Abstract
Purpose
An effective biomarker for the diagnosis of breast cancer (BC) and benign breast diseases (BBD) is crucial for improving the prognosis. We investigated whether N6-methyladenosine (m6A) can be a diagnostic biomarker of BC.
Materials and Methods
We detected the contents of peripheral blood m6A in 62 patients with BC, 41 patients with BBD, and 41 normal controls (NCs) using the colorimetric method. The relative expression of the m6A regulated genes methyltransferase-like 14 (METTL14) and fat mass and obesity-associated (FTO) was analyzed using quantitative real-time polymerase chain reaction.
Results
m6A in peripheral blood RNA was significantly higher in patients with BC than that in patients with BBD (p < 0.001) or the NCs (p < 0.001). m6A was closely associated with the disease stage (from stage 0 to stage I-IV, p=0.003). The receiver operating characteristic curve of m6A contained an area under the curve (AUC) value of 0.887 in BC, which was greater than that of carcinoembryonic antigen (CEA) or carbohydrate antigen 153 (CA153). The combination of m6A, CEA, and CA153 improved the AUC to 0.914. The upregulated and downregulated mRNA expression of METTL14 and FTO, respectively, might contribute to the increase of m6A in patients with BC. m6A combined with METTL14 and FTO improved the AUC to 0.929 with a specificity of 97.4% in the peripheral blood of patients with BC.
Conclusion
The peripheral blood RNA of m6A might be a valuable biomarker for the diagnosis of BC.
Keywords: Breast neoplasms, Benign breast diseases, N6-methyladenosine, Biomarker, Peripheral blood RNA
Introduction
Breast cancer (BC) is one of the most commonly diagnosed malignancies and is the second most common cause of cancer deaths in women [1]. Approximately 2.1 million new cases were diagnosed in 2018, from those of 630,000 women were died of BC, inaccessible and unaffordable treatment often coincides with the tragic consequences of delayed diagnosis [2]. In general, early BC has a 5-year survival rate of > 80% with a good prognosis during the early stages and with basic treatment [3]. In the past few decades, several methods have been used to clinically diagnose BC. These include breast ultrasonography, mammography, exfoliative cytology, and tumor markers that include carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), and carbohydrate antigen 153 (CA153) [4,5]. However, the sensitivity or specificity is not sufficient to screen for BC, or especially, for the early diagnosis of BC. Therefore, a simple, sensitive, and effective diagnostic method is needed to detect BC at an early stage and obtain a better prognosis.
Post-transcriptional modification has become an important regulator in the disease progression of a variety of diseases. Among these, N6-methyladenosine (m6A) is the most abundant mRNA modification. This modification can be installed using methyltransferase and removed using demethylase [6]. Functions of m6A change the expression of target genes and affect the corresponding physiological functions [7]. Additionally, modification of m6A due to methylation in various cancers has been reported recently. The findings have implicated m6A modification as a prognostic target of digestive tract tumors and suggest that m6A modification promotes the brain metastasis of lung cancer [8,9]. More importantly, several articles have suggested that the development of BC may be related to the disharmony of m6A methylation [10,11].
The modification of m6A is a dynamic and reversible process which catalyzed by the methyltransferase complex consisting of the methyltransferase-like 3 and 14 proteins (METTL3 and METTL14) with their cofactors Wilms tumor 1 associated protein (WTAP), RNA binding motif protein 15, and Vir like m6A methyltransferase associated, among others [12]. The reversible process relies on demethylases including fat mass along with obesity-associated protein (FTO) and its homologue AlkB homolog 5 (ALKBH5) to ensure the balance of m6A modification in transcription [7]. In recent years, peripheral blood molecular markers as “liquid biopsies” have gained recognition because of their convenience, good reproducibility, and early detection of cancer. Many blood-based biomarkers have been used in the diagnosis of cancer [13]. m6A had been reported as a novel potential biomarker of type 2 diabetes mellitus in peripheral blood [14]. However, whether the m6A modification in peripheral blood RNA can be a novel diagnostic biomarker for BC has not been investigated.
In this work, we examined m6A that was upregulated in peripheral blood RNA in patients with BC compared to that in patients with benign breast disease (BBD) and normal controls (NCs). The comparison assessed the independent and combined diagnostic values of m6A in peripheral blood RNA, with the aim of exploring whether m6A might be a diagnostic biomarker for BC. We also investigated the upregulation of the METTL14 gene and the downregulation of the FTO gene that accompanied the increase of m6A to explore whether these gene expression patterns contribute to m6A as a biomarker for the early screening and diagnosis of patients with BC.
Materials and Methods
1. Patients and blood samples
Peripheral blood samples from 62 female patients with BC (including 53 tumor resection and 9 without surgery), 41 female patients with BBD (benign fibroadenoma of breast or mastopathy), and 41 female NCs who sought a routine health check-up during the same period and who did not have any breast diseases or other cancerous diseases. All patients were recruited at the Zhongda Hospital of Southeast University from August 2018 to May 2019. All samples were initially diagnosed and untreated (including surgery, chemotherapy, and radiotherapy). There was no history of basic or chronic diseases. Data including age, histologic grade, lymph node metastasis, and TNM stage were recorded. The BC stage was evaluated using the TNM system of the American Joint Committee on Cancer [15]. The background demographic characteristics of the participating patients and NCs are described in S1 Table. Of the 62 patients with BC, 53 underwent surgery, and nine received only chemotherapy or radiotherapy. One milliliter of peripheral blood sample was individually collected in a tube coated with ethylenediamine tetra acetic acid to prevent coagulation. The blood was immediately mixed with 3 mL RNALock Reagent (Tiangen, Beijing, China) to stabilize RNA. The mixed samples were stored at −80°C for no longer than 3 months.
2. Extraction of total RNA in peripheral blood
Total RNA was isolated from 1 mL peripheral whole blood using a commercially available RNAprep Pure Blood Kit (Tiangen) according to the manufacturer’s protocol. RNA quality and yield were measured using spectrophotometry with a NanoDrop 2000 device (Thermo Fisher Scientific, Waltham, MA) at optical densities of 260 and 280 nm. Reverse-transcription from RNA to cDNA was accomplished using PrimeScript RT Master Mix (TaKaRa Bio, Shiga, Japan) according to the manufacturer’s instructions.
3. Colorimetric quantification of RNA m6A
m6A in total RNA was measured using a colorimetric method with EpiQuik m6A RNA Methylation Quantification Kit (Epigentek, Farmingdale, NY) according to the manufacturer’s protocol. Briefly, 200 ng isolated RNA was added to detect wells covered by the binding mixed solution with incubation at 37°C for 90 minutes. After washing three times, the capture antibody solution, detection antibody solution, and enhancer solution were added to detect wells containing various sample dilutions at room temperature according the manufacturer’s instructions. Color developing solution and stop solution were added and the absorbance of each well was measured at a wavelength of 450 nm. A standard curve ranging from 0.02 to 1 ng of m6A was constructed. The m6A levels were calculated based on this standard curve.
4. Analysis of METTL14 and FTO mRNA expression using quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction was performed to quantify the expression levels of METTL14 and FTO using Green Premix Ex Taq II (TaKaRa Bio) and CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The cycle parameters were 95°C for 5 minutes, 40 cycles of 95°C for 10 seconds, 60°C for 30 seconds, and 65°C for 5 seconds. METTL14 quantitative real time polymerase chain reaction primers used were as follows: forward, 5′-AGTGCCGACAGCATTGGTG-3′ and reverse, 5′-GGAG- CAGAGGTATCATAGGAAGC-3′. FTO quantitative real-time polymerase chain reaction primers used were: forward, 5′-TTGGACGGTACAGATATGGAACATTTT-3′ and reverse, 5′-TCTTTTAGTTTCT-TTGCCTTTGGGGAT-3′. mRNA levels were normalized to reference gene sequences of β-actin forward, 5′-CTGGAACGGTGAAGGTGACA-3′ and reverse, 5′-AAGGGACTTCCTGTAACAATGCA-3′. The absolute expression levels of METTL14 and FTO were calculated using the 2−ΔΔCt method.
5. Statistical analysis
All statistical analysis was performed using SPSS Statistics ver. 25.0 software package (IBM Corp., Armonk, NY) and MedCalc ver. 17.0 (MedCalc, Mariakerke, Belgium). Two-tailed unpaired Student’s t tests were used to analyse data unless otherwise indicated. One-Way ANOVA were analyzed for data of three or more groups by Bonferroni test. We report the nominal p-value for each comparison without adjusting for multiple testing. A p-value < 0.05 was considered statistically significant.
Results
1. Upregulation of peripheral blood m6A in patients with BC
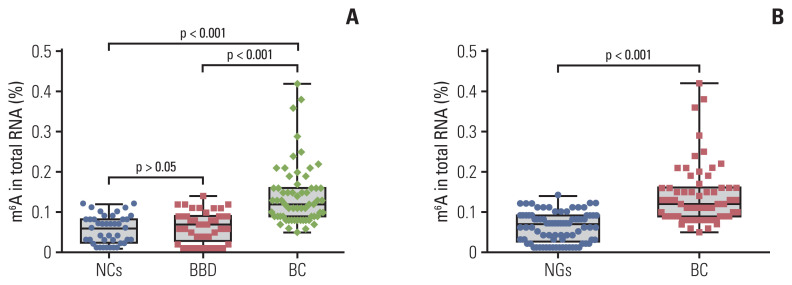
A total of 144 RNA samples (62 patients with BC, 41 patients with BBD, and 41 NCs) were measured using the colorimetric method to determine whether m6A was higher in patients with BC. As shown in Fig. 1A, the mean contents of m6A in RNA from patients with BC, patients with BBD, and NCs were 0.141±0.076%, 0.066±0.039%, and 0.056±0.035%, respectively. m6A in peripheral blood RNA of patients with BBD and the NCs was not significantly different (p > 0.05). The m6A in the peripheral blood RNA of patients with BC was significantly higher than that in the NCs (p < 0.001) and patients with BBD (p < 0.001). In addition, m6A in patients with BC was significantly distinguished from that in the noncancerous groups (NGs; 41 BBD, and 41 NCs), with its expression being significantly upregulated in patients with BC compared to that in the NGs (p < 0.001) (Fig. 1B). These results indicated that the upregulation of peripheral blood m6A in patients with BC can differentiate patients with BC from the patients with BBD or NCs.
Fig. 1.
Quantification and statistical analysis of N6-methyladenosine (m6A) from peripheral blood RNA in patients with breast cancer (BC), patients with benign breast diseases (BBD), normal controls (NCs), and noncancerous groups (NGs). m6A in peripheral blood RNA from 62 patients with BC, 41 patients with BBD and 41 NCs (A), or 82 patients from the NGs (B). Bars represent the mean±standard deviation of the results from replicate measurements.
2. Association between peripheral blood m6A and clinicopathological characteristics in BC
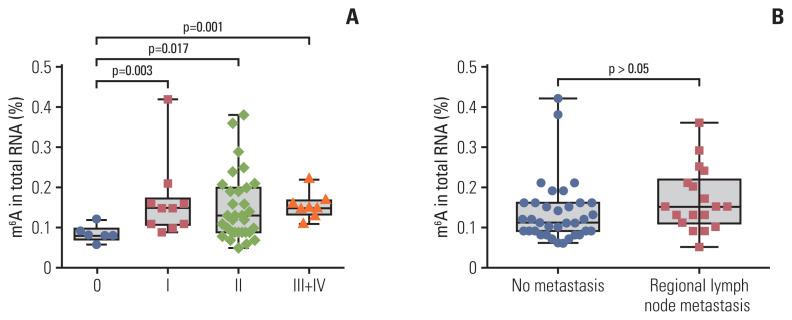
Associations between peripheral blood m6A levels and clinicopathological parameters in patients with BC are presented in Table 1. The m6A level of peripheral blood was closely associated with stage (from stage 0 to stage I–IV, p=0.003). m6A in patients with BC with carcinoma in situ (0 stage, n=6, 0.085±0.020) was significantly different from that in stage I (n=10, 0.166±0.096), II (n=31, 0.152±0.084), and III+IV (n=8, 0.155±0.032) in peripheral blood RNA (Fig. 2A). The m6A level in stage I–IV patients with BC was significantly higher than the level at stage 0 (p=0.005). However, there were no associations in the m6A level with age, T classification, M classification, regional lymph node metastasis, presence/absence of triple-negative breast cancer, and other clinical tumor markers that included α-fetoprotein, CEA, CA125, and CA153 (Table 1, Fig. 2B).
Table 1.
Association between the m6A contents and the clinicopathological characteristics in BC patients
| Variable | No. of patients | Peripheral blood m6A contents mean±SD (%) | p-value |
|---|---|---|---|
| Age (yr) | |||
| < 50 | 22 | 0.141±0.070 | 0.795 |
| ≥ 50 | 40 | 0.141±0.080 | |
| TNM | |||
| 0 | 6 | 0.085±0.020 | 0.968 |
| I | 10 | 0.166±0.096 | |
| II | 31 | 0.152±0.084 | |
| III | 6 | 0.143±0.021 | |
| IV | 2 | 0.190±0.042 | |
| Stage | |||
| 0 | 6 | 0.085±0.020 | 0.003 |
| I–IV | 49 | 0.155±0.080 | |
| T category | |||
| Tis | 6 | 0.085±0.020 | 0.129 |
| T1–T2 | 41 | 0.155±0.086 | |
| T3–T4 | 6 | 0.142±0.029 | |
| N category | |||
| N0 | 35 | 0.135±0.078 | 0.078 |
| N1 | 15 | 0.171±0.085 | |
| N2–N3 | 3 | 0.143±0.031 | |
| M category | |||
| M0 | 53 | 0.146±0.079 | 0.174 |
| M1 | 2 | 0.190±0.042 | |
| TNBC | |||
| Yes | 8 | 0.150±0.068 | 0.72 |
| No | 47 | 0.147±0.081 | |
| AFP (ng/mL) | |||
| < 6 | 59 | 0.142±0.077 | 0.735 |
| ≥ 6 | 3 | 0.127±0.076 | |
| CEA (ng/mL) | |||
| < 5 | 54 | 0.146±0.079 | 0.213 |
| ≥ 5 | 8 | 0.108±0.039 | |
| CA125 (IU/mL) | |||
| < 35 | 55 | 0.141±0.079 | 0.649 |
| ≥ 35 | 7 | 0.140±0.049 | |
| CA153 (IU/mL) | |||
| < 25 | 58 | 0.141±0.077 | 0.985 |
| ≥ 25 | 4 | 0.143±0.078 | |
AFP, α-fetoprotein; BC, breast cancer; CA125, cancer antigen 125; CA153, carbohydrate antigen 153; CEA, carcinoembryonic antigen; m6A, N6-methyladenosine; SD, standard deviation; TNBC, triple-negative breast cancer.
Fig. 2.
Statistical analysis of N6-methyladenosine (m6A) in peripheral blood RNA with different clinical characteristics in patients with breast cancer. m6A in total peripheral blood RNA of patients with breast cancer at different clinical stages (6 stage 0, 10 stage I, 31 stage II, 6 stage III, and 2 stage IV) (A) and those with no metastasis or regional lymph node metastasis (B). Bars represent the mean±standard deviation of the results from replicate measurements.
3. Diagnostic value of peripheral blood m6A in patients with BC
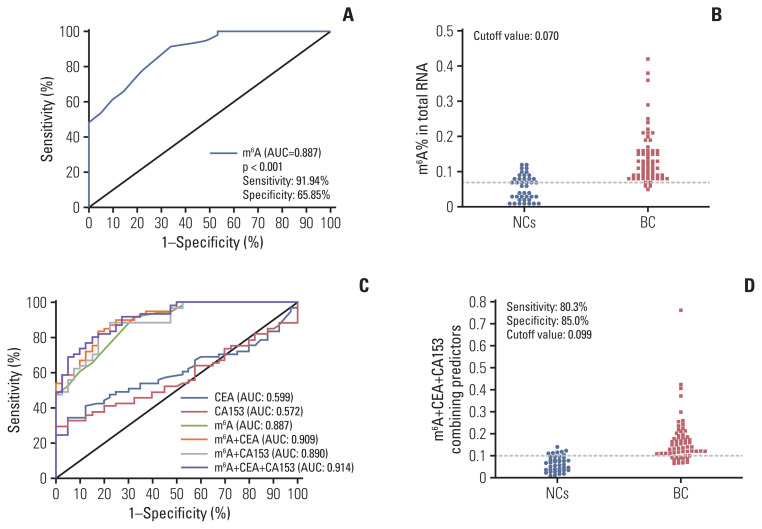
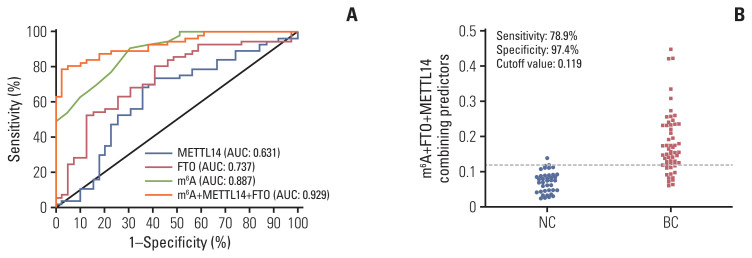
To assess whether the m6A in peripheral blood RNA had diagnostic value for patients with BC, the receiver operating characteristic (ROC) curve was plotted to identify a cutoff value that would differentiate patients with BC from NCs. As shown in Fig. 3A and B and Table 2, the area under the curve (AUC) for m6A was 0.887 (95% confidence interval [CI], 0.826 to 0.948; p < 0.001) and the optimal cutoff value was 0.070 (sensitivity 91.94%, specificity 65.85%), as determined using the highest Youden index. ROC curve analysis of other clinical tumor indicators revealed an AUC of 0.599 (95% CI, 0.489 to 0.709) and 0.572 (95% CI, 0.461 to 0.683) for CEA and CA153, respectively (Table 2, Fig. 3C). These results indicated that the diagnostic value of m6A alone was better than that of CEA or CA153. Additionally, the AUC for the combination of m6A, CEA, and CA153 improved to 0.914 (95% CI, 0.861 to 0.966; p < 0.001) with a specificity of 89.2% specificity with compromised sensitivity 80.3% as the optimal cutoff value for 0.0992 (Fig. 3D). The findings validated the use of m6A alone in peripheral blood RNA as a biomarker to enhance the diagnostic sensitivity for detection of patients with BC and indicated that combining CEA and CA153 with it yielded better specificity for BC.
Fig. 3.
Diagnostic value for peripheral blood RNA of N6-methyladenosine (m6A) alone or combination with carcinoembryonic antigen (CEA) and carbohydrate antigen 153 (CA153) to diagnose patients with breast cancer (BC). Receiver operating characteristic (ROC) curve analysis for m6A in the diagnosis of BC (A) and cutoff value of m6A for normal controls (NCs) and patients with BC (B). ROC curve analysis of m6A compared and combined diagnosis with CEA and CA153 (C) and cutoff value analysis for the combination of m6A, CEA, and CA153 in NCs and patients with BC (D). AUC, area under the curve.
Table 2.
The sensitivities and specificities of diagnostic value about various marker alone and their combination test
| Marker | NCs vs. BC | ||||
|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | AUC | p-value | 95% CI | |
| m6A | 91.94 | 65.85 | 0.887 | < 0.001 | 0.826–0.948 |
| CEA | 34.43 | 95.00 | 0.599 | 0.077 | 0.489–0.709 |
| CA153 | 29.51 | 100.00 | 0.572 | 0.205 | 0.461–0.683 |
| METTL14 | 70.00 | 65.00 | 0.643 | 0.016 | 0.527–0.760 |
| FTO | 52.54 | 85.00 | 0.719 | < 0.001 | 0.615–0.823 |
| m6A+CEA | 83.61 | 82.50 | 0.909 | < 0.001 | 0.855–0.964 |
| m6A+CA153 | 88.52 | 77.50 | 0.890 | < 0.001 | 0.828–0.951 |
| m6A+CEA+CA153 | 80.33 | 89.20 | 0.914 | < 0.001 | 0.861–0.966 |
| m6A+METTL14+FTO | 78.95 | 97.44 | 0.929 | < 0.001 | 0.880–0.977 |
AUC, area under the curve; BC, breast cancer; CA153, carbohydrate antigen 153; CEA, carcinoembryonic antigen; CI, confidence interval; FTO, fat mass and obesity-associated; m6A, N6-methyladenosine; METTL14, methyltransferase-like 14; NC, normal control.
4. Upregulated mRNA expression of the METTL14 gene and downregulated expression of the FTO gene in patients with BC
To explain why m6A increased in patients with BC, we detected the mRNA expression levels of the core regulatory genes (including m6A methyltransferases METTL3, METTL14, and WTAP, and m6A demethylases ALKBH5 and FTO) that might be involved in dynamic m6A modification of RNA in random 15 pair training cohorts using real-time polymerase chain reaction. Only the expression of METTL14 and FTO was significantly different in patients with BC compared with that in the NCs in peripheral blood RNA (p < 0.001) (S2 Fig.). These results demonstrated that along with the dynamic m6A modification of RNA, variants of METTL14 are contributed as did the FTO demethylase in peripheral blood RNA of patients with BC.
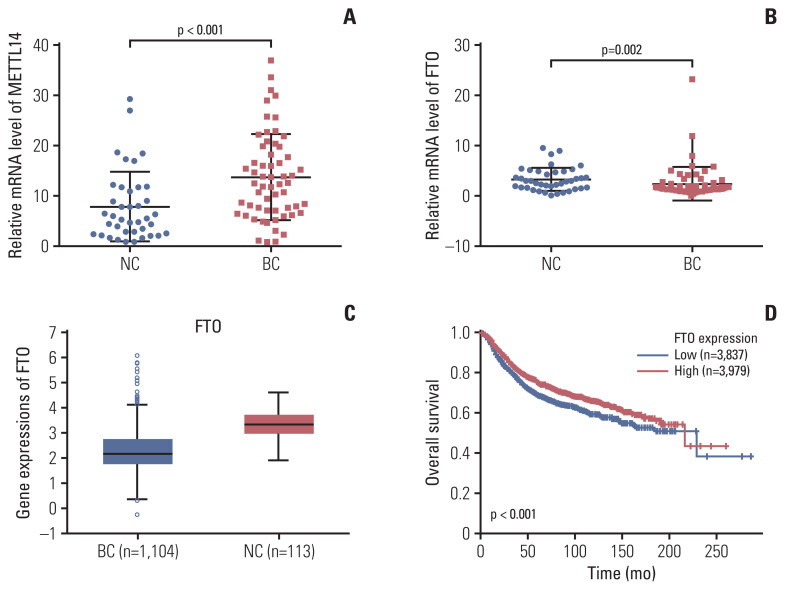
Next, the expression of METTL14 and FTO was checked in the remaining available samples as the validation cohorts. The mRNA expression of METTL14 as the methyltransferases for m6A was prominently upregulated in the peripheral blood RNA of patients with BC compared with that in the NCs (p < 0.001) (Fig. 4A). The average mRNA expression level of METTL14 in BC group was 1.75-fold higher than that in the NCs. Furthermore, the mRNA expression of FTO m6A demethylase in peripheral blood was significantly downregulated in patients with BC (p=0.002) (Fig. 4B). The average mRNA expression level of FTO in BC group was 0.74-fold compared with NCs. This finding was consistent with the lower expression level of FTO in 1,104 tumor tissues than that in 113 normal tissues (Fig. 4C). Moreover, the expression level of FTO in tumor tissues was significantly negatively correlated with the overall survival rate in patients with BC (Fig. 4D). However, both METTL14 and FTO were not significantly different based on other clinicopathological characteristics in patients with BC (S3 Table). The results suggest that the mRNA expression of the METTL14 and FTO genes may be a potential marker for patients with BC.
Fig. 4.
Methyltransferase-like 14 (METTL14) and fat mass and obesity-associated (FTO) mRNA expression in peripheral blood RNA in patients with breast cancer (BC), and the expression of FTO and their association with overall survival in tumor tissue. Quantitative real-time polymerase chain reaction analysis of METTL14 (A) and FTO (B) mRNA expression levels in the peripheral blood of normal controls (NCs) and patients with BC. (C) Analysis of the expression level of FTO from the StarBase database in NCs and patients with BC. (D) Correlation of the expressions with the overall survival analysis from a Kaplan-Meier plot. Bars represent the mean±standard deviation of the results.
5. Combined diagnostic value of m6A, the METTL14 mRNA level, and FTO mRNA level in patients with BC
The performance of m6A and its regulatory gene indicators in detecting BC was assessed in 60 patients with BC. In a ROC curve, the individual AUC of METTL14 and FTO reached 0.631 and 0.737, respectively (Fig. 5A). When m6A was combined with the METTL14 and FTO regulatory genes, the AUC increased to 0.929 (95% CI, 0.880 to 0.977) (Table 2, Fig. 5A). In addition, the specificity reached 97.4% without compromising too much sensitivity (78.9%), and the optimal cutoff value was 0.119 for this combined diagnosis (Fig. 5B). There was no significant correlation between m6A and the relative mRNA levels of METTL14/FTO in peripheral blood of patients with BC (S4 Fig.). m6A combined with METTL14 and FTO in peripheral blood showed the best diagnostic capability for patients with BC (Table 2).
Fig. 5.
Methyltransferase-like 14 (METTL14), fat mass and obesity-associated (FTO), and their combination diagnostic value along with N6-methyladenosine (m6A). Receiver operating characteristic curve analysis for METTL14 and FTO alone or combined diagnosis with m6A (A) and cutoff value analysis for the combination of m6A, METTL14, and FTO in normal controls (NCs) and patients with breast cancer (BC) (B). AUC, area under the curve.
Discussion
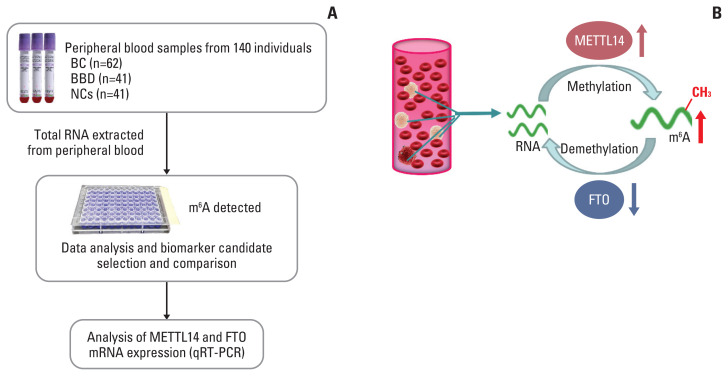
BC is characterized by heterogeneity in the initiation and development of genetic or epigenetic factors, and its sensitivity and specificity in early diagnosis remain the greatest challenge [16]. The low comprehensive predictive value of the main biomarkers for BC tumors in serum, such as CEA, CA153, CA199, and CA125, and their methods of detection lead to the difficulty in the early diagnosis of BC [17]. The m6A modification is one RNA epigenetic modification. Its maladjustment was correlated with tumors, while blood-based DNA methylation biomarkers imply the uncertainty associated with BC detection and risk assessment [18,19]. A schematic diagram of the present study design and the dynamic change of m6A modification in peripheral blood RNA are depicted in the Fig. 6. We found that the upregulation of the m6A level in peripheral blood may potentially be a novel diagnostic biomarker for BC. Simultaneously, m6A combined with its regulated genes METTL14 and FTO as a diagnostic biomarker displayed better diagnostic value in BC compared with current clinically available tumor biomarkers.
Fig. 6.
Study design and the dynamic N6-methyladenosine (m6A) modification of peripheral blood RNA in breast cancer (BC). (A) Flowchart depicting the study design for m6A of peripheral blood RNA biomarker in BC; (B) the schematic illustration for dynamic m6A modification of peripheral blood RNA in BC. BBD, benign breast diseases; FTO, fat mass and obesity-associated; METTL14, methyltransferase-like 14; NC, normal control; qRT-PCR, quantitative real-time polymerase chain reaction.
m6A was first discovered in the 1970s, and many emerging evidences supported that m6A modification can regulate the stability, translation, and splicing process of mRNA [20]. This modification can significantly regulate the occurrence of human cancer, which may be exploited to develop new biomarkers for detection and novel therapeutic targets for clinical studies of malignant tumors. A recent study suggest ed that the m6A level in BC tissues was increased compared with noncancerous tissues [21]. We measured the contents of m6A in the peripheral blood of 62 patients with BC and found that they were significantly higher than the levels in patients with BBD or the NCs. In particular, upregulated stage I–IV BC were significantly different from carcinoma in situ, which may be helpful for the early diagnosis of BC (Figs. 1A and 2A). ROC curve analysis showed m6A could differentiate patients with BC from NCs, with an AUC of 0.887 and high sensitivity of 91.94%, which was markedly greater than the values for CEA or CA153. This means that in patients with early BC, m6A has higher sensitivity and can effectively distinguish patients with BBD or healthy women. Thus, m6A could qualify as an indicator of BC preliminary screening. The combination of CEA and CA153 with m6A enhanced the AUC to 0.914 and improved specificity to 89.20%. We suggest that based on its convenience and diagnostic value, peripheral blood m6A could be a promising biomarker.
We examined the mRNA expression of m6A methylases and demethylases. The METTL14 methylase was significantly increased and the FTO demethylase was decreased in BC peripheral blood relative to NCs (Fig. 4A and B). In BC, promoter DNA hypermethylation has been reported as a common feature and genome-wide hypomethylation can frequently occur in regions of segmental duplications [16]. METTL14 methylase and FTO demethylase contribute to m6A modification in many cancers. Recently, it was reported that METTL14 plays an essential role in normal hematopoiesis. Thus, METTL14 has oncogenic roles in hepatocellular carcinoma and malignant hematopoiesis [8,22]. In addition, FTO was identified as the first RNA demethylase with an eraser function in m6A modification. FTO is critical in the progression of many cancers, including acute myeloid leukemia, glioblastoma, endometrial cancer, gastric cancer, and BC [23,24]. The downregulated expression level of FTO was also detected in 1104 tumor tissues from the StarBase database. Their negative correlation with the overall survival rate in patients with BC from Kaplan-Meier plots suggested that the low expression of FTO is associated with poor prognosis (Fig. 4C and D). Presently, the combination of METTL14 and FTO with m6A in BC peripheral blood improved the specificity to 97.4% and the sensitivity to 78.9%, with a ROC AUC of 0.929. Thus, the regulatory profile of these two genes verified the specific changes of m6A in peripheral blood of patients with BC, laterally. These results indicate that the combination of METTL14, FTO, and m6A has greater diagnostic efficacy for screening BC using peripheral blood.
The upregulation of METTL14 and downregulation of FTO may be related to the increased dynamic m6A modification in peripheral blood RNA of patients with BC. The specific mechanisms for their regulation are unclear. Maladjustment of METTL14 and FTO in BC tumor tissues has been described [25,26]. The RNA extraction protocol from peripheral whole blood mainly extracted RNA from nuclear cells (mainly white blood cells, lymphocytes, and monocytes, as well as reticulocytes). The increased m6A in peripheral blood RNA may have occurred for several reasons. First, the extracellular vesicles secreted by tumor cells may carry some transcriptional information of m6A modification that is pass on to the nuclear cells in peripheral blood RNA [27]. Second, intercellular cross-talk delivers the m6A modified signal to nucleate cells of peripheral blood and modifies their RNA [28]. Third, is the similarity to circulating tumor cells in BC, which act as precursors to metastasis and affect DNA methylation as well as RNA methylation [29]. The specific causes of this phenomenon and whether other cancers also have this characteristic will require further study. Our results were obtained only from peripheral blood. Examination of the relative tumor tissues may verify the results.
In conclusion, m6A in peripheral blood can be a potential novel diagnostic biomarker for BC. Increased mRNA expression of METTL14 and reduced mRNA expression of FTO may laterally underlie the upregulation of m6A, which could be exploited for use as a combination diagnostic biomarker.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81603016, 81773624), the Natural Science Foundation of Jiangsu Province (No. BK20160706, BE2017746), the National Science and Technology Major Project (2018ZX09301026-005, 2020ZX09201015).
Footnotes
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
Ethical Statement
This study was approved by the Ethics Committee of Zhongda Hospital of Southeast University according to the Chinese Ethical Regulations. All blood samples were obtained with informed consent, and ethical approval was granted by the Ethics Committee of Zhongda Hospital of Southeast University.
Author Contributions
Conceived and designed the analysis: Xiao H, Wu G.
Collected the data: Xiao H.
Contributed data or analysis tools: Xiao H, Fan X, Zhang R.
Wrote the paper: Xiao H, Wu G.
Conflicts of Interest
Conflict of interest relevant to this article was not reported.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
References
- 1.Zhang L, Qiang J, Yang X, Wang D, Rehman AU, He X, et al. IL1R2 blockade suppresses breast tumorigenesis and progression by impairing USP15-dependent BMI1 stability. Adv Sci (Weinh) 2020;7:1901728. doi: 10.1002/advs.201901728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang J, Ren J, Cui Q, Zhang D, Kong D, Liao X, et al. A prognostic 10-lncRNA expression signature for predicting the risk of tumour recurrence in breast cancer patients. J Cell Mol Med. 2019;23:6775–84. doi: 10.1111/jcmm.14556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavez-MacGregor M, Mittendorf EA, Clarke CA, Lichtensztajn DY, Hunt KK, Giordano SH. Incorporating tumor characteristics to the American Joint Committee on Cancer breast cancer staging system. Oncologist. 2017;22:1292–300. doi: 10.1634/theoncologist.2017-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Liu M, Shi S, Sun Y, Li M, Zhang M, et al. Diagnostic value of seven biomarkers for breast cancer: an overview with evidence mapping and indirect comparisons of diagnostic test accuracy. Clin Exp Med. 2020;20:97–108. doi: 10.1007/s10238-019-00598-z. [DOI] [PubMed] [Google Scholar]
- 5.Nam SE, Lim W, Jeong J, Lee S, Choi J, Park H, et al. The prognostic significance of preoperative tumor marker (CEA, CA15-3) elevation in breast cancer patients: data from the Korean Breast Cancer Society Registry. Breast Cancer Res Treat. 2019;177:669–78. doi: 10.1007/s10549-019-05357-y. [DOI] [PubMed] [Google Scholar]
- 6.Koh CW, Goh YT, Goh WS. Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat Commun. 2019;10:5636. doi: 10.1038/s41467-019-13561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu BB, Wang XY, Gu XY, Zou C, Gao ZJ, Zhang H, et al. N(6)-methyladenosine (m(6)A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019;18:178. doi: 10.1186/s12943-019-1099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181. doi: 10.1186/s12943-019-1108-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Deng X, Su R, Feng X, Wei M, Chen J. Role of N(6)-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, Tang Q, Cao X, Burwinkel B. Cell-free circulating DNA integrity based on peripheral blood as a biomarker for diagnosis of cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2017;26:1595–602. doi: 10.1158/1055-9965.EPI-17-0502. [DOI] [PubMed] [Google Scholar]
- 14.Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab. 2015;100:E148–54. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 16.Pasculli B, Barbano R, Parrella P. Epigenetics of breast cancer: Biology and clinical implication in the era of precision medicine. Semin Cancer Biol. 2018;51:22–35. doi: 10.1016/j.semcancer.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Di Gioia D, Blankenburg I, Nagel D, Heinemann V, Stieber P. Tumor markers in the early detection of tumor recurrence in breast cancer patients: CA 125, CYFRA 21-1, HER2 shed antigen, LDH and CRP in combination with CEA and CA 15-3. Clin Chim Acta. 2016;461:1–7. doi: 10.1016/j.cca.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Bodelon C, Ambatipudi S, Dugue PA, Johansson A, Sampson JN, Hicks B, et al. Blood DNA methylation and breast cancer risk: a meta-analysis of four prospective cohort studies. Breast Cancer Res. 2019;21:62. doi: 10.1186/s13058-019-1145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Q, Cheng J, Cao X, Surowy H, Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin Epigenetics. 2016;8:115. doi: 10.1186/s13148-016-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722:144076. doi: 10.1016/j.gene.2019.144076. [DOI] [PubMed] [Google Scholar]
- 22.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m(6)A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol. 2019;145:19–29. doi: 10.1007/s00432-018-2796-0. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Liu X, Dong Z, Li J, Yu Y, Chen X, et al. N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10:5447–59. doi: 10.7150/jca.35053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19:326. doi: 10.1186/s12885-019-5538-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–48. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halvorsen AR, Helland A, Gromov P, Wielenga VT, Talman MM, Brunner N, et al. Profiling of microRNAs in tumor interstitial fluid of breast tumors: a novel resource to identify biomarkers for prognostic classification and detection of cancer. Mol Oncol. 2017;11:220–34. doi: 10.1002/1878-0261.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis Seeding. Cell. 2019;176:98–112. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.