Significance
Although cancer immunotherapies have shown potential in inducing durable tumor regression in patients with metastatic melanoma, their efficacy remains highly variable and patient specific. Here, we present one strategy to overcome resistance to T cell–based immunotherapies by pharmacologically modulating the STING pathway, a relatively common suppressed DNA-sensing pathway in human melanomas. We show that rescue of this pathway in human melanoma cells using a clinically available DNA methylation inhibitor can augment their antigenicity and recognition by tumor-infiltrating lymphocytes. These findings have important clinical implications for the development of optimal therapeutic protocols that consider the status of tumor-intrinsic STING activity and provide a strategy to design clinical interventions using T cell–based immunotherapies with appropriate patient selection in melanoma.
Keywords: STING signaling, epigenetic silencing, DNA methylation, antigenicity, immune evasion
Abstract
Lack or loss of tumor antigenicity represents one of the key mechanisms of immune escape and resistance to T cell–based immunotherapies. Evidence suggests that activation of stimulator of interferon genes (STING) signaling in tumor cells can augment their antigenicity by triggering a type I IFN-mediated sequence of autocrine and paracrine events. Although suppression of this pathway in melanoma and other tumor types has been consistently reported, the mechanistic basis remains unclear. In this study, we asked whether this suppression is, in part, epigenetically regulated and whether it is indeed a driver of melanoma resistance to T cell–based immunotherapies. Using genome-wide DNA methylation profiling, we show that promoter hypermethylation of cGAS and STING genes mediates their coordinated transcriptional silencing and contributes to the widespread impairment of the STING signaling function in clinically-relevant human melanomas and melanoma cell lines. This suppression is reversible through pharmacologic inhibition of DNA methylation, which can reinstate functional STING signaling in at least half of the examined cell lines. Using a series of T cell recognition assays with HLA-matched human melanoma tumor-infiltrating lymphocytes (TIL), we further show that demethylation-mediated restoration of STING signaling in STING-defective melanoma cell lines can improve their antigenicity through the up-regulation of MHC class I molecules and thereby enhance their recognition and killing by cytotoxic T cells. These findings not only elucidate the contribution of epigenetic processes and specifically DNA methylation in melanoma-intrinsic STING signaling impairment but also highlight their functional significance in mediating tumor-immune evasion and resistance to T cell–based immunotherapies.
Failure of the immune system in recognizing and eliminating transformed cells leads to cancer development and tumor progression. Although defective T cell function represents one of the key mechanisms of tumor escape, through multiple genetic and epigenetic alterations, tumor cells themselves can directly evade T cell surveillance and destruction (1). Lack of antigenic gene mutations, loss of tumor antigen expression, loss of major histocompatibility complex (MHC) molecules, or other defects in antigen processing machinery are common intrinsic alterations within tumor cells that limit their recognition by immune T cells (2, 3).
Loss of interferon pathway gene function in tumor cells is another escape mechanism by which tumor cells can avoid T cell–mediated immunity. Several studies have indicated that alterations of genes encoding the interferon (IFN)-γ receptor signaling components (IFNGR1, IFNGR2, JAK1, JAK2, and STAT1) in tumor cells contribute to both primary and acquired resistance to immune checkpoint inhibitor therapies in melanoma patients by limiting tumor cell sensitivity to IFN-γ–induced up-regulation of MHC class I and inhibition of growth arrest (4–7).
Type I IFNs also play an important role in the generation of antitumor immunity by triggering a sequence of autocrine and paracrine signaling events within tumor cells that result in the augmentation of MHC class I expression and the induction of CXCR3-binding chemokines CXCL9 and CXCL10 (8, 9).
Studies using both carcinogen-induced and transplantable tumor models have indicated that endogenous induction of type I IFNs is required for the generation of a spontaneous T cell response (10, 11). Therefore, defects in molecules involved in type I IFN-dependent and type I IFN receptor (IFNAR)-transduced signaling pathways in tumor cells may compromise their recognition and elimination by immune T cells.
Among different upstream pathways that trigger type I IFN induction, stimulator of interferon genes (STING) has been identified as a major pathway for the generation of a spontaneous immune response against certain tumors (12). Activation of this pathway occurs through the detection of cytosolic DNA by the sensor cyclic GMP-AMP synthase (cGAS) and the generation of cyclic GMP-AMP (cGAMP) that binds to and activates STING, leading to the activation of the transcription factor IRF3 and induction of type I IFNs (13, 14).
Although the involvement of innate immune components such as antigen-presenting cells (APCs) has been initially reported as a major factor in STING-mediated antitumor immunity (12, 15), now it is becoming more evident that STING activity in tumor cells can also have a functional role in mediating the immune response (16, 17). In fact, we have recently discovered that activation of STING signaling in human melanoma cell lines enhances both their antigenicity and susceptibility to lysis by human melanoma tumor-infiltrating lymphocytes (TIL) through the augmentation of MHC class I molecules (18). However, we have also found that STING signaling is commonly suppressed and dysfunctional in a notable subset of melanomas that limits their antigen presentation and subsequently their sensitivity to T cell–mediated elimination (18, 19). This raises the question whether tumor cells have defects in the components of the STING pathway as a mechanism of immune escape and therefore become resistant to T-cell–based immunotherapies.
Multiple studies have reported the involvement of epigenetic modifications in the suppression of immune-protective signature genes including those of MHC Class I antigen presentation pathway as well as CXCR3-binding chemokines in both preclinical and clinical settings (20–22). Unlike genetic alterations, epigenetic suppressions are potentially reversible by selective epigenetic reprogramming (23).
On the basis of these observations and given the importance of tumor cell–intrinsic STING signaling in antigen presentation and induction of CXCR3-binding chemokines, we have now investigated the extent of promoter hypermethylation-mediated cGAS and STING silencing in clinically-relevant human melanomas and melanoma cell lines and examined whether their therapeutic targeting through pharmacological reprogramming can improve their antigenicity and promote TIL functional activity.
Results and Discussion
Promoter Hypermethylation of STING and cGAS Correlates with Their Silencing in Human Melanoma.
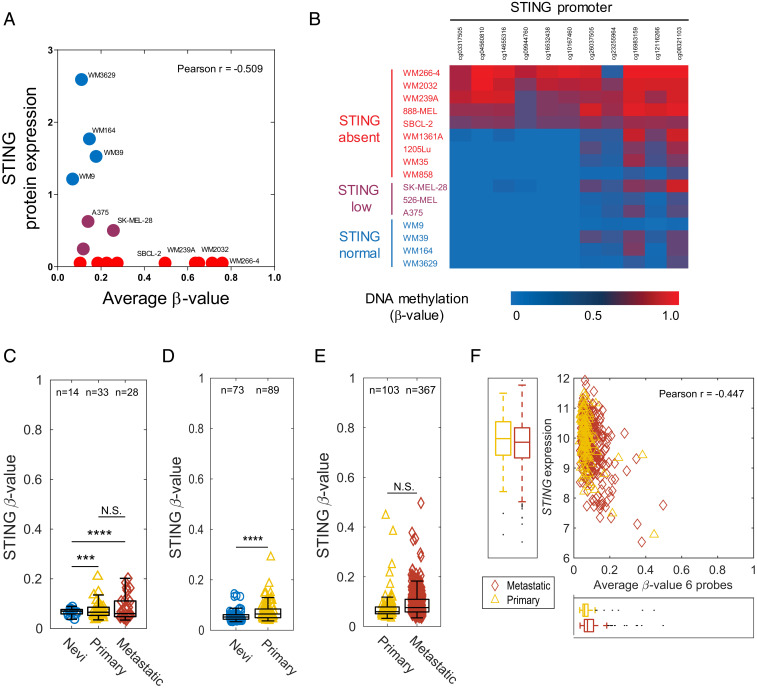
We have previously reported that STING and cGAS protein expression were down-regulated or lost in a large subset of human melanoma cell lines (∼50% for STING and ∼35% for cGAS) (18, 19). To determine the role of DNA methylation in melanoma STING and cGAS silencing, we performed genome-wide DNA methylation profiling using the Illumina MethylationEPIC BeadChip microarray platform across a panel of 16 human melanoma cell lines. We assessed methylation changes in the 18 CpG probes for STING and presented them as β-values (SI Appendix, Fig. S1A). In parallel, we performed immunoblot analysis and quantified STING protein expression relative to β-actin for each cell line and determined the degree of correlation between the β-value in each probe and the STING protein expression. We observed a negative correlation between STING protein expression and DNA methylation in eleven CpG probes (5–15) located in the 5′-untranslated region (5′-UTR) and/or transcription start sites TS1500 and TSS200 (SI Appendix, Fig. S1A). Subsequent correlative analysis using the average β-value for these eleven CpG probes confirmed an inverse association between hypermethylation of STING and the amount of protein expression (Pearson’s r = −0.51) (Fig. 1A). In addition, a β-value heat map of these CpG probes identified three distinct sample subclasses based on their STING protein expression (STINGabsent, STINGlow, and STINGnormal) (Fig. 1B). Among STINGabsent cell lines, WM266-4, WM239A, WM2032, and 888-MEL indicated a high degree of methylation. We also found a high degree of methylation (β-value > 0.7) in two CpG probes (cg16983159 and cg08321103) for WM1361A. Although STING was only expressed in melanoma cell lines showing no promoter methylation (WM3629, WM164, WM39, and WM9), we found one STINGabsent (WM858) and one STINGlow (526-MEL) melanoma cell line that did not indicate promoter hypermethylation. Loss of STING in these cases might be mediated through additional genetic and/or epigenetic alterations such as histone modifications or factors involving microRNAs (24, 25).
Fig. 1.
Promoter hypermethylation of STING correlates with its silencing in human melanoma. Correlative analysis between STING promoter methylation (average β-values for 11 probes) and its protein expression in 16 human melanoma cell lines. Cells are colored based on their STING protein expression (red: no expression, purple: low expression, and blue: normal expression) (A). β-value heat map showing DNA methylation levels for 11 STING probes identifies three distinct sample subclasses based on their STING protein expression (STINGabsent, STINGlow, and STINGnormal) among melanoma cell lines (B). Box plots showing the average β-values (six probes) for STING across nevi, primary, and metastatic melanoma samples in GSE86355 (C), GSE12087 (D), and TCGA SKCM datasets (E). Box plot represents the lower, median, and upper quartile, while whiskers represent the highest and lowest range for the upper and lower quartiles. Each data point represents an individual sample. Numbers above boxplots indicate number of samples in each group. The statistical analysis was performed by Bartlett’s test (***P < 0.001; ****P < 0.0001; N.S., not significant) (C–E). Correlative analysis between STING gene expression and promoter methylation (average β-values for six probes) in primary (n = 102) and metastatic (n = 367) melanoma samples (F).
To validate the clinical relevance of these observations, we next assessed promoter hypermethylation of STING in human nevi, primary, and metastatic melanoma samples using three independent and publicly available gene methylation datasets from The Cancer Genome Atlas (TCGA) skin cutaneous melanoma (SKCM) project and Gene Expression Omnibus (GEO) accessions GSE86355 (24) and GSE120878 (25). While nevi samples consistently displayed low levels of STING promoter hypermethylation, we observed a notable increase in STING β-values both in primary and metastatic samples (Fig. 1 C–E). Furthermore, correlative analysis using TCGA RNA sequencing data indicated that hypermethylation of STING promoter was associated with its transcriptional silencing (Pearson’s r = −0.44) in primary and metastatic melanomas (Fig. 1F). These findings collectively indicate that hypermethylation of STING promoter occurs at the early stage of neoplasia and persists as malignant transformation proceeds, suggesting a possible role for STING silencing in enabling immune evasion in melanoma.
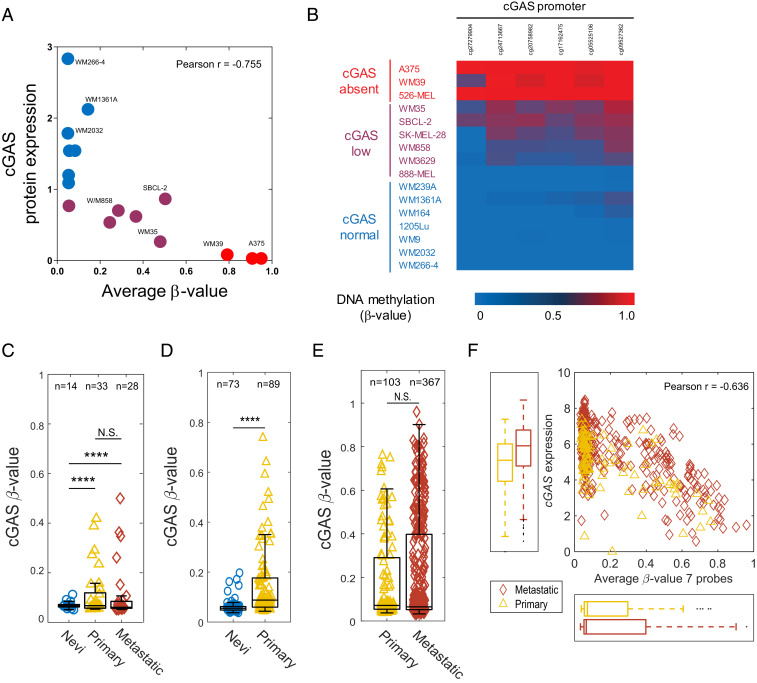
Using a similar genome-wide DNA methylation profiling approach described for STING, we next assessed methylation changes in the eight CpG probes for cGAS and determined the degree of correlation between the β-values and the cGAS protein expression in 16 human melanoma cell lines (SI Appendix, Fig. S1B). We observed a negative correlation between cGAS protein expression and DNA methylation in probes 1 through 6. These hypermethylated CpG probes were located either in the first exon or in the transcription start site TSS200. Using the average β-value for these six CpG probes, we identified a strong inverse correlation between hypermethylation of cGAS and the amount of protein expression (Pearson’s r = −0.75) (Fig. 2A). Similarly, a β-value heat map of these CpG probes clearly demarcated three sample subclasses based on their cGAS protein expression (cGASabsent, cGASlow, and cGASnormal) (Fig. 2B). Notably, all cGASabsent cell lines (526-MEL, A375, and WM39) exhibited high levels of methylation in these CpG probes.
Fig. 2.
Promoter hypermethylation of cGAS is strongly associated with its silencing in human melanoma. Correlative analysis between cGAS promoter methylation (average β-values for six probes) and its protein expression in 16 human melanoma cell lines. Cells are colored based on their cGAS protein expression (red: no expression, purple: low expression, and blue: normal expression) (A). β-value heat map showing DNA methylation levels for six cGAS probes identifies three distinct sample subclasses based on their cGAS protein expression (cGASabsent, cGASlow, and cGASnormal) among melanoma cell lines (B). Box plots showing the average β-values (seven probes) for cGAS across nevi, primary, and metastatic melanoma samples in GSE86355 (C), GSE12087 (D), and TCGA datasets (E). Box plot represents the lower, median, and upper quartile, while whiskers represent the highest and lowest range for the upper and lower quartiles. Each data point represents an individual sample. Numbers above boxplots indicate number of samples in each group. The statistical analysis was performed by Bartlett’s test (****P < 0.0001; N.S., not significant) (C–E). (F) Correlative analysis between cGAS gene expression and promoter methylation (average β-values for seven probes) in primary (n = 102) and metastatic (n = 367) melanoma samples (F).
Consistent with these observations, our gene methylation analysis in human nevi, primary, and metastatic melanoma samples confirmed a significant increase in hypermethylation of cGAS promoter among primary and metastatic melanomas in three independent datasets [GSE86355 (24), GSE120878 (25), and TCGA SKCM] (Fig. 2 C–E). Additionally, using a correlative analysis by incorporating RNA-sequencing data from TCGA SKCM, we identified a negative correlation between hypermethylation of cGAS and its gene expression (Pearson’s r = −0.54) in primary and metastatic melanomas (Fig. 2F). These findings not only reinforce the relevance of promoter hypermethylation-mediated silencing of cGAS in human melanomas but also suggest its involvement in immune evasion mechanisms that facilitate the process of malignant transformation.
Reconstitution of STING Expression through DNA Demethylation Can Rescue STING Signaling in Human Melanoma Cell Lines.
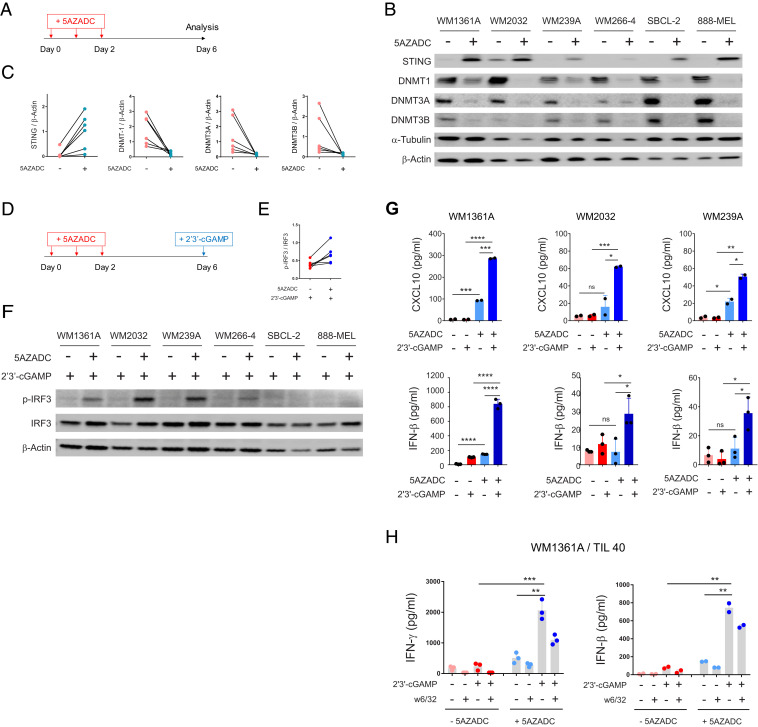
To assess whether DNA demethylation could restore STING expression and functional STING signaling, we next treated six STINGabsent melanoma cell lines that indicated STING promoter hypermethylation (WM1361A, WM2032, WM239A, WM266-4, 888-MEL, and SBCL-2) with a clinically available DNA methyltransferase (DNMT) inhibitor, 5-aza-2’-deoxycytidine (5AZADC) (Fig. 3A). Using immunoblot analysis, we confirmed reconstitution of STING expression in all the cell lines following 5AZADC treatment, although to varying degrees (Fig. 3B). We also observed a marked reduction of DNMT1, DNMT3A, and DNMT3B following reconstitution of STING (Fig. 3 B and C). Along similar lines, genetic depletion of DNMT1 and DNMT3B using small interfering RNA (siRNA) transfection also resulted in re-expression of STING in WM1361A cells (SI Appendix, Fig. S2), further suggesting their involvement in melanoma-STING silencing.
Fig. 3.
Reconstitution of STING expression through DNA demethylation can rescue STING signaling in human melanoma cell lines. Experimental setup for 5AZADC treatment of STINGabsent human melanoma cell lines at indicated time points (A). Immunoblot analysis of STING, DNMT1, DNMT3A, and DNMT3B expression in human melanoma cell lines with or without 5AZADC treatment. α-Tubulin and β-Actin were used as loading controls (B). Ratio of total STING, DNMT1, DNMT3A, and DNMT3B relative to β-Actin for each cell line with or without 5AZADC treatment (C). Experimental setup for 5AZADC treatment and 2’3′-cGAMP stimulation of STINGabsent human melanoma cell lines at indicated time points (D). Ratio of p-IRF3 relative to total IRF3 (E) and immunoblot analysis of p-IRF3 and IRF3 in indicated melanoma cell lines after 4 h stimulation with 2’3′-cGAMP (F). Induction of CXCL10 (Top) and IFN-β (Bottom) in cell culture supernatants measured using ELISA and reported as mean ± SD for two or three biological replicates (G). WM1361A melanoma cells with or without 5AZADC pretreatment were cocultured with HLA-matched human melanoma TIL 40 in the presence or absence of 2’3′-cGAMP. In some experimental conditions, tumor cells were preincubated with an MHC class I blocking Ab (W6/32) to determine whether IFN-γ release was mediated by CD8+ TIL T-cell receptor engagement with peptide/MHC class I. Coculture supernatants were collected after 24 h, and IFN-γ and IFN-β concentrations were measured using ELISA and reported as mean ± SD for two or three biological replicates (H). Statistical significance was determined by unpaired t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
To directly evaluate whether demethylation could restore functional activation of STING signaling following 5AZADC treatment, we next stimulated STINGabsent melanoma cell lines with the STING agonist 2’3′-cGAMP (Fig. 3D). We observed phosphorylation of IRF3, a critical downstream regulatory element for STING-dependent type I IFN induction (26), in four of six 5AZADC-pretreated melanoma cell lines (WM1361A, WM2032, WM239A, and WM266-4) following their stimulation with 2’3′-cGAMP (Fig. 3 E and F). We also determined STING-dependent CXCL10 and IFN-β induction in cell culture supernatants. Three of six 5AZADC-pretreated melanoma cell lines (WM1361A, WM2032, and WM239A) induced CXCL10 and IFN-β expression in response to stimulation with 2’3′-cGAMP, which confirmed activation of STING signaling (Fig. 3G). In contrast, we did not detect STING-dependent induction of CXCL10 and IFN-β in three remaining 5AZADC-pretreated melanoma cell lines (WM266-4, 888-MEL, and SBCL-2) (SI Appendix, Fig. S3), arguing the possibility that additional defective elements downstream of STING may contribute to the impairment of this pathway.
We next examined whether demethylation-mediated rescue of STING signaling in melanoma cells could enhance their antigenicity. To address this possibility, we performed coculture experiments using 5AZADC-pretreated WM1361A [human leukocyte antigen (HLA)-A1/A32] melanoma cell line with HLA-matched human melanoma TIL (TIL 40; HLA-A2/32) in the presence or absence of 2’3′-cGAMP and assessed TIL production of IFN-γ. We also included experimental conditions in which WM1361A cells were preincubated with an MHC class I blocking Ab (W6/32) to determine whether IFN-γ release was mediated by CD8+ TIL T cell receptor engagement with peptide/MHC class I. Indeed, 5AZADC-pretreated WM1361A melanoma cells triggered more than an eightfold higher IFN-γ release by TIL 40 compared to untreated controls in the presence of 2’3′-cGAMP (∼2,000 pg/mL, P < 0.001) (Fig. 3H). We also observed blockade of IFN-γ release in the presence of the MHC class I blocking Ab (W6/32), which confirmed MHC class I–mediated CD8+ reactivity. We also found increased IFN-β induction (P < 0.01) in 5AZADC-pretreated WM1361A coculture groups in response to 2′3′-cGAMP stimulation, which indicated reactivation of STING signaling. Taken together, these data show that reactivation of STING signaling through epigenetic reprogramming enhances antigenicity of melanoma cells.
DNA Demethylation Can Restore cGAS-Dependent STING Activation in cGASabsent Melanoma Cell Lines.
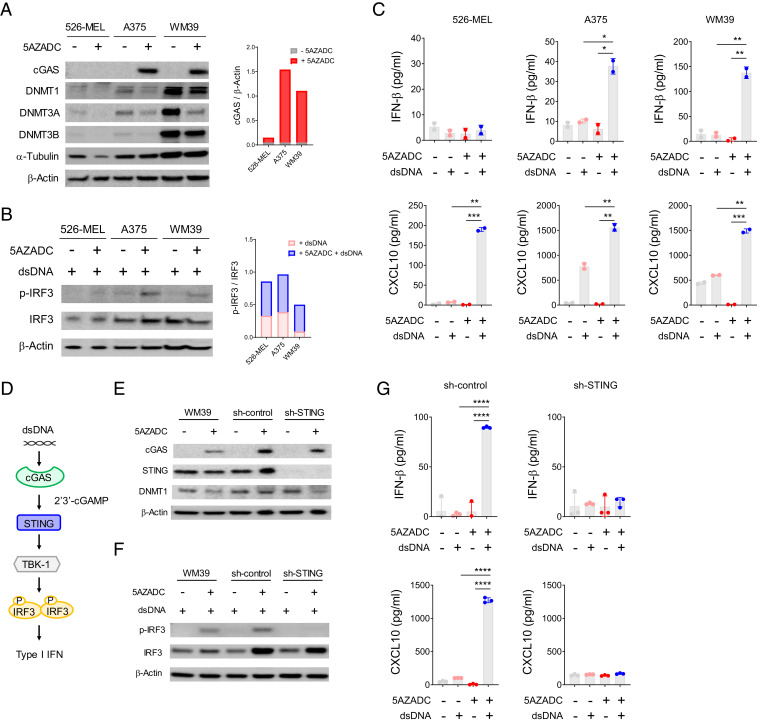
We next examined the effect of DNA demethylation on re-expression of cGAS in three cGASabsent human melanoma cell lines. Reconstitution of cGAS expression was detected in A375 and WM39 but not in 526-MEL following 5AZADC treatment (Fig. 4A). To evaluate cGAS-dependent activation of STING signaling, we next stimulated these melanoma cell lines with double stranded DNA (dsDNA) following 5AZADC treatment. We observed phosphorylation of IRF3 (Fig. 4B) and induction of IFN-β and CXCL10 (Fig. 4C) for 5AZADC-pretreated A375 and WM39 in response to stimulation with dsDNA. As expected, 526-MEL for which 5AZADC treatment failed to restore cGAS expression did not indicate phosphorylation of IRF3 and induction of IFN-β following dsDNA stimulation. Although our DNA methylation microarray data indicated hypermethylation of cGAS promoter, low expression of DNMT1, DNMT3A, and DNMT3B in 526-MEL (Fig. 4A) suggests the involvement of DNMT-independent regulators in its cGAS suppression. In addition, 526-MEL lacked STING expression, and consistent with our previous observation that showed STING loss in this cell line is mediated through DNA methylation–independent processes (Fig. 1B and SI Appendix, Fig. S1A), we did not find reconstitution of STING following 5AZADC treatment (Fig. 4A), further indicating methylation-independent impairment of STING signaling in this cell line.
Fig. 4.
DNA demethylation can restore cGAS-dependent STING activation in cGASabsent melanoma cell lines. Immunoblot analysis of cGAS, STING, DNMT1, DNMT3A, and DNMT3B expression in three cGASabsent human melanoma cell lines (526-MEL, A375, and WM39) with or without 5AZADC treatment. α-Tubulin and β-Actin were used as loading controls. Ratio of total cGAS relative to β-actin for each cell line was quantified using ImageJ software (A). Immunoblot analysis and ratio of p-IRF3 relative to IRF3 in indicated melanoma cell lines after 4 h stimulation with dsDNA (B). Induction of IFN-β (Top) and CXCL10 (Bottom) in indicated melanoma cell lines after stimulation with dsDNA measured using ELISA (C). Schematic of the cGAS-dependent STING signaling activation (D). Immunoblot analysis of cGAS, STING, and DNMT1 in wild-type or stably transduced WM39 cells with a lentiviral shRNA specific for sh-STING or sh-control with or without 5AZADC treatment (E). Immunoblot analysis of p-IRF3 and total IRF3 (F) and induction of IFN-β (Top) and CXCL10 (Bottom) (G) in indicated melanoma cell lines in response to stimulation with dsDNA. Data are mean ± SD of two or three biological replicates. Statistical significance was determined by unpaired t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
In our next experiments, we sought to directly determine whether dsDNA-induced IFN-β production in epigenetically reprogrammed melanoma cells is STING mediated (Fig. 4D). To address this possibility, we stably transduced WM39 cells with a lentiviral short hairpin RNA (shRNA) specific for STING (sh-STING) or nontarget shRNA (sh-control). Despite similar cGAS expression reconstitution following 5AZADC treatment (Fig. 4E), unlike sh-control and WM39 cells, sh-STING cells did not indicate phosphorylation of IRF3 (Fig. 4F) and failed to induce IFN-β and CXCL10 (Fig. 4G) in response to stimulation with dsDNA. These findings confirm that dsDNA-induced downstream effects in 5AZADC-pretreated melanoma cells depend on activation of STING signaling and exclude the possibility of STING-independent mechanisms.
Demethylation-Mediated Reversal of cGAS Silencing Enhances Antigenicity and T Cell Recognition of Melanoma Cells.
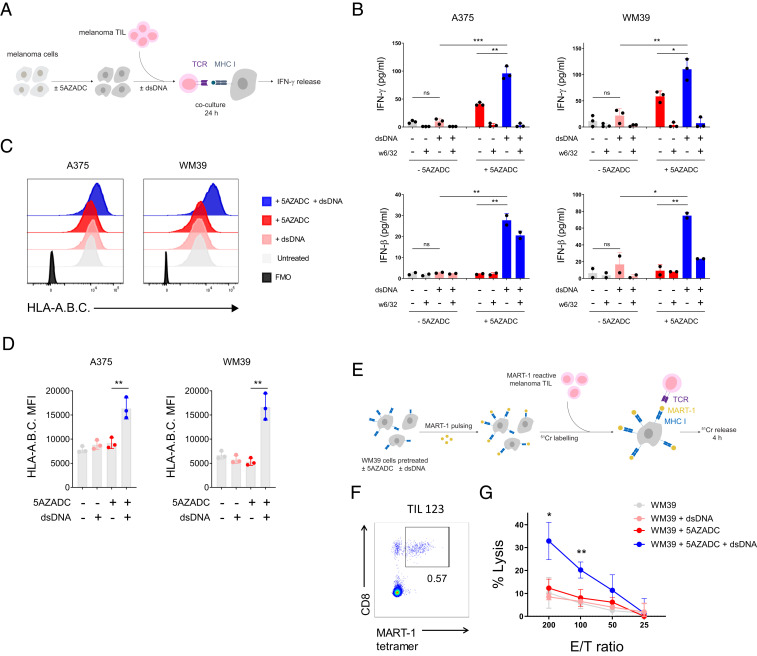
To examine whether demethylation-mediated reconstitution of cGAS and subsequent rescue of STING signaling in cGASabsent melanoma cells could improve their antigenicity, we performed coculture experiments using 5AZADC-pretreated WM39 melanoma cell line (HLA-A2) and HLA-A2–restricted human melanoma TIL (TIL19) in the presence or absence of dsDNA and assessed TIL production of IFN-γ (Fig. 5A). We also included experimental conditions with WM39 cells preincubated with W6/32 (MHC class I blocking Ab) to confirm MHC class I–mediated CD8+ TIL reactivity. We also measured IFN-β expression in coculture supernatants to confirm dsDNA-triggered cGAS-dependent activation of STING signaling. We observed more than a fivefold higher IFN-γ release by TIL 19 for 5AZADC-pretreated WM39 melanoma cells compared to the untreated controls in the presence of dsDNA (P < 0.01) (Fig. 5B), suggesting that rescue of cGAS-dependent STING signaling through epigenetic reprogramming could enhance their antigenicity.
Fig. 5.
Demethylation-mediated reversal of cGAS silencing enhances antigenicity and T cell recognition of melanoma. Schematic of the experimental procedure for the IFN-γ release assay. cGASabsent human melanoma cell lines were pretreated with 5AZADC and cocultured with their HLA-matched human melanoma TIL in the presence or absence of dsDNA for 24 h (A). IFN-γ (Top) and IFN-β (Bottom) concentrations in supernatants were measured using ELISA (B). In some experimental conditions, tumor cells were preincubated with an MHC class I blocking Ab (W6/32) to determine CD8+ TIL reactivity. Representative histograms (C) and mean fluorescence intensity (MFI) of HLA-A.B.C expression on indicated melanoma cell lines (D). Schematic of the experimental procedure for the 51Cr release cytotoxicity assay using MART-1–pulsed WM39 cells (±5AZADC and/or ±dsDNA pretreatment) as target cells and MART-1–reactive TIL as effector cells (E). Frequency of MART-1 tetramer+ T cells in human melanoma TIL 123 (F). Specific lysis of MART-1–pulsed WM39 (±5AZADC and/or ±dsDNA) targets by TIL 123 at the indicated effector/target (E/T) ratios (G). All data are mean ± SD for two or three biological replicates. P values were calculated by one-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant).
For our next epigenetically reprogramed cGAS-recovered model, we selected A375, another HLA-A2–restricted cGASabsent melanoma cell line, and used it in coculture experiments with HLA-A2–restricted TIL 19. Similarly, we found a marked increase (P < 0.01) in IFN-γ secretion by TIL 19 in 5AZADC-pretreated cocultures of A375 compared to the untreated controls in the presence of dsDNA (Fig. 5B). Consistent with the 5AZADC-pretreated WM39 cocultures, we also observed higher IFN-β induction (P < 0.01) for 5AZADC-pretreated cocultures compared to their untreated controls in the presence of dsDNA which indicated cGAS-dependent activation of STING signaling (Fig. 5B).
Following our observation of enhanced antigenicity triggered by the restoration of STING signaling, we next examined MHC class I surface expression on melanoma cell lines with or without 5AZADC pretreatment and dsDNA stimulation. Indeed, we observed increased MHC class I surface expression exclusively for 5AZADC-pretreated A375 and WM39 melanoma cell lines in response to stimulation with dsDNA (Fig. 5 C and D). In contrast, dsDNA stimulation did not change MHC class I surface expression for the 5AZADC-pretreated 526-MEL cell line (SI Appendix, Fig. S4), which indicates intact activation of STING signaling is required for dsDNA-induced up-regulation of MHC class I. Given the importance of MHC class I in tumor antigen presentation and T cell recognition, these findings suggest a functional role for reactivation of melanoma-intrinsic STING signaling in mediating effective antitumor T cell responses.
To better investigate the impact of demethylation-mediated rescue of STING signaling on antigen presentation and immune T cell killing, following 5AZADC pretreatment and dsDNA stimulation, we pulsed WM39 cells with MART-1 [a melanoma-specific peptide recognized by HLA-A2–restricted TILs (26)] and used them as target cells in a 51Cr release cytotoxicity assay (Fig. 5E) with MART-1–reactive human melanoma TIL 123 (Fig. 5F) as effector cells. Despite their MART-1 pulsing, no significant lysis of the control target cells (WM39, WM39+5AZADC, and WM39+dsDNA) was found by TIL 123. However, 5AZADC pretreatment and dsDNA stimulation of WM39 targets resulted in more than a threefold increase (P < 0.05) in their specific lysis by TIL 123 (Fig. 5G). These results highlight how methylation silencing of cGAS in melanoma cells can limit tumor antigen presentation and thereby drive resistance to cytotoxic T cell killing.
In conclusion, we provide evidence that methylation silencing of cGAS and STING is not only a notable mechanism of STING signaling dysfunction in melanoma but also plays a role in tumor antigen presentation and recognition by TIL. By using genome-wide methylation profiling, we show that a strong correlation exists between the loss of STING and cGAS expression and their gene promoter DNA methylation in clinically-relevant human melanomas and melanoma cell lines. Reversal of these repressions using a DNMT inhibitor can restore STING activity and, subsequently, can improve antigenicity in a subset of melanoma cell lines. In demethylation-mediated cGAS-restored melanoma cell lines, activation of STING can augment MHC class I surface expression and improve tumor antigen presentation and tumor cell destruction by immune T cells. Thus, epigenetic silencing of cGAS and STING in melanoma can contribute to tumor immune evasion and can mediate resistance to TIL-based immunotherapies. These findings also have more general clinical implications. In particular, the ability to target and restore these defects using epigenetic modulators provides a rationale to further explore the development of additional therapeutic solutions for patients who do not benefit from current immunotherapeutic interventions, for example, those that not only involve TIL-based adoptive transfers but also checkpoint antibody blockade, among others.
Materials and Methods
Melanoma Cell Lines.
Human melanoma cell lines 1205Lu, A375, SBCL2, SK-MEL-28, WM1361A, WM164, WM2032, WM239A, WM266-4, WM35, WM3629, WM39, WM858, WM9 (provided by Dr. Keiran Smalley, Moffitt Cancer Center, Tampa, FL), and 526-MEL and 888-MEL (established at the Surgery Branch, National Cancer Institute [NCI]/NIH, Bethesda, MD) were maintained as monolayers in complete medium consisting of Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics. HLA typing of melanoma cell lines was performed by the HLA Laboratory (American Red Cross, Dedham, MA). A375, WM39, WM1361A, and 526-MEL were HLA-A typed as A01/02, A01/02, A01/32, and A02/03, respectively. Genomic DNA for all cell lines was extracted using the Blood & Cell Culture DNA Mini Kit (Qiagen), according to the manufacturer’s instructions for cultured cells. For siRNA transfection, tumor cells were transfected with siRNA (20 nM) specific for DNMT1 (catalog no. L-004605-00-0005), DNMT3B (catalog no. L-006395-00-0005) or control siRNA (catalog no. D-001810-10-20) (Dharmacon) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were subjected to further analyses following 48 h incubation at 37 °C. Knockdown of STING in WM39 cells was achieved using lentiviral particles carrying a target gene sequence for human STING (TMEM173) (catalog no. TL307876V) or scrambled control (catalog no. TR30021V) (Origene Technologies). The targeting sequence for STING was 5′-GCAACAGCATCTATGAGCTTCTGGAGAAC. Transduced cells were selected by addition of puromycin (0.5 μg/mL) to the medium 24 h after infection.
Analysis of Methylation Data.
All data preprocessing was performed within the R (version 3.5.2) statistical programming language (27) and computing environment using the minfi package (version 1.28.4) (28, 29), which is available through the Bioconductor (version 3.8) project (30). Preprocessing began with the reading and parsing of the intensity data (IDAT) files containing the signal intensity data from the Infinium MethylationEPIC BeadChip microarray (31). The probe-level detection P values were calculated from the raw intensities using the minfi function detectionP. Utilizing the preprocess FunNorm function, also provided by minfi, the signal intensities were first subjected to dye normalization and background correction, using the NOOB method (32), followed by functional normalization (FunNorm), a between-array normalization (33). The resulting GenomicRationSet object contained the β-values calculated from the corrected methylated (M) and unmethylated (U) intensities using the following formula: β=M/(M+U). The analysis of the normalized methylation data and creation of figures was performed in MATLAB R2019B.
Database Analysis.
Three additional melanoma datasets—The Cancer Genome Atlas (TCGA) skin cutaneous melanoma (SKCM) project, GSE120878 (25), and GSE86355 (24)— all assayed on the Illumina Infinium HumanMethylation450 BeadChip, were also analyzed. IDAT files were available for both SKCM and GSE120878. The raw, Level-1 files for SKCM were downloaded from the Genomic Data Commons (GDC) legacy archive on September 15, 2020 using TCGAbiolinks (34), an R package, while the GSE120878 IDAT files were accessed from the GEO website, also on September 15, 2020. Preprocessing for these two datasets proceeded as described in the previous section. For the GSE86355 dataset, the IDAT files were not available, but the authors supplied beta values, derived from data preprocessed using “preprocessIllumina” from the minfi package (28, 29), as well as the detection P values. The detection P values and the beta values were extracted from the SOFT formatted file, downloaded from the GEO website on September 16, 2020, with a python script, utilizing the regular expression capabilities of the re package.
5AZADC Treatment.
Human melanoma cell lines were treated with 0.1 to 1 μM 5AZADC (Sigma-Aldrich) dissolved in culture medium that was prepared and replaced daily. At day 3, cells were washed and replenished with fresh culture medium (without 5AZADC) and rested for an additional 3 d before assaying (day 6).
STING Agonist Stimulation.
Human melanoma cell lines (4 × 105 cells/well in 24-well plates) were exposed to 2′3′-cGAMP (10 µg/mL) or dsDNA (10 µg/mL) in the presence of Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions as previously described (18). After 4 or 24 h of incubation at 37 °C in a humidified CO2 incubator, the supernatants were collected for detection of CXCL10 and IFN-β release using enzyme-linked immunosorbent (ELISA) assays (Quantikine ELISA Kit, R&D Systems), and cells were scraped, washed, and lysed for assessment of IRF3 phosphorylation by immunoblot.
Immunoblot Analysis.
Proteins were extracted with radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) containing protease inhibitors (Thermo Scientific). Equal amounts of proteins were resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels (Bio-Rad) and transferred to polyvinylidene fluoride membranes (Bio-Rad). After blocking with 5% nonfat dry milk, membranes were incubated with antibodies specific for STING, cGAS, DNMT1, DNMT3A, DNMT3B, p-IRF3, IRF3, α-Tubulin (all from Cell Signaling), and β-actin (Sigma-Aldrich). Following incubation with appropriate secondary antibodies, bands were visualized using an enhanced chemiluminescence detection system.
Preparation of TIL.
Melanoma TIL were established as described previously (35). Briefly, melanomas were minced into 1 to 2 mm3 fragments and plated in 24-well plates with 2 mL TIL culture medium (TIL-CM) containing 6,000 IU/mL IL-2 (Proleukin) per well. The TIL-CM consisted of RPMI 1640, 2.05 mM L-glutamine (HyClone, Thermo Fisher Scientific), 10% heat-inactivated human AB serum (Omega Scientific), 55 μM 2-mercaptoethanol (Invitrogen), 50 μg/mL gentamicin (Invitogen), 100 IU/mL penicillin, 100 μg/mL streptomycin, and 10 mM Hepes Buffer (Mediatech). Half of the medium was replaced every 2 to 3 d or cells were split when 90% confluent. TIL were expanded for 3 to 5 wk. HLA typing of TIL was performed by the HLA Laboratory (American Red Cross, Dedham, MA). TIL 40, TIL 19, and TIL 123 were HLA-A typed as A02/32, A02/26, and A02/11 respectively.
Coculture Assay.
As previously described (18), 1 × 105 of melanoma cells were cultured with TIL at a 1:1 ratio with or without 2′3′-cGAMP (10 µg/mL) in 96-well round-bottom plates. After 24 h of incubation at 37 °C in a humidified CO2 incubator, the supernatant was harvested for detection of IFN-γ release using enzyme-linked immunosorbent assay (Human IFN-γ Quantikine ELISA Kit, R&D Systems). For the MHC class I blocking assay, melanoma cells were incubated with W6/32 (anti-HLA-A,B,C monoclonal antibody, BioLegend) at a final concentration of 50 μg/mL for 1 h at 37 °C prior to the addition of TIL.
51Cr-Release Assay.
Lysis of melanoma cell targets by their HLA-matched TIL cultures was measured in a standard 5lCr release assay, as previously described (36). Briefly, 1 × 106 melanoma cells were labeled with 100 μCi of 51Cr (Amersham Corp) for 2 h at 37 °C. Following three washes with Hanks' balanced salt solution (HBSS), labeled target cells were resuspended in TIL CM at a concentration of 5 × 104 tumor cells/mL and added to the effector cells at different effector-to-target cell ratios in a 96-well plate and incubated at 37 °C. In addition, two control conditions were included in this assay: a minimum release control containing just the target cells and a maximum release control in which target cells were lysed by Triton X-100. After 4 h, supernatant was harvested and measured in TriLux (PerkinElmer). Each point represented the average of quadruplicate wells and percentage of specific lysis was calculated by the following: (experimental release − minimum release) / (maximum release − minimum release) × 100.
Flow Cytometry.
Cells were resuspended in staining buffer [phosphate-buffered saline (PBS) containing 10% FBS and 0.5 M ethylenediaminetetraacetic acid (EDTA) (Ambion)] and stained with HLA-A.B.C–PE antibody (BioLegend, clone W6/32) for 30 min at 4 °C in the dark. Staining of MART-1–specific T cells was performed using MART-1-tetramer-PE (MBL), for 20 min at 37 °C. Following a washing step, cells were stained with CD8-FITC antibody (BioLegend, clone HIT8a) for 30 min at 4 °C in the dark. 4',6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) was used as a viability dye. Sample acquisition was performed on an LSR II flow cytometer (BD Biosciences), and the data were analyzed using FlowJo software (Tree Star).
Statistical Methods.
Statistical analyses were performed using GraphPad Prism7 software as previously described (18). All data are presented as mean ± SD. Means for all data were compared by one-way ANOVA or unpaired t test as described in the figure legends. P values of statistical significance are represented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
This work was supported by the Moffitt Cancer Center Flow Cytometry and Molecular Genomics Core Facilities, all comprehensive cancer center facilities designated by the NCI (P30-CA076292). This work was funded by the NCI-NIH (1R01 CA148995, 1R01 CA184845, P30 CA076292, and P50 CA168536), Cindy and Jon Gruden Fund, Chris Sullivan Fund, V Foundation, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Footnotes
Competing interest statement: J.J.M. is Associate Center Director at Moffitt Cancer Center, has ownership interest in Fulgent Genetics, Inc., Aleta Biotherapeutics, Inc., CG Oncology, Inc., Myst Pharma, Inc., Verseau Therapeutics, Inc., AffyImmune, Inc., and Tailored Therapeutics, Inc., and is a paid consultant/paid advisory board member for ONCoPEP, Inc., CG Oncology, Inc., Morphogenesis, Inc., Mersana Therapeutics, Inc., GammaDelta Therapeutics, Ltd., Myst Pharma, Inc., Tailored Therapeutics, Inc., Verseau Therapeutics, Inc., Iovance Biotherapeutics, Vault Pharma, Inc., Noble Life Sciences, Fulgent Genetics, Inc., UbiVac, LLC, Vycellix, Inc., AffyImmune, Inc., and Aleta Biotherapeutics, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013598118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Schreiber R. D., Old L. J., Smyth M. J., Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Khong H. T., Wang Q. J., Rosenberg S. A., Identification of multiple antigens recognized by tumor-infiltrating lymphocytes from a single patient: Tumor escape by antigen loss and loss of MHC expression. J. Immunother. 27, 184–190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campoli M., Ferrone S., HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance. Oncogene 27, 5869–5885 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaretsky J. M., et al., Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao J., et al., Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manguso R. T., et al., In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin D. S., et al., Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 7, 188–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zitvogel L., Galluzzi L., Kepp O., Smyth M. J., Kroemer G., Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Sistigu A., et al., Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20, 1301–1309 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Diamond M. S., et al., Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med. 208, 1989–2003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuertes M. B., et al., Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J. Exp. Med. 208, 2005–2016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo S.-R., et al., STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa H., Barber G. N., STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corrales L., et al., Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., et al., cGAS/STING axis mediates a topoisomerase II inhibitor-induced tumor immunogenicity. J. Clin. Invest. 129, 4850–4862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitajima S., et al., Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov. 9, 34–45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falahat R., et al., STING signaling in melanoma cells shapes antigenicity and can promote antitumor T-cell activity. Cancer Immunol. Res. 7, 1837–1848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia T., Konno H., Barber G. N., Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 76, 6747–6759 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Peng D., et al., Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527, 249–253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burr M. L., et al., An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell 36, 385–401.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L., Mudianto T., Ma X., Riley R., Uppaluri R., Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti-PD-1 resistance in head and neck cancer. Clin. Cancer Res. 26, 290–300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly T. K., De Carvalho D. D., Jones P. A., Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wouters J., et al., Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med. 15, 101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway K., et al., Identification of a robust methylation classifier for cutaneous melanoma diagnosis. J. Invest. Dermatol. 139, 1349–1361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y., Chen Z. J., STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. C. Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013). [Google Scholar]
- 28.Aryee M. J., et al., Minfi: A flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortin J.-P., Triche T. J. Jr, Hansen K. D., Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 33, 558–560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentleman R. C., et al., Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pidsley R., et al., Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17, 208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triche T. J. Jr, Weisenberger D. J., Van Den Berg D., Laird P. W., Siegmund K. D., Low-level processing of Illumina Infinium DNA methylation beadarrays. Nucleic Acids Res. 41, e90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fortin J.-P., et al., Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 15, 503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colaprico A., et al., TCGAbiolinks: An R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44, e71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pilon-Thomas S., et al., Efficacy of adoptive cell transfer of tumor-infiltrating lymphocytes after lymphopenia induction for metastatic melanoma. J. Immunother. 35, 615–620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu G., et al., Induction of tertiary lymphoid structures with antitumor function by a lymph node-derived stromal cell line. Front. Immunol. 9, 1609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.