Abstract
Background
Mass testing for early identification and isolation of infectious COVID-19 individuals is efficacious for reducing disease spread. Antigen-detecting rapid diagnostic tests (Ag-RDT) may be suitable for testing strategies; however, benchmark comparisons are scarce.
Methods
We used 286 nasopharyngeal specimens from unexposed asymptomatic individuals collected between December 2020 and January 2021 to assess five Ag-RDTs marketed by Abbott, Siemens, Roche Diagnostics, Lepu Medical, and Surescreen.
Results
For the overall sample, the performance parameters of Ag-RDTs were as follows: Abbott assay, sensitivity 38.6% (95%CI 29.1–48.8) and specificity 99.5% (97–100%); Siemens, sensitivity 51.5% (41.3–61.6) and specificity 98.4% (95.3–99.6); Roche, sensitivity 43.6% (33.7–53.8) and specificity 96.2% (92.4–98.5); Lepu, sensitivity 45.5% (35.6–55.8) and specificity 89.2% (83.8–93.3%); Surescreen, sensitivity 28.8% (20.2–38.6) and specificity 97.8% (94.5–99.4%). For specimens with cycle threshold (Ct) <30 in RT-qPCR, all Ag-RDT achieved a sensitivity ≥70%. The modelled negative- and positive-predictive value for 1% prevalence were >99% and <50%, respectively.
Conclusions
When screening unexposed asymptomatic individuals, two Ag-RDTs achieved sensitivity ≥80% for specimens with Ct<30 and specificity ≥96%. The estimated negative predictive value suggests the suitability of Ag-RDTs for mass screenings of SARS-CoV-2 infection in the general population.
Keywords: SARS-CoV-2, Antigen-detecting rapid diagnostic test, Head-to-head comparison, Mass screening
Introduction
Mass testing for early identification and isolation of individuals infected with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), irrespective of symptoms, is potentially an efficacious strategy to reduce disease transmission.1 Recent advances on the validation of Antigen-detecting Rapid Diagnostic Tests (Ag-RDTs) show promise to replace central laboratory techniques for epidemiological control of the SARS-CoV-2 through mass testing.
Reverse transcription-polymerase chain reaction (RT-qPCR) is the current gold standard for identifying the presence of the SARS-CoV-2 in respiratory specimens.2 More recently, transcription-mediated amplification (TMA) of the SARS-CoV-2 genome has been added to the repertoire of nucleic acid amplification tests (NAAT) for SARS-CoV-2 detection.3 Despite their high sensitivity, NAATs are associated with drawbacks that limit their use for community-based testing strategies, including the need for laboratory-processing, high cost, and long turnaround from sampling to results release. Furthermore, there is cumulative evidence indicating that the period of NAAT positivity in infected individuals largely exceeds the time window in which infectious viral particles can be isolated from the respiratory tract, raising doubts about the epidemiological meaning of a NAAT positive result.4
Ag-RDTs, commonly used in diagnosing other infectious diseases, have emerged as an alternative tool that meets the requirements for frequent testing at the point-of-care: rapid turnaround time, low cost, and ease-of-use.5 Overall, Ag-RDTs have lower sensitivity than NAATs; however, clinical validation studies have consistently reported increasing sensitivities in specimens with higher viral loads. These findings, along with the growing body of evidence on the lack of infectivity of cases with low viral load6, 7, 8, 9 and the potential long-tail of positivity when using highly sensitive methods such as PCR, suggest that frequent testing with Ag-RDTs―even those with low sensitivity―may be more effective than less frequent testing with RT-qPCR or TMA for mass screening campaigns to improve SARS-CoV-2 control.9 , 10
The performance parameters of Ag-RDTs are mostly based on testing respiratory specimens from clinically suspected cases11, 12, 13, 14 and contacts after exposure to a positive case.15, 16, 17, 18 However, the sensitivity bias associated with the viral load leads to high heterogeneity in the reported performance parameters, which strongly depend on the disease status and potential exposure (e.g., symptomatic vs. asymptomatic, contact vs. unexposed) of tested individuals. This heterogeneity precludes comparative analyses between tests assessed in different studies and challenges benchmarking of Ag-RDTs. Furthermore, head-to-head comparisons are scarce, particularly in samples from asymptomatic individuals, the target population of community-based screening strategies.19 , 20 In this study, we used fresh nasopharyngeal samples collected in routine mass screening campaigns of unexposed asymptomatic individuals to perform a head-to-head comparison of five Ag-RDTs.
Methods
Study design
As part of the surveillance program for pandemic control in Catalonia (North-East Spain), the local government launched NAAT-based systematic screenings in areas at high risk of an outbreak. The University Hospital Germans Trias i Pujol processed nasopharyngeal specimens collected in a healthcare area in North-East Spain (i.e., Metropolità Nord) with a catchment population of ∼1,400,000 people. These samples enabled us to assess the Ag-RDTs in line with The Foundation for Innovative New Diagnostics (FIND) target product profile for lateral flow assays that directly detect antigens of SARS-CoV-2 antigen assays,21 which recommends at least 100 known negative samples and 100 known positive samples with a documented RT-PCR result. In this study, we used samples collected between December 2020 and January 2021 (i.e., during the third wave of the epidemic in Spain) with RT-qPCR results available (i.e., data on cycle threshold [Ct]) to perform a head-to-head assessment of five Ag-RDTs. Samples with invalid results in any of the assessed Ag-RDTs were excluded from the analysis.
All samples used in this analysis had been collected in the setting of a public health surveillance program, and data were handled according to the General Data Protection Regulation 2016/679 on data protection and privacy for all individuals within the European Union and the local regulatory framework regarding data protection. The study protocol was approved by the ethics committee of Hospital Germans Trias i Pujol (Badalona, Spain).
Procedures
Samples consisted of nasopharyngeal swabs collected by health care workers during mass testing of unexposed asymptomatic individuals living in areas at high risk of an outbreak. Swab specimens were placed into sterile tubes containing viral transport media (DeltaSwab Virus, Deltalab; or UTM Universal Transport Medium, Copan). The reference test (i.e., RT-qPCR) was performed on fresh samples stored at 2–8 °C for up to 24 h; samples were then stored up to 12 h at 2–8 °C until their use for the five Ag-RDTs.
RNA for RT-qPCR tests were extracted from fresh samples using the viral RNA/Pathogen Nucleic Acid Isolation kit for the Microlab Starlet or Nimbus platforms (Hamilton, USA), according to the manufacturer's instructions. PCR amplification was conducted according to the recommendations of the 2019-nCoV RT-qPCR Diagnostic Panel of the Centers for Disease Control and Prevention (CDC) (REF), using the Allplex™ 2019-nCoV assay (Seegene, South Korea) on the CFX96 (Bio-Rad, USA) in line with manufacturer's instruction. Briefly, a 25 μL PCR reaction mix was prepared that contained 8 μL of each sample's nucleic acids, 2019-nCoV positive and negative controls, 5 μL of 2019-nCoV MOM (primer and probe mix) and 2 μL of real-time one-step Enzyme. Thermal cycling was performed at the following conditions: 20 min at 50 °C for reverse transcription, followed by 15 min at 95 °C, and then 45 cycles of 15 s at 94 °C and 30 s at 58 °C. An RT-qPCR was considered positive according to the manufacturer's instructions.22
Index tests included the following Ag-RDTs: PanBioTM COVID-19 Ag Rapid test (Abbott), CLINITEST® Rapid COVID-19 Antigen Test (Siemens), SARS-CoV-2 Rapid Antigen Test (Roche Diagnostics), SARS-CoV-2 Antigen Rapid Test Kit (Lepu Medical), and COVID-19 Coronavirus Rapid Antigen Test Cassette (Surescreen). Supplementary Table 1 provides further details regarding the specifications of each test. All Ag-RDT determinations were performed in parallel by two blinded technicians, who used approximately 100 μL of 1:2 mix of each kit buffer and the sample previously homogenised. Samples were applied directly to the test cassette and incubated for 15 min at room temperature before reading results at the naked eye, according to the manufacturer instructions (i.e., the presence of any test line (T), no matter how faint, indicates a positive result).
Outcomes and statistical analysis
We calculated that a sample size of at least 73 positive specimens and 165 negative specimens would give 80% power to estimate overall sensitivity and specificity of Ag-RDT assays in our study. We based our calculation on the expected sensitivity and specificity in asymptomatic population of 65% and 96%,17 , 23 respectively, fixed precision of the point estimate of 2.5%, and confidence level of 95%. The calculation was in line with FIND recommendations for assessing Ag-RDTs that retrospective assessments should include a minimum of 100 samples per RT-PCR result.21
The primary analysis of the head-to-head comparison was the sensitivity and specificity of each Ag-RDT. Sensitivity and specificity were calculated as defined by Altman and Bland,24 and reported as a percentage and the exact binomial 95% confidence interval (CI). Sensitivity was also analysed in a subset of samples with Ct<30, considered at high risk of transmission.
Secondary analyses were done assessing discordance between results obtained in each Ag-RDTs. Positive and negative-predictive values for each Ag-RDT at population prevalence between 1% and 15% for SARS-CoV-2 infection were modelled25 and plotted with the exact binomial 95% CI.26 All analyses and plots were performed using R version 3.6.27
Role of the funding source
The funders of the study had no role in the study conception, design, conduct, data analysis, or writing of the report. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
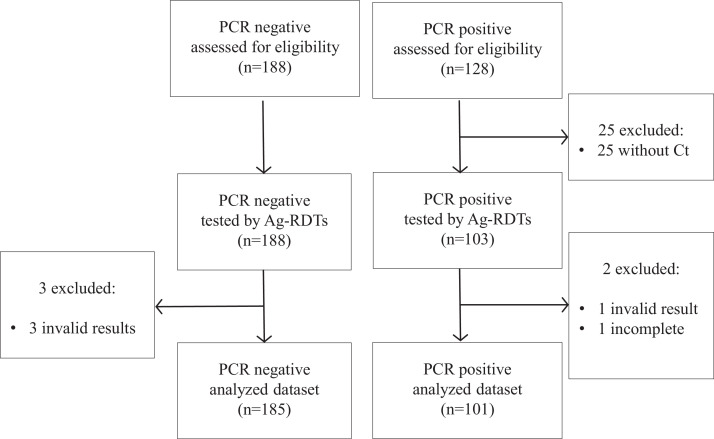
Our sample collection included 316 fresh nasopharyngeal swabs from unexposed asymptomatic individuals who had a RT-qPCR result available. Of these, 30 were excluded because of lack of documented Ct value (n=25), incomplete results due to limited sample volume (n=1), or invalid results in any of the Ag-RDTs (n=4, all of them in the Lepu assay), resulting in a study set of 286 samples: 101 (35.3%) with positive RT-qPCR result and 185 (64.7%) with negative RT-qPCR result (Fig. 1 ).
Fig. 1.
Flow-chart of sample inclusion.
All samples were nasopharyngeal swabs collected from unexposed asymptomatic individuals during screening campaigns.
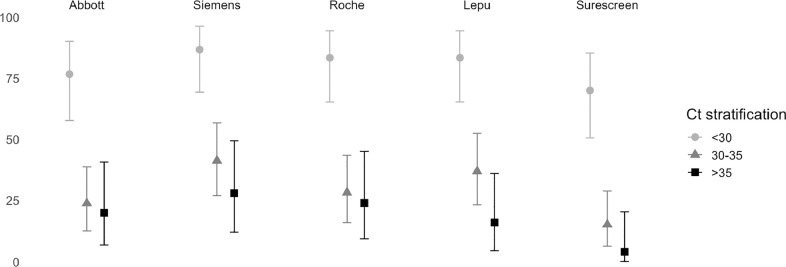
The Ct value of samples with positive RT-qPCR result was <30 in 30 (29.7%) samples, 30-to-35 in 46 (45.5%), and >35 in 25 (24.8%). The overall sensitivity and specificity of the analysed Ag-RDTs ranged from 28.7% to 51.5% and 89.2% to 99.5%, respectively (Table 1 ). When considering only RT-qPCR positive samples with Ct <30 (i.e., indicates a high concentration of viral genetic material which is typically associated with a higher risk of infectivity),28 the sensitivity of Ag-RDTs increased to 76.7% (95% CI 57.7–90.7) for the Abbott assay; 86.7% (69.3–96.3) for the Siemens Assay; 83.3% (65.3–94.4) for the Roche assay; 83.3% (65.3–94.4) for the Lepu assay; and to 70% (50.6–85.3%) for the Surescreen assay (Fig. 2 ).
Table 1.
Sensitivity and specificity of the antigen-detecting rapid diagnostic tests for SARS-CoV-2.
| Abbott | Siemens | Roche | Lepu | Surescreen | |
|---|---|---|---|---|---|
| Overall sensitivity | 38•61% (29•09–48•82) | 51•49% (41•33–61•55) | 43•56% (33•72–53•8) | 45•54% (35•6–55•76) | 28•71% (20•15–38•57) |
| Detected | 39 | 52 | 44 | 46 | 29 |
| Not detected | 62 | 49 | 57 | 55 | 72 |
| Total PCR+ | 101 | 101 | 101 | 101 | 101 |
| Sensitivity in specimens with Ct<30 | 76•67% (57•72–90•07) | 86•67% (69•28–96•24) | 83•33% (65•28–94•36) | 83•33% (65•28–94•36) | 70% (50•6–85•27) |
| Detected | 23 | 26 | 25 | 25 | 21 |
| Not detected | 7 | 4 | 5 | 5 | 9 |
| Total PCR+ | 30 | 30 | 30 | 30 | 30 |
| Specificity | 99•46% (97•03–99•99) | 98•38% (95•33–99•66) | 96•22% (92•36–98•47) | 89•19% (83•8–93•27) | 97•84% (94•56–99•41) |
| Detected | 1 | 3 | 7 | 20 | 4 |
| Not detected | 184 | 182 | 178 | 165 | 181 |
| Total PCR- | 185 | 185 | 185 | 185 | 185 |
All samples were nasopharyngeal swabs collected from unexposed asymptomatic individuals during mass screening campaigns. Sensitivity and specificity results are presented with the 95% confidence interval.
Fig. 2.
Sensitivity of the antigen-detecting rapid diagnostic tests according to the cycle threshold value of the RT-qPCR analysis.
Bars show the 95% confidence interval of the estimated sensitivity.
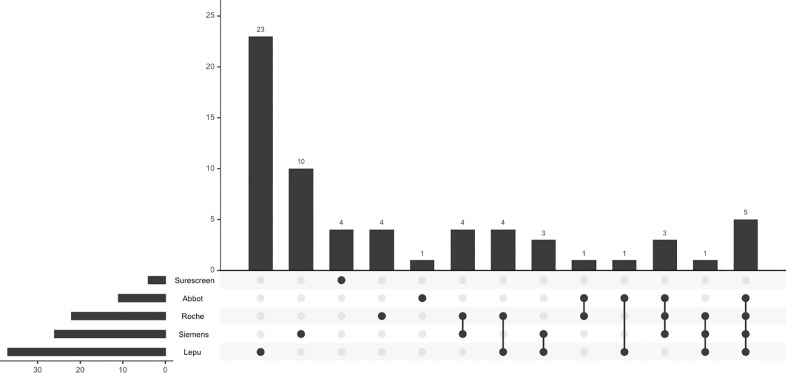
Of the 286 samples analysed by Ag-RDTs, 222 (77.6%) had concordant results across all Ag-RDT assessed. The 29 samples with concordant positive results across Ag-RDTs were all PCR-positive. Conversely, 37 (19.2%) of 193 specimens with negative results in all Ag-RDTs were PCR positive. Fig. 3 shows the distribution of Ag-RDT results in samples with discordant results. The Ag-RDT that most often yielded a positive result in samples with negative results in all other Ag-RDTs was the Lepu assay (n=23; 35.9%), followed by the Siemens assay (n=10, 15.6%). Table S2 summarises the cycle threshold distribution across discordances.
Fig. 3.
Discordance analysis between Ag-RDTs.
Bars show the number of samples for each discordance pattern. Black dots and grey dots indicate the assays showing positive and negative results in each discordance pattern. Table S2 summarizes the cycle threshold distribution across discordances.
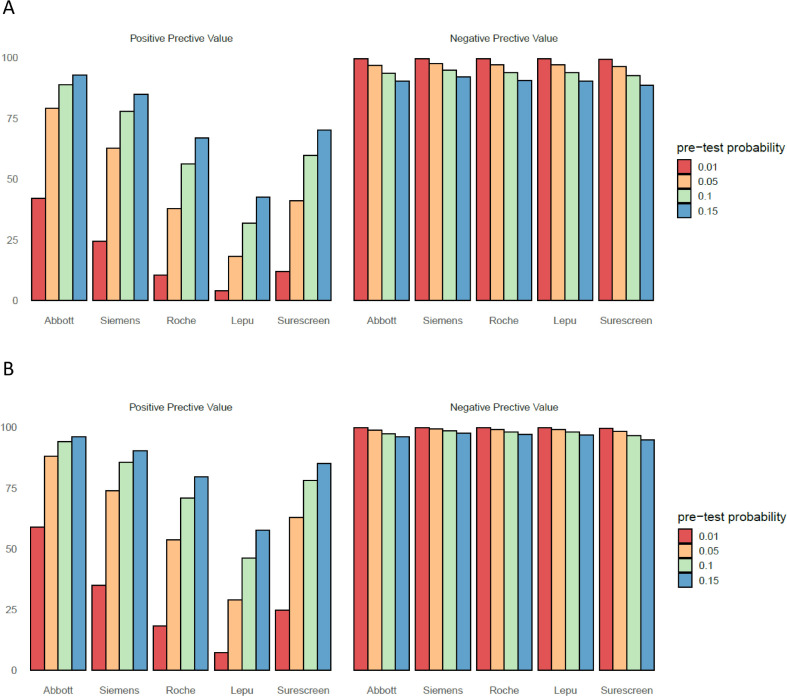
To provide an estimate of misidentified cases―either false-positive or false-negative cases―that can be used for making decisions in the public health setting, we modelled the positive and negative predictive value for a prevalence range consistent with a mass screening of unexposed asymptomatic individuals (Fig. 4 A). For the overall study sample, the estimated positive predictive value (PPV) at a 1% prevalence ranged from 4.1% to 41.9%, with the Lepu assay and the Abbott assay, respectively (Table S3). The estimated PPVs notably increased for the <30 Ct subgroup of samples (Fig. 4B), and when prevalence in the population was higher. The estimated negative predictive value (NPV) at 1% prevalence ranged from 99.3% to 99.5%, with the Surescreen assay and the Siemens assay, respectively.
Fig. 4.
Positive predictive value and negative predictive value according to pre-test probabilities.
A: overall sample (n= 286). B: samples with cycle threshold <30 in the RT-qPCR assay. Table S3 provides detailed values and confidence intervals for predicted false negative and false positives in the investigated prevalence.
Discussion
In this study, we compared head-to-head the sensitivity and specificity of five Ag-RDTs to screen SARS-CoV-2 infected individuals with unknown exposure and no clinical suspicion of COVID-19. Four of the tested Ag-RDTs (i.e., Abbott, Siemens, Roche, and Surescreen assays) showed a specificity higher than 96%. Regarding sensitivity, despite it was low for the overall sample (range 29–51%), the corresponding values for the subset of samples with a RT-qPCR value Ct <30 were higher than 80% for the Siemens, Roche, and Lepu assays. This finding is of particular interest for the proposed use of Ag-RDT as a reliable alternative to RT-qPCR for the rapid detection of individuals with higher risk of infectivity in mass screening of asymptomatic individuals. Pre-clinical studies have persistently reported a very low infectious capacity of respiratory specimens with viral loads below 106 genome copies/mL, which usually correspond to a Ct of approximately 29–31.5 , 8 , 29 These findings align with the significant increase of the secondary attack rate for values of Ct <30,30 indicating higher infectiousness among individuals with viral loads below this Ct threshold.
Although sensitivity and specificity are important intrinsic characteristics of a test, the number of expected errors when using the test for screening purposes strongly depends on the prevalence of the infection in the screened sample. Hence, positive and negative predictive values are a mainstay for making public health decisions regarding the use of a test. The reported prevalence of SARS-CoV-2 infection in PCR-based untargeted screenings of the general population typically ranges between 1% and 3%, depending on the virus transmission context.23 , 31 In low prevalence settings, Ag-RDTs will have a high NPV but a low PPV. According to our estimate, the NPV for SARS-CoV-2 infections at 1% prevalence was higher than 99% for all test, suggesting that a negative test may not require confirmation. In contrast, the PPV at 1% prevalence was lower than 50% in all tests, suggesting that a positive result will need immediate confirmation by RT-qPCR, even for highly specific assays.
Our study has several strengths and limitations. We used the same fresh set of samples for assessing five different Ag-RDTs and the sample size met the FIND recommendation for retrospective assessments of the clinical performance of these tests. Furthermore, to our knowledge, this is the first head-to-head comparison of Ag-RDT in asymptomatic screenings, an intended use proposed by various authors.5 , 10 , 17 , 23 On the other hand, our study was limited by the small number of specimens with Ct <30, a threshold deemed of interest for the use of Ag-RDT in screenings of the general population. In our sample, specimens below this threshold accounted for 30%; however, other authors have reported proportions of nearly 60% in random screenings of the general population.23 Of note, we used specimens in transport medium. This approach is convenient for mass screening strategies in which individuals with positive Ag-RDT results may need further diagnostic confirmation by PCR. However, only one manufacturer (i.e., the Roche assay) provided instructions on how to process samples collected in virus transport medium. The consistency of our results across assays, particularly regarding negative results, suggests that the use of this media had a little or negligible impact on test performance. Finally, it is worth mentioning that all nasopharyngeal swabs in our analysis were collected by trained healthcare professionals. According to a recent report of lateral flow viral antigen detection devices, the positivity rate might be lower in screenings performed by non-trained people.9
Our results provide policymakers with evidence on the use of Ag-RDT for mass screening of unexposed, asymptomatic individuals. Two commercial, widely available assays can be used for SARS-CoV-2 antigen testing to achieve sensitivity in specimens with a Ct<30 and specificity of at least 80% and 96%, respectively. While these tests may overlook SARS-CoV-2 infection with low viral loads, they accurately detect individuals with high viral loads and, therefore, at higher risk of transmission. Our findings also support the idea that Ag-RDTs can be used for mass screening in low prevalence settings and accurately rule out a highly infectious case in such setting. In models according to population prevalence, all Ag-RDTs will have a NPV >99% and a PPV<50% at 1% prevalence. Our results, together with the cumulative evidence on the limited overlapping between PCR positivity and the presence of infectious viral particles in the respiratory tract, encourage the design of public health interventions for containing viral COVID-19 spread that shift from positivity testing to infectivity testing. The low cost and short turnaround time of Ag-RDTs, which ease frequent testing, are additional advantages over assays better suited for diagnostic use like NAATs. In low-income countries with limited laboratory resources, the trade-off between targeted PCR analyses and massive screenings with Ag-RDTs should be carefully considered.
CRediT authorship contribution statement
Bàrbara Baro: Conceptualization, Methodology, Validation, Data curation, Writing – original draft. Pau Rodo: Methodology, Validation, Data curation. Dan Ouchi: Validation, Formal analysis. Antoni E. Bordoy: Data curation. Emilio N. Saya Amaro: Methodology, Data curation. Sergi V. Salsench: Methodology, Data curation. Sònia Molinos: . Andrea Alemany: Data curation. Maria Ubals: Data curation. Marc Corbacho-Monné: Data curation. Pere Millat-Martinez: Writing – review & editing, Data curation. Michael Marks: Writing – review & editing. Bonaventura Clotet: Funding acquisition, Writing – review & editing. Nuria Prat: Writing – review & editing. Oriol Estrada: Funding acquisition, Resources. Marc Vilar: Funding acquisition, Resources. Jordi Ara: Funding acquisition, Writing – review & editing. Martí Vall-Mayans: Writing – review & editing. Camila G-Beiras: Writing – review & editing. Quique Bassat: Funding acquisition, Writing – review & editing. Ignacio Blanco: Conceptualization, Funding acquisition, Writing – review & editing. Oriol Mitjà: Conceptualization, Writing – original draft, Funding acquisition.
Declaration of Competing Interest
We declare no conflicts of interest.
Acknowledgments
Funding
This work was supported by Blueberry diagnostics, Fundació Institut d'Investigació en Ciències de la Salut Germans Trias i Pujol, and #YoMeCorono.org crowdfunding campaign.
Bárbara Baro is a Beatriu de Pinós postdoctoral fellow granted by the Government of Catalonia's Secretariat for Universities and Research, and by Marie Sklodowska-Curie Actions COFUND Programme (BP3, 801370)
ISGlobal receives support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019-2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA Program.
Acknowledgements
The authors would like to thank Gerard Carot-Sans (PhD) for providing professional medical writing support during the preparation of the manuscript. We thank Laia Comí, Josefa Gómez, Maria Pilar Rodríguez and Aida Sanz for technical support with samples selection and storing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.04.009.
Appendix. Supplementary materials
References
- 1.Pavelka M, Van-Zandvoort K, Abbott S, Sherratt K, Majdan M, Jarčuška P, et al. The effectiveness of population-wide, rapid antigen test based screening in reducing SARS-CoV-2 infection prevalence in Slovakia. medRxiv 2020.
- 2.World Health Organization (WHO). Diagnostic testing for SARS-CoV-2. Available at: https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2. Accessed 6 October 2020.
- 3.Gorzalski AJ, Tian H, Laverdure C, Morzunov S, Verma SC, VanHooser S, et al. High-throughput transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sethuraman N, Jeremiah SS, Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA – J Am Med Assoc. 2020;323:2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 5.Mina MJ, Parker R, Larremore DB. Rethinking COVID-19 test sensitivity—a strategy for containment. N Engl J Med. 2020;383 doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 6.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 7.La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quicke K, Gallichote E, Sexton N, Young M, Janich A, Gahm G, et al. Longitudinal surveillance for SARS-CoV-2 RNA among asymptomatic staff in five Colorado skilled nursing facilities: epidemiologic, virologic and sequence analysis. MedRxiv Prepr Serv Heal Sci. 2020 2020.06.08.20125989. [Google Scholar]
- 9.PHE Porton Down & University of Oxford. Preliminary report from the Joint PHE Porton Down & University of Oxford SARS-CoV-2 test development and validation cell: Rapid evaluation of Lateral Flow Viral Antigen detection devices (LFDs) for mass community testing. Available at: https://www.ox.ac.uk/sites/files/oxford/media_wysiwyg/UK evaluation_PHE Porton Down University of Oxford_final.pdf. Accessed 3 January 2020.
- 10.Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2020 doi: 10.1126/sciadv.abd5393. eabd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso AM, et al. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol. 2020;58:0–3. doi: 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected anterior nasal swab versus professional-collected nasopharyngeal swab. medRxiv. 2020;:2003961. [DOI] [PMC free article] [PubMed]
- 14.Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes MÁ, et al. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2020:0. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133 doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerutti F, Burdino E, Milia MG, Allice T, Gregori G, Bruzzone B, et al. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol. 2020;132 doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alemany A, Baró B, Ouchi D, Rodó P, Ubals M, Corbacho-Monné M, et al. Analytical and clinical performance of the panbio COVID-19 antigen-detecting rapid diagnostic test. J Infect. 2021;18:16. doi: 10.1016/j.jinf.2020.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres I, Poujois S, Albert E, Colomina J, Navarro D. Evaluation of a rapid antigen test (PanbioTM COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin Microbiol Infect. 2021:1. doi: 10.1016/j.cmi.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzel T, Legarraga P, Iruretagoyena M, Pizarro G, Vollrath V, Araos R, et al. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Med Infect Dis. 2021;39 doi: 10.1016/j.tmaid.2020.101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman VM, Haage VC, Bleicker T, Schmidt ML, Mühlemann B, Zuchowski M, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests. medRxiv. 2020; 2020.11.12.20230292. [DOI] [PMC free article] [PubMed]
- 21.Foundation for Innovative New Diagnostics. Comparative evaluation of lateral flow assay tests that directly detect antigens of SARS-CoV-2. Available at: https://www.finddx.org/wp-content/uploads/2020/04/20200421-COVID-Ag-RDT-Evaluation-Synopsis.pdf. Accessed 20 December 2020.
- 22.Seegene Inc. AllplexTM 2019-nCoV Assay. Available at: https://www.seegene.com/assays/allplex_2019_ncov_assay. Accessed 20 December 2020.
- 23.Pilarowski G, Lebel P, Sunshine S, Liu J, Crawford E, Marquez C, et al. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. medRxiv 2020; [DOI] [PMC free article] [PubMed]
- 24.Altman DG, Bland JM. Statistics notes: diagnostic tests 1: sensitivity and specificity. BMJ. 1994;308:1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman DG, Bland j.M. Statistics notes: diagnostic tests 2: predictive values. BMJ. 1994;309:102. doi: 10.1136/bmj.309.6947.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collett D. second ed. Chapman and Hall/CRC; 2002. Modelling binary data. [Google Scholar]
- 27.R Core Team. R: A language and environment for statistical com-puting. 2017. Available at: https://www.r-project.org. Accessed 25 May 2020.
- 28.World Health Organization (WHO). Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays. 2020. Available at: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays. Accessed 29 September 2020.
- 29.Singanayagam A, Patel M, Charlett A, Bernal JL, Saliba V, Ellis J, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks M, Millat P, Ouchi D, Roberts C, Alemany A, Corbacho-Monne M, et al. Transmission of Covid-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis. 2021;21(5):629–636. doi: 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.