Abstract
The rapid spread of coronavirus disease has become an example of the worst disruptive disasters of the century around the globe. To fight against the spread of this virus, clinical image analysis of chest CT (computed tomography) images can play an important role for an accurate diagnostic. In the present work, a bi-modular hybrid model is proposed to detect COVID-19 from the chest CT images. In the first module, we have used a Convolutional Neural Network (CNN) architecture to extract features from the chest CT images. In the second module, we have used a bi-stage feature selection (FS) approach to find out the most relevant features for the prediction of COVID and non-COVID cases from the chest CT images. At the first stage of FS, we have applied a guided FS methodology by employing two filter methods: Mutual Information (MI) and Relief-F, for the initial screening of the features obtained from the CNN model. In the second stage, Dragonfly algorithm (DA) has been used for the further selection of most relevant features. The final feature set has been used for the classification of the COVID-19 and non-COVID chest CT images using the Support Vector Machine (SVM) classifier. The proposed model has been tested on two open-access datasets: SARS-CoV-2 CT images and COVID-CT datasets and the model shows substantial prediction rates of 98.39% and 90.0% on the said datasets respectively. The proposed model has been compared with a few past works for the prediction of COVID-19 cases. The supporting codes are uploaded in the Github link: https://github.com/Soumyajit-Saha/A-Bi-Stage-Feature-Selection-on-Covid-19-Dataset
Keywords: Coronavirus, COVID-19 dataset, Deep learning, Feature selection, Dragonfly algorithm, Chest CT image, Convolutional neural network
Introduction
The rapid spread of COVID-19 infection has created an intensive impact on both the physical and mental health of the people throughout the world. The dangerous COVID-19 virus was first observed in December 2019 in Wuhan, China. It then started spreading rapidly by transmitting the virus from China to other countries. This in turn has forced the people to restrict themselves at home, and pulled almost all countries and territories worldwide in a pandemic situation. Each day, thousands of people are getting infected with this dangerous virus [1, 2]. According to the record of WHO (World Health Organization), a total number of people infected by COVID-19 disease is 16,114,449 with 646,641 deaths as of 27 July 2020 [3].
WHO has declared COVID-19 a global health emergency on January 30, 2020. [4]. The general symptoms of COVID-19 disease include fever with cough, fatigue, and breathing problem, loss of taste and smell [5]. Mohamadou et al. in [6] presented a review on COVID-19 that includes various medical images, case reports, management strategies, etc. According to the authors, both mathematical modelling and AI techniques can be considered as reliable tools to fight against COVID-19 disease. According to the authors in [7], chest X-ray and CT-scan images (radiological images) may play an important role for accurate diagnosis of this disease. A few radiologists suggest chest X-ray images to diagnose COVID-19 cases because maximum radiological laboratories and hospitals have X-ray machines through which the chest images of the patients can be achieved easily [8]. However, chest X-ray images can not distinguish the soft tissues perfectly [9]. To overcome this problem, CT-scan images are used which can detect such soft tissues efficiently. Several researchers [10–15] have demonstrated the effectiveness of using CT images for the proper diagnosis of COVID-19 affected person.
The present study attempts to design a classification model which can distinguish the COVID-19 affected people from the normal people when chest CT images are supplied to the model. Initially, a CNN model has been used to generate the features from the chest CT images. However, the dimension of feature vector generated is quite large (800 feature attributes). Hence, we have proposed a bi-stage FS approach to select the optimal feature subset. In the first stage, two different filter methods called Mutual Information (MI) and ReliefF are used to rank the feature attributes. As the ranking of the attributes are diverse and sometimes data dependent, we ensemble the output of filter based techniques by taking the union of top-n ranked features of each method. This ensemble technique combines the information produced by these two rankings into a common subset. we have used a meta-heuristic algorithm called Dragonfly algorithm (DA) [20] based FS method in the second step for the optimization of the feature set generated in first stage. This optimal feature subset is fed to SVM classifier for the prediction purpose.
The key contributions of the proposed work are briefly mentioned below:
An effective hybrid model has been designed to predict COVID-19 disease by combining deep learning (DL) and meta-heuristic-based wrapper FS approaches.
The model uses CNN to generate powerful features and applied a bi-stage FS method to eliminate the redundant and non-informative features, thereby minimizing the computational time for the training of the model.
Efficiency of the proposed model has been evaluated on two publicly available chest CT image datasets namely, SARS-CoV-2 CT image dataset [16] and COVID-CT dataset [17].
The efficiency of the model has been compared with some existing methods, and the significance of the obtained results has been verified through a statistical test.
We have used DA in the second stage of the proposed bi-stage FS procedure for the effective prediction of COVID-19. Mafarja et al. in [18] have introduced DA as an efficient FS method, in which 18 datasets from UCI machine learning repository were used. It was demonstrated that the DA performs better than other FS techniques such as binary grey wolf optimizer (BGWO) [52], binary gravitational search algorithm (BGSA) [53], binary bat algorithm (BBA) [54], particle swarm optimization (PSO) [55], and genetic algorithm (GA) [56] etc. Hammouri et al. in [19] showed the superiority of DA based FS method when considered the same datasets mentioned in [18]. Mirjalili et al. in [20] and Ranolds in [21] demonstrated the merits of DA in FA and other problems. Looking into the effectiveness of DA in various FS applications, we have chosen this algorithm in our proposed work.
The organization of the paper is mentioned as follows: Section 2 describes a few research contributions made by the other authors for the prediction of COVID-19. Section 3 gives the brief description of the used database. Section 4 reports the proposed hybrid model in detail. Feature extraction using the CNN model and a bi-stage FS procedure to select the most relevant features have been discussed in detail in this section. The outcomes observed in the current experiment have been mentioned in Section 5. Finally, the conclusion of the present work along with a few future directions has been reflected in Section 6.
Related work
Recently many researchers across the globe have proposed machine learning (ML) and DL-based image analysis procedures for the prediction of COVID-19 disease, and thereby helping the medical practitioners for proper diagnosis. A number of seminal research works related to the prediction of COVID-19 from X-ray images and chest CT images have been addressed in this section.
Zhang et al. have used a ResNet model to predict COVID-19 cases [1]. They applied their proposed model on chest X-ray image dataset that consists of 1078 images from both COVID-19 and non-COVID patients. Their experiment successfully detects COVID cases with 96% accuracy. In [22], Wang et al. used a CNN for COVID-19 detection and achieved 92.6% test accuracy. In this experiment, they considered X-ray images of COVID-19 patients, Pneumonia patients, and normal people. A detailed analysis on COVID-19 classification has been reported by Narin et al. in [23]. Authors have used three popular DL models namely, ResNet50, Inception-V3 and Inception-ResNetV2 on chest X-ray images. Abbas et al. proposed a CNN based model called DeTraC for the prediction of COVID-19 from chest X-ray (CXR) images in [24]. This system can detect irregularities in the image by observing its class boundaries using class decomposition technique. Their proposed system can detect COVID cases with 95.12% recognition accuracy. Sethy et al. [66] used a DL based method for the extraction of features from CXR images and applied the SVM classifier for the prediction.
In [25], Khan et al. presented CoroNet, a DCNN (Deep CNN) model to apply image processing algorithms on X-ray images for the prediction of COVID-19 . Maghdid et al. [26] demonstrated the effectiveness of DL-based model for the analysis of COVID-19 cases more accurately with fast response. They prepared a dataset comprising CXR and chest CT images from different sources and developed a system for COVID-19 prediction by involving DL and TL (Transfer Learning) techniques. Razzak et al. [27] used pre-trained weights to improve the recognition performance by using TL based approach and compared their results with other CNN models. Authors in [28] proposed a Self-Trans approach for the prediction of COVID-19 disease that combines the power of self-supervised learning algorithm and TL based technique to extract more powerful and unbiased features. Sahlol et al. [30] proposed a hybrid approach for the classification of COVID-19. They employed a CNN model to extract the features from CXR images and MPA (Marine Predators Algorithm [57]) based FS procedure to identify most relevant features for effective recognition.
Some notable research works for the prediction of COVID-19 cases on chest CT images have been briefed here. In [29], Tang et al. considered the chest CT images as an important component for the assessment of COVID-19 severity. The authors proposed an ML-based technique for the assessment of COVID-19 severity from the chest CT images and achieved 87.5% classification accuracy. Farid et al. [33] demonstrated the analysis of Corona disease virus on the basis of a probabilistic model. Their proposed methodology extract features from chest CT images for the prediction of COVID-19 disease. They used a combination of statistical and ML-based methods to extract features from CT images and achieved 96.07% recognition accuracy. He et al [28] proposed a Self-Trans approach that combines self-supervised learning and transfer learning approach to produce powerful and unbiased features that helps to achieved 86% recognition accuracy. Loey et al. [58] presented classical data augmentation techniques with Conditional Generative Adversarial Nets (CGAN) and predicted COVID-19 using a deep transfer learning model with 82.9% correct classification accuracy. Mobiny et al. [60] integrates the power of Capsule Networks with different architectures for the improvement of classification accuracy. Their proposed model classify the COVID-19 cases with 87.6% accuracy. Different DL-based techniques have been adopted by the Authors [61, 62, 64] to improve the prediction capability of COVID-19 cases. Authors in [63] proposed a deep uncertainty-aware TL framework for the prediction of COVID-19 using medical images. Authors have used VGG16, ResNet50, DenseNet121, and InceptionResNetV2 to extract deep features from the images. The features are then fed to different ML and statistical model for final classification purpose and achieved 87.9% accuracy. The approach proposed by Ewen et al. [65] aims to determine whether the self supervision strategy is a good option for COVID-19 prediction. The methodology proposed by the authors [28, 58, 60] for the prediction of COVID-19 has lower recognition rate. Work mentioned in [61] used light CNN for the prediction purpose hence it is less time consuming but recognition accuracy is low. Whereas, works mentioned in [62, 64] achieves better recognition accuracy but consumes more time. To reduce the overall classification time and also to work with small dataset, authors in [63] used TL-based technique and achieved descent outcome.
A few researchers have also tried to identify the most relevant features using different FS-based techniques for better prediction of COVID-19 cases. Al-qaness et al. in [31] proposed an improved ANFI (Adaptive neuro-fuzzy inference) system for forecasting the confirm cases of COVID in China. The proposed system works on SSA (Salp Swarm algorithm) which is an enhanced flower pollination algorithm. Shaban et al. in [32] introduced a novel CPDS (COVID-19 Patients Detection Strategy) methodology for the prediction of COVID-19. They introduced a hybrid FS process that elects most informative features from the features computed from the chest CT images of COVID-19 patients and non COVID-19 person. Their proposed technique combines the information from both filter and wrapper based FS methods. Authors have achieved a recognition accuracy of 96% and also the process is less time consuming as the feature selection technique reduces the features count by tracking the useful features.
From the above discussion about the existing research works, it is clear that many researchers throughout the globe have involved themselves for the automatic prediction of the COVID-19 cases by analyzing the chest X-rays or CT-scans. In this regard, ML, DL and TL based approaches have made a significant contribution to the computer based COVID-19 screening. However, it has been observed that DL-based techniques are slow, and some of the past methods produce lower recognition accuracy. On the other hand, FS-based approaches are able to make the model more robust by eliminating the features which do not contribute much to the classification process. As a result, time required to train the model gets reduced and classification accuracy of the learning model gets enhanced. Keeping these facts in mind, in the current study, a bi-stage classification model has been proposed where CNN is involved for feature extraction, guided-FS is used for the initial screening of the features coming from CNN and lastly, the DA-based FS technique is used to select the most relevant features for the better prediction of COVID-19 cases.
Proposed bi-stage hybrid model
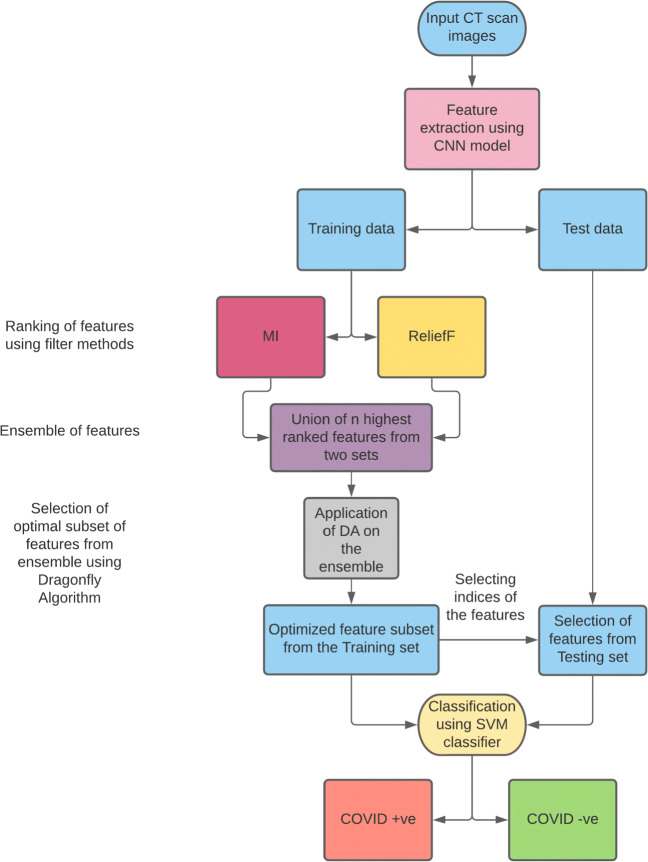
In the present work, we have considered SARS-CoV-2 CT scan image database and COVID-CT database to predict COVID-19 cases. Figure 1 highlights the working procedure of the proposed model. At the beginning of our algorithm, we consider CNN as a feature extractor. The CNN applied on the chest CT images produces 800 feature attributes. As the produced feature set is of high dimension, a bi-stage FS approach is applied to select the most relevant features. In the first stage, a guided-FS technique is applied where the features generated from CNN are ranked by two filter methods namely MI and ReliefF separately. Different filter based techniques rank the feature attributes differently because the rule of assigning the rank is different for the two filter methods. Hence, we utilize a simple ensemble technique by taking the union of the top-n features produced by these two filter based methods. The resultant feature set is then passed through DA for the selection of final and more optimal feature subset in the second stage. This optimized feature subset is then used for the prediction of COVID and non-COVID chest CT images.
Fig. 1.
Flowchart of the proposed model for the prediction of COVID and non-COVID cases from chest CT images
The detailed working procedure of feature extraction using CNN and the bi-stage FS technique used to find most effective optimized feature set (as shown in Fig. 1) are mentioned in the following subsections.
Feature extraction using CNN
CNN is the most utilized architecture where the image is fed into an input layer, which propagates through multiple hidden layers and lastly produces the prediction probability through the output layer. These hidden layers consist of a series of convolutional layers followed by pooling layers that try to detect and learn very complex features and patterns belonging to the images of a particular class. These are the reason for tremendous success of CNN models in field of computer vision and its various applications. Even though CNN performs great in the task of image classification, there are a couple of limitation to this. Firstly, to provide more accurate predictions, the CNN model needs to be trained on a large dataset to include a different kind of possible variations. However, as of now, large sized datasets are not publicly available which can be used for DL based COVID-19 screening. Hence, in the current work, instead of using the CNN model for the prediction of COVID cases, we have used it for extraction of powerful features, and then the feature vector so obtained is passed to the bi-stage FS technique followed by a SVM classifier based classification of the input chest CT images.
The proposed CNN model considers images of size 224x224 in the input layer. The input layer is followed by a Convolutional layer, a Batch Normalization layer and a Maxpooling layer. This set of layers is repeated four times with various number of filters having a size of (3 x 3). All neurons in these layers have the ReLU (Rectified Linear Unit) as their activation functions. The final Convolution layer consists of 8 filters of size (3 x 3) followed by fully connected layers having ReLU activation. For all these layers, stride of size (1x1) is used in Convolutional layers and stride of size (2x2) is used in Maxpooling layers. An overview of this architecture is shown in Table 1. From the last Convolutional layer, a total of 800 features are generated from each image sample. Total parameters of this architecture are 66,079, where trainable parameters are 65,727 and the rest 352 are non-trainable parameters.
Table 1.
Detail of the CNN architecture used for the purpose of feature extraction from the chest CT images
| Layer | Type | Filter size | Number of filters | Strider |
|---|---|---|---|---|
| Input | 224x224x3 | − | − | − |
| Conv_1 | CL+BN+ReLu | 3x3 | 64 | 1x1 |
| MPL_1 | − | 2X2 | − | 2X2 |
| Conv_2 | CL+BN+ReLu | 3x3 | 64 | 1x1 |
| MPL_2 | − | 2X2 | − | 2X2 |
| Conv_3 | CL+BN+ReLu | 3x3 | 32 | 1x1 |
| MPL_3 | − | 2x2 | − | 2x2 |
| Conv_4 | CL+BN+ReLu | 3x3 | 16 | 1x1 |
| MPL_4 | − | 2x2 | − | 2x2 |
| Conv_5 | CL+BN+ReLu | 3x3 | 8 | 1x1 |
| Output | Sigmoid | − | − | − |
Feature selection
A feature extraction procedure may generate a few redundant and irrelevant features in the feature space. It is imperative to eliminate those redundant and irrelevant feature attributes as removal of which not only helps to enhance the overall classification accuracy but also minimizes the computational overheads [35]. This is commonly performed by a FS technique. FS is defined as a procedure to select a subset from a set of candidate features that exhibits the best performance under some classification system [36, 37]. The various FS-based algorithms mentioned in the literature can be categorized into three groups namely, filter-base, wrapper-based, and embedded methods [50]. The filter methods use a statistical or probabilistic approach to score/rank the feature attributes and on the basis of the assigned score, feature attributes are either get selected or removed before entering into the classification model. The wrapper methods are often used in coalition with a ML algorithm, where the impact of the feature attributes are validated by the learning algorithms, which in turn select the optimal feature subsets to augment the classification accuracy. In contrary, an embedded model embeds FS within a specific ML based algorithm. In the proposed work, we have used a bi-stage approach to select most relevant features from the CNN produced feature set so that a near-perfect CT scan image based COVID-19 classification system can be designed. The adopted approach has been mentioned in detail in the following sub-sections.
Guided FS
In the present study, the effectiveness of a bi-stage FS-based procedure has been shown to reduce the feature space for the prediction of COVID-19 cases from the CT scan images. In the first stage, we incorporate a filter based guided FS technique [38] to remove statistically weak features from the search space and considered the reduced feature set as an input to the wrapper based FS module in the next step. In [39], authors have used an efficient guided FS technique where non-negative spectral clustering has been used to recognize cluster labels of the input samples more accurately. To perform our guided FS, we consider two popular filter methods namely, MI [40–42, 51] and ReliefF [43–47].
MI can be defined as a measure of the amount of information that one random variable A owns through the other variable Y https://thuijskens.github.io/2017/10/07/feature-selection/. The MI between two variables A and B is computed by using (1).
| 1 |
where, p(a,b) represents the joint probability density function of A and B. p(a) and p(b) describe the marginal density functions. MI basically determines the similarity between the joint distribution p(a,b) with respect to the products of the factored marginal distributions. If A and B are two independent variables, then the value of p(a,b) and p(a)p(b) will be same and thus the integral value will be zero in this case.
Relief has been proved as very useful and successful attribute selection procedure [44, 47]. This method is capable of estimating the conditional dependencies between attributes very efficiently and thus can deliver an amalgamate view on attribute selection in both regression and classification problems. The basic Relief algorithm measures the quality of feature attributes on the basis of distinguishing capability between instances, those are closer to each other [48].
However, the original Relief algorithm works with nominal and numerical attributes. This algorithm works on two-class problems only and also cannot handle incomplete data. ReliefF, the extension of Relief algorithm, can solve these issues. Like original Relief algorithm, ReliefF also selects an instance x and then finds k nearest neighbors (instead of two neighbors in Relief algorithm) that belong to the same class (also known as nearest hits Hj(x)) and different classes (nearest misses Mj(x)). Next, the algorithm updates the value of Wf (also known as quality estimation) for all attributes f by considering their values for x, Mj(x) and Hj(x). The value of Wf gets decreased if the values of x and Hj(x) are different for attribute f, which is not desirable (means the attribute f separates the two instances of same class). In contrary, the value of Wf gets increased if the values of x and Mj(x) are different for attribute f, which is desirable (means the attribute f separates the two instances of different class). The updating formula, given in (2) is similar to the Relief algorithm, except that we take the average contribution of all the hits and all the misses. This algorithm is repeated for m number of times, where m is a user-defined parameter.
| 2 |
where diff() measures the distance between two samples on the feature f, and p(x) is the probability of a class. The Euclidian distance is used to calculate the inter-class and intra-class distances of the samples and is mentioned in (3).
| 3 |
As mentioned earlier, MI and ReliefF methods has been used here to assign the rank to each attribute of the feature set generated from the CNN model used here. We have selected two feature sets P and Q, each consists of n number of highest ranked feature attributes. Then we have performed a union operation to generate a new feature set Z such that Z=P ∪ Q. The resultant feature set Z has served as input feature vector for DA based FS technique. The steps used in guided FS technique has been mentioned in Algorithm 1. In our case, the values of N and M are set to 300 and 150 respectively. It is to be noted that we have applied this guided FS on the training set only. The feature indices so selected are used to choose the features from the test set.
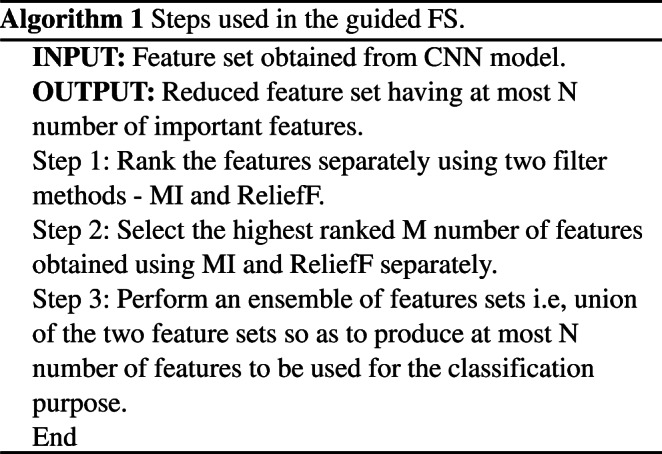
Dragonfly algorithm
In [19], Hammouri et al. have proposed an enhanced meta-heuristic FS algorithm, where dragonfly swarm utilize its static and dynamic swarming behaviours in nature to explore the search space and determine the optimal solution for a given optimization problem. The DA algorithm [20] uses three principals of swarming introduced by Reynolds et al. [21] as follows:
Separation (Si), ensures to avoid the static collision of the individuals from the others belonging to the neighborhood position.
Alignment (Ai), matches the velocity of individuals with respect to the others belongs in neighborhood.
Cohesion (Ci), describes the propensity to move towards the center of mass of the neighborhood.
To implement the above ideas of Separation, Alignment and Cohesion, (4) – (6) have are used in DA [20].
| 4 |
| 5 |
| 6 |
where, X denotes the current position of the dragonfly, Xj represents the position of jth neighbour, Vj represents the velocity of jth neighborhood and N is the neighbourhood size.
According to the natural behavior, each individual dragonfly present in a swarm tends to get attracted towards the food source and repel the enemies. The attraction and repulsion can be represented in mathematical terms as defined in (7) and (8). Where X, X+ and X− represent the current position of the dragonfly, position of the food source, and enemies as follow [20]:
| 7 |
| 8 |
The updating mechanism of the position of the dragonfly in the swarm is governed by δX (termed as step) and the position denoted by X. The step vector determines the direction of the movement of the dragonflies and can be described by (9).
| 9 |
Where, s, a, c, f, e, w and t represent the separation weight, alignment weight, cohesion weight, food factor, enemy factor, inertia weight and iteration number respectively. Si, Ai, Ci, Fi, and Ei specify the separation, alignment, cohesion, food source, position of enemy of ith dragonfly respectively. The main goal of separation, alignment, cohesion, food, and enemy factors(s, a, c, f, and e) is to explore and exploit a wide range of area to obtain the optimal solution. The position vectors are updated by using (10) given below [20].
| 10 |
For our proposed work, we have considered w = 0.9–0.2, s = 0.1, a = 0.1, c = 0.7, f = 1, and e = 1 following the work mentioned in [20].
Some of the above controlling parameters are changed dynamically and adaptively to provide different exploratory and exploitative behaviors for the DA algorithm. The best solution is considered as the food course, and the worst solution is treated as an enemy.
To enhance the stochastic behavior, randomness and exploration, the artificial dragonflies fly around the search space following Le’vy flight random walk when a neighboring solution is absent [20]. In this situation, dragonfly’s position gets updated by following the (11).
| 11 |
where, t represents current iteration, and d is the measurement of the dimension of position vectors.
The Lévy function is defined in (12), where r1 and r2 represent random numbers within the range of 0 to 1, β is a constant value and set to 1.5 for the current experiment, and the value of σ is measured by using (13).
| 12 |
| 13 |
It should be noted that we have slightly modified the DA, where we have incorporate a feasibility checking before the updating the position of the dragonfly. It means the new solution set will be updated if and only if it has the higher fitness value than the previous best food source. Here, the fitness value for a solution vector (in our case it is a feature vector) refers to the classification accuracy achieved by the classifier (for the present experiment SVM classifier) which is used to measure the efficiency of the solution sets generated by the feature selection method. Algorithm 2 shows the basic steps of the DA.
 DA first creates a set of random solutions (in our case it is 20). The position and step vectors of dragonflies are initialized randomly within the range of lower and upper bounds. Then the algorithm proceeds by updating the food sources and enemy which are the best and worst scored feature vectors respectively, and the values of separation, alignment, cohesion, food, and enemy factors (i.e., s, a, c, f, and e) are also updated. The values of S, A, C, F, and E are calculated using (4)–(8) and the radius of the neighborhood to be taken under consideration has also been updated accordingly.
DA first creates a set of random solutions (in our case it is 20). The position and step vectors of dragonflies are initialized randomly within the range of lower and upper bounds. Then the algorithm proceeds by updating the food sources and enemy which are the best and worst scored feature vectors respectively, and the values of separation, alignment, cohesion, food, and enemy factors (i.e., s, a, c, f, and e) are also updated. The values of S, A, C, F, and E are calculated using (4)–(8) and the radius of the neighborhood to be taken under consideration has also been updated accordingly.
After this phase, a checking criterion is set where it is tested whether the current dragonfly has any neighbors, if so, then the position vector and the velocity vector of that dragonfly are updated and stored in temporary position and velocity vectors respectively following the (9) and (10). Otherwise, the position vector of the dragonfly is updated by taking into account Lévy fight as defined in (11) and stored in the temporary position vector. Now, the values in the temporary position vector are checked whether these belong to the feature space, if not then they are brought back to the search space again. Before moving towards to the next iteration, the algorithm checks whether the temporary position vector has better fitness than the existing one (food source), and if so then it replaces the solution vector in the swarm and also updates the velocity. The algorithm then continues until the condition is satisfied. The condition has been set either to obtain a desired solution or maximum number of iterations is reached. At the end of the algorithm, the optimal feature subset is obtained which will be used for the classification purpose.
SVM classifier
In ML based approach, SVM can be termed as the supervised learning model with associated learning algorithm to analyze data for classification and regression purposes. Vapnik [59] proposed the SVM classifier for binary classification problems. The main objective was to find the optimal hyperplane f (w, x) = w ⋅ x + b to separate two classes for a given dataset having features . In other words, for binary classification problem and given a set of training examples, an SVM training algorithm builds a model that assigns new examples to one of two classes. An SVM maps training samples to points in space to maximize the width of the gap between the two classes. New samples are then mapped into the same space and predicted a class based on which side of the gap they fall.
The parameter w is learned by the SVM using the equation mentioned in (14).
| 14 |
where wTw, C, and wTx + b represent the Manhattan or L1 norm, penalty parameter, actual label, and predictor function. Equation (14) represent L1-SVM having standard hinge loss. Its counterpart, L2-SVM (15), provides more stable outcomes. Figure 2 graphically represents the principle of the SVM for a two-class data points separated by a hyperplane.
| 15 |
Fig. 2.
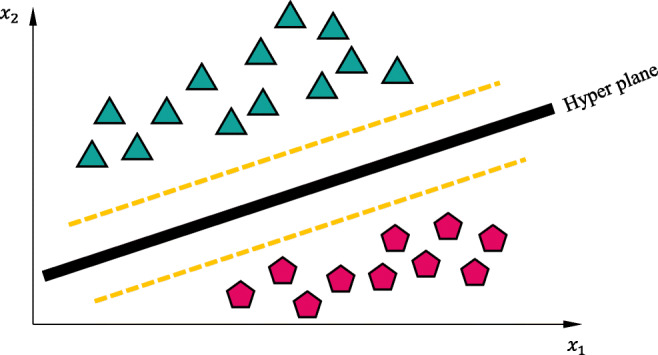
Principle of the SVM for a two-class dataset separated by hyperplane
Database used
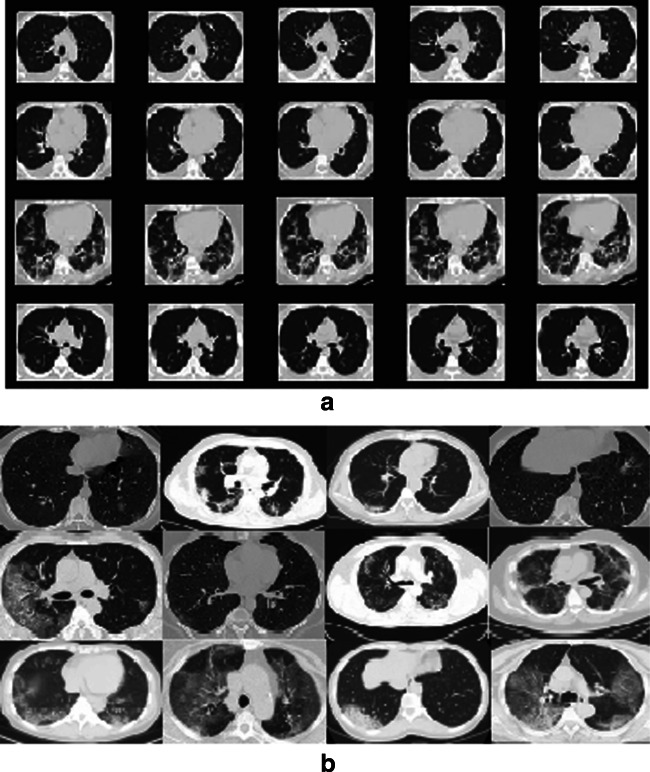
In the present work, we consider SARS-CoV-2 CT scan image database [16] and COVID-CT database [17] to predict of COVID-19 disease by using the proposed model. The SARS-CoV-2 CT image dataset consists of a total of 2,482 chest CT scan images, of which 1,252 images are of COVID-19 infected patients and remaining 1,230 images belong to the non-infected persons [34]. The images of this database have been collected from the hospitalized patients of Sao Paulo, Brazil. Yang et al. have built the open-source COVID-CT dataset that consists of 349 COVID-19 CT images collected from 216 patients and 463 non-COVID-19 CT images [17]. The preliminary goal to build such databases was to provide benchmarks and facilities research in this area. This allows researchers to conveniently identify and test the presence of COVID-19 from the CT scan images by applying various ML/DL based techniques, and therefore to contribute the society for handling the pandemic. Figure 3a and b displays some sample chest CT scan images taken from SARS-CoV-2 CT scan and COVID-CT databases [16, 17].
Fig. 3.
Sample images taken from a SARS-CoV-2 CT scan dataset, and b COVID-CT database that are positive for COVID-19
Experimental results and discussion
As per discussions in the preceding sections, we use SARS-CoV-2 CT scan [16] and COVID-CT dataset [17] for the prediction of COVID-19. We use a CNN model that serves as a feature extractor for the present work, and it produces a feature vector of 800 dimension that is passed through two filter based methods: MI and Relief-F for the initial screening of the features (i.e., the concept of guided population). We have considered first 150 ranked feature attributes, obtained from these two filter based methods separately, to form an ensemble of these feature subsets in which the union of the feature subsets is created. Here, it is found that the produced feature sets are having 284 and 262 feature attributes for SARS-CoV-2 CT and COVID-CT dataset respectively. Then these resultant reduced feature sets have been passed through DA separately for the final feature selection. For measuring and analyzing the performance of the adopted methods, we have considered SVM classifier with five performance metrics, namely accuracy, precision, recall, F1 score and Area Under the Receiver Operating Curve (AUC) values [49]. The Receiver Operating Curve (ROC) is a plot of true positive rate (i.e., TPR or sensitivity) versus false positive rate (FPR or specificity). To measure the value of FPR (14) has been used.
| 16 |
| 17 |
| 18 |
| 19 |
| 20 |
The parameters that have been used in the calculation of these metrics include:
True Positive (TP): When a person is diagnosed for being infected by COVID-19
True Negative (TN): When a person is diagnosed for not being infected by COVID-19.
False Positive (FP): Incorrect detection of a healthy person as infected by COVID-19 (positive).
False Negative (FN): Incorrect detection of an infected person as a healthy (negative).
Table 2 shows the reduced feature dimension, accuracy, precision, recall, F1 score and AUC values for features selected through guided-FS procedure from the feature set originally obtained from CNN model for SARS-CoV-2 CT scan and COVID-CT dataset.
Table 2.
Detail performance measures of the feature set produced by guided-FS procedure applied on features obtained from CNN model for SARS-CoV-2 CT scan and COVID-CT dataset
From Table 2, it can be observed that the performance of guided-FS is quite notable as the procedure successfully reduces the feature dimension from 800 (coming from CNN model) to 284 and 262 respectively for SARS-CoV-2 CT scan and COVID-CT dataset in the first step of our proposed bi-stage FS technique used for the prediction of COVID-19. The proposed guided-FS technique not only reduces the feature dimension substantially by eliminating redundant features, but also increases the classification accuracy of the overall model. From Table 2, it can be observed that the guided-FS procedure produces 95.77% and 85.33% recognition accuracies for the SARS-CoV-2 CT scan and COVID-CT datasets respectively, whereas the features coming from CNN (i.e. 800-element feature vector) produce 94.16% and 82% classification accuracies for the SARS-CoV-2 CT scan and COVID-CT datasets respectively. Hence, guided-FS technique reduces 64.5% and 67.25% feature dimension and increases the prediction accuracy by 1.61% and 3.33% for SARS-CoV-2 CT scan and COVID-CT datasets respectively. For the prediction purpose, we have considered the SVM classifier with polynomial kernel and set the value of the parameter coef0 as 2.0.
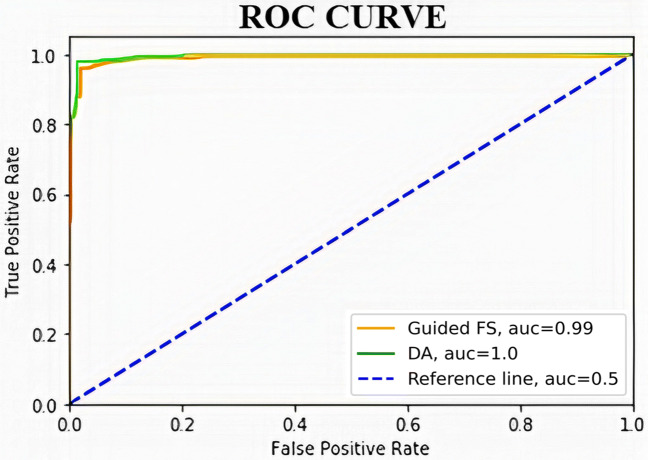
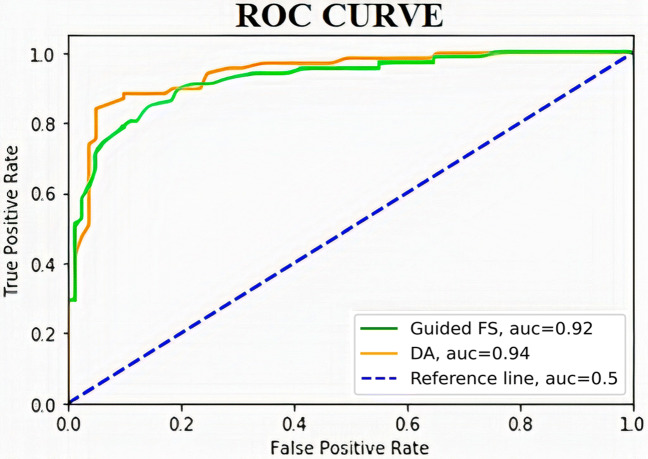
In the second stage, DA is applied on the feature set produced by the guided-FS for the final selection of the optimal feature subset that contains most relevant features. The reduced feature set produced in the second stage has been used for the prediction of COVID-19. Table 3 provides the detailed performance measures of the applied DA for the COVID-19 prediction. Figures 4 and 5 represent the ROC curves for the feature set selected by guided-FS with respect to the features obtained through CNN model and features selected by DA with respect to the features generated using guided-FS technique for SARS-CoV-2 CT scan dataset and COVID-CT scan dataset respectively.
Table 3.
Details of the performance of the feature set produced by applying DA on features obtained from the guided-FS for SARS-CoV-2 CT scan dataset and COVID-CT dataset
Fig. 4.
ROC curves for the feature set selected by guided-FS with respect to the features obtained through CNN model (green line) and features selected by DA with respect to the features generated using guided-FS technique (orange line) for SARS-CoV-2 CT scan dataset
Fig. 5.
ROC curves for the feature set selected by guided-FS with respect to the features obtained through CNN model (green line) and features selected by DA with respect to the features generated using guided-FS technique (orange line) for COVID-CT-Dataset
The measurements of the metrics presented in Table 3 demonstrate the efficacy of applied DA-based FS. After thorough analysis of results in the Tables (2-3), it is observed that the DA is able to increase the accuracy, precision, recall, F1 score and AUC values by 0.0262%, 0.0412, 0.0023, 0.0221 and 0.0096 respectively with respect to guided-FS technique when SARS-CoV-2 CT scan dataset has been considered. Whereas, for COVID-CT dataset, DA-based technique is able to hike the same metrics by 4.67%, 0.0522, 0.0612, 0.0574 and 0.017 respectively in comparison to guided-FS procedure.
For exhaustive analysis and also to justify for choosing SVM polynomial kernel with the value of coef0 as 2, we have tested the same with linear kernel and varied the value of coef0 from 0 to 3. The detailed observed outcomes is mentioned in Table 4. From this table it can be seen that the best result has been observed for polynomial kernel when the value of coef0 has been set to 2.
Table 4.
Observed outcomes after tuning the parameters of SVM classifier on SARS-CoV-2-CT scan dataset and COVID-CT dataset
| Database | Kernel | coef0 | Accuracy | Precision | Recall | F1-score | AUC |
|---|---|---|---|---|---|---|---|
| SARS-CoV-2 | Linear | 0 | 88.93 | 0.8798 | 0.9043 | 0.8919 | 0.9532 |
| 1 | 90.14 | 0.8924 | 0.9106 | 0.9014 | 0.9572 | ||
| 2 | 88.93 | 0.8508 | 0.9214 | 0.8847 | 0.9584 | ||
| 3 | 90.54 | 0.9016 | 0.9053 | 0.9035 | 0.9732 | ||
| Polynomial | 0 | 92.15 | 0.9774 | 0.864 | 0.9172 | 0.9732 | |
| 1 | 95.77 | 0.9736 | 0.9364 | 0.9546 | 0.9848 | ||
| 2 | 98.39 | 0.9821 | 0.9778 | 0.98 | 0.9952 | ||
| 3 | 95.37 | 0.9631 | 0.9438 | 0.9533 | 0.9916 | ||
| COVID-CT-Datbase | Linear | 0 | 76 | 0.7692 | 0.7042 | 0.7353 | 0.8302 |
| 1 | 78.67 | 0.8136 | 0.6957 | 0.75 | 0.86 | ||
| 2 | 80.67 | 0.8169 | 0.7838 | 0.8 | 0.8396 | ||
| 3 | 81.33 | 0.8333 | 0.7639 | 0.7971 | 0.8534 | ||
| Polynomial | 0 | 82 | 0.8158 | 0.8052 | 0.8105 | 0.8820 | |
| 1 | 84.67 | 0.8333 | 0.8267 | 0.8299 | 0.8932 | ||
| 2 | 90 | 0.9355 | 0.8406 | 0.8855 | 0.9414 | ||
| 3 | 87.09 | 0.8309 | 0.8194 | 0.8252 | 0.8878 |
We have also tested the efficiency of the optimized feature vector produced by DA-based FS technique by applying 5-fold cross validation scheme on it. Table 5 reflects the detailed observations obtained for SARS-CoV-2 CT scan dataset and COVID-CT-Dataset.
Table 5.
Detailed outcomes observed for 5-fold cross-validation scheme on SARS-CoV-2 CT scan dataset and COVID-CT-Database
| Dataset | Accuracy | Precision | Recall | F1-score | AUC |
|---|---|---|---|---|---|
| SARS-CoV-2 CT-scan-dataset | 95.32 | 0.953 | 0.953 | 0.953 | 0.953 |
| COVID-CT-Dtabase | 76.01 | 0.761 | 0.760 | 0.759 | 0.834 |
Additional test
To establish the generalizability of the proposed technique, we have also performed experiment on a chest X-ray image database which can be found at: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. The proposed model extracts 800 features applying CNN, then the guided FS technique reduces the feature dimension to 274, finally DA selects 175 most relevant features from the feature set produced by guided FS technique. At the end, the SVM classifier produces 100% recognition accuracy with precision, recall, F1-score, and AUC values 1. , 1.0, 1.0, and 1.0 respectively.
Table 6 provides a comparative study of some of the past works with the current work related to the prediction of COVID-19 disease on SARS-CoV-2 CT scan dataset and COVID-CT dataset. The authors in [16, 34] worked on the SARS-CoV-2 CT scan dataset for the prediction of COVID-19. Authors in [16] have used DL based model and Jaiswal et al. in [34] have used DenseNet201, a pre-trained DL architecture for the prediction of COVID-19. The methods reported in [16, 34] can predict the COVID-19 cases with a recognition accuracy of 97.38% and 96.25% respectively. Authors in [17, 28, 58, 60–65] have used COVID-CT dataset to predict COVID-19 cases.
Table 6.
Comparison of the performances of proposed model with some existing techniques on SARS-CoV-2-CT scan dataset and COVID-CT dataset
| Dataset | References | Accuracy (in %) |
|---|---|---|
| SARS-CoV-2-CT | Soares et al. [16] | 97.38 |
| Jaiswal et al. [34] | 96.25 | |
| Simonyan et al. [68] | 97.4 | |
| He et al. [69] | 95.17 | |
| Chollet et al. [70] | 94.57 | |
| Proposed work | 98.39 | |
| COVID-CT | Yang et al. [17] | 89.1 |
| He et al. [28] | 86 | |
| Mobiny et al. [60] | 87.6 | |
| Polsinelli et al. [61] | 83 | |
| Dan-Sebastian et al. [62] | 87.74 | |
| Shamsi Jokandan et al. [63] | 87.9 | |
| Mishra et al. [64] | 88.34 | |
| Ewen et al. [65] | 86.21 | |
| Loey et al. [58] | 82.91 | |
| Proposed work | 90.0 |
From the results tabulated in Tables 2 and 3, it has been observed that the guided-FS step of the bi-stage FS approach has reduced the feature vector of dimension 800 (extracted from CNN) to dimensions 284 and 262 respectively for SARS-CoV-2 CT scan and COVID-CT datasets. It has rendered accuracy, precision, recall, F1 score and AUC values of 95.77%, 0.9409, 0.9755, 0.9579 and 0.9856 respectively for SARS-CoV-2 CT scan dataset, and 85.33%, 0.8833, 0.7794, 0.8281 and 0.9244 respectively for COVID-CT dataset. Hence, it can be said that the proposed guided-FS technique has efficiently performed the initial screening of the features obtained from the CNN. DA based FS has been applied in the second step on the reduced feature set produced by guided-FS technique. Here, DA further reduces the feature dimension into 179 and 169 for SARS-CoV-2 CT scan and COVID-CT datasets respectively. After the second stage, the obtained accuracy, precision, recall, F1 score and AUC values are 98.39%, 0.9821, 0.9778, 0.98 and 0.9952 respectively for SARS-CoV-2 CT scan dataset, and 90.0%, 0.9355, 0.8406, 0.8855 and 0.9414 respectively for COVID-CT dataset. From the observed outcomes, it is evident that DA can efficiently select the most relevant features after the initial screening by guided-FS. For both the datasets, our proposed model provides better recognition accuracies than the work mentioned in [16, 17, 28, 34, 58, 60–65]. Hence, it can be inferred that the proposed bi-stage FS algorithm has the ability to predict COVID-19 with more perfection then its predecessors.
We have performed Wilcoxon rank-sum test [67] for the statistical significance test of the obtained results. In this non-parametric statistical test pairwise comparison is performed. For each pair, we have considered the accuracy achieved by our proposed method and the results reported by other methods on the same database. The working principle of Wilcoxon signed-rank test is as follows:
Let S denotes the observed value from the proposed model, X1 and X2 denote the predicted value from model 1 and model 2 respectively. Then the absolute error of each prediction is measured using the (21) and (22):
| 21 |
| 22 |
The substantial accuracy of prediction in a model over other models can be determined by conducting a statistical test (e.g. one-tailed hypothesis test). In this test, the null hypothesis is frame in such a way that both population are of the same distribution (E1=E2). The Wilcoxon rank-sum test is performed at significance level of 0.05. If for an observation, we get p-values over 0.05, we cannot reject the null hypothesis. On the other hand, for values less than 0.05 we can reject the null hypothesis with confidence level of 95%. The p-values obtained by this test are 0.007 and 0.04 for SARS-CoV-2-CT and COVID-CT dataset respectively. Hence, this evidences that of the proposed bi-stage FS approach is found to be statistically significant.
Conclusion
Millions of people globally have suddenly become heavily affected by the spread of the novel Coronavirus (COVID-19).As a result, the research community has been trying to harness the power of ML or DL and help the medical personnel to accurately detect this disease. In this work, we proposed a model in which CNN serves as a feature extractor and a bi-stage FS procedure serves as a mechanism to select the features with most relevance for the prediction of COVID-19 accurately from the chest CT images of the patient. Our proposed model was experimented on two publicly available COVID-19 datasets mentioned earlier and can identify the disease with 98.39% and 90.0% classification accuracy. Although our proposed model works better compared to other existing methods as mentioned in Table 6, still there exist some scopes to improve the overall performance by minimizing the error cases. After an exhaustive analysis of the misclassified cases, we observed that the lack of ample historical COVID data and the poor quality of some images may be the probable causes. In our future works, we aim to augment the prediction accuracy of the system by introducing other filter and wrapper combinations. We have also planned to work with other COVID datasets. Another possible target would be to use some well-established pre-trained CNN models to have better features at the initial stage.
Biographies
Shibaprasad Sen
received his B.E. degree in Computer Science and Engineering from Burdwan University in 2003. He received his M.E. degree in Computer Science and Engineering from West Bengal University of Technology in 2009. He received his PhD (Engineering) degrees from Jadavpur University in 2019. He joined Institute of Engineering and Management, Kolkata as a Professor in 2021. He has published more than 25 research articles in various journals and conference proceedings. His areas of current research interest are Pattern Recognition, Document Image Processing, Machine Learning.

Soumyajit Saha
is currently associated with Cognizant as a Program Analyst Trainee. He received his Bachelor of Technology in Computer Science and Engineering from Future Institute of Engineering and Management in the year 2020. His research interests include Feature Selection using optimization techniques, application of optimization techniques in different sectors of Artificial Intelligence, and application of Machine Learning and Deep Learning in Pattern Recognition.

Somnath Chatterjee
is currently pursuing B.Tech in Computer Science & Engineering from Future Institute of Engineering and Management, West Bengal, India. His research interests include handwriting recognition, medical image processing, natural language processing, Ensemble based Learning and other deep learning applications.

Associate Professor Seyedali (Ali) Mirjalili
is the director of the Centre for Artificial Intelligence Research and Optimization at Torrens University Australia. He is internationally recognized for his advances in Swarm Intelligence and Optimization, including the first set of algorithms from a synthetic intelligence standpoint - a radical departure from how natural systems are typically understood - and a systematic design framework to reliably benchmark, evaluate, and propose computationally cheap robust optimization algorithms. Seyedali has published over 200 publications with over 28,000 citations and an H-index of 58. As the most cited researcher in Robust Optimization, he is in the list of 1% highly-cited researchers and named as one of the most influential researchers in the world by Web of Science. Seyedali is a senior member of IEEE and an associate editor of several journals including Neurocomputing, Applied Soft Computing, Advances in Engineering Software, Applied Intelligence, and IEEE Access. His research interests include Robust Optimization, Engineering Optimization, Multi-objective Optimization, Swarm Intelligence, Evolutionary Algorithms, and Artificial Neural Networks. He is working on the application of multi-objective and robust meta-heuristic optimization techniques as well.

Ram Sarkar
received his B. Tech degree in Computer Science and Engineering from University of Calcutta in 2003. He received his M.E. degree in Computer Science and Engineering and PhD (Engineering) degree from Jadavpur University in 2005 and 2012 respectively. He joined the department of Computer Science and Engineering of Jadavpur University as an Assistant Professor in 2008, where he is now working as an Associate Professor. He received Fulbright-Nehru Fellowship (USIEF) for post-doctoral research in University of Maryland, College Park, USA in 2014-15. His areas of current research interest are Image and Video Processing, Optimization Algorithms and Deep Learning. He has published more than 250 research articles in various journals and conference proceedings. He is a senior member of the IEEE, U.S.A.

Footnotes
This article belongs to the Topical Collection: Artificial Intelligence Applications for COVID-19, Detection, Control, Prediction, and Diagnosis
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shibaprasad Sen, Email: shibubiet@gmail.com.
Soumyajit Saha, Email: soumyajitsaha1997@gmail.com.
Somnath Chatterjee, Email: somnathchatterjee796@gmail.com.
Seyedali Mirjalili, Email: ali.mirjalili@gmail.com.
Ram Sarkar, Email: ramjucse@gmail.com.
References
- 1.Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv:2003.12338
- 2.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J, Lang G (2020) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering [DOI] [PMC free article] [PubMed]
- 3.Goel T, Murugan R, Mirjalili S, Chakrabartty D (2020) OptCoNet: an optimized convolutionalneural network for an automatic diagnosis of COVID-19, in the Journal of Applied Intelligence. 10.1007/s10489-020-01904-z [DOI] [PMC free article] [PubMed]
- 4.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) International Journal Surgery. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID-19 Coronavirus Pandemic (2020) Worldometers, Retrieved July 23, 2020,from https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?
- 6.Mohamadou Y, Halidou A, Kapen P. A review ofmathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl Intell. 2020;50:3913–3925. doi: 10.1007/s10489-020-01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh D, Kumar V, Kaur M (2020) Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. European Journal of Clinical Microbiology & Infectious Diseases, pp 1–11. 10.1007/s10096-020-03901-z [DOI] [PMC free article] [PubMed]
- 8.Basavegowda HS, Dagnew G. Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol. 2020;5(1):22–33. doi: 10.1049/trit.2019.0028. [DOI] [Google Scholar]
- 9.Tingting Y, Junqian W, Lintai W, Yong X. Three-stage network for age estimation. CAAI Trans Intell Technol. 2020;4(2):122–126. doi: 10.1049/trit.2019.0017. [DOI] [Google Scholar]
- 10.Wu J, Wu X, Zeng W, Guo D, Fang Z, Chen L, Huang H, Li C. Chest CT findings in patients with corona virus disease 2019and its relationship with clinical features. Invest Radiol. 2020;55(5):257. doi: 10.1097/RLI.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W. Sensitivity of chest CT for COVID19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. Am J Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 13.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Lei P, Zeng B, Li Z, Yu P, Fan B, Wang C, Li Z, Zhou J, Hu S. Coronavirus disease (covid-19): spectrum of CT findings and temporal progression of the disease. Acad Radiol. 2020;27(5):603–608. doi: 10.1016/j.acra.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long C, Xu H, Shen Q, Zhang X, Fan B, Wang C, Zeng B, Li Z, Li X, Li H. Diagnosis of the coronavirus disease (covid-19): rRT-PCRor CT? Eur J Radiol. 2020;126:108961. doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soares E, Angelov P, Biaso S, Froes MH, Abe DK (2020) SARS-Cov-2 CT-scan dataset: a large dataset of real patients CT scans for SARS-cov-2 identification, medRxiv. 10.1101/2020.04.24.20078584
- 17.Yang X, He X, Zhao J, Zhang Y, Zhang S, Xie P (2020) COVID-CT-dataset: a CT scan dataset about COVID-19, pp 1–14. arXiv:2003.13865
- 18.Mafarja M, Aljarah I, Heidari AA, Faris H, Fournier-Viger P, Li X, Mirjalili S. Binary dragonfly optimization for feature selection using time-varying transfer functions. Knowledge Based Systems. 2018;161:185–204. doi: 10.1016/j.knosys.2018.08.003. [DOI] [Google Scholar]
- 19.Hammouri AI, Mafarja M, Al-Betar MA, Awadallah MA, Abu-Doush I. An improved dragonfly algorithm for feature selection. Knowledge-Based Systems. 2020;203:106131. doi: 10.1016/j.knosys.2020.106131. [DOI] [Google Scholar]
- 20.Mirjalili S. Dragonfly algorithm: a new meta-heuristic optimization technique for solving single objective, discrete, and multi-objective problems. Neural Computing and Application. 2016;27(4):1053–1073. doi: 10.1007/s00521-015-1920-1. [DOI] [Google Scholar]
- 21.Reynolds CW (1998) Flocks, herds, and schools: a distributed behavioral model, in Seminal graphics, pp 273–282
- 22.Wang L, Lin ZQ, Wong A (2020) COVID-net: a tailored deep convolutional neural network design for detection of COVID-19cases from chest radiography images. arXiv:2003.09871 [DOI] [PMC free article] [PubMed]
- 23.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks. arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 24.Abbas A, Abdelsamea MM, Gaber MM (2020) Classification of covid-19 in chest x-ray images using detrac deep convolutional neural network. arXiv:2003.13815 [DOI] [PMC free article] [PubMed]
- 25.Khan AI, Shah JL, Bhat M (2020) CORONET: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. arXiv:2004.04931 [DOI] [PMC free article] [PubMed]
- 26.Maghdid HS, Asaad AT, Ghafoor K, Sadiq AS, Khan MK (2020) Diagnosing COVID-19pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv:2004.00038
- 27.Razzak I, Naz S, Rehman A, Khan A, Zaib A (2020) Improving coronavirus (COVID-19) diagnosis using deep transfer learning, medRxiv
- 28.He X, Yang X, Zhang S, Zhao J, Zhang Y, Xing E, Xie P (2020) Sample-efficient deep learning forcovid-19 diagnosis based on ct scans, medRxiv
- 29.Tang Z, Zhao W, Xie X, Zhong Z, Shi F, Liu J, Shen D (2020) Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. arXiv:2003.11988
- 30.Sahlol AT, Yousri D, Ewees AA, Al-qaness MAA, Damasevicius R, Elaziz MA (2020) COVID-19 Image classification using deep features and fractionalorder marine predators algorithm. Scientific reports [DOI] [PMC free article] [PubMed]
- 31.Al-qaness MAA, Ewees AA, Fan H, Abd El Aziz M. Optimization method for forecasting confirmed cases of covid-19 in China. Journal of Clinical Medicine. 2020;9(3):674–689. doi: 10.3390/jcm9030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaban WM, Rabie AH, Saleh AI, Bo-Elsoud M (2020) A new covid-19 patients detectionstrategy (cpds) based on hybrid feature selection and enhanced knn classifier. Knowledge-Based Systems 205. 10.1016/j.knosys.2020.106270 [DOI] [PMC free article] [PubMed]
- 33.Farid AA, Selim GI, Khater HAA. A novel approach of CT images feature analysis and prediction to screen for corona virus disease (COVID-19) International Journal of Scientific Engineeringand Research. 2020;11(3):1–9. [Google Scholar]
- 34.Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M (2020) Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. Journal of Biomolecular Structure & Dynamics, pp 1–8 [DOI] [PubMed]
- 35.Belanche LA, González FF (2011) Review and evaluation of feature selection algorithms in synthetic problems. arXiv:1101.2320
- 36.Jain A. Feature selection: evaluation, application, and small sample performance. IEEE Transaction on Pattern Analysis and Machine Intelligence. 1997;19(2):153–158. doi: 10.1109/34.574797. [DOI] [Google Scholar]
- 37.Jain AK, Chandrasekaran B. Classification pattern recognition and reduction of dimensionality. Handbook of Statistics. 1982;2:835–855. doi: 10.1016/S0169-7161(82)02042-2. [DOI] [Google Scholar]
- 38.Duch W, Winiarski T, Biesiada J, Kachel A (2003) Feature selection and ranking filter. In: International conference on artificial neural networks and international conference on neural information processing, pp 251–254
- 39.Li Z, Liu J, Yang Y, Zhou X, Lu H. Clustering-guided Sparse structural learning for unsupervised feature selection. IEEE Transaction on Knowledgeand Data Engineering. 2014;26(9):2138–2150. doi: 10.1109/TKDE.2013.65. [DOI] [Google Scholar]
- 40.Cover TM, Thomas JA. Elements of information theory. New Jersey: Wiley-Interscience; 2006. [Google Scholar]
- 41.Bell DA, Wang H. Formalism for relevance and its application in feature subset selection. Machine Learning. 2000;41(2):175–195. doi: 10.1023/A:1007612503587. [DOI] [Google Scholar]
- 42.Kojadinovic I. Relevance measures for subset variable selection in regression problems based on k-additive mutual information. Computational Statistics and Data Analysis. 2005;49(4):1205–1227. doi: 10.1016/j.csda.2004.07.026. [DOI] [Google Scholar]
- 43.Spolaôr N, Cherman EA, Monard MC, Lee HD (2013) ReliefF for multi-label feature selection. In: Proceedings of brazilian conference and intelligent systems, pp 6–11. 10.1109/BRACIS.2013.10
- 44.Wang Z, Zhang Y, Chen Z, Yang H, Sun Y, Kang J, Yang Y, Liang X. Application of ReliefF algorithm to selecting feature sets for classification of high resolution remote sensing image. International Geoscience and Remote Sensing Symposium. 2016;2016(41271013):755–758. [Google Scholar]
- 45.Zhang Y, Ding C, Li T. Gene selection algorithm by combining reliefF and mRMR. BMC Genomics. 2008;9(2):1–10. doi: 10.1186/1471-2164-9-S2-S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spola N, Monard MC (2014) Evaluating ReliefF-based multi-label feature selection algorithm. In: Ibero-American conference on artificial intelligence, pp 194–205
- 47.Robnik-Sikonja M, Kononenko I. Theoretical and empirical analysis of Relief and ReliefF. Mach Learn. 2003;53:23–69. doi: 10.1023/A:1025667309714. [DOI] [Google Scholar]
- 48.Kira K, Rendell LA (1992) A practical approach to feature selection, in Machine Learning Proceedings, pp 249–256
- 49.Silva P, Luz E, Silva G, Moreira G, Silva R, Lucio D, Menotti D (2020) COVID-19 detection in CT images with deep learning: a voting-based scheme and cross-datasets analysis, Informatics in Medicine Unlocked 20. 10.1016/j.imu.2020.100427 [DOI] [PMC free article] [PubMed]
- 50.Das S (2001) Filters, wrappers and a boosting-based hybrid for feature selection. In: International conference on machine learning, pp 74–81
- 51.Vergara JR, Estévez PA. A review of feature selection methods based on mutual information. Neural Comput Applic. 2014;24(1):175–186. doi: 10.1007/s00521-013-1368-0. [DOI] [Google Scholar]
- 52.Mirjalili S, Mirjalili SM, Lewis A. Grey wolf optimizer. Advance Engineering Software. 2014;69:46–61. doi: 10.1016/j.advengsoft.2013.12.007. [DOI] [Google Scholar]
- 53.Rashedi E, Nezamabadi-pour H, Saryazdi S. BGSA: binary gravitational search algorithm. Nat Comput. 2010;9:727–745. doi: 10.1007/s11047-009-9175-3. [DOI] [Google Scholar]
- 54.Mirjalili S, Mirjalili SM, Yang X. Binary bat algorithm. Neural Comput Applic. 2014;25:663–681. doi: 10.1007/s00521-013-1525-5. [DOI] [Google Scholar]
- 55.Kennedy J, Eberhart RC (1997) A discrete binary version of the particle swarm algorithm. In: International conference on systems, man, and cybernetics, computational cybernetics and simulation, vol 5, pp 4104–4108
- 56.Kabir MM, Shahjahan M, Murase K. A new local search based hybrid genetic algorithm for feature selection. Neurocomputing. 2011;74:2914–2928. doi: 10.1016/j.neucom.2011.03.034. [DOI] [Google Scholar]
- 57.Faramarzi A, Heidarinejad M, Mirjalili S, Gandomi AH. Marine predators algorithm: a nature-inspired Metaheuristic. Expert Systems with Applications. 2020;152:113377. doi: 10.1016/j.eswa.2020.113377. [DOI] [Google Scholar]
- 58.Loey M, Manogaran G, Khalifa NEM (2020) A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput Applic. 10.1007/s00521-020-05437-x [DOI] [PMC free article] [PubMed]
- 59.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. [Google Scholar]
- 60.Mobiny A, Cicalese PA, Zare S, Yuan P, Abavisani M, Wu CC, Ahuja J, de Groot PM, Van Nguyen H (2020) Radiologist-level COVID-19 detection using CT scans with detail-oriented capsule networks. arXiv:2004.07407
- 61.Polsinelli M, Cinque L, Placidi G (2020) A light CNN for detecting COVID-19 from CT scans of the chest. arXiv:2004.12837 [DOI] [PMC free article] [PubMed]
- 62.Dan-Sebastian B, Delia-Alexandrina M, Sergiu N, Radu B (2020) Adversarial graph learning and deep learning techniques for improving diagnosis within CT and ultrasound images. In: 2020 IEEE 16th international conference on intelligent computer communication and processing (ICCP). IEEE, pp 449– 456
- 63.Shamsi Jokandan A, Asgharnezhad H, Shamsi Jokandan S, Khosravi A, Kebria PM, Nahavandi D, Nahavandi S, Srinivasan D (2020) An uncertainty-aware transfer learning-based framework for Covid-19 diagnosis. arXiv e-prints, arXiv-2007 [DOI] [PMC free article] [PubMed]
- 64.Mishra AK, Das SK, Roy P, Bandyopadhyay S (2020) Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach. Journal of Healthcare Engineering 2020 [DOI] [PMC free article] [PubMed]
- 65.Ewen N, Khan N (2020) Targeted self supervision for classification on a small COVID-19 CT scan dataset. arXiv:2011.10188
- 66.Sethy PK, Behera SK (2020) Detection of coronavirus disease (COVID-19) based on deep features
- 67.Wilcoxon F. Individual comparisons by ranking methods (Springer Series in Statistics) New York: Springer; 1992. pp. 196–202. [Google Scholar]
- 68.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large scale image recognition. arXiv:1409.1556
- 69.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 70.Chollet F (2017) Xception: deep learning with depth-wise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1251– 1258