Significance
Bovine embryonic stem cells and pluripotent stem cells hold the potential to substantially advance biotechnology and agriculture. We report the establishment of bovine expanded potential stem cells (bEPSCs) from preimplantation embryos of both wild-type and somatic cell nuclear transfer (SCNT). EPSCs have broader developmental potential to generate embryonic and extraembryonic cell lineages. bEPSCs express high levels of pluripotency genes, propagate robustly in single cell passaging, are genetically stable, and permit efficient precise gene editing. They differentiate in vitro and in chimeras to both the embryonic and extraembryonic cell lineages. Importantly, genetically modified bEPSCs can be used as donors in SCNT or cloning.
Keywords: bovine, expanded potential stem cell, nuclear transfer
Abstract
Embryonic stem cells (ESCs) and induced pluripotent stem cells have the potential to differentiate to all cell types of an adult individual and are useful for studying development and for translational research. However, extrapolation of mouse and human ESC knowledge to deriving stable ESC lines of domestic ungulates and large livestock species has been challenging. In contrast to ESCs that are usually established from the blastocyst, mouse expanded potential stem cells (EPSCs) are derived from four-cell and eight-cell embryos. We have recently used the EPSC approach and established stem cells from porcine and human preimplantation embryos. EPSCs are molecularly similar across species and have broader developmental potential to generate embryonic and extraembryonic cell lineages. We further explore the EPSC technology for mammalian species refractory to the standard ESC approaches and report here the successful establishment of bovine EPSCs (bEPSCs) from preimplantation embryos of both wild-type and somatic cell nuclear transfer. bEPSCs express high levels of pluripotency genes, propagate robustly in feeder-free culture, and are genetically stable in long-term culture. bEPSCs have enriched transcriptomic features of early preimplantation embryos and differentiate in vitro to cells of the three somatic germ layers and, in chimeras, contribute to both the embryonic (fetal) and extraembryonic cell lineages. Importantly, precise gene editing is efficiently achieved in bEPSCs, and genetically modified bEPSCs can be used as donors in somatic cell nuclear transfer. bEPSCs therefore hold the potential to substantially advance biotechnology and agriculture.
Embryonic stem cells (ESCs) of mouse, rat, human, and nonhuman primate species are established from the inner cell mass in the blastocyst (1–5). A significant advance in mouse ESC derivation is to use small molecule inhibitors, specifically the ones targeting Mek1/2 (PD0325901) and GSK3 (Chir99021), in the 2i/LIF naïve ESC culture condition (6). However, extrapolating the 2i/LIF condition to deriving ESCs of large animals has proven challenging. We and others recently reported establishment of mouse, human, and porcine expanded potential stem cells (EPSCs) (7–10). We posited that deriving stem cells from earlier preimplantation embryos, for example, mouse four-cell or eight-cell cleavage embryos, might help overcome the substantial species differences reported in the blastocyst-stage embryos (11–16). Following this line of reasoning, EPSCs were established by inhibiting molecules and pathways operating in preblastocyst embryos (7, 9, 10). EPSCs are molecularly and functionally similar across species. They possess robust self-renewal capacity in long-term culture, allow efficient genome editing, and generate both embryonic and extraembryonic cell lineages in vitro and in chimeras in terms of mouse and porcine EPSCs (7–9). Mouse EPSCs were reported to self-assemble into blastocyst-like structures (17).
Cattle, or cows, are domesticated bovine farm animals and the most common type of large domesticated ungulates. Bovine ESCs would be expected to substantially facilitate genome editing, to accelerate molecular breeding schemes for economic traits, and to provide a platform for investigating the bovine preimplantation development with potential applications in improving cloning. Intensive efforts have been made to derive bovine ESCs or to reprogram bovine somatic cells to induced plurpotent stem cells (iPSCs) (18–38). These cells, in general, however, lack the standard pluripotent stem cell criteria: poor derivation efficiencies; inability of maintaining pluripotency in long-term culture; limited developmental potential in the in vitro and in vivo assays such as chimera generation. The reported bovine iPSC lines often have leaky expression of the reprogramming genetic factors (28–38). Recently, bovine-primed ESCs were reported (39), which represents a major advance. These cells had the typical mouse- and human-primed ESC properties but morphologically did not form distinct cell colonies, unlike ESCs of other species, and no chimera was generated (39). In this report, we successfully established and characterized bovine EPSCs (bEPSCs). The availability of bEPSCs, which are robustness in culture, permit efficient genome editing, possess expanded developmental potential, are expected to substantially advance bovine stem cell biology to considerably facilitate selecting for superior animals for farming and to open up opportunities for biotechnology.
Results
Identification of a Culture Condition that Maintains Bovine Pluripotency.
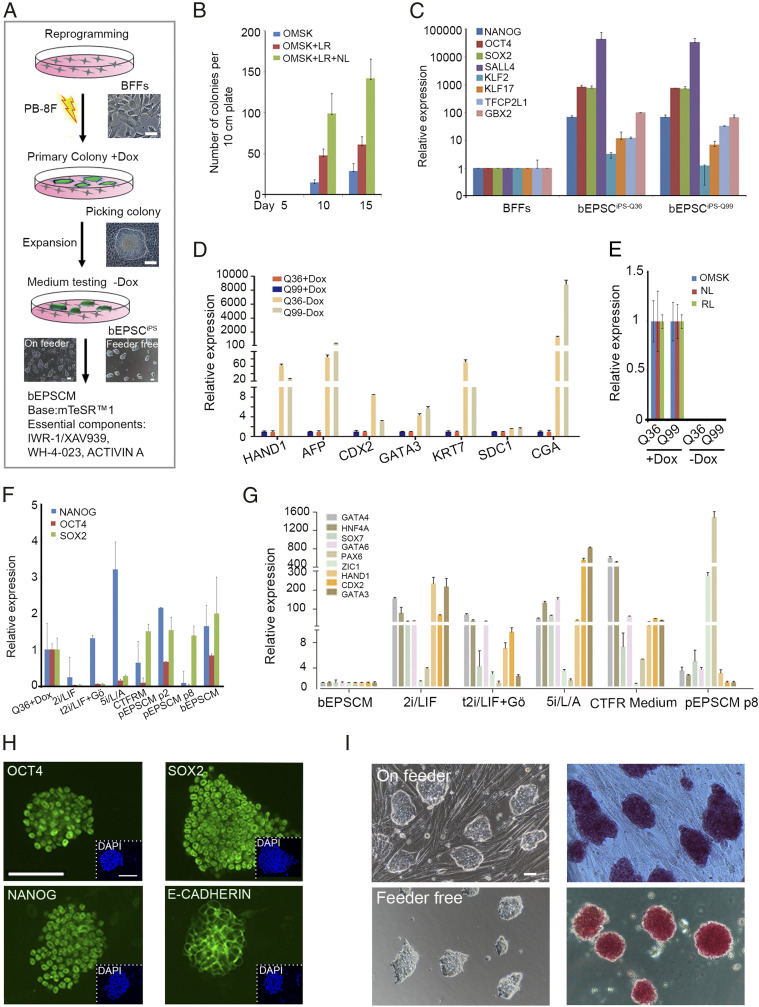
We wished to identify culture conditions under which bona fide bovine pluripotent stem cells could be derived and stably maintained. Due to the limited supply of bovine preimplantation embryos, we chose to initially establish bovine iPSC lines for testing culture conditions. We expressed Dox-inducible eight exogenous reprogramming factors, bOMSK (bovine OCT4, CMYC, SOX2, and KLF4), pNhL (porcine NANOG and human LIN28), and hRL (human RARG and LRH1) in bovine fetal fibroblasts (BFFs) of China Qinchuan bovine, delivered via piggyBac transposition (Fig. 1A). Dox induction reprogrammed ∼0.1% transfected BFFs to primary colonies, which were picked on day 15 through 20 (Fig. 1 A and B). The picked colonies were passaged in single-cell suspension in a serum containing medium (M15) in the presence of Dox. The passaged cells expressed high levels of the endogenous pluripotency genes, such as NANOG, OCT4 (POU5F1), and SOX2 (Fig. 1C), and could be maintained undifferentiated in Dox for at least 50 passages. They were thus named bovine iPSCs. Upon Dox removal, bovine iPSCs were differentiated in 8 d, concomitant with the increased expression of both embryonic and extraembryonic cell-lineage genes and with the loss of pluripotency gene expression (Fig. 1D). Importantly, these Dox-dependent bovine iPSCs did not appear to have detectable leaky expression of the exogenous reprogramming factors once Dox was removed from the culture (Fig. 1E). The pluripotency in these iPSCs thus depended on Dox-induced exogenous factor expression in the serum-containing medium. These bovine iPSCs thus provided a platform for identifying culture conditions that would be able to maintain endogenous pluripotency gene expression, independent of the Dox-induced exogenous factor expression. We tested culture conditions for mouse ESCs and human ESCs including 2i/LIF (6), t2iL+ Gӧ (40), 5i/L/A (41), the recently reported CTFR medium (mTeSR1 supplemented with FGF2 and IWR1) for bovine primed EPSCs (39), and porcine EPSC medium (pEPSCM) (9). Bovine iPSCs Q36 of passage 20 were cultured under these conditions for 8 d without Dox and were morphologically and transcriptionally examined. In 2i, t2iL+ Gӧ, and 5i/L/A, bovine iPSCs lost OCT4 and SOX2 expression (Fig.1F) and expressed high levels of both embryonic and extraembryonic cell-lineage genes (Fig. 1G). Concomitantly, the compact and domed EPSC colonies were now flat and appeared to be differentiated (SI Appendix, Fig. S1A). The CTFR medium for primed bovine ESCs, on the other hand, was able to maintain NANOG and SOX2 expression, but OCT4 expression was substantially decreased (Fig. 1F). Moreover, in CTFR, bovine iPSCs adopted a flat colony morphology (SI Appendix, Fig. S1A) and expressed high levels of the lineage marker genes (GATA4, HNF4A, HAND1, GATA6, CDX2, and GATA3) (Fig. 1G), indicating a primed state and partial differentiation. Porcine EPSC medium (pEPSCM) was able to maintain the iPSCs for up to 7 passages with an undifferentiated morphology and high expression of core pluripotency genes (Fig. 1F). However, under pEPSCM, bovine iPSCs were eventually differentiated in long-term culturing, accompanied with down-regulation of core pluripotency genes and expression of both embryonic and extraembryonic cell-lineage genes, with differentiated cell morphology (Fig. 1 F and G and SI Appendix, Fig. S1A), demanding further adjustment of pEPSCM for bovine stem cells. pEPSCM contains inhibitors targeting GSK3, SRC, and Tanykrases (9). We modified the pEPSCM and developed bEPSCM for bovine stem cells, which contains XAV939 (or IWR-1), CHIR99021, WH-4–023 or A419259, Vitamin C, ACTIVIN A, and LIF in mTeSR basal medium, whereas pEPSCM used N2B27 basal medium (9). Under bEPSC medium (bEPSCM), bovine iPSCs formed compact colonies and expressed high levels of endogenous pluripotency genes (Fig. 1 A, F–H, and SI Appendix, Fig. S1B). Importantly, bovine iPSCs did not have detectable leaky expression of exogenous reprogramming factors (SI Appendix, Fig. S1C), could be maintained for at least 63 passages without noticeable differentiation, and retained a normal karyotype (SI Appendix, Fig. S1D). At passage 63, 2n = 60. 46 out of 52 metaphases or 88%. In vitro and in teratomas, bovine iPSCs could differentiate into cells representative of both embryonic and extraembryonic cell-lineage genes (SI Appendix, Fig. S1 E–G) and were thus named as bEPSCiPS. One remarkable feature of bEPSCiPS was that they remained undifferentiated in feeder-free condition for more than 30 passages and proliferated robustly (Fig. 1I). Similar to porcine EPSCs, bEPSCs were sensitive to Mek1/2 inhibition. When Mek1/2 inhibitor PD0325901 (1.0 µm) was added into bEPSCM, most bEPSCs died in 4 d, and the remaining cells lost OCT4 expression in the OCT4-mCherry reporter bEPSCs (SI Appendix, Fig. S1H). In RT-qPCR analysis, the remaining cells also lost OCT4 and NANOG expression and expressed high levels of SOX2, indicating differentiation (SI Appendix, Fig. S1I). These differentiated cells did not survive the next passaging in bEPSCM. Therefore, porcine and bovine EPSCs require higher levels of Mek1/2 signaling than that in the mouse and human.
Fig. 1.
Reprogramming BFFs to Dox-inducible iPSCs for testing bovine stem cell culture condition. (A) Schematic illustration of reprogramming BFFs to iPSCs. PB-8F: bOMSK+ pN–hLIN + hRL. bOMSK (bovine OCT4, MYC, SOX2, and KLF4 cDNAs), pN–hLIN (porcine NANOG and human LIN28 cDNAs); hRL (human RARG and LRH1 cDNAs). BFFs: bovine fetal fibroblasts; Dox: doxycycline. (Scale bar, 50 μm.) (B) Coexpression of LIN28 (L), NANOG (N), LRH1, and RARG (LR) along with four Yamanaka factors substantially increased the reprogrammed colony numbers. (C) Relative expression of key endogenous pluripotency genes in two iPSC lines cultured in bEPSCM, bEPSCiPS-Q36, and bEPSCiPS-Q99. Data represent the mean ± SD, n = 3 independent experiments. (D) Expression of lineage genes in RT-qPCR of iPSC lines in the presence or absence of Dox in M15 medium. Q36: biPS-Q36 with Dox, Q99: biPS-Q99 with Dox. Data represent the mean ± SD, n = 3 independent experiments. (E) No detectable leaky expression of the exogenous reprogramming factors in iPSCs in RT-qPCR. (F and G) RT-qPCR analysis of pluripotency (F) and lineage genes (G) in bovine iPSCs under several culture conditions in the absence of Dox. These conditions include 2i/LIF, t2iL+Gӧ, and 5i/L/A on day 8; CTFR medium (passage 4); and pEPSCM (cells of passage 2 and passage 8 for analyzing pluripotency genes, and cells of passage 8 for analyzing lineage genes). Cells cultured in bEPSCM for passage 36 were used in the analysis. pEPSCM: porcine expanded potential stem cells medium, bEPSCM: bovine expanded potential stem cells medium, CTFRM: custom TeSR1 base medium supplemented with FGF2 and IWR1. Data represent the mean ± SD, n = 3 independent experiments. (H) Immunostaining of NANOG, OCT4, SOX2, and E-CADHERIN in bovinebEPSCiPS. (Scale bar, 100 μm.) (I) The morphology and alkaline phosphatase (AP) staining of bEPSCiPS-Q36 on feeder cells (Upper) or feeder free (Lower). (Scale bar, 50 μm.)
Establishment of bEPSC Lines from Preimplantation Embryos.
We next investigated deriving EPSC lines from bovine preimplantation embryos (Fig. 2A). From 32 early blastocysts (5 to 6 d postcoitum [dpc]) of Holstein, Angus, and Montbeliarde bovine, nine cell lines (bEPSCES, 3 male and 6 female) were established (Fig. 2B). bEPSCES had high nuclear/cytoplasmic ratios and formed compact domed colonies with smooth colony edges (Fig. 2A). They proliferated robustly and were routinely passaged in 2 to 3 d (1:4 passaging ratio) without the need of the Rho Associated Coiled-Coil Containing Protein Kinase (ROCK) inhibitor Y-27632, and could be maintained in long-term cultures (>82 passages). Cryopreserved bEPSCs could be readily recovered. They were genetically stable and retained a normal karyotype (Fig. 2C) (2n = 60, 38/50, 76%, passage 82 for female bEPSCES, 2n = 60, 36/50, 72%, passage 76 for male bEPSCES). bEPSCES expressed high levels of pluripotency genes but undetectable or minimal levels of lineage genes (Fig. 2 D and E and SI Appendix, Fig. S2 A and B). bEPSCES could also be maintained feeder free in long-term cultures (Fig. 2 E and F) and differentiated via embryoid body formation into cells expressing genes representative of cell types of the three germ layers and trophoblast-like cells (PL-1+) (SI Appendix, Fig. S2 C and D). In vivo, bEPSCES formed mature teratomas that contained cell types of the somatic germ layers (Fig. 2G). After transient expression of the SOX17 transgene, both bEPSCsiPS and bEPSCsES could generate cells in the Embryoid Bodies (EBs) expressing genes highly enriched in primordial germ cells such as OCT4, NANOG, SOX17, TFAP2C, NANOS3, and BLIMP1 (SI Appendix, Fig. S2 E and F), similar to porcine and human EPSCs (9).
Fig. 2.
Establishment of bEPSCs from preimplantation embryos. (A) Schematic diagram of establishment of bEPSCES from bovine day 6 in vivo fertilization embryos. (Scale bar, 50 μm.) (B) bEPSCES lines of three breeds: Holstein, Angus, and Montbéliarde. (C) Karyotyping analysis of bEPSCsES-A6 (female, passage 82) and bEPSCsES-A15 (male, passage 76). (D) Immunostaining of NANOG, OCT4, SOX2, and E-CADHERIN in bEPSCs ES-A15 of passage 38. (Scale bar, 100 μm.) (E) Relative expression of core pluripotency genes OCT4, NANOG and SOX2 in two bEPSC lines (bEPSCsES-A15 on passage 32 and bEPSCiPS-Q36 on passage 36) on feeders or feeder-free. The relative expressions above were normalized to control and housekeeping gene. Data represent the mean ± SD, n = 3 independent experiments. (F) Morphology and AP staining of bEPSCES-A15 on feeder cells (Upper, passage 36) or feeder free (Lower, passage 30). (Scale bar, 50 μm.) (G) Teratoma derived from bEPSCES-A15 (passage 40). (H and E) Analysis revealed the presence of glandular epithelium (endoderm, i), muscle (mesoderm, ii), cartilage (mesoderm, iii), and mature neural tissue (glia and neurons, ectoderm, iv). (Scale bar, 50 μm.)
We determined the colony formation capability from individual bEPSCs and compared to the primed ESCs. Single bEPSCiPS-Q36 and bEPSCES-A6 were picked by microcapillary and seeded into individual 96-wells in bEPSCM. The colony formation efficiency was 47.9% for bEPSCiPS and 40.9% for bEPSCES, whereas much lower efficiencies (2.4% and 1.7%, respectively) were found for the same cells cultured in the primed bovine ESC medium, CTFR (SI Appendix, Fig. S2 G and H).
Transcriptomic and Epigenetic Features of bEPSCs.
Unlike mouse and human naïve ESCs, human and porcine EPSCs have high levels of DNA methylation (9). Whole-genome bisulfite sequencing of bEPSCES and bEPSCiPS revealed that DNA methylation levels in these cells were around 87%, with lower DNA methylation at the promoter regions, including bovine imprinting genes (Fig. 3 A and B and SI Appendix, Fig. S3 A and B). Proper genomic imprinting is required for normal mammalian development (42). Bovine preimplantation embryos have relatively low expression of some genomic imprinted genes (43–45). bEPSCs, in particular bEPSCES, expressed low levels of imprinted genes IGF2, IGF2R, H19, MEST, RTL1, PEG10, DLK1, DIO3, PEG3, and PLAGL1 (SI Appendix, Fig. S3C) (46–54). Compared to BFFs, bEPSCs exhibited comparable DNA methylation in the differential methylation regions at the genomic imprinting loci (Fig. 3B and SI Appendix, Fig. S3B and Table S1). X chromosome reactivation is an important indication in mouse and human naïve ESCs (55, 56). We sought to determine X reactivation state in female bEPSCs as this was not studied in the recently established bovine primed ESCs (39). We stained bEPSCs for H3K27me3 that is enriched on the inactivated X chromosome as a consequence of Xist-dependent recruitment of polycomb repressor complexes PRC2 (57, 58). We analyzed H3K27me3 staining domains or foci in cells of five colonies formed from cells of the two female lines, bEPSCsES-H4 and bEPSCsES-A6. In both cell lines, roughly 40% of the cells had the prominent perinuclear H3K27me3-positive foci (Fig. 3C). Interestingly, in female bovine preimplantation embryo development, X inactivation (XCI) starts as early as in morula, and its onset did not immediately lead to a global down-regulation of X-linked genes (59).
Fig. 3.
Transcriptomic and epigenetic features of bEPSCs. (A) DNA methylation levels in bEPSCs by whole-genome bisulfite sequencing analysis. Boxplots of the averaged DNA methylation levels (CpG sites) of 5 kb tiles in bEPSCiPS-Q36, bEPSCES-A6, and BFFs. The bottom and top of the boxes indicate the first and third quartiles, respectively, and the lines inside the boxes indicate the medians of the data. (B) DNA methylation in DMR of bovine-imprinted genes, including DLK1-DIO3 cluster, H19 cluster, IGF2R, MEG3, PEG3, PEG10, PLAGL1, SNRPPN, and ZIM2 in bEPSCs. (C) Immunostaining detection of H3K27me3 foci in female bEPSCES. The male bEPSCES were the negative control. (Scale bar, 50 μm.) (D) PCA of global gene expression (RNA-seq) of bEPSCs, bovine-primed ESCs, and BFFs. (E and F) Expression of pluripotency genes, lineage genes, and DNA methylation genes in bEPSCs, pEPSCs, and BFFs. Bovine-primed ESCs, n = 10; BFFs, n = 2; bEPSCsiPS-Q36, n = 2; bEPSCsES-A6, n = 2; n represents the number of biologically independent samples. (G) PCA of global gene expression (RNA-seq) of EPSCs and bovine preimplantation embryos (GSE59186) (60). Two replicates in each sample were used. (H) Expression levels of all annotated bovine histone genes in bovine-primed ESCs, EPSCs, and BFFs.
Global gene expression profiling of bEPSCsES and bEPSCsiPS revealed that they were clustered together but distinct from primed bESCs and BFFs (Fig. 3D). Although expression of key pluripotency genes, such as OCT4, SOX2, NANOG, and SALL4, were similar between bEPSCs and primed bESCs (Fig. 3E), DNA methyltransferase genes, DNMT1, DNMT3A, and DNMT3B, were expressed at higher levels in bEPSCs, whereas TET1, TET2, and TET3 were lower in bEPSCs (Fig. 3F), in line with high DNA methylation in bEPSCs. In comparison to bovine fibroblasts, bEPSCs had significantly higher expression of genes functioning in cell cycle and oxidative phosphorylation in Gene Set Enrichment Analysis (GSEA) (SI Appendix, Fig. S3D). Comparing to primed bovine ESCs, bEPSCs had higher gene expression relevant to MYC targets, DNA repair and oxidative phosphorylation in GSEA, whereas those genes in Notch signaling, Wnt signaling, and glycolysis were significantly overrepresented in primed bovine ESCs (SI Appendix, Fig. S3E). Transcriptomic analysis suggested that mouse EPSCs had enriched features of four- to eight-cell blastomeres (7), whereas human expanded potential stem cells (hEPSCs) were more similar to human eight-cell to morula-stage embryos than other developmental stages (9). Similarly, bEPSCs appeared to have enriched transcriptomic features of bovine eight-cell to morula stage embryos (60) in Principal Component Analysis (PCA) (Fig. 3G).
One unique feature of human EPSCs is high expression of certain histone genes (9). Indeed, bEPSCs, but not the primed bESCs, highly expressed many histone genes (Fig. 3H). Across the three species of human, pig, and bovine, EPSCs exhibited similar expression profiles of pluripotency genes and three germ-layer marker genes (SI Appendix, Fig. S3F), extraembryonic cell lineage genes (SI Appendix, Fig. S3G), and genes encoding enzymes for DNA methylation (SI Appendix, Fig. S3H).
bEPSCs In Vivo Developmental Potential.
We next investigated the developmental capacity of bEPSCs in forming chimeras (Fig. 4A). bEPSCES-A15 stably expressing tdTomato were generated by piggyBac transposition. We first tested bEPSCs in the mouse embryo development by injecting tdTomato+ bEPSCs into mouse eight-cell stage embryos and allowed the injected embryos to develop in vitro for 24 to 48 h into blastocysts. tdTomato+ cells were detected in both the trophectoderm and the inner cell mass of the mouse blastocysts (SI Appendix, Fig. S4A). The injected mouse morula embryos were also transferred to pseudopregnant female recipients for postimplantation development. From 62 transferred embryos, only 1 E6.5 chimera embryo was recovered where the donor origin tdTomato+ cells were primarily found in the extraembryonic ectoderm (ExE) region (SI Appendix, Fig. S4B). We also injected tdTomato+ bEPSCs to bovine morula embryos, which were allowed to further develop for 48 h to the blastocyst. tdTomato+ cells were detected in the inner cell mass (ICM) at 88.4% (46/52), 5.7% in the trophectoderm (3/52) (Fig. 4B). Some tdTomato+ cells (5 out of 32) in the trophectoderm (TE) were positive for CDX2 (Fig. 4B). To generate in vivo chimeras, we followed the blastocyst injection practice for producing mouse chimeras. We injected bovine early blastocysts (n = 97) with tdTomato+ bEPSCs and transferred them to pseudopregnant recipient cows (n = 46) (Fig. 4A). Thirteen recipients were found pregnant, and the embryos were harvested on day 38 (n = 4), day 40 (n = 7), and day 72 (n = 2) (Fig. 4A). Whole-mount fluorescence examination detected tdTomato+ cells in 5 of the 13 conceptuses (Fig. 4C and SI Appendix, Table S2). We further confirmed the presence of tdTomato+ cells by carefully dissecting and dissociating tdTomato+ tissues into single cells for fluorescence microscopy examination (Fig. 4C and SI Appendix, Fig. S4C). In two chimeras (No. F1607 and No. F1506, day 40), tdTomato+ cells were detected in both the placenta and embryonic tissues. We performed genomic DNA PCR to detect tdTomato DNA in various tissues of the five chimeras (SI Appendix, Fig. S4D) and genetically confirmed the presence of descendants of the donor tdTomato+ bEPSCs. To identify the descendants of donor bEPSCs in specific tissues, we performed immunofluorescence analysis to detect lineage marker expression in the tdTomato+ cells in chimeras, which revealed that tdTomato+ cells expressed markers of chorionic placenta (PL-1, GATA3, hCGβ, SDC1, and KRT7) and of embryonic cell lineages (SMA, β-TUBULIN III, SOX17, GATA6, and AFP) (Fig. 4D and SI Appendix, Fig. S4E). These results, together with the in vitro differentiation data (Fig. 1D and SI Appendix, Figs. S1 E and F and S2 C and D), indicate that bEPSCs have developmental potential to both the embryonic and extraembryonic cell lineages in chimeras, similar to porcine EPSCs (9).
Fig. 4.
bEPSC’s developmental potential in chimeras. (A) Schematic diagram of chimera experiments using bEPSCs. (B) Contribution of bEPSCs in bovine preimplantation embryo development. The tdTomato+ donor bEPSCsES-A15 (passage 20) were injected into bovine morula embryos, which developed into blastocysts in 48 h in vitro. Panel (i), Injected tdTomato+ bEPSCsES-A15 in the blastocyst; Panel (ii), Several tdTomato+ cells (arrow) expressed trophectoderm factor CDX2. (Scale bar, 100 μm.) TdT, tdTomato. (C) Whole-mount fluorescence and bright field images of 40 d conceptuses from transferred preimplantation embryos. Chimera no. F1506 appeared to have tdTomato+ cells. (Scale bar, 0.5 cm.) (D) Detection of bEPSCsES-A15 tdTomato+ descendants in the chorionic placenta (PL-1+; i) and in smooth muscles (SMA+; ii) in chimera F1506. The control embryos have no tdTomato+ cell injected. DAPI stains nuclei. (Scale bar, 50 μm.) TdT, tdTomato. (E) Genome editing in bEPSCsES-A15. Knock-in of the T2A-H2B-mCherry cassette into the OCT4 locus using the CRISPR/Cas9. The targeting vector with short homology arms from the OCT4 locus flanking the T2A-H2B-mCherry and a Puromycin-resistance cassette was constructed and transfected into bEPSCs. The Puro-resistant transfectant colonies were picked 10 d after transfection. The correctly targeted colonies were identified in genomic DNA PCR. GT primers are for genotyping. (F) Bright field and fluorescence images of the correctly targeted bEPSCs colonies. Eleven out of forty-eight colonies examined were correctly targeted ones. (G and H) tdTomato+ bEPSCES-A15 as the donor in SCNT. Injection of tdTomato+ bEPSCES-A15 (passage 32) donor cells into the perivitelline space of oocytes was shown. (Scale bar, 50 μm.) (I) Derivation of secondary bEPSCs from SCNT (cloned) blastocysts. An outgrowth of day 16 from a SCNT blastocyst with bEPSCs as the donor cell (Upper) was picked for establishing the secondary EPSCs (Lower, Passage 4). (Scale bar, 50 μm.)
Precise Genome Editing in bEPSCs and SCNT Cloning.
Genome editing in bEPSCs would enable dissection of gene functions and advance biotechnology applications. Besides piggyBac transposition, we investigated precision genome editing in bEPSCs using the CRISPR system to knock an mCherry cassette into the bovine OCT4 (POU5F1) locus (Fig. 4E). bEPSCs were transfected with a targeting vector where two short homology arms flanked a T2A-mCherry cassette. At the targeted allele, the mCherry coding sequence replaces the OCT4 stop codon. Out of 48 genotyped colonies, 11 were correctly targeted for 22.9% targeting efficiency. The targeted bEPSCs were mCherry+ under fluorescence microscope (Fig. 4F). The efficient precise genome editing in bEPSCs enables sophisticated genome modifications in the bovine genome. We next explored using bEPSCs as donor cells in somatic cell nuclear transfer (SCNT). Nuclei from tdTomato+ bEPSCsES-A15 were transferred into enucleated oocytes (n = 99), which were allowed to further develop to two-cell embryos (71.7%), eight-cell (41.4%), and the blastocyst (21.2%) (Table 1). These cloned embryo efficiencies were comparable to that of using fibroblasts as the control donors, indicating that bEPSCs could be used in SCNT for producing animals with sophisticated genetic modifications.
Table 1.
bEPSCs as the donor in SCNT in comparison to bovine fibroblasts
| Groups | No. of embryos reconstructed | No. of two-cell (%) | No. of eight-cell (%) | No. of blastocyst (%) |
| SCNT (fibroblast) | 92 | 66 (71.7) | 28 (42.4) | 12 (17.1) |
| bEPSCES-A15-NT | 99 | 70 (71.7) | 29 (41.4) | 14 (21.2) |
The SCNT embryos from bEPSCs provided an embryo source for testing deriving secondary EPSCs. Out of six SCNT blastocysts from bEPSCsES-A15 as the donor, we derived one bEPSC line (Fig. 4 G–I). These secondary bEPSCs had typical EPSC morphology, retained a normal karyotype (2n = 60; 20 out of 25 metaphases at passage 12), and expressed pluripotency factors including SOX2, NANOG, and POU5F1 (SI Appendix, Fig. S4F), demonstrating the robustness of the bEPSC system.
Discussion
Despite major advances in pluripotent stem cell research, establishing bovine ESCs comparable to the mouse and human counterparts is still challenging (18, 20, 26, 29–34, 36–38). In this study, we applied the EPSC technology to establish stem cell lines from bovine preimplantation embryos. We started reprogramming bovine somatic cells to Dox (exogeneous factors)-dependent iPSCs, which expressed high levels of endogenous pluripotency genes and thus allowed interrogating various culture conditions, including our published human and porcine EPSC culture conditions, for bovine stem cells. These experiments revealed that bovine stem cells necessitated a culture condition similar to porcine EPSC medium but demanded modifications and that, identical to porcine cells, bovine EPSCs required proper MEK1/2 activities as even low levels of the inhibitor PD-0325901 caused cell death and differentiation. A recent study reported bovine primed ESCs from the blastocyst (39), which marks a major advance in bovine stem cell research. In this study, we were able to reproduce the derivation of bovine primed ESCs. Compared to the primed ESCs, bEPSCs have several distinct properties that make bEPSCs the first bovine stem cells thatcan substantially facilitate basic and applied research. First, bEPSCs have much higher single-cell subcloning efficiencies in the absence of the ROCK inhibitor, indicating a culture robustness. bEPSCs could even be maintained feeder-free in long-term culture. Second, the culture robustness of bEPSCs enables efficient precise genome editing, which would be challenging in the bovine primed ESCs. Importantly, genetically modified bEPSCs can serve as donor cells in SCNT. One remarkable feature of bEPSCs is the genetic stability. bEPSCs retained a normal karyotype and have high key pluripotency gene expression even after they were single cell–passaged for >82 passages. The secondary EPSCs, which are established from SCNT embryos of genetically modified bEPSCs as the donors, are still karyotypically normal. Third, bEPSCs are able to differentiate to various embryonic and extraembryonic cell lineages in chimeras, whereas primed ESCs are expected to have rare contribution in chimeras, based on mouse primed ESC data (61). Further research is needed to improve procedures including bEPSC culture, bEPSC injection, and embryo transfer for generating high quality live born chimeras. bEPSC’s genetic and epigenetic features, culture properties, efficient precise genome editing, and developmental potential provide a basis for applying bEPSCs in broad biotechnology and agriculture research areas. Importantly, the establishment of EPSCs of multiple mammalian species demonstrate that the EPSC technology could be applicable in additional mammals.
In summary, bEPSCs from preimplantation embryos and by reprogramming somatic cells are established and characterized for their molecular properties and developmental potential. These stem cells propagate robustly in long-term culture, permit precise genome editing, and generate both embryonic and extraembryonic cell lineages in vitro and in chimeras. bEPSCs represent bovine ESCs that are anticipated to have many applications in agriculture and biotechnology.
Materials and Methods
Culturing bEPSCs.
bEPSCs were maintained on BFFs feeder layers, or without feeder cells, and enzymatically passaged every 2 to 3 d by a brief PBS washing followed by treatment for 2 min with TrypLE Select (Gibco, 12563-029). The cells were dissociated and centrifuged (300 g × 5 min) in K10 medium. K10 includes DMEM F12 (Gibco), 10% KSR (Gibco), 1× penicillin-streptomycin, and 1× MEM nonessential amino acids (Gibco). After removing supernatant, the bEPSCs were resuspended and seeded in bEPSCM. bEPSCM is mTeSR1 (STEMCELL, 85850)-based media. bEPSC media (500 mL) was prepared as follows: 485 mL mTeSR1 (STEMCELL), 5.0 mL 100× penicillin-streptomycin (Gibco), 0.1 mM 2-mercaptoethanol (Gibco), and the small molecules and cytokines 1 μM CHIR99021 (GSK3i; Selleck Chemicals, S2924), 0.3 μM WH-4-023 (Selleck Chemicals, S7565), 5 μM XAV939 (Sigma, X3004) or 5 μM IWR-1 (Selleck Chemicals, S7086), 50 μg ⋅ mL−1 Vitamin C (Sigma, 49752-100G), 10 ng ⋅ mL−1 LIF (Millipore, LIF1010), and 20.0 ng ⋅ mL−1 Activin A (R&D, 338-AC).
In Vivo Chimera Assay.
Six to twelve tdTomato+ bEPSCsiPS-Q36 were injected gently into the Institute of Cancer Research (ICR) mice eight-cell stage embryo using a piezo-assisted micromanipulator attached to an inverted microscope (Zeiss, Eppendorf); the protocol was performed as previously described. The injected embryos were cultured in KSOM (Millipore) and bEPSCM mixture medium (1:1) at 37 °C in a 5% CO2 atmosphere overnight and then transferred to the uteri of pseudopregnant ICR mice at 2.5 dpc. The embryos were isolated at embryonic stage E6.5 to check chimeric contribution. Also 5 to 10 bEPSCsES-A15 (tdTomato+) were injected into bovine morulae and early blastocysts with the aid of a piezo-driven micromanipulator in synthetic oviductal fluid (SOF) medium and bEPSCM mixture medium (1:1). After injection, bovine embryos were cultured in the same medium at 38.5 °C in 5% CO2 and 5% O2 for 6 to 48 h. The injected morula embryos were cultured for 24 to 48 h to the blastocysts for in vitro study. In the blastocysts, on average 13.2 cells were tdTomato+ cells. The injected early blastocysts were for the evaluation of postimplantation chimerism. After a short time culture, they were transferred to the uteri of pseudopregnant bovine at 7 dpc. At day 23 to 30 after transplantation, pregnancy was diagnosed by ultrasonography and Rapid Visual Pregnancy Test Kit (IDEXX, 99-41369). The fetuses were isolated at embryonic stage day 38 to 72 to check chimeric contribution.
CRISPR/Cas9-Mediated Genome Editing in bEPSCs.
To target an T2A-H2B–mCherry-EF1a-Puro cassette to the bovine OCT4 locus, OCT4 5′ and 3′ homology arms were amplified by PCR from bEPSCs (837-bp 5′ arm, Chr23: 27,986,458–27,987,294; 734-bp 3′ arm, Chr23: 27,987,203–27,987,937), according to NCBI Reference Sequence NC_037350.1. The sequence 5′- GTGCCTGCT-CACCCCAGGAATGG -3 was designed as the target of gRNA/Cas9.
Production of Nuclear Transfer Embryos Reconstructed with bEPSCsES.
The bEPSCsES within passages 15 through 25 were dispersed to a single-cell suspension by TrypLE select (Invitrogen) and recovered in bEPSCM. They were used as donor cells for nuclear transfer (NT). Single bEPSCs were individually transferred to the perivitelline space of the recipient cytoplasts. Successfully reconstructed embryos were kept in modified SOF (mSOF) (containing 5 mg/mL cytochalasin B) for 2 h until activation. All fused embryos were further activated in 5 mM ionomycin for 5 min, followed by exposure to 2 mM 6-dimethylaminopurine in SOF for 4 h. After the activation, NT embryos were washed and transferred into 500 μL of SOF media covered with mineral oil in a four-well plate, under an atmosphere of 5% CO2, 5% O2, 90% N2. The cleavage rates were determined 48 h after culturing, and the blastocyst rates were determined 7 d after culturing.
SI Appendix, SI Materials and Methods and Tables S1–S4 include further details of the study materials and methods.
Supplementary Material
Acknowledgments
We are grateful to Dr. Wangmei Qi for assistance with cryosectioning and immunofluorescence staining. We also thank the staff members of the Breeding Department of Inner Mongolia Saikexing Institute of Breeding and Reproductive Biotechnology in Domestic Animal for assistance with embryo transferring, recipient feeding, and chimera fetal anatomy. This research was partially supported by the National Natural Science Foundation of China (No. 31560335 and 32070869), the Key Science and Technology Planning Project of Inner Mongolia Autonomous Region (No. 2020ZD0007), and the Science and Technology Planning Project of Inner Mongolia Autonomous Region (No. 201702045). P.L. gratefully acknowledges support from Research Grants Council (RGC) (GRF 17127219) and The University of Hong Kong internal funding schemes.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018505118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Evans M. J., Kaufman M. H., Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). [DOI] [PubMed] [Google Scholar]
- 2.Martin G. R., Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buehr M., et al., Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Thomson J. A., et al., Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Thomson J. A., et al., Isolation of a primate embryonic stem cell line. Proc. Natl. Acad. Sci. U.S.A. 92, 7844–7848 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ying Q. L., et al., The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., et al., Establishment of mouse expanded potential stem cells. Nature 550, 393–397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., et al., Derivation of pluripotent stem cells with in vivo embryonic and extraembryonic potency. Cell 169, 243–257.e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao X., et al., Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 21, 687–699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J., Ryan D. J., Lan G., Zou X., Liu P., In vitro establishment of expanded-potential stem cells from mouse pre-implantation embryos or embryonic stem cells. Nat. Protoc. 14, 350–378 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Blakeley P., et al., Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 142, 3613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boroviak T., et al., Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 145, dev167833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu D., et al., Single-cell RNA-sequencing reveals the existence of naive and primed pluripotency in pre-implantation rhesus monkey embryos. Genome Res. 28, 1481–1493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petropoulos S., et al., Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 167, 285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos-Ibeas P., et al., Pluripotency and X chromosome dynamics revealed in pig pre-gastrulating embryos by single cell analysis. Nat. Commun. 10, 500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X. M., et al., Transcriptome analyses of inner cell mass and trophectoderm cells isolated by magnetic-activated cell sorting from bovine blastocysts using single cell RNA-seq. Reprod. Domest. Anim. 51, 726–735 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Li R., et al., Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell 179, 687–702.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito S., Strelchenko N., Niemann H., Bovine embryonic stem cell-like cell lines cultured over several passages. Rouxs Arch. Dev. Biol. 201, 134–141 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Van Stekelenburg-Hamers A. E., et al., Isolation and characterization of permanent cell lines from inner cell mass cells of bovine blastocysts. Mol. Reprod. Dev. 40, 444–454 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Mitalipova M., Beyhan Z., First N. L., Pluripotency of bovine embryonic cell line derived from precompacting embryos. Cloning 3, 59–67 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Muñoz M., et al., Conventional pluripotency markers are unspecific for bovine embryonic-derived cell-lines. Theriogenology 69, 1159–1164 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Cao S., et al., Isolation and culture of primary bovine embryonic stem cell colonies by a novel method. J. Exp. Zool. A Ecol. Genet. Physiol. 311, 368–376 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Pant D., Keefer C. L., Expression of pluripotency-related genes during bovine inner cell mass explant culture. Cloning Stem Cells 11, 355–365 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Anand T., et al., Buffalo (Bubalus bubalis) embryonic stem cell-like cells and preimplantation embryos exhibit comparable expression of pluripotency-related antigens. Reprod. Domest. Anim. 46, 50–58 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Maruotti J., et al., Efficient derivation of bovine embryonic stem cells needs more than active core pluripotency factors. Mol. Reprod. Dev. 79, 461–477 (2012).Corrected in: Mol. Reprod. Dev.79, 888 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Furusawa T., et al., Characteristics of bovine inner cell mass-derived cell lines and their fate in chimeric conceptuses. Biol. Reprod. 89, 28 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Yuan Y., Capturing bovine pluripotency. Proc. Natl. Acad. Sci. U.S.A. 115, 1962–1963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai C., et al., Melatonin improves reprogramming efficiency and proliferation of bovine-induced pluripotent stem cells. J. Pineal Res. 61, 154–167 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Canizo J. R., et al., Exogenous human OKSM factors maintain pluripotency gene expression of bovine and porcine iPS-like cells obtained with STEMCCA delivery system. BMC Res. Notes 11, 509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao H., et al., Characterization of bovine induced pluripotent stem cells by lentiviral transduction of reprogramming factor fusion proteins. Int. J. Biol. Sci. 8, 498–511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Y., et al., Generation of induced pluripotent stem cells from buffalo (Bubalus bubalis) fetal fibroblasts with buffalo defined factors. Stem Cells Dev. 21, 2485–2494 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Han X., et al., Generation of induced pluripotent stem cells from bovine embryonic fibroblast cells. Cell Res. 21, 1509–1512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaguchi T., et al., Generation of naïve bovine induced pluripotent stem cells using PiggyBac transposition of doxycycline-inducible transcription factors. PLoS One 10, e0135403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar D., et al., Transposon mediated reprogramming of buffalo fetal fibroblasts to induced pluripotent stem cells in feeder free culture conditions. Res. Vet. Sci. 123, 252–260 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Pillai V. V., et al., Induced pluripotent stem cell generation from bovine somatic cells indicates unmet needs for pluripotency sustenance. Anim. Sci. J. 90, 1149–1160 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Sumer H., et al., NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J. Anim. Sci. 89, 2708–2716 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Talluri T. R., et al., Derivation and characterization of bovine induced pluripotent stem cells by transposon-mediated reprogramming. Cell. Reprogram. 17, 131–140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao L., et al., Characterization of the single-cell derived bovine induced pluripotent stem cells. Tissue Cell 49, 521–527 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Bogliotti Y. S., et al., Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc. Natl. Acad. Sci. U.S.A. 115, 2090–2095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takashima Y., et al., Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theunissen T. W., et al., Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 524–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulsen M., Ferguson-Smith A. C., DNA methylation in genomic imprinting, development, and disease. J. Pathol. 195, 97–110 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Driver A. M., Huang W., Kropp J., Peñagaricano F., Khatib H., Knockdown of CDKN1C (p57(kip2)) and PHLDA2 results in developmental changes in bovine pre-implantation embryos. PLoS One 8, e69490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Z., et al., mRNA levels of imprinted genes in bovine in vivo oocytes, embryos and cross species comparisons with humans, mice and pigs. Sci. Rep. 5, 17898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruddock N. T., et al., Analysis of imprinted messenger RNA expression during bovine preimplantation development. Biol. Reprod. 70, 1131–1135 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Gebert C., et al., The bovine IGF2 gene is differentially methylated in oocyte and sperm DNA. Genomics 88, 222–229 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Kim J., Bergmann A., Lucas S., Stone R., Stubbs L., Lineage-specific imprinting and evolution of the zinc-finger gene ZIM2. Genomics 84, 47–58 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Suzuki J. Jr, et al., In vitro culture and somatic cell nuclear transfer affect imprinting of SNRPN gene in pre- and post-implantation stages of development in cattle. BMC Dev. Biol. 9, 9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki J. Jr, et al., Loss of methylation at H19 DMD is associated with biallelic expression and reduced development in cattle derived by somatic cell nuclear transfer. Biol. Reprod. 84, 947–956 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Doherty A. M., O’Shea L. C., Fair T., Bovine DNA methylation imprints are established in an oocyte size-specific manner, which are coordinated with the expression of the DNMT3 family proteins. Biol. Reprod. 86, 67 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Smith L. C., et al., Developmental and epigenetic anomalies in cloned cattle. Reprod. Domest. Anim. 47 (suppl. 4), 107–114 (2012). [DOI] [PubMed] [Google Scholar]
- 52.O’Doherty A. M., et al., DNA methylation dynamics at imprinted genes during bovine pre-implantation embryo development. BMC Dev. Biol. 15, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaneda M., et al., Epigenetic analysis of bovine parthenogenetic embryonic fibroblasts. J. Reprod. Dev. 63, 365–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M., et al., An imprinted long noncoding RNA located between genes Meg8 and Meg9 in the cattle Dlk1-Dio3 domain. Genetica 145, 1–7 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Minkovsky A., Patel S., Plath K., Concise review: Pluripotency and the transcriptional inactivation of the female Mammalian X chromosome. Stem Cells 30, 48–54 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahakyan A., et al., Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20, 87–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva J., et al., Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4, 481–495 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Plath K., et al., Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Yu B., van Tol H. T. A., Stout T. A. E., Roelen B. A. J., Initiation of X Chromosome inactivation during bovine embryo development. Cells 9, 1016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Z., et al., Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genomics 15, 756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brons I. G., et al., Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.