Significance
The outer membrane of gram-negative bacteria is a major deterrent for antibiotic entry, making it difficult to treat these infections. It is composed of both LPS and glycerophospholipids, where the synchronized synthesis between these two components is essential for the outer membrane’s unique permeability barrier. In this report, we identify a previously unidentified association between LPS and glycerophospholipids, where the presence of the complete repertoire of glycerophospholipids is required for efficient transport of LPS. Our results provide insight into how E. coli modifies its lipid composition to maintain the outer membrane’s formidable barrier function.
Keywords: cardiolipin, lipopolysaccharide, MsbA, ClsA, LpxM
Abstract
In Escherichia coli, cardiolipin (CL) is the least abundant of the three major glycerophospholipids in the gram-negative cell envelope. However, E. coli harbors three distinct enzymes that synthesize CL: ClsA, ClsB, and ClsC. This redundancy suggests that CL is essential for bacterial fitness, yet CL-deficient bacteria are viable. Although multiple CL–protein interactions have been identified, the role of CL still remains unclear. To identify genes that impact fitness in the absence of CL, we analyzed high-density transposon (Tn) mutant libraries in combinatorial CL synthase mutant backgrounds. We found LpxM, which is the last enzyme in lipid A biosynthesis, the membrane anchor of lipopolysaccharide (LPS), to be critical for viability in the absence of clsA. Here, we demonstrate that CL produced by ClsA enhances LPS transport. Suppressors of clsA and lpxM essentiality were identified in msbA, a gene that encodes the indispensable LPS ABC transporter. Depletion of ClsA in ∆lpxM mutants increased accumulation of LPS in the inner membrane, demonstrating that the synthetic lethal phenotype arises from improper LPS transport. Additionally, overexpression of ClsA alleviated ΔlpxM defects associated with impaired outer membrane asymmetry. Mutations that lower LPS levels, such as a YejM truncation or alteration in the fatty acid pool, were sufficient in overcoming the synthetically lethal ΔclsA ΔlpxM phenotype. Our results support a model in which CL aids in the transportation of LPS, a unique glycolipid, and adds to the growing repertoire of CL–protein interactions important for bacterial transport systems.
The cell envelope of bacteria allows the cell to cope with a diverse range of environments and harsh conditions (1). In gram-negative bacteria, such as Escherichia coli, the envelope contains a symmetrical inner membrane (IM) consisting of glycerophospholipids (GPLs) and an asymmetrical outer membrane (OM) where GPLs comprise the inner leaflet and primarily lipopolysaccharide (LPS) in the outer leaflet (2) (Fig. 1A). The OM’s topology promotes a formidable barrier to detergents, antibiotics, and to key components of the host innate immune response (1). Hydrophobic packing of acyl chains from GPLs and LPS, coupled with divalent cations promoting strong lateral interactions between LPS molecules, support a barrier with low permeability (3).
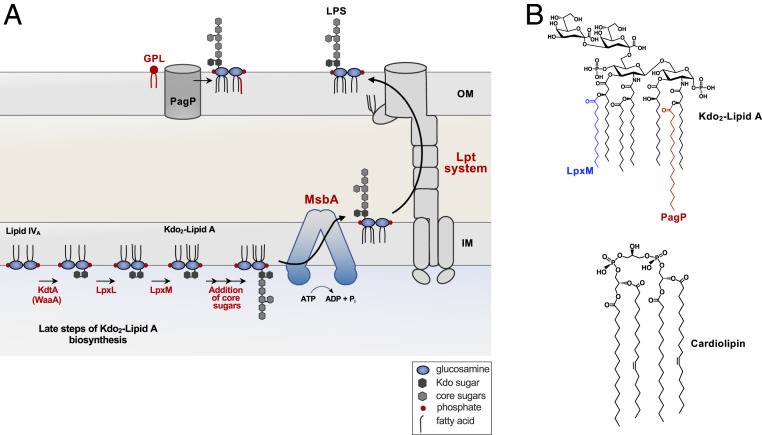
Fig. 1.
Biosynthesis and transport of LPS. (A) The latter steps of Kdo2-lipid A biosynthesis catalyzed by KdtA (WaaA), LpxL, and LpxM are shown. Two Kdo sugars are added to the tetra-acylated lipid A precursor, termed lipid IVA. The Kdo sugars are part of the core oligosaccharide and are required for the ordered addition of last two acyl chains by LpxL and LpxM. First, LpxL adds a laurate (C12:0) followed by LpxM that adds a myristate (C14:0) group forming hexa-acylated Kdo2-lipid A. The remaining core oligosaccharide is extended at the cytoplasmic face of the IM, requiring various glycosyl transferase (not shown). MsbA, an ABC transporter, flips the core–lipid A structure to the periplasmic face of the IM. For simplicity, the O-antigen addition is not shown and is absent in E. coli K-12 strains. The intermembrane translocation of LPS to the OM is conducted by the Lpt system, which forms an envelope-spanning translocation machine. PagP, an OM protein, transfers a palmitate (C16:0) group from mislocalized GPL (red) in the outer leaflet of the OM to LPS. (B, Top) Chemical structure of Kdo2-lipid A with the acyl chain added by LpxM in blue, and the chain added by PagP is in red. (B, Bottom) Structure of cardiolipin.
Disruptions in OM asymmetry compromise the permeability barrier and lead to sensitivity to toxic molecules, including antibiotics. Therefore, the cell must synchronize the synthesis and transport of LPS and GPLs to maintain OM integrity (4). LPS synthesis begins in the cytosol and concludes at the periplasmic face of the IM (5). LPS is divided into three major components: the conserved lipid A domain, the core oligosaccharide, and the distal O-antigen (6). E. coli utilizes nine enzymes for the synthesis of Kdo2-lipid A (5). The enzymes involved in Kdo2-lipid A (lipid A) are essential for bacterial viability, with the exception of the late-step acyltransferases, LpxL and LpxM (Fig. 1A) (7). Although both lpxL and lpxM can be deleted in E. coli, loss of either results in OM permeability defects. Furthermore, suppressor mutations are required for growth of lpxL mutants at temperatures >30 °C (8). The Kdo sugars are the first units of the core oligosaccharide; however, Kdo addition is synonymous with lipid A biosynthesis as the Kdo sugars are required for LpxL and LpxM acyl chain addition in E. coli (9) (Fig. 1). Upon completion of core oligosaccharide assembly onto lipid A, the essential ATPase transporter MsbA flips lipid A–core to the outer leaflet of the IM (Fig. 1A) (6, 10, 11). O-antigen is then appended onto the core region at the periplasmic face of the membrane, although its presence it not essential for viability or present in K-12 strains (6). LPS is then transported to its final destination in the OM by the Lpt system, which is a seven-protein complex forming an envelope-spanning translocating machine (Fig. 1A) (12).
GPL synthesis also begins in the cytosol with the elongation of fatty acids which are then shunted to the IM for synthesis of the three major GPLs: phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL) (SI Appendix, Fig. S1) (1). Interestingly, although CL is the least abundant of the three major GPLs, E. coli has three separate CL synthases: ClsA, ClsB, and ClsC (13). ClsA is responsible for the majority of CL synthesis (13) and is active during logarithmic growth, producing nearly all CL in log phase. During the stationary phase, CL levels increase ∼threefold because of increased ClsA activity (∼10-fold) along with the activation of ClsB and ClsC (13, 14). This redundancy suggests that CL must be critical for the cell, yet CL is not essential for growth under typical laboratory conditions (15). Numerous studies indicate biological interactions between CL and key proteins such as aquaporins, DNA recombination proteins, and essential ATP-binding cassette (ABC) transporters. Despite this, the physiological role of CL remains unclear (16–18), as complete loss of CL does not result in the same phenotypic consequences observed upon loss of function of key CL-interacting proteins (e.g., Sec translocon) (15, 18).
To identify novel CL-mediated functions in E. coli, we utilized transposon sequencing (Tn-seq) in combinatorial clsA, B, and C mutants. We found that the presence of both ClsA and the lipid A acyltransferase LpxM are essential for bacterial fitness. Genetic suppressors in our ∆clsA, ∆lpxM mutant indicated that an accumulation of LPS in the IM is responsible for cell death. We find that lpxM mutants show decreased transport of LPS and that CL levels mitigate penta-acylated LPS transport across the IM. Furthermore, we reveal that lowering total LPS levels through genetic manipulation promotes viability of ∆clsA, ∆lpxM double mutants. We propose a model that CL produced by ClsA aids MsbA in the transport of underacylated LPS.
Results
Loss of ClsA and LpxM Is Synthetically Lethal.
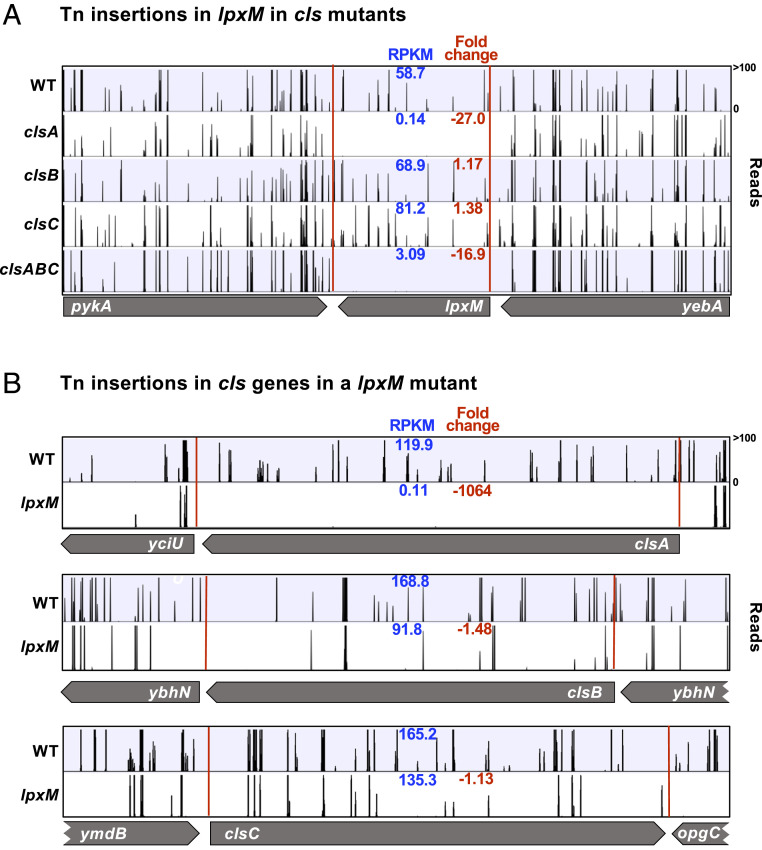
To gain further insight into the redundancy of cls enzymes and the role of CL in the bacterial cell, we wanted to identify genes that impact fitness in combinatorial cls mutants. We constructed an E. coli Tn insertion library in wild-type (WT; W3110), ∆clsA, ∆clsB, ∆clsC, ∆clsAB, ∆clsAC, ∆clsBC, and ∆clsABC backgrounds (∼450,000 mutants per background), utilizing a Tn vector as previously published (Dataset S1) (13, 19). We found that lpxM, the gene encoding the enzyme that catalyzes the last step of lipid A biosynthesis, to be imperative in the absence of clsA (8). In comparison with WT, there were 27-fold fewer Tn insertions in lpxM when clsA was absent (Fig. 2A). Surprisingly, ∆clsB and ∆clsC Tn libraries had a high number of Tn reads in lpxM, suggesting lpxM to be dispensable in these genetic backgrounds (Fig. 2A). Supporting our initial finding that a dual essentiality for clsA and lpxM exist by Tn-seq analysis, we constructed and analyzed a high-density Tn mutant library in a ∆lpxM background (Dataset S1). We observed lpxM mutants to be absent of Tn insertions in clsA, while clsB and clsC had a high number of Tn insertions in lpxM (Fig. 2B). The significant fold change of Tn insertions in clsA and lpxM is illustrated in volcano plots of ΔlpxM and ΔclsABC backgrounds, respectively (SI Appendix, Fig. S2). We were unable to introduce a ∆lpxM::kan allele into ∆clsA, demonstrating that the presence of both clsA and lpxM is essential for cell viability.
Fig. 2.
Identification of lpxM as an essential gene in the absence of clsA by Tn-seq. High-density Tn libraries were generated in WT (W3110) and clsA, clsB, clsC, clsABC, and lpxM mutants. The sites of Tn insertions were identified by deep sequencing and mapped onto the W3110 reference genome. (A) Shown are Tn insertion profiles of the lpxM operon in WT and ∆cls Tn libraries. (B) Tn insertions of the three cls genes in WT and lpxM Tn libraries. The height of each line in the profile represents the number of sequencing reads corresponding to a Tn insertion at the indicated genome position. In blue, the RPKM value for the gene is listed. In red is the fold change of RPKM value of the genes listed relative to WT. A total of three biological replicates where completed. The profile and the RPKM shown is that of a single biological replicate, but the fold change is the average of all replicates.
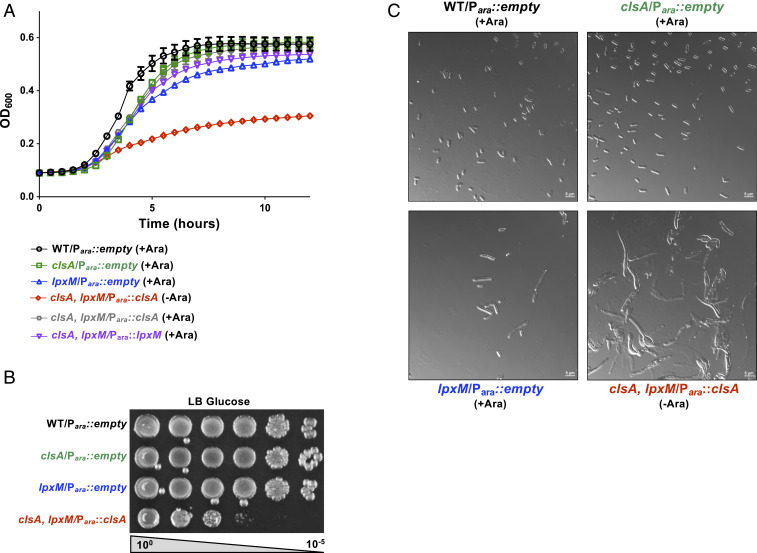
To study the cause of the synthetic lethality of a ∆clsA, ∆lpxM double mutant, we deleted the native clsA gene and introduced a plasmid (pBAD) expressing clsA from an arabinose inducible promoter. The lpxM deletion was then introduced by phage transduction, resulting in the strain ∆clsA, ∆lpxM::kan pBAD::clsA (referred to as clsA, lpxM Para::clsA). W3110 cells harboring a clsA or lpxM deletion have a slight growth defect compared with WT (Fig. 3A); however, ClsA depletion in the absence of the inducer in the clsA, lpxM Para::clsA strain led to a major growth defect (Fig. 3 A and B). Similarly, depletion of lpxM from an arabinose inducible promoter also leads to a severe growth defect of the double mutant (SI Appendix, Fig. S3). Loss of clsA has no major impact on overall cell morphology (Fig. 3C), whereas deletion of lpxM leads to elongated cells, consistent with previous findings (Fig. 3C) (20). Depletion of ClsA in the double mutant, however, exacerbated the ΔlpxM elongated cell morphology, showing gross changes in cell morphology (Fig. 3C).
Fig. 3.
Depletion of ClsA in the absence of LpxM leads to a growth and morphological defect. (A) Growth curve following depletion of ClsA in the absence of lpxM. The strain clsA, lpxM/Para::clsA was grown under inducing conditions with 0.2% arabinose (+Ara, gray) or under repressing conditions with 0.2% glucose (−Ara, red). WT, single clsA, and single lpxM mutants were used as controls. The growth of a clsA, lpxM/Para::lpxM mutant was also observed in inducing conditions. Growth of indicated strains were monitored by OD600 every 30 min. Error bars represent SEM from technical triplicate. (B) Serial dilutions of indicated strains were spotted on LB plates containing 0.2% glucose and grown at 37 °C. (C) Micrographs of cells at the 5 h time point of growth curve shown in A. (Scale bar, 5 μm).
Since ClsA is the predominate CL synthase, we were interested if the overexpression of clsB or clsC could rescue the synthetic lethal phenotype of ∆clsA, ∆lpxM double mutants. To this end, we built plasmids expressing clsB or ymdB-clsC from an arabinose inducible promoter. ymdB is in operon with clsC, and its coexpression with clsC increases ClsC activity (13). Although plasmid expression of either clsB or ymdB-clsC in the triple ΔclsABC mutant leads to CL levels similar to those in WT, obtaining a lpxM mutant in these backgrounds required longer incubation (∼24 to 36 h) and resulted in the formation of smaller colonies. Furthermore, these strains have a growth defect compared with our Para::clsA control (SI Appendix, Fig. S4). Given this data, ClsA may have another functional role outside of CL synthesis that could not be supplied by ClsB or ClsC. To test this, we generated a catalytically inactive ClsA (H224A) by mutating one of the two HKD motifs found in the CL synthases (SI Appendix, Fig. S4). These motifs are characteristic for all members of the phospholipase D super family and are required for enzymatic activity of E. coli CL synthases (15, 21). Expression of both the catalytically inactive clsA and clsB or ymdB-clsC from an arabinose inducible promoter does not phenocopy our Para::clsA control (SI Appendix, Fig. S4B). Together, these results demonstrate that CL synthesized specifically by ClsA must be present to maintain normal growth in the absence of complete lipid A synthesis.
Mutations in msbA Suppress the clsA, lpxM Synthetic Lethal Phenotype.
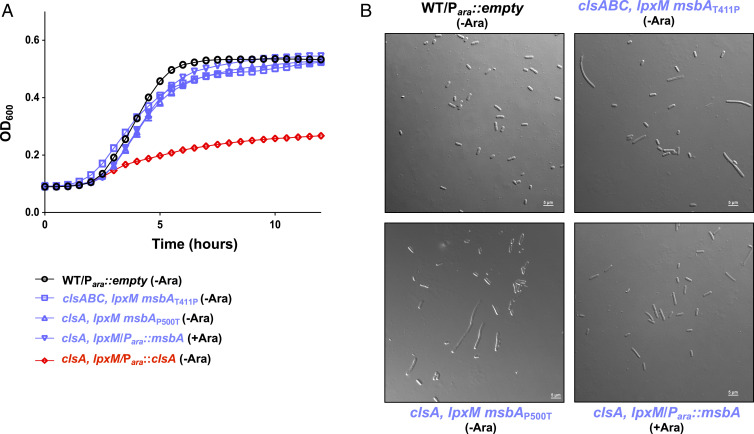
We next turned to suppressor analysis to identify mutations that bypass the synthetic lethal ∆clsA, ∆lpxM phenotype. To this end, both ∆clsA and ∆clsABC mutants were used. The triple mutant was used to avoid suppressors in other CL synthesis enzymes. To select for suppressors, we performed a large-scale transduction and transduced a ΔlpxM::kan mutation into both ΔclsA and ΔclsABC mutants. Additionally, possible transductants were allowed to grow for 36 h at 37 °C as opposed to selection for 16 h. Surviving transductants were isolated and their mutations were mapped by whole genome sequencing. A ∆clsABC ∆lpxM suppressor contained a single missense mutation in msbA encoding a T411P substitution. The ∆clsA ∆lpxM suppressor mutant also contained a single mutation in msbA, with a missense mutation encoding a P500T substitution. MsbA is an ABC transporter that flips the lipid A–core oligosaccharide from the inner leaflet to the periplasmic face of the IM (1). In both suppressors, the substitution is mapped to the nucleotide-binding domain of the protein. The suppressors alleviated the growth and morphological defect of the parent mutant strains (Fig. 4). Since MsbA is essential, we hypothesized that overexpression of MsbA would also rescue the viability of a ∆clsA, ∆lpxM double mutant. Thus, ∆clsA mutants harboring pmsbA were recipients in a transduction using P1 phage ∆lpxM::kan. Kanamycin resistant transductants were obtained on agar containing arabinose to induce the expression of msbA, and these strains had an improved growth (Fig. 4A) and showed WT morphology (Fig. 4B) compared with the ClsA-depleted clsA, lpxM Para::clsA strain.
Fig. 4.
Mutations in msbA suppress the growth defect of cls, lpxM mutants. (A) Growth of msbA suppressors. Listed strains were grown either in the presence 0.2% arabinose (+Ara) to induce plasmid expression or in 0.2% glucose (−Ara) to repress plasmid expression. Cultures were measured by OD600 every 30 min. (B) Micrographs of cells at the 5 h time point of growth curve shown in A. (Scale bar, 5 μm.) Error bars represent SEM from a technical triplicate.
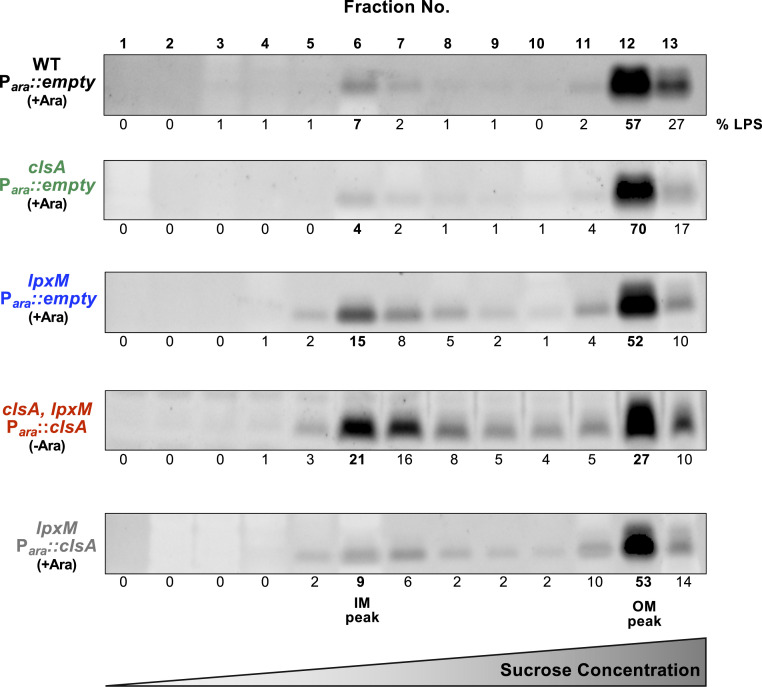
Our initial suppressor analysis suggest LPS transport deficiency is responsible for the lethality of ∆clsA, ∆lpxM double mutants. We therefore examined the LPS distribution between the OM and IM of ClsA-depleted clsA, lpxM Para::clsA and control parent strains. The OM and IM were separated by sucrose density gradient fractionation, and peak OM and IM fractions were identified by quantification of the OM β-barrel protein OmpA (fractions 11, 12, and 13) and NADH oxidase activity (fractions 5, 6, and 7), respectively (SI Appendix, Fig. S5) (21, 22). We establish that lpxM mutants have ∼threefold more LPS in the IM relative to WT (Fig. 5 and SI Appendix, Fig. S6). Furthermore, depletion of ClsA in the absence of lpxM further increases localization of LPS to the IM fraction (∼fourfold more LPS compared with WT) (Fig. 5 and SI Appendix, Fig. S6). Interestingly, LPS levels in the IM of lpxM mutants are lowered when ClsA is overexpressed (Fig. 5 and SI Appendix, Fig. S6). Overall, these data suggest that ClsA levels modulate LPS accumulation in the IM.
Fig. 5.
Depletion of ClsA in the absence of LpxM increases LPS levels in the IM. Indicated strains were grown for 5 h in 0.2% arabinose (+Ara) to induce plasmid expression or in 0.2% glucose to repress plasmid expression (−Ara), and cultures were normalized for OD600. IM and OM fractions from listed cultures were separated by a three-step sucrose gradient. Sodium dodecyl sulphate–polyacrylamide gel electrophoresis stained for LPS was used to quantify LPS concentrations across the isolated fractions. IM) peak fractions are indicated by NADH oxidase, and OM peaks are indicated by OmpA concentration (SI Appendix, Fig. S2). %LPS was calculated by fraction density divided by total LPS density. Gels are representative of three biological experiments.
Depletion of Cardiolipin in the Absence of lpxM Lowers MsbA Transport of LPS.
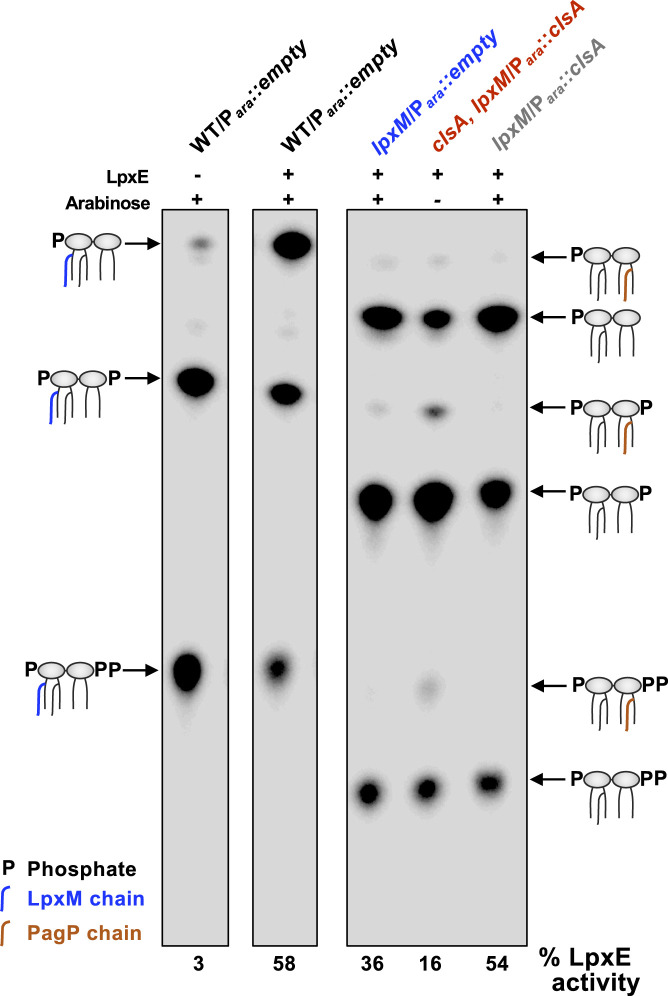
We next wanted to determine if LPS accumulation at the IM occurs in the inner or outer leaflet upon ClsA depletion. LpxE, an LPS modification enzyme present in several gram-negative bacteria (e.g., Francisella), removes the 1-phosphate group from lipid A at the periplasmic face of the IM (23, 24). Although not present in E. coli, heterologous expression of LpxE results in efficient dephosphorylation of E. coli lipid A (25). Thus, LpxE modification of lipid A can be used as a topological reporter of MsbA flippase activity (25). Cells expressing LpxE were grown to early log phase and then labeled with 32Pi for one doubling time prior to lipid A extraction. WT lipid A consists of two major lipid A species: the bulk species (∼70%) is phosphorylated at the 1′ and 4′ positions, and an additional Tris-phosphorylated species contains a diphosphate at the 1′ position that migrates more slowly during thin-layer chromatography (TLC) (Fig. 6) (26). Upon expression of LpxE, 58% of the lipid A is dephosphorylated in the WT background, resulting in lipid A that migrates faster than the two WT lipid A species (Fig. 6). LpxM is responsible for the last acylation step in lipid A synthesis; as expected its absence resulted in penta-acylated lipid A species, reducing the migration of lipid A on a TLC (Fig. 6) (7). The expected dephosphorylated lipid A species makes up 36% of the lipid A, indicating the lipid A is not readily accessible to LpxE compared with the lipid A in a WT background (Fig. 6). Only 16% of the ClsA-depleted clsA, lpxM Para::clsA strain’s lipid A was dephosphorylated by LpxE (Fig. 6), suggesting poor transport of LPS across the IM. However, LpxE activity in the ∆lpxM strain is equivalent to WT if clsA is overexpressed (Fig. 6). Because the absence of CL has been shown to decrease protein transport through the SecYEG system (18), we wanted to demonstrate that the increase in LpxE-modified lipid A when clsA is overexpressed in a ΔlpxM background is independent of increased LpxE levels. Therefore, we measured LpxE-modified lipid A in a WT background overexpressing clsA, and we observed no increase in LpxE activity (SI Appendix, Fig. S7). This suggests that increased LpxE activity in a ΔlpxM mutant overexpressing clsA is independent of SecYEG function. Together, our results indicate that the presence of CL impacts the transport of LPS across the IM.
Fig. 6.
Depletion of ClsA in the absence of LpxM decreases MsbA-mediated LPS transport. Cultures were grown to mid-log phase in either presence 0.2% arabinose (+) to induce plasmid expression or in 0.2% glucose (−) to repress plasmid expression. Cells were then labeled with 32Pi for one doubling, and the lipid A was isolated and separated by TLC and visualized by phosphorimaging. %LpxE-modified lipid A were determined by densitometry. Lipid A species are indicated, and the TLC is representative of three biological experiments.
Depletion of Cardiolipin in lpxM Mutants Exacerbates OM Permeability Defects.
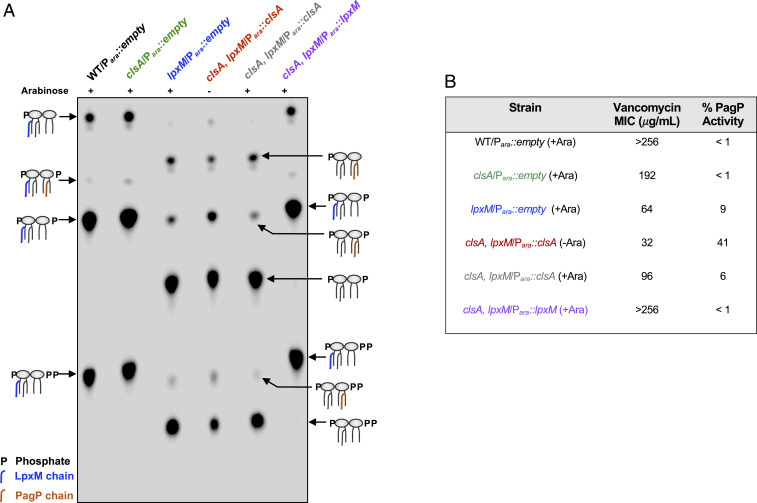
If LPS transport is hindered in our strains, one would expect a disruption of OM asymmetry leading to increased GPLs presented at the bacterial surface (27). One way to monitor this disruption is modification of lipid A by the OM β-barrel acyltransferase PagP. PagP transfers a palmitate (C16:0) from PE, the bulk GPL, to one of the β-hydroxymyristate chains (3-OH-C14:0) of lipid A (Fig. 1) (28). Since the active site of PagP faces the extracellular environment, only PE localized to the outer leaflet of the OM can serve as an acyl donor. Thus, PagP activity functions as a reporter of mislocalized GPLs and OM asymmetry (28). To monitor for PagP modification, cultures were grown in the presence of 32Pi, and lipid A species were extracted and analyzed by TLC (Fig. 7A). The deletion of clsA resulted in no major changes to the lipid A profile compared with WT (Fig. 7A). However, in the lpxM mutant, species are observed migrating similar to hexa-acylated lipid A. These species are PagP-acylated lipid A as determined by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) analysis (Fig. 7A and SI Appendix, Fig. S8) and comprise ∼10% of the total lipid A species, indicating an increase in mislocalized GPLs. In the ClsA-depleted clsA, lpxM Para::clsA strain, PagP-acylated lipid A makes up ∼40% of the lipid A, suggesting that OM asymmetry is severely compromised. This severe reduction in OM asymmetry is not present when an inducible plasmid with clsA or lpxM is grown in the presence of arabinose (Fig. 7A). Interestingly, we observe reduced PagP-acylated lipid A in our msbA suppressors, suggesting LPS flux to the OM is restored (SI Appendix, Fig. S9).
Fig. 7.
Depletion of ClsA in the absence of LpxM increases OM defects. (A) Lipid A profile of ClsA-depleted clsA, lpxM/Para::clsA mutant and control strains. Cultures were grown to mid-log phase in the presence 0.2% arabinose (+) to induce plasmid expression or in 0.2% glucose (−) to repress plasmid expression in the presence of 32Pi. Lipid A was isolated, separated by TLC, and visualized by phosphorimaging. Lipid A species are indicated. (B) Measured OM defects of ClsA-depleted clsA, lpxM/Para::clsA mutant and control strains. Vancomycin MIC was measured in liquid LB cultures with increasing concentrations of vancomycin. Cultures began at an OD600 0.05 and grew at 37 °C for 8 h. MICs were defined as no growth above the starting OD600. The level of PagP-modified lipid A species were determined by densitometry. TLC data and antibiotic sensitivity are representative of three biological experiments.
Additionally, we measured the minimum inhibitory concentration (MIC) of vancomycin for each strain (Fig. 7B). Vancomycin is a large antibiotic that cannot readily pass through the gram-negative OM (29), and increased sensitivity to the antibiotic indicates a disruption in OM asymmetry. The vancomycin MIC mirrored PagP-acylated lipid A levels of each mutant. ΔclsA mutants are slightly less resistant to vancomycin compared with WT cells, 192 and >256 μg/mL, respectively (Fig. 7B). ΔlpxM mutants exhibited a severe decrease in vancomycin resistance, with a MIC of 64 μg/mL, similar to previously published reports (Fig. 7B) (7). The ClsA-depleted clsA, lpxM/Para::clsA strain is even more sensitive with an MIC of 32 μg/mL (Fig. 7B). Induction of clsA or lpxM in the conditional double mutant restored vancomycin phenotypes to the same level or above those of the individual ΔclsA or ΔlpxM strains (Fig. 7B). Additionally, overexpression of clsA increased the vancomycin resistance in lpxM mutants in both W3110 and MG1655 strains (SI Appendix, Table S1). Furthermore, the suppressors mapped to msbA increase vancomycin resistance to 96 μg/mL, above that of ΔlpxM (SI Appendix, Table S1). These data sets suggest that the double mutant has a severe OM defect, and the overexpression of clsA can alleviate OM defects associated with loss of LpxM acyltransferase activity.
Decreasing LPS Synthesis Rescues Growth of CL-Deficient lpxM Mutants.
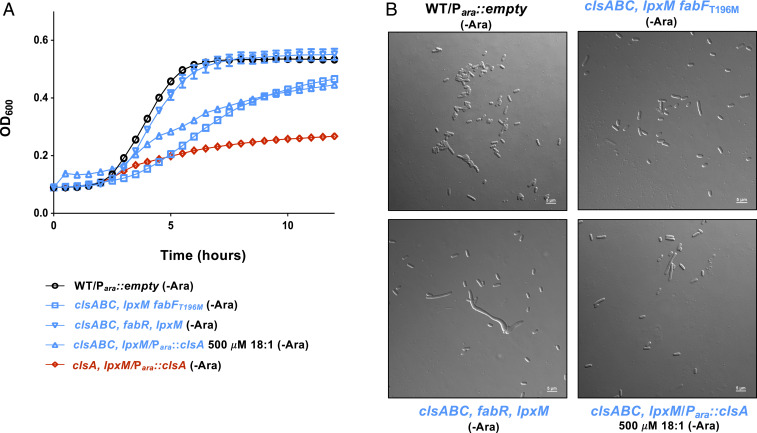
In addition to suppressors in msbA, a single-nucleotide polymorphism in fabF allowed for viable transductants when ∆clsABC mutants were transduced with P1 phage ∆lpxM::kan. The fabFT196M suppressor rescued both growth (Fig. 8A) and cell morphology (Fig. 8B). The deletion of fabF in a ∆clsABC mutant did not allow transductants with ∆lpxM::kan P1 phage to be produced, leading us to speculate that our fabFT196M suppressor is a gain-of-function mutation. FabF is one of three 3-ketoacyl-ACP synthases and has the highest affinity for the elongation of palmitoleic acid (16:1 ∆9) to vaccenic acid (18:1 ∆11) (30). We therefore hypothesized that increasing the level of vaccenic acid within the cell would promote viability in the absence of both clsABC and lpxM. Increasing the cell’s pool of 18:1 fatty acids can be accomplished by adding the fatty acid exogenously or through genetic manipulation. FabR functions as a repressor for fabB, which is an additional 3-ketoacyl-ACP synthase (31). The deletion of fabR results in increased expression of fabB, leading to an increase in the 18:1 fatty acid cellular pool (31). Both the deletion of fabR or the addition of 18:1 fatty acids to the growth media rescued the lethality and aberrant cell morphology exhibited by cells in the absence of ClsA and LpxM (Fig. 8). Altogether, this data indicates that increasing the 18:1 fatty acid pool promotes cell viability in the absence of clsABC and lpxM.
Fig. 8.
Increasing unsaturated fatty acids suppress the growth defect of clsABC, lpxM mutants. (A) Growth of fatty acid synthesis suppressors. Listed strains were grown in 0.2% glucose (-Ara) to repress plasmid expression. Growth of indicated strains were measured by OD600 every 30 min. (B) Micrographs of cells at the 5 h time point of growth curve shown in A. (Scale bar, 5 μm.) Error bars represent SEM from a technical triplicate.
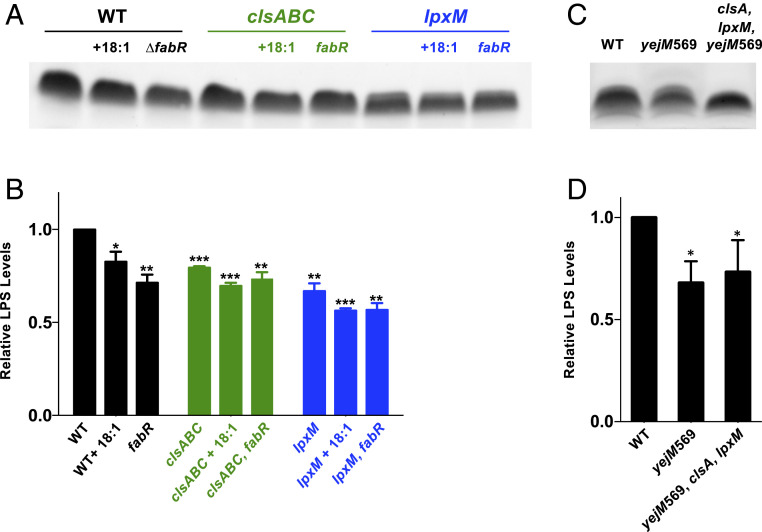
Why does changing the inherent fatty acid pool suppress our synthetic lethal phenotype? We examined if suppressors ΔfabR and fabFT196M restore the OM asymmetry defects seen in the ClsA-depleted clsA, lpxM/Para::clsA strain by performing a vancomycin Etest (SI Appendix, Table S1). The result indicated that the ΔfabR and fabFT196M suppressors failed to restore OM defects as the MICs were even lower than the ∆lpxM parent strain (SI Appendix, Table S1). Furthermore, we see that when fabR is deleted in either a ∆clsABC or ∆lpxM background, vancomycin resistance is decreased (SI Appendix, Table S1). Based on these results, we hypothesize that LPS levels are reduced when 18:1 fatty acid pools increase, alleviating LPS accumulation in the IM observed in ClsA-depleted clsA, lpxM/Para::clsA mutants (Fig. 5) (32). To test this possibility, we measured the overall LPS levels of ∆clsA and ∆lpxM strains containing fabR deletions or grown in the presence of 18:1 fatty acids (Fig. 9 A and B). The addition of 18:1 fatty acids to the growth medium or deletion of fabR led to a decrease in overall LPS levels in WT (Fig. 9 A and B). We also observe that absence of clsA or lpxM also results in an overall decrease in LPS levels.
Fig. 9.
Decreasing LPS levels rescue the growth defect of clsA, lpxM mutants. (A and C) LPS levels were determined by a gel stain of proteinase K–treated whole cell lysates of the indicated strains. (B and D) Densitometry of LPS levels normalized to the WT strain. Error bars represent SEM from a technical triplicate. A t test was used between strains. *P < 0.05, **P < 0.005, and ***P < 0.001.
With the hypothesis that lowering LPS rescues the double mutants, we took advantage of a truncated yejM mutant (yejM569) to lower LPS through genetic means. LpxC catalyzes the committed step for lipid A production, and stability of the enzyme dictates the flux of precursors into lipid A synthesis (33). YejM has recently been identified to regulate LpxC stability, and disruption of YejM synthesis results in lowered LPS levels (34–38). Addition of a yejM569 allele into the clsA strain allows for lpxM to be deleted with no severe growth defect (SI Appendix, Fig. S10). This viability is attributed to the decrease in LPS levels by the yejM569 allele (Fig. 9 C and D). Taken together, our results indicate that lowering LPS levels allows the cell to survive when LPS transport is restricted from loss of CL synthesis.
Discussion
Extensive research on the synthesis and biochemical properties of GPLs has been conducted for over 40 y, yet the biological role of CL remains poorly understood. CL composes ∼5% of the GPL makeup under broad conditions, but CL concentrations can increase threefold during the stationary phase (13). Despite the fluctuating levels of CL and the presence of three separate CL synthase enzymes, ClsA, ClsB, and ClsC, CL is not required for cell growth (13). Previous work has shed light on multiple protein–CL interactions that impact the bacterial cell. Some examples include the activation of respiratory complexes (39), the polar localization of the osmosensory transporter protein ProP (40), and influencing the subcellular distribution of cell division proteins (41–44). More recently, CL has been shown to aid in protein transport through the Sec system and to facilitate drug efflux by binding to a resistance-nodulation-cell division (RND) efflux pump (18, 45). Additionally, previous studies focused on how the absence of CL affects the cell, such as changes in biofilm formation, decreased protein secretion, and cell morphology (15, 46). In this study, we sought to identify major gene networks that become essential in the absence of CL through Tn-seq analysis of cls combinatorial mutants. We identify that the presence of lpxM is essential in the absence of clsA for proper growth, cell division, and OM integrity.
LpxM is the final enzyme in the synthesis of the Kdo2-lipid A substructure transferring a secondary acyl chain, a myristoyl group, to the 3′ position, forming hexa-acylated lipid A (6). The gene encoding LpxM was first described as a multicopy suppressor that rescued the temperature-sensitive growth of an E. coli lpxL mutant (47). Later, Raetz and colleagues characterized the function of LpxM as one of the so-called “late” acyltransferases of lipid A biosynthesis (8, 47) (Fig. 1A). Disruption of the lpxL allele in E. coli results in LPS with tetra-acylated lipid A, termed lipid IVA (Fig. 1A), that is not efficiently transported by MsbA, resulting in a lack of growth at elevated temperatures (10). The poor transport of tetra-acylated LPS results in morphological defects reminiscent of the ClsA-depleted ∆clsA ∆lpxM strain (Fig. 3C). LpxM acylates Kdo2-lipid IVA at a slow rate, and therefore its overexpression increases lipid A acylation, promoting MsbA-mediated flipping and cell viability (8). The transport of lipid A in lpxM mutants has not been studied as extensively as lpxL mutants. However, MsbA’s poor transport of underacylated lipid A, coupled with the sensitivity of lpxM mutants to antibiotics, suggests penta-acylated lipid A may also be poorly transported (7, 10). We have replicated the findings that lpxM mutants are vancomycin sensitive (7) (Fig. 7B and SI Appendix, Table S1) and have observed increased PagP activity in lpxM mutants (Fig. 7B), suggesting disruption of OM asymmetry. Previous lipid A profiles of lpxM mutants did not reveal as high levels of PagP activity as our ∆lpxM mutant, but these experiments characterized lipid A from cells grown at 30 °C in minimal media (7, 8). To prove that OM defects associated with lpxM mutants arise from decreased transport of lipid A, we separated OM and IM fractions. Mutants lacking LpxM had a threefold increase in accumulation of LPS in the IM compared with WT cells (Fig. 5 and SI Appendix, Fig. S7). This accumulation of LPS is located at the inner leaflet of the IM, prior to MsbA transport, as LPS from lpxM mutants were less accessible to periplasmic LpxE modification (Fig. 6). Therefore, MsbA cannot transport penta-acylated lipid A as efficiently as hexa-acylated lipid A. Penta-acylated lipid A produced by lpxM mutants was recently shown to have altered binding to the hydrophobic pockets in the Lpt system, which transports LPS from the periplasmic leaflet of the IM to the OM, further supporting that the acylation state of the substrate affects LPS transport systems (48).
We found that depletion of ClsA in lpxM mutants results in a severe growth and morphological defect (Fig. 3). The depletion of ClsA exacerbated lpxM phenotypes, such as vancomycin sensitivity, disruption of OM asymmetry, and LPS aggregation in the IM (Figs. 5, 6, and 7B). Interestingly, the overexpression of clsA alleviates such associated phenotypes in lpxM mutants (Figs. 5, 6, and 7B and SI Appendix, Table S1). Furthermore, we demonstrate that the synthetic lethal phenotype of ΔclsA and ΔlpxM are associated with MsbA transport (Fig. 6), providing evidence that CL affects MsbA activity. Our first line of evidence that CL affects MsbA activity is that we mapped suppressors to the nucleotide-binding domain of MsbA (Fig. 4A), which powers the transport of LPS through ATP hydrolysis (49). Previous studies have shown that MsbA poorly flips underacylated lipid A because of inefficient stimulation of ATPase activity (11). Together with our results, these findings indicate that the transmembrane regions of MsbA must sense lipid A binding, which triggers conformational changes to stimulate the ATPase, a process in ABC transporters called coupling (50). For MsbA, proper coupling occurs with the completely synthesized hexa-acylated lipid A but must be inefficient with the tetra- or penta-acylated lipid A, suggesting MsbA serves as a checkpoint for whether LPS synthesis has been completed. Notably, the P500T substitution in one of our suppressors is located between the ATPase Walker B motif and the helical subdomain that is involved in coupling conformational movements (51, 52). Thus, our suppressors likely alter coupling of MsbA so that ATPase activity is stimulated by binding of the poor substrate, penta-acylated lipid A, and bypass a normal checkpoint for LPS synthesis. The exacerbation of transport defects in the clsA lpxM double mutant indicates that in bacteria with reduced CL levels, MsbA’s selectivity for hexa-acylated lipid A becomes more stringent. Furthermore, a ∆clsA mutation could not be introduced into a mutant harboring impaired Lpt transport machinery, further supporting that absence of CL in tandem with increased LPS at the IM results in death (53). Overexpression of MsbA rescues the synthetic phenotype (Fig. 4A), possibly in a manner analogous to the rescue of lpxL mutants (54).
Several studies have shown CL–protein interactions. Recent evidence reveals the presence of CL-binding sites in SecY and SecA (18). The SecYEG complex transports proteins through the IM powered by the essential SecA ATPase (55). Absence of CL binding to the Sec system results in lowered ATPase activity of SecA, leading to a reduction in protein translocation activity (18). The protein magnesium transporter A (MgtA), a specialized P-type ATPase, was found to colocalize with CL, but CL also activates the transporter. Absence of CL has also been shown to impact the function of the Acr (acriflavine resistance) multidrug efflux system. In this case, CL may act allosterically to modulate AcrB activity, a transporter belonging to the RND superfamily (45). We speculate that CL may have similar effects during LPS transport, supporting MsbA activity. The bacterial cell can ordinarily tolerate loss of lpxM; however, this comes at the cost of inefficient MsbA-dependent transport, which, when combined with the loss of clsA, results in a synthetic lethal phenotype. Therefore, we propose a model in which MsbA ATPase activity is reduced in the absence of both CL or LpxM, and when both are absent, MsbA is no longer functional leading to cell death. We hypothesize that CL binds to MsbA, and in the absence of CL, MsbA ATPase is lowered, as seen in the SecYEG system (18). Since underacylated LPS forms are poor substrates for MsbA (11), when both CL and LpxM are absent, the likely additive reduction in ATPase activity and LPS transport leads to cell death.
In our Tn-seq analysis, rfaC and rfaF were identified to be essential in both ΔclsABC and ΔlpxM mutants (Dataset S1 and SI Appendix, Fig. S2). RfaC and RfaF are responsible for adding the first and second heptose sugars of the inner core oligosaccharide, respectively (56). Since core addition is sequential, absence of RfaC and RfaF results in a deep-rough LPS phenotype lacking the core region (56). Mutants containing deep-rough LPS are sensitive to antibiotics, and LPS with core truncations decrease MsbA ATPase activity, all suggesting poor transport of deep-rough LPS (11, 57, 58). Furthermore, core modification in Pseudomonas aeruginosa is essential for LPS transport by the Lpt system (59). Therefore, the poor transport of deep-rough LPS coupled with hindered MsbA activity of clsA or lpxM mutants may cause a lethal accumulation of LPS in the IM, decreasing bacterial fitness. We are actively investigating how specific core truncations affect LPS transport.
Through suppressor analysis, we establish that increasing cellular pools of 18:1 fatty acids by exogenous or genetic means rescue the growth defects of ∆clsA ∆lpxM mutants (Fig. 8A). Proteins involved in fatty acid biosynthesis and regulation have recently been identified to interact with and regulate LpxC (60). We speculate that our suppressors modulate LpxC levels to reduce overall LPS levels (Fig. 9). The fabF suppressor does not restore vancomycin resistance, suggesting that OM asymmetry remains disrupted and indicates reduced LPS at the bacterial surface (SI Appendix, Table S1). Our suppressor may decrease LPS levels in one of two ways: 1) FabF overexpression has been shown to decrease LpxC stability in vivo, and therefore FabFT196M may have increased activity or a longer half-life, leading to decreased LpxC levels (60); and 2) FabF is also responsible for increased synthesis of 18:1 fatty acids (31), and previous work in E. coli has shown that mutants with higher 18:1 fatty acid content have decreased amounts of LPS (33). Here, we demonstrate that addition of exogenous 18:1 fatty acid lowers LPS levels (Fig. 9), rescuing our synthetic lethal phenotype. Deletion of the transcriptional regulator FabR also rescues the lpxM, clsA deletion (Fig. 8). Loss of fabR increases transcript levels of FabA and FabB, leading to increased levels of 18:1 fatty acid (61), and fabR mutants produce significantly less LPS (Fig. 9). Notably, FabA overexpression also decreases LpxC stability (60). Regardless, we demonstrate that our mutations in fatty acid synthesis lead to decreased LPS levels and reinforce the critical cross-talk between LPS and fatty acid levels (33, 62, 63).
We observed single ΔclsA and ΔlpxM mutants have lowered LPS levels compared with WT cells. We speculate the cell senses the absence of CL and signals the cell to balance GPL and LPS synthesis by lowering LPS levels. A possible link between CL and LPS synthesis is that LpxK’s catalytic activity is driven by GPLs in vitro, especially CL, as absence of CL reduces LpxK activity drastically (64). Decreased activity of LpxK would lead to an increase in lipid A disaccharide, which has recently been postulated to increase LpxC degradation by FtsH, leading to the observed decrease in LPS (Fig. 9 A and C) (65). We found increased LPS levels in the inner leaflet of the IM in our ΔlpxM strain, leading us to hypothesize the presence of an unknown sensor of LPS accumulation in the IM. The cell may sense this increase in LPS and signal the cell to decrease synthesis to alleviate the poor transport of LPS from the IM to the OM.
Our study demonstrates that CL generated by ClsA promotes the transport of poor lipid A substrates for MsbA. We propose that CL aids in MsbA ATPase activity to transport LPS. Although further studies are required to test this possibility, the identification of CL aiding in maintenance of OM asymmetry represents a novel mechanism of this poorly understood GPL.
Materials and Methods
Bacterial Growth Conditions.
Unless otherwise stated, bacteria were cultured in Luria-Bertani (LB) or on LB agar at 37 °C. LB was supplemented with ampicillin (Amp) (100 μg/mL), kanamycin (Kan) (30 μg/mL), chloramphenicol (Cam) (30 μg/mL), l-arabinose (0.2% [wt/vol]), and/or D-glucose (0.2% glucose [wt/vol]). For growth curves, cultures were grown in 0.2 mL using the BioTek Epoch 2 plate reader in a polystyrene 96-well plate.
Strain Construction in E. coli.
All bacteria strains and plasmids used in this study are listed in SI Appendix, Table S2 in the supplemental material, and oligonucleotides are listed in SI Appendix, Table S3. Chromosomal mutations were introduced into E. coli K-12 strain W3110 using generalized transduction and the Keio collection (66). Strain clsA, lpxM::kan /Para::clsA and clsA, lpxM::kan /Para::lpxM were maintained in medium supplemented with arabinose unless otherwise stated.
Flippase recognition target–flanked resistance cassettes were removed by flippase (FLP)/ FRT) site-specific recombination as previously described (67). pCP20, which expresses FLP from a temperature-sensitive promoter, was electroporated into W3110 containing the Keio allele. Transformants were recovered in LB at 30 °C for 1 h followed by selection on LB agar supplemented with Amp at 30 °C. The following day, single colonies were grown on LB at 37 °C. Colonies were screened on Amp and Kan sensitivity to confirm loss of both pCP20 and removal of the kan allele. PCR was also used to confirm removal of the Kan resistance cassette. The scar region following FLP/FRT recombination contains a single FRT site (68).
Generation of E. coli Tn Mutant Library.
To generate the Tn mutant library in W3110 WT and mutant strains, we modified a previous published method (19). E. coli β-3914, which is a diaminopimelic acid auxotroph, was electroporated with pJNW684 to create the donor strain (19). Mutant Tn libraries were generated by mating β-3914 with our W3110 recipient strains and the exconjugants selected on kanamycin. A total of 450,000 exconjugant colonies were collected by scrapping agar plates, and the colonies were pooled and stored in 30% glycerol at −80 °C.
Growth Challenge Assay and DNA Library Preparation.
Three individual aliquots of the Tn library were thawed and back diluted into 50 mL of plain LB to a starting optical density (OD) at 600 nm of 0.001 and grown at 37 °C until an OD600nm of 0.5. Cells were pelleted, and genomic DNA was extracted using the Easy-DNA Kit from Invitrogen following manufacturer instructions. The DNA was then diluted to 250 ng/μL and sheared by sonication to obtain fragments around 300 base pairs. Poly-C tails were added to 2.5 μg of the sheared DNA using a terminal deoxynucleotidyl transferase (Promega) for 1 h at 37 °C using 9.5 mM deoxycytidine triphosphate/0.5 mM dideoxycytidine triphosphate mix per manufacturer instructions. DNA fragments were purified using AMPure beads (Beckman Coulter) and used as template for a first PCR step using Platinum Pfx Polymerase (Invitrogen) along with primers olj510-Biotin and olj376. PCR products were again purified using AMPure beads, and the biotin-tagged eluted DNA was separated using streptavidin beads (New England Biolabs). Before the DNA purification, the streptavidin beads were equilibrated in 1× bind and wash (B&W) buffer (1 M NaCl, 5 mM Tris HCl, and 0.5 mM ethylenediaminetetraacetic acid [EDTA], pH 7.5) and washed with 1× B&W and two washes in low Tris-EDTA (LOTE) buffer (3 mM Tris HCl and 0.2 mM EDTA, pH 7.5). The biotin-tagged DNA bound to the streptavidin beads was used as a template for a second PCR step, using the Platinum Pfx Polymerase and primers olj511 and BC#. The PCR product was purified using 40 μL AMPure beads, and the concentration of the DNA was quantified using the Qubit double-stranded DNA High Sensitivity assay and the Qubit 3.0 Fluorometer (Life Technologies).
Samples were paired-end sequenced using the Illumina HiSeq 2500 platform at the Georgia Genomics and Bioinformatics Core Facility. Tn-seq data analysis was performed using QIAGEN CLC Genomics Workbench and the E. coli W3110 genome sequence (GenBank accession number NC_007779).
Selection of Suppressors for clsABC and lpxM Essentiality.
Overnight cultures of clsABC or clsA were used as the recipient strain in a transduction using P1 phage with a ∆lpxM::kan allele from the Keio collection. Transductants were plated on LB agar containing Kan and 2.5 mM sodium citrate and incubated until colonies formed. Suppressor mutations in msbA and fabF were identified by whole genome sequencing. Genomic DNA was extracted as previously described, and the DNA was prepped using Nextera DNA Flex Library Prep Kit per manufacturer’s instructions (Illumina). Libraries were sequenced on the Illumina iSq 100, and sequencing analysis was carried out using QIAGEN CLC Genomics Workbench and the E. coli W3110 genome (GenBank accession number NC_007779).
Separation of IM and OM Fractions.
Sucrose density gradient centrifugation was performed as previously described with some modifications (21). For each strain, a 30 mL LB culture was inoculated with a 1/100 dilution of an overnight culture and grown until OD600 ∼0.5. Washed cells were then resuspended in 6 mL 10 mM Tris HCl (pH 8.0) and 20% sucrose (wt/wt) containing 50 μg/mL DNase I and lysed by a single passage through a cell press at 8,000 psi. Unbroken cells were then removed by centrifugation at 10,000 × g for 10 min. A total of 5 mL of cell lysate was layered on a two-step sucrose gradient consisting of 40% (5 mL) and 65% (1.5 mL) sucrose solution (wt/wt) in 10 mM Tris HCl (pH 8.0). To separate the membrane fractions, samples were centrifuged at ∼100,000 × g for 16 h in a Beckman SW41 Rotor in an ultracentrifuge. From the top of each tube, 0.8 mL fractions were then collected and frozen for further analysis. LPS was then visualized with the use of the Pro-Q Emerald 300 Lipopolysaccharide Staining Kit (Molecular Probes, Inc.) after proteinase K treatment. The presence of the OM protein OmpA and the level of NADH oxidase activity were used as OM and IM markers, respectively (SI Appendix, Supplementary Materials and Methods). The fraction containing the highest level of OmpA, including the fraction immediately below and above it, is referred to as our pooled LPS OM fraction. The same method was used to identify our pooled LPS IM fraction, except using fractions with peak NADH oxidase activity. The sum of the LPS density in these pooled membrane fractions were used to determine the fold change in LPS located in the IM and OM between strains.
Supplementary Material
Acknowledgments
This work was funded by NIH Grants AI138576 and AI150098 to M.S.T. We thank Dr. Brent Simpson and Dr. Tori Jeter for their critical reading of the manuscript and for experimental advice.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018329118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Henderson J. C., et al., The power of asymmetry: Architecture and assembly of the gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 70, 255–278 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kamio Y., Nikaido H., Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570 (1976). [DOI] [PubMed] [Google Scholar]
- 3.Schindler M., Osborn M. J., Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry 18, 4425–4430 (1979). [DOI] [PubMed] [Google Scholar]
- 4.Ogura T., et al., Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 31, 833–844 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Raetz C. R. H., et al., Discovery of new biosynthetic pathways: The lipid A story. J. Lipid Res. 50 (suppl.), S103–S108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitfield C., Trent M. S., Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Vorachek-Warren M. K., Ramirez S., Cotter R. J., Raetz C. R. H., A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 277, 14194–14205 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Clementz T., Zhou Z., Raetz C. R. H., Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 272, 10353–10360 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Simpson B. W., Trent M. S., Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z., White K. A., Polissi A., Georgopoulos C., Raetz C. R. H., Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273, 12466–12475 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Doerrler W. T., Raetz C. R. H., ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Sherman D. J., et al., Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan B. K., et al., Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. U.S.A. 109, 16504–16509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiraoka S., Matsuzaki H., Shibuya I., Active increase in cardiolipin synthesis in the stationary growth phase and its physiological significance in Escherichia coli. FEBS Lett. 336, 221–224 (1993). [DOI] [PubMed] [Google Scholar]
- 15.Rowlett V. W., et al., Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199, e00849-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt V., Sidore M., Bechara C., Duneau J.-P., Sturgis J. N., The lipid environment of Escherichia coli Aquaporin Z. Biochim. Biophys. Acta Biomembr. 1861, 431–440 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Sekimizu K., Kornberg A., Cardiolipin activation of dnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem. 263, 7131–7135 (1988). [PubMed] [Google Scholar]
- 18.Corey R. A., et al., Specific cardiolipin-SecY interactions are required for proton-motive force stimulation of protein secretion. Proc. Natl. Acad. Sci. U.S.A. 115, 7967–7972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang N., Ozer E. A., Mandel M. J., Hauser A. R., Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio 5, e01163-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somerville J. E. Jr, Cassiano L., Darveau R. P., Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67, 6583–6590 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy T. J., Kahne D., Walker S., The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrivastava R., Jiang X., Chng S.-S., Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol. Microbiol. 106, 395–408 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Cullen T. W., et al., Helicobacter pylori versus the host: Remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 7, e1002454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran A. X., et al., The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J. Bacteriol. 188, 4531–4541 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Karbarz M. J., McGrath S. C., Cotter R. J., Raetz C. R., MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: Topography of francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 279, 49470–49478 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosner M. R., Tang J., Barzilay I., Khorana H. G., Structure of the lipopolysaccharide from an Escherichia coli heptose-less mutant. I. Chemical degradations and identification of products. J. Biol. Chem. 254, 5906–5917 (1979). [PubMed] [Google Scholar]
- 27.Ruiz N., Gronenberg L. S., Kahne D., Silhavy T. J., Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop R. E., The lipid A palmitoyltransferase PagP: Molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Pootoolal J., Neu J., Wright G. D., Glycopeptide antibiotic resistance. Annu. Rev. Pharmacol. Toxicol. 42, 381–408 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Janßen H. J., Steinbüchel A., Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol. Biofuels 7, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y.-M., Marrakchi H., Rock C. O., The FabR (YijC) transcription factor regulates unsaturated fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 277, 15558–15565 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Emiola A., Andrews S. S., Heller C., George J., Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 113, 3108–3113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson M. S., Raetz C. R., Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J. Biol. Chem. 262, 5159–5169 (1987). [PubMed] [Google Scholar]
- 34.Guest R. L., Samé Guerra D., Wissler M., Grimm J., Silhavy T. J., YejM modulates activity of the YciM/FtsH protease complex to prevent lethal accumulation of lipopolysaccharide. mBio 11, e00598-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen D., Kelly K., Qiu N., Misra R., YejM controls LpxC levels by regulating protease activity of the FtsH/YciM complex of Escherichia coli. J. Bacteriol. 202, JB.00303-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fivenson E. M., Bernhardt T. G., An essential membrane protein modulates the proteolysis of LpxC to control lipopolysaccharide synthesis in Escherichia coli. 11, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clairfeuille T., et al., Structure of the essential inner membrane lipopolysaccharide-PbgA complex. Nature 584, 479–483 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Simpson B. W., Douglass M. V., Trent M. S., Restoring balance to the outer membrane: YejM’s role in LPS regulation. MBio 11, e02624-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arias-Cartin R., et al., Cardiolipin-based respiratory complex activation in bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 7781–7786 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romantsov T., et al., Cardiolipin synthase A colocalizes with cardiolipin and osmosensing transporter ProP at the poles of Escherichia coli cells. Mol. Microbiol. 107, 623–638 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Mileykovskaya E., Dowhan W., Role of membrane lipids in bacterial division-site selection. Curr. Opin. Microbiol. 8, 135–142 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Hsieh C.-W., et al., Direct MinE-membrane interaction contributes to the proper localization of MinDE in E. coli. Mol. Microbiol. 75, 499–512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renner L. D., Weibel D. B., MinD and MinE interact with anionic phospholipids and regulate division plane formation in Escherichia coli. J. Biol. Chem. 287, 38835–38844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vecchiarelli A. G., Li M., Mizuuchi M., Mizuuchi K., Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol. Microbiol. 93, 453–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du D., et al., Interactions of a bacterial RND transporter with a transmembrane small protein in a lipid environment. Structure 28, 625–634.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nepper J. F., Lin Y. C., Weibel D. B., Rcs phosphorelay activation in cardiolipin-deficient Escherichia coli reduces biofilm formation. J. Bacteriol. 201, e00804-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karow M., Georgopoulos C., Isolation and characterization of the Escherichia coli msbB gene, a multicopy suppressor of null mutations in the high-temperature requirement gene htrB. J. Bacteriol. 174, 702–710 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lundstedt E. A., Simpson B. W., Ruiz N., LptB-LptF coupling mediates the closure of the substrate-binding cavity in the LptB2 FGC transporter through a rigid-body mechanism to extract LPS. Mol. Microbiol. 114, 200–213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eckford P. D. W., Sharom F. J., The reconstituted Escherichia coli MsbA protein displays lipid flippase activity. Biochem. J. 429, 195–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Schneider E., Hunke S., ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22, 1–20 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Hollenstein K., Dawson R. J. P., Locher K. P., Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Sutterlin H. A., Zhang S., Silhavy T. J., Accumulation of phosphatidic acid increases vancomycin resistance in Escherichia coli. J. Bacteriol. 196, 3214–3220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voss B. J., Trent M. S., Transport L. P. S., LPS transport: Flipping out over MsbA. Curr. Biol. 28, R30–R33 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Crane J. M., Randall L. L., The Sec system: Protein export in Escherichia coli. Ecosal Plus 7, 10.1128/ecosalplus.ESP-0002-2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z., Wang J., Ren G., Li Y., Wang X., Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli. Mar. Drugs 13, 3325–3339 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konovalova A., Mitchell A. M., Silhavy T. J., A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 5, e15276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Austin E. A., Graves J. F., Hite L. A., Parker C. T., Schnaitman C. A., Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: Insertion mutagenesis of the rfa locus. J. Bacteriol. 172, 5312–5325 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delucia A. M., et al., Lipopolysaccharide (LPS) inner-core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. MBio 2, e00142-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomanek N., et al., Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front. Microbiol. 9, 3285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parsons J. B., Rock C. O., Bacterial lipids: Metabolism and membrane homeostasis. Prog. Lipid Res. 52, 249–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomanek N., Intricate crosstalk between lipopolysaccharide, phospholipid and fatty acid metabolism in Escherichia coli modulates proteolysis of LpxC. Front. Microbiol. 9, 3285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.May K. L., Silhavy T. J., The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio 9, e00379-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ray B. L., Raetz C. R., The biosynthesis of gram-negative endotoxin. A novel kinase in Escherichia coli membranes that incorporates the 4′-phosphate of lipid A. J. Biol. Chem. 262, 1122–1128 (1987). [PubMed] [Google Scholar]
- 65.Emiola A., George J., Andrews S. S., A complete pathway model for lipid A biosynthesis in Escherichia coli. PLoS One 10, e0121216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baba T., et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cherepanov P. P., Wackernagel W., Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.