Significance
The most severe forms of autism spectrum disorder frequently occur in people with large head sizes, many of whom have deletions of a small piece of chromosome 16, known as 16p11.2. We use stem cells derived from people with 16p11.2 deletion syndrome to understand why they exhibit brain overgrowth. We find that CD47 (a protein that signals “don’t eat me”) is overexpressed in the neural brain cells of people with 16p11.2 deletion syndrome, and their unhealthy neural cells are therefore eliminated at lower rates by their immune cells. Blocking CD47 restores the elimination of unhealthy cells to normal rates. These findings suggest that targeting the CD47 pathway may be helpful in identifying treatments for psychiatric disorders associated with brain overgrowth.
Keywords: 16p11.2 deletion, CD47, iPSCs, macrocephaly
Abstract
Copy number variation (CNV) at the 16p11.2 locus is associated with neuropsychiatric disorders, such as autism spectrum disorder and schizophrenia. CNVs of the 16p gene can manifest in opposing head sizes. Carriers of 16p11.2 deletion tend to have macrocephaly (or brain enlargement), while those with 16p11.2 duplication frequently have microcephaly. Increases in both gray and white matter volume have been observed in brain imaging studies in 16p11.2 deletion carriers with macrocephaly. Here, we use human induced pluripotent stem cells (hiPSCs) derived from controls and subjects with 16p11.2 deletion and 16p11.2 duplication to understand the underlying mechanisms regulating brain overgrowth. To model both gray and white matter, we differentiated patient-derived iPSCs into neural progenitor cells (NPCs) and oligodendrocyte progenitor cells (OPCs). In both NPCs and OPCs, we show that CD47 (a “don’t eat me” signal) is overexpressed in the 16p11.2 deletion carriers contributing to reduced phagocytosis both in vitro and in vivo. Furthermore, 16p11.2 deletion NPCs and OPCs up-regulate cell surface expression of calreticulin (a prophagocytic “eat me” signal) and its binding sites, indicating that these cells should be phagocytosed but fail to be eliminated due to elevations in CD47. Treatment of 16p11.2 deletion NPCs and OPCs with an anti-CD47 antibody to block CD47 restores phagocytosis to control levels. While the CD47 pathway is commonly implicated in cancer progression, we document a role for CD47 in psychiatric disorders associated with brain overgrowth.
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by deficits in social interaction and communication. Copy number variation (CNV) at the 16p11.2 locus is associated with ASD (1–8). People who have 16p11.2 deletion syndrome tend to have larger head circumferences (macrocephaly), with disproportionate enlargement in both gray and white matter volume (8–13). Individuals with ASD and macrocephaly have more severe behavioral and cognitive problems and are less responsive to standard medical and therapeutic interventions than those with ASD and normal head circumferences (14). In addition, prior work has documented a very strong cross-sectional and temporal association between macrocephaly and ASD symptoms (8, 9, 11, 12, 14–17). These findings suggest that understanding the underlying mechanisms regulating macrocephaly could provide a window of opportunity for intervention or mitigation of symptoms.
Here, we used patient-derived human induced pluripotent stem cells (hiPSCs) to interrogate the underlying mechanisms contributing to gray and white matter enlargement. We focused on individuals with intellectual disability (IQ < 70) or ASD associated with brain overgrowth in 16p11.2 deletion carriers. We differentiated the iPSCs into neural progenitor cells (NPCs) and oligodendrocyte progenitor cells (OPCs) and investigate the hypothesis that brain enlargement in 16p11.2 deletion carriers may be due to improper cellular elimination. Under normal conditions, classic “eat me” and “don’t eat me” signaling mechanisms associated with phagocytosis maintain cellular homeostasis across diverse tissue types (18, 19). CD47 (a “don’t eat me” signal) protects normal cells from getting cleared (18), but can become overexpressed in many types of cancer cells, preventing tumorigenic cells from getting engulfed or phagocytosed (20–22). In fact, CD47 plays an important role in many pathological disorders associated with an overproduction of cells and cell removal, including cancer (20–22), atherosclerosis (23), and fibrotic diseases (24). NPCs derived from iPSCs of autistic individuals with macrocephaly have increased proliferation relative to controls (25, 26). Therefore, we hypothesized that CD47 may be involved in these disorders.
We find that CD47 is overexpressed in NPCs and OPCs derived from 16p11.2 deletion carriers, leading to reduced phagocytosis by macrophages and microglia. Furthermore, the 16p11.2 deletion NPCs and OPCs have increased cell surface expression of calreticulin (CRT, a prophagocytic “eat me” signal), indicating that these cells should be eliminated but are not due to high levels of CD47 (27). Importantly, treatment with a CD47 blocking antibody restores phagocytosis of 16p11.2 deletion NPCs and OPCs to control levels, particularly in 16p_del NPCs and OPCs that have increased cell surface expression of CRT, indicating that the changes in phagocytosis are mediated by cell surface expression of CD47. We thus identify a role for CD47 in 16p11.2 deletion syndrome and highlight the potential importance of blocking CD47 to promote clearance of unhealthy NPCs and OPCs in 16p11.2 deletion with brain overgrowth.
Results
Increased CD47 on 16p11.2 Deletion NPCs Protects Them from CRT-Mediated Phagocytosis.
We obtained human iPSCs from the Simons Foundation Autism Research Initiative (SFARI) from individuals who are carriers of the 16p11.2 CNV with complete clinical, genetic, and phenotypic data. To better understand and elucidate mechanisms underlying brain overgrowth, we focused our study on 16p11.2 deletion carriers with enlarged head circumferences (mean head circumference of 97 ± 2 percentile; three 16p11.2 deletion subjects had head circumferences in the 99th percentile, and one 16p11.2 deletion subject had a head circumference in the 91st percentile) (Fig. 1A and SI Appendix, Table S1). All four of these 16p11.2 del subjects have been diagnosed with intellectual disability (IQ < 70) or ASD (SI Appendix, Table S1), as well as phonological and language disorders. Two 16p11.2 deletion carriers with normal head circumferences without macrocephaly (SI Appendix, Table S1) were also used as comparison to assess whether our findings are specific to 16p11.2 deletion with brain overgrowth. The 16p11.2 duplication carriers in our study have head circumference percentiles within the normal range of 42.5 ± 30.5 percentile with no diagnosis of macro- or microcephaly (Fig. 1A and SI Appendix, Table S1). iPSCs from control subjects were obtained through the NIH registry with no genetic abnormalities.
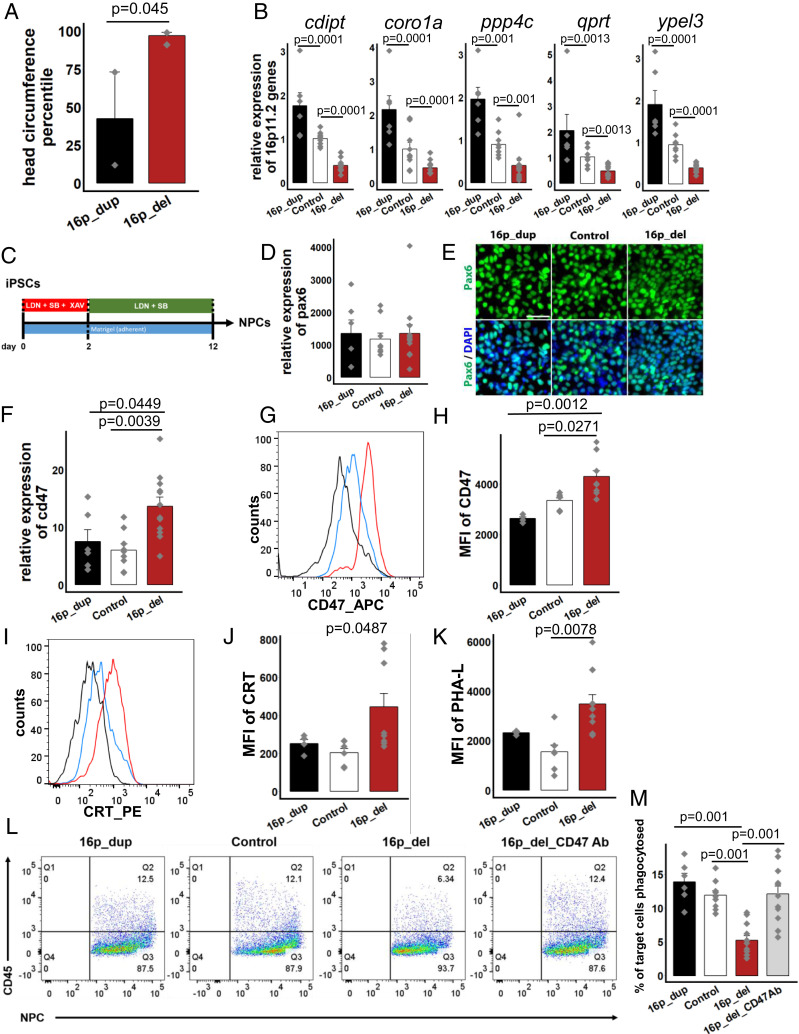
Fig. 1.
CD47 expression is increased on 16p11.2 deletion NPCs protecting them from calreticulin-mediated phagocytosis. (A) Head circumference measurements of 16p11.2 deletion (16p_del, n = 4) and duplication (16p_dup, n = 2) carriers. P value determined by two-tailed equal variance student’s t-test. (B) Quantitative RT-PCR of expression levels of genes within the 16p11.2 locus. Data represent fold change relative to undifferentiated control human iPSCs. (C) Schematic of directed differentiation of iPSCs into neural progenitor cells (NPCs). (D) mRNA expression levels for pax6 after 12 d of differentiation. Data represent fold change relative to undifferentiated control human iPSCs. P values not significant at 5% level. (E) Immunostaining for Pax6 in NPCs. (Scale bar, 50 µm.) (F) Quantitative RT-PCR of cd47 mRNA expression. Data represent fold change relative to undifferentiated control human iPSCs. (G) Flow cytometry histograms of the mean fluorescence intensities (MFI) of CD47 in differentiated NPCs. (H) Quantification of CD47 MFI. (I) Histograms of the MFI of cell-surface expression of Calreticulin (CRT) in NPCs. (J) Quantification of CRT MFI. (K) Quantification of Phaseolus Vulgaris Leucoagglutinin (PHA-L), indicative of asialoglycan binding sites for CRT. (L) Flow cytometry phagocytosis plots showing rates of engulfment of NPCs when cocultured with human derived macrophages (labeled with a CD45 antibody). 16p_del_CD47 Ab, NPCs differentiated from 16p_del lines pretreated with CD47 blocking antibody prior to phagocytosis assessment. (M) Percentages of phagocytosed NPCs. All data are mean ± SE. P values determined by one-way ANOVA followed by post-hoc Tukey HSD Test (B, D, F, H, J, K, M). n = 3 (B, D, F, M) and n = 2 biological replicates (H, J, K) per cell line (16p_dup, n = 2; control, n = 3; 16p_del, n = 4 cell lines).
The extent of CNV insertions and deletions and variant calls for the 16p11.2 CNV iPSC lines were determined from microarray data, whole exome datasets, and molecular inversion probes (MIPs) -targeted sequencing datasets obtained through SFARI (SI Appendix, Fig. S1 and Table S1). We also assessed mRNA levels of a subset of genes spanning the 16p11.2 locus by quantitative real-time PCR (qRT-PCR). Expression levels of many genes within this locus were significantly up-regulated in the 16p11.2 duplication (16p_dup) lines and down-regulated in the 16p11.2 deletion (16p_del) lines relative to control iPSCs (Fig. 1B). Taken together, the MIPs, microarray, and qPCR data confirm the presence of the microdeletions and duplications in the 16p11.2 CNV iPSC lines. Furthermore, expression of pluripotency markers (Nanog and Oct4) were uniformly expressed and comparable in the 16p11.2 CNV iPSCs and control iPSCs (SI Appendix, Fig. S2 A and B).
To begin to investigate the underlying mechanisms, we differentiated the iPSCs into NPCs using an established NPC differentiation protocol that gives rise to cortical neurons (28, 29). Following 12 d of directed differentiation (Fig. 1C), NPCs express Pax6 mRNA at comparable and high levels of >1,000-fold relative to undifferentiated iPSCs (Fig. 1D). Expression of Pax6 at the protein level was also comparable across conditions (Fig. 1E) and was coexpressed with nestin (SI Appendix, Fig. S3A), an intermediate filament protein expressed in neural stem and progenitor cells (30). Expression levels of several other forebrain markers were also increased to a higher extent relative to midbrain and hindbrain markers, indicative of differentiation toward forebrain status (SI Appendix, Fig. S3B). While some degree of variation exists from one differentiation to another even within a cell line, the increased mRNA and protein-expression levels across genes associated with forebrain status, along with concurrent minimal or low expression of genes associated with other brain regions, indicate specificity toward the cortical forebrain lineage as in prior reports (28, 29, 31).
We hypothesized that brain enlargement in 16p11.2 deletion carriers may be due to improper cellular elimination as a result of altered CD47 expression, especially since CD47 is highly expressed in neural and glial cells during brain development (SI Appendix, Fig. S4). To investigate this possibility, we first assessed mRNA expression levels of CD47 in the differentiated NPCs from 16p11.2 CNV carriers and control subjects. Strikingly, CD47 expression was significantly up-regulated in 16p_del carriers relative to control and 16p_dup subjects (Fig. 1F). Flow cytometry analysis for CD47 protein expression at the cell surface also showed a significant shift and up-regulation in the mean fluorescence intensity (MFI) in 16p_del carriers relative to control and 16p_dup subjects (Fig. 1 G and H). CD47 expression of NPCs derived from 16p_del carriers with normal size brains (average head circumference of 50.2 ± 2.2 percentile) was comparable to control and 16p_dup individuals (SI Appendix, Fig. S5 A–E).
Next, we assessed cell surface levels of CRT by flow cytometry in the differentiated NPCs. CRT is a dominant prophagocytic signal (commonly known as an “eat me” signal) that is up-regulated on the surfaces of cancer cells, as well as damaged and aged cells, indicating to the phagocytic system that the cells should be eliminated (32). However, due to high levels of CD47, which dominates the prophagocytic CRT signal, these cells fail to be eliminated (27). Importantly, translocation of CRT to the cell surface of target cells determines cell removal (32).
Within the CD47+ population, NPCs derived from 16p_del iPSCs had a significant up-regulation of CRT relative to control and 16p_dup subjects (Fig. 1 I and J). CRT binds to asialoglycans on the surface of cells; therefore, the more potential binding sites, desialylated glycans, the more potential CRT there is to promote interrogation and clearance by the phagocytic system. The absence of sialic acid on glycans can be determined by using a lectin, phaseolus vulgaris leucoagglutinin (PHA-L) (32). Staining with PHA-L on NPCs derived from control and 16p_CNV iPSCs shows a marked increase in PHA-L binding in 16p_del subjects (Fig. 1K), indicating increased presence of asialoglycan binding sites for CRT. In individuals with 16p11.2 deletion with normal head circumferences (no macrocephaly), expression of CRT was comparable to control subjects (SI Appendix, Fig. S5F).
Given the preceding results, we hypothesized that NPCs with high CD47 and CRT would escape recognition and phagocytosis. To test the functional significance of our findings, we derived macrophages from control human blood samples and cocultured them in vitro with differentiated NPCs from 16p11.2 CNV and control subjects. While NPCs derived from control and 16p_dup subjects had similar rates of phagocytosis as assessed by colabeling with the marker CD45, there was a significant reduction in engulfment of the 16p_del NPCs (Fig. 1 L and M).
Treatment with CD47 blocking antibody can effectively restore phagocytosis of cancer cells that overexpress CD47 without affecting normal cells. In fact, in order for target cells to be phagocytosed upon blockade of an antiphagocytic signal (CD47), the cells must also display a potent prophagocytic signal (e.g., CRT), which is absent on normal cells (27, 33–37). To investigate whether blocking CD47 could restore phagocytic activity, we pretreated 16p_del NPCs with a CD47Ab (anti-CD47 antibody clone B6.H12) and assessed the rate of phagocytosis when cocultured with macrophages. Blockade of CD47 restored phagocytosis to control levels in the 16p_del NPCs (Fig. 1 L and M). Notably, CD47 expression at the mRNA and protein levels, as well as phagocytosis rates, were similar across groups prior to differentiation at the pluripotent stage (SI Appendix, Fig. S2 C–G), emphasizing the role of CD47 in maintaining phagocytic balance during development and highlighting that these findings were specific to the differentiated NPCs. Together, these findings indicate that the NPCs in 16p11.2 deletion syndrome with brain enlargement should be eliminated but are not due to high expression of CD47.
Increased CD47 on 16p11.2 Deletion OPCs Protects Them from CRT-Mediated Phagocytosis.
In addition to increases in gray matter, brain imaging studies have shown increases in white matter volume in 16p11.2 deletion carriers with macrocephaly (11, 13, 17, 38, 39). We next differentiated the iPSCs into OPCs using an established differentiation protocol that enables iPSC-derived OPCs to engraft and function in vivo in the mouse brain (40) (Fig. 2A). Following 12 d of directed differentiation, the cells express pre-OPC markers, Olig2 and Nkx2.2, at the protein level (40) (Fig. 2B). With further differentiation to day 50, mRNA expression of several genes associated with the oligodendrocyte lineage (e.g., Olig1) (40) are up-regulated (Fig. 2C). Following 50 d of differentiation, immunostaining and flow cytometry analyses show that the population of cells also express more mature OPC markers at the protein level, including SRY-box 10 (SOX10) (Fig. 2D), the cell surface marker O4 (Fig. 2E), and the chondroitin sulfate proteoglycan NG2 (SI Appendix, Fig. S6), demonstrating that the majority of the cells are differentiating into the oligodendrocyte lineage (40). In subsequent analyses, we collectively call the day 50 population as OPCs.
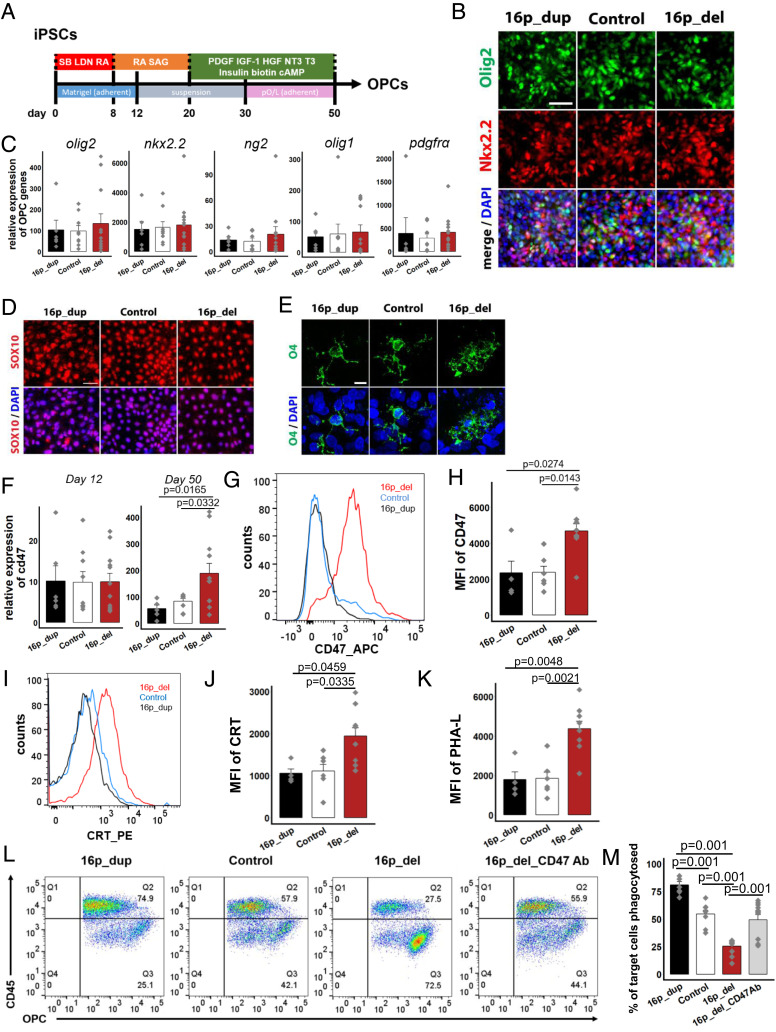
Fig. 2.
CD47 expression is increased on 16p11.2 deletion OPCs protecting them from calreticulin-mediated phagocytosis.(A) Schematic of directed differentiation of iPSCs into oligodendrocyte progenitor cells (OPCs). (B) After 12 days of differentiation, immunostaining for pre-OPC markers Olig2 and Nkx2.2. Nuclei (DAPI, blue). (C) Following 50 days of directed differentiation, quantitative RT-PCR of mRNA expression levels of genes associated with the oligodendrocyte lineage. Data represent fold change relative to undifferentiated control human iPSCs. P values not significant at 5% level. (D) Immunostaining for Sox10 after 50 days. (E) Confocal imaging showing expression of the OPC marker O4 after 50 days. (F) Quantitative RT-PCR of mRNA expression levels of cd47 after 12 days (Left) and 50 days (Right) of directed differentiation. Data represent fold change relative to undifferentiated control human iPSCs. (G) Flow cytometry histograms of the mean fluorescence intensities (MFI) of CD47 in OPCs after 50 d of directed differentiation. (H) Quantification of CD47 MFI. (I) Histograms of the MFI of cell-surface expression of CRT in OPCs. (J)Quantification of CRT MFI. (K) Quantification of PHA-L in OPCs. (L) Flow cytometry phagocytosis plots showing rates of engulfment of OPCs when cocultured with human derived macrophages (labeled with a CD45 antibody). 16p_del_CD47 Ab, OPCs differentiated from 16p_del lines pretreated with CD47 blocking antibody prior to phagocytosis assessment. (M) Percentage of phagocytosed OPCs. All data are mean ± SE. P values determined by one-way ANOVA followed by post-hoc Tukey HSD Test (C, F, H, J, K, M). n = 3 (C, F, M) and n = 2 biological replicates (H, J, K) per cell line (16p_dup, n = 2; control, n = 3; 16p_del, n = 4 cell lines). [Scale bar, 50 µm (B, D, E).]
To investigate the role of CD47 in OPCs, we next assessed the mRNA expression level of CD47 following 12 d of directed differentiation, when the cells are still in the early stages of differentiation at a pre-OPC stage (40). No significant difference in CD47 expression was detectable at this early stage across all 16p CNV and control lines (Fig. 2 F, Left). However, after 50 d of differentiation, there was a significant increase in CD47 expression in the 16p_del OPCs compared with 16p_dup and control OPCs (Fig. 2 F, Right). O4+ OPCs can be isolated through FACS using established protocols (41). In O4+-sorted OPCs, mRNA expression of CD47 was also significantly up-regulated in 16p_del relative to control and 16p_dup conditions (SI Appendix, Fig. S7A). Across all conditions, the O4+ OPCs had high mRNA expression levels of genes associated with the oligodendrocyte lineage (SI Appendix, Fig. S7 B and C), low expression levels of neuronal (Tuj1) and astrocytic (GFAP) genes (SI Appendix, Fig. S7C), and were of cortical origin as genes associated with the spinal cord had low expression (SI Appendix, Fig. S7D). Similar to the NPCs, there was a significant shift and up-regulation of the MFI of CD47 at the protein level at the cell surface of 16p_del OPCs compared to control and 16p_dup subjects after 50 d of differentiation (Fig. 2 G and H). Within the CD47+ population, cell surface levels of the prophagocytic signal CRT and the presence of asialoglycan binding sites marked by an increase in PHA-L binding in 16p_del OPCs were also significantly increased relative to control and 16p_dup subjects (Fig. 2 I–K). This up-regulation in CD47 corresponded with suppressed phagocytosis when the OPC populations were cocultured with macrophages derived from control human blood samples (Fig. 2 L and M). As in NPCs, treatment with the blocking antibody, CD47Ab (anti-CD47 antibody clone B6.H12), restored phagocytosis rates to control levels in 16p_del OPCs (Fig. 2 L and M). Similar to the NPCs, CD47 and CRT expression of OPCs derived from 16p_del carriers with normal size brains and no macrocephaly (average head circumference of 50.2 ± 2.2 percentile) were similar to control and 16p_dup individuals (SI Appendix, Fig. S8 A–D).
Overexpression of CD47 Leads to Suppressed Phagocytosis of 16p11.2 Deletion NPCs and OPCs In Vivo.
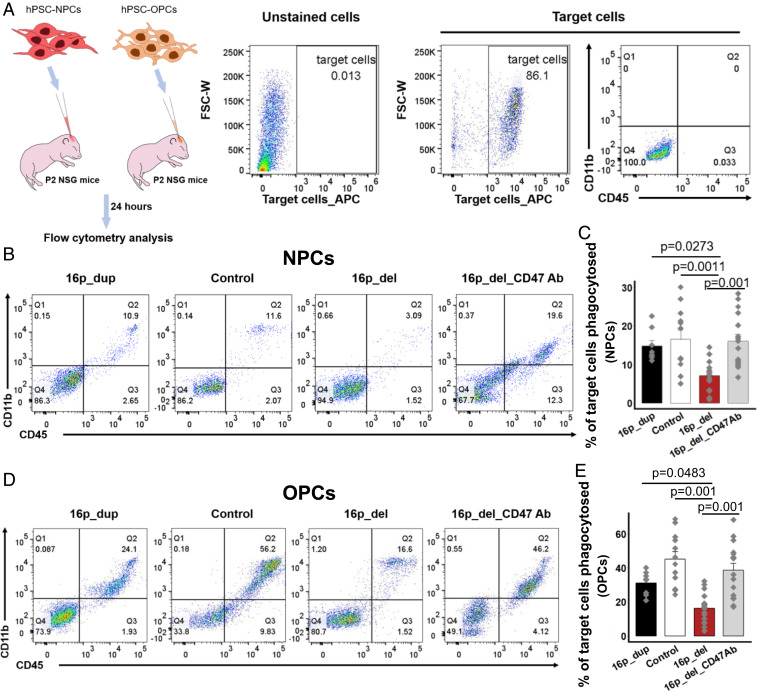
Thus far, our results show that the changes in the balance between CRT and CD47 in the 16p_del NPCs and OPCs are associated with suppressed phagocytosis in vitro. We next investigated whether phagocytosis rates would also be altered and recapitulated in an in vivo setting following intracerebral injections of 16p11.2 CNV-derived NPCs or OPCs. To do so, following in vitro-directed differentiation, hiPSC-derived NPCs and OPCs were labeled with CellTrace red fluorescent dye and injected intracerebrally into individual NOD-scid interleukin (IL) 2γ-null (NSG) postnatal day (P)2 pups (Fig. 3A). Twenty-four hours after injection, pups were killed and brains were dissociated for flow cytometry analyses of phagocytosis. Within the target cell population identified by CellTrace far red, the percentage of cells colabeled with CD45 and CD11b, markers of activated microglia and infiltrating macrophages, were analyzed to determine phagocytosis rates (Fig. 3A) (42). Similar to the in vitro experiments, the percentages of engulfed or phagocytosed NPCs (Fig. 3 B and C) and OPCs (Fig. 3 D and E) were significantly suppressed in vivo in the 16p_del conditions relative to control and 16p_dup conditions. Strikingly, treatment with an anti-CD47 blocking antibody of 16p_del NPCs and OPCs prior to intracerebral injections restored in vivo phagocytosis rates to control levels (Fig. 3 B–E).
Fig. 3.
Overexpression of CD47 suppresses in vivo phagocytosis of 16p 11.2 deletion NPCs and OPCs. (A) Schematic of in vivo phagocytosis assay using NPCs and OPCs differentiated from control, 16p11.2 deletion (16p_del), and 16p11.2 duplication (16p_dup) iPSC lines. NPCs and OPCs differentiated from 16p_del lines were also pretreated with or without a CD47 blocking antibody prior to phagocytosis assessment. CellTrace far red-labeled 12-d differentiated NPCs or 50 d differentiated OPCs were injected into P2 NSG mice brain. Twenty-four hours postinjection, the brains were isolated and disassociated for flow cytometry analysis. Unstained cells show negative expression of CellTrace far red-labeled target cells (NPCs or OPCs). Mouse-CD11b-FITC and mouse-CD45-PE were used to detect mouse microglia and infiltrating macrophages within the population of CellTrace far red-labeled NPC- or OPC-target cells. Within the population of target cells, CD11b and CD45 expression remains negligible prior to in vivo injection. (B) Representative flow cytometry phagocytosis plots showing rates of engulfment of NPCs 24 h postinjection into NSG pup brains. Phagocytosed NPCs are indicated by the coexpression of CD11b and CD45 (Q2). (C) The percentages of phagocytosed NPCs. Rates of phagocytosis when 16p_del NPCs are pretreated with a CD47 blocking antibody (16p_del_CD47Ab) are also shown. (D) Representative flow cytometry phagocytosis plots showing rates of engulfment of OPCs 24 h postinjection into NSG pup brains. Phagocytosed OPCs are indicated by the coexpression of CD11b and CD45 (Q2). (E) The percentages of phagocytosed OPCs. Rates of phagocytosis when 16p_del OPCs are pretreated with a CD47 blocking antibody (16p_del_CD47Ab) are also shown. All data are mean ± SE n = 2 pups per cell line, 2 technical replicates per pup (16p_dup, n = 2; control, n = 3; 16p_del, n = 4 cell lines); P values were determined by one-way ANOVA followed by post hoc Tukey HSD Test.
Anti-CD47 Treatment Restores in Vivo Phagocytosis of 16p11.2 Deletion NPCs and OPCs.
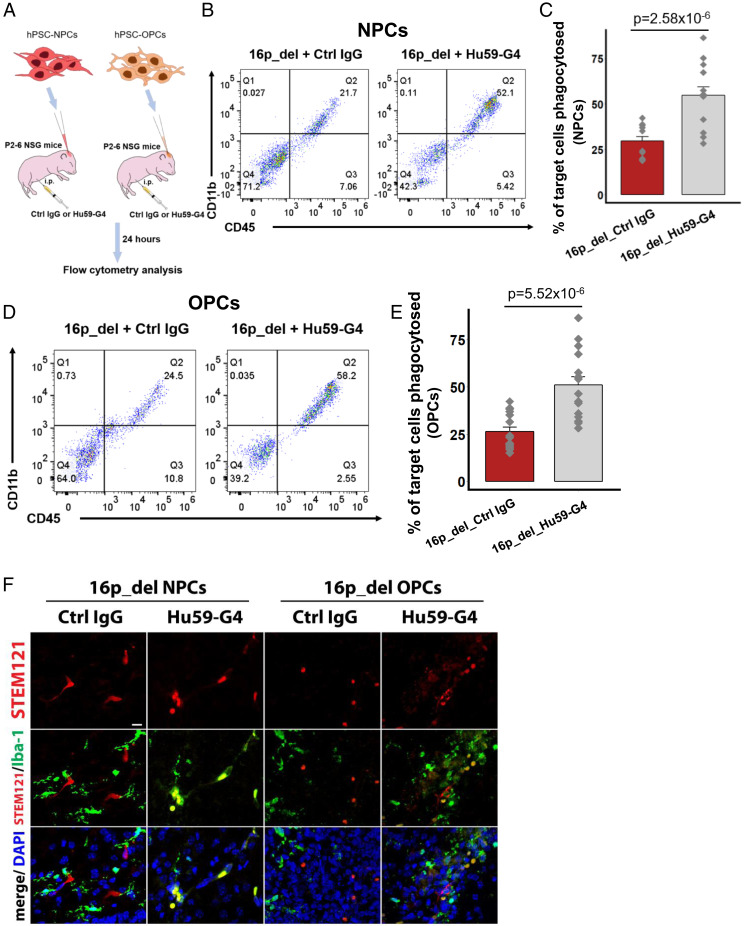
We next evaluated whether intraperitoneal anti-CD47 treatment could restore phagocytosis of 16p11.2_del NPCs and OPCs in vivo, analogous to an approach that has proven to be beneficial in human clinical trials of patients with lymphoma (37). After intracerebrally injecting CellTrace far red-labeled 16p11.2_del NPCs and OPCs into NSG pups, we treated the pups with anti-CD47 (Hu5F9-G4) or human IgG control antibodies intraperitoneally and analyzed phagocytosis by flow cytometry after 24 h (Fig. 4A). Compared to IgG control, treatment with anti-CD47 antibody was associated with a marked increase in phagocytosis, both in 16p_del NPCs (Fig. 4 B and C) and 16p_del OPCs (Fig. 4 D and E). Additionally, we used antibodies to ionizing calcium-binding adaptor molecule (Iba1), a microglial and macrophage-specific calcium-binding protein that is involved in membrane ruffling and phagocytosis (43), to immunolabel and identify phagocytic cells in NSG pup brain tissue sections. Consistent with the flow cytometry analyses, increased Iba1+ cells engulfing target cells marked by the human cytoplasmic marker (STEM121), along with CellTrace far red at the injection site, were evident in 16p_del brains treated with anti-CD47 antibody relative to control IgG treatment (Fig. 4F). In NPCs and OPCs derived from 16p_del individuals without macrocephaly, rates of phagocytosis were comparable between human IgG control and anti-CD47 treatment (SI Appendix, Figs. S5 G and H and S8 E and F), consistent with prior work demonstrating that target cells must display prophagocytic signals, such as CRT, for anti-CD47 antibody-mediated phagocytosis (27, 33–37). Taken together, these results indicate that CD47-blockade could potentially act as an agent in clearing NPCs and OPCs that have high levels of CD47 and CRT in the 16p11.2 deletion syndrome.
Fig. 4.
Anti-CD47 treatment restores phagocytosis of 16p11.2 deletion NPCs and OPCs in vivo. (A) Schematic of in vivo phagocytosis assay using NPCs and OPCs differentiated from 16p11.2 deletion (16p_del) iPSC lines and treated with human IgG control or anti-CD47 antibody (Hu59-G4) intraperitoneally. CellTrace far red-labeled 12 d differentiated NPCs or 50-d differentiated OPCs were injected into P2–P6 NSG mice brain with intraperitoneal treatment of human IgG control or anti-CD47 antibody. Twenty-four hours postinjection, the brains were isolated and disassociated for flow cytometry analysis. Mouse-CD11b-FITC and mouse-CD45-PE were used to detect mouse microglia and infiltrating macrophages within the population of CellTrace far red-labeled NPC- or OPC-target cells. (B) Representative flow cytometry phagocytosis plots showing rates of engulfment of NPCs. Phagocytosed NPCs are indicated by the coexpression of CD11b and CD45 (Q2). (C) Percentages of phagocytosed NPCs derived from 16p_del iPSCs in control IgG-treated or Hu59-G4–treated pups. (D) Representative flow cytometry phagocytosis plots showing rates of engulfment of OPCs. Phagocytosed OPCs are indicated by the coexpression of CD11b and CD45 (Q2). (E) Percentages of phagocytosed OPCs derived from 16p_del iPSCs in control IgG-treated or Hu59-G4–treated pups. (F) Confocal images of CellTrace far red-labeled NPCs or OPCs in NSG pup brains probed for the human cytoplasmic marker STEM121 (red) and Iba1 (green) for phagocytic cells, and nuclei (DAPI, blue) in control IgG or Hu59-G4–treated mice. (Scale bar, 50 μm.) All data are mean ± SE n = 2 pups per cell line, 2 technical replicates per pup (16p_dup, n = 2; control, n = 3; 16p_del, n = 4 cell lines); P values were determined by two-tailed Student’s t test.
Discussion
This study begins to uncover the implications of CD47 for neurodevelopmental disorders associated with brain overgrowth. Recent work has shown that CD47 plays a critical role in regulating synaptic pruning during early brain development (44) and functions as a significant risk locus for bipolar disorder (45), providing further support that the same pathways can go awry in neurodevelopmental and mental disorders. While a number of studies are beginning to show changes in NPC proliferation (25, 26) and neuronal size (46) associated with autism with brain enlargement, these studies focused on changes in gray matter and have only investigated molecular and cellular mechanisms in cultures in vitro (25, 26, 46). We demonstrate that CD47 is up-regulated in two distinct cell types (NPCs and OPCs) at particular stages of development in 16p11.2 deletion syndrome, leading to suppressed phagocytosis both in vitro and in vivo. Moreover, we provide evidence that these findings are more pronounced in individuals with macrocephaly relative to those with normal head circumferences, suggesting that the up-regulation of CD47 and CRT may be specific to individuals with macrocephaly in 16p11.2 deletion syndrome. However, analyses of additional samples are needed to fully understand the role of CD47 in 16p11.2 deletion syndrome without macrocephaly.
We demonstrate that anti-CD47 treatment can act as an agent for clearing NPCs and OPCs with increased cell surface expression of CD47 and CRT in cellular and mouse models. In future work, long-term assessment of changes in phagocytosis in vivo using luciferase expressing reporter cell lines would be valuable. Further work is also needed to determine if anti-CD47 treatment can act as a form of therapy and whether this could be translated to selected autistic individuals with brain overgrowth early in the disease. Importantly, further investigation is needed to determine whether treatment through anti-CD47 treatment or other means would alter behavioral and cognitive deficits or anatomical phenotypes (e.g., in animal models) associated with 16p11.2 deletion syndrome.
While we focused on investigating the changes in CD47 expression in progenitor cells, further work should investigate the role of CD47 in various immature and mature cell types during the different stages of neurodevelopment, given its high expression throughout brain development. Recent work has shown that the critical period during which gene networks become dysregulated in autism with macrocephaly occurs early at the neural stem cell stage prior to further differentiation and maturation into neurons (26). However, prior studies have documented increases in neurogenesis, gliogenesis, and the numbers of progenitor cells as well as mature neurons and glia driving the macrocephaly phenotype in various disorders (25, 44, 47–49). These studies suggest that growth of the embryonic brain may be due to factors acting on cell proliferation and survival of newly born cells. Together with the findings in the present report, these studies call for further investigation of CD47 and other factors of various cell types during the neurodevelopmental process. Further investigating of the phagocytic role of microglia would also provide insight into the pruning processes occurring during these early neurodevelopmental stages.
In acute myeloid leukemia (AML), the hematopoietic stem cell precursors of AML outcompete normal hematopoietic stem cells, enabling the leukemia stem cells to expand beyond by countering signaling associated with programmed cell removal, such as CRT, with overexpression of CD47 (27, 50–52). The imbalance between CD47 and CRT observed in this study may indicate that specific NPCs and OPCs in the 16p deletion syndrome with brain overgrowth undergo clonal expansion and outcompete normal NPCs and OPCs. Future work should evaluate whether expansion of CD47hi NPC or OPC clones contribute to brain overgrowth in psychiatric and neurodevelopmental disorders, such as autism. Interestingly, a surprising number of genes (e.g., PTEN, CHD8) and cellular pathways involved in cancer have been found to overlap with those implicated in autism (53–56). This not only indicates potentially shared mechanisms between autism and cancer, but also that the breadth of therapeutic agents identified for cancer could be beneficial in some types of autism and other neurodevelopmental disorders. Here, we demonstrate the efficacy of anti-CD47 monoclonal antibodies as a potential agent in restoring phagocytosis of 16p11.2 deletion NPCs and OPCs. Within the 16p11.2 locus, ∼29 genes are affected in 16p11.2 CNV carriers (5). This is a region of CNVs with a human specific set of sequences not present in nonhuman primates (57). In future work, it would be interesting to investigate which of these 29 genes directly regulates the CD47 (chr3:107,761,940-107,809,935) pathway and the regulation of asialoglycoproteins (binding sites for CRT [chr7: 93,053,798-93,204,042]), which could have important implications for cancer, neurodevelopment, and other conditions associated with CD47 signaling. Similarly, as in some forms of cancer (58), it would be interesting to investigate whether changes in epigenetics (e.g., H3K27 acetylation) of 16p11.2 deletion NPCs and OPCs influence enhancers and super enhancers to regulate CD47 expression.
The sample sizes in this study are small (though comparable to other iPSC studies modeling subtypes of psychiatric disorders) (25, 26, 46, 59), and it would be useful to analyze more patients with 16p11.2 deletion or other related disorders in future work. Validating these findings in postmortem brain tissue in 16p11.2 deletion individuals with and without brain overgrowth would also be valuable, although this resource is constrained by the fact that most brain banks worldwide do not have head circumference or brain volume data tagged with postmortem human brain tissue. Nonetheless, iPSCs are increasingly proving to be a valuable resource in understanding the mechanisms regulating brain development and growth. In recent work, proliferation of NPCs has been shown to be suppressed in human iPSC models of patients with psychiatric disorders and reduced brain volume with 16p13.11 microduplication (59). Investigating whether imbalances between pro- and antiphagocytic signals, such as CRT and CD47, regulate brain size more generally in other conditions associated with brain overgrowth or undergrowth may lead to novel therapeutic strategies to prevent and treat these neurodevelopmental disorders.
Materials and Methods
Characterization and Maintenance of hiPSCs.
iPSC lines used in this study are listed in SI Appendix, Table S1 and obtained from the SFARI database via Simons Variation in Individuals Project (VIP). Full phenotypic, clinical, and genetic information for each patient were provided by VIP. A total of 11 human iPSC lines were used in the study. Cells were cultured and maintained in mTeSR medium (Stem Cell) at 37 °C and 5% CO2 on Matrigel (BD Biosciences) and with 10 µM ROCK inhibitor (Y27632, Stemgent) 1 d before differentiation. Prior to differentiation, all hiPSC lines were immunostained against OCT3/4 and NANOG for pluripotency analysis.
CNV Validation.
The extent of CNV insertions and deletions for samples 14756.x16, 14756.x9, 14739.x3, 14765.x2, 14799.x1, 14824.x13, 14763.x7, and 14746.x8 was determined from microarray data generated by the Eichler laboratory from the Illumina HumanOmniExpress v1 (104 probands, 280 family members) and v2 (26 probands, 72 family members) microarray platforms. CNV calls were generated by the cnvPartition algorithm, as described by Duyzend et al. (60). Whole-exome datasets from The Simons VIP were downloaded through the SFARI portal, and VCF variant calls were generated with the GATK algorithm. MIPs targeted sequencing data for the above individuals (57) was also obtained through SFARI. The MIPs datasets were aligned to the GRCh37 reference genome using the bowtie2 algorithm. The resulting bam files were filtered to remove reads with sam flags 1,804 as well as reads with quality scores below 30. Bigwig tracks were generated for data visualization in the IGV browser. The script used to process the MIPs datasets is reproduced below:
bowtie2 -x GRCh37 -1 $sample.r1.fastq -2 sample.r2.fastq -p40 | samtools view -F 1804 -q 30 -bS | samtools sort /dev/stdin -o $sample.bam
samtools index $sample.bam
bamCoverage -p16 -v–skipNonCoveredRegions–binSize = 1 -b $sample.bam -o $sample.bw
We also verified that genes within the 16p11.2 locus of the iPSCs have altered gene expression corresponding to either the 16p11.2 deletion or duplication by qPCR. Control lines have been confirmed to have no genetic abnormalities.
CD47 Expression during Brain Development.
Single-cell mRNA sequencing data from primary cortical and medial ganglionic eminence samples across stages of peak neurogenesis were acquired from Nowakowski et al. (61). In brief, cells were captured along the span of neuronal differentiation from progenitor cells to postmitotic neurons across distinct cortical areas. We used weighted gene coexpression network analysis to derive CD47 expression levels log2(TPM) within single-cell expression clusters across the cortex between 5 and 37 wk of fetal development. Using data from the Human Brain Transcriptome Project (62, 63), we also verified microarray expression levels of CD47 during human fetal development across 11 areas of the neocortex and in whole-brain regions.
Regulatory and Institutional Review.
All human pluripotent stem cell experiments were conducted in accord with experimental protocols approved by the Stanford Stem Cell Research Oversight committee.
Generation of NPCs.
Control and 16p11.2 CNV iPSCs were differentiated into cortical NPCs as previously described (28, 29). The iPSC cultures were plated at a high-density monolayer onto GelTrex (ThermoFisher) -coated wells. When cells were confluent, neuroectoderm differentiation began in Essential 6 Media (ThermoFisher) supplemented with small chemical inhibitors of the TGF, SMAD, and Wnt pathways, 500 nM LDN193189 (Tocris) + 10 µm SB431542 (Tocris) + 2 µM of XAV939 (Tocris) for 3 d and then 500 nM LDN193189 + 10 µm SB431542 for the remaining 9 d of differentiation. Medium was changed daily.
Directed Differentiation of OPCs.
OPCs were differentiated from control and 16p11.2 CNV iPSC lines following previously published protocols (40). In brief, iPSCs were plated at a density of 1 × 105 per well on Matrigel-coated six-well plates containing mTeSR and 10 µM ROCK inhibitor. The differentiation medium containing DMEM/F-12 (Life Technologies), 10 μM SB431542 (Stemgent), 250 nM LDN193189 (Stemgent), 100 nM all-trans retinoic acid (RA, Sigma-Aldrich), 1 µM smoothened agonist (SAG, EMD Millipore) was applied for 12 d. Fresh medium was changed daily. Subsequently, cells were detached using a Corning cell lifter (Sigma-Aldrich) and transferred to ultralow-attachment plates for the formation of spheres. From day 12, the differentiation medium was changed every other day. OPC differentiation was induced in the OPC induction media containing DMEM/F-12, 1× MEM nonessential amino acids (NEAA) solution (Life Technologies), 1× GlutaMAX (Life Technologies), 2-mercaptoethanol 1× (Life Technologies), penicillin/streptomycin (PenStrep; Life Technologies), 1× N-2 supplement (Life Technologies), B27 supplement without VitA (Life Technologies), 10 ng/mL recombinant human PDGF-AA, CF (R&D Systems), 10 ng/mL recombinant human IGF-I, CF (R&D Systems), 5 ng/mL recombinant human HGF (R&D Systems), 10 ng/mL neurotrophin 3 (NT3; EMD Millipore), 25 µg/mL insulin solution, human (Sigma-Aldrich), 100 ng/mL biotin (Sigma-Aldrich), 1 µM N6,2′-O-Dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt (cAMP; Sigma-Aldrich), and 60 ng/mL 3,3,5-Triiodo-l-thyronine (T3; Sigma-Aldrich). On day 30, aggregates were plated for attachment on poly-l-ornithine/laminin-coated plates. The oligodendrocytes were continued to be expanded and differentiated in the OPC induction medium until harvested. The cells were immunostained with oligodendroglial specific markers, such as Olig2, Nkx2.2, and O4, to confirm their progression into the oligodendrocyte lineage.
Immunocytochemistry.
Cells were fixed in cold 4% paraformaldehyde in phosphate buffered saline (PBS; Santa Cruz Biotechnology) for 20 min to 1 h under room temperature. Subsequently, the cells were rinsed three times in PBS. For the nuclear staining, cells were permeabilized with 0.3% Triton X-100 (Fisher Bioreagents) for 15 min and washed three times with PBS. Subsequently, blocking solution containing 5% normal donkey serum (Jackson ImmunoResearch)/0.3% Triton X-100 in PBS was applied for 1 h. The primary antibodies and corresponding dilutions used were: rabbit-anti-PAX6 (1:500; BioLegend); mouse-anti-Nestin (1:500; R&D Systems); rabbit-anti-Olig2 (1:100; EMD Millipore); mouse-anti-Nkx2.2 (1:50; DSHB); goat-anti-Sox10 (1:500; R&D Systems); mouse-anti-Oct3/4 (1:500; SantaCruz Biotechnology) and rabbit-anti-NANOG (1:500; Stemgent). After overnight primary antibody incubation at 4 °C, cells were rinsed in PBS/0.3% Triton X-100, incubated with fluorescently tagged secondary antibodies (Alexa-Fluor goat/donkey-antiprimary antibody species IgG 488 or 549; Life Technologies) for 1 h. Nuclear dye, DAPI (Life Technologies), was used to stain all cells. All images were acquired using a Leica Fluorescent Microscope.
The mouse-anti-O4 surface antigen (1:500; clone 81; EMD Millipore) was used to identify hiPSC-derived OPCs. After 15 min of fixation in cold 4% paraformaldehyde/PBS, the cells were blocked in 1% BSA (Sigma-Aldrich) /5% normal donkey/PBS for 1 h at room temperature, and subsequently immunolabeled as described above. The images were taken by confocal microscopy.
RNA Isolation and qRT-PCR.
Total RNA was isolated from control and 16p11.2 CNV hiPSCs, and hiPSC-derived NPCs and OPCs using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. RNA was quantified using the NanoDrop spectrophotometer. Reverse transcription was performed subsequently to synthesize cDNA using SuperScript IV VILO Master Mix with ezDNase (Thermo Fisher) using the same quantity of RNA for all samples.
qRT-PCR was performed using the SYBR green system. One reaction included SYBR green mix (Applied Biosystems), the forward and reverse target gene primers, and 100 to 120 ng cDNA. The StepOnePlus Real-Time PCR System (Applied Biosystems) was used to run the qRT-PCR experiment. The expression of RNA transcripts was analyzed using the ΔΔCT method. Three biological, as well as three technical replicates, were conducted. All samples were normalized to the expression of the housekeeping gene gapdh. Primer sequences used in this study are listed in SI Appendix, Table S2.
Confocal Microscopy.
O4 staining in OPCs derived from control and 16p11.2 CNV iPSC lines, and the mouse brain tissue sections stained with the monoclonal Human Cytoplasmic Marker (STEM121) and microglial/macrophage marker (Iba1) were imaged using Zeiss LSM710 Confocal Microscope with excitation laser lines at 405 (DAPI), 488 (GFP), and 594 nm using a 40× plan-fluor oil-immersion objective.
Flow Cytometry Analysis.
The iPSCs, NPCs, or OPCs were dissociated using Accutase (Life Technologies). The target cells were subsequently suspended in FACS buffer containing 2% bovine serum albumin (BSA; Sigma), and 2 mM EDTA in PBS. Cells were then stained with following antibodies for 30 min on ice: Anti-human-NG2 Chondroitin Sulfate Proteoglycan, Cy3 (1:100; Millipore); Anti-human-CD47-APC (clone: B6H12; 1:100; ThermoFisher); anti-human CRT-PE (1:100; Enzo Life Sciences); anti-mouse/human-CD11b-FITC (clone: M1/70; 1:100; BioLegend); anti-human-CD45-PE (clone: 2D1; 1:100; BioLegend); and anti-mouse-CD45-PE (clone: 30-F11; 1:100; BioLegend). The lectin fluorescein labeled PHA-L (1:200; Vector Laboratories) was used to stain for PHA-L. For O4 staining, the cells were first incubated in primary antibody mouse anti-O4 antibody (clone 81; 1:100; EMD Millipore), followed by secondary antibody incubation, anti-Mouse IgM Alexa Fluor 488 (1:1,000; Life Technologies). The cells were then washed twice with FACS buffer.
DAPI (Sigma) was used to exclude dead cells. Immediately prior to flow cytometric analysis, the cells were strained through a 100-µM filter. Flow cytometry analyses were performed on the BD FACS Aria II (Becton Dickinson) and data were analyzed using FlowJo software (FlowJo). CD47 expression was measured using MFI. The MFI of cell surface CRT expression was obtained from the CD47+ population. Representative unstained controls for CD47 and cell surface CRT MFI are shown in SI Appendix, Fig. S9. The MFI for PHA-L expression was also measured.
O4+ Cell Sorting.
To obtain a pure population of O4-expressing OPCs, the mouse anti-human O4 antibody (clone 81; 1:100; EMD Millipore) was used to stain the 50-d hiPSC-derived OPCs. The cells were incubated in the primary antibody for 40 min on ice, washed with FACS buffer, and subsequently stained with Alexa Fluor 488 goat anti-mouse IgM (μ-chain; Life Technologies) for 30 min on ice. The cells were rinsed twice with FACS buffer. DAPI stain was used to exclude dead cells. Immediately prior to flow cytometric analysis, the cells were strained through a 100-µM filter.
O4+ cell sorting was performed on the BD FACS Aria II (Becton Dickinson) using Accudrop experiments. Cells were sorted into a clean FACS tube containing Buffer RLT (RNeasy Mini Kit, Qiagen) for RNA extractions as described above. For each cell line, 1 × 104 cells were collected.
In Vitro Phagocytosis and Anti-CD47 Treatment.
To generate human peripheral blood-derived macrophages, CD14+ monocytes were enriched from human peripheral blood (purchased from the Stanford Blood Center) and cultured in Iscove’s Modified Dulbecco’s Medium (IMDM; ThermoFisher) with 10% human serum to differentiate to macrophages. After 7-d differentiation, human macrophages are ready for the phagocytosis. The hiPSCs and hiPSC-derived NPCs (at 12-d differentiation) and OPCs (at 50-d differentiation) were fluorescently labeled with CellTrace (far red; ThermoFisher). The human macrophages were lifted using TrypLE (Gibco) and Corning cell lifter, and transferred to 10% human serum/DMEM. In a 3:1 ratio of macrophages (approximately 7.5 × 104 cells) to target cells (approximately 2.5 × 104 cells), the cells were mixed in sterile polystyrene tubes (Fisher Scientific), and incubated at 37 °C for 2 h on a shaker (250 rpm) to promote macrophage and target cell interaction. Subsequently, the cells were washed in cold FACS buffer (2 mM EDTA/2% BSA/PBS) and stained for human marker Human-CD45-PE (clone: 2D1; 1:100; BioLegend) before analysis on the FACS Aria II machine. 7-AAD (BioLegend) stain was used to exclude dead cells.
Treatment with the CD47 blocking antibody was performed immediately after CellTrace labeling. The InVivoMAb anti-human CD47 antibody (clone: B6.H12; BioXcell) was diluted to 10 µg/mL in DMEM and applied to the target cells for 30 min on ice. Samples were then washed in 10% FBS/DMEM and processed for coincubation with human macrophages as described above.
Mice Maintenance.
The NSG mice were kept at a high barrier conditions and under pathogen-free environment in the Lokey Stem Cell Building at Stanford University. All animal handling, surveillance, and experimentation were performed in accordance with approval from the Stanford University Administrative Panel on Laboratory Animal Care.
In Vivo Phagocytosis and Anti-CD47 Treatment.
The target cells were detached using Accutase, counted, stained following the manufacturer’s recommendation with CellTrace (far red; ThermoFisher), and stored on ice until injection. Each P2-6 NSG mouse was anesthetized by hypothermia and received a bilateral injection of 1 L (0.5 L DMEM/F12 and 0.5 L HBSS) containing 30,000 cells of either NPCs or OPCs using a 10-L Hamilton microsyringe (Hamilton). The injection site was determined by injecting at midline, ±1-mm rostral of lambda and injection took place through the skin and soft skull. The pups were gently warmed and then returned back to their mother when active. Twenty-four hours postinjection, pups were killed by decapitation under hypothermia and the fresh brains removed and placed in cold HBSS media on ice as previously described (64). A 2-mm × 2-mm cortical piece of tissue from the midline injection site was subject to neural dissociation (Neural Tissue Dissociation Kit; Miltenyi Biotec). Following dissociation, the cells were strained through a 100-m filter and suspended in FACS buffer. All cells were then stained using Mouse/Human-CD11b-FITC (clone: M1/70; 1:100; BioLegend); Mouse-CD45-PE (clone: 2D1; 1:100; BioLegend). Dead cells were excluded using DAPI, and samples were analyzed on FACS Aria II machine. The targeted NPCs and OPCs population could be determined by APC+ gating. The relative ratio of phagocytosed target cells was determined by coexpression of CD11b-FITC- and CD45-PE+ cells.
Treatment with the CD47 blocking antibody was performed immediately after cell injection. One microiliter of 250 µg/mL of CD47 blocking antibody (Hu5F9-G4, acquired from Forty Seven, Inc.) was injected intraperitoneally into the pups. The pups were gently warmed and then returned back to their mother when active. Twenty-four hours postinjections, brains were processed, stained, and analyzed by flow cytometry as described above.
Serial Sectioning and Staining.
For histological evaluations, brains were extracted and placed in 4% PFA (Electron Microscopy Science) for 24 h at 4 °C. Brains were then placed in 30% sucrose solution (Sigma) dissolved in deionized water for 48 h or until brains sank to the bottom. They were then embedded in optimal cutting temperature compound (OCT; Fisher Scientific). Serial coronal sections were cut at 20-µm-thick using a Cryostat at 200 µm apart for a total of five sections per animal spanning the injection site and then stored at −80 °C. At room temperature, the dried sections were postfixed with 10% formalin, washed three times in 1× PBS, and blocked in 2% donkey serum with 0.1% Triton X-100 for 1 h. Slides were then incubated with primary antibody diluted in blocking solution at 4 °C overnight: mouse-monoclonal human STEM121 (1:500; Takara) for detecting human cytoplasmic protein and Rabbit anti-Iba1 (1:500; Wako) for identifying mouse microglia and infiltrating macrophages. Sections were washed five times with 1× PBS and incubated with fluorescently tagged secondary antibodies (Alexa-Fluor mouse IgG 488 or rabbit 594; Life Technologies) for 1 h at room temperature. Following staining, the slides were then washed 3× in 1× PBS and then mounted in Prolong anti-fade mounting solutions with DAPI.
Statistical Analysis.
All data are graphed as bar blots using the ggplot2 library in RStudio Desktop 1.1.463 environment (https://www.statmethods.net/advgraphs/ggplot2.html). Mean values with SEMs are reported. Student’s unpaired two-tailed t test was used for comparisons of two experimental groups. Multiple comparisons were analyzed using one-way ANOVA followed by post hoc Tukey honest significant difference (HSD) test. P value ≤ 0.05 was considered statistically significant. When P values are not reported in the figures, there was no statistical significance.
Supplementary Material
Acknowledgments
We thank all of the families at the participating Simons Variation in Individuals Project (Simons VIP) sites, as well as the Simons VIP Consortium; the Flow Cytometry Core facilities for their assistance; Rahul Sinha, Danielle Sambo, Kyle Loh, Liang Ma, Mellisa Stafford, Nobuko Uchida, and Theo Palmer for their assistance and feedback on the manuscript; and David Amaral, Joachim Hallmayer, Ruth O’Hara, Christine Nordahl, and members of the I.L.W. laboratory for helpful discussions. We appreciate obtaining access to phenotypic and genetic data on the Simons Foundation Autism Research Initiative Base. Diversity supplement R01CA086065 (to K.M.) provided partial support. This work was supported by grants from the Stanford University School of Medicine, NIH Grant R35CA220434-04 (to I.L.W.), an Autism Center of Excellence grant awarded by the National Institute of Child Health and Development (Grant P50 HD093079 to S.C.), the Ludwig Foundation (I.L.W.), and a Siebel Fellowship (awarded to S.C.)
Footnotes
Competing interest statement: K.M. is a current employee of Forty Seven, Inc.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005483118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Kumar R. A., et al., Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 17, 628–638 (2008). [DOI] [PubMed] [Google Scholar]
- 2.McCarthy S. E.et al.; Wellcome Trust Case Control Consortium , Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 41, 1223–1227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenfeld J. A., et al., Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet. Med. 12, 694–702 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Shinawi M., et al., Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 47, 332–341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss L. A.et al.; Autism Consortium , Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 358, 667–675 (2008). [DOI] [PubMed] [Google Scholar]
- 6.de la Torre-Ubieta L., Won H., Stein J. L., Geschwind D. H., Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leppa V. M., et al., Rare inherited and de novo CNVs reveal complex contributions to ASD risk in multiplex families. Am. J. Hum. Genet. 99, 540–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fombonne E., Rogé B., Claverie J., Courty S., Frémolle J., Microcephaly and macrocephaly in autism. J. Autism Dev. Disord. 29, 113–119 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Bartholomeusz H. H., Courchesne E., Karns C. M., Relationship between head circumference and brain volume in healthy normal toddlers, children, and adults. Neuropediatrics 33, 239–241 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Sacco R., Gabriele S., Persico A. M., Head circumference and brain size in autism spectrum disorder: A systematic review and meta-analysis. Psychiatry Res. 234, 239–251 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Hazlett H. C., et al., Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch. Gen. Psychiatry 68, 467–476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazlett H. C.et al.; IBIS Network; Clinical Sites; Data Coordinating Center; Image Processing Core; Statistical Analysis , Early brain development in infants at high risk for autism spectrum disorder. Nature 542, 348–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maillard A. M.et al.; 16p11.2 European Consortium , The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol. Psychiatry 20, 140–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordahl C. W., et al., Brain enlargement is associated with regression in preschool-age boys with autism spectrum disorders. Proc. Natl. Acad. Sci. U.S.A. 108, 20195–20200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courchesne E., Carper R., Akshoomoff N., Evidence of brain overgrowth in the first year of life in autism. JAMA 290, 337–344 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Redcay E., Courchesne E., When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol. Psychiatry 58, 1–9 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Qureshi A. Y.et al.; Simons VIP Consortium , Opposing brain differences in 16p11.2 deletion and duplication carriers. J. Neurosci. 34, 11199–11211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oldenborg P. A., et al., Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Oldenborg P. A., Gresham H. D., Lindberg F. P., CD47-signal regulatory protein alpha (SIRPalpha) regulates Fcgamma and complement receptor-mediated phagocytosis. J. Exp. Med. 193, 855–862 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majeti R., et al., CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaiswal S., et al., CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138, 271–285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willingham S. B., et al., The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. U.S.A. 109, 6662–6667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojima Y., et al., CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 536, 86–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wernig G., et al., Unifying mechanism for different fibrotic diseases. Proc. Natl. Acad. Sci. U.S.A. 114, 4757–4762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchetto M. C., et al., Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 22, 820–835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schafer S. T., et al., Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 22, 243–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao M. P., et al., Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci. Transl. Med. 2, 63ra94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tchieu J., et al., A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell 21, 399–410.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi Y., et al., Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotechnol. 35, 154–163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elkabetz Y., et al., Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 22, 152–165 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth J. G., et al., 16p11.2 microdeletion imparts transcriptional alterations in human iPSC-derived models of early neural development. eLife 9, e58178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng M., et al., Programmed cell removal by calreticulin in tissue homeostasis and cancer. Nat. Commun. 9, 3194 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao M. P., et al., Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell 142, 699–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D., et al., Anti-CD47 antibodies promote phagocytosis and inhibit the growth of human myeloma cells. Leukemia 26, 2538–2545 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Liu J., et al., Pre-clinical development of a humanized anti-CD47 antibody with anti-cancer therapeutic potential. PLoS One 10, e0137345 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gholamin S., et al., Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci. Transl. Med. 9, eaaf2968 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Advani R., et al., CD47 blockade by Hu5F9-G4 and rituximab in non-Hodgkin’s lymphoma. N. Engl. J. Med. 379, 1711–1721 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owen J. P.et al.; Simons VIP Consortium , Aberrant white matter microstructure in children with 16p11.2 deletions. J. Neurosci. 34, 6214–6223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang Y. S., et al., Reciprocal white matter alterations due to 16p11.2 chromosomal deletions versus duplications. Hum. Brain Mapp. 37, 2833–2848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douvaras P., et al., Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports 3, 250–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douvaras P., Fossati V., Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 1143–1154 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Sedgwick J. D., et al., Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc. Natl. Acad. Sci. U.S.A. 88, 7438–7442 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohsawa K., Imai Y., Kanazawa H., Sasaki Y., Kohsaka S., Involvement of Iba1 in membrane ruffling and phagocytosis of macrophages/microglia. J. Cell Sci. 113, 3073–3084 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Lehrman E. K., et al., CD47 protects synapses from excess microglia-mediated pruning during development. Neuron 100, 120–134.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahl E. A.et al.; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium , Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deshpande A., et al., Cellular phenotypes in human iPSC-derived neurons from a genetic model of autism spectrum disorder. Cell Rep. 21, 2678–2687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caviness V. S. Jr., Takahashi T., Nowakowski R. S., Numbers, time and neocortical neuronogenesis: A general developmental and evolutionary model. Trends Neurosci. 18, 379–383 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Chenn A., Walsh C. A., Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365–369 (2002). [DOI] [PubMed] [Google Scholar]
- 49.McCaffery P., Deutsch C. K., Macrocephaly and the control of brain growth in autistic disorders. Prog. Neurobiol. 77, 38–56 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Jan M., et al., Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 4, 149ra118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chao M. P., Majeti R., Weissman I. L., Programmed cell removal: A new obstacle in the road to developing cancer. Nat. Rev. Cancer 12, 58–67 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Pang W. W., et al., Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc. Natl. Acad. Sci. U.S.A. 110, 3011–3016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Roak B. J., et al., Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernier R., et al., Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler M. G., et al., Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 42, 318–321 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crawley J. N., Heyer W. D., LaSalle J. M., Autism and cancer share risk genes, pathways, and drug targets. Trends Genet. 32, 139–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nuttle X., et al., Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature 536, 205–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Betancur P. A., et al., A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat. Commun. 8, 14802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnstone M., et al., Reversal of proliferation deficits caused by chromosome 16p13.11 microduplication through targeting NFκB signaling: An integrated study of patient-derived neuronal precursor cells, cerebral organoids and in vivo brain imaging. Mol. Psychiatry 24, 294–311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duyzend M. H., et al., Maternal modifiers and parent-of-origin bias of the autism-associated 16p11.2 CNV. Am. J. Hum. Genet. 98, 45–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nowakowski T. J., et al., Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson M. B., et al., Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang H. J., et al., Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brickler T. R., et al., Angiopoietin/Tie2 Axis regulates the age-at-injury cerebrovascular response to traumatic brain injury. J. Neurosci. 38, 9618–9634 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.