Abstract
Long noncoding RNAs (lncRNAs), defined as untranslated and tightly-regulated transcripts with a length exceeding 200 nt, are common outputs of the eukaryotic genome. It is becoming increasingly apparent that many lncRNAs likely serve as important regulators in a variety of biological processes. In particular, some of them accumulate in the nucleus and function in diverse nuclear events, including chromatin remodeling, transcriptional regulation, RNA processing, DNA damage repair, etc. Here, we unite recent progresses on the functions of nuclear lncRNAs and provide insights into the future research directions of this field.
Keywords: MALAT1, NEAT1, XIST, Circular RNA, RNA-Binding protein, Nucleus
1. Introduction
It was long assumed that only a small proportion of the eukaryotic genome is actively transcribed to generate templates for protein translation. However, RNA deep sequencing of eukaryotic transcriptomes has revealed that transcription is pervasively existing across eukaryotic genomes and a diverse group of RNA classes with little or no protein-coding capacity are produced [1,2]. Among these transcripts, long noncoding RNAs (lncRNAs) are ranging from 200 nt to 100 kb in length. Although most lncRNAs (e.g. X-inactive specific transcript, aka XIST) are generated with splicing signals and/or modifications similar to that of protein-coding genes as described by a comprehensive analysis [3], a number of lncRNAs are processed in unexpected ways. For example, the 3’ end of MALAT 1 (Metastasis associated lung adenocarcinoma transcript 1) and NEAT1 (Nuclear enriched abundant transcript 1) are processed by the tRNA biogenesis machinery, generating a stable triple helix structure instead of a poly(A) tail [[4], [5], [6]]. Circular RNAs are generated via back-splicing, a reaction by which a splicing donor is joined to an upstream acceptor [[7], [8], [9]]. Interestingly, some of lncRNAs are highly conserved at the sequence level. The expression levels of certain lncRNAs are higher than most protein-coding genes in a cell- and/or tissue-specific manner. Importantly, the dysregulation of many lncRNAs are associated with various human diseases, such as cancers and brain disorders. For example, H19 is one of the most abundant lncRNAs during fetal development in mammals [10,11]. H19 is robustly induced during embryogenesis at a level comparable to the house-keeping gene β-actin, but is downregulated significantly after birth. Nevertheless, it remains highly expressed in adult skeletal muscle and heart [10,11]. Loss of H19 induces precocious muscle differentiation and defective embryogenesis [10,11].
Similar to mRNAs, lots of lncRNAs display a post-transcriptional role in the cytoplasm as well summarized by Rashid et al. [12]. For example, cytoplasmic lincRNA-p21 has an inhibitory effect on the translation activity of CTNNB1 and JUNB by sterically blocking their transcripts [13]. In addition, circular RNA ND1 and ND5 can promote the mitochondrial entry of nuclear-encoded proteins by interacting with the mitochondrial importation complex [14]. But it is important to note that there are also many lncRNAs accumulating and functioning in the nucleus, which has gained a broad interest in recent years. These nuclear lncRNAs exert critical roles in chromatin remodeling, transcriptional regulation, RNA processing and modification, etc. In this review, we will discuss the functions and underlying mechanisms of several well-studied nuclear lncRNAs and provide insights into the future research directions of nuclear lncRNAs in the field.
2. Nuclear speckle-associated lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1)
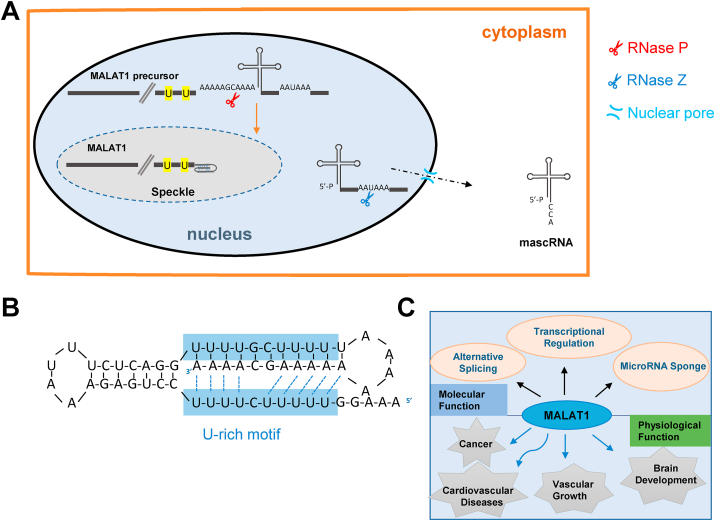
The MALAT1 locus is located on human chromosome 11q13 and is able to generate a long non-coding and nuclear-retained transcript with a length of ~8 kb [[15], [16], [17]]. Despite being transcribed by RNA polymerase II (Pol II) [18,19], mature MALAT1 transcript does not contain a canonical poly(A) tail, since the 3′ end processing of MALAT1 is completely different from the classical mRNA 3′ terminus formation [4,20,21]. The 3′ end of MALAT1 precursor lacks canonical cleavage/polyadenylation signals, but instead consists of a short and highly conserved poly(A)-rich tract [4]. During generation of mature MALAT1, the precursor is first cleaved endonucleolytically by RNase P immediately downstream of the poly(A)-rich tract to simultaneously produce the 3′ end of MALAT1 and the 5′ end of a tRNA-like small RNA (MALAT1-associated small cytoplasmic RNA, aka mascRNA) [4]. The 3′ end of mascRNA is then cleaved by RNase Z, followed by CCA addition (Fig. 1A) [4]. Curiously, MALAT1 accumulates in the nucleus, whereas mascRNA mainly translocates to the cytoplasm (Fig. 1A) [4]. Although MALAT1 lacks a poly(A) tail, it is highly abundant in human and mouse tissues [16,22,23] and expressed at a level higher than a number of house-keeping genes [24], indicating that the 3′ end of MALAT1 is able to resist 3′ degradation by 3′-5′ exonucleases. Indeed, a following study found that the highly conserved A- and U-rich motifs at the 3′ end of MALAT1 can form a triple helix via base-pairing, and disrupting the triple helix triggers the degradation of MALAT1 (Fig. 1B) [5,6,25], demonstrating that this extensive secondary structure is able to protect mature MATAL1 from 3′-5′ exonucleolytic decay.
Fig. 1.
The biogenesis, structure, and functions of MALAT1. (A) The MALAT1 precursor is first cleaved by RNase P immediately downstream of the poly(A)-rich tract, generating the 3′ end of MALAT1 and the 5′ end of mascRNA. The 3′ end of mascRNA is then cleaved by RNase Z and added with CCA. MALAT1 accumulates in nuclear speckles, while mascRNA is exported to the cytoplasm. (B) The A- and U-rich motifs at the 3′ end of MALAT1 form a triple helix via base-pairing, protecting the 3′ end of MALAT1 from degradation. Figure is adapted from Ref. [5]. (C)MALAT1 participates in diverse molecular events and is associated with many diseases.
Different from mascRNA or many other ncRNAs, MALAT1 specifically localizes in nuclear speckles (also called as interchromatin granule cluster) [23,26,27], nuclear domains that are composed of multiple factors involved in transcription activation or repression, pre-mRNA splicing, mRNA modification, and mRNA nuclear export [[28], [29], [30]]. Three nuclear speckle proteins (RNPS1, IBP160, and SRm160) and two distinct motifs on MALAT1 contribute to the specific localization [27]. Such localization suggests that MALAT1 may play important roles in some molecular processes in the nucleus. In fact, MALAT1 is not a structural RNA required for the maintenance of nuclear speckle integrity [31,32], but functions in chromosome segregation during mitosis [32]. Lack of MALAT1 results in aberrant mitosis and the fragmentation of cell nuclei [32]. Intriguingly, Tripathi and colleagues found that MALAT1 interacts with the serine/arginine (SR) splicing factors, controls the distribution and phosphorylation status of SR proteins in nuclear speckles and, in turn, modulates alternative splicing (AS) events of a set of SR-regulated pre-mRNAs in HeLa cells (Fig. 1C) [32]. It is thus likely that the cycling of at least some of SR proteins between speckle domains and sites of transcription is regulated by MALAT1. However, there are several contradictory observations that MALAT1 had almost no effect on the localization of nuclear speckle components or phosphorylation status of SR proteins in knockout mice [24,33,34]. Besides, no obvious connection between MALAT1 and AS regulation was found [24,33]. These inconsistent results might be explained by the possibility that MALAT1 has evolved a function in human cells that is not easily detected in mice or the function of MALAT1 is simply cell/tissue-specific, since MALAT1 expression varies significantly among tissues and cells [16,33].
MALAT1 has also been reported to participate in modulation of transcription (Fig. 1C). Indeed, a comprehensive analysis of genomic binding sites of MALAT1 implied that it exhibits localization to many actively transcribed chromatin sites [35]. The human LTBP3 is a gene downstream of the MALAT1 locus and has an opposite transcription orientation to that of MALAT1. MALAT1 activates the transcription of LTBP3 by promoting the recruitment of the key transcription factor Sp1 to the LTBP3 promotor in mesenchymal stem cells [36]. A large reduction in expression of NEAT1, an lncRNA locus upstream of MALAT1, was observed in MEFs obtained from MALAT1 knockout mice [27]. In addition to the cis effect on neighboring genes, the localization of the polycomb 2 protein (Pc2), a key histone modifier, is controlled by MALAT1, which in turn influences expression of many genes [37]. Yang and colleagues demonstrated that the association between MALAT1 and unmethylated Pc2 promotes E2F1 SUMOylation, resulting in a biochemical switch of the Pc2 chromodomain from interacting with repressive histone marks to preferring active histone marks. Subsequently, a number of growth-control genes are activated by the event [37].
MALAT1 can also act as a microRNA sponge in the nucleus in an AGO2-dependent manner (Fig. 1C) [[38], [39], [40], [41]]. For example, there are two targeting sites for mircoRNA-9 on MALAT1 transcript. The MALAT1 level was significantly reduced in AGO2 immunoprecipitates purified from cells treated with anti-mircoRNA-9 as compared to the control sample [38]. In another case, MALAT1 can modulate c-Myc expression by sponging mircoRNA-34a in melanoma cells [40].
MALAT1 transcript was first discovered in a screening for genes whose expression have a potential relationship to metastasis in early-stage non-small cell lung cancer [16]. Afterwards, MALAT1 was found to be evolutionally conserved from mouse to human and widely expressed in numerous cancer tissues [[42], [43], [44], [45]], implying that the dysregulation of MALAT1 plays crucial roles in cancers and MALAT1 can be used as a valid cancer biomarker as well as a therapeutic target for clinical application (Fig. 1C). For instance, MALAT1 has been found as an important regulator of lung cancer metastasis probably due to its effect on metastasis-associated gene expression [46]. Interestingly, the MALAT1 ASO (antisense oligonucleotide) treated mice was able to resist to EBC-1 tumor metastasis to the lung, indicating a potential application of ASOs against MALAT1 in preventing lung cancer from spreading [46]. In colorectal cancer (CRC), the binding of MATAL1 to the tumor suppressor PSF (PTB-associated splicing factor) dissociates PTB (polypyrimidine-tract-binding protein) from PSF. The released PTB promotes CRC cell proliferation and tumor growth [47]. Two recent studies indicated that the secondary structure of MALAT1 varies in different cancers [48,49]. By dimethyl sulfate-sequencing and RNA structure analysis, MALAT1 is likely to be more unstructured in chronic myeloid leukemia, but undergoes 18 novel structural rearrangements in cervical cancer [49]. This provides new insights into how RNA structure affects its regulatory role under various conditions. Besides cancers, the dysregulation of MALAT1 is also involved in many other physiological processes and pathological diseases, including brain development [26,50], vascular growth [51], cardiovascular diseases [52,53], neurological disorders [[54], [55], [56], [57]], just to name a few. Unfortunately, most of the precise mechanisms underlying the MALAT1-associated diseases still remain unclear. Therefore, further studies are needed to clarify these questions.
3. Nuclear paraspeckle-associated lncRNA NEAT1 (nuclear enriched abundant transcript 1)
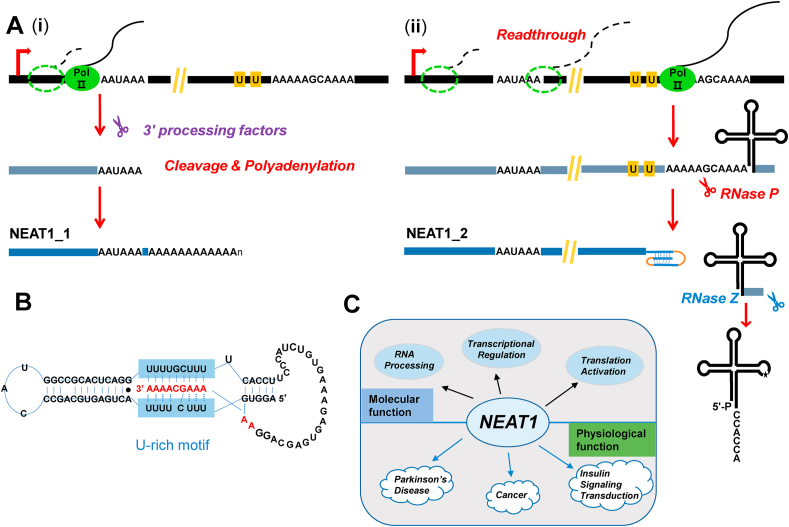
NEAT1 is originally identified as a large and infrequently spliced transcript in a genome-wide screening to search for nuclear ncRNAs that might modulate gene expression [23]. The NEAT1 locus is adjacent to the MALAT1 locus on human chromosome 11q13 and generates two overlapping isoforms [58,59]. NEAT1_1, aka MENε, is the short isoform with a length of ~3700 nucleotides [60]. The 3′ end of NEAT1_1 undergoes the canonical cleavage/polyadenylation steps and, in turn, terminates in a poly(A) tail (Fig. 2A). On the contrary, the long version of NEAT1 (NEAT1_2, aka MEN β) is over 23 kb and stabilized by a triple helix structure following RNase P cleavage at its 3’ end (Fig. 2A and B) [5,6,60], which is similar to MALAT1. Although none of the two isoforms has an open reading frame or a capacity to yield a peptide, they are highly and ubiquitously expressed in a wide range of tissues, such as ovary, prostate, colon, and pancreas [23]. As a type of architectural lncRNA, NEAT1 is essential for organization and integrity of nuclear paraspeckles [[61], [62], [63], [64]], irregularly shaped subnuclear bodies closed to the NEAT1 transcription site [31]. NEAT1 translocates immediately to nuclear paraspeckles once it is transcribed by RNA Pol II [65]. During daughter nuclei reformation, NEAT1 transcription happens prior to the accumulation of the paraspeckle-associated proteins PSP1/p54 in paraspeckles [31]. NEAT1 depletion leads to loss of paraspeckles, whereas overexpression of NEAT1 increases paraspeckle number [31,60,65,66], implying that the paraspeckle-associated proteins might be multifunctional when endogenous NEAT1 expression is altered. In fact, NEAT1 has been found to bind specifically to PSP1/p54 [31]. Interestingly, PSP1 forms a heterodimer with p54 in a NEAT1-independent manner [66]. But PSP1without the RNA recognition motifs no longer exhibits an interaction with NEAT1 or paraspeckles [31]. Furthermore, NEAT1 depletion has no significant effect on the protein level of PSP1 or p54, excluding the possibility that NEAT1 regulates the degradation of some paraspeckle proteins [60]. Therefore, nuclear paraspeckle is built on NEAT1 and the dysfunction of nuclear paraspeckle could be triggered by NEAT1 depletion. For example, the nuclear export of transcripts containing paraspeckle-retention elements (Alu repeats in humans and SINEs in mouse) or hyper-edited A-to-I is enhanced by NEAT1 depletion [66,67].
Fig. 2.
The biogenesis, structure, and functions of NEAT1. (A)NEAT1_1 (the short form of NEAT1) is stabilized by a canonical poly(A) tail following cleavage/polyadenylation. However, NEAT1_2 (the long form of NEAT1) undergoes steps similar to that of MALAT1 biogenesis. (B) The A- and U-rich motifs, present upstream of the RNase P cleavage site, form a triple helix via base-pairing, protecting the 3′ end of NEAT1_2 from degradation. Figure is adapted from Ref. [6]. (C)NEAT1 is involved in diverse molecular events, such as RNA processing and transcriptional regulation. The dysregulation of NEAT1 contributes to many diseases.
NEAT1 not only functions as an architectural component of paraspeckles, but also is important to some other organelles (Fig. 2C). For example, NEAT1 and paraspeckles are crucial for mitochondrial homeostasis [68,69]. A recent study demonstrated that loss of mitochondrial proteins or mitochondrial stress results in abnormal expression of NEAT1 as well as a remarkable increase of elongated nuclear paraspeckles [68]. Moreover, many mRNAs of nuclear-encoded mitochondrial proteins contain paraspeckle-retention elements at their 3’ UTRs and is associated with NEAT1 [68], indicating a strong regulatory communication between nucleus and mitochondria. Abnormal NEAT1 expression leads to mitochondrial dysfunction, such as altered mitochondrial respiration and ATP production rate [68,69].
NEAT1 has strong regulatory effects on many molecular processes (Fig. 2C). Jiang and colleagues found that NEAT1 and its binding partner PSP1/p54 heterodimer can enhance pri-miRNA processing [70]. NEAT1 may provide a binding platform for various pri-miRNAs and microprocessor components that are attracted by the pri-miRNA-like hairpin structures of NEAT1 [70]. Additionally, NEAT1 can regulate the splicing of PPARγ2 (peroxisome proliferator activated receptor) pre-mRNA temporally via the SR protein SRp40 mediated pathway during adipogenesis. In this case, NEAT1 binds to SRp40 and potentiates its phosphorylation status by Clk kinase [71]. Wang and colleagues found that NEAT1 can activate viral gene expression by recruiting STAT3 (signal transducer and activator of transcription 3) to their promoters in response to herpes simplex virus-1 (HSV-1) infection [72]. Similarly, their following study showed that NEAT1 recruits the acetyltransferase CBP/P300 complex to the promoters of the endocytosis-related genes, thereby altering H3K27 acetylation and H3K27 crotonylation in Alzheimer disease [73].
Except for RNA processing or transcriptional regulation, several studies have suggested that NEAT1 has a profound connection with translation activation or protein stability control. For example, during RNA polymerase I inhibition, NEAT1-sequestered p54 and PSF are released from nuclear paraspeckles upon NEAT1 depletion and relocated to c-Myc mRNAs for its translation activation [74]. Curiously, NEAT1 depletion has almost no effect on the protein level of c-Myc under normal conditions [74], indicating that the NEAT1-mediated c-Myc translation only occurs in stressed conditions. In CD4+ T cells, NEAT1 physically binds to STAT3 protein and keeps it from ubiquitination [75]. This was exemplified by the NEAT1 knockdown experiment which led to a large reduction in ubiquitination labeled STAT3 protein [75]. Likewise, the half-life of another NEAT1-binding protein DDX5 is drastically increased upon overexpression of NEAT1 in HT29 cells [76].
Similar to many other ncRNAs, NEAT1 consists of potential targeting sites for various mircoRNAs [[77], [78], [79], [80], [81], [82]]. Importantly, plenty of these mircoRNA-target genes are oncogenes and contribute to tumorigenesis. Disrupting the cross-talk between NEAT1, mircoRNAs, and mircoRNA-target genes may promote proliferation, migration, invasion of cancer cells. Thus NEAT1 could be an effective target for the diagnosis and therapy of many cancer types. In prostate cancer, NEAT1 modulates the expression of the oncogene HMGA2 by sponging mircoRNA-98-5p [81]. In gastric cancer, the binding of microRNA-365a-3p to NEAT1 enhances ABCC4 expression [80]. In esophageal Squamous Cell Carcinoma, mircoRNA-129/CTBP2 axis is controlled by NEAT1 [82]. Nevertheless, it is still elusive how and why cytoplasmic mircoRNAs are controlled by a nuclear lncRNA.
An increasing number of studies focusing on NEAT1's function in physiology have proved that the intensive upregulation of NEAT1 in several human diseases is associated with a drastic decrease in patient survival (Fig. 2C) [[83], [84], [85], [86], [87]]. In regard to Parkinson's disease (PD), a chronic and progressive brain disorder that affects movement, NEAT1 expression was found to be significantly upregulated in PD mice [83,84]. Interestingly, depletion of NEAT1 could reverse apoptosis induced by PD inducers possibly by controlling the expression of some PD contributors, such as α-synuclein and PINK1 [83,84]. Regarding non‐alcoholic fatty liver disease, loss of NEAT1 inhibits the expression of acetyl‐CoA carboxylase and fatty acid synthase through the mTOR/S6K1 signaling pathway, implying that NEAT1 acts as a potent regulator in the biosynthesis of lipid and insulin signaling transduction [88]. With intensive researches of NEAT1-related diseases, we believe that a therapeutic application of NEAT1 is coming soon.
4. Chromatin-associated lncRNA XIST (X-inactive specific transcript)
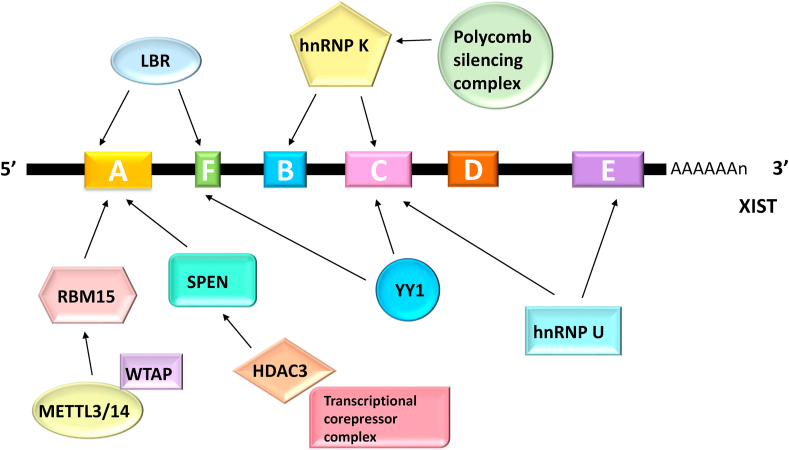
Less than 3% of the ancestral genes survive on the male Y chromosome, while X chromosome retains more than 97% [89,90]. Therefore, males and females have unbalanced genetic content in mammals. To minimize sex chromosome-linked gene expression imbalance between XX females and XY males, a randomly chosen X chromosome is transcriptionally silenced in early female embryogenesis. This dosage compensation process is called X chromosome inactivation (XCI) and is initiated by the lncRNA XIST [[91], [92], [93], [94], [95], [96]]. It is important to note that XCI by no means equal to loss of X chromosome, since 3%–20% of the X-linked genes from the inactive X (Xi) chromosome are still activated and indispensable for female embryogenesis. This is exemplified by Turner syndrome, a genetic disorder disease only occurs in females with only a single X chromosome [97,98]. XIST was first described as a long noncoding transcripts generated exclusively from the Xi chromosome in the early 1990s [[99], [100], [101], [102], [103]]. Once generated, XIST spreads across the entire length of the Xi chromosome, resulting in gene repression chromosome-widely. Further sequence analyses revealed that XIST is a 15–17 kb spliced and polyadenylated RNA which contains blocks of local tandem repeat elements designated A-F (Fig. 3) [102,103]. Significant progresses have elucidated the functional roles of each individual repeat in establishment of X chromosome-linked gene silencing. Hence, we will focus our discussion on these functional elements as well as their RBPs (RNA-binding proteins) in this section.
Fig. 3.
Schematic representation of XIST and its RBPs. As a multi-tasking RNA molecule, XIST functions in a variety of molecular processes through different RBPs. The tandem repeats (denoted in boxes) contribute to the recruitment of different RBPs.
Using a series of inducible XIST transgenes, Wutz and colleagues found that XIST mutations that lack A-repeat, consisting of 7–8 copies of a 24 nt GC-rich core sequence at the 5’ end of XIST, abolish silencing activity and lead to a decrease in stability without influencing localization [104]. Furthermore, the transcription activity of XIST appears to be affected by deletion of the endogenous A-repeat region in DNA [105]. Besides affecting XIST itself, the role of A-repeat in chromosome silencing is also linked to its binding proteins. For example, A-repeat contributes to the interaction with RBM15 (RNA binding motif protein 15) which further targets the methyltransferase complex METTL3/14 to specific RNAs, including XIST, for addition of N6-methyladenosine (m6A modification) [106,107]. Importantly, loss of the m6A catalytic subunit or the m6A reader leads to a major silencing deficiency, indicating a role of m6A in XIST-mediated chromosome silencing [106]. Contradictorily, only a minor decrease in X silencing efficiency was observed in either RBM15 knockdown or knockout cells [106,108]. The phenomena might be explained by the finding of RBM15B, the RBM15 homolog, which shares redundant functions with RBM15 [106,109]. SPEN is another interactor of A-repeat [110]. Depletion of SPEN results in impaired silencing of the X chromosome-linked genes, such as Fbxl17, without disrupting XIST localization [107,108,110,111]. In addition, SPEN might initiate transcriptional repression by actively recruiting the transcriptional corepressor complex and the histone deacetylase HDAC3 [[112], [113], [114]]. Unlike RBM15 and SPEN, LBR (Lamin B receptor) has a widespread distribution across XIST [115,116]. The major binding site encompasses the entire A- and F-repeat as proved by LBR CLIP-seq analysis [115,116]. Losing the interaction between XIST and LBR leads to impaired chromosome silencing [108,116]. B- and C-repeat have been reported to be critical for XIST spreading as well as the recruitment of hnRNP K (heterogeneous nuclear ribonucleoprotein K) and the polycomb silencing complexes to XIST [110,117,118]. Chu and colleagues found that defective chromosome silencing can be induced by depletion of hnRNP K [110]. With further IF-coFISH (combined immunofluorescence and RNA fluorescence in situ hybridization) experiments, they demonstrated that hnRNP K colocalizes with H2AK119ub and H3K27me3, the earliest chromatin modifications occurring in XCI [110]. Depletion of hnRNP K reduces the accumulation of H3K27me3 and H2AK119ub along Xi without influencing the global level of these modifications, suggesting Xi undergoes a specific modification. Indeed, hnRNP K depletion specifically dissociates XIST from the polycomb silencing complexes [110]. C-repeat, together with E-repeat, is also responsible for the binding of hnRNP U (heterogeneous nuclear ribonucleoprotein U) to XIST and, in turn, XIST localization to chromatin [108,110,[119], [120], [121]]. But whether correct XIST localization depends on a combination of additional factors is still elusive, since the effect of hnRNP U cannot be confirmed in certain cell types [122,123]. In fact, an interesting study demonstrated that YY1 protein, capable of binding both RNA and DNA, acts as an adaptor in docking C-repeat of the RNA onto the F-repeat region of the XIST locus [124]. By deleting D-repeat through CRISPR/Cas9 technique in HEK293T cells, a significantly reduction in XIST expression was observed, implying that D-repeat might play an important role in the stability or transcriptional activity of XIST [125]. Consequently, the silencing ability of XIST was abrogated as proved by the phenomenon that levels of some X chromosome-linked genes were upregulated [125].
3D structural reorganization is another critical pathway for establishment of Xi gene repression by XIST, together with SmcHD1 (Structural maintenance of chromosomes flexible hinge domain containing 1) and the polycomb factors [[126], [127], [128], [129], [130], [131], [132], [133], [134]]. In this mechanism, the X inactive genes are relocalized from the periphery to the interior of chromatin during XCI, thereby being isolated from RNA Pol II and other general transcription factors [126].
XCI is not only a dosage compensation process happening during early female embryogenesis and related to X-linked diseases in females, but also a critical modifier of many other diseases including cancers and neurological disorders. For example, XIST could affect cancer development and progression by sponging various microRNAs, thereby influencing the expression of some oncogenes [[135], [136], [137]]. On the other hand, Yue and colleagues found that the level of XIST is significantly increased in AD (Alzheimer's disease) mice models, suggesting that XIST might be used as a diagnostic marker for AD [138].
5. Nuclear circular RNA
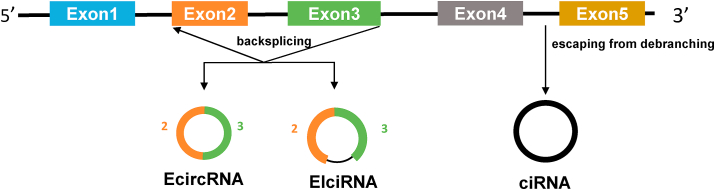
Protein-coding genes of the eukaryotic genome are typically split by one or multiple introns. During the production of a mature mRNA, introns must be removed and exons are joined together in a linear order, which is known as canonical splicing [[139], [140], [141]]. Alternatively, splicing events can also occurs in a way that a splicing donor is joined to an upstream acceptor, thereby generating a covalently-closed RNA (circular RNA) [7,[142], [143], [144], [145]]. Despite the first circular RNA being discovered in 1976 [146], only recently has it been appreciated that these covalently-closed molecules represent a common class of transcriptional outputs from numerous protein-coding genes in the eukaryotic system [147,148]. So far, many labs have been putting a lot of efforts into characterization of circular RNA biology, and thus obtained remarkable progresses as well reviewed in many papers [[7], [8], [9],143,[149], [150], [151], [152], [153], [154]]. In short, the biogenesis of circular RNA is initiated by base-pairing between the flanking intronic repeats as well as some RBPs, including QKI, Mbl, ADAR1, RBM20, FUS, etc [[155], [156], [157], [158], [159]]. The degradation of circular RNA is controlled by different exonucleases and the platform protein gawky (aka GW182) in various context [[160], [161], [162], [163], [164]]. Once generated, most circular RNAs are exported to the cytoplasm in a length-dependent manner with the assistance of the DExH/D-box helicase DDX39A and DDX39B [165,166]; however, a small amount of circular RNAs accumulate in the nucleus. Here, we will focus our discussion on nuclear circular RNAs and their functions in this section.
In searching for lncRNAs involved in transcriptional regulation, Li and colleagues identified a special subclass of Pol II-binding circular RNA, also called EIciRNA (exon-intron circular RNA), whose intron(s) between circularized exons are not spliced out (Fig. 4) [167]. Similar to most EciRNAs (exonic circular RNAs), EiciRNAs are generated via back-splicing reactions in which the flanking Alu repeats facilitate their circularization [167]. Functionally, EIciRNAs have cis regulatory effects on their host gene transcription activity (Table 1), since depletion of EIciRNA leads to a significant reduction in the levels of their nascent mRNAs. Further mechanism analyses indicated that EIciRNA interacts with U1 snRNP directly at the promoter of their host gene. Loss of U1 snRNA or disrupting the U1 snRNA-EIciRNA interaction by antisense morpholino (AMO) impairs the transcription-enhancing effect of EIciRNAs [167]. Notably, EIciRNAs not only localize to their sites of transcription, but also accumulate to other nuclear subdomains, suggesting other potential regulatory roles and mechanisms of EIciRNAs [167]. Another example of nuclear intron-containing circular RNA is ciRNA (intronic circular RNA), a type of lariat RNA which escapes from debranching (Fig. 4) [168]. Like EIciRNAs, ciRNAs (e.g. circANKRD52) are able to interact with the Pol II complex and modulate transcription efficiency of their host genes in cis (Table 1). Mechanistically, depletion of circANKRD52 has almost no effect on Pol II engagement, suggesting that circANKRD52 affects transcriptional rate but not transcription termination [168]. In another case, ciRNAs (circIns2 in rat and circINS in human), derived from the insulin gene, regulate transcription of genes involved in calcium signaling and insulin exocytosis via the RBP TAR DNA-binding protein 43 kDa (TDP43) in the nucleus (Table 1) [169]. Low expression of circIns2 or circINS leads to defective insulin secretion of β-cell to and are associated with type 2 diabetes [169]. EciRNAs are also capable of regulating transcription in the nucleus as exemplified by circFECR1. CircFECR1 can activate its host gene expression by inducing promoter hypomethylation with the demethylase TET1 (Table 1) [170].
Fig. 4.
The biogenesis of circular RNAs. Most circular RNAs are generated from protein-coding genes via back-splicing by which a splicing donor is joined to an upstream acceptor. In other cases, circular RNAs are derived from lariat intronic RNAs that escape from debranching.
Table 1.
The nuclear functions of circular RNAs
| circRNA | Type | Molecular Mechanism | Ref |
|---|---|---|---|
| circEIF3J | EIciRNA | Interacts with RNA Pol II and U1 snRNP to regulate transcription of its host gene | [167] |
| circPAIP2 | EIciRNA | Interacts with RNA Pol II and U1 snRNP to regulate transcription of its host gene | [167] |
| circANKRD52 | ciRNA | Interacts with RNA Pol II to regulate transcription of its host gene | [168] |
| circIns2/INS | ciRNA | Interacts with TDP-43 to regulate transcription | [169] |
| circFECR1 | EciRNA | Induces promoter hypomethylation with the demethylase TET1 | [170] |
| circ2082 | EciRNA | Promotes DICER nuclear translocation via RBM3 | [171] |
| cia-cGAS | EciRNA | Inhibits cGAS as a decoy | [172] |
| circSEP | EciRNA | Regulates alternative splicing of its host gene via R-loop | [173] |
| circCRM1 | Intergenic circRNA | Maintains normal centromeric chromatin organization via R-loop | [174] |
| circSMARCA5 | EciRNA | Downregulates its host gene expression via R-loop | [177] |
Besides transcription, some nuclear circular RNAs can affect the function of their RBPs. For example, a recent study demonstrated that circ2082 has an important role in nucleocytoplasmic distribution of the endoribonuclease DICER, a key component of pre-microRNA processor complex (Table 1) [171]. In this case, circ2082 is recruited to DICER by RNA binding motif protein 3 (RBM3) in the nucleus of glioblastoma cell. The interaction is crucial for nuclear retention of DICER, since depletion of circ2082 results in normalization of DICER localization to the cytoplasm [171]. The change in nucleocytoplasmic distribution of DICER further reestablishes the assortment of cellular microRNAs (microRNAome) [171]. Because the dysregulation of microRNAome is associated with cancers, circ2082 could be used as a therapeutic target for cancer treatment. Circular RNA cia-cGAS act as a protein decoy for the DNA sensor cGAS and inactivates its synthase activity under homeostatic conditions, which in turn inhibits cGAS-mediated autoimmune responses (Table 1) [172].
Interestingly, certain nuclear circular RNAs are able to form an RNA:DNA hybrid (R-loop) with the genomic DNA. In Arabidopsis, nuclear circSEP derived from exon 6 of the SEPALLATA3 gene was reported to affect the splicing of its host gene through an R-loop mechanism (Table 1). Formation of the R-loop leads to an increased level of a linear SEP transcript isoform which lacks exon 6, in turn driving floral homeotic phenotypes [173]. Importantly, circSEP:DNA R-loops are more abundant than their linear RNA:DNA counterparts, indicating the specific role of circSEP in R-loop formation [173]. In Zea mays, circular RNA:DNA R-loops are integral components of centromeric chromatin [174]. The centromeric retrotransposons derived circular RNAs were found to bind to the centromere through R-loops (Table 1). Disrupting the R-loop results in abnormal localization of the centromeric H3 variant (CENH3), in turn affecting centromeric chromatin organization [174]. SMARCA5 is a key component of DNA damage repair pathway and is required for chromatin remodeling [175,176]. In breast cancer cells, circSMARCA5 can downregulate the expression of the SMARCA5 gene via R-loop (Table 1). Therefore, circSMARCA5 has a negative effect on DNA repair capacity [177].
6. Concluding remarks
Eukaryotic nucleus is a membrane-bound organelle contains most genetic information and controls gene expression. Many molecular processes, such as transcription, splicing, replication, histone modification, and DNA repair, occur in the nucleus and are regulated combinatorially by various protein factors. Only until recent 20 years, lncRNAs have been considered important regulators in the processes mentioned above [19,[178], [179], [180]]. Although some of lncRNAs (e.g. XIST) have general effects on nuclear events, the majority of them seem to act uniquely on certain genes, implying that the total numbers of functional nuclear lncRNAs (annotated or unannotated) may be underestimated. Thus defining the novel nuclear roles of lncRNAs will help us understand the complex of nuclear regulation.
Having established an association between a lncRNA and a nuclear event, we could reason a possible mechanism based on the phenotype. Informed by the studies discussed in above sections, we will summarize potential mechanisms for the functions of newly-discovered nuclear lncRNAs. First, lncRNAs might indirectly affect certain events via controlling the expression or subcellular localization of the key protein factors. With the development of high-throughput RNA-seq technique and single molecule tracking system, this possibility could be easily examined. Second, lncRNAs might act as a molecular platform, bringing diverse proteins together into an RNP complex, or a decoy by sponging RNAs or even proteins and preventing them from associating with their targets. In these two cases, deletion analyses can be performed. If the hypothesis is right, then deletion of a protein binding motif in lncRNAs or a RNA binding motif in proteins leads to loss of function. In addition, a variety of high-throughput sequencing methods were developed to identify the interactomes of nuclear lncRNAs in vivo (Table 2). These tools could help us map RNA-DNA [[181], [182], [183], [184], [185], [186], [187], [188], [189]], RNA-protein [[190], [191], [192], [193]], and RNA-RNA interactions [[194], [195], [196], [197], [198]]. Third, lncRNAs might be an organizer of nuclear architecture. Knockdown or knockout of these lncRNAs results in nuclear subdomain breakdown, thereby affecting unique reactions. Fourth, the act of transcription through a lncRNA locus seems to be regulatory, rather than the lncRNA product as discussed by Long et al. and Bassett et al. [178,199]. To answer the question, the lncRNA region in genomic DNA can be replaced by a random sequence without changing its promoter. Combinatorially, the lncRNA transcript can be knockdown by siRNA, ASO, or CRISPR-Cas13a system or functionally blocked by AMO.
Table 2.
The methods used to study RNA interactomes.
| Method | Description | Experimental Purpose | Enhanced Version | Ref |
|---|---|---|---|---|
| CHIRP-seq | A method to pull down and map the RNA-associated chromatin using biotinylated antisense oligos | Identification of RNA-DNA interaction | CHART-seq, CHIRT-seq, hiCHIRP-seq | [[181], [182], [183], [184], [185]] |
| R-CHIP-seq | A method to pull down and map R-loops using catalytic-dead RNase H | Identification of R-loop | [186] | |
| DRIP-seq | A method to pull down and map R-loops using S9.6 antibody | Identification of R-loop | bisDRIP-seq, DRIPc-seq | [[187], [188], [189]] |
| CLIP-seq | A method to pull down and map the protein-binding RNA by immunoprecipitation | Identification of RNA-protein interaction | PAR-CLIP-seq, iCLIP-seq, hiCLIP-seq | [[190], [191], [192], [193]] |
| SHAPE-seq | A method to predict RNA secondary structures using chemical modifiers | Prediction of RNA-RNA interaction | icSHAPE-seq, DMS-seq, SHAPE-seq2.0 | [[194], [195], [196], [197]] |
| Frag-seq | A method to predict RNA secondary structures using endonuclease P1 | Prediction of RNA-RNA interaction | [198] |
In conclusion, the last 20 years have revealed that lncRNAs are not simply rare transcriptional noise, but pervasively exist in the eukaryotic system. Although numerous surprises have been disclosed, they only represent a glimpse of the tip of lncRNA biology. More efforts are required to provide insights into how these special transcripts are controlled and function especially in the nucleus. The era for studies in lncRNA biology is just coming.
Declaration of competing interest
No potential conflict of interest was reported by the authors.
Acknowledgments
This work was sponsored by the National Natural Science Foundation of China (32070633), the Natural Science Foundation of Chongqing, China (cstc2019jcyjmsxmX0085), the Innovation Support Program for Overseas Returned Scholars of Chongqing, China (cx2019142), the Fundamental Research Funds for the Central Universities of China (2020CDJQY-A076), and the 100 Talent Program of Chongqing University (0304001104433). We thank the members of the Shan lab (University of Science and Technology of China) and the Wilusz lab (University of Pennsylvania) for discussions.
References
- 1.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Clark M.B. The reality of pervasive transcription. PLoS Biol. 2011;9(7) doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz J.E., Freier S.M., Spector D.L. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilusz J.E. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26(21):2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J.A. Formation of triple-helical structures by the 3'-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 2012;109(47):19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L. Circular RNAs in eukaryotic cells. Curr. Genom. 2015;16(5):312–318. doi: 10.2174/1389202916666150707161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilusz J.E. A 360 degrees view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA. 2018;9(4):e1478. doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao M.S., Ai Y., Wilusz J.E. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30(3):226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabory A. The H19 gene: regulation and function of a non-coding RNA. Cytogenet. Genome Res. 2006;113(1–4):188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 11.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 12.Rashid F., Shah A., Shan G. Long non-coding RNAs in the cytoplasm. Dev. Reprod. Biol. 2016;14(2):73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon J.H. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 2020;63(10):1429–1449. doi: 10.1007/s11427-020-1631-9. [DOI] [PubMed] [Google Scholar]
- 15.Lin R. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 16.Ji P. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Hamblin M.H., Yin K.J. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–1714. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djebali S. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu B., Shan G. Functions of long noncoding RNAs in the nucleus. Nucleus. 2016;7(2):155–166. doi: 10.1080/19491034.2016.1179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz J.E., Spector D.L. An unexpected ending: noncanonical 3' end processing mechanisms. RNA. 2010;16(2):259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilusz J.E. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim. Biophys. Acta. 2016;1859(1):128–138. doi: 10.1016/j.bbagrm.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tano K. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Hutchinson J.N. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown J.A. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U. S. A. 2016;113(49):14013–14018. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernard D. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyagawa R. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18(4):738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spector D.L., Lamond A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011;3(2) doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamond A.I., Spector D.L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4(8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 30.Galganski L., Urbanek M.O., Krzyzosiak W.J. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017;45(18):10350–10368. doi: 10.1093/nar/gkx759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemson C.M. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathi V. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa S. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eissmann M. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West J.A. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55(5):791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J. Biol. Chem. 2014;289(42):29365–29375. doi: 10.1074/jbc.M114.572693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leucci E. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhuang M. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi: 10.1016/j.ebiom.2018.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F. MALAT1 regulates miR-34a expression in melanoma cells. Cell Death Dis. 2019;10(6):389. doi: 10.1038/s41419-019-1620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao F. miR-211 sponges lncRNA MALAT1 to suppress tumor growth and progression through inhibiting PHF19 in ovarian carcinoma. Faseb. J. 2018 doi: 10.1096/fj.201800495RR. [DOI] [PubMed] [Google Scholar]
- 42.Zhao M. MALAT1: a long non-coding RNA highly associated with human cancers. Oncol. Lett. 2018;16(1):19–26. doi: 10.3892/ol.2018.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z.X. MALAT1: a potential biomarker in cancer. Canc. Manag. Res. 2018;10:6757–6768. doi: 10.2147/CMAR.S169406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Canc. Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez Calle A. Emerging roles of long non-coding RNA in cancer. Canc. Sci. 2018;109(7):2093–2100. doi: 10.1111/cas.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gutschner T. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Canc. Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Canc. 2014;111(4):736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCown P.J. Secondary structural model of human MALAT1 reveals multiple structure-function relationships. Int. J. Mol. Sci. 2019;20(22) doi: 10.3390/ijms20225610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang M.C. Secondary structural model of MALAT1 becomes unstructured in chronic myeloid leukemia and undergoes structural rearrangement in cervical cancer. Noncoding RNA. 2021;7(1) doi: 10.3390/ncrna7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipovich L. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192(3):1133–1148. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michalik K.M. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 52.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115(7):668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Z.H. Long non-coding RNA MALAT1 functions as a mediator in cardioprotective effects of fentanyl in myocardial ischemia-reperfusion injury. Cell Biol. Int. 2017;41(1):62–70. doi: 10.1002/cbin.10701. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J. Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp. Neurol. 2016;277:162–170. doi: 10.1016/j.expneurol.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao J. Long non-coding RNA MALAT1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol. Med. 2016;8(4):346–362. doi: 10.15252/emmm.201505725. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Zhang Q.S. Beta-asarone protects against MPTP-induced Parkinson's disease via regulating long non-coding RNA MALAT1 and inhibiting alpha-synuclein protein expression. Biomed. Pharmacother. 2016;83:153–159. doi: 10.1016/j.biopha.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 57.Ren H. Association of circulating long non-coding RNA MALAT1 in diagnosis, disease surveillance, and prognosis of acute ischemic stroke. Braz. J. Med. Biol. Res. 2020;53(12) doi: 10.1590/1414-431X20209174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prinz F. The implications of the long non-coding RNA NEAT1 in non-cancerous diseases. Int. J. Mol. Sci. 2019;20(3) doi: 10.3390/ijms20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z., Li K., Huang W. Long non-coding RNA NEAT1-centric gene regulation. Cell. Mol. Life Sci. 2020;77(19):3769–3779. doi: 10.1007/s00018-020-03503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki Y.T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. U. S. A. 2009;106(8):2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakagawa S., Yamazaki T., Hirose T. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol. 2018;8(10) doi: 10.1098/rsob.180150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fox A.H., Lamond A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2(7):a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pisani G., Baron B. Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Noncoding RNA Res. 2019;4(4):128–134. doi: 10.1016/j.ncrna.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fox A.H. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 2018;43(2):124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Sunwoo H. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prasanth K.V. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018;20(10):1145–1158. doi: 10.1038/s41556-018-0204-2. [DOI] [PubMed] [Google Scholar]
- 69.Huang Z. Depletion of LncRNA NEAT1 rescues mitochondrial dysfunction through NEDD4L-dependent PINK1 degradation in animal models of alzheimer's disease. Front. Cell. Neurosci. 2020;14:28. doi: 10.3389/fncel.2020.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang L. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017;24(10):816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cooper D.R. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARgamma2 splicing during adipogenesis in 3T3-L1 cells. Genes. 2014;5(4):1050–1063. doi: 10.3390/genes5041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Z. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell. Mol. Life Sci. 2017;74(6):1117–1131. doi: 10.1007/s00018-016-2398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z. NEAT1 regulates neuroglial cell mediating Abeta clearance via the epigenetic regulation of endocytosis-related genes expression. Cell. Mol. Life Sci. 2019;76(15):3005–3018. doi: 10.1007/s00018-019-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen W. Depletion of NEAT1 lncRNA attenuates nucleolar stress by releasing sequestered P54nrb and PSF to facilitate c-Myc translation. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shui X. Knockdown of lncRNA NEAT1 inhibits Th17/CD4(+) T cell differentiation through reducing the STAT3 protein level. J. Cell. Physiol. 2019;234(12):22477–22484. doi: 10.1002/jcp.28811. [DOI] [PubMed] [Google Scholar]
- 76.Zhang M. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 2018;11(1):113. doi: 10.1186/s13045-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan X. Knockdown of lncRNA NEAT1 suppresses hypoxia-induced migration, invasion and glycolysis in anaplastic thyroid carcinoma cells through regulation of miR-206 and miR-599. Canc. Cell Int. 2020;20:132. doi: 10.1186/s12935-020-01222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li S. Long non-coding RNA NEAT1 promotes the proliferation, migration, and metastasis of human breast-cancer cells by inhibiting miR-146b-5p expression. Canc. Manag. Res. 2020;12:6091–6101. doi: 10.2147/CMAR.S252295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luo M. Long noncoding RNA NEAT1 promotes ovarian cancer cell invasion and migration by interacting with miR1321 and regulating tight junction protein 3 expression. Mol. Med. Rep. 2020;22(4):3429–3439. doi: 10.3892/mmr.2020.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao M. Long non-coding RNA NEAT1 serves as sponge for miR-365a-3p to promote gastric cancer progression via regulating ABCC4. OncoTargets Ther. 2020;13:3977–3985. doi: 10.2147/OTT.S245557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo Z. Long non-coding RNA-NEAT1, a sponge for miR-98-5p, promotes expression of oncogene HMGA2 in prostate cancer. Biosci. Rep. 2019;39(9) doi: 10.1042/BSR20190635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y. LncRNA NEAT1 regulates cell viability and invasion in esophageal squamous cell carcinoma through the miR-129/CTBP2 Axis. Dis. Markers. 2017;2017:5314649. doi: 10.1155/2017/5314649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan W. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson's disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 2018;496(4):1019–1024. doi: 10.1016/j.bbrc.2017.12.149. [DOI] [PubMed] [Google Scholar]
- 84.Liu Y., Lu Z. Long non-coding RNA NEAT1 mediates the toxic of Parkinson's disease induced by MPTP/MPP+ via regulation of gene expression. Clin. Exp. Pharmacol. Physiol. 2018;45(8):841–848. doi: 10.1111/1440-1681.12932. [DOI] [PubMed] [Google Scholar]
- 85.Nishimoto Y. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain. 2013;6:31. doi: 10.1186/1756-6606-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sunwoo J.S. Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Mol. Neurobiol. 2017;54(2):1577–1586. doi: 10.1007/s12035-016-9928-9. [DOI] [PubMed] [Google Scholar]
- 87.Bu F.T. LncRNA NEAT1: shedding light on mechanisms and opportunities in liver diseases. Liver Int. 2020;40(11):2612–2626. doi: 10.1111/liv.14629. [DOI] [PubMed] [Google Scholar]
- 88.Wang X. Down-regulation of lncRNA-NEAT1 alleviated the non-alcoholic fatty liver disease via mTOR/S6K1 signaling pathway. J. Cell. Biochem. 2018;119(2):1567–1574. doi: 10.1002/jcb.26317. [DOI] [PubMed] [Google Scholar]
- 89.Soh Y.Q. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159(4):800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mueller J.L. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet. 2013;45(9):1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinheiro I., Heard E. X chromosome inactivation: new players in the initiation of gene silencing. F1000Research. 2017;6 doi: 10.12688/f1000research.10707.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loda A., Heard E. Xist RNA in action: past, present, and future. PLoS Genet. 2019;15(9) doi: 10.1371/journal.pgen.1008333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Galupa R., Heard E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 2018;52:535–566. doi: 10.1146/annurev-genet-120116-024611. [DOI] [PubMed] [Google Scholar]
- 94.Brockdorff N., Bowness J.S., Wei G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020;34(11–12):733–744. doi: 10.1101/gad.337196.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fang H., Disteche C.M., Berletch J.B. X inactivation and escape: epigenetic and structural features. Front. Cell. Dev. Biol. 2019;7:219. doi: 10.3389/fcell.2019.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brockdorff N. Localized accumulation of Xist RNA in X chromosome inactivation. Open Biol. 2019;9(12):190213. doi: 10.1098/rsob.190213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hook E.B., Warburton D. The distribution of chromosomal genotypes associated with Turner's syndrome: livebirth prevalence rates and evidence for diminished fetal mortality and severity in genotypes associated with structural X abnormalities or mosaicism. Hum. Genet. 1983;64(1):24–27. doi: 10.1007/BF00289473. [DOI] [PubMed] [Google Scholar]
- 98.Cockwell A. A cytogenetic and molecular study of a series of 45,X fetuses and their parents. J. Med. Genet. 1991;28(3):151–155. doi: 10.1136/jmg.28.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brockdorff N. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351(6324):329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 100.Borsani G. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351(6324):325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 101.Brown S.D. XIST and the mapping of the X chromosome inactivation centre. Bioessays. 1991;13(11):607–612. doi: 10.1002/bies.950131112. [DOI] [PubMed] [Google Scholar]
- 102.Brockdorff N. The product of the mouse xist gene is a 15 Kb inactive X-specific transcript containing No conserved Orf and located in the nucleus. Cell. 1992;71(3):515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 103.Brown C.J. The human xist gene - analysis of a 17 Kb inactive X-specific rna that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 104.Wutz A., Rasmussen T.P., Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30(2):167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 105.Hoki Y. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development. 2009;136(1):139–146. doi: 10.1242/dev.026427. [DOI] [PubMed] [Google Scholar]
- 106.Patil D.P. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moindrot B. A pooled shRNA screen identifies Rbm15, spen, and Wtap as factors required for xist RNA-mediated silencing. Cell Rep. 2015;12(4):562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McHugh C.A. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Minajigi A. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245) doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chu C. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Monfort A. Identification of spen as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015;12(4):554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guenther M.G., Barak O., Lazar M.A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell Biol. 2001;21(18):6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.You S.H. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 2013;20(2):182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi Y. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15(9):1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cirillo D. Quantitative predictions of protein interactions with long noncoding RNAs. Nat. Methods. 2016;14(1):5–6. doi: 10.1038/nmeth.4100. [DOI] [PubMed] [Google Scholar]
- 116.Chen C.K. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354(6311):468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 117.Pintacuda G. hnRNPK recruits PCGF3/5-PRC1 to the xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell. 2017;68(5):955–969 e10. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Colognori D. Xist deletional analysis reveals an interdependency between xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell. 2019;74(1):101–117 e10. doi: 10.1016/j.molcel.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yamada N. Xist exon 7 contributes to the stable localization of xist RNA on the inactive X-chromosome. PLoS Genet. 2015;11(8) doi: 10.1371/journal.pgen.1005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smeets D. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenet. Chromatin. 2014;7:8. doi: 10.1186/1756-8935-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hasegawa Y. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell. 2010;19(3):469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 122.Kolpa H.J., Fackelmayer F.O., Lawrence J.B. SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell. 2016;39(1):9–10. doi: 10.1016/j.devcel.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sakaguchi T. Control of chromosomal localization of xist by hnRNP U family molecules. Dev. Cell. 2016;39(1):11–12. doi: 10.1016/j.devcel.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 124.Jeon Y., Lee J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146(1):119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lv Q. D-repeat in the XIST gene is required for X chromosome inactivation. RNA Biol. 2016;13(2):172–176. doi: 10.1080/15476286.2015.1137420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chaumeil J. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jansz N. Smchd1 regulates long-range chromatin interactions on the inactive X chromosome and at Hox clusters. Nat. Struct. Mol. Biol. 2018;25(9):766–777. doi: 10.1038/s41594-018-0111-z. [DOI] [PubMed] [Google Scholar]
- 128.Wang C.Y. SMCHD1 Merges chromosome compartments and assists formation of super-structures on the inactive X. Cell. 2018;174(2):406–421 e25. doi: 10.1016/j.cell.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gdula M.R. The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 2019;10(1):30. doi: 10.1038/s41467-018-07907-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Blewitt M.E. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008;40(5):663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 131.Gendrel A.V. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev. Cell. 2012;23(2):265–279. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Denholtz M. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13(5):602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kundu S. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell. 2018;71(1):191. doi: 10.1016/j.molcel.2018.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schoenfelder S. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat. Genet. 2015;47(10):1179–1186. doi: 10.1038/ng.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Du Y. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget. 2017;8(55):94358–94370. doi: 10.18632/oncotarget.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wei W. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J. Cell. Biochem. 2017;118(10):3349–3358. doi: 10.1002/jcb.25988. [DOI] [PubMed] [Google Scholar]
- 137.Zhang Y.L. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol. Sin. 2017;38(3):371–381. doi: 10.1038/aps.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yue D. Silencing of long noncoding RNA XIST attenuated Alzheimer's disease-related BACE1 alteration through miR-124. Cell Biol. Int. 2020;44(2):630–636. doi: 10.1002/cbin.11263. [DOI] [PubMed] [Google Scholar]
- 139.Braunschweig U. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152(6):1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 142.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang C., Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Song Z. Antisense oligonucleotide technology can be used to investigate a circular but not linear RNA-mediated function for its encoded gene locus. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1743-8. [DOI] [PubMed] [Google Scholar]
- 145.Gao L. Circular RNAs from BOULE play conserved roles in protection against stress-induced fertility decline. Sci. Adv. 2020;6(46) doi: 10.1126/sciadv.abb7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sanger H.L. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hansen T.B. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 148.Memczak S. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 149.Kristensen L.S. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 150.Zhou M. New progresses of circular RNA biology: from nuclear export to degradation. RNA Biol. 2020:1–9. doi: 10.1080/15476286.2020.1853977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Patop I.L., Wust S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16) doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen L., Shan G. CircRNA in cancer: Fundamental mechanism and clinical potential. Canc. Lett. 2021;505:49–57. doi: 10.1016/j.canlet.2021.02.004. [DOI] [PubMed] [Google Scholar]
- 153.Li J. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Canc. 2020;6(4):319–336. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 154.Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 155.Conn S.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 156.Ashwal-Fluss R. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 157.Ivanov A. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 158.Khan M.A. RBM20 regulates circular RNA production from the titin gene. Circ. Res. 2016;119(9):996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 159.Errichelli L. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu C.X. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–880 e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 161.Fischer J.W. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell. 2020;78(1):70–84 e6. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jia R. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov. 2019;5:45. doi: 10.1038/s41421-019-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Park O.H. Endoribonucleolytic cleavage of m(6)a-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74(3):494–507 e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 164.Hansen T.B. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang C. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Z., Kearse M.G., Huang C. The nuclear export of circular RNAs is primarily defined by their length. RNA Biol. 2019;16(1):1–4. doi: 10.1080/15476286.2018.1557498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li Z. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 168.Zhang Y. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 169.Stoll L. A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat. Commun. 2020;11(1):5611. doi: 10.1038/s41467-020-19381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen N. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bronisz A. The nuclear DICER-circular RNA complex drives the deregulation of the glioblastoma cell microRNAome. Sci. Adv. 2020;6(51) doi: 10.1126/sciadv.abc0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xia P. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48(4):688–701 e7. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 173.Conn V.M. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 174.Liu Y. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol. 2020;18(1) doi: 10.1371/journal.pbio.3000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Aydin O.Z., Vermeulen W., Lans H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle. 2014;13(19):3016–3025. doi: 10.4161/15384101.2014.956551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Erdel F., Rippe K. Binding kinetics of human ISWI chromatin-remodelers to DNA repair sites elucidate their target location mechanism. Nucleus. 2011;2(2):105–112. doi: 10.4161/nucl.2.2.15209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu X. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Canc. 2020;19(1):128. doi: 10.1186/s12943-020-01246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Long Y. How do lncRNAs regulate transcription? Sci. Adv. 2017;3(9) doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sun Q., Hao Q., Prasanth K.V. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guh C.Y., Hsieh Y.H., Chu H.P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020;27(1):44. doi: 10.1186/s12929-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chu C. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chu H.P. TERRA RNA antagonizes ATRX and protects telomeres. Cell. 2017;170(1):86–101. doi: 10.1016/j.cell.2017.06.017. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mumbach M.R. HiChIRP reveals RNA-associated chromosome conformation. Nat. Methods. 2019;16(6):489–492. doi: 10.1038/s41592-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Simon M.D. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504(7480):465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Simon M.D. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U. S. A. 2011;108(51):20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen L. R-ChIP using inactive RNase H reveals dynamic coupling of R-loops with transcriptional pausing at gene promoters. Mol. Cell. 2017;68(4):745–757 e5. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dumelie J.G., Jaffrey S.R. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. eLife. 2017;6 doi: 10.7554/eLife.28306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ginno P.A. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012;45(6):814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sanz L.A. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell. 2016;63(1):167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hafner M. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Konig J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17(7):909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ule J. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 193.Sugimoto Y. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519(7544):491–494. doi: 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ding Y. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 195.Loughrey D. SHAPE-Seq 2.0: systematic optimization and extension of high-throughput chemical probing of RNA secondary structure with next generation sequencing. Nucleic Acids Res. 2014;42(21) doi: 10.1093/nar/gku909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lucks J.B. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. U. S. A. 2011;108(27):11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Spitale R.C. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519(7544):486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Underwood J.G. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7(12):995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bassett A.R. Considerations when investigating lncRNA function in vivo. eLife. 2014;3 doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]