Lanka E., Roschanski N., Hertwig S.. Binding specificities of the telomere phage ΦKO2 prophage repressor CB and lytic repressor Cro. Viruses. 2016; 8:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stark W.M. Making serine integrases work for us. Curr. Opin. Microbiol. 2017; 38:130–136. [DOI] [PubMed] [Google Scholar]
- 59. Thorpe H.M., Wilson S.E., Smith M.C.M.. Control of directionality in the site-specific recombination system of the streptomyces phage φC31. Mol. Microbiol. 2000; 38:232–241. [DOI] [PubMed] [Google Scholar]
- 60. Ghosh P., Pannunzio N.R., Hatfull G.F., Gottesman M.. Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites. J. Mol. Biol. 2005; 349:331–348. [DOI] [PubMed] [Google Scholar]
- 61. Rutherford K., Van Duyne G.D.. The ins and outs of serine integrase site-specific recombination. Curr. Opin. Struct. Biol. 2014; 24:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mercier R., Petit M.A., Schbath S., Robin S., El Karoui M., Boccard F., Espéli O.. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008; 135:475–485. [DOI] [PubMed] [Google Scholar]
- 63. Dupaigne P., Tonthat N.K., Espéli O., Whitfill T., Boccard F., Schumacher M.A.. Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome. Mol. Cell. 2012; 48:560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lioy V.S., Cournac A., Marbouty M., Duigou S., Mozziconacci J., Espéli O., Boccard F., Koszul R.. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell. 2018; 172:771–783. [DOI] [PubMed] [Google Scholar]
- 65. Lewis D.E.A., Geanacopoulos M., Adhya S. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol. Microbiol. 1999; 31:451–461. [DOI] [PubMed] [Google Scholar]
- 66. Becker N.A., Kahn J.D., Maher L.J.. Effects of nucleoid proteins on DNA repression loop formation in Escherichia coli. Nucleic Acids Res. 2007; 35:3988–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nolis I.K., McKay D.J., Mantouvalou E., Lomvardas S., Merika M., Thanos D. Transcription factors mediate long-range enhancer–promoter interactions. Proc. Natl. Acad. Sci. 2009; 106:20222–20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Deng W., Rupon J.W., Krivega I., Breda L., Motta I., Jahn K.S., Reik A., Gregory P.D., Rivella S., Dean A.et al.. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014; 158:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Morgan S.L., Mariano N.C., Bermudez A., Arruda N.L., Wu F., Luo Y., Shankar G., Jia L., Chen H., Hu J.F.et al.. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 2017; 8:15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.