“What is food to one is bitter poison to another.” —Lucretius (ca. 96 B.C.-55 B.C.), based on his observation of individual variations in adverse reactions caused by food allergy 1
Introduction
Food allergy (FA) is a complex disease of substantial public health concern; it affects ~8% of the pediatric population, and it is a common cause of anaphylaxis.2 FA is the common cause of anaphylaxis in children, affecting up to 15 million people including 5 million children.2–4 FA accounts for 3 million doctor visits, 125,000 to emergency room, 2,000 hospitalization and about150 deaths annually. The economic cost of food allergies is nearly $25 billion per year.5 Food allergy (FA) is an adverse reaction to food (food hypersensitivity) occurring in susceptible individuals, which is mediated by a classical immune mechanism specific for the food itself. The best established mechanism in FA is due to the presence of IgE antibodies against the offending food. Food intolerance (FI) are all non-immune-mediated adverse reactions to food. The subgroups of FI are enzymatic (e.g. lactose intolerance due to lactase deficiency), pharmacological (reactions against biogenic amines, histamine intolerance), and undefined food intolerance (e.g. against some food additives). The diagnosis of an IgE-mediated FA (IgE-FA) is made by a carefully taken case history, supported by the demonstration of an IgE sensitization either by skin prick tests or by in vitro tests, and confirmed by positive oral provocation. For scientific purposes the only accepted test for the confirmation of FA/FI is a properly performed double-blind, placebo-controlled food challenge (DBPCFC). A panel of recombinant allergens, produced as single allergenic molecules, may in future improve the diagnosis of IgE-FA. Due to a lack of causal treatment possibilities, the elimination of the culprit <<food allergen>> from the diet is the only therapeutic option for patients with real food allergy. The basic immunopathology underlying food allergy can be divided into non-IgE-mediated (i.e. cell-mediated slow onset) and IgE-mediated (acute immediate reactions) FA.6 The non-IgE-mediated food hypersensitivity encompasses non-immune mediated adverse reaction with delayed hypersensitivity whose mechanisms are less clear.7 Such reactions include cell-mediated reactions that involve sensitized lymphocytes in tissues rather than antibodies.8 The overall prevalence of cell-mediated reactions remains uncertain.9,10 The most common non-IgE- mediated hypersensitivity reaction affecting all age groups of the population is celiac disease, also known as gluten-sensitive enteropathy, lactose intolerance due to histamine intolerance.11 The IgE-mediated food allergy is the most common and life-threatening type of food allergic disorders.12 It is also called immediate hypersensitivity reactions because of the short onset time between the ingestion of the offending food and the onset of symptoms.3,8,13 Oral exposure to potential food allergens early in life is believed to promote tolerance, whereas cutaneous exposure through an impaired skin barrier promotes sensitization.14 Tolerance is mediated by CD103+ dendritic cells in the gastrointestinal tract that bind to food allergens and migrate to mesenteric lymph nodes where regulatory T (Treg) cells are induced. In IgE-mediated food allergy, the induction and functioning of regulatory T (Treg) cells is believed to be compromised and the immune response is shifted towards the generation of T helper 2 (Th2) cells, leading to IgE class-switching in B cells.15,16 The most common symptoms associated with IgE-mediated reactions involve the skin and may lead to anaphylaxis.17,18 IgE-mediated mechanisms are also responsible for allergic reactions to pollens, mold spores, animal danders, insect venoms and other environmental stimuli.19 Anaphylaxis is a severe and life-threatening systemic allergic reaction characterized by fall of blood pressure, upper airway obstruction, and difficulty breathing. IgE-mediated immunological reactions are the most important type of food allergy because these reactions involve a wide variety of different foods.20More than 170 foods are known to cause IgE-mediated food allergies. In the U.S., eight foods account for 90% of serious allergic reactions: milk, eggs, fish, shellfish, wheat, soy, peanuts, and tree nuts.21,22 Hence, federal law requires food labels to clearly identify the food allergen source of all foods and ingredients that contain any protein derived from these common allergens (Food Allergen Labeling and Consumer Protection Act of 2004, Public L No. 108-282). To date, except strict avoidance, there are no effective curative treatments for IgE-mediated FAs.23,24 This chapter primary addresses the genetics of IgE-mediated food allergy.
FA and the atopic march
The role of FA in the atopic march (progression of allergic diseases starting from infancy and typically includes atopic dermatitis, food allergy, allergic rhinitis, and asthma) is not well understood, but a strong association between FA and AD has been established.25 Although the causal nature of this association has been debated, it is now generally accepted that cutaneous sensitization to food allergens is an important step in the development of FA, whereas exposure to food allergens through the oral route appear to promote tolerance.26–28 Although FA sometimes precedes AD,29 sensitization through an impaired non-lesional skin barrier before manifestation of AD has developed is likely a common route for food allergens. Indeed, skin barrier impairment at birth as measured by transepidermal water loss predicts food allergy at age 2,30 and one study found FLG genotype to be associated with sensitization to peanut allergen regardless of the presence of eczema.31 Several rare monogenic disorders that are caused by genes involved in skin barrier formation and maintenance have been linked to FA. A well-known example is Netherton syndrome, caused by autosomal recessive mutations in the serine protease inhibitor Karzal type 5 (SPINK5).32 The gene product of SPINK5, LEKTI, is involved in the regulation of desquamation. Netherton syndrome is characterized by defective cornification, chronic skin inflammation, impaired skin barrier, and multiple allergies, including FA.33 Furthermore, the desmoglein 1 gene (DSG1) is involved in maintaining the structure of the epidermis, and loss-of-function mutations in this gene are the cause of severe dermatitis, multiple allergies, and metabolic wasting (SAM) syndrome. In addition to skin conditions such as severe psoriasiform dermatitis, ichthyosis and keratosis, elevated IgE levels and multiple food allergies have been observed in patients with SAM syndrome.34,35
The link between FA and monogenic inherited skin disorders further strengthens the case for cutaneous food sensitization through an impaired skin barrier as a causative event in the development of FA.36 There are, however, also several monogenic diseases associated with FA that have been shown to be caused by mutations in genes involved in immune system responses. Immunodysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome is a rare disorder caused by mutations in the FOXP3 gene that leads to impaired development of CD4+CD25+ regulatory T cells. In addition to autoimmunity conditions, IPEX patients have a high risk of food allergies, AD, and elevated IgE levels.37,38 Another example is the connective tissue disorder Loeys–Dietz syndrome, which is usually caused by mutations in TGFBR1 or TGFBR2. The resulting upregulation of TGFβ signaling appear to skew naïve CD4(+) T cells towards TH2 cytokine-producing cells, and patients have a strongly increased risk of all major allergic diseases, including FA.39
Food allergy and racial variation
Pediatric food allergy has increased tremendously over the past two decades and it is on the rise among all demographic groups.40 However, its prevalence rates differ across ethnic groups.41 Results from the NHANES indicated that the prevalence of FA is 4 times higher in African Americans (AA) than European Americans (EA).42 AA children are reported to have an eightfold increase in the prevalence of peanut allergy as compared with the general U.S. pediatric population. AA individuals with FA have elevated levels of immunoglobulin E (IgE), peripheral eosinophilia, T helper 2 cytokines, and epithelial dysfunction as compared with EAs.43–45 Recent studies showed that variants in several cytokine candidate genes that encode Th2-related molecules such as IL-4 and IL-13 show higher allele frequency in AA, suggesting that these alleles have been conserved to combat parasitic infections in Africans but not in Europeans.46 This unique evolutionary trajectory might be the reason for high prevalence of allergic disorders in AAs.47 Although AAs experience higher rates of FA prevalence and mortality than any other racial or ethnic group in America, few studies of FA have focused on this population, and almost all studies have predominantly employed EA samples. A PubMed search demonstrated that EAs are mentioned five times more often in various FA-related literature as compared with AAs. Bringing AA families into research will help to compare their results with those of other groups and to better understand the basis of FA disparities.48,49 It might help to unravel population-specific disease risk in the context of environmental exposure factors.50
Genetics and food allergy
Investigators study the genetics of food allergy for many reasons. First, some investigators study it to understand the evolutionary basis of our current population distribution of FA and its ‘genetic architecture’. For others, such information provides insights into how malleable FA may be and what type of interventions may be most effective in altering population levels of FA. A second reason is the hope of identifying genes associated with increased risk of FA that can be used as prognostic factors to indicate who is likely to become allergic so that they can be given preventive therapy. A third reason is to identify genes that moderate the safety and/or efficacy of treatments. If we can identify such genes, then we are on our way to ‘personalized medicine’.51 Finally, among most biomedical researchers, the most common reason for wishing to identify genes that confer variations in FA is to identify physiologic ‘pathways’ through which FA develops so that such pathways can then be further studied and made targets for potential pharmaceutical intervention. Several experimental designs, including family, twin, and cohort studies, and analytic approaches, such as linkage analysis, candidate gene, genome-wide association, DNA methylation, and microbiome analysis have been used to study important questions such as: Why do some individuals develop FA and others do not? How do environmental and genetics factors interact to increase the risk of FA? A brief history of the search for genetic causes of FA are presented in Figure 1. Investigators screen the genome to locate regions contributing to variance (Box 1) in FA related outcomes.
Figure 1.
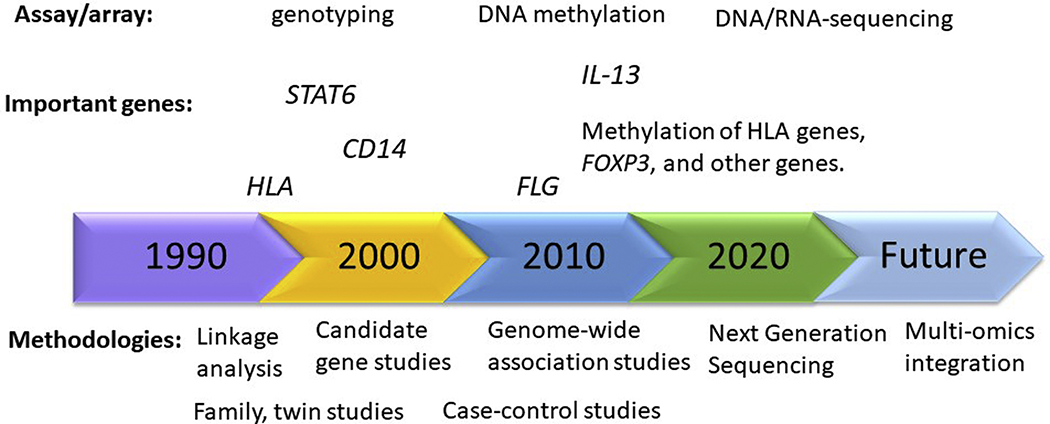
A brief history of the search for genetic causes of food allergy (FA).
Box 1. Definitions of genetic and genetic related terminology.
Variance: a measure of statistical dispersion indicating how far values typically are from the distribution’s mean
Genome: complete set of genetic information for an individual.
Epistatic variance: variance due to interaction among alleles at different loci (gene-gene interactions.
Heritability: the proportion of population variance in a trait attributable to segregation of a gene or genes (overall phenotypic variation/risk that is attributable to genetic factors).
Gene-gene interaction: when two or more DNA variations interact either directly (DNA–DNA or DNA–mRNA interactions), to change transcription or translation levels, or indirectly by way of their protein products, to alter disease risk separate from their independent effects.
Gene-environment interaction: when a DNA variation interacts with an environmental factor, such that their combined effect is distinct from their independent effects. An interaction is indicated when the presence of one factor (eg, diet) affects the influence of the second factor (eg, genetic) on disease risk.
Phenocopy: the presence of a disease phenotype that has a non-genetic (random or environmental) basis.
Trait heterogeneity: when a trait has been defined with insufficient specificity such that it represents at least two distinct underlying traits.
Genetic heterogeneity: the production of the same or similar phenotypes (observed biochemical, physiological, and morphological characteristics of a person determined by his/her genotype) by different underlying genetic mechanisms.
Race: group based on physical attributes, a social construct; no biological basis.
Ethnicity: group based on culture, language, physical attributes, religion, country of origin.
Ancestry: group based on DNA-- line of decent.
Admixture: interbreeding among different ancestry populations.
Candidate gene association studies: Targeted studies of association between selected genes of interest and a phenotype.
Genome- wide association studies: genetics research to associate specific genetic variations with particular trait.
Copy number variation: a sequence of bases within a genome differs in the number of copies among individuals or populations.
Population structure or population stratification: The presence of differences in allele frequencies between subpopulations in a population.
Next- generation sequencing: an entire genome is sequenced from fragmented DNA, producing short (less than 300 bp) sequencing reads at high speed and low cost.
Single- nucleotide variants: a single base within a read or genome differs from the base found at the same position in other individuals or populations.
Meta-analysis: A routine approach to combining smaller GWAs using summary statistics/results to overcome the smaller sample sizes of individual studies.
Fine mapping: A set of statistical and laboratory approaches to determine the causal genetic variant for associated genetic loci.
Family-based studies
Family studies can address whether a disease aggregates in families. Such studies typically compare the prevalence of the disorder among first-degree relatives of affected cases to the prevalence in the population or among relatives of unaffected controls. A higher risk among relatives of cases indicates that the disease may be familial, but it does not necessarily mean that genes are involved; a disease may run in families for nongenetic reasons (i.e., a shared environment). Although environmental factors play a significant role in the onset of FA such as higher rate of food-related-health problems in lower-income children than higher income groups that might indicate the role of environmental exposure factors, the most widely used indicators of FA are familial aggregation and strong genetic component.52–54.54
Twin-based studies
Twin studies compare the concordance rates of a disease between identical / monozygotic (MZ) twins (siblings who are essentially genetically identical) and fraternal /dizygotic (DZ) twins (who share on average half of their genes). Assuming that shared environmental influences on MZ twins are not different from environmental influences on DZ twins (the equal environments assumption), significantly higher concordance rates in MZ twins reflect the action of genes. Nevertheless, an MZ concordance rate less than 100% means that environmental factors influence the phenotype. The importance of genetic variants in FA stems from twin studies. A child with a parent or sibling with peanut allergy, one of the most common forms of FA, has a 7 times higher risk of having the condition than do children without familial risk factors.55 Twin studies have demonstrated that the concordance rate of 82% among monozygotic twins in developing peanut allergy far exceeds the concordance rate of 20% observed among di-zygotic twins .56,57 In general, the heritability estimates for FA is as high as 81%.58
Association-based studies
Successful association studies require adequate sample sizes, and this can be difficult to achieve in GWAS studies in particular, where multiple testing corrections increases the number of participants required to reach significance. It is also important to have well characterized study populations with regard to demographics, and when recruiting from ethnically or racially heterogenous populations, this needs to be adjusted for in the statistical analyses using population stratification.
Accurate and consistent phenotyping of FA is essential to the success of gene association studies since differences in FA definitions and diagnostic methods can have a considerable impact on the results. FA prevalence vary widely depending on the diagnostic criteria, as demonstrated in a systematic review of European studies,59 which found that the point prevalence of self-reported FA was 6-7 times higher than FA confirmed by oral food challenge (OFC). OFC in a clinical setting remains the gold standard for diagnosing FA, but while OFC is considered generally safe, it is not entirely risk-free, and can be associated with considerable discomfort for the patient.60 A positive SPT and elevated specific IgE levels have low predictive value on their own but can confirm the IgE component of possible FA in the presence of a convincing clinical history. Greater SPT wheal size or higher specific IgE levels are associated with increased risk of FA, and a careful evaluation of clinical history together with a positive SPT wheal size or IgE levels above suggested cutoff levels may reduce or eliminate the need for OFC.61
Compared with the genetics of asthma, the genetics of FA is still a somewhat underexplored area. Studies with large and well phenotyped cohorts are needed to confirm results from earlier studies and to identify additional genetic associations, as well as to explore interactions with environmental factors. The results can help increase the understanding of the mechanisms behind the FA, and lead to the development of biomarkers for prediction and monitoring of FA.
Candidate-gene studies
Until recently, studies of FA-gene associations were mainly performed on specific candidate genes based on their known biological function. Hypothesis-driven studies of gene associations with food allergy have targeted well-known immune-related genes, some of which have previously been associated with other allergic diseases. A recent systematic review summarized results from both candidate gene association studies and genome-wide scans with different types of FA as outcomes.62 Some of the genes that have been associated with FA in more than one study will be discussed. The human leukocyte antigen (HLA) system is important for the regulation the immune system and is encoded by a family of genes located in the major histocompatibility complex (MHC) gene complex on chromosome 6p21. Several HLAs belonging to the MHC class II (DP, DM, DO, DQ, and DR), which present antigens from outside of the cell to T-lymphocytes, have been associated with various forms of asthma in a number of studies.63 Several class II genotypes have also been shown to be associated with FA, and PA in particular. In a small study of based on peanut SPT and clinical history of peanut allergy, the genotypes DRB1*08, DRB1*08/12 tyr16, and DQB1*04 were found to be more frequent in peanut-allergic individual compared with controls.64 The association between HLA DQB1 and peanut allergy was also demonstrated in a Canadian study that found associations between it and DQB1*02 and DQB1*06:03P in a study population of European ancestry.
CD14 plays an important role in the innate immunity system as part of the protein complex that binds to lipopolysaccharide, a bacterial cell wall component. Several studies have examined CD14 as a candidate gene for FA. The CD14 SNP rs2569190 was associated with general FA in a racially mixed study population, and the association was found to be stronger when the analysis was restricted to white participants.65 In a study of 53 children with peanut allergy and their peanut-tolerant siblings, rs2569190 was significantly associated with peanut allergy .66 By contrast, the same SNP was not associated with FA in a Japanese study population.67
Interleukin-13 is a cytokine secreted by activated Th2 cells that plays a central role in allergic disease, and the association between the IL-13 gene and asthma is well established. The association between IL-13 and FA was explored in an Australian study of challenge-proven food allergic infants of European ancestry. The IL-13 SNP rs1295686 was associated with FA in the discovery cohort as well as in an independent validation cohort and in a meta-analysis.68 Moreover, the association appeared to be independent on the presence of eczema. Interestingly, no evidence that any of the variants tested increased the risk for FA when comparing food allergic cases to food-sensitized tolerant children, and there was an association with increased plasma IgE levels, which suggests that the association between IL-13 and FA may be mediated by food sensitization. In a Japanese study of associations between FA and 26 loci previously linked to AD and EoE, an association between rs1295686 was seen regardless of eczema comorbidity.69
Several studies have found associations between FA and signal transducer and activator of transcription 6 (STAT6), a gene encoding a transcription factor that plays a role in IL4-mediated responses. STAT6 genotype has been shown to be associated with general nut allergy in a study using participants of European ancestry,70 and the association was later validated at the gene level in a Japanese population. The STAT6 SNPs rs324015 and rs1059513 were found to be associated with challenge-proven FA as well as peanut allergy in a family study of 369 trios that included 262 children with FA. Both SNPs were also associated with more severe FA symptoms. In a recent case-control study of a West Bengal Indian population the STAT6 SNP rs3024974 was not significantly associated with FA. There was, however, an association with food-specific IgE levels in individuals with childhood onset, but not adult onset, of FA.71
As discussed above, entry through an impaired skin barrier is believed to be a common route of sensitizing food allergens, and genes involved in maintenance of the skin barrier have been investigated for associations with FA. The barrier gene filaggrin (FLG),in particular, has been investigated as a candidate gene and found to be significantly associated with peanut allergy72 and general FA.73 In the latter study, path analysis indicated that the effect of FLG-LOF mutations on FA risk was indirect and mediated by eczema and allergic sensitization to foods, suggesting that the association between FLG and FA can be explained by the increased risk of sensitization through a faulty skin barrier in individuals with FLG mutations. Similarly, in a study of peanut allergy, the association between FLG-LOF mutations and FA-sensitized but tolerant children was similar to the association between FLG-LOF mutations and food-sensitized children with FA, though the power to detect a difference was somewhat limited.74
As already mentioned, the gene product of SPINK5, LEKTI, is involved in the maintenance of the skin barrier, and several mutations in the SPINK5 gene have been associated with Netherton syndrome. In addition, the SPINK5 variant rs9325071 has been found to be associated with challenge-proven FA in an Australian candidate gene study of 12-month old infants, and the association was replicated in an independent study population.75 Notably, the association remained in both study populations when the analysis was restricted to children without eczema.
Genome-wide association studies
The strength of genome-wide agnostic approaches is the ability to identify novel loci without an a priori biological hypothesis. Corrections for testing very high numbers of loci can, however, make it difficult to reach statistical significance, and necessitates large study populations. Recently, several GWAS of FA has confirmed some associations with genes previously identified using candidate-gene studies, and in addition, novel candidate loci have been discovered. The first GWAS to specifically investigate FA, as well as peanut, milk, and egg allergy, confirmed the association with HLA-DR and HLA-DQ in peanut allergy and identified specific loci in a region at 6p21.32 tagged by rs7192 and rs9275596.76 These associations were replicated in individuals of European, but not non-European, ancestry. Interestingly, in a genome-wide scan of DNA methylation it was found that differential DNA methylation at 72 different loci were associated with one or both of rs7192 and rs9275596, and, furthermore, there was a difference in methylation levels between peanut allergy cases and controls at 18 of the differentially methylated positions. Further evidence of a role for HLA-DQ in peanut allergy came from a German GWAS of FA diagnosed by OFC in 497 cases and 2387 controls of European ancestry.77 FA was stratified by food-specific allergy (egg, peanut, and milk), and it was found that loci centered on rs9273440 in the HLA-DQB untranslated 3’-region was specifically associated with peanut allergy. By contrast, loci in the epidermal differentiation near the filaggrin genes on 1q21.3 was associated with any FA unstratified for type, and the association was shown to be due to known LOF mutations in FLG. Loci in Serpin Family B Member 7 (SERPINB7), a region centered on rs11949166 between the interleukin 4 gene (IL4) and the kinesin family member 3a gene (KIF3A), and the locus were also associated with risk for any FA. All three loci have previously been implicated in allergic disease. Notably, the associations with FA was observed regardless of the presence of eczema in all 5 loci identified in the study except C11orf30/LRRC32.
To explore novel gene associations with peanut allergy, Asai et al. recently performed a GWAS on a Canadian group of patients as well as a meta-analysis of 7 studies from Canadian, American, Australian, German, and Dutch populations of varied ethnicity.78,79 In the main GWAS analysis, only HLA SNPs and rs115218289, an imputed SNP located close to Integrin α6 (ITGA6), reached genome-wide significance. Conditioning on the top HLA SNP located upstream of HLA-DQB1, rs3134976, identified additional SNPs near the T cell adapter protein Src Kinase Associated Phosphoprotein 1 (SKAP1) and Catenin Alpha 3 (CTNNA3). In the meta-analysis of any FA, rs7936434 near C11orf30 reached genome-wide significance. Loci that were suggestive of significance with both FA and peanut allergy in the meta-analyses included rs115218289, rs523865 in Angiopoietin-4 (ANGPT4), rs144897250 near Matrix metalloproteinase-12 (MMP12) and Matrix metalloproteinase-13 (MMP13), and rs78048444 near Coiled-Coil-Helix-Coiled-Coil-Helix Domain Containing 3 (CHCHD3) and Exocyst complex component 4 (EXOC4). There were variations in phenotype definition ranging from self-report to food challenges among the studies included in the meta-analyses. Notably, the results differed between populations, and the association between FA and rs7936434 when the study using an FA definition based on self-report was excluded.
One genome-wide study of FA associations made use of copy number variations (CNVs), which are common genomic alterations, as molecular markers.80 CNVs in the cell adhesion gene Catenin Alpha 3 (CTNNA3) were significantly associated FA in a discovery cohort of 357 cases with confirmed FA and 3980 controls, and the association was confirmed in an independent replication cohort. CNVs in Fox-1 homolog A (RBFOX1) were significantly associated FA in a meta-analysis of participants of European ancestry. Figure 2 lists variants and genes for which statistically significant (or suggestive) association has been found in several independent studies with food allergies, yet the truth of whether these studies are associated with, let alone cause, FA still remains questionable in most cases.
Figure 2.
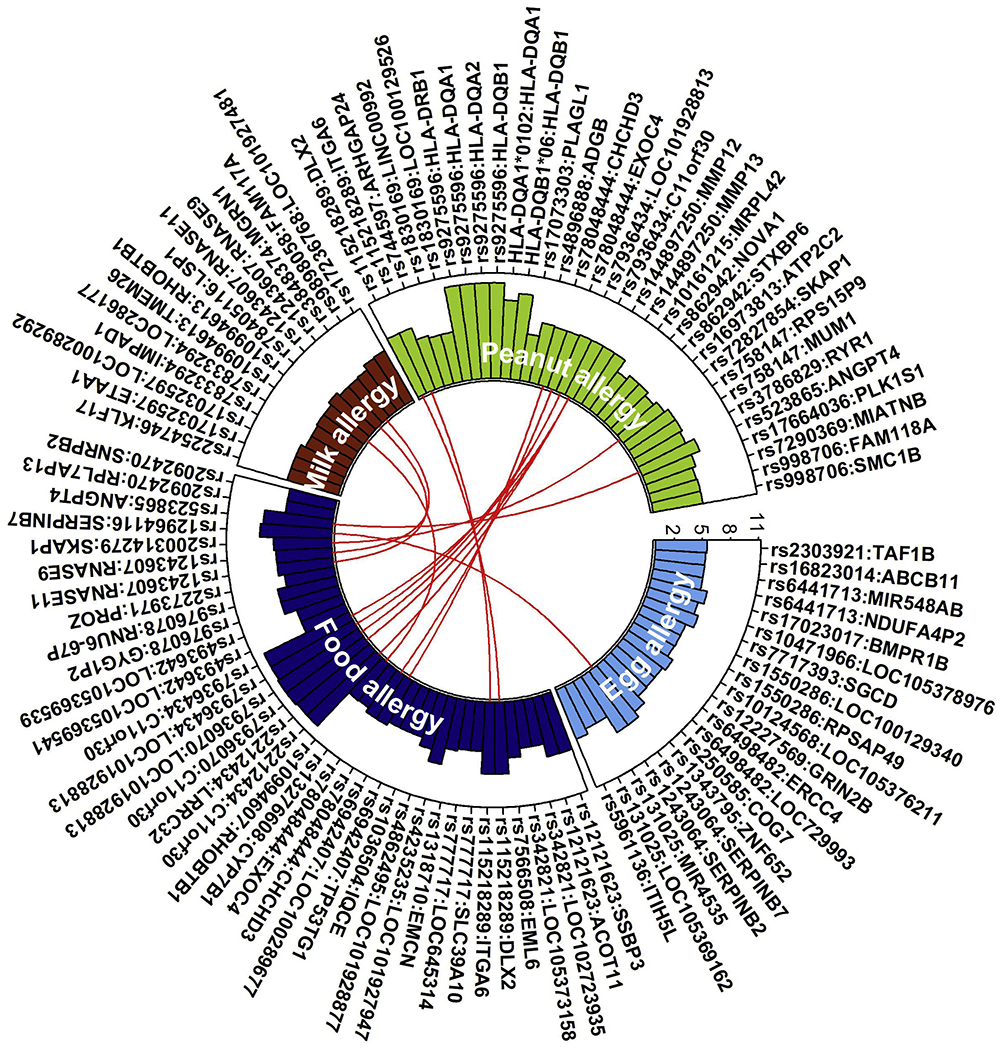
Variants associated with food allergies. Circle plot based on NHGRI-EBI Catalog shows genome-wide association studies (GWAS) variants associated with food allergies. SNPs and the corresponding mapped genes are shown along the outer circle. The rectangles show the log transformed p-value from the SNP-trait association. Y-axis is the −log10(p-value), X-axis is the SNP-gene pair for each disease. Red lines connect the shared gene (regardless of SNP) between traits.
Epigenetics and contributions from the environment in FA
The dramatic rise in FA cases over the past 50 years is more likely due to alterations in the environment rather than to changes in the gene pool. Environmental factors such as the physical environment, the social environment, and the economic environment have all presumably contributed to an increase in the prevalence of FA. The association of putatively influential environment factors on FA has been demonstrated in studies on twins with a high genetic predisposition to FA. Further evidence of environmental effects has come from studies showing that emigrants to the United States usually showed marked differences in the incidence of allergy compared to their counterparts who remained in their native countries. These observations also show the relationship between our genes and the environment.
Among environmental factors implicated in FA are dietary habits, vitamin D intake, exposure to food allergens, pollution, and hygiene-related factors, including pet exposure.60,81 The gut microbiota is part of the total exposome, and is influenced by components in the external environment, such as diet and the environmental microbiome. Early life dysbiosis has been associated with FA,82 and further evidence of a role for the gut microbiota in FA has been provided by studies in mice (40, 41, 42). Environmental factors can modify risk of disease in genetically predisposed individuals, and gene-environment (GxE) interactions are believed to be important for the risk of allergic disease (Figure 3). Although GxE interactions have been studied extensively in asthma,83,84 the literature is more sparse for other allergic diseases, and very few studies on interactions between genes and environmental factors in FA have been published to date. The association between challenge-proven FA and low serum 25-hydroxyvitamin D4 levels have been shown to be modified by a polymorphism in the vitamin D–binding protein (DBP) in such a way that the association was only found in the genotype associated with higher serum DBP levels.85 Since high DBP levels decrease the vitamin D bioavailability, the results are consistent with vitamin D deficiency as a risk factor for FA. Another example of an interaction between genotype and an environmental variable is a study of peanut allergy which demonstrated a modifying effect of peanut allergen levels in dust on the association between FLG and both peanut sensitization and allergy.86 Interestingly, the interaction was significant when adjusted for atopic dermatitis.
Figure 3.
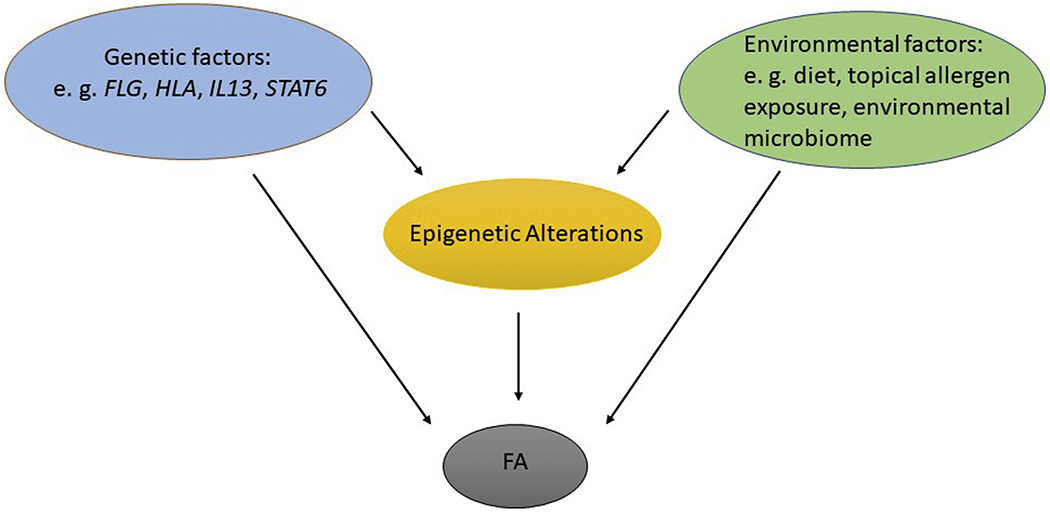
Overview of possible pathways to food allergy (FA), based on genetic and environmental risk factors.
In recent years, evidence has accumulated indicating that environmentally induced epigenetic changes in the DNA of target genes is an important mechanism mediating GxE interactions. In particular, DNA methylation at loci of genetic variation can lead to a variable response to an environmental exposure depending on genotype. Although there are few demonstrated examples of associations between FA and genotypes being modified by environmental factors, several studies have linked DNA methylation to the risk of FA. As already mentioned above, two SNPs in the HLA-DQ and -DR genomic region significantly associated with peanut allergy in a GWAS were also found to be associated with differential methylation at multiple CpG sites.76 In an epigenome-wide association study of methylation in CD4+ T-cells from 12 children with FA to egg or peanut and 12 age-matched controls, 153 CpG loci differentially methylated at birth and 179 differentially methylated at 12 months of age were identified.87 A total of 136 loci were common to both time points. Of these, 44 loci were located within 10 base pairs of single nucleotide polymorphisms, and these loci were annotated to HLA-DQB1. Differentially methylated loci that were not associated with SNPs were annotated to 49 different genes. These were subjected to pathways enrichment analysis, which suggested enrichment of the KEGG pathway “MAPK signaling pathway”, a pro-inflammatory pathway with many different roles, including T cell development and differentiation.
A recent study explored transcriptomic and CD4+ T-cell epigenetic associations with reaction severity in 21 peanut-allergic children during double-blinded peanut challenges.88 Among genes for which expression changes during challenge was significantly associated with reaction severity, 318 were replicated in an independent cohort of 19 peanut-allergic children. Gene ontology analysis on the 318 genes found them to be associated with regulation of nitric oxide processes, neutrophil activation, and macrophage activation, among other processes. Moreover, the association between reaction severity and 203 differentially methylated CpG sites were replicated in the independent cohort, and these CpG sites mapped to 197 unique genes. Four of the severity-associated CpG sites were located within 10Mb of a gene with severity-associated expression, and causal mediation analysis indicated that expression of PHACTR1 and ZNF121 mediates the association between reaction severity and methylation of the associated CpGs. Interaction networks with the differentially expressed genes and the severity associated CpGs identified NFKBIA and ARG1 as key hubs.
DNA methylation has been shown to be associated with immune tolerance in children undergoing oral immunotherapy for peanut allergy. In a study of 23 patients with peanut allergy undergoing oral immunotherapy and 20 controls with peanut allergy receiving standard of care and abstaining from peanut, participants who were immune tolerant to peanut 3 months after therapy withdrawal had decreased methylation of FOXP3 CpG sites in antigen-induced regulatory T cells relative to the baseline before immunotheraphy.89 No change in methylation was seen in controls or participants who remained non-tolerant after the treatment period. The changes in methylation in immune tolerant participants were accompanied by increased expression of FOXP3 in antigen-induced regulatory T cells. The results add support to previous studies showing a role for regulatory T cells in the response to immunotherapy.90
Genome-wide DNA methylation profiling has also been used to develop signatures that can predict clinical outcomes of FA. Using DNA from peripheral blood mononuclear cells in whole blood, a signature of 96 CpG sites was found to predict response to oral food challenge better than skin prick testing and allergen-specific IgE levels in 58 food-sensitized children aged 11-15 months and 13 non-allergic controls.91 The results were validated using CD4+ T cells from an independent cohort. Studies of epigenetic changes in FA can contribute to our understanding of the pathways involved in the development of FA, and the interplay between genes, epigenetics, and environmental factors. More studies are needed to validate and replicate earlier results, and to explore the role of environmental factors in triggering changes in methylation patterns and other epigenetic alterations.
Genomic information is measurable and well established while phenotyping and environmental exposures are less well standardized. As we move forward, deep phenotyping (e.g., oral food challenge (OFC)) will be limiting. In the future, multi-layer and longitudinal data, detailed measures of environmental exposures, detailed and standardized clinical information, combine genetic, race, clinical and environmental information as well as GxE interactions in risk prediction and leveraging big genomic data in racially diverse population could pave the way to personalized genomic medicine in cost effective way.92 Precision medicine in FA requires precise inference of genetic ancestry.
FUTURE RESEARCH AND DIRECTIONS
Rapid progress in molecular genetics has increased our knowledge of the genomes of humans and other organisms and led to more detailed research into the genetic elements of allergy. Common human diseases such as FA are complicated by multiple factors (Box 1): phenocopies, genetic heterogeneity (both locus heterogeneity and allelic heterogeneity), trait heterogeneity, gene-gene interactions, gene-environment interactions, and factors such as admixture.93–95 Future research into FA can be expected to include analyses of gene-gene and gene-environment interactions as well as transcriptome-wide expression studies that estimate the differences in the expression of genes under diverse environmental conditions. Gene-gene interactions, also known as epistasis, refers to the expression of phenotypes only when specific alleles of two or more genes are present. Additionally, genomic studies have been expanded to include variations in DNA structure (Box 1).
Admixture mapping
It is well known that disease does not affect populations equally. This is because different populations are subject to distinct environmental exposure. Natural selection during the out-of-Africa expansion96 may produce population-specific allele frequencies. Assessing variation in the rates of disease according to demographic factors, such as race or ethnicity, is the basis of epidemiologic research and affects clinical and public health practice.97 The association between increased FA risk and African ancestry and the admixed nature of the African-American population suggests that admixture mapping might be an important FA gene-finding strategy to study directly genetically heterogeneous populations of African American nature.98 Admixture mapping involves screening the genome of individuals of mixed ancestry who have a disease for chromosomal regions that have a greater percentage of alleles from the parental population with the higher disease risk.
Multi-omics approach
With the advent of omics-based big data-driven and unbiased approaches to define and characterize endotypes, including more accessible tools for human immunophenotyping, we can now gather detailed molecular information to de-convolute and identify patterns from the data, and gather further insights into the biology of diseases and health states of individual patients.99,100 Statistical methods to integrate multi-omics data are emerging to provide important insights into disease pathophysiology of allergic diseases.101 For example, machine learning approach uses computer algorithms to identify patterns from the systematically collected large molecular profile data, and along with clinical metadata, can assist personalized treatments for effective management of food allergies with similar molecular subtypes.102,103 The increasing focus on multi-omics is expected to play an important role in the development of personalized medicine approaches that takes racial ancestry into account. It needs to be stressed, however, that any racial differences identified using proteomics, transcriptomics, or epigenetics can be expected to be modified or confounded by environmental factors associated with ancestry, given the profound influence of the environment on epigenetics and gene regulation.104 Genomics can provide deep insights into the genetic drivers of food allergy, while transcriptomics sheds light on dysregulated gene regulation, and proteome provides deeper insight into what is going on at the molecular, tissue, and whole-body level.
Although racial disparities have been noted in the prevalence of FA, determining the relative contribution of ancestry-specific genetic risk factors from environmental factors has proved to be challenging because of the limited number of studies performed on African American and other minority populations.105 The delineation and deconstruction of shared and unique biologic and genetic pathways among atopic disorders and ancestry-specific gene-environment interactions can help resolve the clinical complexity and better inform the development of novel therapies.
Future research should include deep phenotyping of diverse ancestral populations, better characterization of environmental determinants, and the application of new technologies utilizing “-omics” tools. These include next-generation sequencing, epigenetics, and eQTL approaches in appropriate tissues/cells along with publicly available bioinformatics and ancestry tools. Systematic integration of “big data” coming from providers (e.g., EMR), from omics (e.g., genomic, proteomic, epigenomic, metabolomic), and from multi-ethnic patients and non-providers (e.g., smart phone, monitoring tools for environmental triggers) can thus provide valuable insights to resolve the clinical complexity and ancestry-specific (or shared) etiology of food allergy.106–109
Synopsis.
The risk factors for food allergy (FA) include both genetic variants and environmental factors. Advances using both candidate-gene association studies and genome-wide approaches have led to the identification of FA-associated genes involved in immune responses and skin barrier functions. Epigenetic changes have also been associated with the risk of FA. Little is known about possible modifying effects of genetic variation on the associations between environmental factors and FA. In this chapter, we outline current understanding of the genetics, epigenetics and the interplay with environmental risk factors associated with FA. Future studies of gene-environment interactions, gene-gene interactions, and multi-omics integration may help shed light on the mechanisms of FA, and lead to improved diagnostic and treatment strategies.
Key Points.
Food allergy (FA) is a growing clinical and public health problem in the U.S. and worldwide.
The development of FA likely results from complex interactions between multiple environmental and genetic factors.
Although the genetics of FA are somewhat understudied, advances have been made in recent years using both candidate gene association studies and agnostic genome-wide approaches.
Most genes found to be associated with FA are involved either in immune responses or in skin/epithelial barrier functioning, with filaggrin and genes in the human leukocyte antigen complex being among the most studied.
Clinics Care Points.
Food allergies are increasing and are a global health problem.
IgE-mediated food allergies are the most common type of allergic reaction to food.
Multiple studies suggest higher rate of food allergen sensitization in African American than European American children.
Risk factors for the development of food allergy include parental history, atopic diseases particularly atopic dermatitis (AD), which can lead to the development of the atopic march through mechanisms of cutaneous sensitization.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
The Authors have nothing to disclose.
References
- 1.Taylor SL. Review of the development of methodology for evaluating the human allergenic potential of novel proteins. Molecular nutrition & food research. 2006;50(7):604–609. [DOI] [PubMed] [Google Scholar]
- 2.Gupta RS, Springston EE, Warrier MR, et al. The prevalence, severity, and distribution of childhood food allergy in the United States. Pediatrics. 2011;128(1):e9–17. [DOI] [PubMed] [Google Scholar]
- 3.Sicherer SH, Sampson HA. 9. Food allergy. J Allergy Clin Immunol. 2006;117(2 Suppl Mini-Primer):S470–475. [DOI] [PubMed] [Google Scholar]
- 4.Sicherer S. Understanding and Managing Your Child’s Food Allergies. Baltimore, MD: The Johns Hopkins University Press. 2006. [Google Scholar]
- 5.Gupta R, Holdford D, Bilaver L, Dyer A, Holl JL, Meltzer D. The economic impact of childhood food allergy in the United States. JAMA pediatrics. 2013;167(11):1026–1031. [DOI] [PubMed] [Google Scholar]
- 6.Sicherer SH. Clinical implications of cross-reactive food allergens. The Journal of allergy and clinical immunology. 2001; 108(6):881–890. [DOI] [PubMed] [Google Scholar]
- 7.Nowak-Wegrzyn A, Sampson HA. Adverse reactions to foods. The Medical clinics of North America. 2006;90(1):97–127. [DOI] [PubMed] [Google Scholar]
- 8.Sampson HA. Peanut anaphylaxis. The Journal of allergy and clinical immunology. 1990;86(1):1–3. [DOI] [PubMed] [Google Scholar]
- 9.Burks AW, Sampson H. Food allergies in children. Current problems in pediatrics. 1993;23(6):230–252. [DOI] [PubMed] [Google Scholar]
- 10.Burks AW, Sampson HA,Buckley RH. Anaphylactic reactions after gamma globulin administration in patients with hypogammaglobulinemia. Detection of IgE antibodies to IgA. The New England journal of medicine. 1986;314(9):560–564. [DOI] [PubMed] [Google Scholar]
- 11.Strober W Gluten-sensitive enteropathy: a nonallergic immune hypersensitivity of the gastrointestinal tract. The Journal of allergy and clinical immunology. 1986;78(1 Pt 2):202–211. [DOI] [PubMed] [Google Scholar]
- 12.Madsen C Prevalence of food allergy: an overview. The Proceedings of the Nutrition Society. 2005;64(4):413–417. [DOI] [PubMed] [Google Scholar]
- 13.Sampson HA. Update on food allergy. The Journal of allergy and clinical immunology. 2004;113(5):805–819; quiz 820. [DOI] [PubMed] [Google Scholar]
- 14.Tordesillas L, Berin MC. Mechanisms of Oral Tolerance. Clin Rev Allergy Immunol. 2018;55(2):107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noval Rivas M, Chatila TA. Regulatory T cells in allergic diseases. J Allergy Clin Immunol. 2016;138(3):639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tordesillas L, Berin MC, Sampson HA. Immunology of Food Allergy. Immunity. 2017;47(1):32–50. [DOI] [PubMed] [Google Scholar]
- 17.Taylor SL. Food allergies and sensitivities. Journal of the American Dietetic Association. 1986;86(5):599–600. [PubMed] [Google Scholar]
- 18.Chehade M IgE and non-IgE-mediated food allergy: treatment in 2007. Current opinion in allergy and clinical immunology. 2007;7(3):264–268. [DOI] [PubMed] [Google Scholar]
- 19.Bruijnzeel-Koomen C, Ortolani C, Aas K, et al. Adverse reactions to food. European Academy of Allergology and Clinical Immunology Subcommittee. Allergy. 1995;50(8):623–635. [DOI] [PubMed] [Google Scholar]
- 20.Werfel T Epicutaneous allergen administration: a novel approach for allergen-specific immunotherapy? The Journal of allergy and clinical immunology. 2009;124(5):1003–1004. [DOI] [PubMed] [Google Scholar]
- 21.Boyce JA, Assa’ad A, Burks AW, et al. Guidelines for the diagnosis and management of food allergy in the United States: summary of the NIAID-sponsored expert panel report. Nutrition research. 2011;31(1):61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyce JA, Assa’ad A, Burks AW, et al. Guidelines for the Diagnosis and Management of Food Allergy in the United States: Summary of the NIAID-Sponsored Expert Panel Report. The Journal of allergy and clinical immunology. 2010;126(6):1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods RK, Stoney RM, Raven J, Walters EH, Abramson M, Thien FC. Reported adverse food reactions overestimate true food allergy in the community. European journal of clinical nutrition. 2002;56(1):31–36. [DOI] [PubMed] [Google Scholar]
- 24.Miles S, Fordham R, Mills C, Valovirta E, Mugford M. A framework for measuring costs to society of IgE-mediated food allergy. Allergy. 2005;60(8):996–1003. [DOI] [PubMed] [Google Scholar]
- 25.Tsakok T, Marrs T, Mohsin M, et al. Does atopic dermatitis cause food allergy? A systematic review. J Allergy Clin Immunol. 2016;137(4):1071–1078. [DOI] [PubMed] [Google Scholar]
- 26.Du Toit G, Sayre PH, Roberts G, et al. Effect of Avoidance on Peanut Allergy after Early Peanut Consumption. N Engl J Med. 2016;374(15):1435–1443. [DOI] [PubMed] [Google Scholar]
- 27.Schmiechen ZC, Weissler KA, Frischmeyer-Guerrerio PA. Recent developments in understanding the mechanisms of food allergy. Curr Opin Pediatr. 2019;31(6):807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brough HA, Nadeau KC, Sindher SB, et al. Epicutaneous sensitization in the development of food allergy: What is the evidence and how can this be prevented? Allergy. 2020;75(9):2185–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paller AS, Spergel JM, Mina-Osorio P, Irvine AD. The atopic march and atopic multimorbidity: Many trajectories, many pathways. J Allergy Clin Immunol. 2019;143(1):46–55. [DOI] [PubMed] [Google Scholar]
- 30.Kelleher MM, Dunn-Galvin A, Gray C, et al. Skin barrier impairment at birth predicts food allergy at 2 years of age. J Allergy Clin Immunol. 2016;137(4):1111–1116 e1118. [DOI] [PubMed] [Google Scholar]
- 31.Johansson EK, Bergstrom A, Kull I, et al. IgE sensitization in relation to preschool eczema and filaggrin mutation. J Allergy Clin Immunol. 2017;140(6):1572–1579 e1575. [DOI] [PubMed] [Google Scholar]
- 32.Chavanas S, Bodemer C, Rochat A, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25(2):141–142. [DOI] [PubMed] [Google Scholar]
- 33.Hannula-Jouppi K, Laasanen SL, Heikkila H, et al. IgE allergen component-based profiling and atopic manifestations in patients with Netherton syndrome. J Allergy Clin Immunol. 2014;134(4):985–988. [DOI] [PubMed] [Google Scholar]
- 34.Samuelov L, Sarig O, Harmon RM, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45(10):1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Has C, Jakob T, He Y, Kiritsi D, Hausser I, Bruckner-Tuderman L. Loss of desmoglein 1 associated with palmoplantar keratoderma, dermatitis and multiple allergies. Br J Dermatol. 2015;172(1):257–261. [DOI] [PubMed] [Google Scholar]
- 36.Carter CA, Frischmeyer-Guerrerio PA. The Genetics of Food Allergy. Curr Allergy Asthma Rep. 2018;18(1):2. [DOI] [PubMed] [Google Scholar]
- 37.Park JH, Lee KH, Jeon B, et al. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome: A systematic review. Autoimmun Rev. 2020;19(6):102526. [DOI] [PubMed] [Google Scholar]
- 38.Torgerson TR, Linane A, Moes N, et al. Severe food allergy as a variant of IPEX syndrome caused by a deletion in a noncoding region of the FOXP3 gene. Gastroenterology. 2007;132(5):1705–1717. [DOI] [PubMed] [Google Scholar]
- 39.Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, et al. TGFbeta receptor mutations impose a strong predisposition for human allergic disease. Sci Transl Med. 2013;5(195):195ra194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arias K, Waserman S, Jordana M. Management of food-induced anaphylaxis: unsolved challenges. Current clinical pharmacology. 2009;4(2):113–125. [DOI] [PubMed] [Google Scholar]
- 41.Celedon JC, Sredl D, Weiss ST, Pisarski M, Wakefield D, Cloutier M. Ethnicity and skin test reactivity to aeroallergens among asthmatic children in Connecticut. Chest. 2004;125(1):85–92. [DOI] [PubMed] [Google Scholar]
- 42.Liu AH, Jaramillo R, Sicherer SH, et al. National prevalence and risk factors for food allergy and relationship to asthma: results from the National Health and Nutrition Examination Survey 2005-2006. The Journal of allergy and clinical immunology. 2010;126(4):798–806 e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vergara C, Caraballo L, Mercado D, et al. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean Coast of Colombia. Human genetics. 2009;125(5-6):565–579. [DOI] [PubMed] [Google Scholar]
- 44.Cardoso BA, Martins LR, Santos CI, et al. Interleukin-4 stimulates proliferation and growth of T-cell acute lymphoblastic leukemia cells by activating mTOR signaling. Leukemia. 2009;23(1):206–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wegienka G, Sitarik A, Bassirpour G, et al. The associations between eczema and food and inhalant allergen-specific IgE vary between black and white children. The journal of allergy and clinical immunology In practice. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Souef PN, Candelaria P, Goldblatt J. Evolution and respiratory genetics. The European respiratory journal. 2006;28(6):1258–1263. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson LA, Gergen PJ, Hoover DR, Rosenstreich D, Mannino DM, Matte TD. Sociodemographic correlates of indoor allergen sensitivity among United States children. The Journal of allergy and clinical immunology. 2001;108(5):747–752. [DOI] [PubMed] [Google Scholar]
- 48.Mahdavinia M, Fox SR, Smith BM, et al. Racial Differences in Food Allergy Phenotype and Health Care Utilization among US Children. The journal of allergy and clinical immunology In practice. 2017;5(2):352–357 e351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartman H, Dodd C, Rao M, et al. Parental timing of allergenic food introduction in urban and suburban populations. Annals of allergy, asthma & immunology: official publication of the American College of Allergy, Asthma, & Immunology. 2016;117(1 ):56–60 e52. [DOI] [PubMed] [Google Scholar]
- 50.Muers M Human disease: Edges, nodes and networks. Nature reviews Genetics. 2010;11(1):4. [DOI] [PubMed] [Google Scholar]
- 51.Kalow W. Pharmacogenetics and pharmacogenomics: origin, status, and the hope for personalized medicine. Pharmacogenomics J. 2006;6(3):162–165. [DOI] [PubMed] [Google Scholar]
- 52.Luccioli S, Ross M, Labiner-Wolfe J, Fein SB. Maternally reported food allergies and other food-related health problems in infants: characteristics and associated factors. Pediatrics. 2008;122 Suppl 2:S105–112. [DOI] [PubMed] [Google Scholar]
- 53.Bonini S, Ruffilli A. Genetics of food allergy. Environmental toxicology and pharmacology. 1997;4(1-2):71–78. [DOI] [PubMed] [Google Scholar]
- 54.Tsai HJ, Kumar R, Pongracic J, et al. Familial aggregation of food allergy and sensitization to food allergens: a family-based study. Clin Exp Allergy. 2009;39(1):101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hourihane JO, Dean TP, Warner JO. Peanut allergy in relation to heredity, maternal diet, and other atopic diseases: results of a questionnaire survey, skin prick testing, and food challenges. BMJ. 1996;313(7056):518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sicherer SH, Furlong TJ, Maes HH, Desnick RJ, Sampson HA, Gelb BD. Genetics of peanut allergy: a twin study. J Allergy Clin Immunol. 2000;106(1 Pt 1):53–56. [DOI] [PubMed] [Google Scholar]
- 57.Liu X, Zhang S, Tsai HJ, et al. Genetic and environmental contributions to allergen sensitization in a Chinese twin study. Clin Exp Allergy. 2009;39(7):991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242(1):10–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muraro A, Werfel T, Hoffmann-Sommergruber K, et al. EAACI food allergy and anaphylaxis guidelines: diagnosis and management of food allergy. Allergy. 2014;69(8):1008–1025. [DOI] [PubMed] [Google Scholar]
- 60.Sicherer SH, Sampson HA. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J Allergy Clin Immunol. 2018;141(1):41–58. [DOI] [PubMed] [Google Scholar]
- 61.Calvani M, Bianchi A, Reginelli C, Peresso M, Testa A. Oral Food Challenge. Medicina (Kaunas). 2019;55(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suaini NHA, Wang Y, Soriano VX, et al. Genetic determinants of paediatric food allergy: A systematic review. Allergy. 2019;74(9):1631–1648. [DOI] [PubMed] [Google Scholar]
- 63.Kontakioti E, Domvri K, Papakosta D, Daniilidis M. HLA and asthma phenotypes/endotypes: a review. Hum Immunol. 2014;75(8):930–939. [DOI] [PubMed] [Google Scholar]
- 64.Howell WM , Turner SJ, Hourihane JO, Dean TP, Warner JO. HLA class II DRB1, DQB1 and DPB1 genotypic associations with peanut allergy: evidence from a family-based and case-control study. Clin Exp Allergy. 1998;28(2):156–162. [DOI] [PubMed] [Google Scholar]
- 65.Woo JG, Assa’ad A, Heizer AB, Bernstein JA, Hershey GK. The -159 C-->T polymorphism of CD14 is associated with nonatopic asthma and food allergy. J Allergy Clin Immunol. 2003;112(2):438–444. [DOI] [PubMed] [Google Scholar]
- 66.Dreskin SC, Ayars A, Jin Y, Atkins D, Leo HL, Song B. Association of genetic variants of CD14 with peanut allergy and elevated IgE levels in peanut allergic individuals. Ann Allergy Asthma Immunol. 2011;106(2):170–172. [DOI] [PubMed] [Google Scholar]
- 67.Campos E, Shimojo N, Inoue Y, et al. No association of polymorphisms in the 5’ region of the CD14 gene and food allergy in a Japanese population. Allergol Int. 2007;56(1):23–27. [DOI] [PubMed] [Google Scholar]
- 68.Ashley SE, Tan HT, Peters R, et al. Genetic variation at the Th2 immune gene IL13 is associated with IgE-mediated paediatric food allergy. Clin Exp Allergy. 2017;47(8):1032–1037. [DOI] [PubMed] [Google Scholar]
- 69.Hirota T, Nakayama T, Sato S, et al. Association study of childhood food allergy with genome-wide association studies-discovered loci of atopic dermatitis and eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140(6):1713–1716. [DOI] [PubMed] [Google Scholar]
- 70.Amoli MM, Hand S, Hajeer AH, et al. Polymorphism in the STAT6 gene encodes risk for nut allergy. Genes Immun. 2002;3(4):220–224. [DOI] [PubMed] [Google Scholar]
- 71.Laha A, Ghosh A, Moitra S, et al. Association of the STAT6 rs3024974 (C/T) Polymorphism with IgE-Mediated Food Sensitization among West Bengal Population in India. Int Arch Allergy Immunol. 2020;181(3):200–210. [DOI] [PubMed] [Google Scholar]
- 72.Brown SJ, Asai Y, Cordell HJ, et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J Allergy Clin Immunol. 2011;127(3):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venkataraman D, Soto-Ramirez N, Kurukulaaratchy RJ, et al. Filaggrin loss-of-function mutations are associated with food allergy in childhood and adolescence. J Allergy Clin Immunol. 2014;134(4):876–882 e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan HT, Ellis JA, Koplin JJ, et al. Filaggrin loss-of-function mutations do not predict food allergy over and above the risk of food sensitization among infants. J Allergy Clin Immunol. 2012;130(5):1211–1213 e1213. [DOI] [PubMed] [Google Scholar]
- 75.Ashley SE, Tan HT, Vuillermin P, et al. The skin barrier function gene SPINK5 is associated with challenge-proven IgE-mediated food allergy in infants. Allergy. 2017;72(9):1356–1364. [DOI] [PubMed] [Google Scholar]
- 76.Hong X, Hao K, Ladd-Acosta C, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun. 2015;6:6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marenholz I, Grosche S, Kalb B, et al. Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat Commun. 2017;8(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Asai Y, Eslami A, van Ginkel CD, et al. A Canadian genome-wide association study and meta-analysis confirm HLA as a risk factor for peanut allergy independent of asthma. J Allergy Clin Immunol. 2018;141(4):1513–1516. [DOI] [PubMed] [Google Scholar]
- 79.Asai Y, Eslami A, van Ginkel CD, et al. Genome-wide association study and meta-analysis in multiple populations identifies new loci for peanut allergy and establishes C11orf30/EMSY as a genetic risk factor for food allergy. J Allergy Clin Immunol. 2018;141(3):991–1001. [DOI] [PubMed] [Google Scholar]
- 80.Li J, Fung I, Glessner JT, et al. Copy Number Variations in CTNNA3 and RBFOX1 Associate with Pediatric Food Allergy. J Immunol. 2015;195(4):1599–1607. [DOI] [PubMed] [Google Scholar]
- 81.Lieberman JA, Greenhawt M, Nowak-Wegrzyn A. The environment and food allergy. Ann Allergy Asthma Immunol. 2018;120(5):455–457. [DOI] [PubMed] [Google Scholar]
- 82.Stephen-Victor E, Crestani E, Chatila TA. Dietary and Microbial Determinants in Food Allergy. Immunity. 2020;53(2):277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johansson H, Mersha TB, Brandt EB, Khurana Hershey GK. Interactions between environmental pollutants and genetic susceptibility in asthma risk. Curr Opin Immunol. 2019;60:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morales E, Duffy D. Genetics and Gene-Environment Interactions in Childhood and Adult Onset Asthma. Front Pediatr. 2019;7:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koplin JJ, Suaini NH, Vuillermin P, et al. Polymorphisms affecting vitamin D-binding protein modify the relationship between serum vitamin D (25[OH]D3) and food allergy. J Allergy Clin Immunol. 2016;137(2):500–506 e504. [DOI] [PubMed] [Google Scholar]
- 86.Brough HA, Simpson A, Makinson K, et al. Peanut allergy: effect of environmental peanut exposure in children with filaggrin loss-of-function mutations. J Allergy Clin Immunol. 2014;134(4):867–875 e861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martino D, Joo JE, Sexton-Oates A, et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics. 2014;9(7):998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Do AN, Watson CT, Cohain AT, et al. Dual transcriptomic and epigenomic study of reaction severity in peanut-allergic children. J Allergy Clin Immunol. 2020;145(4):1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Syed A, Garcia MA, Lyu SC, et al. Peanut oral immunotherapy results in increased antigen-induced regulatory T-cell function and hypomethylation of forkhead box protein 3 (FOXP3). J Allergy Clin Immunol. 2014;133(2):500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ozdemir C, Kucuksezer UC, Akdis M, Akdis CA. Specific immunotherapy and turning off the T cell: how does it work? Ann Allergy Asthma Immunol. 2011;107(5):381–392. [DOI] [PubMed] [Google Scholar]
- 91.Martino D, Dang T, Sexton-Oates A, et al. Blood DNA methylation biomarkers predict clinical reactivity in food-sensitized infants. J Allergy Clin Immunol. 2015;135(5):1319–1328 e1311–1312. [DOI] [PubMed] [Google Scholar]
- 92.Coulson E, Rifas-Shiman SL, Sordillo J, et al. Racial, ethnic, and socioeconomic differences in adolescent food allergy. J Allergy Clin Immunol Pract. 2020;8(1):336–338 e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Savage JH, Lee-Sarwar KA, Sordillo J, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy. 2018;73(1):145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee TD, Gimenez G, Grishina G, Mishoe M, Sampson HA, Bunyavanich S. Profile of a milk-allergic patient who tolerated partially hydrolyzed whey formula. J Allergy Clin Immunol Pract. 2015;3(1):116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jin H, Sifers T, Cox AL, et al. Peanut Oral Food Challenges and Subsequent Feeding of Peanuts in Infants. J Allergy Clin Immunol Pract. 2020. [DOI] [PubMed] [Google Scholar]
- 96.Young JH, Chang YP, Kim JD, et al. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1(6):e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Risch N Dissecting racial and ethnic differences. N Engl J Med. 2006;354(4):408–411. [DOI] [PubMed] [Google Scholar]
- 98.Mersha TB. Mapping asthma-associated variants in admixed populations. Front Genet. 2015;6:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Holzinger A, Dehmer M, Jurisica I. Knowledge Discovery and interactive Data Mining in Bioinformatics--State-of-the-Art, future challenges and research directions. BMC Bioinformatics. 2014;15 Suppl 6:I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu KH, Snyder M. Omics Profiling in Precision Oncology. Mol Cell Proteomics. 2016;15(8):2525–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reinke SN , Gallart-Ayala H, Gomez C, et al. Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J. 2017;49(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Greene CS, Tan J, Ung M, Moore JH, Cheng C. Big data bioinformatics. Journal of cellular physiology. 2014;229(12):1896–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alag A Machine learning approach yields epigenetic biomarkers of food allergy: A novel 13-gene signature to diagnose clinical reactivity. PLoS One. 2019;14(6):e0218253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Watson CT, Cohain AT, Griffin RS, et al. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat Commun. 2017;8(1):1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barnes KC. Genomewide association studies in allergy and the influence of ethnicity. Current opinion in allergy and clinical immunology. 2010; 10(5):427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao W, Ho HE, Bunyavanich S. The gut microbiome in food allergy. Ann Allergy Asthma Immunol. 2019;122(3):276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Irizar H, Kanchan K, Mathias RA, Bunyavanich S. Advancing Food Allergy through Omics Sciences. J Allergy Clin Immunol Pract. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ho HE, Bunyavanich S. Role of the Microbiome in Food Allergy. Curr Allergy Asthma Rep. 2018;18(4):27. [DOI] [PubMed] [Google Scholar]
- 109.Fazlollahi M, Chun Y, Grishin A, et al. Early-life gut microbiome and egg allergy. Allergy. 2018;73(7):1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]


