Abstract
The coronavirus disease 2019 causes a wide degree of organ dysfunction and is associated with bacterial secondary infections. We reported lung microbiota dynamics in a critically ill patient with coronavirus disease 2019, who developed severe Hafnia alvei ventilator-associated pneumonia and required extracorporeal membrane oxygenation support.
Keywords: ARDS, Critical care, Pneumonia, Sepsis, COVID-19, Hafnia Alvei
Abbreviations: ARDS, acute respiratory distress syndrome; BAL, bronchoalveolar lavage; COVID-19, coronavirus disease 2019; MV, mechanical ventilation
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic has been challenging patients, clinicians and healthcare systems worldwide [1]. Although mechanical ventilation (MV) represents a cornerstone of therapy for patients with acute respiratory distress syndrome (ARDS) caused by COVID-19 [2], such an intervention has been identified as a risk factor for ventilator-associated pneumonia (VAP) [3]. Specifically, COVID-19 may cause immune system dysfunction [4] and respiratory tract dysbiosis [5], both fostering the development of secondary infections [6].
In the light of this view, we sought to report lung microbiota dynamics in a critically ill patient with COVID-19, who required MV and developed Hafnia alvei VAP [7].
2. Material and methods
The study was approved by the human research ethics committee of the Fondazione Policlinico A. Gemelli IRCCS (reference number: 1847). The patient provided specific signed permission for the publication.
COVID-19 was diagnosed by reverse transcription-polymerase chain reaction, which was performed on nasopharyngeal swab at hospital admission. Bacterial identification on bronchoalveolar lavage (BAL) culture was performed by MALDI BioTyper® system (Bruker Daltonics, Bremen, Germany) and microbiota analysis was performed by sequencing of amplified 16S rRNA gene V3–V4 and V6 regions. Bacterial richness was assessed by Shannon's diversity index and bacterial equitability was evaluated by Pielou's evenness index.
3. Results
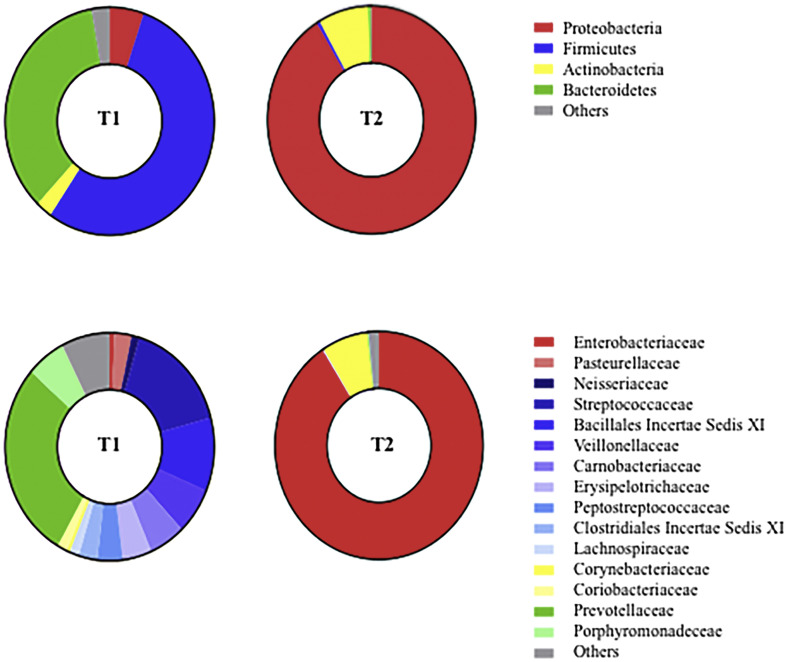
We reported the case of a 52-years-old male who was admitted to the ICU of a tertiary university hospital in Italy for ARDS caused by COVID-19 (eTable1). Although he received Dexamethasone and Remdesivir, invasive MV was necessary to improve life-threatening hypoxemia (eTable1). BAL was performed immediately after endotracheal intubation (T1, Fig. 1 ) and revealed no bacterial growth. Accordingly, empiric antibiotic therapy with Piperacillin-Tazobactam was stopped.
Fig. 1.
Lung microbiota characteristics of the patient. Two bronchoalveolar lavage samples, namely sample T1 (collected after endotracheal intubation) and T2 (collected at the diagnosis of ventilator-associated pneumonia) were comparatively analysed. A total of 41,529 (85% of total) and 32,148 (60% of total) good-quality reads from T1 and T2 were analysed to identify bacterial taxa.
Patient's clinical condition progressively worsened (eTable2) and veno-venous extracorporeal membrane oxygenation was commenced as a rescue organ support for severe ARDS (eFigure 1A). A second BAL (T2, Fig. 1) was performed and allowed the diagnosis of Hafnia alvei VAP (eTable3). Accordingly, empiric antibiotic therapy with intravenous Meropenem was deemed appropriate and continued until clinical improvement and microbiological eradication. Patient's clinical condition progressively improved (eFigure 1B) and he was discharged alive after 114 days of hospital stay (Supplementary eTable4).
The analyses of T1 and T2 (Fig. 1) provided important insight into microbiota dynamics and revealed significant dysbiosis associated with COVID-19 and Hafnia alvei VAP. Bacterial richness and equitability decreased from T1 to T2 (Shannon's diversity index: 3.898 for T1 and 0.522 for T2; Pielou's evenness index: 0.737 for T1 and 0.111 for T2), which corresponded to a transition from Firmicutes (54.2%) and Bacteroidetes (35.0%) predominance in T1 to Proteobacteria (91.1%) in T2.
4. Discussion
We reported clinical characteristics of a critically ill patient admitted to the ICU with ARDS caused by COVID-19, who developed Hafnia alvei VAP. Moreover, we described lung microbiota dysbiosis associated with COVID-19 and Hafnia alvei secondary infection.
Hafnia alvei is a gram-negative bacterium that belongs to Enterobacteriaceae and has been rarely reported as a pathogen that causes respiratory tract infections, especially in immunocompromised patients [7]. Recently, Blonz et al [3] observed a significant incidence of VAP caused by Hafnia alvei (3.9% of respiratory isolates) in critically ill patients with COVID-19. In this study [3], Enterobacteriaceae were isolated in 49.8% of respiratory samples, which may support the pathophysiological hypothesis of COVID-19 induced gut dysbiosis [8] and gut-lung crosstalk. Moreover, male sex represented a risk factor for the development of VAP, that may imply the influence of genetic and hormonal status on immune response. As a results, the emergence of rare infectious diseases in critically ill patients with COVID-19 may be favored by concurrent genetic [3], viral [4] and steroid [9] induced immune dysfunction, in a context of viral [8] and antibiotic induced multi-organ dysbiosis [10].
Our findings implicate that immune dysfunction fosters the development of rare and life-threatening bacterial infections. In this context, microbiota analysis represents a potential monitoring tool of dysbiosis, which may play a role in the prediction and early diagnosis of infectious diseases [10]. Moreover, such analysis may provide important information on the effect of multiple interventions (e.g. pharmacological therapies, MV setting and nutrition) on microbiota balance, in order to orient clinical decision-making towards strategies that allow native microbiota preservation.
This is the first report of Hafnia alvei infection and lung microbiota dysbiosis in a patient with COVID-19. Although our experience is restricted to one patient, Hafnia alvei infections and lung dysbiosis are rarely described in the literature and even a single, detailed, case scenario may be of interest when characterized by these important clinical implications.
5. Conclusion
Hafnia alvei was rarely associated with pneumonia in critically ill patients before COVID-19 pandemic. Immune dysfunction and dysbiosis caused by both COVID-19 and clinical interventions may justify the emergence of rare pathogenic threats. In this context, microbiota analysis represents a potential monitoring tool in order to prevent and early diagnose infectious diseases.
Funding
This work was partially supported by grants from the Italian Ministry for University and Scientific Research, Italy (GR-2018-12367375).
Author contributions
Conceptualization: SLC, GDP, TS, MA; Data curation: SLC, DLG, SC, EST, GP, EP, LC and RX; Formal analysis: SLC, FDM, FRM; Funding acquisition: GDP, MA, BP, TS, MA; Investigation: SLC, FDM, GDP, DLG, FRM, SC, EST, GP, EP, LC and RX, MS, BP, TS, MA; Methodology: SLC, FDM, GDP, MS, BP, TS, MA; Project administration: SLC, FDM, GDP, MS, BP, TS, MA; Resources: SLC, GDP, MS, BP, TS, MA; Software: STATA and R; Supervision: SLC, FDM, GDP, MS, BP, TS, MA; Validation: SLC, FDM, GDP, MS, BP, TS, MA ; Visualization: SLC, FDM, GDP, MS, BP, TS, MA; Writing - original draft: SLC, FDM, GDP, MS, BP, TS, MA; Writing - review & editing: SLC, FDM, GDP, DLG, FRM, SC, EST, GP, EP, LC, RX, MS, BP, TS, MA.
Declaration of Competing Interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcrc.2021.04.008.
Appendix A. Supplementary data
Supplementary data
References
- 1.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region. Italy JAMA. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kassirian S., Taneja R., Mehta S. Diagnosis and Management of Acute Respiratory Distress Syndrome in a time of COVID-19. Diagnostics (Basel) 2020;10(12):1053. doi: 10.3390/diagnostics10121053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blonz G., Kouatchet A., Chudeau N., Pontis E., Lorber J., Lemeur A., et al. Epidemiology and microbiology of ventilator-associated pneumonia in COVID-19 patients: a multicenter retrospective study in 188 patients in an un-inundated French region. Crit Care. 2021;25(1):72. doi: 10.1186/s13054-021-03493-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeannet R., Daix T., Formento R., Feuillard J., François B. Severe COVID-19 is associated with deep and sustained multifaceted cellular immunosuppression. Intensive Care Med. 2020;46(9):1769–1771. doi: 10.1007/s00134-020-06127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao L., Zhang C., Dong J., Zhao L., Li Y., Sun J. Oral Microbiome and SARS-CoV-2: Beware of Lung Co-infection. Front Microbiol. 2020;11:1840. doi: 10.3389/fmicb.2020.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouzé A., Martin-Loeches L., Povoa P., Makris D., Artigas A., Bouchereau M., et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: a European multicenter cohort study. Intensive Care Med. 2021:1–11. doi: 10.1007/s00134-020-06323-9. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janda J., Abbott S. The genus hafnia: from soup to nuts. Clin Microbiol Rev. 2006;19(1):12–18. doi: 10.1128/CMR.19.1.12-28.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rosa S., Cutuli S.L., Ferrer R., Antonelli M., Ronco C. COVID-19 EUPHAS2 Collaborative Group. Polymyxin B hemoperfusion in coronavirus disease 2019 patients with endotoxic shock: Case series from EUPHAS2 registry. Artif Organs. 2020 doi: 10.1111/aor.13900. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Lazzarino M., Orlandi E., Baldanti F., Furione M., Pagnucco G., Astori C., et al. The immunosuppression and potential for EBV reactivation of fludarabine combined with cyclophosphamide and dexamethasone in patients with lymphoproliferative disorders. Br J Haematol. 1999;107(4):877–882. doi: 10.1046/j.1365-2141.1999.01765.x. [DOI] [PubMed] [Google Scholar]
- 10.Fromentin M., Ricard J., Roux D. Respiratory microbiome in mechanically ventilated patients: a narrative review. Intensive Care Med. 2012:1–15. doi: 10.1007/s00134-020-06338-2. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data