ABSTRACT
Background
Human milk contains a diverse community of bacteria that are modified by maternal factors, but whether these or other factors are similar in developing countries has not been explored. Our objective was to determine whether the milk microbiota was modified by maternal age, BMI, parity, lactation stage, subclinical mastitis (SCM), and breastfeeding practices in the first 6 mo of lactation in an indigenous population from Guatemala.
Methods
For this cross-sectional study, Mam-Mayan indigenous mothers nursing infants aged <6 mo were recruited. Unilateral human milk samples were collected (n = 86) and processed for 16S rRNA sequencing at the genus level. Microbial diversity and relative abundance were compared with maternal factors [age, BMI, parity, stage of lactation, SCM, and 3 breastfeeding practices (exclusive, predominant, mixed)] obtained through questionnaires.
Results
Streptococcus was the most abundant genus (33.8%), followed by Pseudomonas (18.7%) and Sphingobium (10.7%) but relative abundance was associated with maternal factors. First, Lactobacillus and Streptococcus were more abundant in early lactation whereas the common oral (Leptotrichia) and environmental (Comamonas) bacteria were more abundant in established lactation. Second, Streptococcus,Lactobacillus,Lactococcus,Leuconostoc, and Micrococcus had a higher abundance in multiparous mothers compared with primiparous mothers. Third, a more diverse microbiota characterized by a higher abundance of lactic acid bacteria (Lactobacillus,Leuconostoc, and Lactococcus), Leucobacter, and Micrococcus was found in mothers with a healthy BMI. Finally, distinct microbial communities differed by stage of lactation and by exclusive, predominant, or mixed breastfeeding practices.
Conclusion
Milk bacterial communities in an indigenous community were associated with maternal factors. Higher microbial diversity was supported by having a healthy BMI, the absence of SCM, and by breastfeeding. Interestingly, breastfeeding practices when assessed by lactation stage were associated with distinct microbiota profiles.
Keywords: indigenous, Mam-Mayan, milk microbiota, 16S rRNA sequencing, microbial diversity, exclusive breastfeeding, predominant breastfeeding
Milk microbiota in an indigenous community showed microbial diversity in adult and multiparous mothers with a healthy BMI and without SCM. Breastfeeding practices were associated with microbial diversity.
Introduction
Human milk has long been acknowledged as the ideal food for infants to meet energy and nutrient needs and for its immune properties (1). Human milk is also recognized as an important source of bacteria for the infant (2, 3). In fact, the human milk microbiota has been described as possibly the most influential postnatal factor in the metabolic and immunological programming of infant growth (4).
Multi-country studies reveal that the most common genera present in the human milk microbiota differ among countries (5, 6). For example, although Lactobacillus is relatively common in Spain (7–11), most of the studies conducted in Africa, Asia, and Latin-America do not report Lactobacillus as a predominant taxon (12–16). There is also evidence that the microbiota differs within a country based on social characteristics including socioeconomic status (14, 17) and rural versus urban setting (4–6, 13, 16, 18), where a more diverse microbiota have been reported in rural agrarian populations. There is evidence that some bacterial taxa in milk vary between hunter-gatherers versus horticulturalist women (19). For instance, Enhydrobacter,Renibacterium, and Lactobacillus were more abundant in horticulturalist women whereas Peptoniphilus,Salinococcus,Planobacterium,Anaerococcus,Granulicatella,Riemerella,Kocuria, and Luteimonas were more abundant in hunter-gatherers. Both populations showed unique bacteria although α-diversity did not vary (19). A study in India reported high Alphaproteobacteria, Betaproteobacteria, Actinobacteria, Clostridia, and Firmicutes in agrarian communities, whereas human milk from urban women had more Proteobacteria and Gammaproteobacteria (16). The rural Ethiopian community differed from urban populations in abundance of the following genera in descending order, Rhizobium,Achromobacter,Streptococcus,Staphylococcus,Propinobacterium, and Rothia (6).
Specific maternal factors including maternal BMI, delivery mode, and lactation stage are known to modify the human milk microbiota (7, 8, 11, 14, 18, 20–24). With regard to BMI, some studies report no association (20, 25) but others report that higher maternal BMI and weight gain during pregnancy impact the diversity of the bacterial community in milk (8, 26). Other studies have correlated human milk microbial composition with lactation stage (27–29). It has been reported that colostrum and transitional milk only have 50% of bacterial genera and 42% of bacterial species in common (12).
In contrast, several important factors that could be associated with the human milk microbiota have not been widely explored; these include maternal age, parity, SCM, and breastfeeding practices. Maternal age, in particular adolescent pregnancies, is associated with increased nutrient requirements (30). Moreover, it is also known that adolescence can impact the development and maturity of the mammary gland and has been associated with changes in immune factors in human milk as well as its microbiota (31). Parity is another factor that could be related to age and might influence the milk microbiota. Assuming retrograde flow, the milk from a multiparous mother will have been previously inoculated by the bacteria transferred by her previous infant(s) during earlier pregnancies (32). The other 2 factors that have been overlooked in the study of the milk microbiota include SCM and breastfeeding practices. The limited research available focuses on mastitis (33) rather than SCM and in relation to breastfeeding practices, most of the literature compares human milk and formula rather than exclusive, predominant, and mixed feeding (27, 29, 34–37).
These overlooked factors may be particularly important in developing countries. Worldwide, 16 M (million) adolescents aged between 15 and 19 y and another 2.5 M under the age of 16 y give birth; moreover, adolescent pregnancies are 3× more frequent in rural and indigenous populations (38). In relation to parity, Central America, Guatemala, Nicaragua, and Panama have the highest adolescent fertility rates (39). Finally, for SCM and breastfeeding practices, previous reports have shown that SCM is common in Guatemala and differs by stage of lactation (40, 41).
On the other hand, developing countries have higher rates of exclusive breastfeeding, whereas mothers in developed countries have lower breastfeeding rates and routinely do not exclusively or predominantly breastfeed for 6 mo (42). Current WHO guidelines recommend either exclusive or predominant breastfeeding for 6 mo (43), and according to recent UNICEF data, rural populations in developing countries are more likely to achieve these goals than other sectors (42). Guatemala reportedly has one of the highest exclusive breastfeeding rates, approaching 76.8%, from which 78.6% of the sample were indigenous women (44). The Mam-Mayan community of Guatemala also has the tradition of introducing ritual fluids called “agüitas” during breastfeeding feeding in order to promote infant health and treat gastrointestinal or other infections (45). The consumption of agüitas in Guatemala has been reported to be 22% during the first 6 mo (46).
Given the maintenance of exclusive and predominant breastfeeding and the ease of obtaining human milk from Mam-Mayan mothers from the Western Highlands of Guatemala, our study provided us with a unique opportunity to measure the milk microbiota in our biobanked human milk samples. The purpose of this cross-sectional study was to explore if genera in the human milk microbiota were associated with maternal age, BMI, parity, lactation stage, the presence of SCM, and various breastfeeding practices.
Methods
Study site and participants
This cross-sectional study was part of a collaboration between McGill University and the Center for Studies of Sensory Impairment, Aging and Metabolism (CeSSIAM) in the Republic of Guatemala. Field studies were conducted from June 2012 through January 2013 in 8 rural Mam-speaking communities of the San Juan Ostuncalco region in Guatemala (47). Lactating mothers of infants aged from 5 to 46 d or 4 to 6 mo postpartum were identified and invited to participate by community health workers. Inclusion criteria included mothers who breastfed for 6 mo, had healthy BMIs, and had vaginal deliveries. Individuals who had been treated with antibiotics during the postpartum period were excluded as the use of antibiotics has been demonstrated to decrease the rate of SCM (48). Ethical approval was obtained from McGill Institutional Review Board and Center for Studies of Sensory Impairment, Aging and Metabolism (CeSSIAM) Human Subjects Committee. All participating mothers provided written informed consent for participation.
Study design
We compared human milk bacterial communities by maternal age (adolescents aged: ≤19 y versus adults: >19 y), weight (healthy: BMI = 18.5–24.9 kg/m2 versus overweight: BMI ≥25), parity (primiparous versus multiparous), stage of lactation [early (5–46 d) versus established (4–6 mo)], SCM (yes: Na/K >0.6 or no: Na/K ≤0.6), and breastfeeding (exclusive or predominant versus mixed). Exclusive breastfeeding (EBF) was defined as providing only human milk to the infant and predominant breastfeeding (PBF) was defined as providing water or agüitas in addition to human milk. Agüitas are ritual fluids; the infusions more commonly used are boiled water, sugar water, chamomile tea, corn paste water, anise water, orange leaf water, mint water, or sage water. They are given for perceived insufficient milk, for irritability and crying and to maintain infant's health, or as herbal medicine to treat gastrointestinal or other infections (45, 47, 49). Breastfeeding was categorized as mixed if mothers provided complementary foods to the infant.
Human milk sample collection
Milk samples from early lactation (5–46 d postpartum) and established lactation (4–6 mo postpartum) were collected. These ranges were chosen to be consistent with our previous studies that measured infant growth (40, 41, 50). Prior to collection, the nipple and areola of the breast were cleaned with 70% ethyl alcohol. Human milk samples were collected during a 3-h time window in the morning from the breast not recently used for breastfeeding via full manual expression by a trained midwife, who used hand sanitizer before and after collection. Milk was collected into 60 mL plastic trace-element-free vials and immediately stored on ice. Samples were partitioned into 15 mL trace-element-free vials at the field laboratory (−30°C) prior to transfer on dry ice to McGill University where they were stored at −80°C until analysis was performed.
16S rRNA pyrosequencing using Illumina MiSeq
DNA extraction was done using 1 mL of whole milk (direct aliquots from the stock) with the DNeasy Blood and Tissue mini kit from Qiagen according to the manufacturer's protocol. A centrifuge was used to spin down the sample and only the pellet was kept for further extraction steps; all the supernatant, including the fat, were discarded. For PCR, a region of ∼526 bp in the 16S rRNA gene, covering V1–V3 was amplified with the universal eubacterial primers 27F (AGAGTTTGATCCTGGCTCAG) and 533R (TTACCGCGGCTGCTGGCAC) (8). The subsequent 16S rRNA sequencing was performed using Illumina MiSeq at McGill University and Genome Quebec Innovation Centre.
Microbial data processing
Using Mothur's MiSeq protocol (51), raw paired-end reads (13.0 M) were assembled into contigs that were aligned together. Unique representative sequences were then extracted and their abundance evaluated. Raw contig counts were used to remove sequences with low abundance (contig count cut-off of 10); 14,742 sequences representing possible unique amplicons were extracted, comprising 10.5 M counts in total (accounting for 97% of initial raw reads).
Metagenomic sequences were analyzed via BLASTn against the nonredundant nucleotide database (nt) with an Expect (E) Value threshold of 1.10−10 and an identity cut-off of 97%. The metatranscriptomic (unconstrained) annotation strategy (52) was employed together with a previously reported annotation selection method used to help select the primary annotation from ambiguous BLASTn returns from metatranscriptomic/metagenomic data (53).
Sequences were identified as a potential genus when their identity cut-off was ≥99%, the BLASTn alignment coverage was ≥95%, and no other competing genera label was present. Taxonomic units (n = 125) were detected at 99% identity (from genus up to phylum level) and were used to bin the remaining sequences at 97%, capturing 5.0 M sequences in total. Sparse taxonomic units (a high proportion of zero counts across samples) and samples with low diversity were removed manually.
The α-diversity of human milk samples, describing the within-community diversity of the microbiota within a milk sample, was measured using 2 different metrics (Shannon and Inverse Simpson) within the Phyloseq R package (54). The α-diversity was compared between groups of samples using a t-test. The β-diversity, describing the microbiota diversity among samples, was estimated based on variation stabilization (VS) normalized counts (DESeq) using Bray–Curtis dissimilarity and the Constrained Analysis of Principal Coordinates (CAP) ordination method. Dispersion ellipses were drawn using the veganCovEllipse function in the Vegan package in R (55). Significant distance was evaluated between groups using nonparametric analysis of similarities (ANOSIM) on normalized counts based on Bray distances (Vegan package). The species richness of samples was also assessed with rarefaction curves.
Diversity relative abundance was represented with Krona (56). Taxonomic profiles were plotted with GraPhlAn (57). Parametric models developed in transcriptomics to characterize differentially abundant taxonomic units between groups of samples perform well when applied to microbiota biomarker data with uneven library sizes, sparsity, and sample representativity (58). From the several programs available, DESeq2 (59) has been found to have the best overall performance (60). The DESeq2 univariate procedure was selected, as implemented in Phyloseq, to calculate differentially abundant taxonomic units. To account for the power to identify differentially abundant genera, the group size was fixed so none of the groups compared had <10 samples and the FC effect size threshold was set at 5 (60). Taxonomic units tested with a Benjamini–Hochberg adjusted P value < 0.05 were considered significant.
Results
Characteristics of the population
Population characteristics are summarized in Table 1 and individual characteristics of participating mothers in Supplemental Table 1. They had a mean age of 23.5 ± 6.4 y and mean maternal BMI was healthy (23.3 ± 3.2); 48% were primiparous and 52% were multiparous. In relation to the breastfeeding practices, 86% exclusively or predominantly breastfed, and 14% breastfed and also provided complementary foods (mixed breastfeeding); 45% were in the early stage of lactation (5–46 d), 55% in established lactation (4–6 mo), and 12% had SCM and 88% did not (non-SCM).
TABLE 1.
Characteristics of the indigenous population of Mam-Mayan mothers (n = 86)
| Maternal factors | Mean ± SD or % |
|---|---|
| BMI, kg/m2 | 23.3 ± 3.2 |
| Healthy weight (18.5–24.9), % | 78 |
| Overweight (>25), % | 22 |
| Stage of lactation | |
| Early 5–46 d, % | 45 |
| Early, x̅ days | 20.2 ± 10.6 |
| Established 4–6 mo, % | 55 |
| Established, x̅ days | 144.1 ± 20.1 |
| Parity | |
| Primiparous, % | 48 |
| Multiparous, % | 52 |
| Age, y | 23.5 ± 6.4 |
| Adolescent ≤19, % | 23 |
| Adult >19, % | 77 |
| Subclinical mastitis | |
| Yes Na/K >0.6, % | 12 |
| No Na/K ≤0.6, % | 88 |
| Breastfeeding practice | |
| Exclusive and predominant, % | 86 |
| Mixed, % | 14 |
Human milk microbiota
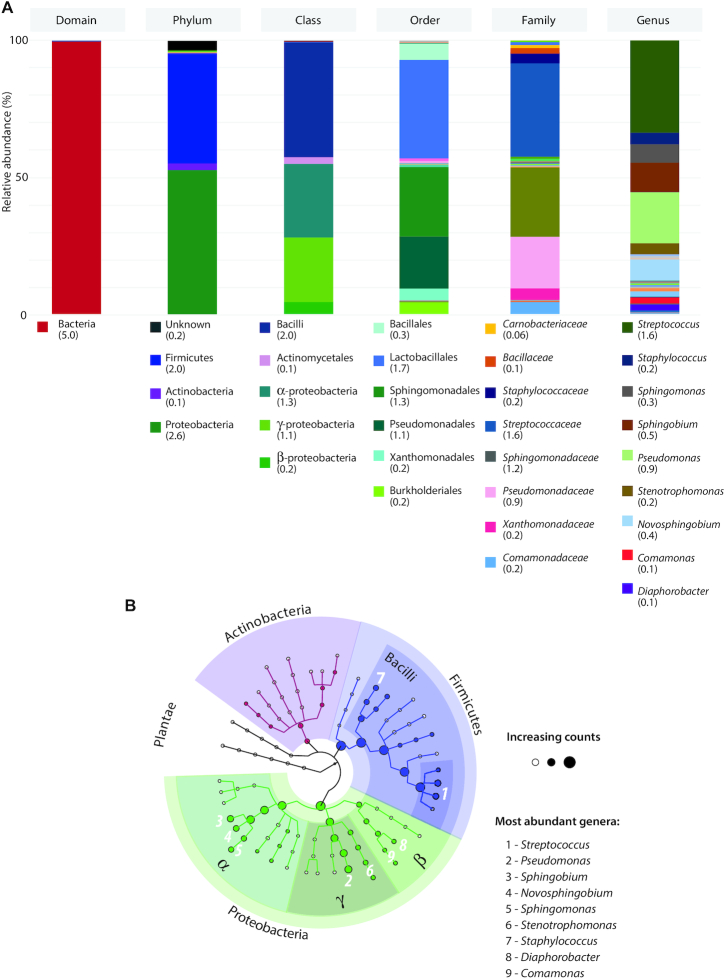
The 16S rRNA analysis of 86 milk samples resulted in 5,053,154 sequences and successfully identified 47 different taxonomic units (from phylum to genera). The total pool of sequences was classified to 3 known phyla, Proteobacteria (52.8%), Firmicutes (40.3%), and Actinobacteria (2.4%). Streptococcus was the most abundant genus making up 33.8% of the sequences, followed by Pseudomonas at 18.7% and Sphingobium at 10.7%. Lactobacillus only accounted for 0.60% of the milk bacterial community (Figure 1). Rarefaction curves are shown in Supplemental Figure 1.
FIGURE 1.
A) Human milk microbiota diversity based on pooled samples from all mothers, illustrated as relative abundance along major taxonomic levels. Total counts for each taxonomic unit are specified between parentheses (in million). Only taxonomic units with a total count >100,000 are accounted for in the legends. B) Major clades including weighted nodes (genera >100,000 counts) as analyzed in the human milk microbiota. The 9 most abundant genera are identified.
Maternal factors
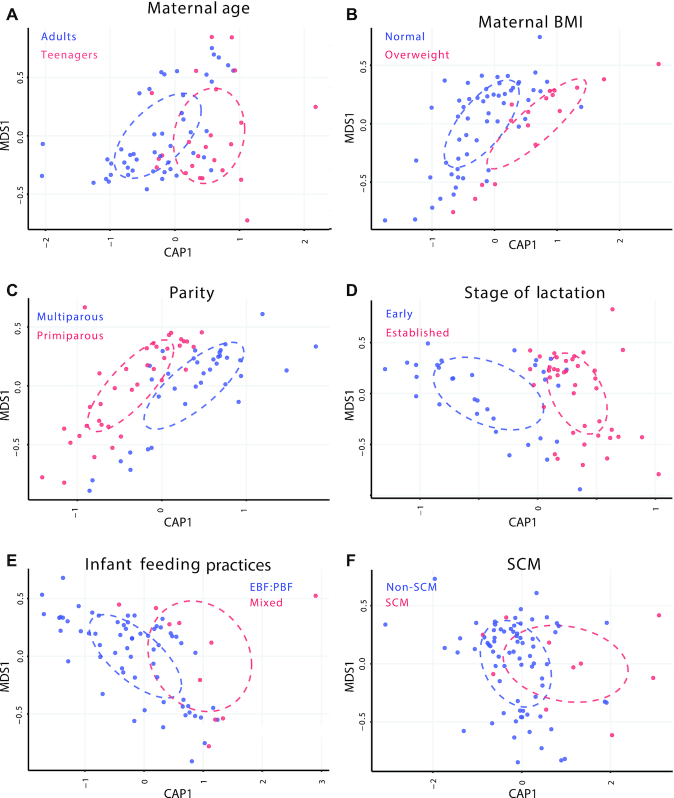
The distance-based redundancy analysis (db-RDA) plots (Figure 2) showed interesting separations in bacterial communities for 3 conditions: maternal BMI, parity, and lactation stage. A lesser separation was observed among communities for maternal age, feeding practices, and SCM. No differences in α-diversities (within-sample diversity) were detected for maternal age, BMI, lactation stage, feeding practices, and SCM, but multiparous mothers had higher richness than primiparous mothers (Shannon, P = 0.01, Inv Simpson, P = 0.03) (Supplemental Figure 2). The β-diversity (between-sample diversity) differed between early and established lactation only (P = 0.001).
FIGURE 2.
Distance-based redundancy analysis (db-RDA or CAP) ordination representations (Bray distance) for A) maternal age; B) maternal BMI; C) parity; D) stage of lactation; E) Infant feeding practices; and F) SCM. Significant clustering of groups was found for maternal BMI, parity, and stage of lactation (ANOVA), P <0.05. EBF, exclusive breastfeeding; PBF, predominant breastfeeding; SCM, subclinical mastitis.
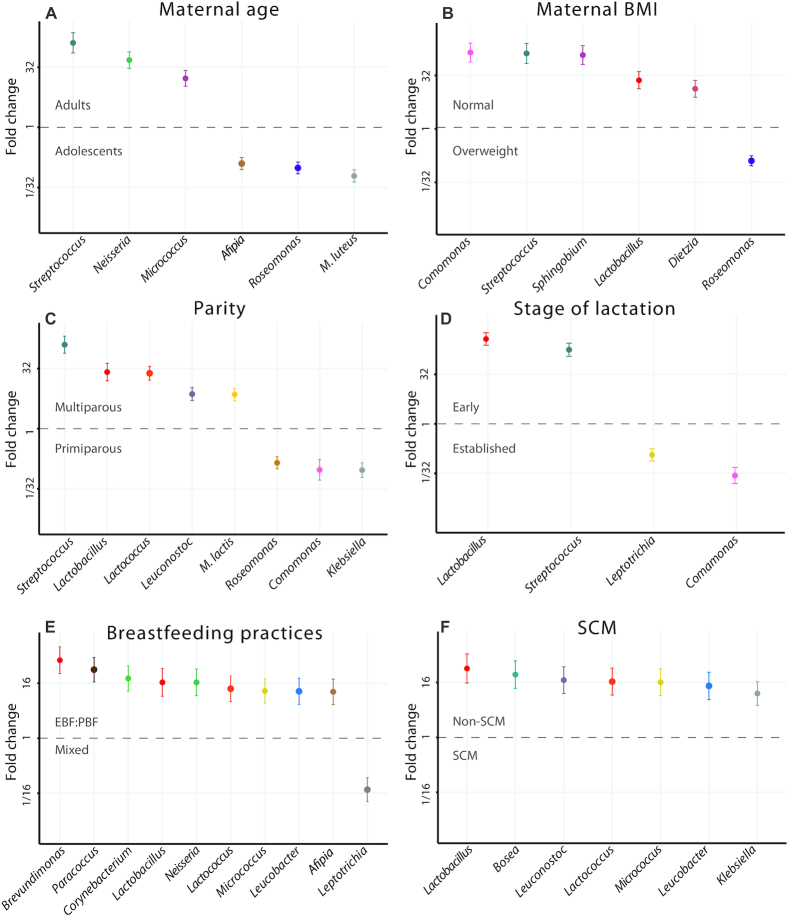
Differential abundance analyses showed that 5 bacterial taxonomic units significantly differed between the milk of adult and adolescent mothers. Adult mothers had a higher abundance of Streptococcus [false discovery rate (FDR) = 3.74 × 10−15; fold change (FC) = 130], Neisseria (FDR = 7.09 × 10−15; FC = 48), and Micrococcus (FDR = 3.40 × 10−9; FC = 17), whereas Roseomonas (FDR = 6.77 × 10−11; FC = 10) and Afipia (FDR = 1.09 × 10−8; FC = 8) were more abundant in adolescent mothers. Compared with adult mothers, adolescent mothers had low mean counts of Streptococcus (Figure 3, Table 2). The relative abundance values of the top 20 genera for maternal age are shown in Supplemental Figure 3.
FIGURE 3.
Differential abundance plots for each maternal factor: A) maternal age (adult versus adolescent); B) maternal BMI (healthy weight versus overweight); C) parity (primiparous versus multiparous); D) stage of lactation (early versus established); E) breastfeeding practices (exclusively/predominant breastfeeding versus mixed feeding); and F) SCM (yes or no). Each plot shows fold change for the differentially abundant taxonomic units (group size fixed to a minimum of 10 and effect size to a fold change of 5). Error bars represent the SE on the estimated fold change. EBF, exclusive breastfeeding; PBF, predominant breastfeeding; SCM, subclinical mastitis.
TABLE 2.
Comparative summary of differentially abundant bacterial genera by maternal factors and infant feeding practices1
| Adult mothers | Healthy BMI | Multiparous >1 childbirth | Early lactation | EBF:PBF | Non-SCM | |
|---|---|---|---|---|---|---|
| Actinobacteria | ||||||
| Actinobacteria; Actinomycetales; Dietziaceae; Dietzia | — | ↑ | — | — | — | — |
| Actinobacteria; Actinomycetales; Micrococcaceae; Micrococcus | ↑ | — | — | — | ↑ | ↑ |
| Actinobacteria; Corynebacteriales; Corynebacteriaceae; Corynebacteriumhis | — | — | — | — | ↑ | — |
| Actinobacteria; Micrococcineae; Microbacteriaceae; Leucobacter | — | — | — | — | ↑ | ↑ |
| Firmicutes | ||||||
| Bacilli; Lactobacillales; Lactobacillaceae; Lactobacillus | — | ↑ | ↑ | ↑ | ↑ | ↑ |
| Bacilli; Lactobacillales; Leuconostocaceae; Leuconostoc | — | — | ↑ | — | — | ↑ |
| Bacilli; Lactobacillales; Streptococcaceae; Lactococcus | — | — | ↑ | — | ↑ | ↑ |
| Bacilli; Lactobacillales; Streptococcaceae; Streptococcus | ↑ | ↑ | ↑ | ↑ | — | — |
| Fusobacteria | ||||||
| Fusobacteria; Fusobacterales; Fusobacteriaceae; Leptotrichia | — | — | — | ↓ | ↓ | — |
| Proteobacteria | ||||||
| Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Brevundimonas | — | — | — | — | ↑ | — |
| Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Afipia | ↓ | — | — | — | ↑ | — |
| Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae; Bosea | — | — | — | — | — | ↑ |
| Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus | — | — | — | — | ↑ | — |
| Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Roseomonas | ↓ | ↓ | ↓ | — | — | — |
| Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium | — | ↑ | — | — | — | — |
| Betaproteobacteria; Burkholderiales; Comamonadaceae; Comamonas | — | ↑ | ↓ | ↓ | — | — |
| Betaproteobacteria; Neisseriales; Neisseriaceae; Neisseria | ↑ | — | — | — | ↑ | — |
| Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Klebsiella | — | — | ↓ | — | — | ↑ |
Group size fixed to a minimum of 10 and effect size to a fold change of 5. Each arrow reflects the higher or lower differential abundance in comparison to the contrasting group (adults versus adolescents, healthy BMI versus overweight, multiparous versus primiparous, early lactation versus established lactation, exclusive and predominant breastfeeding versus mixed feeding and Non-SCM versus SCM). The symbol (—) represents ''not found''.
For maternal BMI, 6 differentially abundant taxonomic units also emerged. Healthy weight mothers had a higher abundance of Comamonas (FDR = 3.47 × 10−14; FC = 142), Streptococcus (FDR = 8.47 × 10−13; FC = 134), Sphingobium (FDR = 8.14 × 10−14; FC = 120), Lactobacillus (FDR = 1.37 × 10−7; FC = 23), and Dietzia (FDR = 1.31 × 10−5; FC = 13) than overweight mothers. The mean counts of Comamonas and Streptococcus of overweight mothers were very low. Only Roseomonas (FDR = 1.93 × 10−9; FC = 8) was more abundant in the milk of overweight mothers than healthy weight mothers (Figure 3, Table 2). The relative abundance values of the top 20 genera for maternal BMI are shown in Supplemental Figure 3.
For parity, 8 bacterial taxonomic units significantly differed between the milk of primiparous and multiparous mothers. Multiparous mothers had a higher abundance of Streptococcus (FDR = 3.92 × 10−21; FC = 127), Lactobacillus (FDR = 1.29 × 10−9; FC = 26), Lactococcus (FDR = 1.73 × 10−14; FC = 24), Leuconostoc (FDR = 5.81 × 10−7; FC = 7), and Micrococcus (FDR = 1.49 × 10−7; FC = 7) in milk than primiparous mothers, whereas Klebsiella (FDR = 1.49 × 10−7; FC = 11), Comamonas (FDR = 3.03 × 10−4; FC = 11), and Roseomonas (FDR = 1.49 × 10−7; FC = 7) were more abundant in the milk of primiparous mothers (Figure 3, Table 2). The relative abundance values of the top 20 genera for parity are shown in Supplemental Figure 3.
Lactation characteristics
Differential abundance analyses showed that 4 bacterial taxonomic units significantly differed between early and established lactation. During early lactation, milk had a higher abundance of Lactobacillus (FDR = 3.73 × 10−40; FC = 378) and Streptococcus (FDR = 4.61 × 10−28; FC = 178), whereas during established lactation, milk had a higher abundance of Comamonas (FDR = 1.47 × 10−9; FC = 37) and Leptotrichia (FDR = 3.18 × 10−6; FC = 9). Lactobacillus spp. were not detected in most milk samples collected during established lactation (Figure 3, Table 2). The relative abundance values of the top 20 genera for stage of lactation are shown in Supplemental Figure 3.
For breastfeeding practices, 10 bacterial taxonomic units significantly differed between EBF/PBF and mixed feeding mothers. EBF/PBF mothers had a higher abundance of Brevundimonas (FDR = 2.11 × 10−7; FC = 51), Paracoccus (FDR = 4.84 × 10−7; FC = 32), Corynebacterium (FDR = 3.13 × 10−5; FC = 20), Lactobacillus (FDR = 2.82 × 10−4; FC = 17), Neisseria (FDR = 1.72 × 10−4; FC = 17), Lactococcus (FDR = 5.05 × 10−4; FC = 12), Micrococcus (FDR = 5.05 × 10−4; FC = 11), Leucobacter (FDR = 9.81 × 10−4; FC = 11), and Afipia (FDR = 8.24 × 10−4; FC = 10) in milk. Only Leptotrichia (FDR = 1.34 × 10−4; FC = 13) was more abundant in the milk of mixed feeding mothers (Figure 3, Table 2). The relative abundance values of the top 20 genera for breastfeeding practices are shown in Supplemental Figure 3.
When we further categorized breastfeeding practices (exclusive, predominant, and mixed) by stage of lactation (early and established), distinct separations in microbiota among each of these groups were observed. The microbiota of exclusive differed from predominant by stage of lactation and both differed compared with mixed feeding (Figure 4).
FIGURE 4.
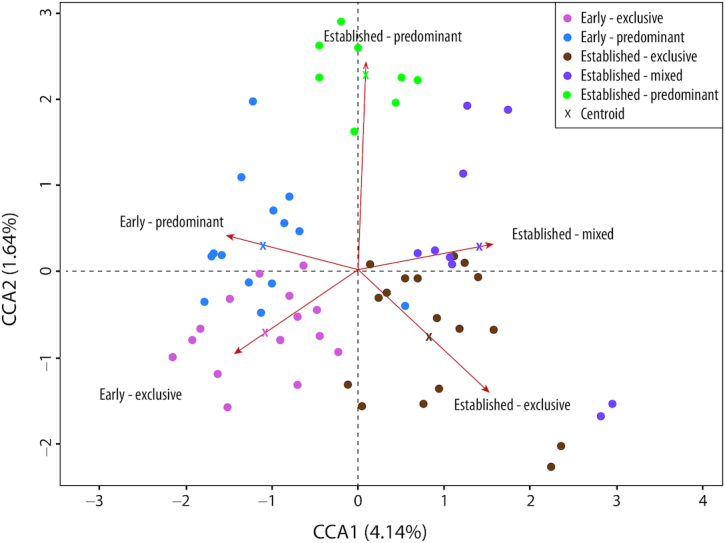
Comparison at genera level by lactation stage and breastfeeding practices. Centroide crosses represent the shift of each group and their distinct cluster. Early lactation clusters differently than established; predominant and mixed feeding cluster higher on the y-axis scale than exclusive breastfeeding.
With regards to SCM, 7 bacterial taxonomic units were significantly more abundant in the milk of mothers without SCM than mothers with SCM. Mothers without SCM had a higher abundance of Lactobacillus (FDR = 4.53 × 10−5; FC = 32), Bosea (FDR = 7.37 × 10−5; FC = 24), Leuconostoc (FDR = 1.63 × 10−4; FC = 18), Lactococcus (FDR = 2.27 × 10−4; FC = 17), Micrococcus (FDR = 2.27 × 10−4; FC = 16), Leucobacter (FDR = 8.73 × 10−4; FC = 13), and Klebsiella (FDR = 9.67 × 10−4; FC = 9). Bosea,Lactococcus, and Leucobacter were absent in all SCM samples and Lactobacillus was only detected in 1 SCM sample (Figure 3, Table 2). The relative abundance values of the top 20 genera for SCM are shown in Supplemental Figure 3.
Discussion
Our study is the first to investigate variations of bacterial communities in the human milk of the Mam-Mayan indigenous community in Guatemala, where mothers routinely comply with WHO recommendations to exclusively or predominantly breastfeed for 6 mo. In addition to BMI, the impact of understudied maternal factors including adolescent pregnancy, parity, late-stage lactation, the presence of SCM, and breastfeeding practices on the human milk microbiota were also explored. Six findings emerged. First, the human milk microbiota in our study population differed between early (<6 wk) and established lactation (4–6 mo). Lactobacillus and Streptococcus were more abundant in early lactation, whereas the common oral (Leptotrichia) and environmental (Comamonas) bacteria were more abundant in established lactation. Second, the milk microbiota differed between adolescent and adult mothers. Streptococcus,Neisseria, and Micrococcus were higher in adult milk whereas, Roseomonas and Afipia were predominant in the milk of adolescent mothers. Third, a significantly higher abundance of Streptococcus,Lactobacillus,Lactococcus,Leuconostoc, and Micrococcus were observed in multiparous mothers, whereas the milk of primiparous mothers had a higher abundance of Comamonas,Klebsiella, and Roseomonas. Fourth, the milk of mothers with a healthy BMI was higher in Comamonas,Streptococcus, Sphingobium,Lactobacillus, and Dietzia, whereas in mothers with obesity only Roseomonas was higher. Fifth, in the absence of SCM, mothers had a more diverse microbiota that was characterized by a higher abundance of lactic acid bacteria (Lactobacillus,Leuconostoc, and Lactococcus), Leucobacter, and Micrococcus compared with mothers with SCM, suggesting that the absence of lactic-acid-producing bacteria may be associated with human milk dysbiosis and the emergence of SCM. Finally, greater microbial diversity was associated with exclusive and predominant breastfeeding practices compared with mixed feeding. Moreover, the microbiota differed between exclusive and predominant at each stage of lactation. Collectively, these findings showed that indigenous communities have several distinct maternal factors that shift the milk microbiota. Based on our findings, we propose that evidence from developed countries regarding the milk microbiota may not be transferable to developing countries due to differing environments and population characteristics.
Characteristics of predominant phyla and genera
Our identification of 3 phyla in human milk, Proteobacteria (52.8%), Firmicutes (40.3%), and Actinobacteria (2.4%), aligns with the latest systematic review that described Firmicutes and Proteobacteria as the most predominant phyla with Actinobacteria present at lower relative abundances (24). The most abundant genera were Streptococcus (33.8%), Pseudomonas (18.7%), and Sphingobium (10.7%). On one hand, Streptococcus spp. have been commonly reported as one of the most abundant genera in human milk (15, 18–20, 23, 25, 61–63). Pseudomonas is also a commonly detected genus in a range of relative abundance from <1 to 17% (24). On the other hand, Sphingobium spp. have been previously observed in the human milk microbiota (10, 64); however, it is not generally considered as an abundant or frequent genus.
Maternal factors
Stage of lactation
It is well-established that human milk volume and nutritional immunological composition progressively change over the lactation period (36, 65). There is now evidence that the milk microbiota also differs by stage of lactation (8, 11, 37, 66), although not all studies report differences (18, 20, 23). However, none of these aforementioned studies were conducted in agrarian indigenous communities. In our study, we were able to take advantage of the fact that all of the indigenous mothers complied with WHO guidelines to breastfeed for 6 mo. Under these conditions Streptococcus and Lactobacillus were higher in early lactation. Lactobacillus has been reported to be more abundant in the early stage of lactation (8) and Streptococcus is considered to be part of the “core” microbiota in human milk (6, 8, 22, 62). Additionally, Streptococcus, the most abundant genus observed in our samples, has been reported as a putative organism (67) that is capable of suppressing the growth of pathogenic bacteria such as Staphylococcus aureus. Both Lactobacillus and Streptococcus, which were more abundant in early lactation in our study, have immunomodulatory and anti-inflammatory properties and functions that contribute to the stability of bacterial populations (68–70). On the other hand, Lactobacillus was absent in 55% of milk samples collected during established lactation, which contrasts with previous research that reports an increasing prevalence of Lactobacillus as lactation progresses (8, 11). However, these studies were conducted in developed countries and other factors could be intervening as well. We observed that both Comamonas and Leptotrichia were higher in established lactation. Leptotrichia is a common oral bacteria previously reported in established lactation, whereas Comamonas is a common environmental bacteria that has been associated on occasion with human disease (71), suggesting dynamic differences in the milk microbiota during lactation associated with other factors that also differed by stage of lactation in our study.
Maternal age
To date, the only study to consider whether maternal age was associated with the human milk microbiota reported no association between adult maternal age (33.0 ± 4.2 y) and the human milk microbiota; all participants were adults aged between 20 and 40 y (22). Our study is the first to perform a differential abundance analysis of the human milk bacterial taxonomy between adolescent and adult mothers. Our findings show that Streptococcus,Neisseria, and Micrococcus had a higher abundance in milk samples of adult mothers. Streptococcus and Neisseria are normally present in the infant oral cavity and saliva (72, 73). Micrococcus, which is a common human colonizer that rarely causes infections, is found in the skin, mucosa, and oropharynx and is likely transmitted through retrograde flow (66). In contrast, the genera Roseomonas and Afipia, of which some species are known to be opportunistic bacteria, had a higher abundance in the milk of adolescents (74, 75).
It is possible that these differences in the human milk microbiota of adolescent and adult mothers could be linked to the development of breast milk immunity, which occurs during adolescence (76). The immature mammary gland undergoes 3 age-related developmental phases; embryonic, adolescent, and adult, with the majority of its alveolar branching taking place during adolescence (31). The alveolar epithelium is responsible for secreting immune modulators including IL-6, TNF, and IgA (77), and these concentrations in milk can be affected during adolescence (78). Perhaps the immaturity of the adolescent breast and its associations with lower concentrations of immune factors might underscore a higher relative abundance of opportunistic bacteria in adolescents, but this requires further investigation.
In addition to the immune system, adolescence is a period characterized by its own requirements for growth and physical development. In the developing country context where the prevalence of adolescence pregnancy is high (39) and where nutrient deficiency and malnutrition are common (79), human milk nutrient composition might modulate the human milk microbiota. There is evidence that adolescents’ milk is lower in copper, zinc, retinol, and α-tocopherol compared with adult milk (80–84). In these Mam-Mayan mothers, we have evidence that several human milk minerals and trace elements associated with human SCM were altered (40). Thus, variation in the nutrient composition of human milk might contribute to further alterations in the human milk microbiota.
Parity
Parity is another underexplored maternal factor. To date, only 1 study has reported an association of milk microbiota composition with multiparity but did not characterize the microbiota (22). In our study, a significantly higher abundance of Streptococcus, Lactobacillus,Lactococcus,Leuconostoc, and Micrococcus were observed in our multiparous mothers. By contrast, primiparity was associated with a significant abundance of Comamonas,Klebsiella, and Roseomonas. Interestingly, Roseomonas is an example of bacteria that showed a higher relative abundance in both adolescent and primiparous mothers. Presumably primiparous mothers were more likely to be adolescents, further supporting a link with immaturity of the human milk immune system during adolescence, as previously reported (76).
Maternal BMI
Most studies that have considered the impact of BMI on the human milk microbiota have been conducted in urban populations and developed countries, and the results are contradictory. Some studies do not find any association (20, 25), whereas according to other studies a high maternal BMI impacts the bacterial composition in human milk (85); in some cases the diversity is less, with higher amounts of Staphylococcus and Akkermansia and lower numbers of Bifidobacterium in mature milk at 6 mo observed in women of a higher maternal BMI (8, 26). Among those conducted in developing countries, some have also reported no association (14, 20). In our study, only Roseomonas was more abundant in the milk of obese than healthy weight mothers. Roseomonas, which is considered an opportunistic bacteria to humans, and although it is a lactic acid producer, it has been isolated from aquatic environments and has been found in human blood (86). It has not been reported in previous studies in human milk but due to its environmental origin it might colonize milk after the infant's consumption of agüitas or directly via consumption by the mother. Further research is needed to understand its higher abundance in obese mothers.
SCM
SCM is a common, asymptomatic, inflammatory condition of the lactating mammary gland (87, 88). Due to the lack of symptoms, SCM is commonly underdiagnosed and it has been suggested that human milk bacterial dysbiosis may underscore SCM (48) and lead to impaired growth (40, 41). Previous reports describing the microbiota composition in both acute and subacute mastitis have identified Aeromonas,Staphylococcus,Ralstonia,Klebsiella,Serratia,Enterococcus, and Pseudomonas (33). Specifically, in subacute mastitis, human milk is reportedly dominated by the genus Staphylococcus (33, 89, 90). Erwinia,Bacillus,Pantoea,Cronobacter, and Pseudomonas may be present but Acinetobacter, Ruminococcus,Clostridium,Faecalibacterium, and Eubacterium are reportedly scarce (91). In general, previous studies have associated subacute (different from subclinical) mastitis with a lower microbial diversity, a greater abundance of pathogenic bacteria, and a depletion of commensal bacteria in human milk (33, 63, 91). In our SCM mothers, a lower microbial diversity was also found. Of note, Bosea,Lactococcus, and Leucobacter were absent in all our SCM samples and Lactobacillus was only rarely detected. In contrast, mothers who did not have SCM had a much higher abundance of lactic acid bacteria (Lactobacillus,Leuconostoc,Lactococcus), Leucobacter and Micrococcus compared with mothers with SCM. Lactic acid bacteria isolated from human milk have previously been shown to prevent proliferation of pathogenic bacteria such as Staphylococcus aureus (92) and more recently, against a wider range of human pathogens such as Salmonella enterica, Klebsiella pneumonia, Listeria monocytogenes, Vibrio cholerae, and Shigella flexneri (69). In addition, Lactobacillus gasseri,Lactobacillus salivarius, and Lactobacillus fementum have been used to treat mastitis with positive results; a lower recurrence of mastitis (93), lower staphylococcal count, and absence of symptoms at 14 d posttreatment (94) have been reported in mothers in Spain. In our study, the presence of Lactobacillus and Lactococcus in non-SCM milk and the complete absence of these bacteria in SCM milk would suggest that the lack of lactic-acid-producing bacteria is associated with human milk dysbiosis and the emergence of SCM.
Bosea and Klebsiella, were identified in samples from non-SCM mothers in our study. Klebsiella normally colonizes the gut although it may also be found on the skin and nasopharynx and transmission is probably fecal-oral or via direct contact (95). Both Klebsiella and Bosea have also been isolated from soil and water (96, 97). Recent studies highlight the potential for environmental factors to modify the human milk microbiome (98). These observations may suggest that lactating women in our study acquired such microbes from the environment or from the infant through the retrograde flow of bacteria from agüitas to the mammary gland and would suggest that this linkage might be an important contributor to the microbial diversity in human milk (98).
Interestingly, although Bifidobacteria is normally found in human milk (61, 99, 100), we did not detect it in our milk samples. Other studies have also reported its absence in human milk samples (8, 25, 62, 101). Several other studies also report low concentrations ranging from 2 to 11% (100, 102–104). Others have reported low concentrations at birth that substantially increase within the first month (105). Remarkably, a longitudinal study concluded that Bifidobacteria first appears in infant feces rather than in human milk and suggests that Bifidobacteria is more likely to be transferred from the infant gut to human milk, contrary to what was previously believed (102). There is also some limited evidence that the absence of Bifidobacterium in the mouse gut has been associated with micronutrient deficiencies (106). We have previously reported a range of mineral and trace element concentrations in breast that fail to meet nutrient requirements for breastfeeding infants (88). Thus, the absence of Bifidobacteria in our population may be associated with selection of the primer (98) but could also be associated with low concentrations of micronutrients in the milk; however, this requires further investigation.
Breastfeeding practices
Breastfeeding provides the infant with optimal nutrition and protects against morbidity and mortality associated with infection especially in developing countries (107). Furthermore, exclusive breastfeeding has demonstrated protection against infant deaths attributable to respiratory infections and diarrhea in developing countries (108, 109), and exclusive breastfeeding is associated with healthier bacterial communities in the infant's gut (110). It has been suggested that this protection is related to the colonization of the infant gastrointestinal tract with hundreds of bacterial phylotypes transmitted through human milk (62, 111).
When we compared EBF/PBF and mixed feeding mothers, 10 bacterial taxonomic units significantly differed. Comparing our findings with others, previous reports show the presence of similar bacterial genera in the exclusively breastfed group including Corynebacterium,Lactobacillus, and Micrococcus (36). Corynebacterium has been previously identified in human milk (6, 25, 62) and has been reported in infant gut and feces, as well as maternal feces, which might suggest that Corynebacterium colonizes the infant gut through human milk (100, 112, 113). On the other hand, Brevundiomonas,Afipia, and Paracoccus, which were also found in our milk samples, are Gram-negative bacteria that have been associated with human infections (74, 97). Brevundimonas have previously been identified in human milk and in the maternal gut. Leptotrichia, which was the only bacteria predominantly abundant in the mixed feeding group, is an oral bacteria where certain species have been associated with clinical diseases (114). Because it has been reported previously in mature milk (8), these observations highlight the importance of promoting further research at species level.
In our study, we used CAP plots to further differentiate the milk microbiota by breastfeeding practices, noting distinct differences among those who exclusively breastfed in early or established lactation, those that were classified as predominantly breastfeeding because of the added agüitas during early or established lactation, and those who added solid foods while continuing to breastfeed during established lactation (mixed breastfeeding). The introduction of solid foods is known to be associated with the development of an adult-like microbiota (115). However, there are no reports in the scientific literature describing the addition of ritual fluids or herbal teas, commonly used in this population (49), on the microbiota. In this population, we were interested to know even though prepared with boiled water, if shifts in the composition of the milk microbiota might occur with the consumption of agüitas. Our analysis of exclusive and predominant breastfeeding by stage of lactation and by breastfeeding practice revealed variation of the milk microbiota due to the introduction of agüitas in predominantly breastfed infants. Together, our identification of subtle changes in the milk microbiota is novel but does require further investigation.
Strengths and limitations
The strengths of our study include the observation that the majority of Mam-Mayan mothers complied with WHO recommendations to exclusively breastfeed for 6 mo (43, 49). We are among the first studies to explore the influence of parity, maternal age, and SCM on human milk microbiota composition in an indigenous community, where there is evidence that growth faltering occurs within the first 6 mo postpartum (50). The method of extraction is also a strength, by doing it manually we avoided the use of a pump that could be a source of contamination (22).
We recognize our study had some limitations. In this study we used the 27F/533R primer, which can amplify the core human milk genus Cutibacterium (10, 62, 100), but cannot amplify species from the genus Bifidobacterium (116), whereas the other commonly used primer 515F/806R amplifies Bifidobacterium but not Cutibacterium (formerly Propriobacterium) (98). Yet, Bifidobacterium spp. are often (22) but not always identified in human milk studies using such primers (14, 117) if Bifidobacterium abundance and prevalence is low (105).
Conclusion
The WHO recommends EBF for the first 6 mo of life, followed by breastfeeding in combination with the introduction of complementary foods until at least the age of 24 mo (118). Our data supports this global recommendation as we showed that 9 bacterial genera including Lactobacillus and Lactococcus, had a higher abundance in the milk of mothers who exclusively and/or predominantly breastfed, had a healthy BMI, and who did not have SCM. The current study provides new insights into the composition of microbiota in human milk by establishing those maternal factors that impact the human milk microbiota in the Mam-Mayan indigenous community in Guatemala. Breastfeeding practices can shift the composition of the milk microbiota during early and in established lactation. These findings highlight the importance of promoting exclusive and predominant breastfeeding to enhance bacterial diversity during lactation. Even though we were able to identify similarities in the major phyla and genera with studies from developed countries, our differential abundance analyses revealed that understudied maternal factors, including adolescent pregnancies, parity, and SCM, which are common characteristics of developing countries and rural populations, modified the differential abundance of the human milk microbiota. Finally, we caution against the direct application of findings in developed countries to mothers living in remote agrarian settings in developing countries. More evidence is needed from developing countries and rural populations in order to confirm our findings.
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks to McGill University and Genome Quebec Innovation Centre for technical assistance with 16S rRNA sequencing. We thank HM Wren-Atilola, AM Chomat, and the CeSSIAM field team for sample collection and the participating mothers for providing valuable milk samples.
The authors’ responsibilities were as follows—KGK, CL, and EG: designed the study; CL: prepared samples for the 16S rRNA sequencing; EG: analyzed 16S rRNA sequencing data, created figures, and performed bioinformatics analyses; NWS: supervised field studies and milk collection; KGK, LBA, and NWS: provided financial support; LLL, CL, TA, MES, EG, and KGK: contributed to writing the final manuscript; and all authors: read and approved the final manuscript.
Notes
This study was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant #RGPIN-2016-04961.
Author disclosures: The authors report no conflicts of interest.
Supplemental Table 1 and Supplemental Figures 1–3 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/cdn/.
LLL and EG are principle coauthors.
Abbreviations used: CAP, constrained analysis of principal coordinates; db-RDA, distance-based redundancy analysis; EBF, exclusive breastfeeding; FC, fold change; FDR, false discovery rate; PBF, predominant breastfeeding; SCM, subclinical mastitis.
Contributor Information
Lilian Lopez Leyva, School of Human Nutrition, McGill University, Ste-Anne-de-Bellevue, QC, Canada.
Emmanuel Gonzalez, Canadian Centre for Computational Genomics (C3G), Department of Human Genetics, McGill University and Genome Quebec Innovation Centre, Montréal, QC, Canada; Microbiome Research Platform, McGill Interdisciplinary Initiative in Infection and Immunity (MI4), Genome Centre, McGill University, Montréal, QC, Canada.
Chen Li, School of Human Nutrition, McGill University, Ste-Anne-de-Bellevue, QC, Canada.
Tamara Ajeeb, School of Human Nutrition, McGill University, Ste-Anne-de-Bellevue, QC, Canada.
Noel W Solomons, Center for Studies of Sensory Impairment, Aging and Metabolism (CeSSIAM), Guatemala City, Guatemala.
Luis B Agellon, School of Human Nutrition, McGill University, Ste-Anne-de-Bellevue, QC, Canada.
Marilyn E Scott, Institute of Parasitology, McGill University, Ste-Anne-de-Bellevue, QC, Canada.
Kristine G Koski, Email: kristine.koski@mcgill.ca, School of Human Nutrition, McGill University, Ste-Anne-de-Bellevue, QC, Canada.
Data Availability
The data for this article may be shared following a written request to the corresponding author.
References
- 1. WHO . Breastfeeding. [Internet]. 2020; [cited 2020 May 18]. Available from: https://www.who.int/westernpacific/health-topics/breastfeeding. [Google Scholar]
- 2. Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, Rodríguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69(1):1–10. [DOI] [PubMed] [Google Scholar]
- 3. McGuire MK, McGuire MA. Human milk: mother nature's prototypical probiotic food?. Adv Nutr. 2015;6(1):112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomez-Gallego C, Garcia-Mantrana I, Salminen S, Collado MC. The human milk microbiome and factors influencing its composition and activity. Seminars in Fetal and Neonatal Medicine. 2016;21(6):400–5. [DOI] [PubMed] [Google Scholar]
- 5. Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, Nicol MP, Isolauri E, Yang B, Collado MCet al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. 2016;7:1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, Foster JA, Sellen DW, Kamau-Mbuthia E, Kamundia EWet al. What's normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE Study. Front Nutr. 2019;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016;7(1):54–60. [DOI] [PubMed] [Google Scholar]
- 8. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96(3):544–51. [DOI] [PubMed] [Google Scholar]
- 9. Collado MC, Delgado S, Maldonado A, Rodríguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48(5):523–8. [DOI] [PubMed] [Google Scholar]
- 10. Jiménez E, de Andrés J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, Codoñer FM, Ramón D, Fernández L, Rodríguez JM. Metagenomic analysis of milk of healthy and mastitis-suffering women. J Hum Lact. 2015;31(3):406–15. [DOI] [PubMed] [Google Scholar]
- 11. Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014;34(8):599–605. [DOI] [PubMed] [Google Scholar]
- 12. Chen P-W, Lin Y-L, Huang M-S. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. Journal of Food and Drug Analysis. 2018;26(4):1235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corona-Cervantes K, García-González I, Villalobos-Flores LE, Hernández-Quiroz F, Piña-Escobedo A, Hoyo-Vadillo C, Rangel-Calvillo MN, García-Mena J. Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns. PeerJ. 2020;8:e9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davé V, Street K, Francis S, Bradman A, Riley L, Eskenazi B, Holland N. Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr Res. 2016;79(6):846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drell T, Štšepetova J, Simm J, Rull K, Aleksejeva A, Antson A, Tillmann V, Metsis M, Sepp E, Salumets Aet al. The influence of different maternal microbial communities on the development of infant gut and oral microbiota. Sci Rep. 2017;7(1):9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vaidya YH, Patel SH, Patel RJ, Pandit RJ, Joshi CG, Kunjadia AP. Human milk microbiome in urban and rural populations of India. Meta Gene. 2017;13:13–22. [Google Scholar]
- 17. Carvalho-Ramos II, Duarte RTD, Brandt KG, Martinez MB, Taddei CR. Breastfeeding increases microbial community resilience. J Pediatr (Rio J). 2018;94(3):258–67. [DOI] [PubMed] [Google Scholar]
- 18. Sakwinska O, Moine D, Delley M, Combremont S, Rezzonico E, Descombes P, Vinyes-Pares G, Zhang Y, Wang P, Thakkar Set al. Microbiota in breast milk of Chinese lactating mothers. PLoS One. 2016;11(8):e0160856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meehan CL, Lackey KA, Hagen EH, Williams JE, Roulette J, Helfrecht C, McGuire MA, McGuire MK. Social networks, cooperative breeding, and the human milk microbiome. Am J Hum Biol. 2018;30(4):e23131. [DOI] [PubMed] [Google Scholar]
- 20. Li S-W, Watanabe K, Hsu C-C, Chao S-H, Yang Z-H, Lin Y-J, Chen C-C, Cao Y-M, Huang H-C, Chang C-Het al. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and mainland China. Front Microbiol. 2017;8:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McGuire MK, McGuire MA. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr Opin Biotechnol. 2017;44:63–8. [DOI] [PubMed] [Google Scholar]
- 22. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza R, Becker AB, Mandhane PJet al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host & Microbe. 2019;25(2):324–35..e4. [DOI] [PubMed] [Google Scholar]
- 23. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016;4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zimmermann P, Curtis N. Breast milk microbiota: a complex microbiome with multiple impacts and conditioning factors. J Infect. 2020;81(1):17–47. [DOI] [PubMed] [Google Scholar]
- 25. Williams JE, Carrothers JM, Lackey KA, Beatty NF, York MA, Brooker SL, Shafii B, William JP, Settles ML, McGuire MAet al. Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J Nutr. 2017;147(9):1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88(4):894–9. [DOI] [PubMed] [Google Scholar]
- 27. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xia H, Zhong Het al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host & Microbe. 2015;17(5):690–703. [DOI] [PubMed] [Google Scholar]
- 28. Lawrence AR. Biochemistry of human milk. [Internet]. 6th ed. 2005, [cited 2020 Aug 8]. Available from: https://www.sciencedirect.com/book/9780323028233/breastfeeding#book-info. [Google Scholar]
- 29. Le Huërou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. 2010;23(1):23–36. [DOI] [PubMed] [Google Scholar]
- 30. Marvin-Dowle K, Burley VJ, Soltani H. Nutrient intakes and nutritional biomarkers in pregnant adolescents: a systematic review of studies in developed countries. BMC Pregnancy Childbirth. 2016;16:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sternlicht MD, Kouros-Mehr H, Lu P, Werb Z. Hormonal and local control of mammary branching morphogenesis. Differentiation. 2006;74(7):365–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nasioudis D, Forney LJ, Schneider GM, Gliniewicz K, France M, Boester A, Sawai M, Scholl J, Witkin SS. Influence of pregnancy history on the vaginal microbiome of pregnant women in their first trimester. Sci Rep. 2017;7(1):10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Angelopoulou A, Field D, Ryan CA, Stanton C, Hill C, Ross RP. The microbiology and treatment of human mastitis. Med Microbiol Immunol. 2018;207(2):83–94. [DOI] [PubMed] [Google Scholar]
- 34. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Yet al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castanys-Muñoz E, Martin MJ, Vazquez E. Building a beneficial microbiome from Birth. Adv Nutr. 2016;7(2):323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Collado MC, Cernada M, Baüerl C, Vento M, Pérez-Martínez G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. 2012;3(4):352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi Ket al. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. 2016;11(6):e0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. WHO . WHO | Global Strategy for Women's, Children's and Adolescent's Health 2016–2030. [Internet]. WHO. World Health Organization; 2015; cited 2020 Aug 8. Available from: http://www.who.int/life-course/publications/global-strategy-2016-2030/en/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. PAHO, WHO . PAHO/WHO | Latin America and the Caribbean have the second highest adolescent pregnancy rates in the world. [Internet]. Pan American Health Organization /World Health Organization; 2018; [cited 2020 Jan 25]. Available from: https://www.paho.org/hq/index.php?option=com_content&view=article&id=14163:latin-america-and-the-caribbean-have-the-second-highest-adolescent-pregnancy-rates-in-the-world&Itemid=1926&lang=en#:∼:targetText=Adolescent%20pregnancy%20by%20the%20numbers%3A&targetText=The%20adolescent%20fertility%20rate%20in,1000%20women%20during%202010%2D2015.&targetText=Approximately%2016%20million%20girls%20aged,each%20year%20around%20the%20world. [Google Scholar]
- 40. Li C, Solomons NW, Scott ME, Koski KG. Anthropometry before day 46 and growth velocity before 6 months of Guatemalan breastfed infants are associated with subclinical mastitis and milk cytokines, minerals, and trace elements. J Nutr. 2019;149(9):1651–9. [DOI] [PubMed] [Google Scholar]
- 41. Wren-Atilola HM, Solomons NW, Scott ME, Koski KG. Infant growth faltering linked to subclinical mastitis, maternal faecal-oral contamination, and breastfeeding. Matern Child Nutr. 2019;15(3):e12756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. UNICEF . Infant and young child feeding. [Internet]. UNICEF DATA. 2019, [cited 2020 Jan 25], Available from: https://data.unicef.org/topic/nutrition/infant-and-young-child-feeding/. [Google Scholar]
- 43. WHO . WHO | The optimal duration of exclusive breastfeeding: report of the expert consultation. [Internet]. WHO. World Health Organization; 2001, [cited 2020 Aug 5], Available from: http://www.who.int/nutrition/publications/infantfeeding/WHO_NHD_01.09/en/. [Google Scholar]
- 44. Colombara DV, Hernández B, Gagnier MC, Johanns C, Desai SS, Haakenstad A, McNellan CR, Palmisano EB, Ríos-Zertuche D, Schaefer Aet al. Breastfeeding practices among poor women in Mesoamerica. J Nutr. 2015;145(8):1958–65. [DOI] [PubMed] [Google Scholar]
- 45. Doak CM, van der Starre RE, van Beusekom I, Campos Ponce M, Vossenaar M, Solomons NW. Earlier introduction of aguitas is associated with higher risk of stunting in infants and toddlers in the Western Highlands of Guatemala. Am J Clin Nutr. 2013;97(3):631–6. [DOI] [PubMed] [Google Scholar]
- 46. van Beusekom I, Vossenaar M, Montenegro-Bethancourt G, Doak CM, Solomons NW. Estimates of exclusive breastfeeding rates among mother-infant dyads in Quetzaltenango, Guatemala, vary according to interview method and time frame. Food Nutr Bull. 2013;34(2):160–8. [DOI] [PubMed] [Google Scholar]
- 47. Chomat AM, Solomons NW, Koski KG, Wren HM, Vossenaar M, Scott ME. Quantitative methodologies reveal a diversity of nutrition, infection/illness, and psychosocial stressors during pregnancy and lactation in rural Mam-Mayan mother-infant dyads from the Western Highlands of Guatemala. Food Nutr Bull. 2015;36(4):415–40. [DOI] [PubMed] [Google Scholar]
- 48. WHO . WHO | Mastitis: causes and management. [Internet]. WHO. World Health Organization; 2000; [cited 2020 Aug 8]. Available from: https://www.who.int/maternal_child_adolescent/documents/fch_cah_00_13/en/. [Google Scholar]
- 49. Wren HM, Solomons NW, Chomat AM, Scott ME, Koski KG. Cultural determinants of optimal breastfeeding practices among indigenous Mam-Mayan women in the Western Highlands of Guatemala. J Hum Lact. 2015;31(1):172–84. [DOI] [PubMed] [Google Scholar]
- 50. Li C, Solomons NW, Scott ME, Koski KG. Minerals and trace elements in human breast milk are associated with Guatemalan infant anthropometric outcomes within the first 6 months. J Nutr. 2016;146(10):2067–74. [DOI] [PubMed] [Google Scholar]
- 51. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJet al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. AEM. 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gonzalez E, Brereton NJB, Marleau J, Guidi Nissim W, Labrecque M, Pitre FE, Joly S. Meta-transcriptomics indicates biotic cross-tolerance in willow trees cultivated on petroleum hydrocarbon contaminated soil. BMC Plant Biol. 2015;15(1):246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brereton NJB, Gonzalez E, Desjardins D, Labrecque M, Pitre FE. Co-cropping with three phytoremediation crops influences rhizosphere microbiome community in contaminated soil. Sci Total Environ. 2020;711:135067. [DOI] [PubMed] [Google Scholar]
- 54. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O'Hara R, Simpson G, Solymos Pet al. vegan: community ecology package, [Internet]. . 2020, [cited 2021 Mar 9]. Available from:, https://cran.r-project.org/web/packages/vegan/index.html. [Google Scholar]
- 56. Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a web browser. BMC Bioinformatics. 2011;12(1):385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ. 2015;3:e1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham Aet al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jonsson V, Österlund T, Nerman O, Kristiansson E. Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics. 2016;17(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. González R, Mandomando I, Fumadó V, Sacoor C, Macete E, Alonso PL, Menendez C. Breast milk and gut microbiota in African mothers and infants from an area of high HIV prevalence. PLoS One. 2013;8(11):e80299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MAet al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6(6):e21313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murphy K, Curley D, O'Callaghan TF, O'Shea C-A, Dempsey EM, O'Toole PW, Ross RP, Ryan CA, Stanton C. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. 2017;7:40597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ding M, Qi C, Yang Z, Jiang S, Bi Y, Lai J, Sun J. Geographical location specific composition of cultured microbiota and Lactobacillus occurrence in human breast milk in China. Food Funct. 2019;10(2):554–64. [DOI] [PubMed] [Google Scholar]
- 65. Gueimonde M, Laitinen K, Salminen S, Isolauri E. Breast milk: a source of Bifidobacteria for infant gut development and maturation?. Neonatology. 2007;92(1):64–6. [DOI] [PubMed] [Google Scholar]
- 66. Ramsay DT, Kent JC, Owens RA, Hartmann PE. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics. 2004;113(2):361–7. [DOI] [PubMed] [Google Scholar]
- 67. Damaceno QS, Souza JP, Nicoli JR, Paula RL, Assis GB, Figueiredo HC, Azevedo V, Martins FS. Evaluation of potential probiotics isolated from human milk and colostrum. Probiotics & Antimicro Prot. 2017;9(4):371–9. [DOI] [PubMed] [Google Scholar]
- 68. Delorme C, Abraham A-L, Renault P, Guédon E. Genomics of Streptococcus salivarius, a major human commensal. Infect Genet Evol. 2015;33:381–92. [DOI] [PubMed] [Google Scholar]
- 69. Mohammadi F, Eshaghi M, Razavi S, Sarokhalil DD, Talebi M, Pourshafie MR. Characterization of bacteriocin production in Lactobacillus spp. isolated from mother's milk. Microb Pathog. 2018;118:242–6. [DOI] [PubMed] [Google Scholar]
- 70. Toscano M, De Grandi R, Stronati L, De Vecchi E, Drago L. Effect of Lactobacillus rhamnosus HN001 and Bifidobacterium longum BB536 on the healthy gut microbiota composition at phyla and species level: a preliminary study. WJG. 2017;23(15):2696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Willems A. The Family Comamonadaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. [Internet]. Berlin (Heidelberg): Springer Berlin Heidelberg; 2014. p. 777–851., Available from: http://link.springer.com/10.1007/978-3-642-30197-1_238. [Google Scholar]
- 72. Liu G, Tang CM, Exley RM. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiol Read Engl. 2015;161(7):1297–312. [DOI] [PubMed] [Google Scholar]
- 73. Rodríguez JM. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation?. Adv Nutr. 2014;5(6):779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weyant RS, Whitney AM. Afipia. In: Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S, . Bergey's manual of systematics of archaea and bacteria. [Internet]. Chichester (UK): John Wiley & Sons, Ltd; 2015. p. 1–11., Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781118960608.gbm00798. [Google Scholar]
- 75. Dé I, Rolston KVI, Han XY. Clinical significance of Roseomonas species isolated from catheter and blood samples: analysis of 36 cases in patients with cancer. Clin Infect Dis. 2004;38(11):1579–84. [DOI] [PubMed] [Google Scholar]
- 76. Bartlett JA, Schleifer SJ, Demetrikopoulos MK, Delaney BR, Shiflett SC, Keller SE. Immune function in healthy adolescents. Clin Diagn Lab Immunol. 1998;5(1):105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Garofalo R. Cytokines in human milk. J Pediatr. 2010;156(2 Suppl):S36–40. [DOI] [PubMed] [Google Scholar]
- 78. Flores N, Villegas E, Villacís D, Fornasini M, Cifuentes SG, Narváez L, Baldeón ME. Concentrations of immunoglobulin A, interleukin-6, and tumor necrosis factor in breastmilk of adolescent and adult mothers in Quito, Ecuador: a cohort study. Breastfeeding Medicine. 2014;9(2):107–8. [DOI] [PubMed] [Google Scholar]
- 79. Galicia L, Grajeda R, de Romaña DL. Nutrition situation in Latin America and the Caribbean: current scenario, past trends, and data gaps. Rev Panam Salud Pública. 2016;40(2):104–13. [PubMed] [Google Scholar]
- 80. Al-Awadi FM, Srikumar TS. Trace-element status in milk and plasma of Kuwaiti and non-Kuwaiti lactating mothers. Nutrition. 2000;16(11–12):1069–73. [DOI] [PubMed] [Google Scholar]
- 81. Almeida AA, Lopes C, Silva AMS, Barrado E. Trace elements in human milk: correlation with blood levels, inter-element correlations and changes in concentration during the first month of lactation. J Trace Elem Med Biol. 2008;22(3):196–205. [DOI] [PubMed] [Google Scholar]
- 82. Bolognini Pereira K, Blondet de Azeredo V, Barros da Sileira C, Magnago Pedruzzi L. Composición de la leche de madres lactantes adolescentes en relación con el tiempo de lactancia. Nutr Hosp. 2013;28(6):1971–6., Available from: http://scielo.isciii.es/pdf/nh/v28n6/28originalpediatria06.pdf. [PubMed] [Google Scholar]
- 83. de Azeredo VB, Trugo NMF. Retinol, carotenoids, and tocopherols in the milk of lactating adolescents and relationships with plasma concentrations. Nutrition. 2008;24(2):133–9. [DOI] [PubMed] [Google Scholar]
- 84. Silvestre MD, Lagarda MJ, Farré R, Martínez Costa C, Brines J. Copper, iron and zinc determinations in human milk using FAAS with microwave digestion. Food Chem. 2000;68(1):95–9. [Google Scholar]
- 85. Lundgren SN, Madan JC, Karagas MR, Morrison HG, Hoen AG, Christensen BC. Microbial communities in human milk relate to measures of maternal weight. Front Microbiol. 2019;10::2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Han XY, Pham AS, Tarrand JJ, Rolston KV, Helsel LO, Levett PN. Bacteriologic characterization of 36 strains of Roseomonas species and proposal of Roseomonas mucosa sp nov and Roseomonas gilardii subsp rosea subsp nov. Am J Clin Pathol. 2003;120(2):256–64. [DOI] [PubMed] [Google Scholar]
- 87. Willumsen J, Filteau S, Coutsoudis A, Uebel K, Newell M, Tomkins A. Subclinical mastitis as a risk factor for mother-infant HIV transmission. In: Short and long term effects of breast feeding on child health. USA: Springer; 2002. p. 211–23. [DOI] [PubMed] [Google Scholar]
- 88. Li C, Solomons NW, Scott ME, Koski KG. Subclinical mastitis (SCM) and proinflammatory cytokines are associated with mineral and trace element concentrations in human breast milk. J Trace Elem Med Biol. 2018;46:55–61. [DOI] [PubMed] [Google Scholar]
- 89. Delgado S, Arroyo R, Martín R, Rodríguez JM. PCR-DGGE assessment of the bacterial diversity of breast milk in women with lactational infectious mastitis. BMC Infect Dis. 2008;8(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Delgado S, Collado MC, Fernández L, Rodríguez JM. Bacterial analysis of breast milk: a tool to differentiate Raynaud's phenomenon from infectious mastitis during lactation. Curr Microbiol. 2009;59(1):59–64. [DOI] [PubMed] [Google Scholar]
- 91. Patel SH, Vaidya YH, Patel RJ, Pandit RJ, Joshi CG, Kunjadiya AP. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep. 2017;7:7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Heikkilä MP, Saris PEJ. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95(3):471–8. [DOI] [PubMed] [Google Scholar]
- 93. Arroyo R, Martín V, Maldonado A, Jiménez E, Fernández L, Rodríguez JM. Treatment of infectious mastitis during lactation: antibiotics versus oral administration of lactobacilli isolated from breast milk. Clin Infect Dis. 2010;50(12):1551–8. [DOI] [PubMed] [Google Scholar]
- 94. Jiménez E, Fernández L, Maldonado A, Martín R, Olivares M, Xaus J, Rodríguez JM. Oral administration of lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. AEM. 2008;74(15):4650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fox J, Anderson L, Lowe F, Quimby F . Laboratory animal medicine. In: Laboratory animal medicine. [Internet]. 2nd ed. San Diego: Academic Press; 2002. p. 1325, [cited 2020 Aug 8], Availabe from: https://www.elsevier.com/books/laboratory-animal-medicine/fox/978-0-12-263951-7. [Google Scholar]
- 96. De Meyer SE, Willems A. Multilocus sequence analysis of Bosea species and description of Bosea lupini sp. nov., Bosea lathyri sp. nov. and Bosea robiniae sp. nov., isolated from legumes. Int J Syst Evol Microbiol. 2012;62(Pt 10):2505–10. [DOI] [PubMed] [Google Scholar]
- 97. Lamoth F, Greub G. Amoebal pathogens as emerging causal agents of pneumonia. FEMS Microbiol Rev. 2010;34(3):260–80. [DOI] [PubMed] [Google Scholar]
- 98. Lopez Leyva L, Brereton NJB, Koski KG. Emerging frontiers in human milk microbiome research and suggested primers for 16S rRNA gene analysis. Computational and Structural Biotechnology Journal. 2021;19:121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Biagi E, Aceti A, Quercia S, Beghetti I, Rampelli S, Turroni S, Soverini M, Zambrini AV, Faldella G, Candela Met al. Microbial community dynamics in mother's milk and infant's mouth and gut in moderately preterm infants. Front Microbiol. 2018;9:2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110(7):1253–62. [DOI] [PubMed] [Google Scholar]
- 101. Ward TL, Hosid S, Ioshikhes I, Altosaar I. Human milk metagenome: a functional capacity analysis. BMC Microbiol. 2013;13:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Makino H, Martin R, Ishikawa E, Gawad A, Kubota H, Sakai T, Oishi K, Tanaka R, Ben-Amor K, Knol Jet al. Multilocus sequence typing of bifidobacterial strains from infant's faeces and human milk: are Bifidobacteria being sustainably shared during breastfeeding?. Beneficial Microbes. 2015;6(4):563–72. [DOI] [PubMed] [Google Scholar]
- 103. Solís G, de los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. Establishment and development of lactic acid bacteria and Bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe. 2010;16(3):307–10. [DOI] [PubMed] [Google Scholar]
- 104. Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and Bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J Pediatr Gastroenterol Nutr. 2014;59(1):78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sakwinska O, Bosco N. Host microbe interactions in the lactating mammary gland. Front Microbiol. 2019;10:1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pachikian BD, Neyrinck AM, Deldicque L, De Backer FC, Catry E, Dewulf EM, Sohet FM, Bindels LB, Everard A, Francaux Met al. Changes in intestinal Bifidobacteria levels are associated with the inflammatory response in magnesium-deficient mice. J Nutr. 2010;140(3):509–14. [DOI] [PubMed] [Google Scholar]
- 107. Duijts L, Jaddoe VWV, Hofman A, Moll HA. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics. 2010;126(1):e18–25. [DOI] [PubMed] [Google Scholar]
- 108. Arifeen S, Black RE, Antelman G, Baqui A, Caulfield L, Becker S. Exclusive breastfeeding reduces acute respiratory infection and diarrhea deaths among infants in Dhaka slums. Pediatrics. 200;108(4):e67. [DOI] [PubMed] [Google Scholar]
- 109. Turin CG, Ochoa TJ. The role of maternal breast milk in preventing infantile diarrhea in the developing world. Curr Trop Med Rep. 2014;1(2):97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, Bender JM, Azad MB, Thompson AL, Weiss Set al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. 2018;9:4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sanz Y. Gut microbiota and probiotics in maternal and infant health. Am J Clin Nutr. 2011;94(6 Suppl):2000S–5S. [DOI] [PubMed] [Google Scholar]
- 112. Bezirtzoglou E, Stavropoulou E. Immunology and probiotic impact of the newborn and young children intestinal microflora. Anaerobe. 2011;17(6):369–74. [DOI] [PubMed] [Google Scholar]
- 113. Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, Faldella G, Candela M, Brigidi P, Corvaglia L. The bacterial ecosystem of mother's milk and infant's mouth and gut. Front Microbiol. 2017;8:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Eribe ERK, Olsen I. Leptotrichia species in human infections II. J Oral Microbiol. 2017;9(1):1368848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci. 2011;108(Supplement_1):4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ramani S, Stewart CJ, Laucirica DR, Ajami NJ, Robertson B, Autran CA, Shinge D, Rani S, Anandan S, Hu Let al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat Commun. 2018;9(1):5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. WHO . Infant and young child feeding, [Internet]. World Health Organization. 2020, [cited 2020 Dec 2]. Available from: https://www.who.int/news-room/fact-sheets/detail/infant-and-young-child-feeding. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this article may be shared following a written request to the corresponding author.