Abstract
Objectives
The present study compared the performance of the Lumipulse G Sars-CoV-2 Ag kit with the TaqPath COVID-19 RT-PCR CE IVD kit.
Methods
The study was conducted on 4266 naso-oropharyngeal swabs. Samples were subjected to antigen RT-PCR tests for the detection of Sars-CoV-2 and related variants. Statistical analyses were conducted in R software.
Results
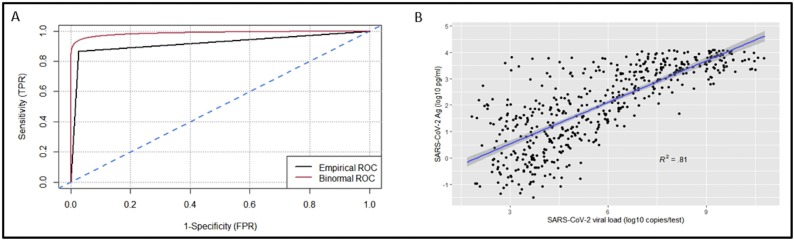
We found 503 positives (including 138 H69-V70 deletion carriers) and 3763 negatives by RT-PCR, whereas 538 positives and 3728 negatives were obtained by antigen testing. We achieved empirical and binormal AU-ROCs of 0.920 and 0.990, accuracy of 0.960, sensitivity of 0.866, specificity of 0.973, positive and negative predictive values of 0.810 and 0.980. We obtained a positive correlation between viral loads and antigen levels (R2 = 0.81), finding a complete concordance for high viral loads (log10 copies/mL > 5.4). Antigen levels > 222 pg/mL were found to be reliable in assigning positive samples (p < 0.01). Concerning variant carriers, antigen test detected them with the same accuracy as other positive samples.
Conclusions
Molecular and antigen tests should be evaluated regarding the prevalence of the area. In case of low prevalence, antigen testing can be employed as a first-line screening for the timely identification of affected individuals with high viral load, also if carriers of Sars-CoV-2 variants.
Keywords: COVID-19, Naso-oropharyngeal swabs, Antigen test, Viral load, Sars-CoV-2 variants, Screening
The pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (Sars-CoV-2) dramatically affects health and quality of life worldwide (Strafella et al., 2020a, Strafella et al., 2020b). To date, the gold standard test for Coronavirus-19 disease (COVID-19) diagnosis is represented by Real Time PCR (RT-PCR), which detects viral genome with high-sensitivity in naso-oropharyngeal swab fluids (Pascarella et al., 2020, Wiersinga et al., 2020). However, preanalytical and analytical issues, including long turnaround time, limit its use in specialized laboratories. Thus, novel laboratory and point-of-care tests for the detection of viral antigens have been developed in order to provide faster responses to a large number of individuals and to counteract the diffusion of infection. On this subject, a chemiluminescence-based assay has been recently developed for the quantitative measurement of SARS-CoV-2 N protein in swab and saliva (Lumipulse G SARS-CoV-2 Ag kit, Fujirebio). To date, comparative studies showed its high reliability (Gili et al., 2021, Hirotsu et al., 2020, Hirotsu et al., 2021), although the general application of antigen testing deserves to be further investigated and validated on larger cohorts, taking into account also the current spreading of novel, more infectious Sars-CoV-2 variants.
Hence, the study aimed to compare the performance of the quantitative antigen test Lumipulse G SARS-CoV-2 Ag kit (Fujirebio) and of the molecular test TaqPath COVID-19 RT PCR CE IVD kit (ThermoFisher Scientific).
The study was conducted on 4266 samples obtained from 2426 individuals, which were enrolled in the Scientific Institute for Research, Hospitalization and Healthcare (IRCCS) Santa Lucia Foundation from December 2020 to February 2021.
The research was approved by the local Ethics Committee and was performed according to the Declaration of Helsinki. All the recruited individuals signed written informed consents. Naso-oropharyngeal samples were obtained and tested by antigen and molecular tests in parallel (materials and methods are available in the Supplementary materials).
As a result, we detected 503 positive and 3763 negative samples by RT-PCR. This analysis allowed finding 138 samples with the H69-V70 deletion within the S gene, which may be suggestive of Sars-CoV-2 variants, particularly VOC 202012/01 (Bal et al., 2021).
Antigen testing, instead, detected 538 positive and 3728 negative samples, of which 436 were true positives and 102 false positives, whereas 3661 were true negative and 67 false negative results. We obtained an accuracy of 0.960, a specificity of 0.973, a sensitivity of 0.866, with positive and negative predictive values of 0.810 and 0.980, respectively. ROC curve analysis achieved an antigen test binormal and empirical AU-ROCs of 0.920 and 0.990, respectively (Figure 1 A).
Figure 1.
Comparison between antigen and molecular tests. (A) ROC curve analysis. (B) Correlation between antigen levels (pg/mL) and viral loads obtained by RT-PCR (log10 copies/mL).
We further analyzed the correlation among the viral loads obtained by RT-PCR and the antigen level reported as pg/mL, finding a positive correlation (R2 = 0.81, Figure 1B). Given these data, we subdivided our samples according to viral load ranges (Table 1 ) and assessed the accuracy of antigen testing for each range. A complete concordance was observed for samples with high viral load (log10 copies/mL > 5.4), whereas samples with lower concentrations of virus showed decreased accuracy (Table1). Subsequently, we splitted the samples according to the antigen level in order to identify a reliable range in which positive samples are correctly assigned by antigen testing.
Table 1.
Accuracy rates of antigen tests for different ranges of viral load. The viral load is reported in log10 (copies/mL). For each range, log10 (copies/mL), the total number of samples and the number of concordant and discordant samples are reported. The number of concordant and discordant variants, included in the total cohort, are extrapolated. N.: number.
| Viral load (log10 copies/mL) | N | N. of concordant samples (%) | N. of discordant samples (%) | Accuracy rate | N. of variants | N. of concordant variants | N. of discordant variants |
|---|---|---|---|---|---|---|---|
| >7.2 | 154 | 154 (100%) | 0 (0%) | 1.0 | 51 | 51 (100%) | 0 (0%) |
| 5.4–7.2 | 106 | 106 (100%) | 0 (0%) | 1.0 | 38 | 38 (100%) | 0 (0%) |
| 3.7–5.4 | 144 | 122 (85%) | 22 (15%) | 0.85 | 38 | 31 (82%) | 7 (18%) |
| 2.0–3.7 | 99 | 54 (54%) | 45 (46%) | 0.54 | 11 | 5 (46%) | 6 (54%) |
| Negatives | 3763 | 3661 (97%) | 102 (3%) | 0.97 | / | / | / |
As a result, antigen levels > 222 pg/mL showed the highest concordance rate (99%), indicating thereby the ability to correctly assign positive samples (p < 0.01). At lower antigens levels, instead, a decreased concordance rate (1.87–222 pg/mL = 76%; <1.87 pg/mL = 52%) was reported.
Interestingly, samples carrying the H69-V70 deletion were detected by antigen test with the same accuracy as other positive non-variant samples (Table1).
Overall, these results showed that samples with high viral load, including variant carriers, were successfully identified by antigen test, which reported high concentration results (Figure 1B). On the other hand, antigen detection revealed a reduced accuracy for samples with lower viral load, requiring the confirmation by RT-PCR.
Considering these data and evidence from literature (Hirotsu et al., 2020, Hirotsu et al., 2021), the use of molecular and antigen tests should be carefully evaluated regarding the prevalence of infection in a specific area. As shown by the high reliability in correctly identifying negative samples, quantitative antigen testing may be suitable as a first-line screening in low prevalence communities. In fact, antigen testing may allow a faster identification of individuals infected with a high viral load and/or carrying viral variants, which need to be identified in a timely manner and isolated in order to control viral spreading. Conversely, antigen testing has a limited utility in monitoring infection in positive patients and in high prevalence communities. As several positive individuals with variable viral loads are expected in these areas, molecular testing for Sars-CoV-2 infection should be recommended to avoid false negative results.
Authors’ contributions
Conception and design of the study: Valerio Caputo, Cristina Bax, Giulia Sancesario, Emiliano Giardina.
Acquisition, analysis and interpretation of data: Valerio Caputo, Cristina Bax, Luca Colantoni, Cristina Peconi, Andrea Termine, Carlo Fabrizio, Giulia Calvino, Laura Luzzi, Giorgia Panunzi, Claudia Fusco, Claudia Strafella, Raffaella Cascella.
Draft of the article: Valerio Caputo, Cristina Bax, Luca Colantoni, Cristina Peconi, Giulia Calvino, Laura Luzzi.
Critical revision of the manuscript: Luca Battistini, Carlo Caltagirone, Antonino Salvia, Giulia Sancesario and Emiliano Giardina.
All authors read and approved the final version of the manuscript.
Funding
The authors declare no funding.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.04.048.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gili A., Paggi R., Russo C., Cenci E., Pietrella D., Graziani A. Evaluation of automated test Lumipulse® G SARS-CoV-2 antigen assay for detection of SARS-CoV-2 nucleocapsid protein (NP) in nasopharyngeal swabs for community and population screening. Int J Infect Dis. 2021;105:391–396. doi: 10.1016/j.ijid.2021.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K. Prospective study of 1,308 nasopharyngeal swabs from 1,033 patients using the LUMIPULSE SARS-CoV-2 antigen test: comparison with RT-qPCR. Int J Infect Dis. 2021;105:7–14. doi: 10.1016/j.ijid.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascarella G., Strumia A., Piliego C., Bruno F., Del Buono R., Costa F. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strafella C., Caputo V., Termine A., Barati S., Caltagirone C., Giardina E. Investigation of genetic variations of IL6 and IL6R as potential prognostic and pharmacogenetics biomarkers: implications for COVID-19 and neuroinflammatory disorders. Life (Basel) 2020;10 doi: 10.3390/life10120351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strafella C., Caputo V., Termine A., Barati S., Gambardella S., Borgiani P. Analysis of ACE2 genetic variability among populations highlights a possible link with COVID-19-related neurological complications. Genes (Basel) 2020;11 doi: 10.3390/genes11070741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.