Abstract
Argonaute proteins are programmable nucleases that are found in both eukaryotes and prokaryotes and provide defense against invading genetic elements. Although some prokaryotic argonautes (pAgos) were shown to recognize RNA targets in vitro, the majority of studied pAgos have strict specificity toward DNA, which limits their practical use in RNA-centric applications. Here, we describe a unique pAgo nuclease, KmAgo, from the mesophilic bacterium Kurthia massiliensis that can be programmed with either DNA or RNA guides and can precisely cleave both DNA and RNA targets. KmAgo binds 16–20 nt long 5′-phosphorylated guide molecules with no strict specificity for their sequence and is active in a wide range of temperatures. In bacterial cells, KmAgo is loaded with small DNAs with no obvious sequence preferences suggesting that it can uniformly target genomic sequences. Mismatches between the guide and target sequences greatly affect the efficiency and precision of target cleavage, depending on the mismatch position and the nature of the reacting nucleic acids. Target RNA cleavage by KmAgo depends on the formation of secondary structure indicating that KmAgo can be used for structural probing of RNA. These properties of KmAgo open the way for its use for highly specific nucleic acid detection and cleavage.
INTRODUCTION
Prokaryotic argonautes (pAgos) form a diverse family of proteins that are found in both bacteria and archaea and are related to eukaryotic Ago proteins involved in RNA interference (1–4). pAgos can be divided into three clades, including long active, long inactive pAgos and short pAgos (5). The conserved core structure of long pAgo proteins includes the N-terminal, L1, PAZ, L2, MID and PIWI domains, while short pAgos consist of only MID and PIWI domains. Long active pAgos have a catalytic tetrad DEDX (X is D, H or K) in the PIWI domain, which binds divalent metal ions and participates in catalysis. When programmed with small nucleic acid guides, active pAgos cleave complementary targets between the 10th and 11th positions of the guide nucleic acid (1,3,4,6).
Most previously characterized catalytically active pAgos are DNA-guided nucleases that target DNA. They were initially isolated from thermophilic prokaryotes, including TtAgo from Thermus thermophilus, PfAgo from Pyrococcus furiosus and MjAgo from Methanocaldococcus jannaschii (7–13). More recently, pAgo nucleases from mesophilic bacteria have also been described including CbAgo from Clostridium butyricum (14,15), LrAgo from Limnothrix rosea (15) and SeAgo from Synechococcus elongatus (6). Among them, CbAgo shows robust activity toward single-stranded DNA substrates in the range between 20 and 55°C and can also precisely cleave double-stranded DNA at elevated temperature (14,15). Recognition of double-stranded DNA by pAgos requires its prior melting, which can be modulated by variations in the GC/AT-content, temperature, the action of accessory proteins or cellular processes of DNA replication, transcription and repair (8,15–19). In the absence of guides, some pAgos including TtAgo, MjAgo, CbAgo and SeAgo, were shown to perform non-specific processing of target DNA in vitro, possibly resulting in autonomous generation of guide molecules, but the role of this activity in vivo remains to be established (6,13,15,20).
Similarly to eukaryotic Agos, pAgos have been implicated in the cell's defense against mobile genetic elements (1,4,21). Recently, CbAgo has been shown to protect bacterial cells from phage infection (16). CbAgo as well as thermophilic TtAgo and PfAgo can also decrease plasmid DNA content and/or transformation efficiency (7,8,16). RsAgo from Rhodobacter sphaeroides, a long inactive pAgo, preferentially targets plasmid DNA, transposons and prophages (19). In addition, TtAgo has been implicated in chromosomal DNA decatenation and completion of replication (22), suggesting that pAgos may have a wider range of cellular functions.
Ago proteins can be used for specific and precise cleavage of nucleic acid targets, thus providing a potential tool for nucleic acid manipulations. However, most previously studied pAgos act on DNA and there are only a few known examples of pAgo nucleases that can be loaded with RNA guides or recognize RNA targets. In particular, MpAgo from thermophilic Marinitoga piezophila binds RNA guides to cleave DNA and, with a somewhat lower efficiency, RNA targets (23). TtAgo from thermophilic T. thermophilus uses DNA guides to cleave DNA but can also cleave RNA (8,10,24). No mesophilic pAgos with the ability to recognize both types of target nucleic acids, DNA and RNA, have been described to date, and none of the previously studied pAgos could be guided by both DNA and RNA. The search for other mesophilic pAgo proteins with relaxed specificity, which could be used to manipulate both DNA and RNA, is therefore of a great importance. In this work, we characterize a novel mesophilic pAgo nuclease, KmAgo from Kurthia massiliensis. We show that, similarly to other mesophilic pAgos, KmAgo is active at ambient temperatures. However, in contrast to them, KmAgo can be programmed with both DNA and RNA guides of any sequence to efficiently cleave both types of nucleic acids which paves the way for its use for site-specific cleavage and structural probing of DNA and RNA.
MATERIALS AND METHODS
Protein expression and purification
The gene of KmAgo was amplified by PCR from genomic DNA of K. massiliensis JC30 and cloned into the pET28b expression vector in frame with the N-terminal His6-tag using the NheI–XhoI restriction sites. Escherichia coli BL21(DE3) was transformed with the expression plasmid and the cells were cultivated in the LB medium with 50 μg/ml kanamycin at 25°C until OD600 = 0.4, cooled down to 16°C, induced with 0.2 mM IPTG and grown at 16°C overnight with aeration. The cells were collected by centrifugation and stored at –70°C. The cell pellet was resuspended in buffer A (30 mM Tris–HCl pH 7.9, 0.3 M NaCl, 5% glycerol) supplemented with 1 mM of PMSF and disrupted using Cell Disruptor CF (Constant Systems). The lysate was cleared by centrifugation, and loaded onto Co2+-charged TALON Metal Affinity Resin (Clontech) for 1.5 h with agitation. The beads were washed with buffer A containing 5 mM of imidazole, then with the same buffer with 20 mM of imidazole, and eluted with buffer containing 300 mM imidazole. The eluted protein was diluted 3-fold with a buffer containing 20 mM Tris–HCl pH 7.9 and 5% glycerol. The protein sample was loaded onto a Heparin FF column (GE Healthcare) equilibrated with 20 mM Tris–HCl pH 7.9, 100 mM NaCl and 5% glycerol, washed with 10 volumes of the same buffer and eluted with a linear NaCl gradient (0.1–1.0 M). The purity of the final protein samples was assessed by denaturing PAGE with Coomassie staining. Fractions containing KmAgo were concentrated using Amicon Ultra 50K (Merck Millipore), placed in a storage buffer (40 mM Tris–HCl, 0.3 M NaCl, 50% glycerol, 1 mM DTT, 0.5 mM EDTA, pH 7.9), aliquoted and frozen in liquid nitrogen. The protein concentration was determined by the Qubit protein assay kit (Thermo Fischer Scientific). Catalytically dead mutant variant (D527A; D596A) was obtained by site-directed mutagenesis using QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent), expressed and purified in the same way.
Analysis of KmAgo-associated smDNAs
Small nucleic acids were extracted by phenol-chloroform treatment from KmAgo after the first purification step (Co2+-resin). Nucleic acids were treated with DNase I or RNase A and analyzed by denaturing PAGE as described previously (15). Small DNA libraries were prepared as described in (16). Briefly, extracted nucleic acids were ethanol-precipitated, treated with RNase A (ThermoFisher), ligated with adaptor oligonucleotides and sequenced using the HiSeq2500 platform (Illumina) in the rapid run mode (50-nucleotide single-end reads). Analysis of smDNA sequences was performed as described previously (16).
Analysis of cell growth
Overnight culture prepared from E. coli BL21(DE3) transformed with the expression plasmid pET28-KmAgo or the empty vector was diluted to OD 0.015 in the LB medium containing 50 μg/ml kanamycin and 0.2 mM IPTG or 0.2% glucose and aliquoted in a 24-well plate, 1 ml per well. The plate was incubated at 30°C with agitation at 200 rpm and OD was measured each 10 min in a Tecan microplate reader. Data from three independent biological replicates were used to plot the growth curves.
Single strand nucleic acid cleavage
The cleavage assays were performed using synthetic guide and target DNAs and RNAs (see Supplementary Table S1 for oligonucleotide sequences). For some experiments 5′-Cy3- or 5′-32P-labeled targets were used. The 6S RNA gene was PCR-amplified from the genomic DNA of E. coli strain MG1655 including the T7 RNA polymerase promoter in the forward primer. 6S RNA was synthesized by T7 RNA polymerase (Thermo Fisher Scientific), purified by 8% denaturing PAGE, treated with DNase I and ethanol precipitated. The cleavage reactions were performed in low adhesion tubes at 37°C in a buffer containing 20 mM Tris–HCl pH 7.4, 100 mM NaCl, 10% glycerol and 10 mM MnCl2. To analyze the effect of various divalent cations 10 mM MgCl2, CoCl2, CuCl2 or ZnCl2 were added instead of MnCl2. 500 nM of KmAgo was mixed with 500 nM guide DNA or RNA and incubated for 10 min at 37°C for guide loading. All guides were 5′-phosphorylated using T4 PNK (New England Biolabs) except for experiments with 5′-OH guides. Target DNA or RNA was then added to the final concentration of 100 nM. For analysis of temperature dependence of ssDNA cleavage, KmAgo was loaded with guide DNA for 10 min at 37°C, the samples were transferred to indicated temperatures in a dry block heater (BioSan, CH-100), target DNA or RNA was added and the samples were incubated for 4 minutes with G-DNA and T-DNA, 40 min with G-DNA and T-RNA and 60 minutes for G-RNA with T-DNA or T-RNA. For experiments on multiple turnover cleavage (Supplementary Figure S4A), an excess of target DNA (200 nM) was incubated with 100 nM KmAgo-guide complex (equimolar KmAgo and guide ratio) at 37°C or 60°C. For experiments in Supplementary Figure S4B, 50 or 100 nM of the KmAgo-guide complex was incubated for 30 min with 200 nM of label-free target DNA and then 5′-Cy3 labeled target DNA was added to 200 nM concentration. For analysis of 6S RNA cleavage, the reactions were performed for 30 minutes with corresponding DNA guides. All reactions were carried out at 37 or 60°C as indicated. The reactions were stopped after indicated time intervals by mixing the samples with equal volumes of stop solution (8 M urea, 20 mM EDTA, 0.005% Bromophenol Blue, 0.005% Xylene Cyanol). The cleavage products were resolved by 19% denaturing PAGE, stained with SYBR Gold (Invitrogen) or visualized with a Typhoon FLA 9500 scanner (GE Healthcare) in the case of 5'-32P-labeled targets, and analyzed by the ImageQuant (GE Healthcare) software. For reactions containing Cy3-labels, the gels with the reaction products were first scanned in the Cy3 channel and then stained with SYBR Gold. In the case of 6S RNA, the cleavage products were resolved by 10% denaturing PAGE and stained with SYBR Gold.
Plasmid DNA cleavage
For analysis of plasmid DNA cleavage, the final pAgo and guide concentrations were 500 nM. When using two or four guide molecules, the samples of KmAgo were independently loaded with the guides (at the 1:1 KmAgo:guide ratio) and then mixed together to the final concentration of 500 nM. The sequences of all guide oligonucleotides are shown in Supplementary Table S1. The target plasmids used in the assays are pJET1.2 derivatives (Thermo Fisher Scientific) containing short inserts shown in Supplementary Table S1. The plasmid was added to the reaction mixtures to the final concentration of 2 nM, followed by incubation for indicated time intervals at 37 or 60°C. SSB proteins (E. coli SSB purified as described before (6) or thermostable ET SSB purchased from New England BioLabs) were added to the reaction mixtures to the final concentration of 1 μM 10 min prior to the addition of KmAgo, when indicated. The reactions were stopped by treatment with Proteinase K for 20 min at 25°C, the samples were mixed with 6× SDS-free Purple Loading Dye (New England Biolabs) supplemented with SYBR Gold and the cleavage products were resolved by native 1.2% agarose gel electrophoresis. The relaxed plasmid in control samples was generated by treatment of the supercoiled plasmid with RNase H2 (New England Biolabs) for 2 h at 37°C, the linear plasmid was obtained by treatment with a single cut restriction endonuclease (XhoI, Thermo Fisher Scientific).
RESULTS
Expression of KmAgo in E. coli and analysis of associated nucleic acids
KmAgo (accession number WP_010289662.1 in the NCBI protein database) is distantly related to CbAgo and contains the canonical catalytic tetrad in the PIWI domain (residues D527, E562, D596 and D713). To study the biochemical properties and in vivo functions of KmAgo, we amplified the KmAgo gene from the genomic DNA of K. massiliensis and cloned it in an expression vector. We obtained E. coli strains with plasmid-encoded KmAgo and used them for in vivo assays and purification of KmAgo. The expression of KmAgo in E. coli led to a significant delay in cell growth in comparison with control strains grown without the addition of the inducer or containing an empty plasmid, suggesting that KmAgo protein is toxic for the cells (Figure 1A). We purified KmAgo using metal-affinity and heparin chromatography steps, resulting in >98% pure preparations (Supplementary Figure S1, Materials and Methods) and used it for further functional analysis (see next sections).
Figure 1.
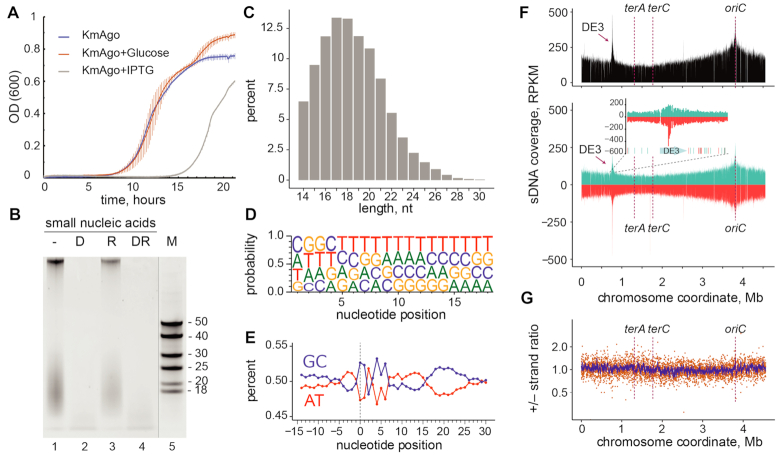
KmAgo binds small DNAs from genomic sequences during expression in E. coli. (A) Effect of KmAgo expression on cell growth. The experiment was performed in BL21(DE3) strains containing the expression vector (pET28-KmAgo) in the absence of IPTG, in the presence of Glc (to fully suppress possible leaky expression of KmAgo) and in the presence of the inducer (IPTG) (mean values and standard deviations from three biological replicates are shown). (B) Purification of KmAgo-associated nucleic acids after its expression in E. coli. Nucleic acids were treated with DNaseI (lane 2), RNase A (lane 3), both nucleases (lane 4), or left untreated (lane 1), separated by denaturing PAGE and stained with SYBR Gold. DNA length markers are shown in lane 5. (C) Length distribution of small DNAs in the sequenced libraries. (D) Frequencies of individual nucleotides at different positions of smDNAs associated with KmAgo. (E) AT/GC-content along the smDNA sequences. Nucleotide positions relative to the smDNA 5′-end are shown below the plot (negative numbers correspond to genomic DNA adjacent to the 5′-end of smDNA). (F) Genomic distribution of KmAgo-bound smDNAs mapped to the chromosome of the BL21(DE3) strain (top, the total number of smDNAs; bottom, strand-specific distribution of smDNAs; turquoise, plus strand; red, minus strand). The inset shows the genomic neighborhood of the DE3 prophage, with distribution of Chi sites in this region (turquoise, Chi sites in the plus strand; red, Chi sites in the minus strand). The numbers of smDNAs are shown in RPKM (reads per kilobase per million reads in the smDNA library). Positions of the replication origin (ori) and termination sites (terA and terC) are indicated. (G) Ratio of the smDNA densities for the plus and minus genomic strands. The ratio of RPKM values in each 1 kb window is shown in orange; the averaged ratio in the 10 kb window is shown in blue.
Previously studied pAgo proteins were shown to bind small guide nucleic acids when expressed in their native hosts or in the heterologous E. coli system (8,13,14,16,19,22). To explore if KmAgo is associated with guide nucleic acids when expressed in E. coli cells, we extracted and labeled nucleic acids from the KmAgo samples purified by metal-affinity chromatography. KmAgo was bound with 13–25 nt small guide DNAs (smDNAs), as could be concluded from their sensitivity to DNase I treatment and resistance to RNase A (Figure 1B). Next, we cloned KmAgo-associated smDNAs and performed their high-throughput sequencing. As expected, the length distribution of the resulting sequences corresponded to the lengths of smDNAs isolated from KmAgo, with the peak at 16–19 nucleotides (Figure 1C). We observed large diversity of associated guides with no nucleotide biases along the guide sequences, with comparable frequencies of all four nucleotides at all positions (Figure 1D). Thus, unlike many of previously studied pAgos, KmAgo does not have a preference for specific residue at the first position of the guide. In addition, mapping of guide sequences to the E. coli genome showed no sequence bias in the genomic sequence upstream of the guide 5′-end (Figure 1E).
To analyze the origin of KmAgo-associated smDNAs, we mapped them to the E. coli chromosome and the expression plasmid. A significant portion of smDNAs corresponded to plasmid sequences (3.9 and 6.8% in the two replicas). However, when normalized to the length of corresponding replicons, the ratio of smDNAs mapping to the plasmid and chromosomal DNA (24 and 44 in the two replicas) roughly equaled the known copy number of the plasmid vector (pET28, about 20 copies per cell). Thus, unlike previously studied pAgos (8,16,19), KmAgo-bound guides are not enriched in sequences derived from plasmid DNA.
Chromosomal mapping of KmAgo-bound smDNAs revealed their almost uniform distribution along the genome with no preferences for specific genomic regions, apart from a shallow enrichment around the origin of replication, which likely results from the higher DNA content in this region due to ongoing replication (Figure 1F and Supplementary Figure S2). The smDNA reads were evenly distributed along the plus and minus genomic strands, with about equal ratio of the strands along the whole genome (Figure 1F, G and Supplementary Figure S2). In addition to the origin region, a smaller peak of smDNAs around the 0.77 Mb genomic coordinate was detected, which corresponded to the DE3 prophage in the genome of the BL21(DE3) strain used for KmAgo expression (Figure 1F). This indicated some kinds of genomic rearrangements in this region (see Discussion). In summary, these experiments demonstrated that KmAgo is loaded with smDNA guides when expressed in vivo, and that these DNAs are generated by uniform sampling of chromosomal and plasmid sequences by KmAgo.
KmAgo can use both DNA and RNA guides to target DNA and RNA in vitro
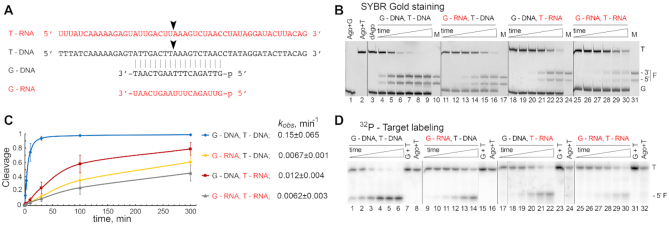
Most previously characterized pAgo nucleases have strict specificity for DNA guides and DNA targets (see Introduction). To test whether KmAgo have a distinct specificity for DNA or RNA in vitro, we analyzed its activity using a set of synthetic guide and target oligonucleotides (Figure 2A, Supplementary Table S1). KmAgo was first loaded with guide DNA or RNA and both types of binary complexes were incubated with single-stranded DNA or RNA targets. After incubation the cleavage products were analyzed by denaturing gel-electrophoresis (Figure 2B). Surprisingly, it was found that KmAgo is active as guide-directed endonuclease with all combinations of guide and target nucleic acids, with the highest activity observed for DNA-guided DNA cleavage, and the lowest for RNA-guided RNA cleavage. Importantly, in all cases the cleavage products corresponded to the single expected cleavage site (the target position between the 10th and 11th nt of the guide, the canonical cleavage site observed for all previously studied Ago proteins), demonstrating that KmAgo retains high level of precision with various combinations of guides and targets. No cleavage activity was detected for a catalytically dead KmAgo variant with substitutions of two of the tetrad residues (D527A and D596A) (Figure 2B, lane 3).
Figure 2.
Guide and target specificity of KmAgo in vitro. (A) Scheme of the guide and target nucleic acids (DNA or RNA); the cleavage position is indicated with an arrow. (B) Analysis of nucleic acid cleavage by KmAgo. KmAgo pre-loaded with guide DNA or RNA was incubated with single-stranded DNA or RNA targets for indicated time intervals, nucleic acids were separated by denaturing PAGE and visualized by SYBR Gold staining. Positions of the targets (T), guides (G) and the 5′- and 3′-cleavage fragments (F) are indicated. The lengths of the synthetic RNA and DNA marker (M) oligonucleotides correspond to the expected cleavage site between the 10th and 11th guide positions. (C) and (D) Kinetics of nucleic acid cleavage by KmAgo measured with radiolabeled target DNA or RNA. The data were fitted to a single-exponential equation. For each guide-target pair, the resulting kobs value is shown (means and standard deviations from three independent measurements). All experiments were performed at 37°C. Representative gels from three independent measurements are shown.
To determine the catalytic parameters of KmAgo, we measured the kinetics of the reaction using 5′-32P-labeled target molecules and calculated the observed rates (kobs) of target cleavage under single-turnover conditions (i.e. in excess of the KmAgo-guide complex over the target) (Figure 2C and D). In agreement with the previous experiment, the fastest cleavage was observed for DNA guide and DNA target (kobs = 0.15 min−1, corresponding to the reaction half-time t1/2 of about 4.6 min), followed by DNA-guided RNA cleavage (kobs = 0.012 min−1), and RNA-guided DNA and RNA cleavage (kobs = 0.0067 and 0.0062 min−1, respectively) (Figure 2C). Importantly, although the reaction rates varied significantly for different guide/target combinations, at least half of the substrate was processed during the reaction time in all cases, thus making KmAgo the first Ago nuclease that is active with all variants of guide and target nucleic acids.
KmAgo is active under a wide range of reaction conditions and has no strict sequence requirements
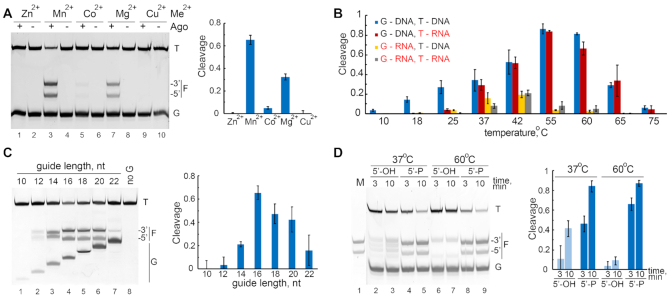
Divalent metal ions bound in the catalytic center of Ago proteins play the key role in the cleavage reaction. We observed that, similarly to other pAgos, the most efficient cleavage occurs with Mn2+ and, less efficiently, with Mg2+ cofactors (Figure 3A). A low level of activity was observed with Co2+ and no activity was detected with Zn2+ and Cu2+. Therefore, all further experiments were performed in the presence of Mn2+ ions.
Figure 3.
Characteristics of nuclease activity of KmAgo. (A) DNA-guided DNA cleavage by KmAgo with various divalent cation cofactors. The reaction was performed for 10 min at 37°C. The fraction of cleaved target for each guide-target combination is shown. (B) Temperature dependence of the cleavage reaction with various combinations of guide and target nucleic acids. (C) Dependence of the efficiency of DNA cleavage on the length of guide DNA. The reaction was performed for 4 min at 37°C. (D) The requirement of the 5′-phosphate group in guide DNA for precise target DNA cleavage. The reaction was performed for 3 or 10 min at 37 or 60°C. The marker (M) lane contains DNA fragments corresponding to the canonical cleavage site. For each experiment, representative gels and means and standard deviations from three to four independent measurements are shown.
Most previously studied pAgos were isolated from thermophilic bacteria and were active only in a high temperature range (7,8,11,13,23). Recently, several DNA-guided pAgos from mesophilic bacteria were demonstrated to cleave DNA targets at ambient temperatures (6,14,15,25,26). We therefore determined optimal temperature conditions for nucleic acid cleavage by KmAgo. When loaded with the DNA guide, KmAgo was able to precisely cleave the corresponding DNA target at temperatures between 18 and 65°C, with the most efficient cleavage at 37–60°C (Figure 3B). A similar temperature dependence was observed for the corresponding RNA target between 37 and 65°C; however, the RNA target was cleaved less efficiently at low temperatures (18–25°C). In the case of the RNA guide, KmAgo was active in a much narrower range of temperatures, between 37 and 42°C, with no strong differences observed for the DNA and RNA targets (Figure 3B). Therefore, DNA guides can be used by KmAgo to cleave both DNA and RNA targets in a wider temperature range in comparison with RNA guides.
We further studied the role of guide structure such as its length and the presence of the 5′-phosphate on target DNA cleavage. KmAgo was most active with 16–20 nt guide DNAs, with a lower cleavage efficiency observed with shorter or longer guides (Figure 3C). Interestingly, the cleavage position was shifted by one nucleotide if shorter (12 and 14 nt) guides were used, so that the cleavage occurred between the 9th and 10th guide positions (Figure 3C). Another factor that affects the precision of target processing is the presence of the 5′-phosphate in guide DNA. KmAgo can use 5′-OH guide DNA to cleave targets, however, the reaction efficiency was decreased and the cleavage positions were shifted relative to the canonical cleavage site (the cleavage occurred between 9th and 10th or 11th and 12th guide nucleotides at 37°C, or between 11th and 12th nucleotides at 60°C) (Figure 3D).
Many studied Ago proteins from both prokaryotes and eukaryotes have certain specificity for the first nucleotide in the guide strand, which is bound in the MID pocket of Ago (19,27). In contrast, no changes in the cleavage rate and efficiency were observed when KmAgo was loaded with guides with different 5′-nucleotides but otherwise identical sequences (Supplementary Figure S3A). KmAgo was also able to cleave target sites with completely different sequences with comparable efficiency (except for one of the tested guides that was less efficient) (Supplementary Figure S3B). This correlates with the absence of strong nucleotide preferences at any guide position in smDNA associated with KmAgo in vivo (see above). Therefore, KmAgo can potentially be programmed for specific recognition and cleavage of any target sequence.
To test whether KmAgo is a single-turnover enzyme or a multiple-turnover enzyme that can dissociate from the target after cleavage, we analyzed the cleavage kinetics in reactions containing a 2-fold excess of target DNA over the binary guide-KmAgo complex. It was found that at 37°C only a fraction of the target was cleaved during the 100 min course of the reaction (Supplementary Figure S4A, left panel). In contrast, when the reaction was performed at 60°C almost all DNA was cleaved during 10 min, indicative of multiple-turnover reaction (right panel). To directly detect exchange of the reacting target molecules, we analyzed cleavage of identical DNA targets, unlabeled or labeled with a Cy3 dye, sequentially added to the reaction with the KmAgo-guide complex. The reactions were started with adding an excess of unlabeled target to the KmAgo-guide complex (2–4-fold molar ratio). After 30 min, Cy3-labeled target was added to the reaction. The cleavage of the labeled secondary target was detected after 3 min incubation at 60°C, but not at 37°C (Supplementary Figure S4B). Extended (3-h) incubation led to processing of the labeled target at both temperatures. Thus, KmAgo acts as a multiple-turnover endonuclease, and exchange of target molecules is facilitated at high temperatures.
Effects of guide-target mismatches on target cleavage
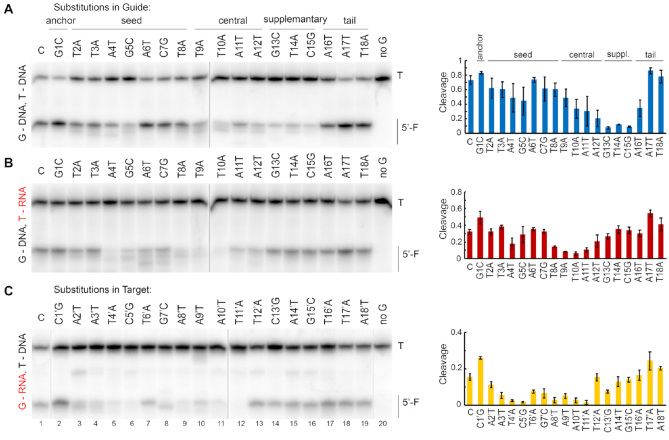
Precise recognition and cleavage of target nucleic acids is essential for the activities of genome defense systems that employ programmable nucleases, including CRISPR–Cas systems in prokaryotes and RNA interference systems in eukaryotes. To determine the sequence specificity of KmAgo, we analyzed the effects of mismatches between the guide and target strands on its nuclease activity, using three variants of guide and target nucleic acids: guide DNA and target DNA (Figure 4A and Supplementary Figure S5A), guide RNA and target DNA (Figure 4B and Supplementary Figure S5B), and guide DNA and target RNA (Figure 4C and Supplementary Figure S5C). In the effector complexes of Ago proteins, the guide molecule can be functionally divided into several regions, including the 5′-nucleotide bound in the MID pocket of Ago, the 5′-seed region (positions 2–8), the central region surrounding the cleavage site (positions 9–12), the 3′-supplementary region (positions 13–15), and the 3′-tail region (position 16 and downstream of it) (Figure 4). We found that mismatches in each of these regions had various effects on the efficiency and precision of target cleavage by KmAgo, depending on the nature of the reacting molecules. In reactions with DNA guides and targets (G-DNA/T-DNA), mismatches in the 3′-supplementary region of guides had the strongest effects on DNA cleavage (Figure 4A). In addition, mismatches in the central region and at positions 4–5 in the seed region also decreased the efficiency of cleavage. In contrast, when the same set of mismatched guide DNA was tested with target RNAs (G-DNA/T-RNA), no changes in cleavage were observed for mismatches in the 3′-suppplementary region (Figure 4B). Instead, the strongest decrease in cleavage efficiency was observed for mismatches at the cleavage site and in the middle of the seed region. Finally, for reactions containing guide RNA and target DNA (G-RNA/T-DNA) the target cleavage was strongly effected by mismatches at most positions in the seed and central regions (Figure 4C). This correlated with the lower reaction rate for this combination of guide and target nucleic acids (see above), likely making the reaction more dependent on correct base-pairing.
Figure 4.
Effects of guide-target mismatches on DNA and RNA cleavage by KmAgo. Reactions with guide DNA and target DNA (A), guide DNA and target RNA (B) and guide RNA and target DNA (C) were performed at 37°C for 15, 60 and 100 minutes, respectively, using 5′- P32-labeled targets. Positions of the 5′-cleavage fragments (F) are indicated. In panels A and B, a series of different DNA guides contained substitutions at each guide position, shown above the panels. In panel C, a series of DNA targets contained substitutions at each position in the region of complementarity to the guide RNA, shown above the panel (see Supplementary Table S1 for full guide and target sequences). Means and standard deviations from three independent measurements.
Unexpectedly, in addition to changing the efficiency of target cleavage, mismatches also changed its pattern. The appearance of additional products resulting from cleavage at noncanonical sites was observed not only for mismatches at the site of cleavage (Figure 4, positions 9–12) but also for mismatches in the seed region (e.g. at positions 2–5 in the G-DNA/T-DNA reactions, positions 2, 4, 5–8 in the G-DNA/T-RNA reactions, positions 2–4, 7, 8 in the G-RNA/T-DNA reactions) and even more surprisingly in the 3′-supplementary region (position 14 in the G-DNA/T-DNA reactions). The pattern of noncanonical cleavage fragments was highly reproducible in three independent experiments (compare Figure 4 and Supplementary Figure S5), indicating that mismatches at individual positions have distinct effects on the conformation of the effector complex. Overall, these experiments demonstrated that the complete base-pairing between the guide and target molecules is important for precise target cleavage and that even single mismatches at certain positions can disrupt proper DNA and RNA processing by KmAgo.
Plasmid DNA cleavage by KmAgo
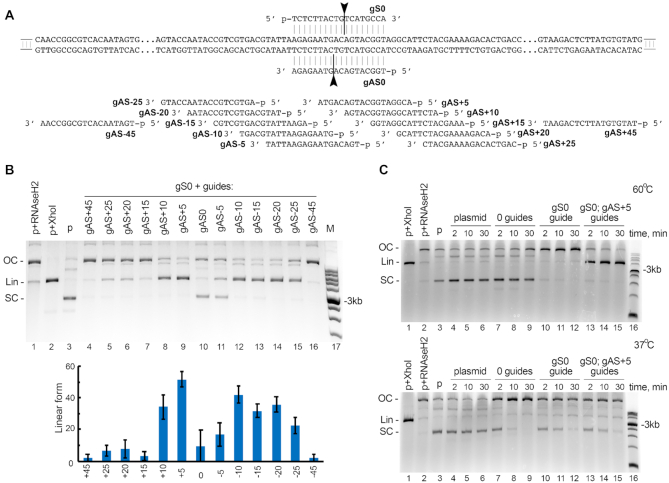
In the experiments described above, the activity of KmAgo was tested on single-stranded DNA targets, which are preferred substrates for most previously studied pAgos (see Introduction). To determine the ability of KmAgo to process double-stranded DNA, we analyzed cleavage of supercoiled plasmid DNA with KmAgo loaded with guides corresponding to the two plasmid strands. To test whether the relative positions of the guide molecules targeting opposite DNA strands influence the efficiency of dsDNA cleavage, we used a set of guide pairs in which the position of the guides targeting the bottom ‘antisense’ DNA strand was shifted relative to the position of the guide targeting the top ‘sense’ strand by 5, 10, 15, 20, 25 or 45 nucleotides in both directions (sense guide gS0, antisense guides gAS-5, gAS+5, gAS-10, gAS+10 etc.) (Figure 5A). To facilitate melting of double-stranded DNA, the experiments were performed at 60°C. In all cases, we observed relaxation and linearization of the plasmid, resulting in disappearance of the supercoiled form and increase in the amounts of the relaxed (OC, open circle) and linear (Lin) plasmid forms (Figure 5B). Remarkably, shifting the position of the antisense guide by 5 and 10 nt to the right (Figure 5B, lanes 8, 9) or by 10, 15, 20 and 25 nt to the left (lanes 12–15) relative to the sense guide significantly increased the efficiency of plasmid linearization in comparison with oppositely located guides (lane 10). This suggests that the effector complexes of KmAgo interfere with each other during recognition of the two DNA strands if they are located directly opposite each other.
Figure 5.
Cleavage of plasmid DNA by KmAgo. (A) Scheme of the plasmid target used in the cleavage assay (pJET1.2 containing insert 1 shown in Supplementary Table S1). Positions of guide DNAs corresponding to the sense (gS) and antisense (gAS) strands of the target are shown along the plasmid sequence. (B) Plasmid linearization with pairs of guide DNAs with different lengths between the cleavage sites in the two DNA strands. The reactions were performed for 2 min at 60°C. The cleavage efficiency is shown as a fraction of linear DNA form relative to the sum of supercoiled, relaxed and linear DNA (means and standard deviations from three measurements). (C) The target plasmid was incubated in the absence of KmAgo (lanes 4–6), with empty KmAgo (lanes 7–9), or with KmAgo loaded with one or two guide DNAs (gS0 and gAS+5; lanes 10–15) at 60°C (top) or 37°C (bottom) for indicated time intervals. Control samples containing linear (Lin) plasmid obtained by treatment with XhoI (lane 1), relaxed open circle (OC) plasmid obtained by treatment with RNase H2 (lane 2), and supercoiled (SC) plasmid (lane 3) were incubated in the absence of KmAgo. Representative gels from three independent experiments are shown.
We further programmed KmAgo with two pairs of guide DNAs to observe whether it can cleave the corresponding DNA segment out of the target plasmid (Supplementary Figure S6A). For both target sites, the guides targeting opposite DNA strands were shifted by five nucleotides relative to each other, to increase target cleavage. Incubation of the plasmid with the mixture of four effector complexes of KmAgo resulted not only in the linearization of the plasmid but also in the appearance of the two plasmid fragments corresponding to its cleavage at the expected sites, indicating that KmAgo can potentially be used for site-specific plasmid DNA cleavage.
To better understand processing of plasmid DNA by KmAgo in vitro, we compared plasmid cleavage with empty KmAgo or with KmAgo loaded with one or two DNA guides (gS0 and gAS+5, Figure 5A) at 60°C and 37°C. At 60°C, KmAgo rapidly linearized the plasmid with two guides (Figure 5C, upper panel, lanes 13–15) and relaxed it with one guide (lanes 10–12), as expected. Empty KmAgo did not target the plasmid under these conditions (lanes 7–9, compare with lanes 4–6 in which the plasmid was incubated in the absence of KmAgo). In contrast, incubation of the plasmid with empty KmAgo at 37°C led to its relaxation (Figure 5C, bottom panel, compare lanes 7–9 with the control samples in lanes 4–6). This may result from non specific guide-independent activity of KmAgo, which is suppressed at higher temperatures. Several other pAgo proteins were also shown to cleave DNA in the absence of guides under certain conditions (13–15,20). At 37°C, loading of KmAgo with one or two plasmid-specific guides had only minor effects on the distribution of plasmid isoforms in comparison with empty KmAgo (Figure 5C, bottom panel, lanes 10–15). Previously, DNA cleavage by TtAgo was shown to be stimulated by SSB proteins, which may help to facilitate target melting (18). However, we did not observe any stimulatory effect of SSB on plasmid DNA cleavage by KmAgo (Supplementary Figure S6C). Thus, KmAgo cannot efficiently cleave double-stranded DNA at 37°C but can be used for programmable cleavage of double-stranded substrates at elevated temperatures.
Several previously studied pAgos were shown to decrease plasmid content and/or plasmid transformation efficiency in the host cells (including RsAgo, CbAgo, TtAgo, PfAgo) (7,8,16,19). We therefore tested whether KmAgo could have a similar effect on plasmid maintenance in E. coli. For this purpose, we compared the rate of plasmid loss in E. coli strains containing or lacking KmAgo. Under the conditions of our experiments, the expression of KmAgo did not increase the fraction of plasmid-free cells after 10 passages (Supplementary Figure S7). This is consistent with the absence of preferential loading of KmAgo with smDNAs derived from plasmid DNA (see above).
Probing of the secondary RNA structure with KmAgo
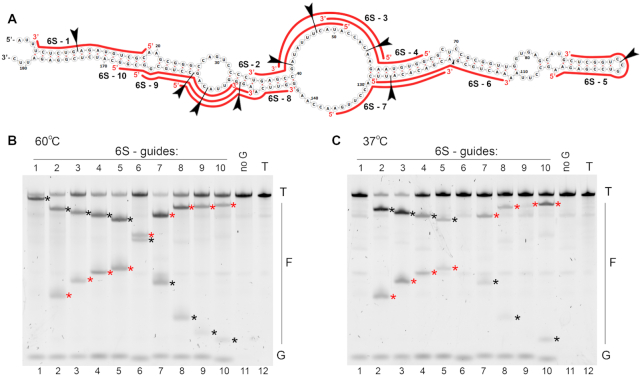
To explore if processing of target RNA by KmAgo depends on its secondary structure, we analyzed cleavage of highly structured 6S RNA, which binds to bacterial RNA polymerase and serves as a regulator of gene expression at the stationary phase (Figure 6A) (28). KmAgo was loaded with guide DNAs corresponding to different regions of 6S RNA that either maintain single-stranded conformation or are involved in extensive base-pairing (6S-1 through 6S-10; Figure 6A). The cleavage products were observed with all guide DNAs when reactions were performed at 60°C (Figure 6B). In contrast, at 37°C the cleavage efficiency greatly varied for different target positions. The most efficient cleavage was observed with guides 6S-2 and 6S-3 corresponding to the central single-stranded bubble in 6S RNA (Figure 6C, lanes 2 and 3). In contrast, no cleavage could be detected with guides 6S-1 and 6S-6, corresponding to fully double-stranded target regions. The efficiency of cleavage was also significantly lower for other guides corresponding to partially double-stranded target regions. For some of these guides (6S-4, 6S-5, 6S-9) the RNA region corresponding to the seed segment of guide DNA was directly involved in base-pairing with other segments of 6S RNA, while for others (6S-7, 6S-8) the seed region was partially exposed but the target site could not be cleaved efficiently. Taken together, these experiments indicate that base-pairing of a target RNA site determines the efficiency of its processing by KmAgo, while destabilization of the secondary structure at elevated temperatures allows for efficient processing at all sites. Thus, KmAgo can potentially be used for precise processing and structural probing of RNA targets.
Figure 6.
RNA probing with KmAgo. (A) Secondary structure of 6S RNA used in the experiments. Positions of small guide DNA loaded into KmAgo are shown with red lines (see Supplementary Table S1 for full guide sequences). Positions of the expected cleavage sites for each guide smDNA are indicated with arrowheads. (B) Analysis of the cleavage products obtained after incubation of KmAgo with 6S RNA for 30 min at 60°C. (C) The same experiment performed at 37°C. Representative gels from two independent experiments are shown. Positions of the 5′-terminal and 3′-terminal cleavage fragments are indicated with red and black asterisks, respectively.
DISCUSSION
In this study, we have characterized a novel pAgo nuclease from the mesophilic bacterium K. massiliensis. In comparison with known pAgos KmAgo has an unusually broad specificity toward guide and target nucleic acid molecules and can target both DNA and RNA at physiological temperatures. Previously, individual pAgo nucleases were shown to have strict specificity for either DNA or RNA guides, including AaAgo, TtAgo, PfAgo, CbAgo, LrAgo, SeAgo (6,7,10,14,15,29,30) (all requiring DNA guides) or MpAgo (23) (using RNA guides). Most of these nucleases strongly prefer DNA targets, although thermophilic AaAgo, TtAgo and MpAgo can also cleave RNA in vitro (10,23,29–31). Mesophilic KmAgo is therefore the first example of an Ago protein which is able to utilize all combinations of nucleic acids as its guides and targets. Furthermore, we have shown that KmAgo can target single-stranded DNA and RNA under a wide range of reaction conditions, with the efficiency and precision of cleavage being modulated by temperature, divalent ion cofactors, the length and phosphorylation of the small nucleic acid guide and its complementarity to the target. While our study was being prepared for publication, another manuscript describing the in vitro activities of KmAgo was published (32). The authors had come to similar conclusions about the biochemical activities of KmAgo. The most important complementary observations made in our study are discussed below.
We have shown that in bacterial cells KmAgo is associated with small DNAs of diverse sequences suggesting that DNA is used as the guide molecule in vivo. Since in vitro KmAgo is more active on DNA targets, it is natural to suppose that DNA is also its preferred substrate in vivo. Accordingly, small RNAs are not detected in KmAgo samples purified from bacterial cells; however, it cannot be excluded that RNA may also be targeted by KmAgo, with potential effects on gene expression, which requires further investigation. Analysis of KmAgo-bound smDNAs has revealed that they are derived from the whole genome with no obvious preferences for specific regions, except for moderate enrichment around the replication origin, likely as a result of a higher DNA content in this region in the dividing bacterial population. Interestingly, we have also observed a peak in smDNAs corresponding to the DE3 prophage in the BL21(DE3) chromosome. A similar peak in smDNAs and in genomic DNA around DE3 was observed previously during analysis of CbAgo in E. coli in the stationary phase of growth (16). This suggests that this chromosomal region may be amplified in E. coli cells, possibly as a result of partial prophage excision during the stationary stage, which may promote local DNA repair and replication.
Unlike CbAgo, SeAgo and TtAgo (16,22), KmAgo-associated smDNAs are not enriched at the sites of replication termination. Furthermore, KmAgo has no preference for smDNAs derived from the expression plasmid and does not affect plasmid maintenance in E. coli. This contrasts previously studied pAgos, including TtAgo, RsAgo, PfAgo, CbAgo and MjAgo, which all were shown to preferentially target plasmid DNA and decrease plasmid content and/or transformation efficiency (7,8,13,16,19). CbAgo, but not KmAgo, can also induce DNA interference between homologous plasmid and chromosomal regions resulting in targeted chromosomal DNA processing (16). In contrast, KmAgo-associated smDNAs are uniformly distributed along the chromosomal and plasmid sequences. It therefore remains to be established whether KmAgo can directly participate in DNA processing or be programmed for specific DNA recognition in vivo.
In vitro, KmAgo preferentially uses 16–20 nt long 5′-phosphorylated guide DNAs and is mostly active as endonuclease in the presence of manganese or magnesium ions, similarly to previously studied pAgos. However, in contrast to several other pAgos, including TtAgo, MjAgo and RsAgo (8,11,19,33), KmAgo has no strong sequence preferences for the 5′-guide nucleotide or for any other position within the guide sequence. This is consistent with the broad spectrum of smDNAs associated with KmAgo in bacterial cells and potentially allows specific targeting of any desired sequence. Importantly, even single-nucleotide mismatches with the target molecule greatly affect the nuclease activity of KmAgo. We have shown that the efficiency and the position of cleavage can be modulated by mismatches in the seed, central and 3′-supplementary regions of the guide. Interestingly, cleavage of DNA and RNA targets is differentially affected by mismatches. The presence of mismatches also changes the precision of target cleavage by KmAgo. Surprisingly, these effects have been observed not only for mismatches immediately at the site of cleavage but also for many positions inside the seed or 3′-supplementary region, suggesting that these mismatches strongly affect the conformation of the catalytic complex and dislodge the target molecule from its correct position in the active site. In comparison, mismatches between the guide and target nucleic acids did not affect the cleavage site for TtAgo or the structure of the ternary complex of RsAgo (9,33).
Given the strong effects of mismatches on KmAgo-dependent cleavage, accurate design of guide oligonucleotides may allow discrimination of closely related target sequences by KmAgo in various applications. In particular, degradation of one of the target variants can be used for enrichment of rare nucleic acid variants, both DNA and RNA, for their further detection. Thermophilic TtAgo and PfAgo were previously applied for precise recognition and cleavage of complementary targets and their closely related variants depending on the presence of mismatches (34,35). Similarly, thermophilic MpAgo, programmed with RNA guides, was used for selective binding of rare RNA variants with single-nucleotide changes without their cleavage (31). In comparison with thermophilic proteins, KmAgo offers a more flexible tool for the detection of single-nucleotide variations in the target nucleic acids because it is active in a wider range of temperatures. The specificity of KmAgo can likely be further increased by using RNA guides that are more sensitive to the presence of mismatched nucleotides.
All analyzed pAgo proteins, including KmAgo, are significantly less active on dsDNA than on ssDNA substrates. TtAgo, CbAgo and RsAgo can interact with double-stranded DNA only if its melting is facilitated by various factors, including low GC-content, increased temperature, or supercoiling (8,14,15,17,20). Similarly, specific cleavage of double-stranded plasmid DNA by KmAgo is also enhanced at higher temperatures. Previously, SSB proteins were shown to stimulate cleavage of both single-stranded and double-stranded target DNA by TtAgo but had no effect on plasmid DNA cleavage by SeAgo (6,18). The ability of KmAgo to cut plasmid DNA was not stimulated by SSB, however, since KmAgo retains high level of activity up to 60°C, it can be used for site-specific double-stranded DNA cleavage under these conditions.
Finally, we have demonstrated that KmAgo can be used for specific cleavage and structural probing of RNA targets. We have found that the efficiency of cleavage depends on the secondary structure of RNA target and that the presence of double-stranded regions prevents processing by KmAgo. Thus, KmAgo-dependent RNA cleavage can be used to detect the conformational state of RNA targets. Previously, eukaryotic Ago protein from the budding yeast Kluyveromyces polysporus was used for site-specific RNA cleavage (36). Similar to our findings, it was found that the efficiency of cleavage depended on the secondary RNA structure. Importantly, in comparison with previous works we have shown that KmAgo can also perform efficient RNA cleavage at elevated temperatures that promote melting of the secondary structure and thus make possible unbiased RNA processing. Therefore, KmAgo can also be used for precise cleavage of complex RNA targets independently of the secondary structure formation. Furthermore, KmAgo can potentially be applied in other RNA-centric methods such as recently developed Ago-based fluorescence in situ hybridization (Ago-FISH) (37). In conclusion, we demonstrate that KmAgo is a unique programmable nuclease that has a broad specificity for guide and target nucleic acids and can potentially be used in a wide range of applications in the field of nucleic acid biotechnology.
DATA AVAILABILITY
The smDNA sequencing datasets generated in this study are available from the Gene Expression Omnibus (GEO) database under the accession number GSE168010.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sergei Ryazansky for help with bioinformatic analysis of pAgo proteins and Denis Yudin for initial analysis of smDNA libraries.
Contributor Information
Ekaterina Kropocheva, Institute of Molecular Genetics, National Research Centre ‘Kurchatov Institute’, Moscow 123182, Russia.
Anton Kuzmenko, Institute of Molecular Genetics, National Research Centre ‘Kurchatov Institute’, Moscow 123182, Russia; Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA 91125, USA.
Alexei A Aravin, Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA 91125, USA.
Daria Esyunina, Institute of Molecular Genetics, National Research Centre ‘Kurchatov Institute’, Moscow 123182, Russia.
Andrey Kulbachinskiy, Institute of Molecular Genetics, National Research Centre ‘Kurchatov Institute’, Moscow 123182, Russia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research [18-29-07086 to A.K.; biochemical analysis of KmAgo]; Russian Science Foundation [19-14-00359 to D.E.; analysis of in vivo activities of KmAgo]. Funding for open access charge: Russian Foundation for Basic Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lisitskaya L., Aravin A.A., Kulbachinskiy A.. DNA interference and beyond: structure and functions of prokaryotic argonaute proteins. Nat. Commun. 2018; 9:5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Makarova K.S., Wolf Y.I., van der Oost J., Koonin E.V.. Prokaryotic homologs of argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct. 2009; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Swarts D.C., Makarova K., Wang Y., Nakanishi K., Ketting R.F., Koonin E.V., Patel D.J., van der Oost J.. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014; 21:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Willkomm S., Makarova K., Grohmann D. DNA-silencing by prokaryotic argonaute proteins adds a new layer of defence against invading nucleic acids. FEMS Microbiol. Rev. 2018; 42:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ryazansky S., Kulbachinskiy A., Aravin A.A.. The expanded universe of prokaryotic argonaute proteins. mBio. 2018; 9:e01935–01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olina A., Kuzmenko A., Ninova M., Aravin A.A., Kulbachinskiy A., Esyunina D. Genome-wide DNA sampling by Ago nuclease from the cyanobacterium Synechococcus elongatus. RNA Biol. 2020; 17:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swarts D.C., Hegge J.W., Hinojo I., Shiimori M., Ellis M.A., Dumrongkulraksa J., Terns R.M., Terns M.P., van der Oost J.. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 2015; 43:5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J.J.et al.. DNA-guided DNA interference by a prokaryotic argonaute. Nature. 2014; 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y., Juranek S., Li H., Sheng G., Wardle G.S., Tuschl T., Patel D.J.. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009; 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Willkomm S., Oellig C.A., Zander A., Restle T., Keegan R., Grohmann D., Schneider S.. Structural and mechanistic insights into an archaeal DNA-guided argonaute protein. Nature Microbiology. 2017; 2:17035. [DOI] [PubMed] [Google Scholar]
- 12. Zander A., Holzmeister P., Klose D., Tinnefeld P., Grohmann D. Single-molecule FRET supports the two-state model of argonaute action. RNA Biology. 2014; 11:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zander A., Willkomm S., Ofer S., van Wolferen M., Egert L., Buchmeier S., Stockl S., Tinnefeld P., Schneider S., Klingl A.et al.. Guide-independent DNA cleavage by archaeal argonaute from Methanocaldococcus jannaschii. Nat. Microbiol. 2017; 2:17034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hegge J.W., Swarts D.C., Chandradoss S.D., Cui T.J., Kneppers J., Jinek M., Joo C., van der Oost J.. DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum argonaute. Nucleic Acids Res. 2019; 47:5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuzmenko A., Yudin D., Ryazansky S., Kulbachinskiy A., Aravin A.A.. Programmable DNA cleavage by Ago nucleases from mesophilic bacteria Clostridium butyricum and Limnothrix rosea. Nucleic Acids Res. 2019; 47:5822–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuzmenko A., Oguienko A., Esyunina D., Yudin D., Petrova M., Kudinova A., Maslova O., Ninova M., Ryazansky S., Leach D.et al.. DNA targeting and interference by a bacterial argonaute nuclease. Nature. 2020; 587:632–637. [DOI] [PubMed] [Google Scholar]
- 17. Lisitskaya L., Petushkov I., Esyunina D., Aravin A., Kulbachinskiy A.. Recognition of double-stranded DNA by the Rhodobacter sphaeroides argonaute protein. Biochem. Biophys. Res. Commun. 2020; 533:1484–1489. [DOI] [PubMed] [Google Scholar]
- 18. Hunt E.A., Evans T.C. Jr, Tanner N.A. Single-stranded binding proteins and helicase enhance the activity of prokaryotic argonautes in vitro. PLoS One. 2018; 13:e0203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A.. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013; 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swarts D.C., Szczepaniak M., Sheng G., Chandradoss S.D., Zhu Y., Timmers E.M., Zhang Y., Zhao H., Lou J., Wang Y.et al.. Autonomous generation and loading of DNA guides by bacterial argonaute. Mol. Cell. 2017; 65:985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olina A.V., Kulbachinskiy A.V., Aravin A.A., Esyunina D.M.. Argonaute proteins and mechanisms of RNA interference in eukaryotes and prokaryotes. Biochemistry. 2018; 83:483–497. [DOI] [PubMed] [Google Scholar]
- 22. Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D.. Thermus thermophilus argonaute functions in the completion of DNA replication. Cell. 2020; 182:1545–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial argonaute with noncanonical guide RNA specificity. PNAS. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheng G., Zhao H., Wang J., Rao Y., Tian W., Swarts D.C., van der Oost J., Patel D.J., Wang Y.. Structure-based cleavage mechanism of Thermus thermophilus argonaute DNA guide strand-mediated DNA target cleavage. PNAS. 2014; 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao Y., Sun W., Wang J., Sheng G., Xiang G., Zhang T., Shi W., Li C., Wang Y., Zhao F.et al.. Argonaute proteins from human gastrointestinal bacteria catalyze DNA-guided cleavage of single- and double-stranded DNA at 37 degrees C. Cell Discovery. 2019; 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia-Quintans N., Bowden L., Berenguer J., Mencia M.. DNA interference by a mesophilic argonaute protein, CbcAgo. F1000Research. 2019; 8:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wassarman K.M. 6S RNA, a global regulator of transcription. Microbiol. Spectrum. 2018; 6:doi:10.1128/microbiolspec.RWR-0019-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J.. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008; 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J.. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005; 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lapinaite A., Doudna J.A., Cate J.H.D. Programmable RNA recognition using a CRISPR-associated Argonaute. PNAS. 2018; 115:3368–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y., Li W., Jiang X., Wang Y., Zhang Z., Liu Q., He R., Chen Q., Yang J., Wang L.et al.. A programmable omnipotent argonaute nuclease from mesophilic bacteria Kurthia massiliensis. Nucleic Acids Res. 2021; 49:1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y., Esyunina D., Olovnikov I., Teplova M., Kulbachinskiy A., Aravin A.A., Patel D.J.. Accommodation of helical imperfections in Rhodobacter sphaeroides argonaute ternary complexes with guide RNA and target DNA. Cell Rep. 2018; 24:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J.et al.. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 2020; 48:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He R., Wang L., Wang F., Li W., Liu Y., Li A., Wang Y., Mao W., Zhai C., Ma L.. Pyrococcus furiosus argonaute-mediated nucleic acid detection. Chem. Commun. 2019; 55:13219–13222. [DOI] [PubMed] [Google Scholar]
- 36. Dayeh D.M., Cantara W.A., Kitzrow J.P., Musier-Forsyth K., Nakanishi K.. Argonaute-based programmable RNase as a tool for cleavage of highly-structured RNA. Nucleic Acids Res. 2018; 46:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin S., Jung Y., Uhm H., Song M., Son S., Goo J., Jeong C., Song J.J., Kim V.N., Hohng S.. Quantification of purified endogenous miRNAs with high sensitivity and specificity. Nat. Commun. 2020; 11:6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The smDNA sequencing datasets generated in this study are available from the Gene Expression Omnibus (GEO) database under the accession number GSE168010.