Abstract
An important bottleneck for non-viral gene transfer commonly relates to translocation of nucleic acids into the nuclear compartment of target cells. So-called 3NFs are optimized short nucleotide sequences able to interact with the transcription factor nuclear factor κB (NF-κB), which can enhance the nuclear import of plasmid DNA (pDNA) carrying such motifs. In this work, we first designed a consistent set of six pDNAs featuring a common backbone and only varying in their 3NF sequences. These constructions were then transfected under various experimental settings. In vitro, cationic polymer-assisted pDNA delivery in five human-derived cell lines showed the potential advantage of 3NF carrying pDNA in diverse cellular contexts. In vivo, naked pDNAs were hydrodynamically delivered to muscle hindlimbs in healthy mice; this direct accurate comparative (in the absence of any gene carrier) revealed modest but consistent trends in favor of the pDNAs equipped with 3NF. In summary, the results reported emphasize the implications of various parameters on NF-κB-mediated pDNA nuclear import; under specific conditions, 3NF can provide modest to substantial advantages for pDNA gene transfer, in vitro as well as in vivo. This study thus further underscores the potential of optimized nuclear import for more efficient non-viral gene transfer applications.
Keywords: hydrodynamic limb vein injection, NF-κB, nuclear import, plasmid DNA, skeletal muscle, transfection
Graphical Abstract
Nucleic acid translocation into the nucleus is a well-recognized obstacle for non-viral gene transfer. In this study, the authors worked out a consistent set of plasmid DNAs that allowed underscoring the potential of short cis-DNA sequences (3NFs) interacting with NF-κB to enhance nuclear import under various in vitro and in vivo experimental conditions.
Introduction
Gene therapy consists in bringing therapeutic nucleic acids (NAs) to target cells in order to supply and/or to correct a genetic defect responsible for an inherited or acquired disease.1 For such delivery, several methods, usually classified as viral and non-viral, can be used.2,3 Viral gene delivery strategies can be particularly efficient to transduce quiescent or non-quiescent cells and were used in most of the gene therapy clinical trials conducted so far.4 However, viral-based carriers are also usually characterized with some drawbacks, notably (1) limited NA encapsidation capacity, (2) costly production process with extensive quality assessments, and (3) induction of a host immune response against the vector.5 Much less concerned by such limitations, non-viral gene transfer strategies, making use of cationic lipids/polymers or amphiphilic triblock copolymers,6, 7, 8, 9, 10 have, however, a much lower transfection yield. This is partly due to a non-optimal entry of the NA into the nuclear compartment and a limited access to the transcriptional machinery of target cells. In vitro experiments indeed showed that 24–36 h post-transfection, only 1%–10% of the intracytoplasmic copies of a given plasmid DNA (pDNA) effectively entered into the nucleus.11
DNA nuclear targeting sequences (DTSs) are short consensus motifs recognized by specific transcription factors (TFs). They can be used to actively shuttle, upon activation and following an importin-mediated process, a pDNA from the cytosol to the nucleus through nuclear pores. For instance, the SV40 enhancer can be used as a DTS since it is recognized by various TFs such as Oct1.11 Other DTSs bound by the nuclear factor κB (NF-κB) TF have also been described, e.g., a NF-κB-optimized DTS motif named 3NF consisting of three 10-bp κB sites interspaced with optimized spacers.12,13 The insertion of two 3NFs upstream of the promoter and downstream of the cDNA sequence triggered a NF-κB-mediated nuclear import of pDNA, as demonstrated through qPCR and confocal microscopy.12,14 It was shown that, compared with control pDNA, 3NF-pDNA mediated a longer transgene expression in mouse liver upon hydrodynamic tail vein (HTV) injection.14 Such DTSs also proved to enhance pDNA transfection in mouse skeletal muscle after intramuscular injection followed or not by electroporation.13 Beyond these findings, 3NF motifs could be beneficial in inflammatory diseases such as myopathies in which the NF-κB pathway is constitutively activated.15 Alternatively, the latter could be triggered by the transfection procedure itself, in a way to enhance the nuclear import of 3NF-pDNA.
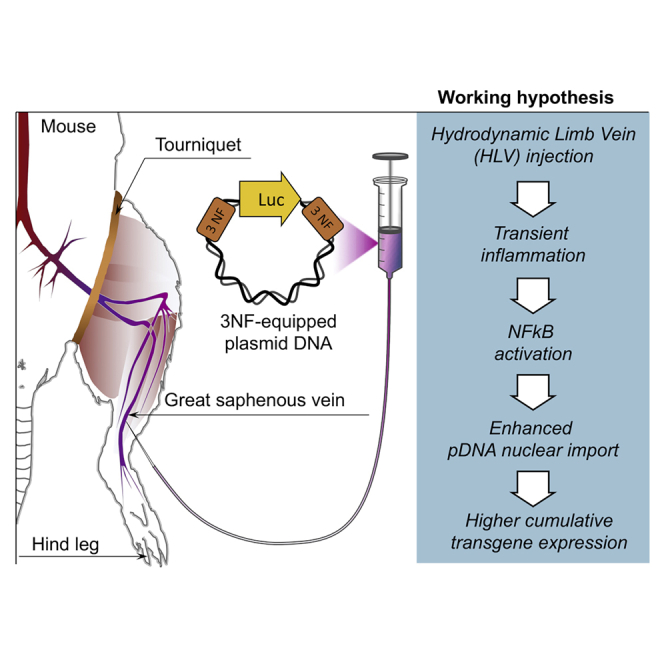
Hydrodynamic limb vein (HLV) injection is a loco-regional gene delivery method originally developed by Wolff and colleagues.16 This safe and clinically relevant procedure allows transfection of the whole musculature of a given limb isolated by a tourniquet, thanks to the rapid injection of a great volume of solution.17,18 Thus, naked pDNA can be delivered with some efficacy.19 This makes HLV injection particularly suitable for accurately studying the effect of DTSs under in vivo conditions, in the absence of any adjunct (carrier) that would interfere with the transfection process and complicate interpretation of results. In addition, the target tissue is ideal for such study since skeletal myofibers (which have peripheral nuclei) are non-dividing cells. Thus, access to the nuclear compartment cannot rely on nuclear envelope breakdown and can only occur by translocation through nuclear pores. Furthermore, skeletal muscle represents both a challenging study model and a relevant tissue for non-viral gene therapy since it can be affected by monogenic disorders such as dystrophies.
This study was first devoted to design fully comparable pDNAs varying only according to their equipment in 3NF motif(s). These pDNAs were tested in a series of experimental conditions, both in vitro (using cell lines originating from diverse organs and tissues) and in vivo (using HLV injection as a clinically relevant delivery method to target skeletal muscle). All combined, the data obtained unveiled the potential specific contribution of 3NF to non-viral gene transfer under various conditions, especially in skeletal myofibers.
Results
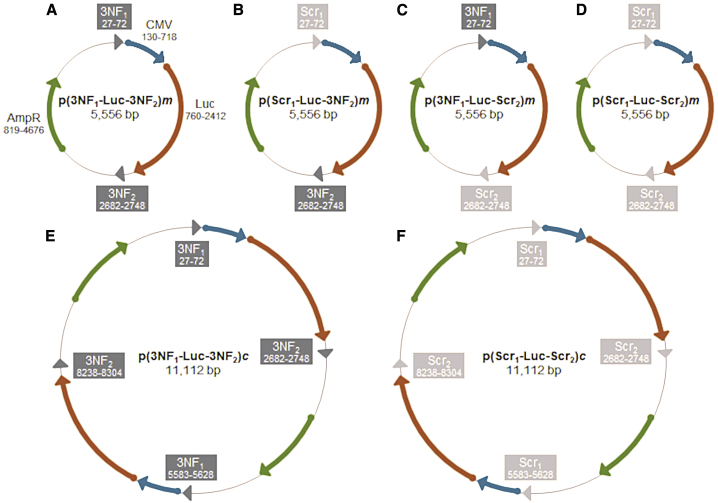
The parental monomer pDNA p(3NF1-Luc-3NF2)m (5,556 bp) carries (1) a luciferase cassette under control of the strong immediate early (IE) cytomegalovirus (CMV) promoter (allowing transgene expression in a wide range of cell types) and (2) 3NF1 and 3NF2 respectively upstream and downstream of the luciferase gene (Figures 1A and S1). To accurately evaluate the contribution of 3NF to transfection efficacy, three other pDNAs were derived from p(3NF1-Luc-3NF2)m by replacing 3NF1 and/or 3NF2 by scramble sequences (named Scr1 and Scr2) while keeping unchanged plasmid size and backbone. Scr1 and Scr2 feature exactly the same length as 3NF1 and 3NF2, respectively, and they show no similarity with any known consensus sequence, according to NCBI BLAST (Table S1). In practice, p(Scr1-Luc-3NF2)m and p(3NF1-Luc-Scr2)m (Figures 1B, 1C, and S2) were first obtained, and p(Scr1-Luc-Scr2)m was then derived from p(3NF1-Luc-Scr2)m (Figures 1D and S2). In addition, concatemers (i.e., forward fusions of two identical pDNAs) were constructed following a straightforward (“enzymatic cut then ligation”) protocol. Thus, p(3NF1-Luc-3NF2)c and p(Scr1-Luc-Scr2)c exactly twice the size of monomers (i.e., 11,112 bp) were obtained (Figures 1E and 1F). Overall, sequence homology of pDNAs (either monomers or concatemers) was ≥97% (see Supplemental information for details).
Figure 1.
Simplified maps of the parental monomer pDNA p(3NF1-Luc-3NF2)m and its monomer or concatemer derivatives
(A–F) Optimized NF-κB DNA nuclear targeting sequences (DTSs) named 3NF1 and 3NF2 sequences, respectively, upstream and downstream of the luciferase gene are shown as dark arrowheads. Scramble sequences named Scr1 and Scr2 are shown as light gray arrowheads. The ampicillin resistance gene AmpR, the luciferase cassette, and the strong ubiquitous CMV promoter are shown as green, orange, and blue arrows.
In vitro investigations
The in vitro transfection of the pDNAs designed (Figure 1) was then evaluated under various experimental conditions, using as gene carrier either a commercial branched polyethylenimine (bPEI; 25 kDa) or a histidinylated linear PEI (His-lPEI; 22 kDa).10 Gel retardation assays were first performed to determine the cationic polymer-to-pDNA mass ratio (MR) from which full DNA retardation was obtained. For bPEI, this was 0.5, whereas it was 5.0 for His-lPEI (Figure S3). Accordingly, for comparative purposes, bPEI and His-lPEI polyplexes formed at a same MR (i.e., MR = 5; see Table S2 for size and zeta potential measurements) were used in all subsequent in vitro experiments that aimed to address a first series of questions.
What benefit can 3NF motifs provide for in vitro pDNA transfection?
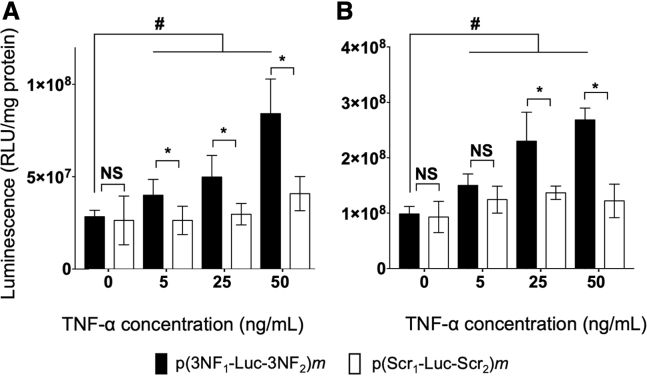
As a first proof of principle of 3NF-assisted transfection, p(3NF1-Luc-3NF2)m and p(Scr1-Luc-Scr2)m were transfected in HeLa, SK-Mel-28, and 16HBE cells, using His-lPEI as a DNA carrier. Transfections were done in the presence of the pro-inflammatory cytokine tumor necrosis factor (TNF)-α (which triggers the NF-κB pathway) for concentrations in the same range as in other related studies.12,20 Reporter gene expression (i.e., luminescence) was evaluated shortly (i.e., 24 h) after transfection to limit the possibility for pDNA to be internalized in nuclei thanks to cell mitosis. In the absence of TNF-α, the luciferase activities obtained with both pDNAs were almost the same (Figures 2A and 2B). In contrast, following addition of TNF-α, higher luminescence signals were obtained with p(3NF1-Luc-3NF2)m than with p(Scr1-Luc-Scr2)m. Differences were greater for higher TNF-α concentrations, being statistically significant in most instances in every cell line considered (Figures 2A, 2B, and S4). For the highest cytokine concentration tested, a nearly 3-fold statistically significant enhancement of luminescence was obtained with p(3NF1-Luc-3NF2)m compared to basal condition in HeLa and SK-Mel-28 cells. Of note, a slight luminescence increase could also be measured with p(Scr1-Luc-Scr2)m when increasing the TNF-α concentration, arguing also for some transfection benefit of NF-κB motifs (see Discussion).
Figure 2.
In vitro transfection using His-lPEI polyplexes incorporating either p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m in the presence of increasing concentrations of TNF-α
(A and B) Transfections were carried out in (A) HeLa and (B) SK-Mel-28 cells using His-lPEI polyplexes. Results are expressed as relative light units per milligram of protein (RLU/mg protein) as mean values of six wells ± SD. ∗p ≤ 0.05, with a two-tailed unpaired t test used to compare p(3NF1-Luc-3NF2)m with p(Scr1-Luc-Scr2)m at a given TNF-α dose; #p ≤ 0.05, with a two-tailed unpaired t test used to compare p(3NF1-Luc-3NF2)m without TNF-α versus with TNF-α at the indicated doses. NS, not significant.
Can a gene carrier modulate the in vitro gene transfection of 3NF-pDNA?
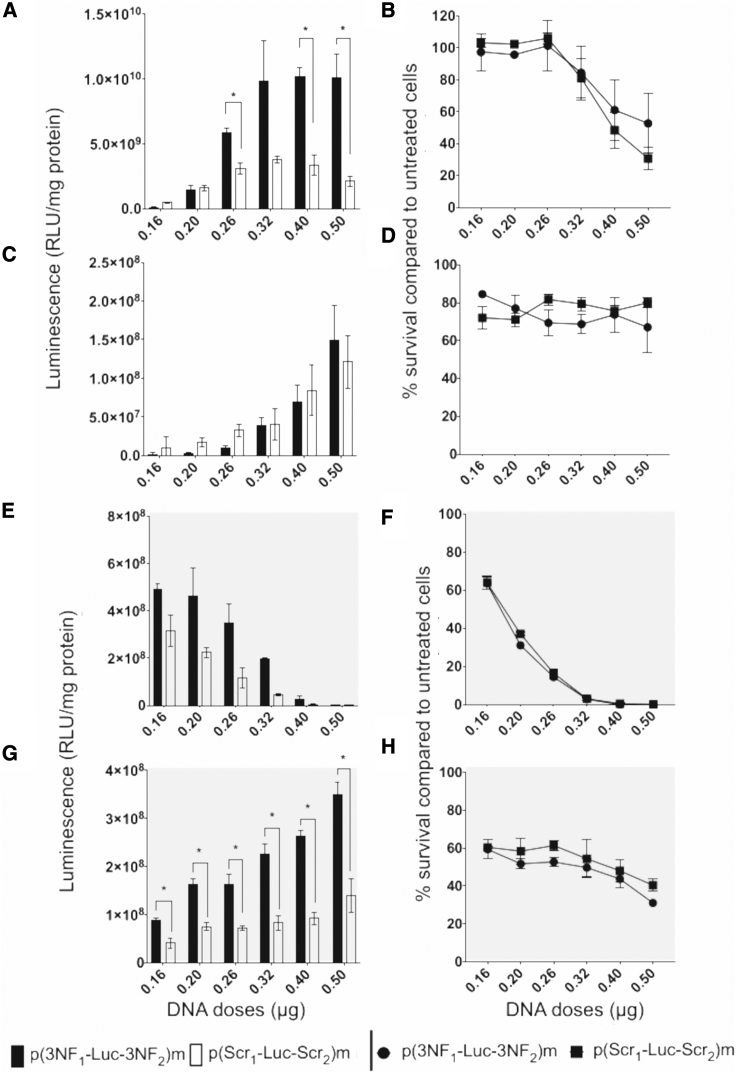
A dose escalation study was performed in SK-Mel-28, C2C12 (at myoblast stage), and A549 cells for comparing His-lPEI- and bPEI-based polyplexes. The latter were deposited for quantities corresponding to 0.16, 0.20, 0.26, 0.32, 0.40, and 0.50 μg of pDNA per well. Results with p(3NF1-Luc-3NF2)m and p(Scr1-Luc-Scr2)m are given in Figure 3 (see also Figures S5 and S6 for the results obtained with p(Scr1-Luc-3NF2)m and p(3NF1-Luc-Scr2)m). (1) When using His-lPEI to transfect pDNA in SK-Mel-28 cells, for a dose of 0.16 μg of pDNA/well, p(Scr1-Luc-Scr2)m provided slightly higher luminescence than did p(3NF1-Luc-3NF2)m. For a dose of 0.20 μg of pDNA/well, luminescence was almost the same with all pDNAs (Figures 3A and S5A). From 0.26 μg of pDNA/well, higher luminescence was obtained with p(3NF1-Luc-3NF2)m than with p(Scr1-Luc-Scr2)m. Differences increased for higher pDNA dose, being statistically significant at 0.26, 0.40, and 0.50 μg of pDNA/well (Figure 3A). Cell viabilities were similar for all pDNAs, being 100% for the smallest pDNA dose assayed but dropping down to 40% at 0.50 μg of pDNA/well (Figures 3B and S5B). (2) When using His-lPEI to transfect pDNA in C2C12, p(Scr1-Luc-Scr2)m yielded slightly higher luminescence than did p(3NF1-Luc-3NF2)m for 0.16–0.26 μg of pDNA/well. When increasing the pDNA dose, transfection efficiencies were roughly similar (Figure 3C). Cell viability remained above 80% in every experimental condition (Figure 3D). The same experiment with A549 provided similar findings (Figures S6A and S6B). (3) When using bPEI to transfect pDNA in SK-Mel-28 cells, the highest luminescence signals were noticeably obtained with p(3NF1-Luc-3NF2)m in every case (Figures 3E and S5E). Cell viability was 60% for the lowest pDNA dose and decreased quickly for higher doses (Figure 3F). (4) When using bPEI to transfect pDNA in C2C12 cells, p(3NF1-Luc-3NF2)m provided significantly higher luminescence signals in every case, which progressively increased when increasing the pDNA dose delivered (Figures 3G and S5G). From 60% for the lowest pDNA dose, cell viability progressively decreased to 40% at the highest pDNA dose assayed (Figure 3H). Similar transfection profiles were observed in A549 cells but with higher cell viabilities (Figures S6C and S6D). Additionally, consistent results were obtained in a time course study using bPEI to deliver p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m in C2C12 and A549 cells (Figures S7A and S7B). In almost every condition considered, p(Scr1-Luc-3NF2)m and p(3NF1-Luc-Scr2)m yielded reporter gene activity at intermediate levels between those of p(3NF1-Luc-3NF2)m and p(Scr1-Luc-Scr2)m (Figures S5 and S6). Altogether, these findings highlight that, depending on the DNA carrier used and the induced cytotoxicity, the in vitro transfection of 3NF-pDNA can actually vary and be more efficient compared with the same pDNA devoid of such motifs.
Figure 3.
In vitro transfection using ascending doses of His-lPEI or bPEI polyplexes incorporating either p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m
(A–H) Experiments were carried out in SK-Mel-28 (A, B, E, and F) and C2C12 (C, D, G, and H) cells using His-lPEI (A–D) or bPEI (E–H) polyplexes. Results are mean values of three wells ± SD. ∗p ≤ 0.05 (multiple t test Holm-Sidak method) between p(3NF1-Luc-3NF2)m and p(Scr1-Luc-Scr2)m. Cell survival is given as a percentage compared to untreated cells. See Figure S5 for complementary results obtained with p(3NF1-Luc-Scr2)m and p(Scr1-Luc-3NF2)m.
In vivo investigations
In vivo transfection experiments were then carried out to further investigate the potential of 3NF under experimental conditions relevant for some therapeutic applications. The clinically approved HLV injection procedure was used considering it allows to efficiently deliver naked pDNA in myofibers. Thus, p(3NF1-Luc-3NF2)m and its derivatives p(Scr1-Luc-Scr2)m, p(3NF1-Luc-3NF2)c, and p(Scr1-Luc-Scr2)c were assayed for addressing additional questions.
Can 3NF provide an advantage for pDNA transfection following HLV injection?
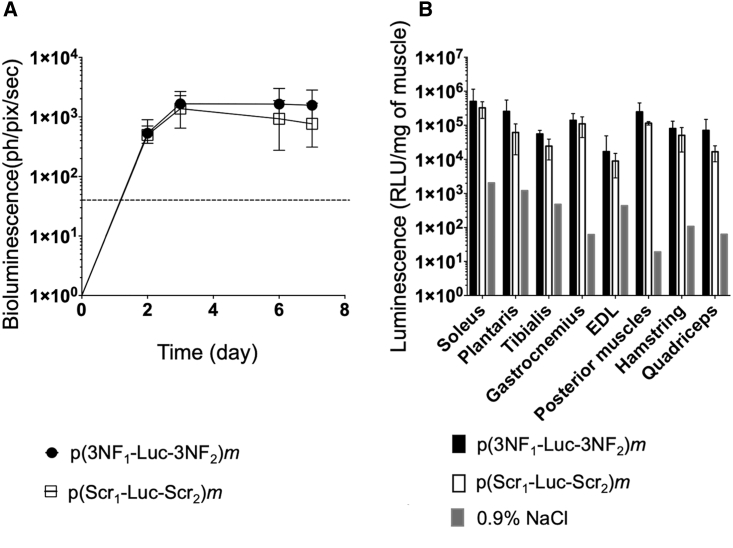
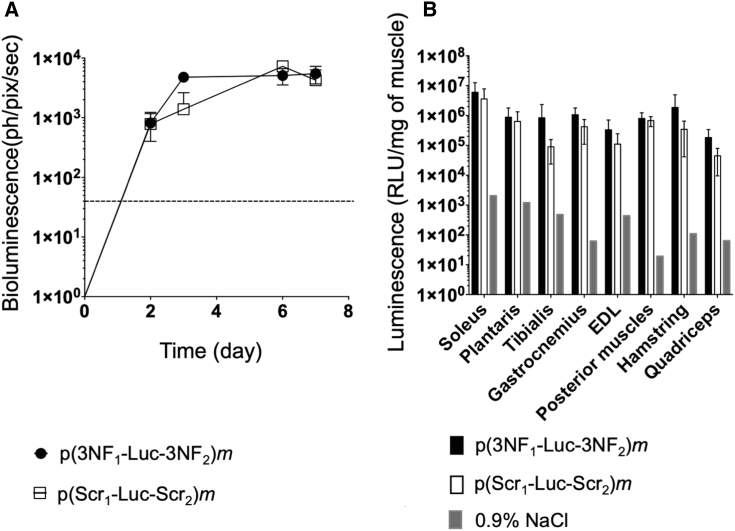
Swiss mice, exempt of any known inflammation, were administrated via HLV injection with either p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m. Each hindlimb of animals was injected with a suboptimal (according to our expertise) pDNA dose (i.e., 0.5 μg per leg). Luciferase expression was monitored in living animals via bioluminescence imaging (BLI) (Figure S8A) for 1 week. At the steady state (i.e., from day 3 after injection), p(3NF1-Luc-3NF2)m yielded higher bioluminescence compared with p(Scr1-Luc-Scr2)m (Figure 4A). Mice were then sacrificed and eight muscles per limb were harvested for further analyses (see Figure S8B for a simplified anatomy of the mouse leg showing the various muscles harvested). The higher bioluminescence obtained with the 3NF-pDNA was confirmed with luminescence measurements performed on muscle homogenates. Although no statistically significant difference was reached, the mean of the luminescence ratios (p(3NF1-Luc-3NF2)m/p(Scr1-Luc-Scr2)m) in every muscle homogenate was 2.39 (Figure 4B; see also Table S3 for detailed values and statistical analyses). In other experiments, a 10-fold higher dose was assayed, i.e., mice received 5 μg of either p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m per hindlimb. Compared to the dose previously injected, BLI signals and luminescence in muscle homogenates were about 1 log higher (Figures 4A, 4B, 5A, and 5B), indicating a linear range dose/response following HLV injection.21 Although not statistically significant, a trend in favor of the 3NF-pDNA was again observed in every muscle considered. The mean of the luminescence ratios (p(3NF1-Luc-3NF2)m/p(Scr1-Luc-Scr2)m) in every muscle homogenate was here on average 3.57 (Table S3).
Figure 4.
In vivo HLV injection in Swiss mice using 0.5 μg of either naked p(3NF1-Luc-3NF2)m or naked p(Scr1-Luc-Scr2)m
(A) In vivo bioluminescence measured in treated legs (mean values ± SD of n = 4 limbs per group) until 7 days after injection is expressed in photons per pixel per second (ph/pix/sec). The dashed line indicates the luminescence background. (B) Ex vivo luminescence measured in the homogenates of eight types of muscle (with extensor digitorum longus [EDL]) (Figure S8B). For each treatment, the value for each type of muscle is the average of n = 4 legs. Results are mean values ± SD given as RLU/mg of muscle. The background for each type of muscle was determined using a limb injected with 0.9% NaCl.
Figure 5.
In vivo HLV injection in Swiss mice using 5 μg of either naked p(3NF1-Luc-3NF2)m or naked p(Scr1-Luc-Scr2)m
(A) In vivo bioluminescence measured in treated legs (mean values ± SD of n = 5–6 limbs per group) until 7 days after injection is expressed in ph/pix/sec. The dashed line indicates the luminescence background. (B) Ex vivo luminescence measured in the homogenates of eight types of muscle. For each treatment, the value for each type of muscle is the average of n = 5–6 legs. Results are mean values ± SD given as RLU/mg of muscle. The background for each type of muscle was determined using a limb injected with 0.9% NaCl.
Can 3NF rescue the transfection of long-size pDNA following HLV injection?
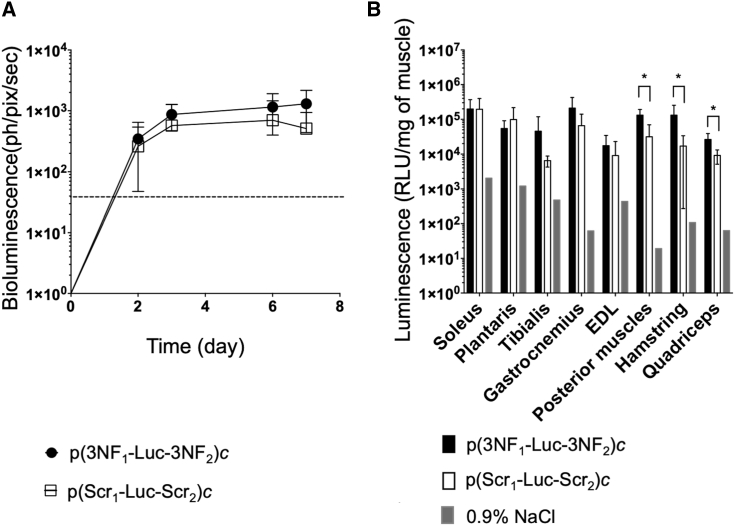
Generally speaking, transfection is sensitive to the size of the pDNA delivered, which has been demonstrated notably following HLV injection.21 Given the relatively small size of our so-called pDNA monomers (5,556 bp), they could easily move through the cytoplasm to reach the nucleus. Furthermore, the hydrodynamic effect of HLV injection may promote their nuclear entry independently of any DTS-mediated import. Thus, under these experimental conditions, 3NF may be useless (providing no substantial beneficial effect) for transfection of such pDNA, and the 3NF benefit would be better observed for transfection of pDNA with much greater size.22,23 To test this hypothesis, we compared p(3NF1-Luc-3NF2)c and p(Scr1-Luc-Scr2)c, both being 11,112 bp in length. Since no noticeable effect of the pDNA dose was observed in previous in vivo assays (Figures 4 and 5), pDNA concatemers were injected for a dose of 0.5 μg per limb. First, compared with respective monomers, concatemers consistently showed lower luminescence signals, as observed via BLI (on average 35%) and in muscle homogenates (40%). When comparing concatemers, BLI showed a trend in favor of p(3NF1-Luc-3NF2)c compared with p(Scr1-Luc-Scr2)c, without reaching any statistical significance (Figure 6A) but with greater differences than when comparing corresponding monomers (Figure 4A). This was further detailed with luminescence measurements performed in muscle homogenates (Figure 6B). The mean of luminescence ratios (p(3NF1-Luc-3NF2)c/p(Scr1-Luc-Scr2)c) in every muscle homogenate was on average 3.56. Importantly, compared with p(Scr1-Luc-Scr2)c, p(3NF1-Luc-3NF2)c allowed obtaining significantly (2.9- to 7.8-fold) higher luminescence signals in posterior muscles, hamstring, and quadriceps (Table S3).
Figure 6.
In vivo HLV injection in Swiss mice using 0.5 μg of either naked p(3NF1-Luc-3NF2)c or naked p(Scr1-Luc-Scr2)c
(A) In vivo bioluminescence measured in treated legs (mean values ± SD of n = 5 limbs per group) until 7 days after injection is expressed in ph/pix/sec. The dashed line indicates the luminescence background. (B) Ex vivo luminescence measured in the homogenates of eight types of muscle. For each treatment, the value for each type of muscle is the average of n = 5 legs. Results are mean values ± SD given as RLU/mg of muscle. The background for each type of muscle was determined using a limb injected with 0.9% NaCl. ∗p ≤ 0.05 (Mann-Whitney test) between p(3NF1-Luc-3NF2)c and p(Scr1-Luc-Scr2)c.
Discussion
DNA translocation from the cytoplasm to the nucleus is a well-known limiting step for non-viral gene transfer, especially in terminal non-dividing cells such as skeletal muscle fibers. Considering this bottleneck, we investigated the effect of NF-κB-optimized DTSs termed 3NF on pDNA transfection in various cellular contexts. For this purpose, fully comparable pDNAs (either monomers or concatemers) varying only as regards their content in 3NF were first designed and thoroughly characterized (Figures 1 and S2). It is noteworthy that the 3NF motifs used in this work were previously shown to enhance transfection by promoting pDNA nuclear translocation.12,14 Of note, the pDNAs assayed in those previous studies contained a luciferase cassette under control of the weak TAL promoter, whereas in the present work, pDNAs incorporated the IE-CMV promoter.
In vitro transfections were carried out using the set of pDNAs (Figure 1) in various cell lines. Considering that a gene carrier was required, two types of PEI were used, i.e., the widely used bPEI and the histidinylated lPEI derivative His-lPEI (which is known to be less cytotoxic).10 In a first series of in vitro experiments, His-lPEI was used to assay 3NF-assisted pDNA transfection under TNF-α-induced NF-κB activation (Figure 2). The increasing luciferase signals obtained with p(3NF1-Luc-3NF2)m for growing doses of that cytokine demonstrated the role of 3NF in active nuclear import of 3NF-pDNA in the various cell lines considered (Figures 2A and 2B). A slight enhancement of luminescence signals was also observed with p(Scr1-Luc-Scr2)m when increasing TNF-α concentration, which was likely due to non-optimized NF-κB sites present in the CMV enhancer/promoter (Figure S9).24 Next, a dose escalation of polyplexes was performed to test the hypothesis that the gene carrier could also enhance by itself, through some NF-κB activation, the transfection efficiency of 3NF-pDNA (Figures 3, S5, and S6). Using low doses of His-lPEI-based polyplexes, p(Scr1-Luc-Scr2)m yielded higher luciferase signals than did p(3NF1-Luc-3NF2)m. However, when increasing the dose of such polyplexes, p(3NF1-Luc-3NF2)m gave luciferase activities similar to, or better than, those of p(Scr1-Luc-Scr2)m, coincidentally to cell toxicity due to pDNA transfection (Figures 3, S5, and S6). Using bPEI-based polyplexes, a lower cell viability than with His-lPEI was expectedly observed. However, compared with p(Scr1-Luc-Scr2)m, p(3NF1-Luc-3NF2)m always yielded higher luciferase expressions, which were enhanced when increasing the dose of polyplexes (in correlation with the cytotoxicity induced). All combined, these results underscore that the DNA carrier used can indeed impact the transfection efficacy of 3NF-pDNA, notably likely through its ability to activate the NF-κB pathway. Indeed, it has already been reported that bPEI could activate the nuclear translocation of NF-κB. Additionally, compared with His-lPEI, bPEI should have a better ability to trigger the NF-κB pathway for several reasons. (1) Histidinylation has been shown to reduce PEI toxicity (which could trigger some inflammatory chemokine release) while providing amorphous complexes and enhancing the buffering capacity, thus facilitating endosomal escape of NA, resulting in a better pDNA release into the cytoplasm.10,25 (2) Otherwise, the NF-κB activation induced by the gene carrier may be inversely correlated to the endosomal buffering capacity of the latter.26,27 (3) Furthermore, polyamines have already been shown to be able to activate the NF-κB pathway.28 It should be stressed that compared with bPEI, His-lPEI has a lower secondary amine content, since 16% of the ethylenimine motifs in this polymer are substituted with an histidine moiety (which likely explains the lesser pDNA condensation ability of His-lPEI compared with bPEI; Figure S3). (4) PEI toxicity is known to trigger the AKT kinase, a protein involved in NF-κB activation, although this is still a matter of debate (it has also been described that bPEI could downregulate NF-κB gene expression).29,30 Considering altogether our in vitro results and the above-mentioned comments, a simple hypothetical mechanistic model can be proposed, as follows (Figure S10): In the absence of activation of NF-κB (e.g., when using an almost non-toxic dose of polyplexes), this TF is sequestrated in the cytoplasm with its inhibitor IκBα, with the latter hiding the NF-κB nuclear localization signal (NLS).31 In this situation, NF-κB DTSs may be bound to the inactive form of NF-κB, thus sequestrating some 3NF-pDNA copies in the cytoplasm. As a result, p(3NF1-Luc-3NF2)m luciferase activity is inferior to that of p(Scr1-Luc-Scr2)m (Figure S10; comparison of Figures S10A, S10C, and S10D). When a stimulus triggers the NF-κB pathway (e.g., a gene carrier or a chemokine such as TNF-α at a sufficient dose), IκBα is phosphorylated and no longer interacts with NF-κB, which can thus shuttle from the cytoplasm to the nucleus. If bound to a 3NF-pDNA, the latter can be actively imported into the nucleus. As a result, p(3NF1-Luc-3NF2)m luciferase activity is superior to that of p(Scr1-Luc-Scr2)m (Figure S10; comparison of Figures S10B–S10D). For intermediate situations (i.e., when the NF-κB pathway is subactivated), the pDNA entry into the nucleus can vary, due to various exogenous (e.g., experimental parameters, pDNA design [see below]) and/or endogenous, cell-dependent (confluency, sensitivity, NF-κB basal activity, and activation threshold) parameters or stimuli. This may explain the variability observed in some of our results, especially in the dose escalation study (Figures 3, S5, and S6). In addition to the DNA carrier, other parameters may participate in 3NF-pDNA transfection ability, notably corticotherapy treatments (which may inhibit NF-κB) given in some chronic human diseases.32 A dexamethasone dose escalation study showed a non-specific increase of the luciferase signal, irrespective of the pDNA used (Figures S11 and S12), in accordance with data reported in another study.33 This was more probably due to some transcriptional process rather than an active pDNA nuclear import (no glucocorticoid response element [GRE] DTS was present in the sequence of the pDNAs assayed in our study).34 This finding thus suggests that, depending on the targeted cell type, corticoids may not preclude the potential interest of 3NF-pDNA. However, further evaluations should be done considering other cell types and corticoid drugs (e.g., deflazacort that is given in Duchenne muscular dystrophy [DMD] patients).
In vivo transfection experiments in mice were then conducted using the HLV injection procedure. First, compared with p(Scr1-Luc-Scr2)m, p(3NF1-Luc-3NF2)m yielded consistent (although not statistically significant) higher luminescence signals, irrespective of the pDNA dose administered (Figures 4 and 5). Concatemers p(3NF1-Luc-3NF2)c and p(Scr1-Luc-Scr2)c were then assayed, which both featured a size (11,112 bp) closer to that of therapeutic pDNA that would be used for gene therapy of muscular diseases such as DMD (for which large transgenes can be required). Compared to respective monomers injected at a given dose (i.e., 0.5 μg of pDNA), clearly lower luminescence signals were measured with concatemers. This finding is in accordance with previous observations by Wooddell et al.,21 who noticed that the transgene expression decreased when delivering larger pDNA via HLV injection. It has been shown that the molecular weight of pDNA impacts their ability to diffuse through the cytosol and their entry into the nucleus, both being strongly decreased for pDNA >2 kb.22 As a result, a lower pDNA copy number in nuclei yields lower luciferase expression.21 Noticeably, when comparing the transfection efficiency of p(3NF1-Luc-3NF2)c and p(Scr1-Luc-Scr2)c, a reproducible trend in favor of the former was obtained, in accordance with results obtained when comparing p(3NF1-Luc-3NF2)m and p(Scr1-Luc-Scr2)m (Figures 4 and 6). However, the 3NF benefit was more obvious with concatemers than with monomers, reaching statistical significance in three large muscles after injection of p(3NF1-Luc-3NF2)c (Figure 6B; Table S3). Thus, as well as HTV administration can activate the NF-κB pathway, HLV injection may trigger a transient NF-κB-assisted nuclear import of 3NF-pDNA; this effect would be all the more noticeable with larger pDNA for which the hydrodynamic effects of the procedure on cytoplasmic diffusion and non-assisted nuclear entry are less efficient. Additionally, other non-physical gene delivery methods that do not enhance cytosolic diffusion may allow to unveil greater benefits when using smaller DTS-carrying pDNA.35
Optimizations of the pDNA sequence can be achieved by modulating many parameters.36 It is noteworthy here that the IE-CMV strong promoter used in this study allows to mediate high (although short-lived) transgene expression in most cell lines. In vivo, a long-term expression can be obtained in skeletal muscle after HLV injection (notably due to the long lifespan of myocytes).21 It was reported earlier that such a promoter could mitigate the potential benefits provided with DTS.24 In spite of this, we show in the present study the potential of 3NF for enhanced pDNA transfection. In ongoing studies, the use of a tissue (e.g., muscle)-specific promoter, weaker than the IE-CMV (but more physiologically relevant), could allow obtaining greater benefits with 3NF-pDNA. This would be desired, as it was suggested that optimization at each step of the gene delivery process, even leading to modest (as small as 2-fold) improvements, could lead to a cumulative enhancement as high as 212-fold of the transgene expression.37
In conclusion, this study further demonstrates that optimized NF-κB-DTSs termed 3NF can enhance the efficacy of pDNA gene transfer in various in vitro and in vivo settings. It is noteworthy that our in vivo evaluations were performed in healthy mice, devoid of any known inflammation. The higher gene transfer obtained with 3NF-pDNAs was likely attributable to some NF-κB activation triggered by the HLV injection procedure itself. Thus, it can be hypothesized that a NF-κB nuclear import strategy might be relevant in chronic inflammatory diseases such as DMD or cystic fibrosis. Additionally, multiple administrations required for treating long-term genetic disorders could also provide cumulative DTS-related benefits. Finally, further optimized gene transfer protocols could consist in combining DTS-pDNA with non-ionic gene carriers.38 In particular, amphiphilic triblock copolymers would be good candidates since some can improve in vivo skeletal muscle gene transfer and/or activate the NF-κB pathway.7,39,40
Materials and methods
pDNAs
The luciferase-encoding pDNA p(3NF1-Luc-3NF2)m (5,556 bp; designed by C.P. and P.M.) (Figures 1A and S1) contains two optimized NF-κB DTSs named 3NFs respectively upstream (3NF1) and downstream (3NF2) of a luciferase cassette. The latter is under transcriptional control of the strong IE-CMV promoter. 3NF1 and 3NF2 sequences are as follows: 3NF1, 5′-CTGGGGACTTTCCAGCCTGGGGACTTTCCAGCTGGGACTTTCCAGG-3′; 3NF2, 5′-CTGGGGACTTTCCAGCTGGGGACTTTCCAGCTGGGACTTTCCAGGAG-3′. Each contains three NF-κB binding sites (underlined motifs) interspaced with optimized spacers. Each 3NF motif could thus simultaneously interact with up to three NF-κB proteins. Five other pDNAs were derived from p(3NF1-Luc-3NF2)m (Figure 1). Briefly, p(Scr1-Luc-3NF2)m, p(3NF1-Luc-Scr2)m, and p(Scr1-Luc-Scr2)m (each 5,556 bp; Figures 1B–1D) were obtained by replacing 3NF1 and/or 3NF2 by scramble sequences (respectively Scr1 and Scr2) using KpnI and EcoRI or BamHI and SalI (New England Biolabs, Evry, France). Two other pDNAs, named p(3NF1-Luc-3NF2)c and p(Scr1-Luc-Scr2)c (each 11,112 bp), were fusions of two copies of either p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m (Figures 1E and 1F). They were obtained by digestion of the latter with KpnI followed by ligation with the T4 DNA ligase (Invitrogen, Paris, France). An agarose gel electrophoresis was run and the bands corresponding to the concatemerized pDNA forms were isolated and purified. In every case, plasmids were transformed and amplified in E. coli DH5α. They were purified using the Macherey-Nagel NucleoBond PC 10000 kit (Macherey-Nagel, Düren, Germany) and checked by gel electrophoresis, PCR (Table S1), and Sanger sequencing. See Supplemental information for more details.
Cell culture
HeLa (human epithelioid cervix carcinoma cells) (CCL-2, ATCC, Rockville, MD, USA), C2C12 (mouse myoblasts) (CRL-1772, ATCC, Rockville, MD, USA), and SK-Mel-28 (human melanoma cells) (HTB-72, ATCC, Rockville, MD, USA) cell lines (DGRI approval no. 6670) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Ozyme, Saint-Cyr-l’École, France) containing 10% heat-inactivated serum (Lonza, Levallois-Perret, France), 1% antibiotics (10,000 U/mL penicillin, 10,000 μg/mL streptomycin) (PAA Laboratories, Les Mureaux, France) and a supplementation of 1% l-glutamine (Ozyme, Saint-Cyr-l’École, France). All incubations were carried out at 37°C in a humidified atmosphere containing 5% CO2. Typically, 24 h before transfection experiments, cells were seeded in 96-well plates (Sarstedt, Marnay, France) at a density of 25,000 cells in 200 μL of DMEM per well.
Transfection reagents
The cationic polymers bPEI (25 kDa) and His-lPEI (22-kDa lPEI grafted with 16% histidine) were obtained from Sigma-Aldrich (Saint-Quentin-Fallavier, France) and Polytheragène (Evry, France; synthesis reported previously),10 respectively. Stock solution of each polymer was prepared at 20 mg/mL in 20 mM HEPES (Sigma-Aldrich, Saint-Quentin-Fallavier, France).
Preparation of polyplexes
Unless otherwise stated, polyplexes were formed for cationic polymer/pDNA MR of 5, by gently mixing in 20 mM HEPES, a volume of cationic polymer with the same volume of pDNA.
In vitro transfection upon TNF-α activation
Prior to transfection, the culture medium of cells was replaced by fresh medium supplemented with 0, 5, 25, or 50 ng/mL TNF-α (ImmunoTools, Friesoythe, Germany). Cells were then incubated for 30 min at 37°C prior to be transfected with 0.25 μg of p(3NF1-Luc-3NF2)m or p(Scr1-Luc-Scr2)m complexed with His-lPEI. A further incubation was then performed (without any change of the medium) for 24 h until reporter gene expression assessment.
In vitro ascending dose of His-lPEI and bPEI polyplexes
Polyplexes were deposited in the culture medium of cells for quantities corresponding to 0.16, 0.20, 0.26, 0.32, 0.40, or 0.50 μg of pDNA per well. Cells were incubated (without any change of the medium) for 24 h until reporter gene expression assessment.
In vitro transfection efficiency and cell toxicity assays
The culture medium was carefully discarded and then 75 μL of 0.5× passive lysis buffer (PLB) (Promega, Charbonnières-les-Bains, France) was introduced into each well. Luminescence was determined using a luciferase assay system kit (Promega, Charbonnières-les-Bains, France). The protein content in each well was determined using a bicinchoninic acid (BCA) protein assay kit (Interchim, Montluçon, France). Luciferase activity was then expressed as relative light units (RLU) per mg of protein (RLU/mg protein). Cell viability was determined with the ViaLight kit (Lonza, Levallois-Perret, France), with results expressed in survival percentage compared to untreated control cells.
Animals
Swiss mice from Janvier Labs (Le Genest-Saint-Isle, France) were used for HLV injection experiments. They were housed at the Animal Facility of the University of Brest. All protocols (APAFIS no. 2125) were approved by the Laboratory Animal Care Guidelines and by the Institutional Animal Care and Research Advisory Committee of the Faculty of Brest.
HLV injection
Prior to HLV injection, mice were anesthetized by intraperitoneal (i.p.) injection of a ketamine/xylazine mixture (100 and 10 mg/kg body weight, respectively). When complete narcosis was obtained, the two hindlimbs were shaved and then a tourniquet was placed to the proximal part of one leg. A small incision of the skin was performed to expose the distal segment of the great saphenous vein. A catheterized 30G needle was carefully introduced into the vein and held in place during the time of the procedure. Injection was done using a 5-mL syringe (Terumo, Leuven, Belgium) and a Harvard PHD 2000 syringe pump (Harvard Apparatus, Les Ulis, France), delivering 1 mL of solution per hindlimb at a rate of 6.290 mL/min. The tourniquet was removed about 2 min after the end of the injection. The second leg was then processed following the same protocol. The skin incised was closed with absorbable suture (Covidien, Mansfield, MA, USA). Animals were monitored until they recovered from anesthesia.
In vivo bioluminescence imaging
Mice received an i.p. injection of 200 μL of luciferin (Interchim, Montluçon, France; 0.16 g/kg body weight). Two minutes later, anesthesia was induced using isoflurane inhalation (Abbvie, Rungis, France; 4% isoflurane/air blend). BLI images were acquired with a NightOWL NC320 device (Berthold, Thoiry, France). In general, luminescence images were taken for an exposure time of 2×30 sec and 4×4 binning. During image acquisition, narcosis of animals was maintained with a 2% isoflurane/air blend. Luminescence background was determined by imaging a non-treated limb.
Ex vivo luminescence measurement
Eight to nine days after HLV injection, mice were sacrificed by cervical dislocation. Eight muscles were sampled from each leg, i.e., soleus, plantaris, gastrocnemius, tibialis anterior, extensor digitorum longus (EDL), quadriceps, posterior muscles, and hamstring (Figure S8B). They were flash-frozen in liquid nitrogen and kept at −80°C until analysis. Each muscle was weighed prior to being crushed in 1 mL of 0.5× PLB using a gentleMACS dissociator (Miltenyi Biotec, Paris, France). Homogenates were centrifuged at 2,000 rpm, 4°C for 10 min to collect the supernatants. The latter were analyzed using a Promega luciferase assay system kit, as described above. Results are given in RLU/mg of muscle. The luminescence background for each muscle was determined using a limb injected with 0.9% NaCl.
Statistical analyses
Various tests were used for assessing statistical significance. For datasets from in vitro experiments, we used the unpaired two-tailed Student’s t test, a multiple t test using the Holm-Sidak method, or two-way ANOVA (GraphPad Prism 7). For datasets from in vivo experiments, the Mann-Whitney test was preferred. Results were considered significant for p values ≤0.05.
Acknowledgments
The authors greatly acknowledge Prof. Pierre Lehn for constant support. They pay homage to Prof. Jon Wolff, thanks to whom the expertise to perform HLV injection was acquired. Julian Ravel is also thanked for technical support. This work was funded by grants from the AFM – Association Française contre les Myopathies (project no. AFM2017#20609; AFM, Evry, France); the ANR – Agence Nationale de la Recherche (project no. ANR-17-CE18-0015-03, “monopDNA-Nanoparticules Virus-Inspirées pour transfert de gènes”; ANR, Paris, France); the Association de Transfusion Sanguine et de Biogénétique Gaétan Saleün (Brest, France); and the Région Bretagne (France). The Service Général des Plateformes, Animalerie Commune (Brest, France) and SynNanoVect (Brest-Rennes, France) are also acknowledged. Y.T.L.G. was a recipient of a PhD fellowship from the French Ministry of Higher Education, Research and Innovation.
Author contributions
Y.T.L.G., T.L.G., P.G., C.P., P.M., and T.M. designed the experiments; Y.T.L.G., T.L.G., K.P., T.H., and S.Q. conducted the experiments; P.G. synthetized the His-lPEI; Y.T.L.G., T.L.G., and T.M. analyzed the data; and Y.T.L.G., T.L.G., P.G., J.R., C.P., P.M., and T.M. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.03.012.
Contributor Information
Tony Le Gall, Email: tony.legall@univ-brest.fr.
Tristan Montier, Email: tristan.montier@univ-brest.fr.
Supplemental information
References
- 1.Montier T., Delépine P., Pichon C., Férec C., Porteous D.J., Midoux P. Non-viral vectors in cystic fibrosis gene therapy: Progress and challenges. Trends Biotechnol. 2004;22:586–592. doi: 10.1016/j.tibtech.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Nayerossadat N., Maedeh T., Ali P.A. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012;1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jayant R.D., Sosa D., Kaushik A., Atluri V., Vashist A., Tomitaka A., Nair M. Current status of non-viral gene therapy for CNS disorders. Expert Opin. Drug Deliv. 2016;13:1433–1445. doi: 10.1080/17425247.2016.1188802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018;20:e3015. doi: 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan E.E., DeGiulio J.V., Dean D.A. Intracellular trafficking of plasmids for gene therapy: Mechanisms of cytoplasmic movement and nuclear import. Curr. Gene Ther. 2006;6:671–681. doi: 10.2174/156652306779010688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomel C., Leborgne C., Cheradame H., Scherman D., Kichler A., Guégan P. Synthesis and evaluation of amphiphilic poly(tetrahydrofuran-b-ethylene oxide) copolymers for DNA delivery into skeletal muscle. Pharm. Res. 2008;25:2963–2971. doi: 10.1007/s11095-008-9698-9. [DOI] [PubMed] [Google Scholar]
- 7.Richard P., Bossard F., Desigaux L., Lanctin C., Bello-Roufai M., Pitard B. Amphiphilic block copolymers promote gene delivery in vivo to pathological skeletal muscles. Hum. Gene Ther. 2005;16:1318–1324. doi: 10.1089/hum.2005.16.1318. [DOI] [PubMed] [Google Scholar]
- 8.Bouraoui A., Ghanem R., Berchel M., Vié V., Le Guen Y., Paboeuf G., Deschamps L., Le Gall T., Montier T., Jaffrès P.-A. Bis-thioether-containing lipid chains in cationic amphiphiles: Physicochemical properties and applications in gene delivery. ChemPhysChem. 2019;20:2187–2194. doi: 10.1002/cphc.201900626. [DOI] [PubMed] [Google Scholar]
- 9.Bouraoui A., Berchel M., Ghanem R., Vié V., Paboeuf G., Deschamps L., Lozach O., Le Gall T., Montier T., Jaffrès P.-A. Substitution of unsaturated lipid chains by thioether-containing lipid chains in cationic amphiphiles: Physicochemical consequences and application for gene delivery. Org. Biomol. Chem. 2019;17:3609–3616. doi: 10.1039/c9ob00473d. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand E., Gonçalves C., Billiet L., Gomez J.P., Pichon C., Cheradame H., Midoux P., Guégan P. Histidinylated linear PEI: A new efficient non-toxic polymer for gene transfer. Chem. Commun. (Camb.) 2011;47:12547–12549. doi: 10.1039/c1cc15716g. [DOI] [PubMed] [Google Scholar]
- 11.Dean D.A., Strong D.D., Zimmer W.E. Nuclear entry of nonviral vectors. Gene Ther. 2005;12:881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breuzard G., Tertil M., Gonçalves C., Cheradame H., Géguan P., Pichon C., Midoux P. Nuclear delivery of NFκB-assisted DNA/polymer complexes: Plasmid DNA quantitation by confocal laser scanning microscopy and evidence of nuclear polyplexes by FRET imaging. Nucleic Acids Res. 2008;36:e71. doi: 10.1093/nar/gkn287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahindhoratep S., Bouda H.A., El Shafey N., Scherman D., Kichler A., Pichon Ch., Midoux P., Mignet N., Bureau M.F. NF-kB related transgene expression in mouse tibial cranial muscle after pDNA injection followed or not by electrotransfer. Biochim. Biophys. Acta. 2014;1840:3257–3263. doi: 10.1016/j.bbagen.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Gonçalves C., Ardourel M.-Y., Decoville M., Breuzard G., Midoux P., Hartmann B., Pichon C. An optimized extended DNA kappa B site that enhances plasmid DNA nuclear import and gene expression. J. Gene Med. 2009;11:401–411. doi: 10.1002/jgm.1312. [DOI] [PubMed] [Google Scholar]
- 15.Evans N.P., Misyak S.A., Robertson J.L., Bassaganya-Riera J., Grange R.W. Dysregulated intracellular signaling and inflammatory gene expression during initial disease onset in Duchenne muscular dystrophy. Am. J. Phys. Med. Rehabil. 2009;88:502–522. doi: 10.1097/PHM.0b013e3181a5a24f. [DOI] [PubMed] [Google Scholar]
- 16.Hagstrom J.E., Hegge J., Zhang G., Noble M., Budker V., Lewis D.L., Herweijer H., Wolff J.A. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004;10:386–398. doi: 10.1016/j.ymthe.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Fan Z., Kocis K., Valley R., Howard J.F., Chopra M., An H., Lin W., Muenzer J., Powers W. Safety and feasibility of high-pressure transvenous limb perfusion with 0.9% saline in human muscular dystrophy. Mol. Ther. 2012;20:456–461. doi: 10.1038/mt.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Z., Kocis K., Valley R., Howard J.F., Jr., Chopra M., Chen Y., An H., Lin W., Muenzer J., Powers W. High-pressure transvenous perfusion of the upper extremity in human muscular dystrophy: A safety study with 0.9% saline. Hum. Gene Ther. 2015;26:614–621. doi: 10.1089/hum.2015.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegge J.O., Wooddell C.I., Zhang G., Hagstrom J.E., Braun S., Huss T., Sebestyén M.G., Emborg M.E., Wolff J.A. Evaluation of hydrodynamic limb vein injections in nonhuman primates. Hum. Gene Ther. 2010;21:829–842. doi: 10.1089/hum.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mesika A., Grigoreva I., Zohar M., Reich Z. A regulated, NFκB-assisted import of plasmid DNA into mammalian cell nuclei. Mol. Ther. 2001;3:653–657. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- 21.Wooddell C.I., Hegge J.O., Zhang G., Sebestyén M.G., Noble M., Griffin J.B., Pfannes L.V., Herweijer H., Hagstrom J.E., Braun S. Dose response in rodents and nonhuman primates after hydrodynamic limb vein delivery of naked plasmid DNA. Hum. Gene Ther. 2011;22:889–903. doi: 10.1089/hum.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukacs G.L., Haggie P., Seksek O., Lechardeur D., Freedman N., Verkman A.S. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G., Wooddell C.I., Hegge J.O., Griffin J.B., Huss T., Braun S., Wolff J.A. Functional efficacy of dystrophin expression from plasmids delivered to mdx mice by hydrodynamic limb vein injection. Hum. Gene Ther. 2010;21:221–237. doi: 10.1089/hum.2009.133. [DOI] [PubMed] [Google Scholar]
- 24.van Gaal E.V.B., Oosting R.S., van Eijk R., Bakowska M., Feyen D., Kok R.J., Hennink W.E., Crommelin D.J.A., Mastrobattista E. DNA nuclear targeting sequences for non-viral gene delivery. Pharm. Res. 2011;28:1707–1722. doi: 10.1007/s11095-011-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maury B., Gonçalves C., Tresset G., Zeghal M., Cheradame H., Guégan P., Pichon C., Midoux P. Influence of pDNA availability on transfection efficiency of polyplexes in non-proliferative cells. Biomaterials. 2014;35:5977–5985. doi: 10.1016/j.biomaterials.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Ma Y.-F., Yang Y.-W. Delivery of DNA-based cancer vaccine with polyethylenimine. Eur. J. Pharm. Sci. 2010;40:75–83. doi: 10.1016/j.ejps.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Saito Y., Higuchi Y., Kawakami S., Yamashita F., Hashida M. Immunostimulatory characteristics induced by linear polyethyleneimine-plasmid DNA complexes in cultured macrophages. Hum. Gene Ther. 2009;20:137–145. doi: 10.1089/hum.2008.013. [DOI] [PubMed] [Google Scholar]
- 28.Shah N., Thomas T., Shirahata A., Sigal L.H., Thomas T.J. Activation of nuclear factor κB by polyamines in breast cancer cells. Biochemistry. 1999;38:14763–14774. doi: 10.1021/bi991291v. [DOI] [PubMed] [Google Scholar]
- 29.Kafil V., Omidi Y. Cytotoxic impacts of linear and branched polyethylenimine nanostructures in a431 cells. Bioimpacts. 2011;1:23–30. doi: 10.5681/bi.2011.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dan H.C., Cooper M.J., Cogswell P.C., Duncan J.A., Ting J.P.-Y., Baldwin A.S. Akt-dependent regulation of NF-κB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs M.D., Harrison S.C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 32.Auphan N., DiDonato J.A., Rosette C., Helmberg A., Karin M. Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 33.Braun S., Jenny C., Thioudellet C., Perraud F., Claudepierre M.-C., Längle-Rouault F., Ali-Hadji D., Schughart K., Pavirani A. In vitro and in vivo effects of glucocorticoids on gene transfer to skeletal muscle. FEBS Lett. 1999;454:277–282. doi: 10.1016/s0014-5793(99)00818-2. [DOI] [PubMed] [Google Scholar]
- 34.Dames P., Laner A., Maucksch C., Aneja M.K., Rudolph C. Targeting of the glucocorticoid hormone receptor with plasmid DNA comprising glucocorticoid response elements improves nonviral gene transfer efficiency in the lungs of mice. J. Gene Med. 2007;9:820–829. doi: 10.1002/jgm.1082. [DOI] [PubMed] [Google Scholar]
- 35.Belmadi N., Midoux P., Loyer P., Passirani C., Pichon C., Le Gall T., Jaffres P.-A., Lehn P., Montier T. Synthetic vectors for gene delivery: An overview of their evolution depending on routes of administration. Biotechnol. J. 2015;10:1370–1389. doi: 10.1002/biot.201400841. [DOI] [PubMed] [Google Scholar]
- 36.Lindberg M.F., Le Gall T., Carmoy N., Berchel M., Hyde S.C., Gill D.R., Jaffrès P.-A., Lehn P., Montier T. Efficient in vivo transfection and safety profile of a CpG-free and codon optimized luciferase plasmid using a cationic lipophosphoramidate in a multiple intravenous administration procedure. Biomaterials. 2015;59:1–11. doi: 10.1016/j.biomaterials.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 37.Ramamoorth M., Narvekar A. Non viral vectors in gene therapy—An overview. J. Clin. Diagn. Res. 2015;9:GE01–GE06. doi: 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Guen Y.T., Le Gall T., Midoux P., Guégan P., Braun S., Montier T. Gene transfer to skeletal muscle using hydrodynamic limb vein injection: Current applications, hurdles and possible optimizations. J. Gene Med. 2020;22:e3150. doi: 10.1002/jgm.3150. [DOI] [PubMed] [Google Scholar]
- 39.Alimi-Guez D., Leborgne C., Pembouong G., Van Wittenberghe L., Mignet N., Scherman D., Kichler A. Evaluation of the muscle gene transfer activity of a series of amphiphilic triblock copolymers. J. Gene Med. 2009;11:1114–1124. doi: 10.1002/jgm.1396. [DOI] [PubMed] [Google Scholar]
- 40.Gaymalov Z.Z., Yang Z., Pisarev V.M., Alakhov V.Y., Kabanov A.V. The effect of the nonionic block copolymer pluronic P85 on gene expression in mouse muscle and antigen-presenting cells. Biomaterials. 2009;30:1232–1245. doi: 10.1016/j.biomaterials.2008.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.