Abstract
Abstract
Gluten is a mixture of storage proteins in wheat and occurs in smaller amounts in other cereal grains. It provides favorable structure to bakery products but unfortunately causes disease conditions with increasing prevalence. In the human gastrointestinal tract, gluten is cleaved into proline and gluten rich peptides that are not degraded further. These peptides trigger immune responses that might lead to celiac disease, wheat allergy, and non-celiac gluten sensitivity. The main treatment option is a gluten-free diet. Alternatively, using enzymes or microorganisms with gluten-degrading properties might alleviate the disease. These components can be used during food production or could be introduced into the digestive tract as food supplements. In addition, natural food from the environment is known to enrich the microbial communities in gut and natural environmental microbial communities have high potential to degrade gluten. It remains to be investigated if food and environment-induced changes in the gut microbiome could contribute to the triggering of gluten-related diseases.
Key points
• Wheat proteins, gluten, are incompletely digested in human digestive tract leading to gluten intolerance.
• The only efficient treatment of gluten intolerance is life-long gluten-free diet.
• Environmental bacteria acquired together with food could be source of gluten-degrading bacteria detoxifying undigested gluten peptides.
Keywords: Celiac disease, Peptidases, Food, Environment
Introduction
Gluten is a mixture of grain storage proteins in cereal grains. The major gluten source in the human diet is wheat, domesticated approximately 10,000 years ago when hunter-gatherers settled in the Middle East. Since then, wheat has been an integral part of the Western type of diet. The dough made from wheat flour has unique baking qualities which provides it with advantages over other crops in temperate regions. The properties of wheat dough depend on the major grain endosperm storage proteins, which together form the “gluten” protein fraction. The major components of gluten are responsible for providing “viscoelasticity” while making leavened bread, but also unleavened bread, cookies, pasta, and many other food products. As a consequence, the total protein concentration in the grain is the most widely applied criteria for wheat baking quality, and an important factor in the price of wheat (Barneix 2007).
Despite 10,000 years of coexistence, gluten-rich food is new for humans—we lack the enzymes that can fully degrade gluten proteins. As a consequence, consumption of wheat products may trigger an autoimmune enteropathy, celiac disease (CD), in genetically susceptible individuals. The genetic predisposition depends on the human leukocyte antigen (HLA) type, specifically the DQ2 or DQ8 alleles (Van Heel et al. 2007; Dubois and Van Heel 2008). However, it has been shown that only 3% of genetically susceptible persons develop the disease; therefore, possessing a predisposing HLA risk allele is necessary but not sufficient to develop CD (Pozo-Rubio et al. 2012; Chibbar and Dieleman 2019). A constant increase in the prevalence of CD has been well documented, and while some of this increase can be attributed to improvements in the complex diagnostic procedures (Kelly et al. 2015), it is also evident that the prevalence of CD is growing worldwide. This can be related to changes in our diet and environmental factors: the increased quantity of ingested gluten, infant feeding patterns, the spectrum of intestinal infections, and gut microbiota colonization (Mustalahti et al. 2010; Dydensborg et al. 2012; Chan et al. 2017; Dydensborg Sander et al. 2019). Moreover, new diseases related to wheat consumption have emerged such as wheat allergy (WA) and non-celiac gluten sensitivity (NCGS) (Inomata 2009; Sapone et al. 2011; Pietzak 2012). Together with CD, these disorders are referred to as gluten intolerance.
The only effective way for CD treatment is a life-long gluten-free diet. The products that have been specially processed to reduce the gluten content can be labeled “gluten-free” if the total gluten content is below 20 mg/kg. Maintaining a gluten free diet is problematic, because it is often not commercially accessible to everyone (Newberry 2019). In addition, small amounts of the gluten components can still be present in food due to either inefficient processing or cross-contamination. Therefore, alternative strategies are needed.
Recent research has shown that gluten and gluten-derived peptides can be degraded by peptidases from different sources. These peptidases can either be used to produce gluten-free foods from gluten-containing raw materials (Rizzello et al. 2007), or they have been suggested as an oral therapy for CD, in which dietary gluten is hydrolyzed by coingested peptidases already in the mouth or stomach, thus preventing CD-specific immune reactions in the small intestine. Oral enzyme therapy is considered a promising candidate to assist gluten-free diet (Plugis and Khosla 2015). Glutenases used for this purpose originate from environmental species of bacteria, fungi, plants, and insects.
A promising approach involves relying on living microorganisms to alleviate the gluten-triggered conditions. Here, we review potential microbes that could be used for this purpose and discuss the sources that could be relevant for enrichment of food with gluten-degrading microbes.
Gluten
Wheat is currently the most important crop in the world and together with maize and rice accounts for over 70% of the total production of cereals. About 95% of the wheat grown worldwide is hexaploid, with most of the remaining 5% being tetraploid durum wheat, also termed pasta wheat, which is mostly cultivated in the dry Mediterranean region. Hexaploid bread wheat (Triticum aestivum L. ssp. aestivum) derives its three genomes (A, B, and D) from three diploid wild ancestors: Triticum urartu (AA), an unknown close relative of Aegilops speltoides (BB), and Ae. tauschii (DD) (Marcussen et al. 2014). The initial allopolyploidization event is hypothesized to have involved the A and B genome donors, giving rise to tetraploid emmer wheat (T. turgidum; AABB). This species subsequently hybridized with the D genome donor Ae. tauschii to form modern hexaploid bread wheat (AABBDD). Durum wheat Triticum turgidum L. spp. dicoccum, genome AABB is derived from the ancient T. turgidum (AABB) (Mefleh et al. 2019). Therefore, the wild emmer wheat is the progenitor of both cultivated hexaploid bread wheat and tetraploid pasta wheat.
Although the protein content in cereal grains is relatively low (about 10–20% of dry weight), in modern wheat, 80–90% of the total protein is gluten (Bromilow et al. 2017). The precise number of different gluten proteins present in mature seeds can vary, but is probably between 50 and 100. Traditionally, gluten proteins have been classified into roughly equal fractions according to their solubility in alcohol–water solutions (e.g., 60% ethanol): the soluble gliadins and the insoluble glutenins (Obsborn 1924). However, based on their composition, both gliadins and glutenins can be referred to as prolamins. The gliadins comprise monomeric subunits which are further classified into α-, γ-, and ω-gliadin fractions. The glutenins comprise two groups of subunits, high molecular weight (HMW) and low molecular weight (LMW) glutenin subunits, which form alcohol-insoluble polymers stabilized by inter-chain disulfide bonds. However, amino acid sequences show that gliadins and glutenin subunits are related. Therefore, a new classification divides all prolamins into three groups in relation to structural and evolutionary relationships: sulfur-rich (S-rich); sulfur-poor (S-poor); and high molecular weight (HMW) prolamins (Shewry and Tatham 1990; Shewry and Halford 2002) (Table 1.). A characteristic of all prolamins is the presence of large domains of short repetitive peptide sequence motifs dominated by proline and glutamine residues. An important feature is the presence of cysteine residues that link prolamin proteins through intra- or intermolecular disulfide bonds, forming a continuous network in dough.
Table 1.
Classification of gluten prolamins from wheat, barley, rye, and oat, based on Shewry and Halford (2002), Wieser and KoeHler (2012), and Shewry (2019)
| Type of gluten prolamins | Partial amino acid composition (mol %) | Components | State | Repetitive unit | Proportion of prolamin fraction (%) | Grain species |
|---|---|---|---|---|---|---|
| HMW prolamins | 30–35% glutamine, 10–16% proline, 15–20% glycine, 0.5–1.5% cysteine, 0.7–1.4% lysine | HMW subunit of glutenin | Polymeric | QQPGQG; GYYPTSPQQ | 6–10% | Wheat |
| D-hordein | Polymeric | QQPGQG | 2–4% | Barley | ||
| HMW secalin | Polymeric | QQPGQG | 2% | Rye | ||
| S-rich prolamins | 30–40% glutamine, 15–20% proline, 2–3% cysteine, <1% lysine | γ-gliadin | Monomeric | PQQPFPQ | 70–80% | Wheat |
| α-gliadin | Monomeric | QPQPFP; PQQPY | ||||
| B- and C-type LMW subunit of glutenin | Polymeric | QPQQPFP | ||||
| B-hordein | Monomeric | QQPFPQ | 80% | Barley | ||
| γ-hordein | Polymeric | QPQQPFP | ||||
| γ-secalins | Poly/Monomeric | (Q)QPQQPFP | 80% | Rye | ||
| S-poor prolamins | 40–50% glutamine, 20–30% proline, 0–0.5% lysine, 0 cysteine, 1 cysteine residue in D-type LMW subunit | ω-gliadin | Monomeric | PQQPFPQQ | 10–20% | Wheat |
| D-type LMW subunits of glutenin | Polymeric | PQQPQQ | ||||
| C-hordein | Monomeric | PQQPFPQQ | 10–15% | Barley | ||
| ω-secalin | Monomeric | QPQQPFP | 10–15% | Rye | ||
| Other gluten prolamins | 23–29% glutamine, 8–10% proline, 6.5–8.5% valine, 3.5–5% cysteine | Avenin | Monomeric | PFVQQQQ | a | Oat |
S-rich, sulfur-rich; S-poor, sulfur-poor; HMW, high molecular weight; LMW, low molecular weight; a, 10–20% of total proteins (Anderson 2014)
In addition to wheat, a prolamin fraction also exists in related cereal grains that belong to the family Poaceae such as rye (Secale cereale) (secalin), barley (Hordeum vulgare) (hordein), and oat (Avena sativa) (avenin) albeit in much lower quantities. The sequences within these proteins are similar, but unique bread making properties are provided only by wheat gluten and to a limited extent rye gluten.
Gluten-related disorders
The extensive proline- and glutamine-rich repeated sequences of wheat seed storage proteins are responsible for bread quality, yet unfortunately play a crucial role in triggering hypersensitivity reactions such as celiac disease (CD), wheat allergy (WA), and non-celiac gluten sensitivity (NCGS) (Scherf et al. 2016).
Celiac disease
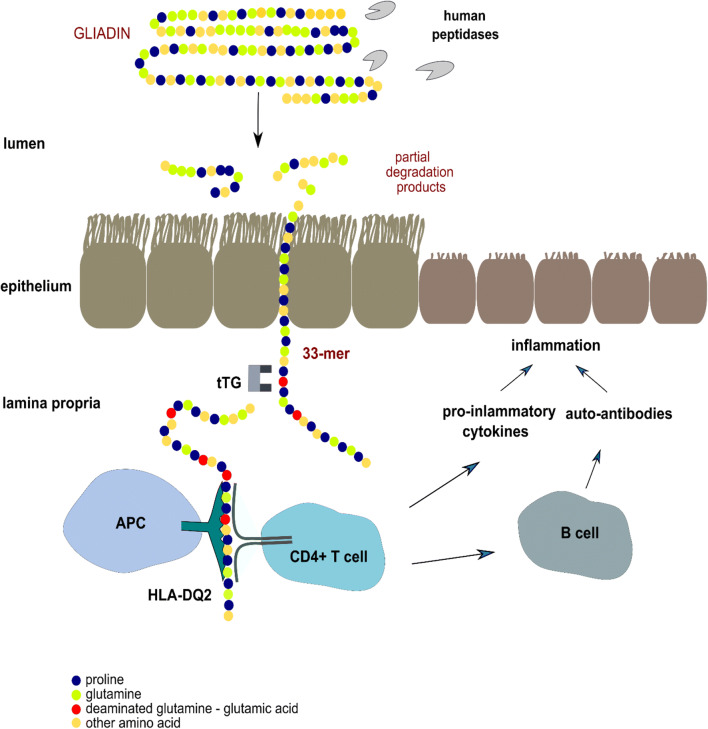
Celiac disease (CD) is an autoimmune enteropathy caused by genetic and environmental factors with an estimated worldwide prevalence of about 1% of the global population (Mustalahti et al. 2010). The symptoms are malabsorption, steatorrhea, weight loss, or growth failure (Kelly et al. 2015). Ingested food proteins are usually digested into amino acids, dipeptides, and tripeptides by gastric, pancreatic, and brush border proteases. Proline-rich sequences of prolamines are partially resistant to enzymatic degradation in the gastrointestinal tract that results in relatively long peptide fragments. These undigested proline- and glutamine repetitive peptide fragments pass through the epithelial barrier of the small intestine and reach the lamina propria where transglutaminase (tTG) deaminates the selected glutamine residues, thereby enhancing their affinity to the HLA receptors, DQ2, and DQ8 (Schuppan 2000). The presentation of peptides to HLA-DQ2/DQ8 protein subunits on the surface of antigen-presenting cells (APC) in the presence of gluten-specific T cells induces both adaptive as well as an innate immune response in CD patients. The characteristics of CD are crypt hyperplasia, increased infiltration villous atrophy, increased lymphocyte infiltration and the stimulation of CD4+ T cells against gluten epitopes, and the production of tTG targeting autoantibodies (Dewar et al. 2004).
All gluten proteins (gliadin and glutenin from wheat, hordein from barley, secalin from rye, and avenin from oat) possess their own sets of toxic and immunogenic peptides (or epitopes) with distinct immunogenicity. However, gliadin peptides are known to be the most toxic and numerous, specifically derived from α- and γ-gliadin: the strongest and most common adaptive response to gluten is directed toward an α2-gliadin fragment of 33 amino acids in length containing six partially overlapping immunodominant CD epitope regions (Shan et al. 2002; Camarca et al. 2009) (Figs. 1 and 2). α-gliadins genes from diploid Aegilops tauschii (DD) introduced six types of α a-gliadins (named 1–6) into the currently used hexaploid wheat (AABBDD) (Ozuna et al. 2015; Schalk et al. 2017). Distinct types of α-gliadins differ mainly in the number of repeat blocks that contain the interspersed motifs PFPPQQ and PYPQPQ. Only type 1 α-gliadins contain the full immunodominant 33-mer epitopes.
Fig. 1.
The resistance of gliadin to human proteolytic enzymes causes inflammation in human small intestine. Partially digested gliadin fragments cross the epithelium and reach the lamina propria. The most toxic fragment contains 33 amino acid residues mostly proline and glutamine. This peptide can bind to DQ2 molecules directly on the surface of antigen presenting cells (APC) (Qiao et al. 2005). Tissue transglutaminase (tTG) deaminates the glutamine residues to glutamic acid, thereby enhancing their affinity to the HLA receptor DQ2, that activates gliadin-specific T-cells. The result is chronic inflammation of the intestinal mucosa
Fig. 2.
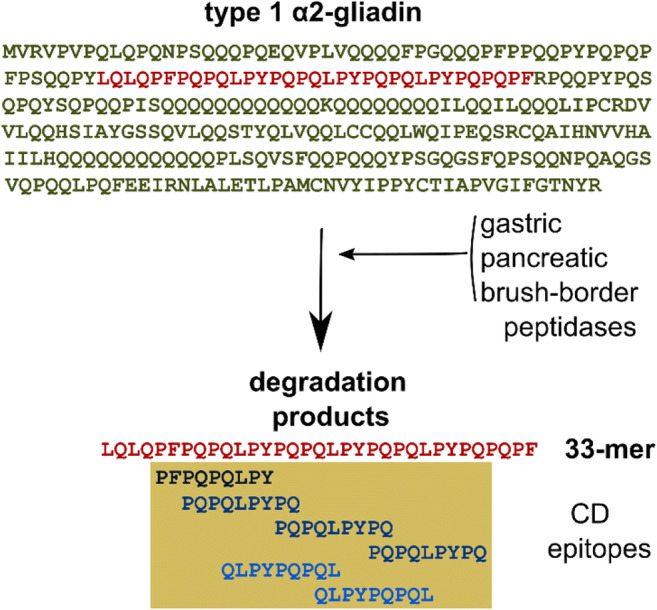
Proteolytic processing of the type 1 α2-gliadin. A 33-mer peptide fragment is resistant to the proteolytic enzymes in the human gastrointestinal tract. This peptide is the largest and most toxic degradation product. It contains six partially overlapping immunodominant CD epitopes that induce gluten-specific immune responses and intestinal inflammation
Wheat allergy
Wheat flour-triggered IgE-mediated food allergy is one of the top eight food allergies reported 0.2–1.0% globally (Cianferoni 2016). Wheat allergy (WA) is classified as a classic food allergy that is induced by wheat (not only gluten) intake and involves subtypes such as immediate wheat allergy, baker’s asthma, wheat contact dermatitis, and wheat-dependent exercise-induced anaphylaxis (Roszkowska et al. 2019). WA typically develops in childhood during early infancy and is frequently outgrown between the ages of 3 and 5 (Patel and Volcheck 2015).
IgE-mediated food allergies have been associated with sensitization to particular cereal storage prolamins or the non-gluten proteins within wheat, α-amylase inhibitors, thioredoxin, lipid transfer protein, profilin, and serpin, among others (Matsuo et al. 2015). A major allergenic protein from wheat is the ω5-gliadin repetitive region that consists almost entirely of the peptides FPQQQ and QQIPQQ (Battais et al. 2005; Denery-Papini et al. 2011). Other prolamins have been described to contain IgE binding epitopes including ω1, 2-gliadins, α-gliadins, γ- gliadins, LMW, and HMW subunits of glutenin (Denery-Papini et al. 2011; Battais et al. 2005).
Non-celiac gluten sensitivity
Non-celiac gluten sensitivity (NCGS) may be defined as a gluten (wheat)-dependent disorder with symptoms similar to CD (abdominal pain, discomfort, bloating, altered bowel habit, and fatigue) but with mostly normal small intestinal histology and lack of anti-tTG autoantibodies. Prevalence of NGCS is highly variable 0.63–6.0% (although it is difficult to estimate because the condition lacks specific biomarker(s)) (Sapone et al. 2012). Its pathogenesis is still not clear; gluten triggers an innate immune response in NCGS and provokes an additional adaptive immune response with increased expression of IL-6, IL-21, IL-17, and IFN-γ. NCGS is more frequently diagnosed in adults rather than in children (Volta et al. 2014). Symptoms disappear after starting on a gluten-free diet and appear again after a gluten challenge within a few hours or a couple of days (Catassi et al. 2013; Barbaro et al. 2018).
Peptidases able to degrade gluten
There is no cure for gluten intolerance disorders and the only treatment option is avoiding gluten in food, although even this does not eliminate all the symptoms. Providing enzymes able to degrade gluten and the immunogenic peptides is being investigated as a complementary treatment option. Peptidases can be used to eliminate gluten from food material (review Scherf et al. 2018). In addition, peptidases given as oral therapy have been tested to alleviate gluten-related conditions (Cerf-Bensussan et al. 2007; Tye-Din et al. 2010; König et al. 2017).
Enzymes that cleave the peptide bond are called peptidases or proteases. These enzyme names are often used as synonyms. Here, we call this class of enzymes peptidases. In the digestive tract, peptidases are responsible for cleavage of ingested proteins. Gluten-related health problems arise from the inefficiency of peptidases to degrade the immunogenic epitopes. Most peptidases, even those with low specificity, are unable to hydrolyze peptide bonds formed by proline residues due to the cyclic structure and the special constraints it exerts on the peptide backbone structure. The N-terminal amine of proline lacks a hydrogen, and the side chains are typically in the cis conformation. These properties impair the susceptibility of X↓P bonds (between any amino acid “X” and proline residue “P”) to proteolytic cleavage.
There are several peptidases from different classes that have been shown to degrade proline and gluten-rich prolamine molecules with varying efficiently. Peptidases can either act only near the ends of polypeptide chains (exopeptidases EC 3.4.11-19), or amidst polypeptide chains (endopeptidases EC 3.4.21-24 and EC 3.4.99). Here, we focus on endopeptidases that are more effective at gluten degradation than exopeptidases (Table 2). We use the classification of peptidases employed by MEROPS (http://merops.sanger.ac.uk.), a comprehensive database containing information on peptidase sequence, structure, substrates, and inhibitors (Rawlings et al. 2018).
Table 2.
Most studied microbial endopeptidases cleaving proline- and glutamine-rich prolamine molecules
| Enzyme family | Cleavage site | Substrate | Subcellular location | pH optimum | Temp. optimum (°C) | Microbe; peptidase | Origin | Reference |
|---|---|---|---|---|---|---|---|---|
| Metallopeptidase | ||||||||
| M4 elastase | Q↓L | Intact molecule | Extracellular | 2.0–7.0 | 37 | Bacteria; Pseudomonas aeruginosa; LasB | Human feces | (Wei et al. 2015; Caminero et al. 2016) |
| Serine peptidase | ||||||||
| S8 subtilisin | XPQ↓; LPY↓ | Intact molecule | Extracellular | 3–10 | 37 | Bacteria; Rothia spp. | Human oral cavity | (Zamakhchari et al. 2011) |
| S9A prolyl-oligopeptidase | P↓F; P↓Y; P↓Q | Oligopeptide | Cytosolic | 6.0–7.0 | 37 | Bacteria; Myxococcus xanthus | Soil | (Shan et al. 2004; Kocadag Kocazorbaz and Zihnioglu 2017) |
| P↓Q; P↓Y | Oligopeptide | Periplasmic | 6.0–7.0 | 43 | Bacteria; Sphingomonas capsulata | Distilled water | (Kabashima et al. 1998; Shan et al. 2004) | |
| GP↓; AP↓ | Oligopeptide | Extracellular | 6.0–8.0 | Bacteria; Chryseobacterium taeanense | Rhizosphere of cereal crops | (de Amador et al. 2019) | ||
| SAP↓; GGP↓; TP↓G; GP↓ | ND | Cytosolic | 6.0–9.0 | 37 | Bacteria; Cellulomonas sp. | Human gut | (Kumar et al. 2018) | |
| P↓Q; P↓F; P↓Y | Oligopeptide | Cytosolic | 5.0–8.0 | 63 | Bacteria; Sphaerobacter thermophilus | Sewage sludge | (Pati et al. 2010; Shetty et al. 2017) | |
| GP↓; AP↓: AA↓ | Intact molecule, oligopeptide | Not exported | ∼7 | 85–90 | Archaea; Pyrococcus furiosus | (Harwood et al. 1997; Harris et al. 2001) | ||
| S28 prolyl endopeptidase | XP↓ | Intact molecule, oligopeptide | Extracellular | 4.0–5.0 | 15–60 | Fungi; Aspergillus niger | (Edens et al. 2005) | |
| S53 serine-carboxyl peptidase | F↓P; Q↓L | Intact molecule | Extracellular | 3.0–6.0 | 37 | Bacteria; Actinoallomurus A8 | Soil pH 4.2 | (Cavaletti et al. 2019) |
Metallopeptidases
Family M4. Elastase (E.C.3.4.24.26)
Among metallopeptidases that degrade gluten, elastase LasB has gained significant attention because it is an extracellular metallopeptidase excreted from Pseudomonas aeruginosa that has been isolated from human feces (Wei et al. 2015) and the duodenum of CD patients (Caminero et al. 2016). P. aeruginosa is an opportunistic human pathogen that requires elastase to degrade mucins and surfactant proteins (Kuang et al. 2011) and exogenous flagellin which acts to prevent flagellin-mediated immune recognition (Casilag et al. 2015). Elastase LasB also efficiently hydrolyses gluten molecules, but produces a multitude of shorter immunogenic peptides that could activate gluten-specific T-cells in CD patients. It has been shown in germ-free mice that Lactobacillus rhamnosus and Lactobacillus fermentum isolated from the duodenum of non-CD humans degrade gluten peptides produced by P. aeruginosa peptidases thereby reducing their immunogenicity (Caminero et al. 2016). The peptidase activity was provided by the exopeptidases (of other peptidase families, not metallopeptidases) that Lactobacillus spp. often produce.
Serine peptidases
Most of the gluten-degrading bacterial peptidases described so far belong to the serine family of proteases.
Family S8. Subtilisin
Peptidase family S8 contains the serine endopeptidase subtilisin and its homologs that cleave after the XPX↓ motif. Most of these enzymes are inefficient against many immunogenic gluten sequences.
However, the enzyme from Rothia spp. is active against the 33-mer immunogenic peptide. Rothia aeria is a commensal bacterium that inhabits the human oral cavity and is active over much of the intestinal pH range (pH 3–10) (Zamakhchari et al. 2011). Rothia subtilisins are unique in that they cleave both XPQ↓ and LPY↓.
Two S8 subtilisins from Bacillus licheniformis, subtilisin A (sold by Sigma) and the food-grade Nattokinase (extracted from a dietary food supplement), degrade the immunogenic gliadin-derived 33-mer peptide and the immunodominant epitopes recognized by the R5 (QQPFP and related sequences) and G12 (QPQLPY) antibodies. Nattokinase abolishes the R5 epitopes but is less effective in eliminating the G12 epitopes (Wei et al. 2016).
Family S9, Serine-carboxyl peptidase (sedolisin, E.C.3.4.21.100)
Soil Actinomycete Actinoallomurus A8 produces extracellular sedolisin that has optimal pH around 3. This enzyme cleaves both F↓P and Q↓L in the 33-mer as well as intact gliadin proteins. Its recombinant active form has been produced in Streptomyces lividans TK24, a strain generally regarded as a safe (GRAS) and a source of proteins for human alimentary use (Cavaletti et al. 2019).
Many natural sedolisins are not very efficient in degrading gluten; however, it has been demonstrated that their properties can be improved by engineering. Kumamolisin from Alicyclobacillus sendaiensis, with an optimal pH around 3, is a serine-carboxyl peptidase (S53) with collagenase activity (Tsuruoka et al. 2003). After molecular engineering of kumamolisin to shift its cleavage specificity from P↓R or P↓K to P↓Q, a mutant termed Kuma030 with high proteolytic activity against gluten was obtained (Gordon et al. 2012).
Family S9, subfamily S9A, prolyl oligopeptidase (POP, E.C.3.4.21.26)
These enzymes are specific for cleavage after proline residues, thus making this group of peptidases very synergistic with human digestive proteases. The S9A subfamily of enzymes are also referred to as prolyl endopeptidases (PEP). These enzymes have been extensively studied and are already being developed for medicinal claims. POP from the bacterium Sphingomonas capsulata (SC-PEP) (cleaves after P) has been combined with the barley cysteine endopeptidase PEP-B2 (cleaves after glutamine) to form a combined enzymatic activity that attacks the immunogenic peptides after the most frequent amino acid residues (P and Q). This formulation, developed under the name Latiglutanase, is expected to provide synergy and create a more active medication (Lähdeaho et al. 2014).
Generally, POP hydrolyses oligopeptides that consist of less than 30 residues (Camargo et al. 1997). However, POP from the thermophilic archaeon Pyrococcus furiosus has been shown to cleave larger substrates (Harwood et al. 1997; Harris et al. 2001). The Sphaerobacter thermophilus enzyme also seems to cleave full intact gluten molecules, although the enzymatic assay was performed in a complex mixture that also contains malt (Shetty et al. 2017). The POP studied so far originate from a wide range of environmental bacteria and archaea from phyla/class: Actinobacteria (Cellulomonas sp.), Bacteroidetes (Flavobacterium meningosepticum, Chryseobacterium taeanense), Alphaproteobacteria (Sphingomonas capsulata), Deltaproteobacteria (Myxococcus xanthus), Chloroflexi (Sphaerobacter thermophilus), and Euryarchaeota (Pyrococcus furiosus) (Table 2). All these enzymes can detoxify the 33-mer peptide but differ in their hydrolytic activity and stability under in vitro artificial gastrointestinal conditions (acidic pH, pancreatic proteases, and membrane peptidases of the small intestinal mucosa).
Based on a bioinformatic analysis, Kaushik and Sowdhamini (2014) showed that POP are widely distributed in all classes of bacteria and archaea with diverse domain architectures. Several of the proteins contain signaling peptides for export, which suggests their periplasmic or extracellular location (Table 2).
Unfortunately, the strains that produce promising POP enzymes cannot be used in the food industry because of their environmental origin. Lactic acid bacteria are food-grade and have beneficial gut properties and resistance to harsh gut conditions. Recombinant Lactobacillus casei strains were constructed to deliver POP from M. xanthus to the gut. It was found that the strain that secretes POP into the extracellular medium more effectively degrades the 33-mer oligopeptide compared to the strain that retained POP to the intracellular environment. The probable reason is that the extracellular enzymes can directly attack the peptide and do not require uptake events prior to cleavage (Alvarez-Sieiro et al. 2014). Another group used Lactococcus lactis as delivery vector for secreting F. meningosepticum POP into the growth medium (Lim et al. 2017).
Other enzymes of nonhuman origin
It is expected that gluten degradation occurs in environments where gluten occurs naturally, i.e., in cereal plant grains and in organisms who use it as a food source. Endogenous cereal proteases that hydrolyze storage proteins, including gluten, are synthesized during germination. The well-studied cysteine endopeptidase B2 of barley cuts at glutamine residues and prefers intact proteins as a substrate (Bethune et al. 2006; Osorio et al. 2019). A combination of the barley cysteine endopeptidase EP-B2 with the bacterial POP from F. meningosepticum-PEP has been proven to be effective in detoxifying gluten proteins. Site saturation mutagenesis effectively increased the thermostability of F. meningosepticum-PEP, which should allow it to maintain its enzymatic activity in the core of bread during baking, where temperatures generally do not exceed 100°C (Osorio et al. 2020).
Gluten-degrading peptidases are also found in organisms involved in the decomposition of organic matter. Family S28 prolyl endopeptidases from the fungus Aspergillus niger and edible mushroom in the family Physalacriaceae, Flammulina velutipes, are unique in the serine peptidase family because they have endopeptidase activity that cleaves after XP (Edens et al. 2005; Stepniak et al. 2006; Kang et al. 2013; Schulz et al. 2018). Aspergillus niger AN-PEP is active between a pH of 2 and 8, with an optimal activity between pH 4 and 5. It is not degraded by pepsin, and thereby remains functional in the stomach (Stepniak et al. 2006; Mitea et al. 2008). It has been shown that AN-PEP significantly degrades most gluten in the stomach before it enters the duodenum (Salden et al. 2015). Capsules containing the AN-PEP enzyme have been marketed for over a decade, although the health effects have been disputed (Krishnareddy et al. 2017).
For Tenebrio molitor, Rhizopertha dominica, and related cereal-feeding insects, the main dietary proteins are the storage proteins of cereal grains. Post-glutamine cleaving peptidases were isolated from the midgut of the pest, Tenebrio molitor (yellow mealworm) (Goptar et al. 2013). POP from the gastrointestinal system of the beetle R. dominica cleaves proline-rich peptides from wheat and barley in P↓Q and P↓Y (Mika et al. 2015).
Gluten-degrading bacteria in human digestive tract
There are thousands of bacterial species that inhabit the human digestive tract. Several of these bacteria can potentially degrade gluten and a healthy microbiome composition could modulate the symptoms of gluten-related diseases.
It ought to be noted that the microbiome composition is mostly investigated by the 16S rRNA gene marker. Because different strains of the same bacterial species can have very different proteolytic activities, 16S-based studies must be complemented with cultivation and metagenomics-based approaches to assess proteolytic activity. In addition, the 16S rRNA-based approaches have shown that the bacterial communities are very different in different parts of the digestive tract. Here, we discuss the gluten-degrading potential of these different communities.
Oral cavity
The digestion of food starts in the oral cavity and it has been shown that bacteria from the oral cavity have the capacity to degrade gluten. There are approximately 800 prokaryotic species in the oral cavity, of which 70% were found to be cultivable (Verma et al. 2018). Rothia spp., Actinomyces odontolyticus, Neisseria mucosa, and Capnocytophaga sputigena were found to have the highest activity against gliadin or the 33-mer immunogenic peptide (Fernandez-Feo et al. 2013). Rothia spp. enzymes have been studied in more detail (see above). Although their activities are quite low, the high abundance of Streptococcus sp. could still substantially contribute to the overall gliadin-degrading capacity in the human mouth (Aas et al. 2005; Fernandez-Feo et al. 2013). The bacterial strains studied to date have considerably different peptidase activities, which suggests that efficient gluten degradation can be achieved in combination. Approximately, 1 l of saliva, which contains a diverse collection of aerobic and anaerobic bacteria, is produced on a daily basis (Maukonen et al. 2008). Saliva contains a high amount of proline-rich proteins; the degradation of which most likely starts by the subtilisins produced by oral bacteria (Messana et al. 2008; Helmerhorst et al. 2008). Therefore, it is probable that some of these bacteria can also contribute to the degradation of gluten. Saliva is swallowed and thereby also provides substrates for bacteria in the lower digestive tract.
Stomach
The pH of the stomach after a meal is in the range of 2–4. This vastly restricts the bacterial groups that can be active in the stomach or even pass its harsh conditions alive. In stomach, the microbial density is ca 102 to 104 cfu/ml. The gastric microbiota is diverse and includes species from the genera Streptococcus, Propionibacterium, Lactobacillus, Staphylococcus, Prevotella, Veillonella, Rothia, and Haemophilus (Delgado et al. 2013; Nardone and Compare 2015). Although there are no studies on the capacity of stomach bacteria to degrade gluten, it has to be noted that Rothia spp. can have activity at low pH and therefore could be involved in gluten degradation in the stomach.
Small intestine
The degradation of nutrients continues in the duodenum, jejunum, and ileum of the small intestine. Many degradation products are absorbed in the small intestine, which also performs strong immunomodulatory functions. In line with this, CD is triggered in the small intestine. The conditions in the duodenum can also be harsh for certain bacteria because it contains high levels of human proteases and bile acids. This is probably the reason why the bacterial count is lower in duodenum (101–105 CFU/ml) and rises in the jejunum (104–107 CFU/ml) and ileum (103–108 CFU/ml) (O’Hara and Shanahan 2006; Leser and Mølbak 2009; Zoetendal et al. 2012). The physiological bacterial communities within the small intestine are difficult to study because of the need to take biopsy is needed. Therefore, only one study has been published to date that assesses the gluten degradation potential of bacterial communities in small intestine. This work is restricted to the duodenum.
Bacteria from biopsy samples from the duodenum were investigated for their activity when degrading intact gluten and the 33-mer peptide (Herrán et al. 2017). This study showed that 15 bacterial species from genera Actinomyces, Bacillus, Lactobacillus, Prevotella, Pseudomonas, and Stenotrophomonas were able to hydrolyze the 33-mer. Additionally, bacteria that showed extracellular gluten-degrading activity were as follows: Bacillus licheniformis, B. subtilis, B. subtilis/amyloliquefaciens, Bacillus pumilus, L. casei/paracasei, Pseudomonas aeruginosa, S. aureus, S. epidermidis, Stenotrophomonas maltophilia, Streptococcus salivarius/thermophilus, and Virgibacillus pantothenticus. Overall, 60% of the bacteria identified were Lactobacilli. Because of their auxotrophy for numerous amino acids, Lactobacilli hydrolyze proteins through the action of their proteolytic system in order to get the amino acids required for their growth. Lactobacilli mostly produce cell wall attached exopeptidases that are relatively inefficient in eliminating gluten and gluten-derived peptides (Scherf et al. 2016). Still, L. helveticus has serine endopeptidases that belong to the subtilisin family that break down larger proteins into oligopeptides that are transported into the cytoplasm (Griffiths and Tellez 2013). Subsequently, the internalized peptides, 2 to 9 amino acid residues, are degraded into amino acids by the combined action of numerous internal peptidases that vary depending on the species (Savijoki et al. 2006; Raveschot et al. 2018). No single Lactobacillus strain possesses all the peptidases required to degrade toxic gluten oligomers. Gluten, and its shorter immunogenic peptides, can be degraded by a combination of bacteria with complementary peptidase activities (Francavilla et al. 2017).
Colon
Colonic bacteria are characterized by studying feces. The estimated bacterial count in feces is ca 1010–1012 CFU/ml, and the number of cultivable species is estimated to be over 1500 (O’Hara and Shanahan 2006; Lagier et al. 2016).
Early work with human gut contents by Macfarlane identified fecal bacteria with proteolytic activity: Propionibacterium, Clostridium, Streptococcus, Bacillus, and Staphylococcus. Extracellular proteases were formed by Streptococcus faecalis ST6, Propionibacterium acnes P6, Clostridium perfringens C16, C. bifermentans C21, and C. sporogenes C25 (Macfarlane and Allison 1986; Macfarlane et al. 1988; Gibson et al. 1989).
In 2014, Caminero et al. (2014) identified gluten-degrading activity in strains from Bacillus licheniformis, B. subtilis, B. pumilus, Bifidobacterium longum, Clostridium sordellii, C. perfringens, C. botulinum/sporogenes, C. butyricum/beijerinckii, Enterococcus faecalis, E. faecium, Propionibacterium acnes, Pediococcus acidilactici, Paenibacillus jamilae, Staphylococcus epidermidis, S. hominis, and Stenotrophomonas maltophilia.
The importance of the prolamin degrading activity in colon is questionable because the damaging activity of gluten-derived epitopes already occurs in the duodenum.
Probiotics in CD
Currently, there is no clear understanding of the role of the gut microbiota in CD. Several studies have reported changes in the composition of gut microbiota, “intestinal dysbiosis,”,in CD patients compared to healthy subjects (Nadal et al. 2007; de Palma et al. 2012; Cheng et al. 2013). A balanced composition of the human commensal microbiota has been considered crucial for the development of a healthy immune system; however, it is not clear if intestinal dysbiosis is the cause or effect of CD (Chibbar and Dieleman 2019). Probiotics have been considered as a strategy to modulate the gut microbiome in CD patients. Probiotics can influence gluten intolerance by digesting the gluten proteins to non-immunogenic small polypeptides, maintain the intestinal barrier by preventing immunogenic polypeptides from accessing the mucosa, and by regulating the immune system. The potential of several probiotics has been investigated. De Angelis and others reported the potential benefits of a probiotic cocktail with eight strains (VSL#3), Bifidobacterium breve, Bifidobacterium infantis, acidophilus, Lactobacillus plantarum, Lactobacillus casei, Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus, and Bifidobacterium longum, which decreased wheat-induced discomfort (De Angelis et al. 2006). It ought to be noted that the probiotic cocktail was added during the food-processing step and could produce predigested and thus tolerable gliadins that increase the palatability of gluten-free products. Importantly, as mentioned above, individual Lactobacillus strains are not capable of complete detoxification of gluten-derived peptides (Francavilla et al. 2017). The same is true for VSL#3; individual probiotic strains were inadequate to break down gliadin compared to the efficiency with a pool of strains (De Angelis et al. 2006; Harnett et al. 2016).
Bifidobacterium spp. has been shown to modulate a proinflammatory milieu of CD in several in vitro studies (Medina et al. 2008; Lindfors et al. 2008; Laparra et al. 2012). Bifidobacterium spp. also helped to restore the healthy percentage of the main microbial components (Quagliariello et al. 2016). It would be prudent to study the effects of Bifidobacterium spp. on CD in further clinical studies.
It is expected that probiotics will not provide a rapid cure for complex diseases such as CD, but rather alleviate the severity and symptoms. More studies are needed that specifically address how the gut microbiome can modulate or alter the course of the disease. We expect that probiotics will probably need to be combined with long-term dietary changes.
Natural sources of gluten-degrading bacteria
CD is an autoimmune disease triggered by gluten and most probably other environmental factors not yet defined. CD prevalence has increased over time in geographical regions characterized by a Western lifestyle (Catassi 2015). In Western society, the gut microbiome has changed when compared to the traditional societies. The “Biodiversity hypothesis” states that interaction with diverse and abundant environmental microbiota is important for the prevention of immune-mediated non-communicable diseases (von Hertzen et al. 2011; Hanski et al. 2012). There are a rising number of studies that show that a living environment, and changes therein, likely shape the skin as well as the composition of gut microbiota and are important for the development of a normal immune system and protection from immune-mediated diseases (Seiskari et al. 2007; Stein et al. 2016; Lehtimäki et al. 2017; Hui et al. 2019). For example, diverse vegetation around homes, particularly shrubs and non-woody flowering plants, and the coverage of built areas are associated with health-related changes in gut microbiota composition (Parajuli et al. 2020). The effect of environmental bacteria on allergies has been associated with their immunomodulatory effects (Nurminen et al. 2018; Ottman et al. 2019; Roslund et al. 2020).
Empirical studies have implicated bacteria as the dominant contributor to proteolytic activity in soils (Watanabe and Hayano 1993; Nguyen et al. 2019). Bacterial isolates from Bacillus, Pseudomonas, and Flavobacterium-Cytophaga have been shown to be important agents of proteolysis and act as the main sources of soil peptidase activity (Bach and Munch 2000; Vranova et al. 2013). Given the nature of nitrogen limitation in most soils and aquatic environments, the ability to readily break down high molecular weight proteinaceous material into amino acid precursors for cell growth or energy generation would be highly favorable (Geisseler et al. 2010; Kolton et al. 2013).
Root vegetables are in direct contact with soil and share some of the surrounding microbiome. We have recently shown in an in vitro study that bacteria that originate from root vegetables can degrade gluten and detoxify the toxic immunogenic CD epitopes at 37°C under microoxic conditions (Kõiv et al. 2020). Bacteria with strong extracellular protease activity were identified as Bacillus pumilus, Bacillus cereus, Bacillus subtilis, Bacillus circulans, Bacillus licheniformis, Bacillus psychrosaccharophilus, Clostridium bifermentans, Clostridium sporogenes, and Clostridium subterminale. Four bacterial strains belonging to the species Bacillus pumilus, Clostridium subterminale, and Clostridium sporogenes produce peptidases with post-proline cleaving activity that successfully neutralized the toxic immunogenic epitopes. The bacterial groups identified as 33-mer and gluten degrades are similar or even identical to the bacteria degrading these substrates in duodenum and feces (described above in the “Gluten-degrading bacteria in human digestive tract” section). Therefore, it is possible that these strains are introduced to the gastrointestinal tract together with food. Currently, it is not possible to estimate if these bacteria inhabit the gastrointestinal tract stably, perhaps with specific required dietary substrates, or are constantly introduced with food.
It is not known if the observed peptidase activity is provided by one enzyme or a mixture of several enzymes. As mentioned above, B. pumilus and C. sporogenes isolated from duodenum and feces hydrolyze both the 33-mer, and have also proteolytic activity against intact gluten proteins (Macfarlane et al. 1988; Caminero et al. 2014; Herrán et al. 2017). Both bacteria produce several peptidases that degrade other proline and glutamine-rich proteins such as keratin (7.5% proline and 12% glutamine) (Ionata et al. 2008; Fellahi et al. 2016). The complete degradation of keratin to single amino acids requires the synergistic action of (at least) three kinds of peptidases, namely endo-, exo-, and oligo-peptidase (Qiu et al. 2020). It is probable that the natural degradation of gluten also occurs by a combination of different enzymes although this hypothesis requires further investigation. It is also probable that gluten degradation activity can be achieved by enzymes that usually degrade certain proline-rich proteins such as keratin, collagen, and elastin. It is described above that elastase from P. aeruginosa can degrade gluten. The gluten-degrading activity of human gastrointestinal elastases has also been described (Gutiérrez et al. 2017).
The human digestive tract is saturated with easily accessible sources of nitrogen for bacteria. Therefore, only a few microbes that colonize the digestive tract “waste” energy on the production of extracellular proteases. Environmental bacteria are more active in using extracellular proteases and, when introduced into the gut, could provide beneficial proteolytic activity.
In addition, opportunistic pathogens sometimes produce high levels of peptidases that could also degrade gluten (P. aeruginosa, Stenotrophomonas maltophilia, C. perfringens, Enterococcus faecalis, E. faecium) (Caminero et al. 2014; Caminero et al. 2016). CD is largely a contemporary disease. It has been noted that oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet: species found preferentially in hunter-gatherers included microbes often considered as oral pathogens, despite their hosts’ apparent good oral health (Lassalle et al. 2018). It is possible that also in contemporary diets, microbes considered as opportunistic pathogens can contribute to degradation of gluten-derived peptides.
In conclusion, the enzymatic food supplements and probiotics are being developed to alleviate the symptoms of gluten intolerance. In addition, natural bacterial communities from food could also facilitate detoxification of gluten. Complete gluten degradation is a complex process that can be achieved with assistance of various combinations of enzymes produced by a wide range of microorganism, including bacteria. These bacterial communities could facilitate adaptation to our consumption of crop products that were introduced rather recently to humankind.
Acknowledgements
Special thanks to Elin Org and Marta Putrinš for critical reading of the manuscript. The authors acknowledge David Schryer for reviewing English language usage.
Author contribution
VK and TT wrote the manuscript.
Funding
The work was supported by European Union from the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering (2014-2020.4.01.15-0013) and grant from the Estonian Research Council (PRG335).
Declarations
Ethics declarations and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Sieiro P, Martin MC, Redruello B, Del Rio B, Ladero V, Palanski BA, Khosla C, Fernandez M, Alvarez MA. Generation of food-grade recombinant Lactobacillus casei delivering Myxococcus xanthus prolyl endopeptidase. Appl Microbiol Biotechnol. 2014;98:6689–6700. doi: 10.1007/s00253-014-5730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach HJ, Munch JC. Identification of bacterial sources of soil peptidases. Biol Fertil Soils. 2000;31:219–224. doi: 10.1007/s003740050648. [DOI] [Google Scholar]
- Barbaro MR, Cremon C, Stanghellini V, Barbara G (2018) Recent advances in understanding non-celiac gluten sensitivity [version 1; peer review: 2 approved]. F1000Research 7 [DOI] [PMC free article] [PubMed]
- Barneix AJ. Physiology and biochemistry of source-regulated protein accumulation in the wheat grain. J Plant Physiol. 2007;164:581–590. doi: 10.1016/j.jplph.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Battais F, Mothes T, Moneret-Vautrin DA, Pineau F, Kanny G, Popineau Y, Bodinier M, Denery-Papini S. Identification of IgE-binding epitopes on gliadins for patients with food allergy to wheat. Allergy Eur J Allergy Clin Immunol. 2005;60:815–821. doi: 10.1111/j.1398-9995.2005.00795.x. [DOI] [PubMed] [Google Scholar]
- Bethune MT, Strop P, Tang Y, Sollid LM, Khosla C. Heterologous expression, purification, refolding, and structural-functional characterization of EP-B2, a self-activating barley cysteine endoprotease. Chem Biol. 2006;13:637–647. doi: 10.1016/j.chembiol.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Bromilow SNL, Gethings LA, Langridge JI, Shewry PR, Buckley M, Bromley MJ, Mills ENC. Comprehensive proteomic profiling of wheat gluten using a combination of data-independent and data-dependent acquisition. Front Plant Sci. 2017;7:2020. doi: 10.3389/fpls.2016.02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarca A, Anderson RP, Mamone G, Fierro O, Facchiano A, Costantini S, Zanzi D, Sidney J, Auricchio S, Sette A, Troncone R, Gianfrani C. Intestinal T cell responses to gluten peptides are largely heterogeneous: implications for a peptide-based therapy in celiac disease. J Immunol. 2009;182:4158–4166. doi: 10.4049/jimmunol.0803181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo ACM, Gomes MD, Reichl AP, Ferro ES, Jacchieri S, Hirata IY, Juliano L. Structural features that make oligopeptides susceptible substrates for hydrolysis by recombinant thimet oligopeptidase. Biochem J. 1997;324:517–522. doi: 10.1042/bj3240517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminero A, Herrán AR, Nistal E, Pérez-Andrés J, Vaquero L, Vivas S, Ruiz de Morales JMG, Albillos SM, Casqueiro J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. 2014;88:309–319. doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- Caminero A, Galipeau HJ, McCarville JL, Johnston CW, Bernier SP, Russell AK, Jury J, Herran AR, Casqueiro J, Tye-Din JA, Surette MG, Magarvey NA, Schuppan D, Verdu EF. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology. 2016;151:670–683. doi: 10.1053/J.GASTRO.2016.06.041. [DOI] [PubMed] [Google Scholar]
- Casilag F, Lorenz A, Krueger J, Klawonn F, Weiss S, Häussler S. LasB elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect Immun. 2015;84:162–171. doi: 10.1128/IAI.00939-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catassi C. Gluten Sensitivity. Ann Nutr Metab. 2015;67:15–26. doi: 10.1159/000440990. [DOI] [PubMed] [Google Scholar]
- Catassi C, Bai JC, Bonaz B, Bouma G, Calabrò A, Carroccio A, Castillejo G, Ciacci C, Cristofori F, Dolinsek J, Francavilla R, Elli L, Green P, Holtmeier W, Koehler P, Koletzko S, Meinhold C, Sanders D, Schumann M, Schuppan D, Ullrich R, Vécsei A, Volta U, Zevallos V, Sapone A, Fasano A. Non-celiac gluten sensitivity: the new frontier of gluten related disorders. Nutrients. 2013;5:3839–3853. doi: 10.3390/nu5103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaletti L, Taravella A, Carrano L, Carenzi G, Sigurtà A, Solinas N, De Caro S, Di Stasio L, Picascia S, Laezza M, Troncone R, Gianfrani C, Mamone G. E40, a novel microbial protease efficiently detoxifying gluten proteins, for the dietary management of gluten intolerance. Sci Rep. 2019;9:13147. doi: 10.1038/s41598-019-48299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Matysiak-Budnilc T, Cellier C, Heyman M. Oral proteases: a new approach to managing coeliac disease. Gut. 2007;56:157–160. doi: 10.1136/gut.2005.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Mack DR, Manuel DG, Mojaverian N, De Nanassy J, Benchimol EI. Validation of an algorithm to identify children with biopsy-proven celiac disease from within health administrative data: an assessment of health services utilization patterns in Ontario. Canada PLoS One. 2017;12:e0180338. doi: 10.1371/journal.pone.0180338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kalliomäki M, Heilig HGHJ, Palva A, Lähteenoja H, de Vos WM, Salojärvi J, Satokari R. Duodenal microbiota composition and mucosal homeostasis in pediatric celiac disease. BMC Gastroenterol. 2013;13:113. doi: 10.1186/1471-230X-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibbar R, Dieleman LA (2019) The gut microbiota in celiac disease and probiotics. Nutrients 11 [DOI] [PMC free article] [PubMed]
- Cianferoni A. Wheat allergy: diagnosis and management. J. Asthma Allergy. 2016;9:13–25. doi: 10.2147/JAA.S81550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Amador MLM, Arévalo-Rodríguez M, Durán EM, JCM R, Martín CS, de Moreno Amador ML, Arévalo-Rodríguez M, Durán EM, Martínez Reyes JC, Sousa Martín C. A new microbial gluten-degrading prolyl endopeptidase: potential application in celiac disease to reduce gluten immunogenic peptides. PLoS One. 2019;14:e0218346. doi: 10.1371/JOURNAL.PONE.0218346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M, Rizzello CG, Fasano A, Clemente MG, De Simone C, Silano M, De Vincenzi M, Losito I, Gobbetti M. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for Celiac Sprue probiotics and gluten intolerance. Biochim Biophys Acta Mol basis Dis. 2006;1762:80–93. doi: 10.1016/J.BBADIS.2005.09.008. [DOI] [PubMed] [Google Scholar]
- de Palma G, Capilla A, Nova E, Castillejo G, Varea V, Pozo T, Garrote JA, Polanco I, López A, Ribes-Koninckx C, Marcos A, García-Novo MD, Calvo C, Ortigosa L, Peña-Quintana L, Palau F, Sanz Y. Influence of milk-feeding type and genetic risk of developing coeliac disease on intestinal microbiota of infants: the PROFICEL study. PLoS One. 2012;7:e30791. doi: 10.1371/journal.pone.0030791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65:763–772. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- Denery-Papini S, Bodinier M, Pineau F, Triballeau S, Tranquet O, Adel-Patient K, Moneret-Vautrin DA, Bakan B, Marion D, Mothes T, Mameri H, Kasarda D. Immunoglobulin-E-binding epitopes of wheat allergens in patients with food allergy to wheat and in mice experimentally sensitized to wheat proteins. Clin Exp Allergy. 2011;41:1478–1492. doi: 10.1111/j.1365-2222.2011.03808.x. [DOI] [PubMed] [Google Scholar]
- Dewar D, Pereira SP, Ciclitira PJ. The pathogenesis of coeliac disease. Int J Biochem Cell Biol. 2004;36:17–24. doi: 10.1016/S1357-2725(03)00239-5. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Van Heel DA. Translational mini-review series on the immunogenetics of gut disease: immunogenetics of coeliac disease. Clin Exp Immunol. 2008;153:162–173. doi: 10.1111/j.1365-2249.2008.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dydensborg Sander S, Nybo Andersen A-M, Murray JA, Karlstad Ø, Husby S, Størdal K. Association between antibiotics in the first year of life and celiac disease. Gastroenterology. 2019;156:2217–2229. doi: 10.1053/J.GASTRO.2019.02.039. [DOI] [PubMed] [Google Scholar]
- Dydensborg S, Toftedal P, Biaggi M, Lillevang ST, Hansen DG, Husby S. Increasing prevalence of coeliac disease in Denmark: a linkage study combining national registries. Acta Paediatr. 2012;101:179–184. doi: 10.1111/j.1651-2227.2011.02392.x. [DOI] [PubMed] [Google Scholar]
- Edens L, Dekker P, Van Der Hoeven R, Deen F, De Roos A, Floris R. Extracellular prolyl endoprotease from Aspergillus niger and its use in the debittering of protein hydrolysates. J Agric Food Chem. 2005;53:7950–7957. doi: 10.1021/jf050652c. [DOI] [PubMed] [Google Scholar]
- Fellahi S, Chibani A, Feuk-Lagerstedt E, Taherzadeh MJ. Identification of two new keratinolytic proteases from a Bacillus pumilus strain using protein analysis and gene sequencing. AMB Express. 2016;6:42. doi: 10.1186/s13568-016-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Feo M, Wei G, Blumenkranz G, Dewhirst FE, Schuppan D, Oppenheim FG, Helmerhorst EJ (2013) The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin Microbiol Infect 19:E386–E394. 10.1111/1469-0691.12249 [DOI] [PMC free article] [PubMed]
- Francavilla R, De Angelis M, Rizzello CG, Cavallo N, Dal Bello F, Gobbetti M. Selected probiotic lactobacilli have the capacity to hydrolyze gluten peptides during simulated gastrointestinal digestion. Appl Environ Microbiol. 2017;83:83. doi: 10.1128/AEM.00376-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisseler D, Horwath WR, Joergensen RG, Ludwig B. Pathways of nitrogen utilization by soil microorganisms - a review. Soil Biol Biochem. 2010;42:2058–2067. doi: 10.1016/j.soilbio.2010.08.021. [DOI] [Google Scholar]
- Gibson SAW, McFarlan C, Hay S, Macfarlane GT. Significance of microflora in proteolysis in the colon. Appl Environ Microbiol. 1989;55:679–683. doi: 10.1128/aem.55.3.679-683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goptar IA, Shagin DA, Shagina IA, Mudrik ES, Smirnova YA, Zhuzhikov DP, Belozersky MA, Dunaevsky YE, Oppert B, Filippova IY, Elpidina EN. A digestive prolyl carboxypeptidase in Tenebrio molitor larvae. Insect Biochem Mol Biol. 2013;43:501–509. doi: 10.1016/j.ibmb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Gordon SR, Stanley EJ, Wolf S, Toland A, Wu SJ, Hadidi D, Mills JH, Baker D, Pultz IS, Siegel JB. Computational design of an α-gliadin peptidase. J Am Chem Soc. 2012;134:20513–20520. doi: 10.1021/ja3094795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths MW, Tellez AM (2013) Lactobacillus helveticus: the proteolytic system. Front Microbiol 4 [DOI] [PMC free article] [PubMed]
- Gutiérrez S, Pérez-Andrés J, Martínez-Blanco H, Ferrero MA, Vaquero L, Vivas S, Casqueiro J, Rodríguez-Aparicio LB. The human digestive tract has proteases capable of gluten hydrolysis. Mol Metab. 2017;6:693–702. doi: 10.1016/j.molmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, Karisola P, Auvinen P, Paulin L, Mäkelä MJ, Vartiainen E, Kosunen TU, Alenius H, Haahtela T. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnett J, Myers SP, Rolfe M (2016) Probiotics and the microbiome in celiac disease: a randomised controlled trial. Evidence-based Complement Altern Med 2016:1–16. 10.1155/2016/9048574 [DOI] [PMC free article] [PubMed]
- Harris MN, Madura JD, Ming LJ, Harwood VJ. Kinetic and mechanistic studies of prolyl oligopeptidase from the hyperthermophile Pyrococcus furiosus. J Biol Chem. 2001;276:19310–19317. doi: 10.1074/jbc.M010489200. [DOI] [PubMed] [Google Scholar]
- Harwood VJ, Denson JD, Robinson-Bidle KA, Schreier HJ. Overexpression and characterization of a prolyl endopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1997;179:3613–3618. doi: 10.1128/jb.179.11.3613-3618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst EJ, Sun X, Salih E, Oppenheim FG. Identification of Lys-Pro-Gln as a novel cleavage site specificity of saliva-associated proteases. J Biol Chem. 2008;283:19957–19966. doi: 10.1074/jbc.M708282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrán AR, Pérez-Andrés J, Caminero A, Nistal E, Vivas S, Ruiz de Morales JM, Casqueiro J. Gluten-degrading bacteria are present in the human small intestine of healthy volunteers and celiac patients. Res Microbiol. 2017;168:673–684. doi: 10.1016/J.RESMIC.2017.04.008. [DOI] [PubMed] [Google Scholar]
- Hui N, Parajuli A, Puhakka R, Grönroos M, Roslund MI, Vari HK, Selonen VAO, Yan G, Siter N, Nurminen N, Oikarinen S, Laitinen OH, Rajaniemi J, Hyöty H, Sinkkonen A. Temporal variation in indoor transfer of dirt-associated environmental bacteria in agricultural and urban areas. Environ Int. 2019;132:105069. doi: 10.1016/j.envint.2019.105069. [DOI] [PubMed] [Google Scholar]
- Inomata N. Wheat allergy. Curr Opin Allergy Clin Immunol. 2009;9:238–243. doi: 10.1097/ACI.0b013e32832aa5bc. [DOI] [PubMed] [Google Scholar]
- Ionata E, Canganella F, Bianconi G, Benno Y, Sakamoto M, Capasso A, Rossi M, La Cara F. A novel keratinase from Clostridium sporogenes bv. pennavorans bv. nov., a thermotolerant organism isolated from solfataric muds. Microbiol Res. 2008;163:105–112. doi: 10.1016/j.micres.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Kabashima T, Fujii M, Meng Y, Ito K, Yoshimoto T. Prolyl endopeptidase from Sphingomonas capsulata: isolation and characterization of the enzyme and nucleotide sequence of the gene. Arch Biochem Biophys. 1998;358:141–148. doi: 10.1006/abbi.1998.0836. [DOI] [PubMed] [Google Scholar]
- Kang C, Yu XW, Xu Y. Gene cloning and enzymatic characterization of an endoprotease Endo-Pro-Aspergillus niger. J Ind Microbiol Biotechnol. 2013;40:855–864. doi: 10.1007/s10295-013-1284-4. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Sowdhamini R. Distribution, classification, domain architectures and evolution of prolyl oligopeptidases in prokaryotic lineages. BMC Genomics. 2014;15:985. doi: 10.1186/1471-2164-15-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CP, Bai JC, Liu E, Leffler DA. Advances in diagnosis and management of celiac disease. Gastroenterology. 2015;148:1175–1186. doi: 10.1053/j.gastro.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocadag Kocazorbaz E, Zihnioglu F. Purification, characterization and the use of recombinant prolyl oligopeptidase from Myxococcus xanthus for gluten hydrolysis. Protein Expr Purif. 2017;129:101–107. doi: 10.1016/j.pep.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Kõiv V, Adamberg K, Adamberg S, Sumeri I, Kasvandik S, Kisand V, Maiväli Ü, Tenson T. Microbiome of root vegetables—a source of gluten-degrading bacteria. Appl Microbiol Biotechnol. 2020;104:8871–8885. doi: 10.1007/s00253-020-10852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolton M, Sela N, Elad Y, Cytryn E. Comparative genomic analysis indicates that niche adaptation of terrestrial Flavobacteria is strongly linked to plant glycan metabolism. PLoS One. 2013;8:76704. doi: 10.1371/journal.pone.0076704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Holster S, Bruins MJ, Brummer RJ. Randomized clinical trial: effective gluten degradation by Aspergillus niger-derived enzyme in a complex meal setting. Sci Rep. 2017;7:13100. doi: 10.1038/s41598-017-13587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnareddy S, Stier K, Recanati M, Lebwohl B, Green PH. Commercially available glutenases: a potential hazard in coeliac disease. Ther Adv Gastroenterol. 2017;10:473–481. doi: 10.1177/1756283X17690991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Hao Y, Walling BE, Jeffries JL, Ohman DE, Lau GW. Pseudomonas aeruginosa elastase provides an escape from phagocytosis by degrading the pulmonary surfactant protein-A. PLoS One. 2011;6:e27091. doi: 10.1371/journal.pone.0027091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Verma MK, Kumar T, Gupta S, Pandey R, Yadav M, Chauhan NS. S9A serine protease engender antigenic gluten catabolic competence to the human gut microbe. Indian J Microbiol. 2018;58:294–300. doi: 10.1007/s12088-018-0732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, Dubourg G, Durand G, Mourembou G, Guilhot E, Togo A, Bellali S, Bachar D, Cassir N, Bittar F, Delerce J, Mailhe M, Ricaboni D, Bilen M, Dangui Nieko NPM, Dia Badiane NM, Valles C, Mouelhi D, Diop K, Million M, Musso D, Abrahão J, Azhar EI, Bibi F, Yasir M, Diallo A, Sokhna C, Djossou F, Vitton V, Robert C, Rolain JM, La Scola B, Fournier PE, Levasseur A, Raoult D. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:1–8. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- Lähdeaho ML, Kaukinen K, Laurila K, Vuotikka P, Koivurova OP, Kärjä-Lahdensuu T, Marcantonio A, Adelman DC, Mäki M. Glutenase ALV003 attenuates gluten-induced mucosal injury in patients with celiac disease. Gastroenterology. 2014;146:1649–1658. doi: 10.1053/j.gastro.2014.02.031. [DOI] [PubMed] [Google Scholar]
- Laparra JM, Olivares M, Gallina O, Sanz Y. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS One. 2012;7:30744. doi: 10.1371/journal.pone.0030744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassalle F, Spagnoletti M, Fumagalli M, Shaw L, Dyble M, Walker C, Thomas MG, Bamberg Migliano A, Balloux F. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol Ecol. 2018;27:182–195. doi: 10.1111/mec.14435. [DOI] [PubMed] [Google Scholar]
- Lehtimäki J, Karkman A, Laatikainen T, Paalanen L, von Hertzen L, Haahtela T, Hanski I, Ruokolainen L. Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Sci Rep. 2017;7:45651. doi: 10.1038/srep45651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser TD, Mølbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol. 2009;11:2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- Lim PY, Tan LL, Ow DS-W, Wong FT. A propeptide toolbox for secretion optimization of Flavobacterium meningosepticum endopeptidase in Lactococcus lactis. Microb Cell Factories. 2017;16:221. doi: 10.1186/s12934-017-0836-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindfors K, Blomqvist T, Juuti-Uusitalo K, Stenman S, Venäläinen J, Mäki M, Kaukinen K. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol. 2008;152:552–558. doi: 10.1111/j.1365-2249.2008.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Allison C. Utilisation of protein by human gut bacteria. FEMS Microbiol Lett. 1986;38:19–24. doi: 10.1111/j.1574-6968.1986.tb01934.x. [DOI] [Google Scholar]
- Macfarlane GT, Allison C, Gibson SAW, Cummings JH. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol. 1988;64:37–46. doi: 10.1111/j.1365-2672.1988.tb02427.x. [DOI] [PubMed] [Google Scholar]
- Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, Jakobsen KS, Wulff BBH, Steuernagel B, Mayer KFX, Olsen OA, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, Sourdille P, Endo TR, Kubaláková M, Šíhalíková J, Dubská Z, Vrána J, Šperková R, Šimková H, Febrer M, Clissold L, McLay K, Singh K, Chhuneja P, Singh NK, Khurana J, Akhunov E, Choulet F, Alberti A, Barbe V, Wincker P, Kanamori H, Kobayashi F, Itoh T, Matsumoto T, Sakai H, Tanaka T, Wu J, Ogihara Y, Handa H, Maclachlan PR, Sharpe A, Klassen D, Edwards D, Batley J, Sandve SR, Lien S, Wulff B, Caccamo M, Ayling S, Ramirez-Gonzalez RH, Clavijo BJ, Wright J, Martis MM, Mascher M, Chapman J, Poland JA, Scholz U, Barry K, Waugh R, Rokhsar DS, Muehlbauer GJ, Stein N, Gundlach H, Zytnicki M, Jamilloux V, Quesneville H, Wicker T, Faccioli P, Colaiacovo M, Stanca AM, Budak H, Cattivelli L, Glover N, Pingault L, Paux E, Sharma S, Appels R, Bellgard M, Chapman B, Nussbaumer T, Bader KC, Rimbert H, Wang S, Knox R, Kilian A, Alaux M, Alfama F, Couderc L, Guilhot N, Viseux C, Loaec M, Keller B, Praud S (2014) Ancient hybridizations among the ancestral genomes of bread wheat. Science (80- 345(6194). 10.1126/science.1250092 [DOI] [PubMed]
- Matsuo H, Yokooji T, Taogoshi T. Common food allergens and their IgE-binding epitopes. Allergol Int. 2015;64:332–343. doi: 10.1016/j.alit.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Maukonen J, Mättö J, Suihko ML, Saarela M. Intra-individual diversity and similarity of salivary and faecal microbiota. J Med Microbiol. 2008;57:1560–1568. doi: 10.1099/jmm.0.47352-0. [DOI] [PubMed] [Google Scholar]
- Medina M, De Palma G, Ribes-Koninckx C, Calabuig M, Sanz Y. Bifidobacterium strains suppress in vitro the pro-inflammatory milieu triggered by the large intestinal microbiota of coeliac patients. J Inflamm. 2008;5:19. doi: 10.1186/1476-9255-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefleh M, Conte P, Fadda C, Giunta F, Piga A, Hassoun G, Motzo R. From ancient to old and modern durum wheat varieties: interaction among cultivar traits, management, and technological quality. J Sci Food Agric. 2019;99:2059–2067. doi: 10.1002/jsfa.9388. [DOI] [PubMed] [Google Scholar]
- Messana I, Cabras T, Pisano E, Sanna MT, Olianas A, Manconi B, Pellegrini M, Paludetti G, Scarano E, Fiorita A, Agostino S, Contucci AM, Calò L, Picciotti PM, Manni A, Bennick A, Vitali A, Fanali C, Inzitari R, Castagnola M. Trafficking and postsecretory events responsible for the formation of secreted human salivary peptides. Mol Cell Proteomics. 2008;7:911–926. doi: 10.1074/mcp.M700501-MCP200. [DOI] [PubMed] [Google Scholar]
- Mika N, Zorn H, Rühl M. Prolyl-specific peptidases for applications in food protein hydrolysis. Appl Microbiol Biotechnol. 2015;99:7837–7846. doi: 10.1007/s00253-015-6838-0. [DOI] [PubMed] [Google Scholar]
- Mitea C, Havenaar R, Wouter Drijfhout J, Edens L, Dekking L, Koning F, Dekking EHA. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for coeliac disease. Gut. 2008;57:25–32. doi: 10.1136/gut.2006.111609. [DOI] [PubMed] [Google Scholar]
- Mustalahti K, Catassi C, Reunanen A, Fabiani E, Heier M, McMillan S, Murray L, Metzger M-H, Gasparin M, Bravi E, Mäki M. The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann Med. 2010;42:587–595. doi: 10.3109/07853890.2010.505931. [DOI] [PubMed] [Google Scholar]
- Nadal I, Donant E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56:1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- Nardone G, Compare D. The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol J. 2015;3:255–260. doi: 10.1177/2050640614566846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry C. The gluten-free diet: use in digestive disease management. Curr Treat Options Gastroenterol. 2019;17:554–563. doi: 10.1007/s11938-019-00255-0. [DOI] [PubMed] [Google Scholar]
- Nguyen TTH, Myrold DD, Mueller RS. Distributions of extracellular peptidases across prokaryotic genomes reflect phylogeny and habitat. Front Microbiol. 2019;10:413. doi: 10.3389/fmicb.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminen N, Lin J, Grönroos M, Puhakka R, Kramna L, Vari HK, Viskari H, Oikarinen S, Roslund M, Parajuli A, Tyni I, Cinek O, Laitinen O, Hyöty H, Sinkkonen A (2018) Nature-derived microbiota exposure as a novel immunomodulatory approach. 13:737–744 . 10.2217/fmb-2017-0286 [DOI] [PubMed]
- O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obsborn T. The vegetable proteins. 2. London: Longmans Green & Co; 1924. [Google Scholar]
- Osorio CE, Wen N, Mejias JH, Liu B, Reinbothe S, von Wettstein D, Rustgi S. Development of wheat genotypes expressing a glutamine-specific endoprotease from barley and a prolyl endopeptidase from Flavobacterium meningosepticum or Pyrococcus furiosus as a potential remedy to celiac disease. Funct Integr Genomics. 2019;19:123–136. doi: 10.1007/s10142-018-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio CE, Wen N, Mejías JH, Mitchell S, von Wettstein D, Rustgi S. Directed-mutagenesis of Flavobacterium meningosepticum prolyl-oligopeptidase and a glutamine-specific endopeptidase from barley. Front Nutr. 2020;7:11. doi: 10.3389/fnut.2020.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottman N, Ruokolainen L, Suomalainen A, Sinkko H, Karisola P, Lehtimäki J, Lehto M, Hanski I, Alenius H, Fyhrquist N. Soil exposure modifies the gut microbiota and supports immune tolerance in a mouse model. J Allergy Clin Immunol. 2019;143:1198–1206.e12. doi: 10.1016/J.JACI.2018.06.024. [DOI] [PubMed] [Google Scholar]
- Ozuna CV, Iehisa JCM, Giménez MJ, Alvarez JB, Sousa C, Barro F. Diversification of the celiac disease α-gliadin complex in wheat: a 33-mer peptide with six overlapping epitopes, evolved following polyploidization. Plant J. 2015;82:794–805. doi: 10.1111/tpj.12851. [DOI] [PubMed] [Google Scholar]
- Parajuli A, Hui N, Puhakka R, Oikarinen S, Grönroos M, Selonen VAO, Siter N, Kramna L, Roslund MI, Vari HK, Nurminen N, Honkanen H, Hintikka J, Sarkkinen H, Romantschuk M, Kauppi M, Valve R, Cinek O, Laitinen OH, Rajaniemi J, Hyöty H, Sinkkonen A. Yard vegetation is associated with gut microbiota composition. Sci Total Environ. 2020;713:136707. doi: 10.1016/j.scitotenv.2020.136707. [DOI] [PubMed] [Google Scholar]
- Patel BY, Volcheck GW. Food allergy: common causes, diagnosis, and treatment. Mayo Clin Proc. 2015;90:1411–1419. doi: 10.1016/j.mayocp.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Pati A, la Butti K, Pukall R, Nolan M, del Rio TG, Tice H, Cheng JF, Lucas S, Chen F, Copeland A, Ivanova N, Mavromatis K, Mikhailova N, Pitluck S, Bruce D, Goodwin L, Land M, Hauser L, Chang YJ, Jeffries CD, Chen A, Palaniappan K, Chain P, Brettin T, Sikorski J, Rohde M, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lapidus A. Complete genome sequence of Sphaerobacter thermophilus type strain (S 6022 T) Stand Genomic Sci. 2010;2:49–56. doi: 10.4056/sigs.601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzak M. Celiac disease, wheat allergy, and gluten sensitivity. J Parenter Enter Nutr. 2012;36:68S–75S. doi: 10.1177/0148607111426276. [DOI] [PubMed] [Google Scholar]
- Plugis NM, Khosla C. Therapeutic approaches for celiac disease. Best Pract Res Clin Gastroenterol. 2015;29:503–521. doi: 10.1016/j.bpg.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo-Rubio T, Olivares M, Nova E, De Palma G, Mujico JR, Ferrer MD, Marcos A, Sanz Y. Immune development and intestinal microbiota in celiac disease. Clin Dev Immunol. 2012;2012:12–12. doi: 10.1155/2012/654143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S-W, Bergseng E, Molberg Ø, Jung G, Fleckenstein B, Sollid LM. Refining the rules of gliadin T cell epitope binding to the disease-associated DQ2 molecule in celiac disease: importance of proline spacing and glutamine deamidation. J Immunol. 2005;175:254–261. doi: 10.4049/jimmunol.175.1.254. [DOI] [PubMed] [Google Scholar]
- Qiu J, Wilkens C, Barrett K, Meyer AS. Microbial enzymes catalyzing keratin degradation: classification, structure, function. Biotechnol Adv. 2020;44:107607. doi: 10.1016/j.biotechadv.2020.107607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliariello A, Aloisio I, Bozzi Cionci N, Luiselli D, D’ Auria G, Martinez-Priego L, Pérez-Villarroya D, Langerholc T, Primec M, Mičetić-Turk D, Di Gioia D. Effect of Bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. 2016;8:660. doi: 10.3390/nu8100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveschot C, Cudennec B, Coutte F, Flahaut C, Fremont M, Drider D, Dhulster P (2018) Production of bioactive peptides by Lactobacillus species: from gene to application. Front Microbiol 9 [DOI] [PMC free article] [PubMed]
- Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzello CG, De Angelis M, Di Cagno R, Camarca A, Silano M, Losito I, De Vincenzi M, De Bari MD, Palmisano F, Maurano F, Gianfrani C, Gobbetti M. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: new perspectives for celiac disease. Appl Environ Microbiol. 2007;73:4499–4507. doi: 10.1128/AEM.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslund MI, Puhakka R, Grönroos M, Nurminen N, Oikarinen S, Gazali AM, Cinek O, Kramná L, Siter N, Vari HK, Soininen L, Parajuli A, Rajaniemi J, Kinnunen T, Laitinen OH, Hyöty H, Sinkkonen A. Environmental studies biodiversity intervention enhances immune regulation and health-associated commensal microbiota among daycare children. Sci Adv. 2020;6:2578. doi: 10.1126/sciadv.aba2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszkowska A, Pawlicka M, Mroczek A, Bałabuszek K, Nieradko-Iwanicka B (2019) Non-celiac gluten sensitivity: a review. Medicina. 55 [DOI] [PMC free article] [PubMed]
- Salden BN, Monserrat V, Troost FJ, Bruins MJ, Edens L, Bartholomé R, Haenen GR, Winkens B, Koning F, Masclee AA. Randomised clinical study: Aspergillus niger-derived enzyme digests gluten in the stomach of healthy volunteers. Aliment Pharmacol Ther. 2015;42:273–285. doi: 10.1111/apt.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapone A, Lammers KM, Casolaro V, Cammarota M, Giuliano MT, De Rosa M, Stefanile R, Mazzarella G, Tolone C, Russo MI, Esposito P, Ferraraccio F, Cartenì M, Riegler G, de Magistris L, Fasano A. Divergence of gut permeability and mucosal immune gene expression in two gluten-associated conditions: celiac disease and gluten sensitivity. BMC Med. 2011;9:23. doi: 10.1186/1741-7015-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapone A, Bai JC, Ciacci C, Dolinsek J, Green PHR, Hadjivassiliou M, Kaukinen K, Rostami K, Sanders DS, Schumann M, Ullrich R, Villalta D, Volta U, Catassi C, Fasano A. Spectrum of gluten-related disorders: consensus on new nomenclature and classification. BMC Med. 2012;10:13. doi: 10.1186/1741-7015-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savijoki K, Ingmer H, Varmanen P. Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol. 2006;71:394–406. doi: 10.1007/s00253-006-0427-1. [DOI] [PubMed] [Google Scholar]
- Schalk K, Lang C, Wieser H, Koehler P, Scherf KA (2017) Quantitation of the immunodominant 33-mer peptide from α-gliadin in wheat flours by liquid chromatography tandem mass spectrometry. Sci Rep 7. 10.1038/srep45092 [DOI] [PMC free article] [PubMed]
- Scherf KA, Koehler P, Wieser H. Gluten and wheat sensitivities - an overview. J Cereal Sci. 2016;67:2–11. doi: 10.1016/j.jcs.2015.07.008. [DOI] [Google Scholar]
- Scherf KA, Wieser H, Koehler P. Novel approaches for enzymatic gluten degradation to create high-quality gluten-free products. Food Res Int. 2018;110:62–72. doi: 10.1016/j.foodres.2016.11.021. [DOI] [PubMed] [Google Scholar]
- Schulz K, Giesler L, Linke D, Berger RG. A prolyl endopeptidase from Flammulina velutipes for the possible degradation of celiac disease provoking toxic peptides in cereal proteins. Process Biochem. 2018;73:47–55. doi: 10.1016/j.procbio.2018.07.019. [DOI] [Google Scholar]
- Schuppan D. Current concepts of celiac disease pathogenesis. Gastroenterology. 2000;119:234–242. doi: 10.1053/gast.2000.8521. [DOI] [PubMed] [Google Scholar]
- Seiskari T, Kondrashova A, Viskari H, Kaila M, Haapala AM, Aittoniemi J, Virta M, Hurme M, Uibo R, Knip M, Hyöty H, Ilonen J, Reunanen A, Ludvigsson J, Marciulionyte D, Hermann R, Soltesz G, Fuechtenbusch M, Ziegler A, Romanov A. Allergic sensitization and microbial load - a comparison between Finland and Russian Karelia. Clin Exp Immunol. 2007;148:47–52. doi: 10.1111/j.1365-2249.2007.03333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Molberg Ø, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004;383:311–318. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty R, Vestergaard M, Jessen F, Hägglund P, Knorr V, Koehler P, Prakash HS, Hobley TJ. Discovery, cloning and characterisation of proline specific prolyl endopeptidase, a gluten degrading thermo-stable enzyme from Sphaerobacter thermophiles. Enzym Microb Technol. 2017;107:57–63. doi: 10.1016/J.ENZMICTEC.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Shewry P (2019) What is gluten—why is it special? Front. Nutr. 6:101 [DOI] [PMC free article] [PubMed]
- Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. In: Journal of Experimental Botany. Oxford University Press, pp 947–958 [DOI] [PubMed]
- Shewry PR, Tatham AS. The prolamin storage proteins of cereal seeds: structure and evolution. Biochem J. 1990;267:1–12. doi: 10.1042/bj2670001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, Ledford JG, Marques dos Santos M, Anderson RL, Metwali N, Neilson JW, Maier RM, Gilbert JA, Holbreich M, Thorne PS, Martinez FD, von Mutius E, Vercelli D, Ober C, Sperling AI. Innate immunity and asthma risk in Amish and Hutterite farm children. N Engl J Med. 2016;375:411–421. doi: 10.1056/nejmoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepniak D, Spaenij-Dekking L, Mitea C, Moester M, De Ru A, Baak-Pablo R, Van Veelen P, Edens L, Koning F. Highly efficient gluten degradation with a newly identified prolyl endoprotease: implications for celiac disease. Am J Physiol Gastrointest Liver Physiol. 2006;291:291–G629. doi: 10.1152/ajpgi.00034.2006. [DOI] [PubMed] [Google Scholar]
- Tsuruoka N, Nakayama T, Ashida M, Hemmi H, Nakao M, Minakata H, Oyama H, Oda K, Nishino T. Collagenolytic serine-carboxyl proteinase from Alicyclobacillus sendaiensis strain NTAP-1: purification, characterization, gene cloning, and heterologous expression. Appl Environ Microbiol. 2003;69:162–169. doi: 10.1128/AEM.69.1.162-169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye-Din JA, Anderson RP, Ffrench RA, Brown GJ, Hodsman P, Siegel M, Botwick W, Shreeniwas R. The effects of ALV003 pre-digestion of gluten on immune response and symptoms in celiac disease in vivo. Clin Immunol. 2010;134:289–295. doi: 10.1016/j.clim.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MCNM, Bethel G, Holmes GKT, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JRF, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Garg PK, Dubey AK (2018) Insights into the human oral microbiome [DOI] [PubMed]
- Volta U, Bardella MT, Calabrò A, Troncone R, Corazza GR, Bagnato C, Belcari C, Bellantoni A, Caio G, Calella F, Cappello M, Ciacci C, D’Agate C, De Vitis I, Di Sabatino A, Fava G, Frau MR, Fugazza A, Grassi SA, Larcinese G, Latella G, Lauri A, Lauria A, Lenoci N, Lopez Rios NB, Macchia D, Minelli M, Molinari B, Morelli O, Mumolo MG, Niccoli G, Pacenza C, Pallone F, Parente F, Pulitanò R, Pumpo R, Riegler G, Rispo A, Rispoli FR, Tedone F, Valiante F, Viviani G, Satta PU. An Italian prospective multicenter survey on patients suspected of having non-celiac gluten sensitivity. BMC Med. 2014;12:85. doi: 10.1186/1741-7015-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hertzen L, Hanski I, Haahtela T. Natural immunity. EMBO Rep. 2011;12:1089–1093. doi: 10.1038/embor.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranova V, Rejsek K, Formanek P. Proteolytic activity in soil: a review. Appl Soil Ecol. 2013;70:23–32. doi: 10.1016/j.apsoil.2013.04.003. [DOI] [Google Scholar]
- Watanabe K, Hayano K. Distribution and identification of proteolytic Bacillus spp. in paddy field soil under rice cultivation. Can J Microbiol. 1993;39:674–680. doi: 10.1139/m93-097. [DOI] [PubMed] [Google Scholar]
- Wei G, Tian N, Valery AC, Zhong Y, Schuppan D, Helmerhorst EJ. Identification of pseudolysin (lasB) as an aciduric gluten-degrading enzyme with high therapeutic potential for celiac disease. Am J Gastroenterol. 2015;110:899–908. doi: 10.1038/ajg.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Tian N, Siezen R, Schuppan D, Helmerhorst EJ. Identification of food-grade subtilisins as gluten-degrading enzymes to treat celiac disease. Am J Physiol Gastrointest Liver Physiol. 2016;311:G571–G580. doi: 10.1152/ajpgi.00185.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser H, KoeHler P. Detoxification of gluten by means of enzymatic treatment. J AOAC Int. 2012;95:356–363. doi: 10.5740/jaoacint.SGE_Wieser. [DOI] [PubMed] [Google Scholar]
- Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG, Helmerhorst EJ. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS One. 2011;6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CCGM, Troost FJ, Bork P, Wels M, de Vos WM, Kleerebezem M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–1426. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OD (2014) The spectrum of major seed storage genes and proteins in oats (Avena sativa). PLoS One 9. 10.1371/journal.pone.0083569 [DOI] [PMC free article] [PubMed]