Abstract
Rationale
Targeted cystic fibrosis (CF) therapy with lumacaftor/ivacaftor partly restores chloride channel function and improves epithelial fluid transport in the airways. Consequently, changes may occur in the microbiome, which is adapted to CF lungs.
Objectives
To investigate the effects of lumacaftor/ivacaftor on respiratory microbial composition and microbial metabolic activity by repeatedly sampling the lower respiratory tract.
Methods
This was a single-centre longitudinal observational cohort study in adult CF patients with a homozygous Phe508del mutation. Lung function measurements and microbial cultures of sputum were performed as part of routine care. An oral and nasal wash, and a breath sample, were collected before and every 3 months after starting therapy, for up to 12 months.
Results
Twenty patients were included in this study. Amplicon 16S RNA and metagenomics sequencing revealed that Pseudomonas aeruginosa was most abundant in sputum and seemed to decrease after 6 months of treatment, although this did not reach statistical significance after correction for multiple testing. Two types of untargeted metabolomics analyses in sputum showed a change in metabolic composition between 3 and 9 months that almost returned to baseline levels after 12 months of treatment. The volatile metabolic composition of breath was significantly different after 3 months and remained different from baseline until 12 months follow-up.
Conclusions
After starting CF transmembrane conductance regulator (CFTR) modulating treatment in CF patients with a homozygous Phe508del mutation, a temporary and moderate change in the lung microbiome is observed, which is mainly characterised by a reduction in the relative abundance of Pseudomonas aeruginosa.
Short abstract
Lumacaftor/ivacaftor in adult cystic fibrosis patients with homozygous Phe508del results in temporal and moderate changes in lung microbiome and metabolome, that are mainly characterised by a reduction in the relative abundance of Pseudomonas aeruginosa https://bit.ly/3pcPUfX
Introduction
Cystic fibrosis (CF) is a recessively inherent, progressive multi-organ disease that results from mutations in both copies of the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which encodes for a chloride ion channel [1]. CF-related lung disease results in mucus retention, pathogen accumulation, pulmonary inflammation and loss of lung function with subsequent chronic respiratory failure when left untreated. Treatment with CFTR modulators became available during the last decade and improves lung function, body mass index (BMI) and quality of life in specific genotypes [2–7].
The homozygous Phe508del genotype is the most common worldwide and is now routinely treated with lumacaftor/ivacaftor or ivacaftor/tezacaftor combination therapy throughout the Western world [2, 4, 7, 8]. This therapy improves CFTR function significantly, but only results in small and highly variable improvement in lung function [9]. Besides lung function, pulmonary microbial changes might be equally important in shaping the trajectory of disease in CF. Small studies in other CF genotypes suggest changes in the pulmonary microbiome after starting CFTR modulators, but these alterations have been small and transient [10–14]. In addition, microbial ecology studies in CF suggest that not only is bacterial composition important, but also the metabolic behaviour of the microbes present [15–18].
In this study, we investigated the effects of lumacaftor/ivacaftor on respiratory microbial composition and microbial metabolic activity in adult CF patients with the homozygous Phe508del genotype, in an observational cohort study in which we repeatedly sampled the lower respiratory tract. We hypothesised that starting lumacaftor/ivacaftor in CF patients with a homozygous Phe508del mutation may change the respiratory microbiome and metabolome.
Methods
Design, inclusion criteria and ethical considerations
This was a single-centre longitudinal observational cohort study, performed at the Amsterdam University Medical Center, Location AMC, after the Dutch government started insurance coverage for lumacaftor/ivacaftor in November 2017. Adult CF patients with a homozygous Phe508del mutation were sampled at five routine 3-monthly visits over the course of 12 months. The first visit was just before starting lumacaftor/ivacaftor. There were no exclusion criteria. The ethical board of the Amsterdam UMC approved the study protocol. All patients provided written informed consent.
Study procedures
The study visits were planned to be concurrent with the scheduled outpatient clinic visits. Lung function measurements and microbial cultures of sputum were performed as part of routine care. An oral and nasal wash, and an exhaled breath sample, were obtained for research purposes (see online supplement).
Measurements and processing
Microbiota analysis
Semi-quantitative bacterial cultures were performed for clinical purposes. Microbial composition was analysed by 16S rRNA gene V4–V5 sequencing and metagenomic sequencing performed at the CGEB–Integrated Microbiome Resource and Department of Pharmacology, Dalhousie University, Halifax, Canada, as previously described [19, 20] (see online supplement).
Processing of metagenomic sequences was performed with PRINSEQ'S prinseq-lite 0.20.4 to remove low-quality reads and Bowtie's bowtie2-2.2.7 to remove reads that align to the human genome [21]. The parameters for prinseq-lite were set as min_qual_read 30 and ns_max_n 0. Next, we used bowtie2 to remove quality filtered reads that aligned to the human genome hg38. The remainder of the reads were used as input for MIDAS (Metagenomic Intra-species Diversity Analysis System) version 1.2.1 [22].
Metabolomic analysis
Two types of untargeted metabolomics analyses were performed on the sputum supernatant by the West Coast Metabolomics Center (WCMC, UC Davis, Davis, CA, USA) [23]. Gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) was used to separate primary metabolite classes. Hydrophilic interaction chromatography quadrupole time-of-flight mass spectrometry (HILIC-QTOF-MS) was used for analysis of biogenic amines, after extraction in sequential ethyl acetate solvation followed by methanol solvation. For volatile metabolites, exhaled breath samples were analysed using thermal desorption (TD100; Markes) coupled to GC-MS (Shimadzu QP2010, Kyoto, Japan).
Statistical analysis
Statistical analysis was performed in R (version 3.6.1) through the R studio interface. An R-markdown file with the code and output that outlines all analytical steps is available as a supplementary file for full transparency. For all analysis, the baseline visit, before the start of lumacaftor/ivacaftor, was used as the reference. Patients with missing samples/data were not included in the individual analyses. Microbiome data were analysed using the vegan package for diversity measurements and comparisons of composition. PERMANOVA was used to compare microbial composition between visits. The paired Wilcoxon signed rank test was used to compare univariate, non-compositional data between visits. The most important changes in metabolomics data were identified using sparse partial least square discriminant analysis (sPLS-DA) via the mixOmics package [24, 25] after principal components analysis was used for outlier detection. The parameters of the sPLS-DA model were tuned for the optimal number of variables in two predefined projections. Linear mixed effects models were used to evaluate the association between the relative abundance of Pseudomonas aeruginosa in sputum and changes in metabolites (package lme4, relative abundance as dependent variable, 10-log transformed metabolite concentration as independent variable, visit number as fixed effect and a random intercept per patient). A pathway was further explored when multiple metabolites in it had a strong association with the most important changes in lung microbiome composition and if the metabolites could be linked to the micro-organism via the KEGG database. A p-value below 0.05 was considered significant. The p-value was adjusted for multiple comparison (false discovery rate according to the Benjamini–Hochberg method).
Results
The included patients did not improve in lung function or BMI
Of the 45 patients that were eligible to participate, the first 20 patients scheduled for a routine visit were included in this study. Nine patients were excluded because of compassionate use of lumacaftor/ivacaftor, and five were not interested in participating in the study (figure E1). Table 1 shows the patient characteristics. The median patient age was 25 years (IQR: 22.0–28.5). The included patients all started lumacaftor/ivacaftor treatment and one patient stopped due to side effects after 3 months follow-up. For this patient, only data collected up to 3 months after starting treatment were included in the analyses. Antibiotic use was patient-specific (table E5). After a year of treatment, forced expiratory volume in 1 s (FEV1) did not change (mean at start: 77.5±23.1% predicted, mean after a year: 76.9±27.4%, p=0.61, figure E2) and BMI did not change (mean at start: 21.6±3.1, mean after a year: 21.1±3.3, p=0.51, figure E3). Sweat chloride decreased from a mean±sd of 95±12.2 mmol·L−1 before start of treatment to a mean of 79±19.2 mmol·L−1 (p=0.00089) after treatment.
TABLE 1.
Baseline patient characteristics
| Age years | 25 (22.0–28.5) |
| Male | 12 (60) |
| BMI kg·m−2 | 21.6 (20.2–23.3) |
| Colonisation | |
| Pseudomonas aeruginosa | 10 (50) |
| Staphylococcus aureus | 15 (75) |
| FEV1 L | 2.92 (2.19–3.77) |
| FEV1 % pred | 76 (60–93) |
| FVC L | 4.44 (3.30–5.59) |
| FVC % pred | 98 (83–107) |
| Exacerbation in 3 months before inclusion | 2 (10) |
Data are presented as median (interquartile range) or n (%). BMI: body mass index; FEV1: forced expiratory volume in 1 s; FVC: forced vital capacity.
Microbial composition of the respiratory tract
Sixteen patients were able to expectorate sputum at the first visit, before start of therapy (figure E1). 10 patients (50%) were chronically colonised with Pseudomonas aeruginosa and 15 (75%) with Staphylococcus aureus based on sputum or cough swab culture results (figure E5 and E6).
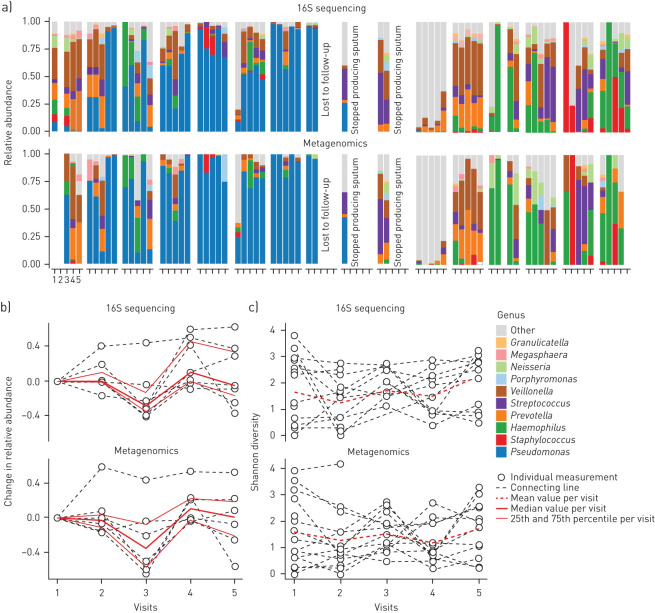
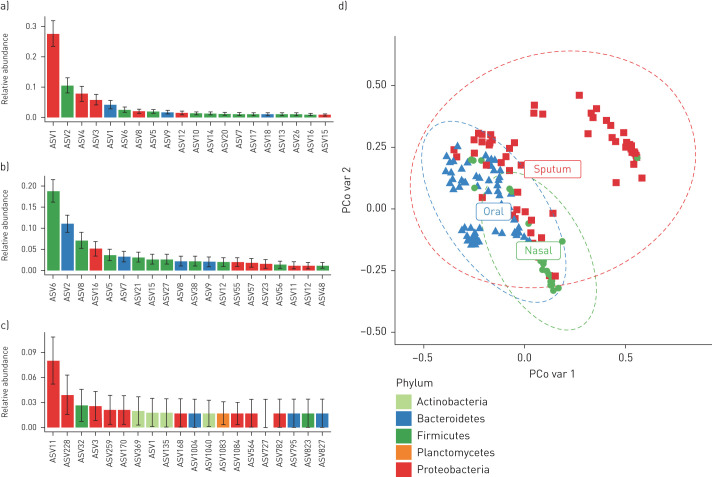
An overview of all the collected samples and performed analyses for each individual patient can be found in table E1. Sequence data have been submitted to the National Center for Biotechnology Information's Sequence Read Archive under the following accession number: BioProject PRJNA599290. 16S rRNA sequencing revealed that the most abundant amplicon sequence variant (ASV) in sputum was Pseudomonas, followed by oral bacteria such as Veillonella, Prevotella and Streptococcus (figure 1). The composition of the sputum microbiome was different from the oral and nasal microbiome (figure 2; p<0.0001). The microbial composition was unique per patient (figure 1a) and the genus Pseudomonas appears to decrease after 6 months of treatment (figure 1b), although this did not reach statistical significance after correction for multiple testing (unadjusted PERMANOVA p=0.001; adjusted p=0.48) and was not accompanied by an increase in alpha-diversity (figure 1c; p=0.63).
FIGURE 1.
Change in the lung microbiome during lumacaftor/ivacaftor treatment. a) Composition of the lung microbiome measured in sputum by 16S rRNA and metagenomics sequencing. The x-axis indicates the visits, with visit 1 before start of treatment and subsequent visits 3 months apart. The y-axis indicates relative abundance. The upper panel shows results from 16S sequencing and the lower panel from metagenomics analysis. Fill colours correspond to the genus level annotation. b) Cumulative relative abundance of Pseudomonas aeruginosa based on ASVs/metagenomes matched to this particular species. Only patients with a non-zero Pseudomonas aeruginosa count at baseline are included in the graph. The x-axis indicates the visits, with visit 1 before start of treatment and subsequent visits 3 months apart. The y-axis indicates absolute change in relative abundance. The red line indicates the median value per visit with the thinner lines indicating the quantile intervals. The upper panel shows results from 16S sequencing and the lower panel from metagenomics analysis. c) Shannon diversity. The x-axis indicates the visits, with visit 1 before start of treatment and subsequent visits 3 months apart. The y-axis indicates Shannon diversity. The red line indicates the mean value per visit. The upper panel shows results from 16S sequencing and the lower panel from metagenomics analysis.
FIGURE 2.
Most abundant amplicon sequence variants (ASVs) in sputum, nasal wash and oral wash. The mean relative abundance of the top 20 most prominent ASVs found in a) sputum, b) oral wash and c) nasal wash of the included patients. The bar indicates the mean relative abundance while the error bar gives the standard deviation of the mean. d) Microbial composition between sample materials, with the x-axis indicating principal coordinate 1 (PCo var 1) and the y-axis principal coordinate 2 (PCo var 2) on the Bray–Curtis dissimilarity measure of 16S microbiome data. The sputum samples are distinct from the oral and nasal samples (p<0.0001).
Compositional analysis of metagenomes of non-human DNA showed similar trends for Pseudomonas aeruginosa dynamics as with 16S rRNA analysis (figure 1; bottom graphs for each part; unadjusted PERMANOVA p=0.001; adjusted p=0.52).
Change in metabolic composition of sputum
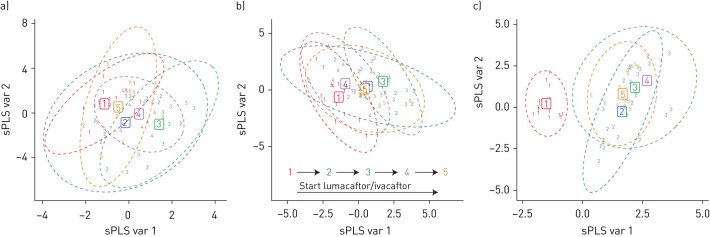
Sixty-nine sputum samples were successfully analysed by GC-TOF-MS, whereas 79 sputum samples were analysed by HILIC-TOF-MS. sPLS-DA showed a change in metabolic composition between 3 and 9 months after treatment that almost returned to baseline levels after 12 months of follow-up (figure 3). In dataset 1, the difference was mainly driven by ribitol and phenylpyruvate concentrations (table E2). In dataset 2, cortodoxone and diazepam concentrations were the most important contributors to the changing metabolic composition of sputum (table E3). None of the molecules that were selected by sPLS-DA as important variables to change during treatment are linked to metabolism of Pseudomonas aeruginosa based on the KEGG database and the currently available literature.
FIGURE 3.
Change in metabolic composition of sputum and breath during lumacaftor/ivacaftor treatment. The x-axis is the sPLS projected variable 1 (sPLS var 1, containing 9, 9 and 23% of variation) and the y-axis sPLS projected variable 2 (sPLS var 2, containing 12, 6 and 8% of variation). a and b) Change in metabolic composition of sputum after the start of lumacaftor/ivacaftor treatment. There is a significant change from visit 1 (before start of treatment) to visit 3 (6 months after treatment; p=0.0015 for dataset 1 and p=0.004 for dataset 2) with tendency to return to baseline after 12 months (p=0.031 for dataset 1 and p=0.014 for dataset 2). a) Data obtained by GC-TOF-MS (n=69). b) Data obtained by HILIC-TOF-MS (n=79). c) Change in metabolomic composition of exhaled breath (n=68) after the start of lumacaftor/ivacaftor. After start of treatment there is a significant difference in metabolic composition (p<0.0001) that does not return to baseline at the end of the observation period (p=0.0002).
Change in metabolic composition of breath
Sixty-eight breath samples were successfully analysed by TD-GC-MS. Most samples from visit 4 were missing due to sampling error; therefore, this visit was not evaluated in statistical analysis. The metabolic composition of breath was significantly different after 3 months of treatment and remained different from baseline until 12 months after treatment (figure 3). The breath concentration of 4-ethylbenzanoic acid 2-pentyl ester was the most important variable in the sPLS-DA model (table E4). Fifty-one volatile organic compounds (VOCs) (48.6%) changed in concentration 3 months after start of treatment. The majority (90.2%) of these VOCs remained significantly different at 12 months after treatment.
Reflection of change in the microbiome in metabolomics datasets
The metabolites in the three datasets were correlated with the relative abundance of Pseudomonas aeruginosa using mixed-model analysis to account for between-patient variation. After accounting for multiple testing, the false discovery rate for the association between the change in relative abundance of Pseudomonas aeruginosa and metabolite datasets 1, 2 and 3 was 8.7, 10.5 and 71.6%, respectively. Dataset 3 was disregarded for further analysis due to the high probability of chance findings.
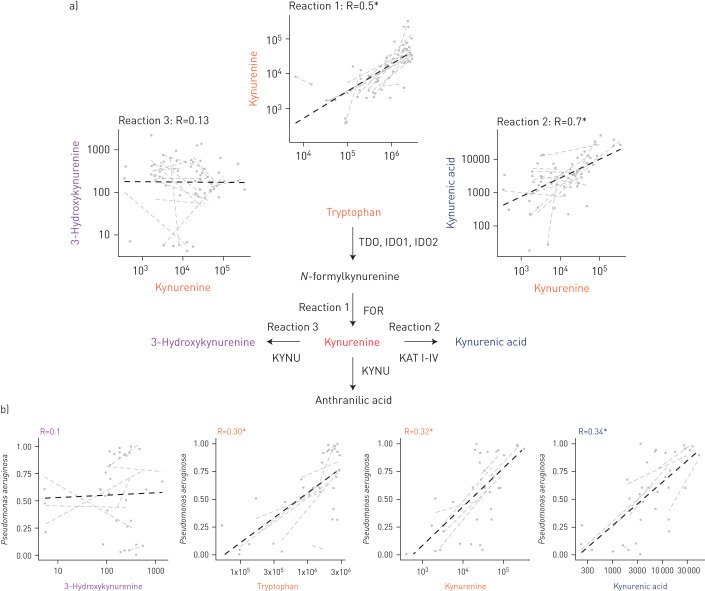
Several metabolites in the pathway of tryptophan–kynurenic acid metabolism (figure 4) were consistently associated with changes in the microbiome. This pathway is known to be of particular importance to Pseudomonas aeruginosa and was therefore further evaluated (figure 4 shows the relationships between metabolites and microbes). Pseudomonas aeruginosa abundance was significantly associated with tryptophan (B-fixed: 0.44 [95% CI: 0.27–0.61]), kynurenine (B-fixed: 0.33 [95% CI: 0.21–0.47]) and kynurenic acid (B-fixed: 0.38 [95% CI: 0.24–0.53]). In contrast, 3-hydroxykynurenine was not associated with Pseudomonas aeruginosa (B-fixed: 0.008 [95% CI: −0.19–0.21]).
FIGURE 4.
Tryptophan and its metabolites in sputum and the association with Pseudomonas aeruginosa. a) Tryptophan metabolism in Pseudomonas aeruginosa. There was a strong association between tryptophan and kynurenine and between kynurenine and kynurenic acid concentration in sputum. There was no association between kynurenine and 3-hydroxykynurenine concentration in sputum. This suggests that reaction 2 predominated over reaction 3 in the lungs of CF patients. b) Tryptophan, kynurenine and kynurenic acid are associated with relative abundance of Pseudomonas aeruginosa in sputum, but 3-hydroxykynurenine is not. FOR: formamidase; IDO: indoleamine-2,3-dioxygenase; KAT 1–IV: kynurenine aminotransferase; KYNU: kynureninase; TDO: tryptophan-2,3-dioxygenase.
Discussion
This study suggests that after starting lumacaftor/ivacaftor in CF patients with a homozygous Phe508del mutation, a temporary and moderate change in the lung microbiome is observed, mainly characterised by a reduction in the relative abundance of Pseudomonas aeruginosa. Metabolites from the tryptophan–kynurenic acid pathway follow a similar trajectory and seem to be related to the abundance of Pseudomonas aeruginosa. In contrast, the breath metabolome is distinct after start of treatment and is persistently altered after 12 months of follow-up. The identity of the molecules suggests a relationship with inflammation and oxidative stress rather than microbial metabolism. Taken together, we reject the hypothesis that starting lumacaftor/ivacaftor in patients with homozygous Phe508del has important effects on microbiological composition that would complement the moderate effects on clinical end-points such as lung function or BMI.
This is the first study to evaluate the lung microbiome and metabolome after starting lumacaftor/ivacaftor. Previous studies in patients with class III mutations treated with ivacaftor monotherapy have shown temporary reduction in Pseudomonas aeruginosa abundance in sputum [10–13]. We see a heterogeneous response across the cohort with an apparent temporary reduction in Pseudomonas aeruginosa abundance in sputum that does not persist for 12 months. Despite this difference, the studies consistently show a transient reduction followed by normalisation towards baseline levels of Pseudomonas aeruginosa, irrespective of the CF-related mutations and the treatments that have been studied so far [12].
There are several ecological explanations for this phenomenon. First, one could hypothesise that the elimination of pathogens by improved mucociliary clearance due to CFTR-targeted treatment is only temporary. Therefore, the forces of immigration and elimination quickly fall, to the advantage of elimination of pathogens like Pseudomonas. There is evidence that ivacaftor improves mucociliary clearance in class III mutations at least up to 3 months after start of treatment [26]. Data for lumacaftor/ivacaftor and longer durations of treatment are lacking, but based on the pharmacology of the drugs and the persistent improvement in sweat chloride concentrations it is likely that improved mucociliary clearance will persist after 3 months of treatment. A second hypothesis is that the airways are irreversibly damaged and that normalisation of CFTR function will not change the regional growth circumstances sufficiently to eliminate Pseudomonas aeruginosa. This could certainly partly explain the persistence of Pseudomonas, but does not explain the temporal decrease. Lastly, it can be postulated that Pseudomonas aeruginosa is challenged by the changing regional environment due to improved CFTR function [9] and improved killing capacity of macrophages [27], but adapts to the new circumstances [17, 28]. Previous studies did not identify a replacement of Pseudomonas aeruginosa by different subtypes but rather persistence of the same strain [12]. Importantly, multilocus sequence typing (MLST) analysis could miss adaptive changes to the genome and we speculate that the organism indeed adapted. Our data suggest that the metabolic environment does indeed change after 3–6 months of treatment but that the original composition is restored after 12 months. This is exemplified by the association of molecules in the tryptophan–kynurenic acid pathway and Pseudomonas aeruginosa abundance. Indeed, Pseudomonas is known to catalyse tryptophan to kynurenic acid and utilises this pathway to influence the host response [29, 30]. One could speculate that Pseudomonas aeruginosa plays a role in the restoration of regional growth factors promoting its own growth.
This study has several strength and weaknesses. The most important weaknesses of the study are the lack of a control group that did not receive lumacaftor/ivacaftor and the limited sample size. Another weakness is the lack of multiple samples prior to lumacaftor/ivacaftor, because the sputum microbiome in CF patients may also vary over time in the absence of treatment [31]. The inclusion of a control group without medication was not considered to be ethical due to the effectiveness (albeit moderate) of the medication [7]. Nevertheless, it is important to evaluate other effects besides clinical end-points, in order to gain more insight into the mechanisms underlying the beneficial effect of CFTR-modulating treatment and that is the pivotal novelty of this study. Patients who had already started lumacaftor/ivacaftor treatment in a compassionate use programme could not participate in this study due to the absence of a pre-treatment sample. This resulted in exclusion of the most severely affected CF patients with a homozygous Phe508del mutation in our hospital. Therefore, interpretation and extrapolation of these results to this specific group of patients has to be performed with caution. Thirdly, we did not investigate any changes in medication that occurred during the 12 months of follow-up other than changes in antibiotic exposure. Changes in the use of medication could partly explain the changes in metabolic composition after start of lumacaftor/ivacaftor therapy. The major strength of this paper is the combination of serial measurements: two techniques for evaluation of microbial composition and three techniques for metabolomic profiling [15]. In this way we obtained a comprehensive picture of the microbial and metabolic landscape and how it changes during lumacaftor/ivacaftor treatment. Our analytical approach was in line with the recently published recommendation from the American Thoracic Society's Assembly on Allergy, Immunology and Inflammation [32]. Furthermore, breath analysis allowed us to also evaluate patients who were unable to expectorate sputum.
The two methods for determining the microbial composition of sputum and the two metabolomics analyses of sputum showed similar trends after the start of lumacaftor/ivacaftor treatment. Breath metabolomics, however, revealed a more persistent change that was characterised by an increase in oxidative stress markers. Because the samples were collected at different visits ranging over the course of a year but require analysis within 2 weeks, batch effects should be investigated as an explanation for these differences. Retention time shift and changes in the sensitivity of the mass spectrometer were evaluated but did not explain the sudden change in metabolic profile. Alternatively, the patients could have changed their behaviour (e.g. eating behaviour and/or physical activity behaviour), which might bias the metabolic content of their breath. In order to explain our findings, the patients would have had to do so unanimously. We can disregard this explanation because patients responded very differently; some exacerbated frequently (figure E4) and had to decrease activities, while others improved and became more active. The metabolites of the drugs were also not identified in breath. Therefore, we might attribute the change in metabolic composition of the breath to the effects of treatment, possibly on the inflammatory response in the lung. Unfortunately, the sputum sample was of insufficient volume to perform additional analyses after the predefined analyses were performed, so a stronger link with inflammation could not be established.
The results presented in this manuscript have several important implications. As Pseudomonas aeruginosa possibly adapts to the improved CFTR activity and the regional growth conditions are restored, these types of treatment have particular potential in cases where Pseudomonas aeruginosa has not yet colonised the lung. Furthermore, there might be a window of opportunity to attempt to eliminate pathogens after 3–6 months of treatment, when they seem to struggle to persist. It is important to test this speculation in randomised controlled trials that coincide with the initiation of CFTR-targeted treatment. Finally, our study provides an overview of sampling and analysis procedures that might be copied to patients starting on triple therapy. Transient changes can only be evaluated when treatment is started and the approval of the newest generation of drugs provides a unique opportunity for the scientific community to better understand the influence of these therapies on the airway microbiome.
In conclusion, after starting lumacaftor/ivacaftor in CF patients with homozygous Phe508del, a temporary and moderate change in the lung microbiome is observed that is mainly characterised by a reduction in the relative abundance of Pseudomonas aeruginosa. Permanent changes to the exhaled breath metabolome were also found. These findings may provide a starting point to better understand the mechanisms through which CFTR-modulating treatment exerts its beneficial effect in CF patients.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00731-2020.SUPPLEMENT (702.6KB, pdf)
Footnotes
This article has supplementary material available from openres.ersjournals.com.
Members of the Amsterdam Mucociliary Clearance Disease Research Group: E. Haarman (Amsterdam UMC, Vrije Universiteit Amsterdam, Department of Pediatric Medicine, Amsterdam, The Netherlands); N.W. Rutjes (Amsterdam UMC, University of Amsterdam, Department of Pediatric Respiratory Medicine and Allergy, Amsterdam, The Netherlands); S.W.J. Terheggen-Lagro (Amsterdam UMC, University of Amsterdam, Department of Pediatric Respiratory Medicine and Allergy, Amsterdam, The Netherlands); D. Seljogi (Amsterdam UMC, University of Amsterdam, Department of Pediatric Respiratory Medicine and Allergy, Amsterdam, The Netherlands); E.M. Kemper (Amsterdam UMC, University of Amsterdam, Department of Clinical Pharmacology, Amsterdam, The Netherlands); R. Lutter (Amsterdam UMC, University of Amsterdam, Department of Experimental Immunology, Amsterdam, The Netherlands); S.J. Vijverberg (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); S.E.M. Vonk (Amsterdam UMC, University of Amsterdam, Department of Clinical Pharmacology, Amsterdam, The Netherlands); N. Adriaens (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); R. Lub (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); M. van Brederode (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); L. van der Schaaf (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); M. Verkleij (Amsterdam UMC, University of Amsterdam, Department of Pediatric Psychology, Amsterdam, The Netherlands); N.A. van Gilst (Amsterdam UMC, University of Amsterdam, Department of Pediatric Respiratory Medicine and Allergy, Amsterdam, The Netherlands); G.H. Hofsteenge (Amsterdam UMC, University of Amsterdam and Vrije Universiteit Amsterdam, Department of Nutrition and Dietetics, Amsterdam, The Netherlands); C.L.H. Brackel (Amsterdam UMC, Vrije Universiteit Amsterdam, Department of Pediatric Medicine, Amsterdam, The Netherlands); P. Lakeman (Amsterdam UMC, University of Amsterdam, Department of Clinical Genetics, Amsterdam, The Netherlands); I.C.M. Bon (Amsterdam UMC, Vrije Universiteit Amsterdam, Department of Pediatric Respiratory Medicine, Amsterdam, The Netherlands);and S.P.M. Tanner (Amsterdam UMC, Location VUmc, Department of Pediatric Respiratory Medicine, Amsterdam, The Netherlands).
Members of the Amsterdam UMC Breath Research Group: P.J. Sterk (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); C. Longo (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); A. Sinha (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); D. Fenn (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); A. Lammers (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); L.B. Richards (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); J.M. van Bragt (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); R. Kos (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); J.W.F. Dagelet (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); S.J.A. Lone-Latif (Amsterdam UMC, University of Amsterdam, Department of Respiratory Medicine, Amsterdam, The Netherlands); M.J. Schultz (Amsterdam UMC, University of Amsterdam, Department of Intensive Care, Amsterdam, The Netherlands); M.R. Smit (Amsterdam UMC, University of Amsterdam, Department of Intensive Care, Amsterdam, The Netherlands); L.A. Hagens (Amsterdam UMC, University of Amsterdam, Department of Intensive Care, Amsterdam, The Netherlands).
Author contributions: Substantial contributions to the conception or design of the work were made by A.H. Neerincx, K. Whiteson, C.J. Majoor, A.H. Maitland-van der Zee and L.D.J. Bos. The acquisition and analysis of the data were performed by A.H. Neerincx, J.L. Phan, P. Brinkman, M.I. Abdel-Aziz, E.J.M. Weersink, J. Altenburg, C.J. Majoor and L.D.J. Bos. Interpretation of data for the work was performed by A.H. Neerincx, K. Whiteson, J.L. Phan, P. Brinkman, M.I. Abdel-Aziz, A.H. Maitland-van der Zee and L.D.J. Bos. Drafting the work or revising it critically for important intellectual content was performed by all authors. Final approval of the version to be published and agreement to be accountable were given by all authors.
Data availability: Sequence data have been submitted to the National Center for Biotechnology Information's Sequence Read Archive under the following accession number: BioProject PRJNA599290.
Conflict of interest: A.H. Neerincx has nothing to disclose.
Conflict of interest: K. Whiteson has nothing to disclose.
Conflict of interest: J.L. Phan has nothing to disclose.
Conflict of interest: P. Brinkman has nothing to disclose.
Conflict of interest: M.I. Abdel-Aziz reports an Egyptian Government PhD Scholarship outside the submitted work.
Conflict of interest: E.J.M. Weersink has nothing to disclose.
Conflict of interest: J. Altenburg has nothing to disclose.
Conflict of interest: C.J. Majoor has nothing to disclose.
Conflict of interest: A.H. Maitland-van der Zee reports an Innovation Grant from Vertex outside the submitted work.
Conflict of interest: L.D.J. Bos has nothing to disclose.
Support statement: L.F.J. Bos is supported by the Dutch Lung Foundation (Longfonds) Young Investigator Award. The authors have no additional financial interests. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Stoltz DA, Meyerholz DK, Welsh MJ. Origins of cystic fibrosis lung disease. N Engl J Med 2015; 372: 351–362. doi: 10.1056/NEJMra1300109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies JC, Moskowitz SM, Brown C, et al. . VX-659–tezacaftor–ivacaftor in patients with cystic fibrosis and one or two Phe508del alleles. N Engl J Med 2018; 379: 1599–1611. doi: 10.1056/NEJMoa1807119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman LR, Ramsey BW. Cystic fibrosis therapeutics: the road ahead. Chest 2013; 143: 207–213. doi: 10.1378/chest.12-1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keating D, Marigowda G, Burr L, et al. . VX-445–tezacaftor–ivacaftor in patients with cystic fibrosis and one or two Phe508del alleles. N Engl J Med 2018; 379: 1612–1620. doi: 10.1056/NEJMoa1807120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey BW, Davies J, McElvaney NG, et al. . A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med 2011; 365: 1663–1672. doi: 10.1056/NEJMoa1105185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor-Cousar JL, Munck A, McKone EF, et al. . Tezacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508del. N Engl J Med 2017; 377: 2013–2023. doi: 10.1056/NEJMoa1709846 [DOI] [PubMed] [Google Scholar]
- 7.Wainwright CE, Elborn JS, Ramsey BW, et al. . Lumacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med 2015; 373: 220–231. doi: 10.1056/NEJMoa1409547 [DOI] [PubMed] [Google Scholar]
- 8.Bobadilla JL, Macek M Jr, Fine JP, et al. . Cystic fibrosis: a worldwide analysis of CFTR mutations—correlation with incidence data and application to screening. Hum Mutat 2002; 19: 575–606. doi: 10.1002/humu.10041 [DOI] [PubMed] [Google Scholar]
- 9.Gräber S, Dopfer C, Naehrlich L, et al. . Effects of Lumacaftor-Ivacaftor Therapy on CFTR Function in Phe508del Homozygous Patients with Cystic Fibrosis. Eur Respiratory Soc 2018; 197: 1433–1442. [DOI] [PubMed] [Google Scholar]
- 10.Bernarde C, Keravec M, Mounier J, et al. . Impact of the CFTR-potentiator ivacaftor on airway microbiota in cystic fibrosis patients carrying a G551D mutation. PLoS One 2015; 10: e0124124. doi: 10.1371/journal.pone.0124124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris JK, Wagner BD, Zemanick ET, et al. . Changes in airway microbiome and inflammation with ivacaftor treatment in patients with cystic fibrosis and the G551D mutation. Ann Am Thorac Soc 2020; 17: 212–220. doi: 10.1513/AnnalsATS.201907-493OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hisert KB, Heltshe SL, Pope C, et al. . Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am J Respir Crit Care Med 2017; 195: 1617–1628. doi: 10.1164/rccm.201609-1954OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peleg AY, Choo JM, Langan KM, et al. . Antibiotic exposure and interpersonal variance mask the effect of ivacaftor on respiratory microbiota composition. J Cyst Fibros 2018; 17: 50–56. doi: 10.1016/j.jcf.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 14.Rogers GB, Taylor SL, Hoffman LR, et al. . The impact of CFTR modulator therapies on CF airway microbiology. J Cyst Fibros 2020; 19: 359–364. doi: 10.1016/j.jcf.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bevivino A, Bacci G, Drevinek P, et al. . Deciphering the ecology of cystic fibrosis bacterial communities: towards systems-level integration. Trends Mol Med 2019; 25: 1110–1122. doi: 10.1016/j.molmed.2019.07.008 [DOI] [PubMed] [Google Scholar]
- 16.Quinn RA, Lim YW, Mak TD, et al. . Metabolomics of pulmonary exacerbations reveals the personalized nature of cystic fibrosis disease. PeerJ 2016; 4: e2174. doi: 10.7717/peerj.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn RA, Lim YW, Maughan H, et al. . Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung. MBio 2014; 5: e00956-13. doi: 10.1128/mBio.00956-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whiteson KL, Meinardi S, Lim YW, et al. . Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2, 3-butanedione fermentation. ISME J 2014; 8: 1247–1258. doi: 10.1038/ismej.2013.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comeau AM, Douglas GM, Langille MG. Microbiome helper: a custom and streamlined workflow for microbiome research. MSystems 2017; 2: e00127-16. 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan BJ, McMurdie PJ, Rosen MJ, et al. . DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13: 581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27: 863–864. doi: 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nayfach S, Rodriguez-Mueller B, Garud N, et al. . An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 2016; 26: 1612–1625. doi: 10.1101/gr.201863.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cajka T, Fiehn O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal Chem 2016; 88: 524–545. doi: 10.1021/acs.analchem.5b04491 [DOI] [PubMed] [Google Scholar]
- 24.Lê Cao K-A, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 2011; 12: 253. doi: 10.1186/1471-2105-12-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohart F, Gautier B, Singh A, et al. . mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 2017; 13: e1005752. doi: 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donaldson SH, Laube BL, Corcoran TE, et al. . Effect of ivacaftor on mucociliary clearance and clinical outcomes in cystic fibrosis patients with G551D-CFTR. JCI insight 2018; 3: e122695. doi: 10.1172/jci.insight.122695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnaby R, Koeppen K, Nymon A, et al. . Lumacaftor (VX-809) restores the ability of CF macrophages to phagocytose and kill Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol 2018; 314: L432–L438. doi: 10.1152/ajplung.00461.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol 2017; 7: 39. doi: 10.3389/fcimb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bortolotti P, Hennart B, Thieffry C, et al. . Tryptophan catabolism in Pseudomonas aeruginosa and potential for inter-kingdom relationship. BMC Microbiol 2016; 16: 1–10. doi: 10.1186/s12866-016-0756-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirthgen E, Hoeflich A, Rebl A, et al. . Kynurenic acid: the Janus-faced role of an immunomodulatory tryptophan metabolite and its link to pathological conditions. Front Immunol 2018; 8: 1957. doi: 10.3389/fimmu.2017.01957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caverly LJ, Lu J, Carmody LA, et al. . Measures of cystic fibrosis airway microbiota during periods of clinical stability. Ann Am Thorac Soc 2019; 16: 1534–1542. doi: 10.1513/AnnalsATS.201903-270OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carney SM, Clemente JC, Cox MJ, et al. . Methods in lung microbiome research. Am J Respir Cell Mol Biol 2020; 62: 283–299. doi: 10.1165/rcmb.2019-0273TR [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material 00731-2020.SUPPLEMENT (702.6KB, pdf)