Abstract
Entamoeba histolytica is the only pathogenic species of the Entamoeba genus and is morphologically identical to E. dispar/E. moshkovskii (Entamoeba complex) hence cannot be microscopically differentiated. The other Entamoeba spp. found in humans (E. hartmanni, E. polecki, and E. coli) can be differentiated morphologically from this Entamoeba complex. However, some of their morphologic features overlap making differential diagnosis difficult. This study aimed at determining the occurrence of Entamoeba spp. in patients seeking treatment for diarrhea and/or abdominal discomfort at two clinics in Mukuru informal settlement in Nairobi, Kenya. Faecal samples were collected from 895 patients, examined microscopically following direct wet smear and formal-ether concentration methods. Entamoeba spp. positive faecal samples were subjected to DNA extraction and species-specific nested polymerase chain reaction of the 18S ribosomal RNA (rRNA). By microscopy, Entamoeba spp. cysts or trophozoites were detected in 114/895 (12.7%, 95% Confidence Interval (CI) 10.6–15.1) faecal samples. By nested PCR, the prevalence was: E. histolytica (7.5%, 95% CI 5.9–9.4, 67/895) and E. dispar (8.2%, 95% CI 6.5–10.2, 73/895). Among the Entamoeba spp. complex positive samples, nested PCR detected E. coli and E. hartmanni DNA in 63/114 (55.3%) and 37/114 (32.5%), samples respectively. Among the E. histolytica/E. dispar PCR negative samples (32.5%), 21 (18.4%) contained cysts of either E. coli (19) or E. hartmanni (2) by nested PCR. Entamoeba spp. infections were most common among participants aged 21–30 years; however it was not significant (P = 0.7). Entamoeba spp. infections showed an inverse relationship with diarrhea being most common among participants without diarrhea (P = 0.0). The difference was significant for E. histolytica (P = 0.0) but not significant for E. dispar (P = 0.1). Only E. dispar infections were significantly associated with sex (P = 0.0). This study highlights the need for differentiation of E. histolytica from other Entamoeba spp. by molecular tools for better management of amoebiasis.
Keywords: Entamoeba species, Diarrhea, Abdominal discomfort, Mukuru informal settlement, Nairobi, Kenya
Highlights
-
•
Identified four Entamoeba species (E. histolytica, E. dispar, E. coli and E. hartmanni)
-
•
Entamoeba spp. infection were most common in participants aged 21–30 years.
-
•
Entamoeba spp. infection were inversely related to diarrhea.
-
•
E. coli and E. hartmanni were detected in faecal samples containing Entamoeba spp. complex.
1. Introduction
The protozoan Entamoeba histolytica is the causative agent of amoebiasis, where patients are asymptomatic or may present with amoebic dysentery and liver abscess among other symptoms. The burden of amoebiasis is high in developing countries due to poor sanitary conditions, low socioeconomic status and non-hygienic practices (Shirley et al., 2018). An estimated 50 million people worldwide are infected with E. histolytica annually leading to death of 100,000 people (WHO, 1997). The genus Entamoeba consists of many species, seven of which colonize the human intestinal lumen, namely Entamoeba histolytica, Entamoeba dispar, Entamoeba moshkovskii, Entamoeba polecki, Entamoeba coli, Entamoeba hartmanni, and Entamoeba bangladeshi (Fotedar et al., 2007; Royer et al., 2012). Entamoeba histolytica cause amoebic colitis and extra-intestinal disease in humans, while patients infected with either E. dispar and/or E. moshkovskii have presented with gastrointestinal symptoms (Fotedar et al., 2008; Ximenez et al., 2010). The other Entamoeba species are commensal parasites in humans (Stanley Jr., 2003).
Microscopy has been the traditional method for diagnosis of E. histolytica/E. dispar in stool despite having low sensitivity (Gonzalez-Ruiz et al., 1994). The sensitivity of microscopy is further reduced by the periodic release of cysts that necessitates the examination of multiple faecal samples in subsequent days (Nazer et al., 1993). Additionally, pathogenic E. histolytica is indistinguishable in its cysts and trophozoite stages from non-pathogenic E. dispar and E. moshkovskii (WHO, 1997; Ali et al., 2003). The diagnosis is also complicated by the presence of other commensal Entamoeba spp. (E. hartmanni, E. coli, and E. polecki) with overlapping morphologic features (Tanyuksel and Petri Jr., 2003). Therefore, there is need for sensitive and accurate diagnostic tools to inform the management of amoebiasis and reduce unnecessary treatment for non-pathogenic Entamoeba spp. (WHO, 1997; Pritt and Clark, 2008). Polymerase chain reaction (PCR), is one of such tools that can differentiate E. histolytica from E. dispar/E. moshkovskii and other species of the Entamoeba genus. The Entamoeba spp. 18S rRNA gene exists as a multicopy loci and exhibits genetic variation and therefore a good target for detection and differentiation of members of this genus (Cruz-Reyes et al., 1992; Troll et al., 1997; Bhattacharya et al., 1998). The development and application of a nested multiplex PCR based on 18S rRNA has contributed greatly into epidemiology of amoebiasis in many regions of the world (Khairnar and Parija, 2007).
Worldwide more than half of the population live in urban areas and this has resulted in exponential growth of informal settlements. In Kenya the informal settlements are home to more than 71% of the urban population (WHO and UN-Habitat, 2010). They are densely populated, lack clean water and have inadequate sanitation, poor waste management and drainage (APHRC, 2014). These conditions favour parasite transmission and put the residents at risk of acquiring infectious diseases with high morbidity and mortality (Brooker et al., 2006). Indeed the Mukuru informal settlements are hot spots for infectious diseases such as multidrug resistant nontyphoidal Salmonella and intestinal protozoans according to findings of Kariuki et al. (2019) and Mbae et al. (2013), respectively.
Although E. histolytica/E. dispar is one of the most common parasitic cause of diarrheal diseases in Kenya, besides Cryptosporidium species and Giardia lamblia (Gatei et al., 2006; Mbae et al., 2013). The epidemiology of amoebiasis is poorly understood owing to the fact that previous studies employed microscopy for diagnosis and could not distinguish pathogenic E. histolytica from other commensal Entamoeba spp. Therefore, the actual prevalence of E. histolytica in those studies is still unknown since microscopy was shown to overestimate the prevalence of E. histolytica when molecular tools were applied in studies across African countries (Kebede et al., 2003; Ben Ayed et al., 2008; Efunshile et al., 2015; Yimer et al., 2017). In addition, there is limited data on the occurrence of other Entamoeba spp. (E. hartmanni, E. coli, E. polecki) (Matey et al., 2016) and how their presence could easily mislead the diagnosis of E. histolytica. The present study reports the prevalence of Entamoeba spp. in patients seeking treatment for diarrhea and/or abdominal discomfort at two clinics in Mukuru informal settlement in Nairobi, Kenya.
2. Materials and methods
2.1. Study design and study sites
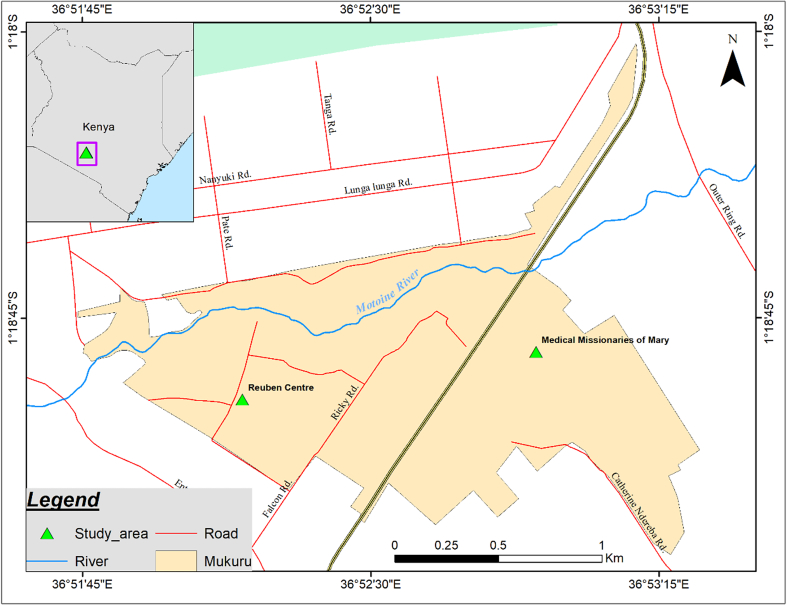
This was a cross-sectional study carried out at two clinics (Reuben Centre and Medical Missionaries of Mary) within Mukuru informal settlement in Nairobi, Kenya (Fig. 1). Mukuru slum is one of the urban informal settlements in Nairobi city and is subdivided into 8 villages. The Reuben Centre (1° 18′ 57” S, 36° 52′ 10″ E) and Medical Missionaries of Mary clinics (MMM) (1° 18′ 50” S, 36° 52′ 55″ E) are located in Mukuru Kwa Reuben and Mukuru Kwa Njenga villages respectively. The villages are characterized by poor sanitary conditions, poor drainage systems, shortage of clean drinking water and improper waste management factors that are likely to enhance transmission of enteric infections such as amoebiasis.
Fig. 1.
A map of Mukuru informal settlement showing the Reuben Centre and Medical Missionaries of Mary clinics.
2.2. Specimen collection, processing and microscopy examination
The majority of the patients seeking treatment in these clinics reside within Mukuru informal settlement. Fresh faecal samples were collected (February – May 2013) from individual patients of all ages presenting with gastrointestinal symptoms (diarrhea, abdominal pain, bloody stool). The faecal samples were examined macroscopically for consistency, mucus and blood and processed by direct wet smear and formal-ether sedimentation techniques, iodine staining and examined microscopically for the presence of cysts or trophozoites of Entamoeba spp. and other intestinal parasites (Cheesbrough, 2009). Stool samples found positive for Entamoeba spp. cysts or trophozoites were aliquoted and preserved in 2.5% potassium dichromate at 4 °C until further analysis. For detection of Cryptosporidium and other protozoa, faecal samples were subjected to Modified Ziehl-Neelsen staining technique and examined by microscopy (Casemore, 1991).
2.3. Extraction of Entamoeba spp. genomic DNA
Genomic DNA was extracted from 114 stool samples microscopically positive for E. histolytica/E. dispar. Briefly 200 μl of faecal suspension from each Entamoeba spp. positive sample was washed five times with triple-distilled water by centrifugation. DNA was purified using QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany) with slight modifications of the manufacturer's protocol. The cysts were lysed by adding 1.4 ml of ASL buffer and subjected to five cycles of freeze (−80 °C for 30 min) and thaw (80 °C for 15 min). The DNA was eluted in 50 μl of elution buffer and stored at −20 °C before used in PCR reactions. Entamoeba spp. control DNA was obtained from the Department of Parasitology, Medical School, Kanazawa University, Japan courtesy of Dr. Tokoro Masaharu.
2.4. Nested polymerase chain reaction based on 18S ribosomal RNA gene
A nested PCR targeting 18S ribosomal RNA gene was performed for detection and differentiation of E. histolytica, E. dispar and E. moshkovskii according to Khairnar and Parija (2007) with slight modifications. The primary PCR which is Entamoeba genus-specific used forward primer E-1 and reverse primer E-2 (Table 1). The PCR assay was carried out in a 25 μl reaction volume consisting of 1 × 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 2 mM MgCl2, 200 μM of each dNTPs, 0.625 units of DreamTaq™ Green DNA Polymerase, (Thermo Fisher Scientific, Massachusetts, USA), 0.25 μM of each primer and 2.5 μl of DNA template. The cycling conditions included an initial denaturation at 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min and a final extension at 72 °C for 7 min. The nested PCR for the Entamoeba complex differentiation were done individually instead of using multiplex PCR. Amplification was achieved using primer sets EH-1 and EH-2 for E. histolytica; ED-1 and ED-2 for E. dispar; Mos-1 and Mos-2 for E. moshkovskii (Table 1) as described by Khairnar and Parija (2007). In the secondary amplification reagent concentrations and cycling conditions were similar to the first PCR except that 2.5 μl of the primary PCR product was used as DNA template and different annealing temperatures were used for each species, 52 °C for E. dispar, 50 °C for E. histolytica and 48 °C for E. moshkovskii (Table 1). To illustrate how other Entamoeba spp. beside E. histolytica/E. dispar/E. moshkovskii complex might interfere with microscopy diagnosis of amoebiasis, another nested PCR was performed on the faecal samples containing this complex but targeting E. coli and E. hartmanni DNA as previously described by Matey et al. (2016). For the primary PCR universal Entamoeba genus-specific forward (TN21) and reverse (TN14) primers were used. In the secondary PCR species-specific forward primers MA67 and MA113 for E. hartmanni and E. coli respectively, the reverse primer TA28 was used for both species (Table 1). The secondary PCR products were subjected to electrophoresis in 2% agarose gels stained with ethidium bromide and visualized on a UV-transilluminator.
Table 1.
Primers for detection of Entamoeba species.
| Primer | Genus/species | Sequence (5′-3′) | Annealing temperature | Reference |
|---|---|---|---|---|
| E-1 | Entamoeba | TAAGATGCAGAGCGAAA | 58 °C | (Khairnar and Parija, 2007) |
| E-2 | Entamoeba | GTACAAAGGGCAGGGACGTA | 58 °C | (Khairnar and Parija, 2007) |
| EH-1 | E. histolytica | AAGCATTGTTTCTAGATCTGAG | 50 °C | (Khairnar and Parija, 2007) |
| EH-2 | E. histolytica | AAGAGGTCTAACCGAAATTAG | 50 °C | (Khairnar and Parija, 2007) |
| ED-1 | E. dispar | TCTAATTTCGATTAGAACTCT | 52 °C | (Khairnar and Parija, 2007) |
| ED-2 | E. dispar | TCCCTACCTATTAGACATAGC | 52 °C | (Khairnar and Parija, 2007) |
| Mos-1 | E. moshkovskii | GAAACCAAGAGTTTCACAAC | 48 °C | (Khairnar and Parija, 2007) |
| Mos-2 | E. moshkovskii | CAATATAAGGCTTGGATGAT | 48 °C | (Khairnar and Parija, 2007) |
| TN21 | Entamoeba | AAGATTAAGCCATGCATGTSKA | 58 °C | (Matey et al., 2016) |
| TN14 | Entamoeba | GATACCTTGTTACGACTTCTY | 58 °C | (Matey et al., 2016) |
| MA67 | E. hartmanni | TTGGATGTAGAGATACATTC | 54 °C | (Matey et al., 2016) |
| T28 | E. hartmanni | CACTATTGGAGCTGGAATTAC | 54 °C | (Matey et al., 2016) |
| MA113 | E. coli | GCCAAGAGAATTGTAGAAATCG | 54 °C | (Matey et al., 2016) |
| T28 | E. coli | CACTATTGGAGCTGGAATTAC | 54 °C | (Matey et al., 2016) |
2.5. Ethical approval
The study protocol (SSC. No. 2258) was reviewed and approved by the Scientific and Ethics Review Unit (SERU) at KEMRI and by the management of Mukuru clinics (Reuben Centre and Medical Missionaries of Mary). Verbal consent was sought from all study participants before they were enrolled.
2.6. Data analysis
The data was entered in Microsoft Excel 2013 and transferred into STATA version 12.0 (STATA Corporation, College Station, Texas, USA) for analysis. The chi square (χ2) test was used to determine the potential correlation between age, sex of the study participants, centre and stool constituency with Entamoeba infections. The difference was considered significant when the P value was ≤0.05.
3. Results
3.1. Demographic information
A total of 895 participants were recruited into the study, 470 from the Medical Missionaries of Mary (MMM) and 425 from the Reuben Centre. Among the participants 439 (49.1%) were males, 426 (47.6%) females and 30 (3.4%) whose gender was not recorded (Table 2), the latter were excluded from analysis (Table 3). The ages of the study participants ranged from 2 weeks to 83 years and were stratified into 8 groups (0–5, 6–10, 11–20, 21–30, 31–40, 41–50, 51–60 and ≥ 61 years). The majority of the participants were aged 21–30 years (34.8%), followed by infants under 5 years at 16.0% while only 2 participants (0.2%) were aged 61 and above. The ages of 11 (1.2%) participants were not recorded (Table 2) and were excluded from analysis (Table 3).
Table 2.
Demographic data and prevalence of Entamoeba species by age, sex and centre.
| Variables | No. of subjects (%) n = 895 |
Microscopy (%) n = 114 (12.7) |
E. histolytica (%) n = 67 (7.5) |
E. dispar (%) n = 73 (8.2) |
E. coli (%) n = 63 (55.3) |
E. hartmanni (%) n = 37 (33.0) |
|---|---|---|---|---|---|---|
| Age groups (years) | ||||||
| ≤5 | 143 (16.0) | 14 | 9 | 11 | 8 | 5 |
| 6-10 | 119 (13.3) | 14 | 5 | 6 | 6 | 5 |
| 11-20 | 114 (12.7) | 13 | 9 | 9 | 11 | 6 |
| 21-30 | 311 (34.8) | 44 | 23 | 25 | 25 | 16 |
| 31-40 | 131 (14.6) | 17 | 14 | 13 | 6 | 4 |
| 41-50 | 54 (6.0) | 7 | 5 | 6 | 6 | 1 |
| 51-60 | 10 (1.1) | 2 | 1 | 1 | 0 | 0 |
| 6≥ | 2 (0.2) | 1 | 0 | 1 | 0 | 0 |
| Unknown | 11 (1.2) | 2 | 1 | 1 | 1 | 1 |
| Gender | ||||||
| Male | 439 (49.1) | 50 (5.6) | 27 (3.0) | 28 (3.1) | 26 (22.8) | 15 (13.2) |
| Female | 426 (47.6) | 62 (6.9) | 39 (4.4) | 44 (4.9) | 36 (31.6) | 22 (19.3) |
| Unknown | 30 (3.4) | 2 (0.2) | 1 (0.1) | 1 (0.1) | 1 (0.9) | 0 (0.0) |
| Centre | ||||||
| Reuben | 425 (47.5) | 54 (12.7) | 31 (7.3) | 36 (8.5) | 27 (23.7) | 17 (14.9) |
| MMM | 470 (52.5) | 60 (12.8) | 36 (7.7) | 37 (7.7) | 36 (31.6) | 20 (17.5) |
Table 3.
Association of Entamoeba species infection with age, sex, centre and stool consistency.
| Variables | No. of subjects (%) n = 884 |
Microscopy (%) n = 112 (13.0) |
E. histolytica (%) n = 66 (7.6) |
E. dispar (%) n = 72 (8.0) |
E. coli (%) n = 63 (56.3) |
E. hartmanni (%) n = 37 (32.5) |
|---|---|---|---|---|---|---|
| Age groups (years) | ||||||
| ≤5 | 143 (16.2) | 14 | 9 | 11 | 8 | 5 |
| 6-10 | 119 (13.5) | 14 | 5 | 6 | 6 | 5 |
| 11-20 | 114 (12.9) | 13 | 9 | 9 | 11 | 6 |
| 21-30 | 311 (35.2) | 44 | 23 | 25 | 25 | 16 |
| 31-40 | 131 (14.8) | 17 | 14 | 13 | 6 | 4 |
| 41-50 | 54 (6.1) | 7 | 5 | 6 | 6 | 1 |
| 51-60 | 10 (1.1) | 2 | 1 | 1 | 0 | 0 |
| 61≥ | 2 (0.2) | 1 | 0 | 1 | 0 | 0 |
| χ2 = 5.0 P = 0.7 |
χ2 = 4.6 P = 0.7 |
χ2 = 7.5 P = 0.4 |
||||
| Gender | n = 865 | |||||
| Male | 439 (50.8) | 50 (11.4) | 27 (6.2) | 28 (6.4) | 26 (41.3) | 15 (40.5) |
| Female | 426 (49.3) | 62 (14.6) | 39 (9.2) | 44 (10.3) | 36 (57.1) | 22 (59.5) |
| χ2 = 1.9 P = 0.2 |
χ2 = 2.8 P = 0.1 |
χ2 = 4.4 P = 0.0⁎ |
||||
| Centre | n = 865 | |||||
| Reuben | 420 (48.6) | 53 (12.6) | 31 (7.4) | 36 (8.6) | 27 (24.1) | 17 (15.2) |
| MMM | 445 (51.6) | 59 (13.3) | 35 (7.9) | 36 (8.1) | 36 (32.1) | 20 (17.9) |
| χ2 = 0.1 P = 0.8 |
χ2 = 0.1 P = 0.8 |
χ2 = 0.1 P = 0.8 |
||||
| Consistency | n = 865 | |||||
| Diarrhoeic | 534 (61.7) | 58 (51.8) | 32 (48.5) | 38 (52.8) | 24 (21.4) | 16 (14.3) |
| Non-diarrhoeic | 331 (38.3) | 54 (48.2) | 34 (51.5) | 34 (47.2) | 39 (34.8) | 21 (18.8) |
| χ2 = 5.4 P = 0.0⁎ |
χ2 = 5.3 P = 0.0⁎ |
χ2 = 2.7 P = 0.1 |
P-value statistically significant.
3.2. Microscopy examination
Entamoeba cysts or trophozoites were detected in 114 (12.7% 95% CI 10.6–15.1) participants. The prevalence of Entamoeba spp. infection was higher in MMM than in Reuben Centre (13.3 vs 12.6% respectively), however the difference was not significant (χ2 = 0.1, P = 0.8). Entamoeba spp. infection were more common in female participants compared to males (14.6% vs 11.4%), however this difference was not statistically significant (χ2 = 1.9, P = 0.2) (Table 3). Other intestinal parasites detected included Giardia lamblia (2.0%), Ascaris lumbricoides (1.2%), Hymenolepis nana (0.5%), Chilomastix mesnili (0.2%), Cryptosporidium spp., (0.2%), H. diminuta (0.1%) and Cyclospora spp. (0.1%).
3.3. Nested PCR assays
Entamoeba spp. were detected in 98/114 (86.0%) faecal samples by nested PCR. The prevalence of Entamoeba spp. was (7.5%, 95% CI 5.9–9.4, 67/895) for E. histolytica and (8.2%, 95% CI 6.5–10.2, 73/895) for E. dispar (Table 2). The prevalence of E. histolytica (7.9 vs 7.4) and E. dispar (8.1 vs 8.6) was comparable between the two clinics (Table 3). Among the Entamoeba spp. complex positive samples (114), nested PCR detected E. coli and E. hartmanni DNA in 63/114 (55.3%) and 37/114 (32.5%) samples respectively (Table 2). This study did not detect E. moshkovskii, while E. histolytica/E. dispar mixed infections were detected in 7.0% of the faecal samples. Mixed infection with all four Entamoeba spp. was detected in 15 (16.8%) faecal samples. Among the E. histolytica/E. dispar PCR negative samples (32.5%), 21 (18.4%) of them contained cysts of either E. coli (19) or E. hartmanni (2) by nested PCR. Entamoeba spp. infections, including E. histolytica were highest in the age group 21–30 years. The age groups 0–5 and 31–40 years also recorded high infections for all the Entamoeba spp., while participants aged over 41 years had the least infections (Table 2). None of the Entamoeba spp. infections were significantly associated with the age of the participants (Table 3). Female participants were more infected by all the Entamoeba spp. compared to males: E. histolytica (9.2% vs 6.2%) and E. dispar (10.3% vs 8.3%). The difference was only significant for E. dispar (χ2 = 4.4, P = 0.0) but not for E. histolytica (χ2 = 2.8, P = 0.1) (Table 3).
3.4. Correlation of faecal samples consistency with the presence of Entamoeba species
Majority of the participants presented with diarrhea (watery, mucoid, loose stool) 534 (61.7%) against 331 (38.2%) without (formed stool). Entamoeba spp. infection showed an inverse relationship with diarrhea, being most common among participants without diarrhea (χ2 = 5.4, P = 0.0). The difference was significant for E. histolytica (χ2 = 5.3, P = 0.0), but not significant for E. dispar (χ2 = 2.7, P = 0.1). A total of 31 (3.5%) faecal samples contained blood, among these 4 bloody mucoid samples were positive for E. histolytica by PCR.
4. Discussion
This study reports the presence of E. histolytica, E. dispar, E. coli and E. hartmanni in patients seeking treatment for diarrhea and/or abdominal discomfort in two clinics in Mukuru informal settlement in Nairobi, Kenya. Several microscopy-based epidemiological studies in Kenya failed to differentiate E. histolytica from E. dispar/E. moshkovskii complex and other Entamoeba spp. (Chunge et al., 1991; Joyce et al., 1996; Gatei et al., 2006; Nyarango et al., 2008; Nguhiu et al., 2009; Kamau et al., 2012; Kipyegen et al., 2012; Mbae et al., 2013; Obala et al., 2013). As it is the usual practice in Kenya, these patients were treated indiscriminately using antiamoebic drugs. This study therefore, highlights the importance of differentiating of E. histolytica from non-pathogenic Entamoeba spp. before treatment of patients to avoid drug resistance.
Accurate identification and differentiation of Entamoeba spp. is a critical step in the management of amoebiasis as recommended by WHO (WHO, 1997). It is necessary to distinguish pathogenic E. histolytica infections from those of the Entamoeba spp. complex and other non-pathogenic species such as E. coli, E. hartmanni and E. polecki (Ali et al., 2008; Gomes Tdos et al., 2014). In this study, the presence of these commensal Entamoeba spp. was determined in faecal samples containing the Entamoeba spp. complex to illustrate how their presence complicates the diagnosis of E. histolytica microscopically. Entamoeba coli and E. hartmanni were identified in faecal samples containing Entamoeba spp. complex microscopically and from those negative for this complex by nested PCR. The detection of E. coli or E. hartmanni DNA in samples originally identified microscopically to contain cysts or trophozoites for Entamoeba complex but negative for this complex by nested PCR clearly shows the limitation of microscopy in diagnosis of E. histolytica. Entamoeba coli and E. hartmanni were also common among HIV-infected and HIV-uninfected children in western Kenya (Matey et al., 2016).
The prevalence of Entamoeba spp. by microscopy in the present study was in agreement with those reported previously from Kenya in children (Obala et al., 2013; Njambi et al., 2020), food-handlers (Kamau et al., 2012) and people of all age groups (Nguhiu et al., 2009), as well as in other African countries including Ethiopia, Cameroon and Nigeria (Aribodor et al., 2012; Assob et al., 2012; King et al., 2013). However, elsewhere in Kenya high prevalence of Entamoeba spp. was recorded in children under 5 years of age from Maasailand (Joyce et al., 1996), as a co-infection in sleeping sickness patients (Kagira et al., 2011) and among HIV-patients in Baringo County (Kipyegen et al., 2012). The true prevalence of Entamoeba spp. in this study and previous ones could be underestimates due to the limited sensitivity of microscopy (Gonzalez-Ruiz et al., 1994). However, over the last two decades, the application of PCR on E. histolytica/E. dispar samples initially identified by microscopy has led to the conclusion that the prevalence of E. histolytica in Africa was indeed overestimated (Kebede et al., 2003; Ben Ayed et al., 2008; Efunshile et al., 2015; Yimer et al., 2017).
The prevalence of E. histolytica in this study (7.5%) was higher than in most studies from African countries such as in Uganda (1.5%) (Morawski et al., 2017), Tanzania (2.9%) (Beck et al., 2008), Ethiopia (1.7%) (Yimer et al., 2017), Sudan (5.0%) (Saeed et al., 2015) and South Africa (4.1%) (Samie et al., 2020). However, the prevalence was lower than those reported from patients with gastrointestinal complaints in South Africa (15.6%) (Samie et al., 2006) and Egypt (10.3%) (Roshdy et al., 2017). In Kenya the prevalence of E. histolytica in this study was lower than (14.4–15%) reported previously (Easton et al., 2016; Mwendwa et al., 2017) but higher than (0.4–4.5%) in other studies (Matey et al., 2016; Kyany'a et al., 2019). The difference in prevalence could be due to the diagnostic tool applied for example Easton et al. (2016) used Real-time PCR which is more sensitive than conventional or nested PCR for detection and differentiation of Entamoeba spp. complex (Lau et al., 2013). Although the study by Mwendwa et al. (2017) shared the same study site with the current one, all the faecal samples were tested for E. histolytica by PCR while the present study applied E. histolytica specific PCR on Entamoeba spp. positive faecal samples following microscopic examination. The different study populations, geographical sites, environmental conditions, their socio-economic status and immune status could also explain the difference prevalence with other studies in Kenya.
The age group 21–30 years was the most infected with Entamoeba spp. including E. histolytica and is in agreement with previous studies in Kenya (Kamau et al., 2012; Kimosop et al., 2018). Populations sandwiching this age group recorded high E. histolytica infections in different countries for example 18–40 years in Yemen (Al-Areeqi et al., 2017) and 20–46 years in South Africa (Samie et al., 2006; Samie et al., 2020). This age-related prevalence of E. histolytica was also reported in Malaysia, where infections were more common in children below the age of 15 years (Shahrul Anuar et al., 2012). The age-dependency prevalence of E. histolytica is influenced by acquired immunity (Haque et al., 2006) and the interaction of this Entamoeba spp. and with host microbiome (Ngobeni et al., 2017; Leon-Coria et al., 2020). The age group 21–30 years was also shown to harbour the highest burden of other intestinal parasites in Kenya (Kipyegen et al., 2012). This age group in Kenya comprises of young adults who are likely buy food from the streets with low standards of hygiene and therefore exposing them to foodborne diseases. A recent study reported that buying food from the street as a common behaviour for residents of Mukuru slums and found significant association of this habit with salmonella disease (Mbae et al., 2020). This study found an inverse relationship between E. histolytica infections and diarrhea. The finding of positive association between E. histolytica infections and diarrhea (Samie et al., 2006; Yimer et al., 2017; Samie et al., 2020) or lack of association (Wumba et al., 2010) has been reported previously. A similar inverse relationship has been reported for G. lamblia in case-controls studies paired by age and sex in other sub-Saharan Africa counties including Côte d'Ivoire, Central African Republic, and Tanzania (Becker et al., 2015; Tellevik et al., 2015; Breurec et al., 2016). The cause of the inverse relationship between Entamoeba spp. infections and diarrhea could be due to infections with other parasitic agents. In this study 19 participants who had diarrhea but no E. histolytica infections were infected with G. lamblia (12), A. lumbricoides (5), a co-infection of H. nana and H. diminuta (1), Cryptosporidium spp. (1) and Cyclospora spp. (1). The cause of diarrhea in those negative for E. histolytica could also be other agents besides parasites such as viruses (Gikonyo et al., 2017) and bacterial infections (Kariuki et al., 2019) as reported before in residents of Mukuru informal settlement.
Previous studies in Kenya recorded lower prevalence of E. dispar than the present study, but were in agreement with this study that E. dispar was more common in patients than E. histolytica (Matey et al., 2016; Kyany'a et al., 2019). Although E. dispar is considered common among asymptomatic patients, it has been reported in symptomatic patients (Ximenez et al., 2010; Oliveira et al., 2015; Samie et al., 2020) including those with liver abscess (Dolabella et al., 2012). The proportion of participants infected with E. dispar and presented with or without diarrhea were comparable in this study (not statistically significant), however, it should be noted that majority of the E. dispar infections occurred as a co-infection with E. histolytica (86.3%).
This study failed to detect E. moshkovskii in patients with gastrointestinal symptoms and is consistent with previous findings in Mukuru slums (Mwendwa et al., 2017) and elsewhere in Kenya (Easton et al., 2016; Matey et al., 2016). However, E. moshkovskii was the most prevalent Entamoeba spp. detected in symptomatic and asymptomatic participants in Kenya (Kyany'a et al., 2019). The failure to detect E. moshkovskii in this study could be due to the inability by this nested PCR to amplify DNA from limited number of cysts of this species compared to those of E. histolytica and E. dispar (Khairnar and Parija, 2007; Lau et al., 2013). Indeed Hamzah et al. (2010) showed that a ten-fold amount of E. dispar and E. moshkovskii DNA (2 pg) was detectable by real-time PCR compared to that of E. histolytica (0.2 pg). Furthermore, a single round PCR assay needed double amount of DNA for the detection of E. histolytica and E. moshkovskii compared to that of E. dispar (Hamzah et al., 2006).
The failure by nested PCR to detect Entamoeba spp. DNA in microscopically-positive faecal samples could be due to the presence of PCR inhibitors that were not completely eliminated during DNA extraction. Another possible reason is limited amount of cysts or trophozoites in those samples that fell below the nested PCR detection limit. It is also postulated that faecal samples containing trophozoites alone compared to those with cysts are likely to fail on PCR due to their fast degradation. Lastly the cysts from those samples could belong to other Entamoeba spp. such as E. polecki which was not tested in this study, and confusion of macrophages with trophozoites or polymorphonuclears with cysts (Fotedar et al., 2007; Ngui et al., 2012).
The limitations of this study included 1) The examination of a single faecal sample instead of three-days consecutive samples is likely to underestimate the prevalence of Entamoeba complex because cysts or trophozoites are shed periodically (Nazer et al., 1993). 2) The use of microscopy for detection of Entamoeba spp. in faecal samples instead of nested PCR could also lower the actual prevalence of Entamoeba species given the limited sensitivity of microscopy. 3) The cause of diarrhea and abdominal discomfort could have been associated with other infections such viruses (Rotavirus, adenoviruses) and bacteria which were not diagnosed in this study. 4) The data on patients presenting with abdominal pain was not recorded and analyzed in the current study and this has been reported as a major symptom of amoebiasis (Samie et al., 2020).
In conclusion, this study identified E. histolytica, E. dispar, E. coli and E. hartmanni in patients seeking treatment for diarrhea and/or abdominal discomfort from two clinics in Mukuru informal settlements in Nairobi, Kenya. Further studies are needed on the occurrence of Entamoeba species in asymptomatic individuals who might act as reservoirs of E. histolytica and account for majority of amoebiasis infections. The study also highlights the need for differentiation of E. histolytica from other Entamoeba spp. by molecular tools for better management of amoebiasis to avoid unnecessary treatment for infections with non-pathogenic Entamoeba spp.
Financial support
This study was funded through the Kenya Medical Research Institute, Internal Research Grants L0407.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors sincerely thank the residents of Mukuru informal settlements who consented and submitted faecal samples for this study. The clinical and laboratory staff of both clinics who assisted in recruitment of participants and collection of faecal samples are highly appreciated. Many thanks to Dr. Tokoro Masaharu of the Department of Parasitology, Medical School, Kanazawa University for providing the Entamoeba spp. control DNA. This research article is published with permission from the Director General, Kenya Medical Research Institute.
References
- Al-Areeqi M.A., Sady H., Al-Mekhlafi H.M., Anuar T.S., Al-Adhroey A.H., Atroosh W.M., Dawaki S., Elyana F.N., Nasr N.A., Ithoi I., Lau Y.L., Surin J. First molecular epidemiology of Entamoeba histolytica, E. dispar and E. moshkovskii infections in Yemen: different species-specific associated risk factors. Tropical Med. Int. Health. 2017;22:493–504. doi: 10.1111/tmi.12848. [DOI] [PubMed] [Google Scholar]
- Ali I.K., Hossain M.B., Roy S., Ayeh-Kumi P.F., Petri W.A., Jr., Haque R., Clark C.G. Entamoeba moshkovskii infections in children. Bangladesh. Emerg. Infect. Dis. 2003;9:580–584. doi: 10.3201/eid0905.020548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I.K., Clark C.G., Petri W.A., Jr. Molecular epidemiology of amebiasis. Infect. Genet. Evol. 2008;8:698–707. doi: 10.1016/j.meegid.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHRC . African Population and Health Research Center; Nairobi: 2014. Population and Health Dynamics in Nairobi's Informal Settlements: Report of the Nairobi Cross-Sectional Slums Survey (NCSS) 2012. [Google Scholar]
- Aribodor D., Anidebe A., Eneanya O., Emelummadu O. Entamoeba histolytica infection in children aged 0-12 years in rural communities in Anambra state, Nigeria. Nigerian J. Parasitol. 2012;33 [Google Scholar]
- Assob J.C., Nde P.F., Nsagha D.S., Njimoh D.L., Nfor O., Njunda A.L., Kamga H.L. The incidence of feco-oral parasites in street-food vendors in Buea, south-west region Cameroon. Afr. Health Sci. 2012;12:376–380. doi: 10.4314/ahs.v12i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D.L., Dogan N., Maro V., Sam N.E., Shao J., Houpt E.R. High prevalence of Entamoeba moshkovskii in a Tanzanian HIV population. Acta Trop. 2008;107:48–49. doi: 10.1016/j.actatropica.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S.L., Chatigre J.K., Gohou J.P., Coulibaly J.T., Leuppi R., Polman K., Chappuis F., Mertens P., Herrmann M., N'Goran E.K., Utzinger J., von Müller L. Combined stool-based multiplex PCR and microscopy for enhanced pathogen detection in patients with persistent diarrhoea and asymptomatic controls from Côte d'Ivoire. Clin. Microbiol. Infect. 2015;21(591) doi: 10.1016/j.cmi.2015.02.016. e591-510. [DOI] [PubMed] [Google Scholar]
- Ben Ayed S., Ben A.R., Mousli M., Aoun K., Thellier M., Bouratbine A. Molecular differentiation of Entamoeba histolytica and Entamoeba dispar from Tunisian food handlers with amoeba infection initially diagnosed by microscopy. Parasite (Paris, France) 2008;15:65–68. doi: 10.1051/parasite/2008151065. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Som I., Bhattacharya A. The ribosomal DNA plasmids of Entamoeba. Parasitol. Today. 1998;14:181–185. doi: 10.1016/s0169-4758(98)01222-8. [DOI] [PubMed] [Google Scholar]
- Breurec S., Vanel N., Bata P., Chartier L., Farra A., Favennec L., Franck T., Giles-Vernick T., Gody J.C., Luong Nguyen L.B., Onambélé M., Rafaï C., Razakandrainibe R., Tondeur L., Tricou V., Sansonetti P., Vray M. Etiology and epidemiology of diarrhea in hospitalized children from low income country: a matched case-control study in Central African Republic. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker S., Clements A.C., Bundy D.A. Global epidemiology, ecology and control of soil-transmitted helminth infections. Adv. Parasitol. 2006;62:221–261. doi: 10.1016/S0065-308X(05)62007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casemore D.P. ACP Broadsheet 128: June 1991. Laboratory methods for diagnosing cryptosporidiosis. J. Clin. Pathol. 1991;44:445–451. doi: 10.1136/jcp.44.6.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesbrough M. Cambridge University Press; Cambridge: 2009. District laboratory practice in tropical countries. Part 2. [Google Scholar]
- Chunge R.N., Nagelkerke N., Karumba P.N., Kaleli N., Wamwea M., Mutiso N., Andala E.O., Gachoya J., Kiarie R., Kinoti S.N. Longitudinal study of young children in Kenya: intestinal parasitic infection with special reference to Giardia lamblia, its prevalence, incidence and duration, and its association with diarrhoea and with other parasites. Acta Trop. 1991;50:39–49. doi: 10.1016/0001-706x(91)90071-q. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J.A., Spice W.M., Rehman T., Gisborne E., Ackers J.P. Ribosomal DNA sequences in the differentiation of pathogenic and non-pathogenic isolates of Entamoeba histolytica. Parasitology. 1992;104(Pt 2):239–246. doi: 10.1017/s0031182000061679. [DOI] [PubMed] [Google Scholar]
- Dolabella S.S., Serrano-Luna J., Navarro-García F., Cerritos R., Ximénez C., Galván-Moroyoqui J.M., Silva E.F., Tsutsumi V., Shibayama M. Amoebic liver abscess production by Entamoeba dispar. Ann. Hepatol. 2012;11:107–117. [PubMed] [Google Scholar]
- Easton A.V., Oliveira R.G., O'Connell E.M., Kepha S., Mwandawiro C.S., Njenga S.M., Kihara J.H., Mwatele C., Odiere M.R., Brooker S.J., Webster J.P., Anderson R.M., Nutman T.B. Multi-parallel qPCR provides increased sensitivity and diagnostic breadth for gastrointestinal parasites of humans: field-based inferences on the impact of mass deworming. Parasit. Vectors. 2016;9:38. doi: 10.1186/s13071-016-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efunshile M.A., Ngwu B.A., Kurtzhals J.A., Sahar S., Konig B., Stensvold C.R. Molecular detection of the carriage rate of four intestinal protozoa with real-time polymerase chain reaction: possible overdiagnosis of Entamoeba histolytica in Nigeria. Am. J. Trop. Med. Hyg. 2015;93:257–262. doi: 10.4269/ajtmh.14-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R., Stark D., Beebe N., Marriott D., Ellis J., Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin. Microbiol. Rev. 2007;20:511–532. doi: 10.1128/CMR.00004-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R., Stark D., Marriott D., Ellis J., Harkness J. Entamoeba moshkovskii infections in Sydney. Australia. Eur. J. Clin. Microbiol. Infect. Dis. 2008;27:133–137. doi: 10.1007/s10096-007-0399-9. [DOI] [PubMed] [Google Scholar]
- Gatei W., Wamae C.N., Mbae C., Waruru A., Mulinge E., Waithera T., Gatika S.M., Kamwati S.K., Revathi G., Hart C.A. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am. J. Trop. Med. Hyg. 2006;75:78–82. [PubMed] [Google Scholar]
- Gikonyo J.N., Nyangao J., Mbae C., Sang C., Njagi E., Ngeranwa J., Esona M., Seheri M.L., Gitau G.W., Raini K., Kariuki S. Molecular characterization of group A rotaviruses in Mukuru slums Kenya: detection of novel strains circulating in children below 5 years of age. BMC Res. Notes. 2017;10:290. doi: 10.1186/s13104-017-2611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes Tdos S., Garcia M.C., de Souza Cunha F., Werneck de Macedo H.P., Peralta J.M., H R. Differential diagnosis of Entamoeba spp. in clinical stool samples using SYBR green real-time polymerase chain reaction. Sci. World J. 2014;2014:645084. doi: 10.1155/2014/645084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ruiz A., Haque R., Aguirre A., Castanon G., Hall A., Guhl F., Ruiz-Palacios G., Miles M.A., Warhurst D.C. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J. Clin. Pathol. 1994;47:236–239. doi: 10.1136/jcp.47.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzah Z., Petmitr S., Mungthin M., Leelayoova S., Chavalitshewinkoon-Petmitr P. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J. Clin. Microbiol. 2006;44:3196–3200. doi: 10.1128/JCM.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamzah Z., Petmitr S., Mungthin M., Leelayoova S., Chavalitshewinkoon-Petmitr P. Development of multiplex real-time polymerase chain reaction for detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii in clinical specimens. Am. J. Trop. Med. Hyg. 2010;83:909–913. doi: 10.4269/ajtmh.2010.10-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque R., Mondal D., Duggal P., Kabir M., Roy S., Farr B.M., Sack R.B., Petri W.A., Jr. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect. Immun. 2006;74:904–909. doi: 10.1128/IAI.74.2.904-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce T., McGuigan K.G., Elmore-Meegan M., Conroy R.M. Prevalence of enteropathogens in stools of rural Maasai children under five years of age in the Maasailand region of the Kenyan Rift Valley. East Afr. Med. J. 1996;73:59–62. [PubMed] [Google Scholar]
- Kagira J.M., Maina N., Njenga J., Karanja S.M., Karori S.M., Ngotho J.M. Prevalence and types of coinfections in sleeping sickness patients in Kenya (2000/2009) J. Trop. Med. 2011;2011:248914. doi: 10.1155/2011/248914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau P., Aloo-Obudho P., Kabiru E., Ombacho K., Langat B., Mucheru O., Ireri L. Prevalence of intestinal parasitic infections in certified food-handlers working in food establishments in the City of Nairobi. Kenya. J. Biomed. Res. 2012;26:84–89. doi: 10.1016/S1674-8301(12)60016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariuki S., Mbae C., Onsare R., Kavai S.M., Wairimu C., Ngetich R., Ali M., Clemens J., Dougan G. Multidrug-resistant nontyphoidal Salmonella hotspots as targets for vaccine use in management of infections in endemic settings. Clin. Infect. Dis. 2019;68:S10–s15. doi: 10.1093/cid/ciy898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebede A., Verweij J., Dorigo-Zetsma W., Sanders E., Messele T., van Lieshout L., Petros B., Polderman T. Overdiagnosis of amoebiasis in the absence of Entamoeba histolytica among patients presenting with diarrhoea in Wonji and Akaki. Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 2003;97:305–307. doi: 10.1016/s0035-9203(03)90153-2. [DOI] [PubMed] [Google Scholar]
- Khairnar K., Parija S.C. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7:47. doi: 10.1186/1471-2180-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimosop R., Mulambalah C., Ngeiywa M. Prevalence of enteric parasitic diseases among patients referred at a teaching hospital in Kenya. J. Health Res. Rev. 2018;5:78–85. [Google Scholar]
- King J.D., Endeshaw T., Escher E., Alemtaye G., Melaku S., Gelaye W., Worku A., Adugna M., Melak B., Teferi T., Zerihun M., Gesese D., Tadesse Z., Mosher A.W., Odermatt P., Utzinger J., Marti H., Ngondi J., Hopkins D.R., Emerson P.M. Intestinal parasite prevalence in an area of Ethiopia after implementing the SAFE strategy, enhanced outreach services, and health extension program. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipyegen C.K., Shivairo R.S., Odhiambo R.O. Prevalence of intestinal parasites among HIV patients in Baringo. Kenya. Pan. Afr. Med. J. 2012;13:37. [PMC free article] [PubMed] [Google Scholar]
- Kyany'a C., Eyase F., Odundo E., Kipkirui E., Kipkemoi N., Kirera R., Philip C., Ndonye J., Kirui M., Ombogo A., Koech M., Bulimo W., Hulseberg C.E. First report of Entamoeba moshkovskii in human stool samples from symptomatic and asymptomatic participants in Kenya. Trop. Dis. Travel Med. Vaccines. 2019;5:23. doi: 10.1186/s40794-019-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y.L., Anthony C., Fakhrurrazi S.A., Ibrahim J., Ithoi I., Mahmud R. Real-time PCR assay in differentiating Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii infections in Orang Asli settlements in Malaysia. Parasit. Vectors. 2013;6:250. doi: 10.1186/1756-3305-6-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Coria A., Kumar M., Chadee K. The delicate balance between Entamoeba histolytica, mucus and microbiota. Gut Microbes. 2020;11:118–125. doi: 10.1080/19490976.2019.1614363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matey E.J., Tokoro M., Nagamoto T., Mizuno T., Saina M.C., Bi X., Oyombra J.A., Okumu P., Langat B.K., Sang W.K., Songok E.M., Ichimura H. Lower prevalence of Entamoeba species in children with vertically transmitted HIV infection in Western Kenya. AIDS (London, England) 2016;30:803–805. doi: 10.1097/QAD.0000000000001002. [DOI] [PubMed] [Google Scholar]
- Mbae C., Mwangi M., Gitau N., Irungu T., Muendo F., Wakio Z., Wambui R., Kavai S., Onsare R., Wairimu C., Ngetich R., Njeru F., Van Puyvelde S., Clemens J., Dougan G., Kariuki S. Factors associated with occurrence of salmonellosis among children living in Mukuru slum, an urban informal settlement in Kenya. BMC Infect. Dis. 2020;20:422. doi: 10.1186/s12879-020-05134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbae C.K., Nokes D.J., Mulinge E., Nyambura J., Waruru A., Kariuki S. Intestinal parasitic infections in children presenting with diarrhoea in outpatient and inpatient settings in an informal settlement of Nairobi. Kenya. BMC Infect. Dis. 2013;13:243. doi: 10.1186/1471-2334-13-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski B.M., Yunus M., Kerukadho E., Turyasingura G., Barbra L., Ojok A.M., DiNardo A.R., Sowinski S., Boulware D.R., Mejia R. Hookworm infection is associated with decreased CD4+ T cell counts in HIV-infected adult Ugandans. PLoS Negl. Trop. Dis. 2017;11 doi: 10.1371/journal.pntd.0005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwendwa F., Mbae C.K., Kinyua J., Mulinge E., Mburugu G.N., Njiru Z.K. Stem loop-mediated isothermal amplification test: comparative analysis with classical LAMP and PCR in detection of Entamoeba histolytica in Kenya. BMC Res. Notes. 2017;10:142. doi: 10.1186/s13104-017-2466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazer H., Greer W., Donnelly K., Mohamed A.E., Yaish H., Kagalwalla A., Pavillard R. The need for three stool specimens in routine laboratory examinations for intestinal parasites. Br. J. Clin. Pract. 1993;47:76–78. [PubMed] [Google Scholar]
- Ngobeni R., Samie A., Moonah S., Watanabe K., Petri W.A., Jr., Gilchrist C. Entamoeba species in South Africa: correlations with the host microbiome, parasite burdens, and first description of Entamoeba bangladeshi outside of Asia. J. Infect. Dis. 2017;216:1592–1600. doi: 10.1093/infdis/jix535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguhiu P.N., Kariuki H.C., Magambo J.K., Kimani G., Mwatha J.K., Muchiri E., Dunne D.W., Vennervald B.J., Mkoji G.M. Intestinal polyparasitism in a rural Kenyan community. East Afr. Med. J. 2009;86:272–278. doi: 10.4314/eamj.v86i6.54138. [DOI] [PubMed] [Google Scholar]
- Ngui R., Angal L., Fakhrurrazi S.A., Lian Y.L., Ling L.Y., Ibrahim J., Mahmud R. Differentiating Entamoeba histolytica, Entamoeba dispar and Entamoeba moshkovskii using nested polymerase chain reaction (PCR) in rural communities in Malaysia. Parasit. Vectors. 2012;5:187. doi: 10.1186/1756-3305-5-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njambi E., Magu D., Masaku J., Okoyo C., Njenga S.M. Prevalence of intestinal parasitic infections and associated water, sanitation, and hygiene risk factors among school children in Mwea irrigation scheme, Kirinyaga County. Kenya. J. Trop. Med. 2020;2020:3974156. doi: 10.1155/2020/3974156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyarango R.M., Aloo P.A., Kabiru E.W., Nyanchongi B.O. The risk of pathogenic intestinal parasite infections in Kisii municipality, Kenya. BMC Public Health. 2008;8:237. doi: 10.1186/1471-2458-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obala A.A., Simiyu C.J., Odhiambo D.O., Nanyu V., Chege P., Downing R., Mwaliko E., Mwangi A.W., Menya D., Chelagat D., Nyamogoba H.D., Ayuo P.O., O'Meara W.P., Twagirumukiza M., Vandenbroek D., Otsyula B.B., de Maeseneer J. Webuye health and demographic surveillance systems baseline survey of soil-transmitted helminths and intestinal protozoa among children up to five years. J. Trop. Med. 2013;2013:734562. doi: 10.1155/2013/734562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira F.M., Neumann E., Gomes M.A., Caliari M.V. Entamoeba dispar: could it be pathogenic. Trop. Parasitol. 2015;5:9–14. doi: 10.4103/2229-5070.149887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt B.S., Clark C.G. Amebiasis. Mayo Clin. Proc. 2008;83:1154–1159. doi: 10.4065/83.10.1154. (quiz 1159-1160) [DOI] [PubMed] [Google Scholar]
- Roshdy M.H., Abd El-Kader N.M., Ali-Tammam M., Fuentes I., Mohamed M.M., El-Sheikh N.A., Rubio J.M. Molecular diagnosis of Entamoeba spp. versus microscopy in the great Cairo. Acta Parasitol. 2017;62:188–191. doi: 10.1515/ap-2017-0022. [DOI] [PubMed] [Google Scholar]
- Royer T.L., Gilchrist C., Kabir M., Arju T., Ralston K.S., Haque R., Clark C.G., Petri W.A., Jr. Entamoeba bangladeshi nov. sp., Bangladesh. Emerg. Infect. Dis. 2012;18:1543–1545. doi: 10.3201/eid1809.120122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A., Abd H., Sandstrom G. Microbial aetiology of acute diarrhoea in children under five years of age in Khartoum. Sudan. J. Med. Microbiol. 2015;64:432–437. doi: 10.1099/jmm.0.000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie A., Obi L.C., Bessong P.O., Stroup S., Houpt E., Guerrant R.L. Prevalence and species distribution of E. Histolytica and E. Dispar in the Venda region, Limpopo, South Africa. Am. J. Trop. Med. Hyg. 2006;75:565–571. [PubMed] [Google Scholar]
- Samie A., Mahlaule L., Mbati P., Nozaki T., ElBakri A. Prevalence and distribution of Entamoeba species in a rural community in northern South Africa. Food. Waterborne Parasitol. 2020;18 doi: 10.1016/j.fawpar.2020.e00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrul Anuar T., H M.A.-M., Abdul Ghani M.K., Osman E., Mohd Yasin A., Nordin A., Nor Azreen S., Md Salleh F., Ghazali N., Bernadus M., Moktar N. Prevalence and risk factors associated with Entamoeba histolytica/dispar/moshkovskii infection among three Orang Asli ethnic groups in Malaysia. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley D.T., Farr L., Watanabe K., Moonah S. A review of the global burden, new diagnostics, and current therapeutics for amebiasis. Open Forum Infect. Dis. 2018;5 doi: 10.1093/ofid/ofy161. ofy161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S.L., Jr. Amoebiasis. Lancet (London, England) 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- Tanyuksel M., Petri W.A., Jr. Laboratory diagnosis of amebiasis. Clin. Microbiol. Rev. 2003;16:713–729. doi: 10.1128/CMR.16.4.713-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellevik M.G., Moyo S.J., Blomberg B., Hjøllo T., Maselle S.Y., Langeland N., Hanevik K. Prevalence of Cryptosporidium parvum/hominis, Entamoeba histolytica and Giardia lamblia among young children with and without diarrhea in Dar es Salaam. Tanzania. PLoS Negl. Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0004125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll H., Marti H., Weiss N. Simple differential detection of Entamoeba histolytica and Entamoeba dispar in fresh stool specimens by sodium acetate-acetic acid-formalin concentration and PCR. J. Clin. Microbiol. 1997;35:1701–1705. doi: 10.1128/jcm.35.7.1701-1705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO World Health Organization/pan American health organization/UNESCO report. A consultation with experts on amoebiasis. Epidemiol. Bull. 1997;18:13–14. [PubMed] [Google Scholar]
- WHO, UN-Habitat . World Health Organization; Geneva: 2010. Hidden Cities: Unmasking and Overcoming Health Inequities in Urban Settings. [Google Scholar]
- Wumba R., Longo-Mbenza B., Mandina M., Odio W.T., Biligui S., Sala J., Breton J., Thellier M. Intestinal parasites infections in hospitalized AIDS patients in Kinshasa, Democratic Republic of Congo. Parasite (Paris, France) 2010;17:321–328. doi: 10.1051/parasite/2010174321. [DOI] [PubMed] [Google Scholar]
- Ximenez C., Cerritos R., Rojas L., Dolabella S., Moran P., Shibayama M., Gonzalez E., Valadez A., Hernandez E., Valenzuela O., Limon A., Partida O., Silva E.F. Human amebiasis: breaking the paradigm? Int. J. Environ. Res. Public Health. 2010;7:1105–1120. doi: 10.3390/ijerph7031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer M., Zenebe Y., Mulu W., Abera B., Saugar J.M. Molecular prevalence of Entamoeba histolytica/dispar infection among patients attending four health centres in north-West Ethiopia. Trop. Dr. 2017;47:11–15. doi: 10.1177/0049475515627236. [DOI] [PubMed] [Google Scholar]