Significance
The recent emergence and spread of zoonotic viruses, including Ebola virus and severe acute respiratory syndrome coronavirus 2, demonstrate that animal-sourced viruses are a very real threat to global public health. Virus discovery efforts have detected hundreds of new animal viruses with unknown zoonotic risk. We developed an open-source risk assessment to systematically evaluate novel wildlife-origin viruses in terms of their zoonotic spillover and spread potential. Our tool will help scientists and governments assess and communicate risk, informing national disease prioritization, prevention, and control actions. The resulting watchlist of potential pathogens will identify targets for new virus countermeasure initiatives, which can reduce the economic and health impacts of emerging diseases.
Keywords: emerging infectious disease, wildlife, zoonotic virus, disease ecology, public health
Abstract
The death toll and economic loss resulting from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic are stark reminders that we are vulnerable to zoonotic viral threats. Strategies are needed to identify and characterize animal viruses that pose the greatest risk of spillover and spread in humans and inform public health interventions. Using expert opinion and scientific evidence, we identified host, viral, and environmental risk factors contributing to zoonotic virus spillover and spread in humans. We then developed a risk ranking framework and interactive web tool, SpillOver, that estimates a risk score for wildlife-origin viruses, creating a comparative risk assessment of viruses with uncharacterized zoonotic spillover potential alongside those already known to be zoonotic. Using data from testing 509,721 samples from 74,635 animals as part of a virus discovery project and public records of virus detections around the world, we ranked the spillover potential of 887 wildlife viruses. Validating the risk assessment, the top 12 were known zoonotic viruses, including SARS-CoV-2. Several newly detected wildlife viruses ranked higher than known zoonotic viruses. Using a scientifically informed process, we capitalized on the recent wealth of virus discovery data to systematically identify and prioritize targets for investigation. The publicly accessible SpillOver platform can be used by policy makers and health scientists to inform research and public health interventions for prevention and rapid control of disease outbreaks. SpillOver is a living, interactive database that can be refined over time to continue to improve the quality and public availability of information on viral threats to human health.
We now live in an era in which threats posed by viral pandemics are a daily reality. A single lethal virus can emerge suddenly and spread rapidly to every household and every community without regard to national borders or to social and economic standing. Recognizing the importance of emerging infectious diseases (EIDs), the World Health Organization (WHO) highlighted Disease X, a currently unknown pathogen capable of causing a serious human epidemic, as a target for research and development in their 2018 Blueprint of Priority Diseases (1). However, despite increased investment in pandemic prevention and knowledge gained from previous EID outbreaks, such as severe acute respiratory syndrome coronavirus (SARS-CoV), Ebola virus (Zaire ebolavirus), and Zika virus, we were unprepared for the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2) that emerged and rapidly spread around the world in 2019 to 2020 with a devastating death toll and an estimated global economic loss of ∼28 trillion (3).
SARS-CoV-2 is one of many potential viral threats to humans. There are just over 250 known zoonotic viruses—viruses that have previously spilled over from animals to humans and caused disease in people (4). While these viruses are of ongoing concern to human health, as repeated Ebola epidemics demonstrate, the yet to be identified viruses pose an equal if not more serious threat to humanity. Approximately 1.67 million undescribed viruses are thought to exist in mammals and birds, up to half of which are estimated to have the potential to spill over into humans (5).
Virus discovery efforts have initiated the process of investigating potential viral threats. Our team sampled wildlife at high-risk human disease transmission interfaces in over 30 countries (6), resulting in the discovery of hundreds of previously undetected viruses (SI Appendix). Although interesting, virus discovery creates a plethora of data without direction on where to focus efforts to prevent viral spillovers that could lead to future epidemics and pandemics. A strategy is needed to evaluate viruses and identify those that are most important for further investigation and surveillance.
The risk each virus poses to human health is not equal. Two viruses may be nearly identical, one zoonotic and the other not. However, several factors about the virus, host (the organism in which a virus can live and multiply), environment (the location and ecology where the host lives), and related human behavior influence the likelihood that a virus will become zoonotic and spread within human populations (7, 8). Risk assessments, such as those used by lenders, account for variances among risk factors in order to determine a comparative risk score. Previous studies and tools using this approach for viruses have been insightful but limited in scope by focusing on narrow groups of viruses or few risk factors (7–9). Here, we present an innovative relative risk assessment and interactive web application to systematically evaluate viruses of wildlife origin in terms of their potential for zoonotic spillover and spread in people (henceforth, “spillover risk”).
Using literature reviews and input from experts, we identified risk factors that are most likely to contribute to spillover risk. We then created a risk ranking framework and web tool called SpillOver: Viral Risk Ranking (https://spillover.global) that uses data for these risk factors to calculate a comparative “risk score” for each virus, much like a credit report. We used SpillOver to rank viruses detected during our viral discovery efforts and compared them with viruses that are already known to be zoonotic. The SpillOver tool also allows scientists, clinicians, policy makers, and the public to input data or explore and customize the output to their needs and interests while providing an adaptive crowdsourcing platform for uploading viruses as they are discovered.
Risk Ranking Methods
Selection of Risk Factors.
We conducted an extensive literature review to identify risk factors suggested to contribute to spillover risk. Since zoonotic spillover risk inherently is characterized by the ability of an animal-sourced virus to infect humans and cause disease, we looked for factors that covered topics relating to the likelihood of virus transmission from animals to humans and the ability of the virus to cause disease and spread within human populations. We identified 50 potential risk factors, including all those that could be identified in the peer-reviewed literature, as well as those identified through our viral discovery field work and surveys (6). A selection of 150 experts from 20 countries were asked to evaluate each of these factors in terms of influence on animal-origin virus spillover risk to humans (SI Appendix).
Experts were identified from relevant literature, Google searches, and conference attendance lists. The majority were affiliated with academic institutions (66%), and the remainder (33%) were from government and private or international organizations/foundations (SI Appendix, Table S32). A panel of 65 experts participated, each with a median of 19 y (interquartile range 12 to 27 y) of experience in virology, epidemiology, ecology, public health, molecular and microbiology, bioinformatics, and/or environmental science (SI Appendix, Figs. S1 and S2). Experts assigned a Spillover Risk (options = high [3], medium [2], low [1], or not relevant for spillover [0]) to each of the 50 identified risk factors and also characterized their own Level of Expertise (options = novice [1], competent [2], proficient [4], expert [8], or master [16]) for each risk factor (SI Appendix, Fig. S3 and Table S33).
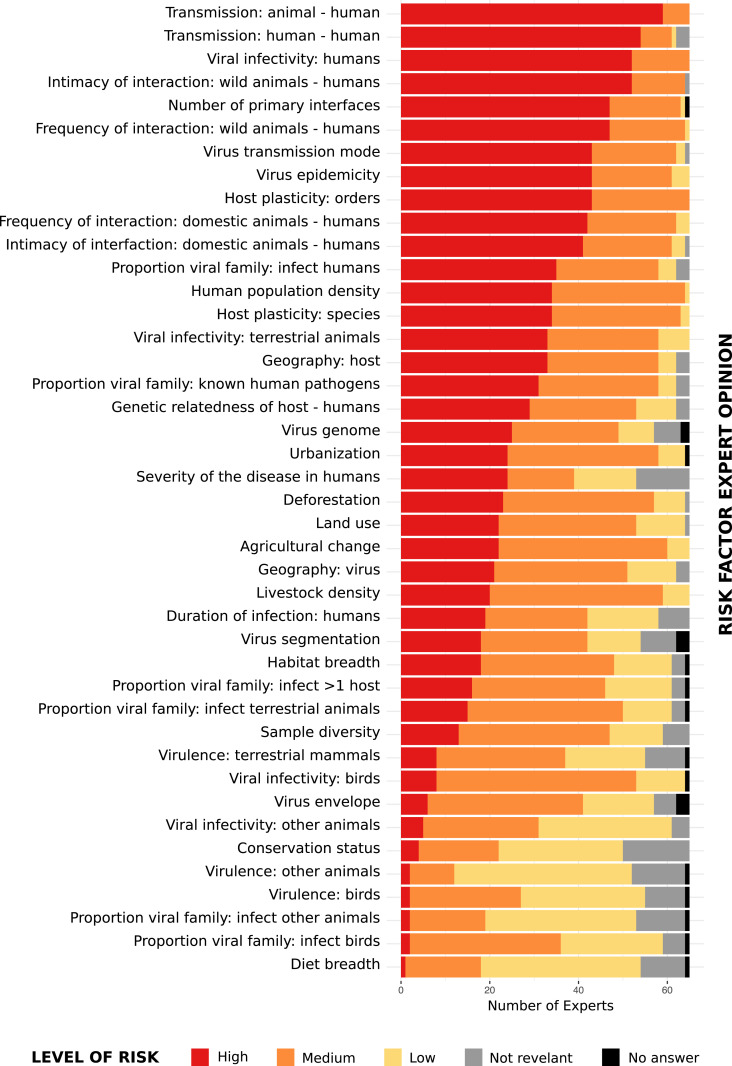
Expectedly, opinion with regard to Spillover Risk varied among experts (Fig. 1). To account for uncertainty, we calculated a weighted average score for each risk factor from the sum of expert responses to Spillover Risk, accounting for the Level of Expertise of each expert within each subject. If a participant did not declare a Spillover Risk or Level of Expertise, a value of zero or eight was assigned, respectively. A weighted average score (Risk Factor Influence) was then calculated for each risk factor from the sum of expert responses (i) using the following formula:
Fig. 1.
Expert opinion (n = 65) of the level of risk each factor included in the risk ranking assessment (n = 42) plays in the risk of a new virus spillover from animals to humans.
In addition to factors inherently linked to viruses already known to be zoonotic, factors deemed as high contribution to spillover risk were frequency and intimacy of human interaction with wild and domestic animal hosts, mode of transmission, and host plasticity (Fig. 1 and SI Appendix, Fig. S4). Seven risk factors including host mass, the percentage of host range in protected areas, and virus phylogenetics (virus to human pathogens distance, virus to animal pathogens distance, virus distance to a virus that is known to infect humans, virus distance to a virus that is known to infect animals, virus distance to a virus that is not known to infect humans) were assessed by experts as worthy of consideration but were eliminated due to the insufficiency of available data sources. One risk factor “Virus associated with unknown cause of illness in humans” was merged with the risk factor “Severity of disease in humans.”
The Risk Factor Influence was used as basis for selection of factors for inclusion in the risk assessment. Factors assessed by experts as having low or no contribution to spillover risk, evidenced by a Risk Factor Influence score of below two, were not included in the ranking with the exception of two risk factors, virus segmentation and virus envelope, that were included in the assessment due to the publication of a risk assessment supporting them having an important role in human-to-human transmission of viruses (7). The resulting risk assessment was based on 31 risk factors; however, methodology and results for the 42 risk factors for which data are available can be found in SI Appendix.
A Database of Wildlife-Origin Viruses.
We then created a database of virus detections in wildlife, confirmed by PCR and/or genetic sequencing and the necessary metadata to characterize the viruses according to the risk factors described above. Host species were limited to classes of Mammalia, Aves, Reptilia, and Amphibia. The database contains 35,751 rows of unique detections with metadata for 887 viruses from 19 virus families (viruses often were detected in several animals at several locations and interfaces) (SI Appendix).
Initially, virus detection data were collated from multiple sources, including viruses detected during our viral discovery project, the National Center for Biotechnology Information (NCBI) virus nucleotide database, and a literature review of zoonotic viruses (SI Appendix). These data included, when available, virus taxonomy; an identifier (i.e., accession number); host species taxonomy; sample type in which the virus was detected; primary disease transmission interface; and longitude and latitude where the animal was sampled or at minimum, the country of detection.
Between 2009 and 2019, our team collected and tested 509,721 samples from 74,635 wild animals at high-risk pathogen transmission interfaces in 28 countries (https://ohi.vetmed.ucdavis.edu/programs-projects/predict-project). Consensus PCR protocols and Sanger/Next-Generation sequencing were used to screen samples for viruses belonging to 15 viral families of concern to human health (SI Appendix). The resulting dataset used in this study comprises 855 unique viruses from 7,726 individual animals belonging to 251 mammalian species (plus 30 identified only to genus) within 26 countries (SI Appendix). Of the viruses detected, 721 were newly discovered, 133 were previously identified in other animals prior to the project, and 6 were known zoonotic viruses. In addition, virus detection data for 38 International Committee on Taxonomy of Viruses (ICTV)-recognized known zoonotic viruses identified from wildlife (including the 6 detected by the project) and the project-detected 133 previously identified viruses were extracted from the NCBI virus nucleotide database server (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/) and characterized via a review of the literature (SI Appendix).
Additional metadata necessary to characterize risk for each virus detection were derived from 19 publicly available resources on the host [International Union for Conservation of Nature (IUCN) Red List (10), Birdlife International (11), Catalogue of Life (12), TimeTree (13), diet database (14)], the virus [ICTV (15), ViralZone database (16), reference virology textbooks (17–19), published human/zoonotic virus databases (7, 16, 20, 21), the NCBI nucleotide database], and the environment [Land Degradation Assessment in Dryland (LADA) land use systems (22), NASA Sedac Gridded population of the World United Nations (UN) Adjusted population density v4 2015 (23), Global Grid of Probabilities of Urban Expansion to 2030 (24), adapted history database of the global environment (HYDE) land conversion 2005 (25, 26), Global Forest Change 2017 (27)] (SI Appendix).
Virus Risk Assessment.
The above data were used to qualitatively rank the spillover risk of wildlife-origin viruses by calculating a relative Spillover Risk Score for each virus (Fig. 2).
Fig. 2.
Schematic outline of the SpillOver risk ranking tool framework and website (https://spillover.global/).
Each risk factor had multiple categorical Risk-Level options (e.g., DNA or RNA for “Virus Genome” risk factor). These options were assigned a Risk-Level Score on a scale between zero and five, with five being the riskiest, based on scientific evidence and expert opinion. For example, the Risk Levels for the risk factor “Livestock density” were high (5), medium (3), low (1), none (0), or unknown (2.5). A central Risk-Level Score of 2.5 was assigned to all Risk Levels categorized as unknown. Further details on Risk Levels for each factor and justifications are provided in SI Appendix. For viruses with more than one detection and thus, more than one row of data, the assessment uses the principle of the worst case scenario (i.e., the most severe possible outcome is assigned using the highest Risk-Level Score among all rows for that virus). For example, a virus may have been detected in two locations, one that had high livestock densities and the other that had low livestock density. The assigned Risk Level for this virus’ risk factor would be “High livestock density” or five. The highest risk score for each risk factor was then combined with the expert assigned weight for that risk factor. Using the livestock density example, the calculation would be
For each virus, a relative Spillover Risk Score was then calculated as the sum of the 31 Virus Data Risk scores. Finally, virus ranking positions were assigned in descending order according to each virus’ Spillover Risk Score, thus relative spillover potential.
Development of the SpillOver: Viral Risk Ranking Tool.
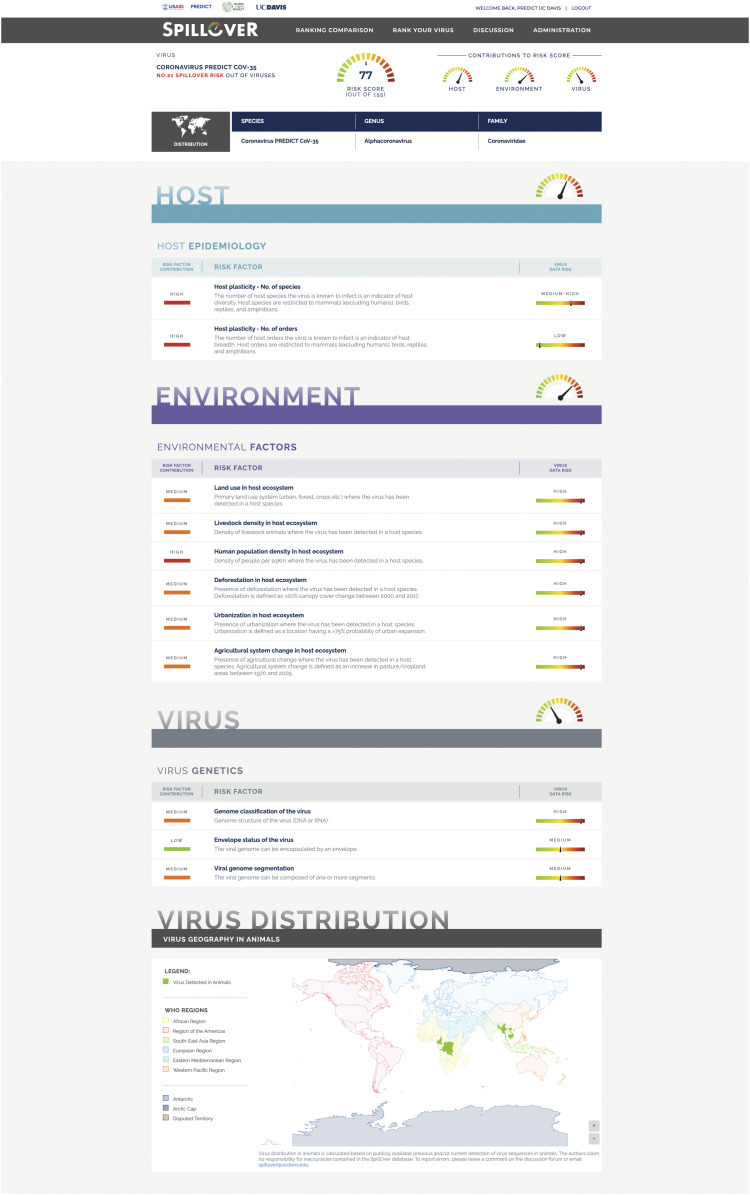
To provide a living platform for ongoing ranking of spillover risk from newly discovered wildlife-origin viruses, we created the SpillOver: Viral Risk Ranking tool in a browser-based application (https://spillover.global/) designed to increase accessibility to evidence-based risk ranking of viruses for scientists, policy makers, public health managers, and communities, especially in areas with higher risk of EIDs. SpillOver produces a detailed spillover risk report for each virus (Fig. 3), and the “Risk Comparison” tool allows users to compare and contrast ranked viruses and to filter viruses on a selection of key attributes, including virus-specific factors, host species, and country of detection.
Fig. 3.
Virus spillover ranking estimate for PREDICT-CoV-35, including the contribution of a selection of risk factors and its associated data toward the overall spillover risk assessment. Full details are in https://spillover.global.
SpillOver is a crowdsourcing platform on which people can choose to privately or publicly rank a new virus or add data to existing viruses using the “Rank Your Virus” tool. Currently, the application is designed for wildlife-origin viruses with host species in the classes Mammalia, Reptilia, Amphibia, and Aves, and data can autopopulate for 26 virus families of concern to human health (5). It is not intended for viruses that only infect humans, vector-borne viruses, or viruses of domestic species (animals that have been domesticated to be kept as pets or used as food) origin. Users are encouraged to submit their data for public sharing, which are reviewed by website administrators prior to integration. The minimum set of data required for ranking a virus includes virus taxonomy with sequence or reference number for quality control, host species, sample type, the estimated timeline of host species divergence from humans (13), host longitude/latitude or nearest town, and the primary high-risk disease transmission interface where the host was sampled. Variables for other host, virus, and environmental factors to complete the information on all 31 factors are autopopulated using data sources detailed above. Users can further edit automated entries from these publicly available datasets to improve accuracy of information. The innovative design is fully customizable for future developments, such as adding new risk factors and incorporating evolving scientific information to adjust the weighting of existing variables.
Data used for the current analyses were restricted to genetic evidence of virus infection (e.g., PCR, genome sequencing) and exclude experimental evidence of infection and serological studies of antibody response. However, SpillOver does not rely on specific detection protocols or methods; instead, we place the responsibility of robust virus data on the submitter. This approach facilitates future integration of all data-sourcing technologies, especially in high-risk countries where access to advanced methods may be limited but where surveillance is most needed.
SpillOver Risk Ranking Results.
Of 887 ranked viruses, the top 12 viruses were all known zoonotic viruses, as expected (Table 1). SARS-CoV-2 ranked second behind between known zoonotic viruses Lassa virus and Ebola virus. It may seem surprising that SARS-CoV-2 did not rank in first position given it has caused a pandemic (2). This slightly lower ranking is the result of much less data being available for SARS-CoV-2, as well as the more frequent documentation of spillover events for known viruses. For example, the risk ranking for SARS-CoV-2 is currently based solely on virus detections in zoo tigers, lions, and mink (28, 29). At the time of publication, we do not yet have key information about the natural wildlife host(s) that would have allowed the tool to more accurately estimate the spillover risk of SARS CoV-2, including number and range of host species, geographic distribution of hosts, and types of environments in which the hosts live. The initial paucity of data on SARS-CoV-2 reminds us how important it is to identify these data ahead of virus spillover, so that ranking tools may be populated and refined to create more comprehensive watchlists for surveillance and study. The more we study SARS-CoV-2, the higher it will likely rank within the tool, but the real goal is to collect and use such important data for novel viruses in advance of a pandemic to enable prevention and preparedness. In fact, two severe acute respiratory syndrome (SARS)-related coronaviruses detected in bats during our surveillance were ranked highly by SpillOver, with SARS-related betacoronavirus Rp3 in 15th and SARS-related bat coronavirus RsSHC014 in 51st position (Table 1).
Table 1.
A virus spillover risk ranking comparison of the top 50 wildlife viruses in the spillover database, including viruses known to be zoonotic and those with unknown zoonotic potential, which were detected in a broad-scale virus discovery effort in Africa and Southeast and South Asia from 2009 to 2019
| Risk ranking position | Risk ranking score* | Virus | |||
| Virus | Genus | Family | Detection in hosts | ||
| 1 | 91.18 | Lassa virus† | Mammarenavirus | Arenaviridae | Regional |
| 2 | 87.14 | SARS-CoV-2† | Betacoronavirus | Coronaviridae | Semiglobal |
| 3 | 87.00 | Ebola virus† | Ebolavirus | Filoviridae | Regional |
| 4 | 86.49 | Seoul virus† | Hantavirus | Bunyaviridae | Global |
| 5 | 86.49 | Nipah virus† | Henipavirus | Paramyxoviridae | Semiglobal |
| 6 | 86.38 | Hepatitis E virus† | Orthohepevirus | Hepeviridae | Global |
| 7 | 85.70 | Marburg virus† | Marburgvirus | Filoviridae | Regional |
| 8 | 85.04 | SARS-CoV† | Betacoronavirus | Coronaviridae | National—large |
| 9 | 84.78 | Simian immunodeficiency virus† | Lentivirus | Retroviridae | Semiglobal |
| 10 | 84.69 | Rabies virus† | Lyssavirus | Rhabdoviridae | Global |
| 11 | 84.61 | Lymphocytic choriomeningitis virus† | Mammarenavirus | Arenaviridae | Global |
| 12 | 83.99 | Simian foamy virus† | Spumavirus | Retroviridae | Global |
| 13 | 80.98 | Coronavirus 229E (bat strain) | Alphacoronavirus | Coronaviridae | Regional |
| 14 | 80.01 | Rousettus bat coronavirus HKU9 | Betacoronavirus | Coronaviridae | Global |
| 15 | 79.71 | SARS-related betacoronavirus Rp3 | Betacoronavirus | Coronaviridae | National—large |
| 16 | 78.97 | European bat lyssavirus 1† | Lyssavirus | Rhabdoviridae | Regional |
| 17 | 78.81 | Andes virus† | Hantavirus | Bunyaviridae | National—small |
| 18 | 78.63 | Murine coronavirus | Betacoronavirus | Coronaviridae | Global |
| 19 | 78.57 | Puumala virus† | Hantavirus | Bunyaviridae | Regional |
| 20 | 78.03 | Chaerephon bat coronavirus/Kenya/KY22/2006 | Alphacoronavirus | Coronaviridae | Regional |
| 21 | 77.32 | Coronavirus PREDICT_CoV-35 | Alphacoronavirus | Coronaviridae | Semiglobal |
| 22 | 77.21 | Borna disease virus† | Bornavirus | Bornaviridae | Semiglobal |
| 23 | 76.42 | Longquan Aa mouse coronavirus | Betacoronavirus | Coronaviridae | Semiglobal |
| 24 | 76.14 | Monkeypox virus† | Orthopoxvirus | Poxviridae | Semiglobal |
| 25 | 75.78 | European bat lyssavirus 2† | Lyssavirus | Rhabdoviridae | Regional |
| 26 | 75.51 | Laguna Negra virus† | Hantavirus | Bunyaviridae | Regional |
| 27 | 75.05 | Eidolon bat coronavirus/Kenya/KY24/2006 | Betacoronavirus | Coronaviridae | Regional |
| 28 | 74.65 | Cowpox virus† | Orthopoxvirus | Poxviridae | Regional |
| 29 | 74.64 | Coronavirus PREDICT CoV-24 | Betacoronavirus | Coronaviridae | Semiglobal |
| 30 | 74.60 | Macaque Foamy virus | Spumavirus | Retroviridae | Global |
| 31 | 73.80 | Rodent coronavirus | Alphacoronavirus | Coronaviridae | Regional |
| 32 | 73.36 | Sin Nombre virus† | Hantavirus | Bunyaviridae | Regional |
| 33 | 73.23 | Human mastadenovirus G | Mastadenovirus | Adenoviridae | Semiglobal |
| 34 | 72.94 | Coronavirus PREDICT CoV-22 | Betacoronavirus | Coronaviridae | Semiglobal |
| 35 | 72.91 | Reston virus† | Ebolavirus | Filoviridae | Semiglobal |
| 36 | 72.49 | Bombali virus | Ebolavirus | Filoviridae | Regional |
| 37 | 72.46 | Coronavirus HKU1 | Betacoronavirus | Coronaviridae | National—small |
| 38 | 72.17 | Kenya bat coronavirus/BtKY56/BtKY55 | Betacoronavirus | Coronaviridae | Regional |
| 39 | 72.08 | Paramyxovirus PREDICT PMV-10 | Unassigned | Paramyxoviridae | Regional |
| 40 | 71.73 | Bat coronavirus 1 | Alphacoronavirus | Coronaviridae | Semiglobal |
| 41 | 71.64 | BtVs-BetaCoV/SC2013 | Betacoronavirus | Coronaviridae | National—large |
| 42 | 71.54 | Australian bat lyssavirus† | Lyssavirus | Rhabdoviridae | National—large |
| 43 | 71.37 | Bat coronavirus Hipposideros/GhanaKwam/20/2008 | Betacoronavirus | Coronaviridae | Regional |
| 44 | 71.24 | Coronavirus PREDICT CoV-68 | Betacoronavirus | Coronaviridae | Regional |
| 45 | 71.14 | Mamastrovirus 1 | Mamastrovirus | Astroviridae | Semiglobal |
| 46 | 71.13 | Dobrava-Belgrade virus† | Hantavirus | Bunyaviridae | Regional |
| 47 | 71.06 | Scotophilus bat coronavirus 512 | Alphacoronavirus | Coronaviridae | Regional |
| 48 | 80.00 | Paramyxovirus PREDICT PMV-13 | Unassigned | Paramyxoviridae | Semiglobal |
| 49 | 70.98 | Paramyxovirus PREDICT PMV-15 | Unassigned | Paramyxoviridae | Regional |
| 50 | 70.96 | Coronavirus PREDICT CoV-16 | Betacoronavirus | Coronaviridae | Regional |
Out of a maximum score of 155 from 31 risk factors.
Zoonotic virus.
Several newly discovered viruses had higher spillover risk estimates than some known zoonotic pathogens (Table 1 and SI Appendix, Fig. S4). Some newly detected coronaviruses, including the Alphacoronavirus provisionally named PREDICT_CoV-35 ranked within the top 20. Broad host and virus geography combined with detection in bats at high-risk disease transmission interfaces, including hunting and within human dwellings, suggests that PREDICT_CoV-35 is of high public health relevance. Although these newly detected coronaviruses have not yet been found in humans, spillover events may be going unrecognized due to lack of diagnostic capabilities and underreporting, as well as the propensity of coronaviruses to cause only mild symptoms or asymptomatic cases.
A Global Resource for Zoonotic Threats.
SpillOver is a comprehensive publicly accessible risk assessment tool to systematically facilitate evaluation of novel viruses in terms of their zoonotic spillover and pandemic potential. The framework is not intended to predict the origins, impact, or timing of future pandemics. However, the tool begins to address the need for platforms to help interpret global infectious disease data on a broader scale (30). The simple design of the SpillOver tool requires only a few pieces of readily available information to be uploaded by the submitters of new viruses in order to source a rich set of risk data on each virus from many publicly available sources, producing comparative risk estimates.
The ability of SpillOver to meaningfully rank animal–human transmission risk from wildlife-origin viruses is inherently limited by the data available, or lack thereof, for each virus, especially when a novel virus is initially identified. If we are to use the tool to its full utility and continue to improve it over time, the paucity of collated, validated public virus information must be rectified. The fact that many attributes are unknown for newly detected viruses could cause an initial under- or overestimate of spillover risk since we have to assign a central or “unknown value” to some of these variables until more data become available: for example, whether or not the virus has the capability to infect human cells, the mode(s) of transmission, and the full host and geographic range. In addition to simply too few detections of viruses, limiting our ability to rigorously assess their host and geographic scope and associated risk factors, we most often identified inconsistent reporting of host species and the location of sampling, as well as nonstandardized naming of viruses in public datasets. Therefore, we advocate for the improvement of data reporting for viruses, as well as an expansion of large-scale viral detection and characterization to improve the available data for risk ranking efforts.
Making those viruses that may rank highly available to responsible virologists to further assess receptor binding and other virological risk factors is also crucial, as we could not yet incorporate some likely important virological risk factors into the tool because there are almost no publicly available data on such parameters for anything beyond those viruses that have already made many people sick. While the ranking includes a wide range of risk factors judged to be important by the global experts, there remain complexities and undocumented risk factors that are not yet incorporated. In fact, insufficient evidence or availability of broad-scale data prevented inclusion of several risk factors in this study that may have influence on virus spillover and spread, such as the role virus phylogeny, host mass, and remoteness of host home ranges. Additionally, data deficiencies in vector distributions and knowledge relating to competent hosts limited our ability to estimate relative risk in a meaningful way for vector-borne viruses.
The power of this tool lies in the fact that it is open source and adaptable—the more virus detection data that are entered, the more robust the rankings. New risk factors can and will be added when evidence in the peer-reviewed literature provides insight into newly identified risk factors. Therefore, we hope future virus detections in animals will be reported into the system so that risk comparisons can be refined. The authors also commit to regular review of publicly available resources to incorporate new discoveries into the SpillOver database.
Existing tools to rank influenza virus strains are increasingly being used by public health communities and researchers (e.g., the Centers for Disease Control and Prevention Influenza Risk Assessment Tool). Because there is a disproportionate availability of diverse and high-quality data for influenza viruses, we have intentionally not included them in this ranking of viruses for which much less is known. The goal of the SpillOver tool is to identify those viruses that should be prioritized for surveillance and studied as intensively as influenzas have been. Knowledge regarding existing and novel subtype and strain diversity and how those vary and impact the potential risk for zoonotic emergence, disease, and spread is already being generated for influenza viruses. In the future, however, the foundations on which the SpillOver tool has been developed could be duplicated and customized with virus-specific risk factors within a group of related and well-studied viruses for a deeper dive into cross-species transmission risk.
Collaboration Is Key to Success.
Despite the propensity of viruses to influence ecosystems and especially, the health of hosts within them, data on virus diversity and transmission characteristics are lacking relative to cellular pathogens. We aimed to begin to address this gap in knowledge by conducting virus detection and discovery in regions forecasted to be hot spots for emergence of disease. In addition to expanding the general knowledge of our world, virus discovery efforts have the potential to allow characteristics of viruses, hosts, environmental factors, and their associated interactions to be analyzed and acted upon to target surveillance, improving cost-effectiveness, as well as epidemic preparedness and prevention activities to reduce impacts of spillover events. As more and more viruses are discovered, we must establish a scientific dialogue that will result in tools that can be used to prioritize these activities, informed by data on spillover risk and the potential for further amplification and spread in humans. We ranked known zoonotic viruses in relation to a large body of novel virus findings and built a “straw” tool to begin this dialogue. The impact of the tool on public health will be dependent on global participation and collaboration. By crowdsourcing and integrating additional virus discovery data from scientists around the world into the SpillOver database and with continued risk ranking refinement, we could advance regional and global understanding of animal-origin viruses and be better prepared to prevent future viral spillovers that could lead to the next pandemic by investing in mitigation activities for the most risky viruses at the most likely locations and interfaces. Without such efforts, we are doomed to wait for the next Disease X and once again, be ill prepared.
The SpillOver tool is an essential first step that begins to address the burden of uncertainty raised while new viruses are being discovered. By creating a comprehensive watchlist of the top potential pathogens, our tool helps scientists and governments assess and communicate risk from an evidence-based perspective, informing national-level zoonotic disease prioritization, prevention, and action frameworks such as enhanced surveillance of people and wildlife at key locations and interfaces. After high-risk viruses have been characterized, further investigation can distinguish candidates for Disease X (1), assess likely relative impacts, and mitigate spillover risk in advance of devastating consequences. These investigations will require collaboration among experts in virology, epidemiology, ecology, computing technologies, synthetic biology, sociology, politics, and user intention to improve data sourcing in order to develop preventive and controlling countermeasures, including the prioritized development of vaccines and targeted multivalent therapies, as well as public health interventions. Specifically, changing human behavior at interfaces for the riskiest viruses can limit interactions with wildlife and may make the most immediate impact. By learning to live safely with wildlife, we not only can mitigate future disease outbreaks in humans but also, can conserve species that are essential to our life on the planet.
New initiatives including the WHO Research and Development (R&D) Blueprint (1), the Coalition for Epidemic Preparedness Innovations (31), the Trinity Challenge (https://www.thetrinitychallenge.org/), and proposed collaborative efforts among the International Barcode of Life’s program BIOSCAN, the Earth Biogenome Project, and the Global Virome Project (32) are making strides forward by building vaccine technology and collaborative networks to address virus threats. SpillOver will complement these programs by providing a resource for collating massive global datasets across multiple disciplines (virology, epidemiology, ecology, etc.) and producing a meta-analysis that can focus research and future countermeasure development, which together could reduce the economic and public health impacts of Disease X.
The research protocol was approved by the University of California Animal Care and Use Committee (permit nos. 17504 and 19300).
Supplementary Material
Acknowledgments
We thank P. Pandit and C. Zambrana-Torellio for analytical support and D. Carroll, A. Clements, and Murray Trostle for their vision and support. This study was made possible by the generous support of the American people through US Agency for International Development (USAID) Emerging Pandemic Threats PREDICT Project Awards GHN-A-00-09-00010-00 and AID-OAA-A-14-00102. USAID had no involvement in the design and content of this manuscript. The contents of this manuscript and associated materials are the responsibility of the authors and do not necessarily reflect the views of USAID or the US Government.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.D. is a guest editor invited by the Editorial Board.
3The complete lists of Expert Panel and PREDICT Consortium can be found in SI Appendix.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002324118/-/DCSupplemental.
Contributor Information
Collaborators: Kevin Anderson, Prasert Auewarakul, Lark Coffey, Ronald Corley, Gwenaelle Dauphin, Jonathan Epstein, Keiji Fukuda, Simon Goodman, Barbara Han, James Hughes, Martin Jeggo, William Karesh, Rudovick Kazwala, Terra Kelly, Gerald Keusch, Micheal Kurilla, John Mackenzie, Wanda Markotter, Corina Monagin, David Morens, Vincent Munster, Elke Muhlberger, Pranav Pandit, Alison Peel, Dirk Pfeiffer, Olivier Restif, Oyewale Tomori, Jonathan Towner, Sylvie Van Der Werf, Sophie VonDobschetz, Supaporn Wacharapluesadee, Micheal Ward, Lidewij Weirsma, Mary Wilson, David Wolking, Kachen Wongsathapornchai, Liam Brierley, University of Edinburgh Epigroup members, Carlos Tambrana-Torellio, those who wish to remain anonymous, Arif Islam, Shariful Islam, Zia Raman, Vibol Hul, Veasna Duong, Moctar Mouiche, Julius Nwobegahay, Kalpy Coulibaly, Charles Kumakamba, Eddy Kambale Syaluha, Jean-Paul Lukusa, Desalegn Belay, Nigatu Kebede, William Ampofo, Sammuel Bel-Nono, Richard Suu-Ire, Kalivogui Douokoro, Huda Dursman, Imung Pamungkas, Novie Rachmitasari, Suryo Saputro, Wirda Damanik, Tina Kusumaningrum, Maya Rambitan, Beounly Rey, Dodi Safari, Amin Soebandrio, Juliana Triastuti, Ehab Abu-Basha, Kwallah Allan, Kamau Joseph, Mutura Samson, Bouaphanh Khamphaphonphane, Watthana Theppanga, Jim Desmond, Sandra Samules, Mei Ho Lee, Jimmy Lee, Batchuluun Damdinjav, Enkhtuvshin Shiilegdamba, Ohnmar Aung, Manisha Bista, Dibesh Karmacharya, Rima Shrestha, Julius Nziza, Jean-Claude Tumushime, Modou Moustapha Lo, Amadou Ndiaye, Mame Cheikh Seck, James Bangura, Edwin Lavalie, Grace Mwangoka, Zikankuba Sijali, Ricky Okwir Okello, Benard Ssebide, Supaporn Wacharpluesadee, Nga Nguyen, Jon Epstein, Emily Hagan, William Karesh, Alice Latinne, Anne Laudisoit, Hongying Li, Catherine Machalaba, Stephanie Martinez, Noam Ross, Ava Sullivan, Carlos Zambrana Torrelio, John Mackenzie, Ron Waldman, Subash Morzaria, Wantanee Kalpravidh, Yilma Makonnen, Sophie Von Dubscheutz, Filip Claes, Katie Pelican, Casie Barton Behravesh, Elizabeth Mumford, John Paul Clark, Trong Duoc Vu, Karen Saylors, Bethany Edison, Jason Euren, Amethyst Gillis, Christian Lange, Mat LeBreton, David McIver, Daniel O’Rourke, Marc Valitutto, Dawn Zimmerman, Jaber Belkhiria, Brian Bird, Hannah Chale, Eunah Preston, Nicole Gardner, Brooke Genovese, Kevin Gonzalez, Lucy Keatts, Terra Kelly, Elizabeth Leasure, Corina Monagin, Pranav Pandit, Nistara Randhawa, Brett Smith, Woutrina Smith, Alex Tremeau-Bravard, David Wolking, Carolina Churchill, Sarah Olson, Chris Walzer, and Amanda Fine
Data Availability
All datasets along with the R code and R package dependencies needed to fully replicate and evaluate these analyses have been deposited in Open Source Framework (https://osf.io/mb6qn/?view_only=f6326d48d7d941afa7af02714819a1a2) (33).
Change History
September 16, 2021: The text of this article and SI Appendix have been updated; please see accompanying Correction for details.
References
- 1.Mehand M. S., Al-Shorbaji F., Millett P., Murgue B., The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res. 159, 63–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization , WHO Director-General’s opening remarks at the media briefing on COVID-19–11 March 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020. Accessed 11 March 2021.
- 3.International Monetary Fund (IMF) , World Economic Outlook, October 2020: A Long and Difficult Ascent (International Monetary Fund, Washington, DC, 2020). [Google Scholar]
- 4.Mollentze N., Streicker D. G., Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl. Acad. Sci. U.S.A. 117, 9423–9430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll D., et al., The global virome project. Science 359, 872–874 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kelly T. R.et al.; PREDICT Consortium , One Health proof of concept: Bringing a transdisciplinary approach to surveillance for zoonotic viruses at the human-wild animal interface. Prev. Vet. Med. 137, 112–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geoghegan J. L., Senior A. M., Di Giallonardo F., Holmes E. C., Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl. Acad. Sci. U.S.A. 113, 4170–4175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olival K. J., et al., Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trock S. C., Burke S. A., Cox N. J., Development of framework for assessing influenza virus pandemic risk. Emerg. Infect. Dis. 21, 1372–1378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.IUCN , The IUCN red list of threatened species. http://www.iucnredlist.org. Accessed 1 October 2020.
- 11.BirdLife International , Species distribution data request. http://datazone.birdlife.org/species/requestdis. Accessed 1 October 2017.
- 12.Roskov Y., et al., Species 2000 & ITIS Catalogue of Life, 2018 Annual Checklist (Species 2000, Naturalis, Leiden, the Netherlands, 2018). [Google Scholar]
- 13.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Wilman H., et al., EltonTraits 1.0: Species-level foraging attributes of the world’s birds and mammals. Ecology 95, 2027 (2014). [Google Scholar]
- 15.Lefkowitz E. J., Adams M. J., Davison A. J., Siddell S. G., Simmonds P., Virus taxonomy: The classification and nomenclature of viruses. https://talk.ictvonline.org/ictv-reports/ictv_online_report/introduction/. Accessed 1 October 2017.
- 16.Hulo C., et al., ViralZone: A knowledge resource to understand virus diversity. Nucleic Acids Res. 39, D576–D582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubovi E. J., Maclachlan N. J., Eds., Fenner’s Veterinary Virology (Academic Press, Boston, MA, ed. 5, 2016). [Google Scholar]
- 18.Tidona C., Darai G., The Springer Index of Viruses (Springer, New York, NY, ed. 2, 2011). [Google Scholar]
- 19.Burrell C. J., Howard C. R., Murphy F. A., Fenner and White’s Medical Virology (Academic Press, ed. 5, 2017), p. 604. [Google Scholar]
- 20.Woolhouse M. E. J., Brierley L., Epidemiological characteristics of human-infective RNA viruses. Sci. Data 5, 180017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson C. K., et al., Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 5, 14830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nachtergaele F., Pertri M., “Mapping land use systems at global and regional scales for land degradation assessment analysis” in Land Degradation Assessment in Drylands, Woodfine A., Ed. (FAO, Rome, Italy, 2011), pp. 1–19. [Google Scholar]
- 23.Center for International Earth Science Information Network–CIESIN–Columbia University , Gridded Population of the World, Version 4 (GPWv4): UN-Adjusted Population Density Adjusted to Match 2015 Revision UN WPP Country Totals. NASA Socioeconomic Data and Applications Center (SEDAC). https://beta.sedac.ciesin.columbia.edu/data/set/gpw-v4-population-density-adjusted-to-2015-unwpp-country-totals. Accessed 8 October 2017.
- 24.Seto K., Güneralp B., Hutyra L. R., Global grid of probabilities of urban expansion to 2030, v1 (2000–2030). https://sedac.ciesin.columbia.edu/data/set/lulc-global-grid-prob-urban-expansion-2030. Accessed 17 January 2019.
- 25.Kees K. G., Arthur B., Gerard D., Martine V., The HYDE 3.1 spatially explicit database of human-induced global land-use change over the past 12,000 years. Glob. Ecol. Biogeogr. 20, 73–86 (2011). [Google Scholar]
- 26.Klein Goldewijk K., Beusen A., Janssen P., Long-term dynamic modeling of global population and built-up area in a spatially explicit way: HYDE 3.1. Holocene 20, 565–573 (2010). [Google Scholar]
- 27.Hansen M. C., et al., High-resolution global maps of 21st-century forest cover change. Science 342, 850–853 (2013). [DOI] [PubMed] [Google Scholar]
- 28.OIE , Events in animals. https://www.oie.int/en/scientific-expertise/specific-information-and-recommendations/questions-and-answers-on-2019novel-coronavirus/events-in-animals/ Accessed 1 December 2020.
- 29.McAloose D., et al., From people to Panthera: Natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. MBio 11, e02220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han B. A., Drake J. M., Future directions in analytics for infectious disease intelligence: Toward an integrated warning system for emerging pathogens. EMBO Rep. 17, 785–789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brende B., et al., CEPI-a new global R&D organisation for epidemic preparedness and response. Lancet 389, 233–235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kress W. J., Mazet J. A. K., Hebert P. D. N., Opinion: Intercepting pandemics through genomics. Proc. Natl. Acad. Sci. U.S.A. 117, 13852–13855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grange Z. L., et al., Ranking the risk of animal-to-human spillover for newly discovered viruses. Open Science Framework. https://osf.io/mb6qn/?view_only=f6326d48d7d941afa7af02714819a1a2. Deposited 12 November 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets along with the R code and R package dependencies needed to fully replicate and evaluate these analyses have been deposited in Open Source Framework (https://osf.io/mb6qn/?view_only=f6326d48d7d941afa7af02714819a1a2) (33).