Abstract
The nucleoprotein (NP) of influenza virus is the core component of the ribonucleoprotein (RNP) and performs multiple structural and functional roles. Structures of the influenza A, B and D NP molecules have been solved previously, but structural information on how NP interacts with RNA remains elusive. Here we present the crystal structure of an obligate monomer of H5N1 NP in complex with RNA nucleotides to 2.3 Å, and a C-terminal truncation of this mutant, also in complex with RNA nucleotides, to 3 Å. In both structures, three nucleotides were identified near two positive grooves of NP suggested to be important for RNA binding. Structural evidence supports that conformational changes of flexible loops and the C-terminal tail both play important roles in the binding of RNA. Based on the structure, we propose a mechanism by which NP captures RNA by flexible loops and transfers it onto the positive binding grooves. Binding of RNA by NP is a crucial step for template re-encapsidation during transcription and replication and cRNP formation. Our structures thus provide insights into the molecular virology of the influenza virus.
INTRODUCTION
The ribonucleoprotein (RNP) of influenza virus is the structural unit where the negative-sense genome segments are encompassed. Each RNP contains the heterotrimeric polymerase complex at one end, and multiple copies of nucleoprotein (NP) forming a double helical rod shape architecture. The structure of RNP is maintained by NP-NP homo-oligomerization, which is mediated by the insertion of the tail-loop from one NP protomer into the groove of its neighbour (1,2). Genomic viral RNA (vRNA) is predicted to wrap along a helical path constituted by positive residues on NP (3–5). During viral replication when a positive-sense cRNA is synthesized, NP is recruited to stabilize nascent cRNA and forms a replicative intermediate named complementary RNP (cRNP) (6,7). In the vRNA synthesis stage that follows, NP is again recruited for its encapsidation and formation of new progeny vRNP.
The crystal structures of H1N1 and H5N1 NP have been solved previously (1,2). The NP molecule shows a crescent shape with a head domain (residues 150–276 and 429–452) and a body domain (residues 1–149 and 277–386) comprised of mainly helices and beta strands. The tail-loop structure spans from residues 402 – 428 and protrudes outwards the head domain. It demonstrates considerable degree of flexibility so that tail-loop insertion at different angles is possible, leading to the formation of oligomers of various sizes. Later, monomeric structures of H1N1 NP harbouring R416A mutation, or with the tail-loop structure deleted, have also been resolved (8,9). Crystal structures of influenza B and D NP both show conserved overall fold highly similar to influenza A NP (10,11). NP is a basic molecule with pI of about 9.4. Conserved positive residues have been identified on the surface of NP. By mutagenesis, contribution of these residues towards RNA binding has been analysed. By Surface Plasmon Resonance assay, Ng et al. (2) have found that mutation of the positive groove formed by R74, R75, R174, R175, R221 (NP-G1) completely abolished RNA binding, while mutation of a nearby groove formed by R150, R152, R156, R162 (NP-G2) led to a 4.7-fold decrease in RNA affinity. Also implicated in RNA-binding is a flexible loop spanning E73-K91, deletion of which rendered a 6.4-fold drop in RNA affinity.
NP binds RNA without sequence specificity, although it has higher affinity towards C and U (12,13). Each NP roughly binds 24 nucleotides (14), while more recent evidence supports the presence of RNA secondary structures which remain unbound by NP, and is proposed for RNA–RNA interaction during genome packaging (15,16). The affinity of NP towards RNA is dependent on RNA length. Yamanaka et al. (17) have showed that NP binds RNA of at least 15 nucleotides in length. RNA binding to NP is mediated by arginines on NP (18), consistent with recent findings that RNA binding affinity is also dependent on salt-concentration (19).
Here, by solving the crystal structures H5N1 NP–RNA complexes, we specify the importance of R65 and K87 during the capture of RNA molecule. Our structures not only demonstrate previously uncharacterized amino acids in close contact with incoming RNA, but also describe residues reported to be important for RNA binding. We also demonstrate the regulatory role of the C-terminal residues of NP on RNA binding. Based on crystallographic and microscale thermophoresis data, we propose a mechanism by which RNA is recruited to NP molecules.
MATERIALS AND METHODS
Plasmids and reagents
cDNA of A/HK/483/97 (H5N1) NP was described previously (2). pET28a vector was obtained from Invitrogen. RNA oligos for crystallization and Kd determination were obtained from Ribobio, Guangzhou, China. C-terminal peptide of NP was obtained from GL Biochem, Shanghai, China.
Protein expression and purification
NP was subcloned onto pET28a vector and mutations were introduced where appropriate by overlapping extension PCR. NP and variant inserts were double-digested with EcoRI and HindIII and ligated using T4 DNA ligase (ThermoFisher). NP and variant proteins were overexpressed in Escherichia coli BL21(DE3)pLysS cells as recombinant proteins tagged with hexahistidine at the N-terminus. Cells were grown in LB medium supplemented with kanamycin and chloramphenicol. Isopropyl β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM was added at OD600 = 0.6–0.8 to induce protein expression. Cells were cultured at 21°C with agitation at 210 rpm for 12–16 h and harvested by centrifugation. Cells were then resuspended and sonicated in cold NP lysis buffer [20 mM sodium phosphate pH7, 1.5 M sodium chloride, 1 mM phenylmethylsulfonyl fluoride (PMSF)]. Whole cell lysate was cleared by centrifugation at 21 000 g for 1 h at 4 °C. Supernatant was loaded onto a nickel affinity column. After initial washing with NP lysis buffer, ribonuclease A (RNase A) from bovine pancreas (Sigma) was added at 1 U/ml to remove residual endogenous RNA from E. coli. The nickel affinity beads were incubated at room temperature for 1 h with shaking and then washed extensively with NP lysis buffer supplemented with 50 mM imidazole. Proteins were eluted with NP elution buffer (20 mM sodium phosphate pH7, 150 mM NaCl, 500 mM imidazole). Eluted proteins were further purified with a heparin column and eluted with 1.5 M NaCl in 20 mM sodium phosphate pH 7. For proteins used for crystallization screenings, an additional step of gel filtration was performed with Superdex 200 (GE Healthcare) in 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS) pH 7, 150 mM NaCl. NP mutant proteins used in this study is described in Table 1.
Table 1.
Influenza A NP mutants generated
| NPA mutant | Mutations | NPA mutant | Mutations |
|---|---|---|---|
| NP-DL | Δ402–428 | DL87 | Δ402–428, K87E |
| DLDC2 | Δ402–428, Δ497–498 | DL174 | Δ402–428, R174E |
| DLDC3 | Δ402–428, Δ496–498 | DL87–74 | Δ402–428, R74E, K87E |
| DLDC4 | Δ402–428, Δ495–498 | DL87–75 | Δ402–428, R75E, K87E |
| DLDC8 | Δ402–428, Δ491–498 | DL87–65 | Δ402–428, R65E, K87E |
| DL65 | Δ402–428, R65E | DL65–75 | Δ402–428, R65E, R75E |
| DL72 | Δ402–428, D72K | DL65–174 | Δ402–428, R65E, R174E |
| DL74 | Δ402–428, R74E | DL3A | Δ402–428, Y148A, R152A, R156A |
| DL75 | Δ402–428, R75E |
Crystallization of NP-RNA complexes
NP from gel filtration was pooled and concentrated to 8–10 mg/ml using Amicon-ultra centrifugal filter unit at MWCO 10,000 kDa (Millipore). NP was mixed with 2′-O-methylated RNA oligos (Ribobio) at molar ratio of 1:1.5 and incubated in 16 °C for 1 h or overnight. Protein-RNA mixture was centrifuged to remove possible precipitation before used for crystallization screening. Initial crystallization conditions were picked after surveying through commercially available crystallization kits. RNA with length of 5, 8, 9 and 24 nucleotides were tested, but only the 8-mer and 9-mer oligoes led to diffractable crystals. Crystals for NP tail-loop deletion mutant (NP-DL) and a 9-mer poly-uridine RNA were obtained in 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.0, 0.1 M NaCl and 11% (w/v) PEG2000. Crystals for NP with two extreme C-terminal residues truncated (NP-DLDC2) and an 8-mer poly-uridine RNA were obtained in 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5 and 5% propan-2-ol.
Structure determination, processing and refinement
Crystals were harvested into a 1:1 mixture of reservoir buffer and sucrose (final concentration 17% as cryo-protectant) and then flash-frozen in liquid nitrogen. All datasets were collected at Shanghai Synchrotron Radiation Facility, Shanghai (SSRF, Shanghai, China) beamline BL17U1. Data reduction was performed within the CCP4 suite (20,21). Structure of NP-DL/RNA was determined by molecular replacement using an unpublished H5N1 NP tail-loop deletion mutant as search model (available upon request). Structure of NP-DLDC2/RNA was determined by molecular replacement using NP-DL/RNA complex with RNA moiety manually removed as search model. All molecular replacement calculations were performed in Phaser-MR (22). Model building was carried out in Coot (23) with the RNA nucleotides built and fit manually. Structures were iteratively refined with Refmac/CCP4 suite (24). Structures were validated with MolProbity (25). Protein structures were visualized in PyMOL (DeLano 2002, The PyMOL Molecular Graphics System, Schrödinger, LLC). Protein-RNA interaction was analyzed by PISA (26) and LigPlot+ (27).
Dissociation constant determination using microscale thermophoresis
9-nucleotide poly-uridine RNA oligos were obtained from Ribobio with a Cy5 tag at the 5′ end and were methylated at the 2′-O positions. NPA or variants were concentrated to concentrations as indicated in the text or figures and buffer-exchanged to MST buffer (PBS pH 7.4, 0.1% Tween-20). MST experiments were performed on a Monolith NT.115 machine (Nanotemper, Munich, Germany). 10 μl of RNA at 50 nM was titrated against a serial dilution of proteins so that each sample was at a total volume of 20 μL. RNA-protein mixtures were briefly centrifuged before loading into Nanotemper's Standard-Treated capillaries. Measurements were made in 23°C using 15% LED power and 20% MST power.
Competition assay
9-Nucleotide poly-uridine RNA oligos methylated at the 2′-O positions was also obtained from Ribobio. The DLDC8 variant was concentrated to 2.2 uM and buffer-exchanged to MST buffer (PBS pH 7.4, 0.1% Tween-20). A short peptide, corresponding to the C-terminal eight residues of NPA, with the sequence DNAEEYDN, was mixed with DLDC8 at a molar ratio of 1:20. MST experiments were performed on a Monolith NT.LabelFree machine (Nanotemper, Munich, Germany). 10 μl of protein/peptide mixture was titrated against a serial dilution of 9-mer RNA so that each sample was at 20 μl. RNA-protein mixtures were briefly centrifuged before loading into Nanotemper's Standard-Treated capillaries for LabelFree machine. Measurements were made in 23°C using 10% LED power and 40% MST power.
Microscale thermophoresis data analysis
Data analysis was performed on Nanotemper's MO.Affinity V2.3 software using thermophoresis signal at MST-on-time = 2.5 s (for NT115 machine) or 1.5 s (for LabelFree machine). Curves of Fnorm against concentration were fitted with Kd model, assuming 1:1 binding, using data points from at least three independent measurements and with target concentration fixed. Fnorm is defined as (Fhot/Fcold) × 1000. Obvious outlier points due to protein aggregation or capillary contamination were removed. Error bars represent mean ± standard deviation.
RESULTS
Structure of NP bound with RNA
Structure of NP
Despite various NP structures are available by crystallography, NP–RNA complexes are recalcitrant to crystallization. Eventually, we were able to co-crystallize the obligated monomeric mutant of NP, NP-DL, with a 9-mer RNA. Crystal structure of NP-DL/9-mer RNA complex was solved to 2.3 Å in P1 space group. There were two NP molecules in an asymmetric unit, while three nucleotides could be modelled to chain B. We also serially truncated NP-DL at the C-terminus to give DLDC2, DLDC3, DLDC4 and DLDC8 variants. Nevertheless, only DLDC2 produced diffraction quality crystals when co-crystallized with RNA nucleotides. Hence we also solved the crystal structure of DLDC2/8-mer RNA complex and refined it to 3 Å in P1 space group, with also two NP molecules in one asymmetric unit. Data collection and refinement statistics for both complexes are detailed in Table 2.
Table 2.
Data collection and refinement statistics for NP–RNA complexes. Values in brackets are for the highest-resolution shell. RMSD: root-mean-square-deviation from ideal values. Ramachandran analysis was done by MolProbity (25)
| NP-DL-RNA | DLDC2-RNA | |
|---|---|---|
| Crystallization condition | 0.1 M MES, pH 6.0, 0.1 M NaCl and 11% PEG2000 | 0.1 M HEPES, pH 7.5 and 5% propan-2-ol |
| Ligand | 9-mer poly-uridine RNA | 8-mer poly-uridine RNA |
| Beamline | SSRF BL17U1 | SSRF BL17U1 |
| Data collection | ||
| Space Group | P1 | P1 |
| Cell dimensions a, b, c (Å) α, β, γ (degrees) | 54.0, 60.6, 82.7 106.7, 109.0, 96.6 | 53.8, 60.1, 81.4 107.3, 106.7, 96.1 |
| Resolution (Å) | 25.52–2.30 (2.42–2.30) | 38.58–3.00 (3.18–3.00) |
| Rmerge | 10.2% (25.3%) | 16.8% (17.1%) |
| I/σI | 7.1 (2.5) | 5.8 (3.6) |
| Completeness | 90.4 (91.6) | 86.1 (88.2) |
| Multiplicity | 1.9 (1.9) | 1.9 (2.0) |
| Total observations | 69 016 (10198) | 30 372 (5137) |
| Total unique | 37106 (5475) | 15710 (2610) |
| Refinement | ||
| No. of atoms | 6727 (overall), 6498 (macromolecules), 74 (ligands), 155 (waters) | 6421 (overall), 6309 (macromolecules), 63 (ligands), 49 (waters) |
| Resolution (Å) | 24.50–2.30 | 36.62–3.00 |
| R work/Rfree | 0.193/0.248 | 0.289/0.332 |
| RMSD bond length (Å) | 0.0102 | 0.007 |
| RMSD angle (°) | 1.710 | 1.312 |
| MolProbity score | 2.23 | 2.17 |
| Ramachandran | Favored 96.6%, allowed 2.1%, outlier 1.25% | Favored 96.7%, allowed 3.2%, outlier 0.13% |
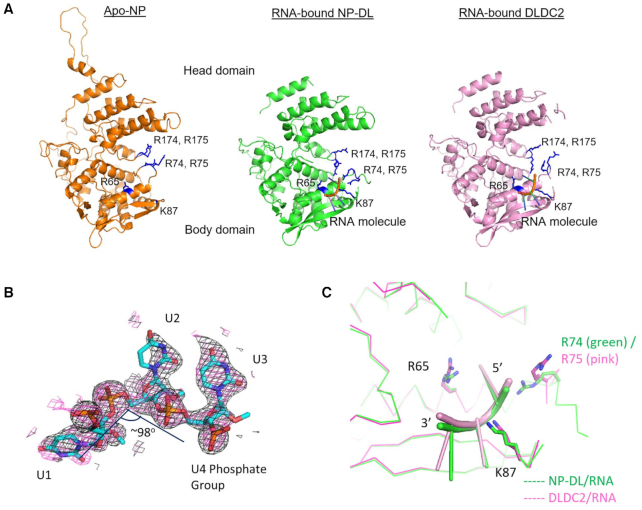
The overall fold of the protein core of NP-DL was highly similar to the wild-type apo-NP (PDB 2Q06), indicating that the structure was not significantly distorted upon deletion of the tail-loop (Figure 1A). The root-mean-square deviation (RMSD) between NP-DL and apo-NP (PDB 2Q06) was 0.78 Å (over 386 Cα atoms in chain A). Residues 23–75, 87–201, 206–207, 211–388, 438–452, 462–497 of chain A and 22–78, 85–388, 437–453, 458–497 of chain B were modelled unambiguously. Deletion of the tail-loop (S402–A428) renders the flanking N- and C- terminal sequences flexible. Also, only part of region I204–R214 could be modelled in our structures. This region encompasses a large portion of a nuclear localization signal (K198–R216) on NP and is exposed on the surface. On the other hand, residues K91–K112 in our structures formed a well-structured beta-sheet motif which was more ordered when compared to wild-type NP. In the DLDC2 structure, residues 23–77, 87–200, 213–388, 438–452, 462–496 of chain A and 21–76, 85–202, 204–208, 215–389, 437–453, 459–496 of chain B were modeled. RMSD between NP-DL and DLDC2 were 0.46 (over 387 Cα atoms in chain A), indicating high degree of resemblance between the two variants.
Figure 1.
(A) Apo-NP (PDB 2Q06) and the obligate monomeric NP-DL and DLDC2 shown in the same orientation. Three RNA nucleotides are modeled onto the body domain near the 74–88 loop. The proteins share highly similar tertiary folds demonstrating neither RNA binding nor tail-loop deletion distorts the overall structure. G1 residues (R74, R75, R174, R175), R65 and K87 are marked in blue in the structures. (B) 2Fo – Fc omit maps were generated for the nucleotides modeled on NP-DL (black: simple omit map; pink: simulated-annealing omit map) and contoured at 1σ as shown. (C) The RNA moieties on NP-DL and DLDC2 are situated in a cavity defined by R65, R74/75 and K87. The conformations of two RNA moieties do not deviate significantly. Protein cores are shown as ribbons (NP-DL: green; DLDC2: magenta). RNA is shown as cartoon in green (NP-DL/RNA) and pink (DLDC2/RNA).
Structure of RNA bound on NP
9-mer and 8-mer RNA oligo was used, respectively, in co-crystallization with NP-DL and DLDC2, while three nucleotides could be modelled to chain B at 1.0σ in each structure (Figure 1B). Only residual density which was too weak to be assigned was observed at the equivalent position at the chain A. These nucleotides were bound to a cavity on the NP surface at the body domain defined by positive residues R65, R74/75 and K87 (Figure 1C). The interacting interface spanned an area of 235 and 255 Å2, respectively, between the RNA and the protein core in NP-DL and DLDC2 complexes. The position and orientation of the three nucleotides were both highly similar in the NP-DL-RNA and DLDC2-RNA structures, suggesting the interaction is reproducible crystallographically. These nucleotides were in close proximity to the flexible 74–88 loop previously proposed for RNA capture (2).
The RNA backbone adopted a special conformation so that in the NP-DL complex, the 5′ (U1) and the middle (U2) nucleotides were twisted to give an angle of 98o (Figure 1B). Similarly, a backbone twist of 96o was also observed for the three equivalent nucleotides in the DLDC2 complex. In fact, conformations of these three nucleotides in the two structures were very similar (Figure 1C). All three nucleotides adopted the anti- conformation. The two uracil bases of U2 and the 3′ nucleotide (U3) were almost co-planar to one another, as a result base stacking between them was only minimal. The RNA molecule was mainly held by polar contacts (Table 3 and Supplementary figure S1). Extensive hydrogen bonding was identified between phosphate groups and NP side chains. U1 phosphate was in contact with R74, U2 phosphate was held by S69 and T92, while U3 phosphate interacted with K87 (Table 3 and Figure 2A).
Table 3.
Interactions between RNA and the protein core identified in the NP-RNA complexes
| Side-chain interactions | Interacting RNA moiety on NP-DL (shortest distance in Å) | Interacting RNA moiety on DLDC2 (shortest distance in Å) |
|---|---|---|
| R65 | U1/O2 (2.96) | U1/O2 (3.41) |
| R74 | U1/PO4 (2.51) | Not applicable |
| R75 | Not applicable | U1/PO4 (4.16) |
| K87 | U3/PO4 (3.33) | U2/PO4 (3.64), U3/PO4 (3.52) |
| S69 | U2/PO4 (2.50) | U2/PO4 (3.03) |
| T92 | U2/PO4 (2.63) | U2/PO4 (3.05) |
| R175 | U1/O4 (3.26) | U1/O4 (3.98) |
| S367 | U3/O3′ (3.33) | U3/O3′ (3.29) |
| Main-chain interactions | Interacting RNA moiety on NP-DL (shortest distance in Å) | Interacting RNA moiety on DLDC2 (shortest distance in Å) |
| D88/N | U2/O4 (2.66) | U2/O4 (3.00) |
Figure 2.
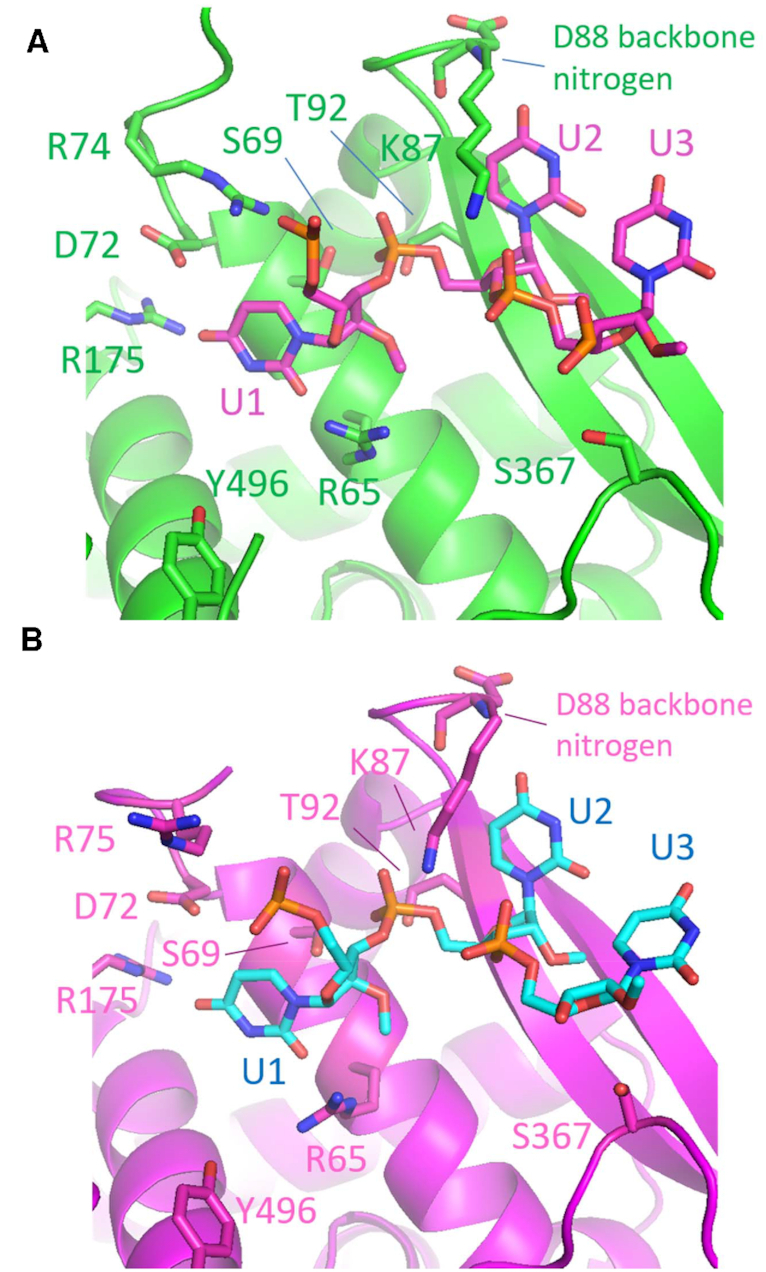
Three nucleotides built onto (A) NP-DL and (B) DLDC2, respectively, with side chains of nearby interacting residues shown. Numerous side-chain interactions can be identified as detailed in Table 3, including R65 and K87 which form hydrogen bonds with RNA and are previously uncharacterized. The RNA can possibly interact with both R74 or R75 depending on loop flexibility.
To sustain the characteristic conformation of the bases, side chain of R65 was within less than 3 Å to O2 of U1 uracil base. R175 side chain was also in close proximity to O4 position of U1 base at around 4 Å. O4 of U2 uracil base interacted with main chain nitrogen of D88. O3′ of U3 ribose was held by side chain of S367. These polar contacts were conserved in the DLDC2 complex, except that phosphate of U1, which interacted with R74 in the NP-DL complex, formed hydrogen bond with R75 instead (Figure 2B and Supplementary figure S1). In both structures, a salt bridge between D72 and R175 was possible. R74, R75 and R174, R175 formed a positive groove and were located respectively on two flexible loops (Figure 1). The 74–88 loop had been suggested to capture RNA from ambient environment while the R174, R175 had been proposed to be important for correct positioning of the RNA (2). The changes in local interaction environment should be implicated in the mechanism of RNA binding.
Microscale thermophoresis identified important elements for RNA binding
Single-point and double-point mutants on NP-DL were designed based on the two NP structures. We then utilized microscale thermophoresis (MST) to determine the dissociation constants (Kd) of NP variants against a Cy5-tagged 9-mer RNA oligo (Table 4 and Supplementary Figures S2 and S3). NP-DL bound to 9-mer RNA with Kd of 4.2 μM. DLDC2 bound to this RNA with a similar affinity, with Kd of 4.5 μM. Single-point variants DL65, DL74 and DL87 each decreased affinity to RNA for about 2-fold, with Kd of 9.2 μM, 10.5 μM and 10.9 μM respectively. Effect of mutation on R75 was more prominent, where a decrease in affinity of about 4-fold was observed (Kd = 17.6 μM). We considered these fold-changes reasonable taking into account that the RNA relied on multiple contact points for binding. Double mutants DL87–65, DL87–75, DL65–75 and DL87–74 were then generated which showed around 5- to 6-fold decrease in RNA-binding affinity. Taken together, the MST results were consistent with the binding models observed in the two NP structures. DL174 only demonstrated a Kd of ∼52 μM, which was more than 10-fold decrease than NP-DL. The DL65–174 double mutant interacted with RNA very weakly at a Kd of ∼92 μM, equivalent to a >20-fold drop.
Table 4.
K d values of NP mutants against 9-mer RNA measured by MST
| NP variants | K d value (μM) | Fold-change (relative to NP-DL) |
|---|---|---|
| NP-DL (Δ402–428) | 4.17 ± 0.26 | 1 |
| DL65 (Δ402–428, R65E) | 9.22 ± 0.89 | –2.2 |
| DL72 (Δ402–428, D72K) | 11.96 ± 1.08 | –2.9 |
| DL74 (Δ402–428, R74E) | 10.49 ± 0.76 | –2.5 |
| DL75 (Δ402–428, R75E) | 17.64 ± 1.07 | –4.2 |
| DL87 (Δ402–428, K87E) | 10.94 ± 1.03 | –2.6 |
| DL174 (Δ402–428, R174E) | 52.53 ± 4.08 | –12.6 |
| DL87–65 (Δ402–428, R65E, K87E) | 26.92 ± 1.83 | –6.5 |
| DL65–75 (Δ402–428, R65E, R75E) | 21.57 ± 1.98 | –5.2 |
| DL75–87 (Δ402–428, R75E, K87E) | 19.16 ± 0.74 | –4.6 |
| DL74–87 (Δ402–428, R74E, K87E) | 27.32 ± 2.63 | –6.5 |
| DL65–174 (Δ402–428, R65E, R174E) | 91.86 ± 15.46 | –22.0 |
| DLDC2 (Δ402–428, Δ497–498) | 4.54 ± 0.40 | –1.1 |
| DL-3A (Δ402–428, Y148A, R152A, R156A) | 18.47 ± 1.23 | –4.4 |
Structural analysis of the NP-RNA complexes
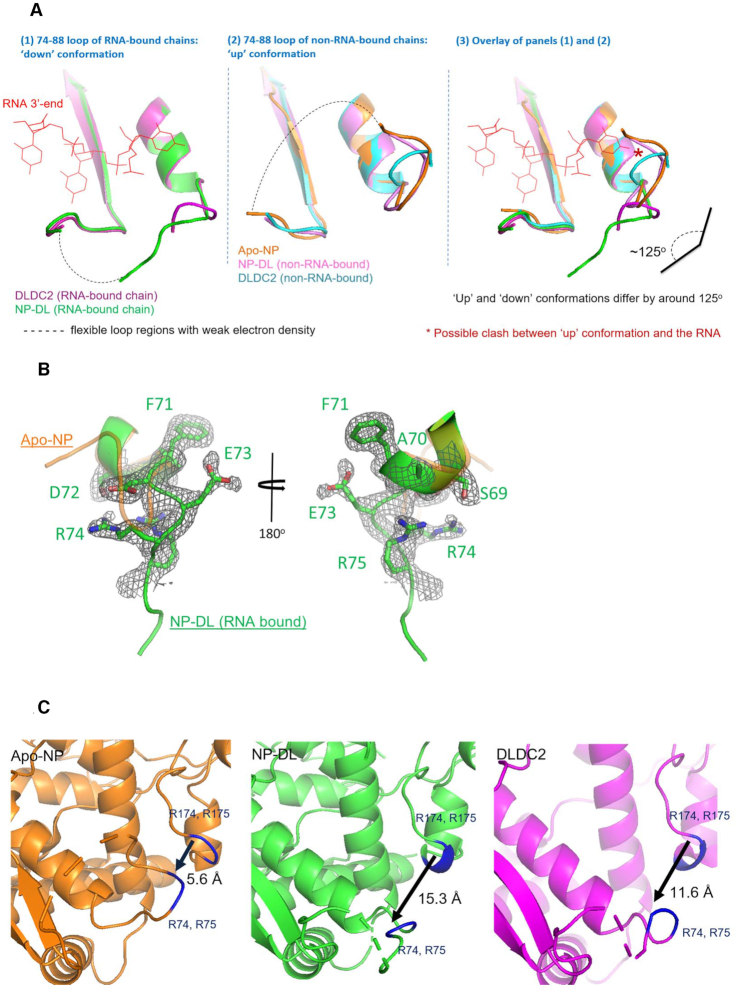
By comparing our structures with the wild-type H5 NP (PDB 2Q06), we observed different conformations within the loop regions of NP. As shown in Figure 3A and 3B, the 74–88 loop adopted two major orientations. The C-terminal end, roughly after K87 of the loop, remained at about the same orientation in all protomers. The N-terminal portion (from D72 onwards) in the RNA-bound chains adopted a ‘down’ conformation whereas the non-RNA-bound chains had 74–88 loops resembling wild-type NP, adopting an ‘up’ conformation. The two conformations differ by about 125o. It should however be mentioned that also because of loop flexibility, electron density for the tip portion of the loop was weak despite effort on map sharpening. As a result, a complete loop could not be modeled. The ‘down’ conformation could be stabilized by the salt bridge between R175 and D72. By MST analysis, a D72K NP variant (DL72) recorded a drop in RNA affinity by ∼3-fold. We also surveyed all influenza A NP structures in the Protein Data Bank and observed both conformations in the RNA-free state. However, it was obvious from our structures that only the ‘down’ conformation can accommodate the RNA, because side-chains of N76, R77 and Y78 would clash with the 5′-end of the RNA moiety if arranged in ‘up’ conformation. Taken together, we conclude that during the capture of RNA, flexibility of the 74–88 loop leads to dynamic changes of local loop conformations. The overall result of this flexibility is the opening and closure of positive G1 groove (defined by R74, R75, R174 and R175) (Figure 3C).
Figure 3.
(A) Here we align the 74–88 loops on all RNA-bound chains in panel (1) and non-RNA bound chains from our structures to apo-NP (PDB 2Q06) in panel (2). Peptides are colored according to colors of labels. When bound with RNA, the loop adopts a ‘down’ conformation, in contrast to the ‘up’ conformation when RNA-free. Due to the weak density, part of the loops could not be modeled confidently and is indicated by dashes here. Panel (3) is an overlay of panels (1) and (2) and the possible clash between the loop in ‘up’ conformation with the 5′ RNA is indicated. (B) 2mFo – DFc map (black, contoured at 1σ) for residues F71-R75 (green) on NP-DL complex is shown. The peptide is overlaid with the corresponding residues from apo-NP (PDB 2Q06, orange). The two peptides align with each other until D72 where they deviate to give two different conformations. (C) The 74–88 loop from apo-NP (PDB 2Q06), NP-DL and DLDC2 are shown with positive residues R74, R75, R174 and R175 marked blue. The ‘down’ conformation of the 74–88 loop in NP-DL and DLDC2 lead to opening up of G1 groove as indicated by increased distances between R174 to R74.
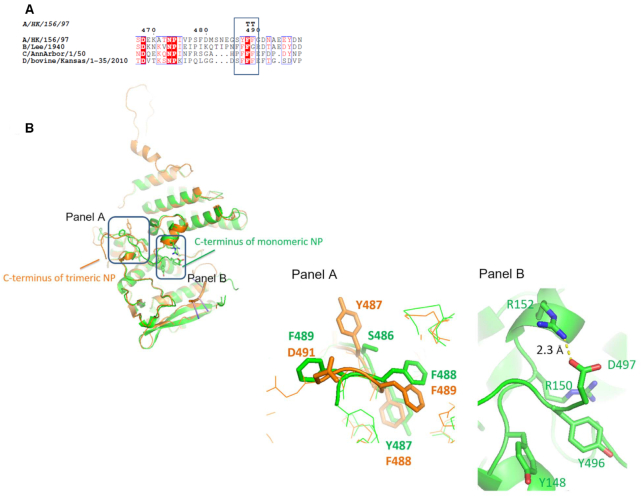
Consistent with the H1 NP tail-loop deletion and R416A monomer structures, the G2 binding site (defined by R150, R152, R156 and R162) was blocked by the C-terminal residues of NP protein. We confirmed the contribution of G2 groove to RNA binding by demonstrating a drop of RNA affinity to the DL3A variant, harboring triple mutations Y148A–R152A–R156A, by >4-fold. Our structures showed that the C-terminal tail in H5 NP made more extensive polar contacts with the protein core, compared with the H1 NP. We identified three pairs of salt bridges, namely D491–R355, E494–R361 and D497–R152. The phenyl ring of Y496 was adjacent to aromatic Y148 with the two rings roughly perpendicular to each other. R150 was also <4 Å apart to Y496 making cation- π interaction possible. Side chain of E495 interacted with the main chain atoms of Q149 and T151. As the G2 groove is defined by positive residues, covering of this groove by a highly acidic tail (DNAEEYDN) inevitably rendered the region less basic compared to the wild-type H5 NP (PDB 2Q06) where the C-terminal residues point to the opposite direction out of the molecule (Supplementary Figure S4). The tail residues span ∼22 Å on G2 groove, which can roughly accommodate around four RNA bases. The re-orientation of the C-terminal tail also led to a disrupted arrangement of a 487-YFF-489 motif which is highly conserved among NPA, NPB, NPC and NPD (Figure 4A and 4B). In the trimeric wild-type H5 NP, Y487 points outwards from the oligomerization groove and is 2.7 Å from E421 from the insertion-loop of the incoming protomer, facilitating a hydrogen bond pair which stabilizes the NP trimer. Whereas in the monomeric structures we solved, Y487 was pointing towards an opposite direction and was not available for interaction with the incoming protomer. Instead it formed hydrogen bond with main chain oxygen of A146 as a compensating stabilization. Altogether, our structures hinted that RNA-binding and NP-NP oligomerization is coupled.
Figure 4.
(A) Alignment of NP sequences near the C-terminal region extracted from influenza A, B, C and D nucleoproteins. Conserved YFF motif is highlighted in box. (B) In H5 NP, the C-terminal tail of obligate monomers flips onto the protein core to cover the RNA-groove and as such Y496 is brought to vicinity to both Y148 and R150 of the positive G1 groove. D497-R152 salt-bridge is one of the stabilizing forces for this conformation. Shown in Figure 4B is an overlay of wild-type NP (orange) and the obligate monomer NP-DL (green). Panel A highlights the rearrangement of the YFF motif during this conformation change while panel B shows the local environment around the D497-R152 salt-bridge when NP is in monomeric state.
Tail-peptide blocks RNA binding in vitro
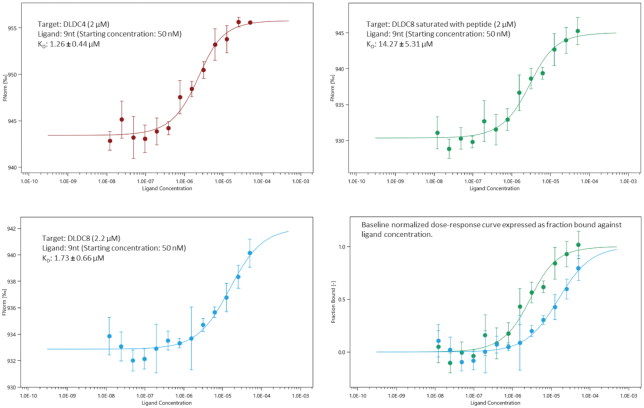
Since the C-terminal residues covered the positive binding groove, we decided to investigate the effect of this C-terminal tail on RNA-binding in vitro. We designed further truncations of four and eight residues at the C-terminal end resulting in the DLDC4 and DLDC8 variants. Utilizing label-free microscale thermophoresis, we determined the Kd of these variants towards a 9-mer RNA to be 1.26 and 1.73 μM, representing a roughly 2- to 3-fold increase in affinity. Then we attempted to attenuate RNA binding using a synthetic peptide DNAEEYDN, corresponding to the last 8 residues on NP. Before the assay, we confirmed that there was no binding detected between this peptide and a tagged 9-mer RNA (Supplementary figure S5). When saturated by 20-fold molar excess of peptide, affinity of DLDC8 to 9-mer RNA was significantly decreased by around 8-fold (Figure 5). Taken together, the tail peptide played a role in regulating RNA binding.
Figure 5.
Label-free MST was performed to determine the affinity between DLDC4 and DLDC8 NP variants towards 9-mer RNA (top-left and bottom-left panels, respectively). By saturating DLDC8 mutant with a synthetic tail peptide, RNA binding was attenuated by 8-fold. MST curve is expressed as Fnorm (0/00) versus concentration. Data represent mean values from at least three independent measurements. Error bars represent standard deviation. In the bottom right panel, for comparison, MST curves were expressed as fraction of protein bound vs ligand (RNA) concentration (Blue: DLDC8; Green: peptide-saturated DLDC8).
DISCUSSION
We present here two complex structures formed between RNA oligo and H5 NP. Previously, we demonstrated that the Kd of a 24-mer RNA to NP monomers was within the range of 0.4 to 1 μM (28) which represents around 17- to 43-fold weaker binding compared to wild-type trimeric NP with Kd of 23 nM (2). Here, with an RNA oligo which was around one-third in length, the Kd between monomeric NP-DL versus a 9-mer RNA was further decreased to 4 μM. Based on structural analysis, we have confirmed residues R74, R75, R175 are important for RNA binding, and showed R65 and K87, two previously uncharacterized residues, to be important for RNA binding. Indeed, the positive charge at position 65 is conserved among NPA, NPB, NPC and NPD, further signifying its functional importance. We also demonstrate that flexibility of 74–88 loop allows it to adopt at least two major conformations, with only one of them (the ‘down’ conformation) allowing it to capture an RNA molecule. The presence of a highly conserved aspartic acid at position 72, and the fact that its side chain being spatially close to the side chains of R174/175 tempted us to postulate that the flexibility of the 74–88 loop is regulated by the two positively charged arginine residues at 174 and 175. Previous studies by Li et al. (29) could not rescue D72A mutant virus, highlighting the biological relevance of this residue. Changes in loop conformations are therefore crucial to RNA binding. Our MST results show that deleting four or more C-terminal residues are sufficient to increase RNA affinity for 2- to 3-fold. Hence, deletions on the tail likely open up an additional binding pocket on the protein. Blockage of RNA-binding site by the tail likely accounts for the discrepancy in RNA affinity between trimeric and monomeric NP. Therefore, we have also provided additional evidence for the role of NP loops and tail elements in regulating RNA binding.
Further the above evidence, following the electron density of phosphorous atoms we can attempt to extend the nucleotide chain and trace the RNA phosphate backbone based on the DLDC2 complex which allows two more nucleotides to be modelled at the 5′ end. The nucleotides thus built would resemble a distorted hook with the 5′ end right next to the G2 groove (Supplementary figure S6A). These two extra nucleotides exhibit nice interaction with the protein. (Supplementary figure S6B and Supplementary table S1). Displacement of the C-terminal tail would likely facilitate the correct positioning of these nucleotides.
In our structures, positive arginine or lysine residues are highly relevant for RNA binding. Intriguingly, base stacking is minimal in our NP-RNA model, in contrast to the extensive base stacking in vesicular stomatitis virus (VSV) and rabies NP. Only 3–5 nucleotides could be recorded in our NP-RNA structures, despite that 9-mer or 8-mer RNAs were used during crystallization. The remaining nucleotides likely hover over the protein core flexibly, implying that significant degree of RNA disorder can be tolerated in influenza NP, which is not the case in VSV and rabies where RNA is encapsidated in the interior of a ring of NP oligomers (30,31).
Based on our current structural and biochemical data, we can propose a mechanism by which NP binds RNA (Supplementary Figure S7). In the first instance, the G1 groove, formed mainly by R74/75 and R174/175 on two flexible loops, is able to open and close. This movement is dependent on the N-terminal portion of the 74–88 loop. The opening and closure of this region packages RNA to the site near R65, K87, as shown in our structures. The C-terminal tail must displace from its current position. Currently there is no direct evidence as to whether this displacement is dependent of NP-NP oligomerization. However, based on the rearrangement near the YFF motif, we postulate that NP-NP oligomerization should aid this movement. Finally, the 5′-end of the RNA shifts into the G2 groove which has positive charges exposed after the departure of the acidic tail residues. This model is in line with our biochemical analysis in this and our previous work (2) which showed the importance of G1 and G2 residues. Besides, molecular dynamics (32) had suggested several loop regions including 73–90 and 360–373 for RNA capture. We have now showcased the importance of loop 74–88 on RNA-binding. Also, as observed in our structure, S367 interacts with 3′end of the RNA and R361 forms a salt bridge with E494 at the C-terminal tail. The importance of loop 360–373 on RNA binding is thus substantiated.
Our work here not only deepens the understanding on the molecular basis of influenza NP-RNA binding but also offers new insights to RNA binding mechanism by NP. We propose that the tail peptide could be an attractive RNA binding inhibitor to be tested further.
DATA AVAILABILITY
CCP4 suite is downloaded under academic licenses at https://www.ccp4.ac.uk/download/.
Crystal structures of NP-DL and NP-DLDC2 complexed with RNA are deposited with the Protein Data bank under accession number 7DXP and 7DKG, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof Paul KS Chan (Department of Microbiology, The Chinese University of Hong Kong) for the influenza H5N1 NP cDNA. We also thank Dr Andy KL Ng and Dr Edwin CY Lo for useful discussions on experimental design and the manuscript.
Contributor Information
Yun-Sang Tang, Centre for Protein Science and Crystallography, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China.
Shutong Xu, Department of Medical Oncology and Department of Cancer Biology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA; College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China.
Yu-Wai Chen, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hong Kong SAR, China.
Jia-Huai Wang, Department of Medical Oncology and Department of Cancer Biology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA.
Pang-Chui Shaw, Centre for Protein Science and Crystallography, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong SAR, China; Li Dak Sum Yip Yio Chin R & D Centre for Chinese Medicine, The Chinese University of Hong Kong, Hong Kong SAR, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Theme-based Research Grant under Hong Kong Research Grant Council (HKRGC) [T11-705/14N to P.C.S.]; NIH [HL103526 to J.H.W.]; Claudia Adams Barr Award [to J.H.W.]. Funding for open access charge: The Chinese University of Hong Kong.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ye Q., Krug R.M., Tao Y.J.. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006; 444:1078–1082. [DOI] [PubMed] [Google Scholar]
- 2. Ng A.K.L., Zhang H., Tan K., Li Z., Liu J.H., Chan P.K.S., Li S.M., Chan W.Y., Au S.W.N., Joachimiak A.et al.. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008; 22:3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coloma R., Valpuesta J.M., Arranz R., Carrascosa J.L., Ortín J., Martín-Benito J.. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009; 5:e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arranz R., Coloma R., Chichon F.J., Conesa J.J., Carrascosa J.L., Valpuesta J.M., Ortin J., Martin-Benito J.. The structure of native influenza virion ribonucleoproteins. Science. 2012; 338:1634–1637. [DOI] [PubMed] [Google Scholar]
- 5. Moeller A., Kirchdoerfer R.N., Potter C.S., Carragher B., Wilson I.A.. Organization of the influenza virus replication machinery. Science. 2012; 338:1631–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vreede F.T., Ng A.K.L., Shaw P.C., Fodor E.. Stabilization of influenza virus replication intermediates is dependent on the RNA-binding but not the homo-oligomerization activity of the viral nucleoprotein. J. Virol. 2011; 85:12073–12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. York A., Hengrung N., Vreede F.T., Huiskonen J.T., Fodor E.. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E4238–E4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chenavas S., Crépin T., Delmas B., Ruigrok R.W., Slama-Schwok A.. Influenza virus nucleoprotein: structure, RNA binding, oligomerization and antiviral drug target. Future Microbiol. 2013; 8:1537–1545. [DOI] [PubMed] [Google Scholar]
- 9. Ye Q., Guu T.S.Y., Mata D.A., Kuo R.L., Smith B., Krug R.M., Tao Y.J.. Biochemical and structural evidence in support of a coherent model for the formation of the double-helical influenza A virus ribonucleoprotein. MBio. 2012; 4:e00467–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng A.K.L., Lam M.K.H., Zhang H., Liu J., Au S.W.N., Chan P.K.S., Wang J., Shaw P.C.. Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein. J. Virol. 2012; 86:6758–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Donchet A., Oliva J., Labaronne A., Tengo L., Miloudi M.C.A., Gerard F., Mas C., Schoehn G.W.H., Ruigrok R., Ducatez M.et al.. The structure of the nucleoprotein of influenza D shows that all Orthomyxoviridae nucleoproteins have a similar NPCORE, with or without a NPTAIL for nuclear transport. Sci. Rep. 2019; 9:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baudin F., Bach C., Cusack S., Ruigrok R.W.. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994; 13:3158–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Albo C., Valencia A., Portela A.. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J. Virol. 1995; 69:3799–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ortega J., Martín-Benito J., Zürcher T., Valpuesta J.M., Carrascosa J.L., Ortín J.. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 2000; 74:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee N., Le Sage V., Nanni A.V, Snyder D.J., Cooper V.S., Lakdawala S.S.. Genome-wide analysis of influenza viral RNA and nucleoprotein association. Nucleic Acids Res. 2017; 45:8968–8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dadonaite B., Gilbertson B., Knight M.L., Trifkovic S., Rockman S., Laederach A., Brown L.E., Fodor E., Bauer D.L.V.. The structure of the influenza A virus genome. Nat. Microbiol. 2019; 4:1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamanaka K., Ishihama A., Nagata K.. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J. Biol. Chem. 1990; 265:11151–11155. [PubMed] [Google Scholar]
- 18. Elton D., Medcalf L., Bishop K., Harrison D., Digard P.. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 1999; 73:7357–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labaronne A., Swale C., Monod A., Schoehn G., Crépin T., Ruigrok R.W.H.. Binding of RNA by the nucleoproteins of influenza viruses A and B. Viruses. 2016; 8:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W.. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. D. Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kovalevskiy O., Nicholls R.A., Long F., Carlon A., Murshudov G.N.. Overview of refinement procedures within REFMAC 5: Utilizing data from different sources. Acta Crystallogr. Sect. D Struct. Biol. 2018; 74:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B.et al.. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018; 27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 27. Wallace A.C., Laskowski R.A., Thornton J.M.. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein. Eng. 1995; 8:127–134. [DOI] [PubMed] [Google Scholar]
- 28. Chan W.H., Ng A.K.L., Robb N.C., Lam M.K.H., Chan P.K.S., Au S.W.N., Wang J.H., Fodor E., Shaw P.C.. Functional analysis of the influenza virus H5N1 nucleoprotein tail loop reveals amino acids that are crucial for oligomerization and ribonucleoprotein activities. J. Virol. 2010; 84:7337–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z., Watanabe T., Hatta M., Watanabe S., Nanbo A., Ozawa M., Kakugawa S., Shimojima M., Yamada S., Neumann G.et al.. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J. Virol. 2009; 83:4153–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Green T.J., Zhang X., Wartz G.W., Luo M.. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006; 313:357–360. [DOI] [PubMed] [Google Scholar]
- 31. Albertini A.A.V., Wernimont A.K., Muziol T., Ravelli R.B.G., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W.H.. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006; 313:360–363. [DOI] [PubMed] [Google Scholar]
- 32. Tarus B., Chevalier C., Richard C.A., Delmas B., Di Primo C., Slama-Schwok A.. Molecular dynamics studies of the nucleoprotein of influenza A virus: role of the protein flexibility in RNA binding. PLoS One. 2012; 7:e30038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CCP4 suite is downloaded under academic licenses at https://www.ccp4.ac.uk/download/.
Crystal structures of NP-DL and NP-DLDC2 complexed with RNA are deposited with the Protein Data bank under accession number 7DXP and 7DKG, respectively.