Significance
Clostridium perfringens causes gas gangrene and is a leading cause of bacterial food poisoning, with 1,000,000 US cases annually. Food-poisoning strains produce an enterotoxin (CpE) that breaks apart tight junctions, protein assemblies composed of claudins that fortify the gut barrier. CpE selectively targets claudins via its C-terminal domain (cCpE) to disrupt the gut barrier—the basis for targeting was unknown. Here, we determine the structure of cCpE bound to human claudin-4, discover the structural origin of CpE targeting, and reveal that the primary CpE receptors differ in mice and humans. These insights elucidate CpE’s mechanism for selective targeting of claudins in mammalian gut and can be applied to aid design of new CpE-based therapeutics to treat claudin-linked diseases.
Keywords: claudin, tight junction, membrane protein, X-ray structure, enterotoxin
Abstract
The bacterium Clostridium perfringens causes severe, sometimes lethal gastrointestinal disorders in humans, including enteritis and enterotoxemia. Type F strains produce an enterotoxin (CpE) that causes the third most common foodborne illness in the United States. CpE induces gut breakdown by disrupting barriers at cell–cell contacts called tight junctions (TJs), which are formed and maintained by claudins. Targeted binding of CpE to specific claudins, encoded by its C-terminal domain (cCpE), loosens TJ barriers to trigger molecular leaks between cells. Cytotoxicity results from claudin-bound CpE complexes forming pores in cell membranes. In mammalian tissues, ∼24 claudins govern TJ barriers—but the basis for CpE’s selective targeting of claudins in the gut was undetermined. We report the structure of human claudin-4 in complex with cCpE, which reveals that enterotoxin targets a motif conserved in receptive claudins and how the motif imparts high-affinity CpE binding to these but not other subtypes. The structural basis of CpE targeting is supported by binding affinities, kinetics, and half-lives of claudin–enterotoxin complexes and by the cytotoxic effects of CpE on claudin-expressing cells. By correlating the binding residence times of claudin–CpE complexes we determined to claudin expression patterns in the gut, we uncover that the primary CpE receptors differ in mice and humans due to sequence changes in the target motif. These findings provide the molecular and structural element CpE employs for subtype-specific targeting of claudins during pathogenicity of C. perfringens in the gut and a framework for new strategies to treat CpE-based illnesses in domesticated mammals and humans.
Type F isolates of the pathogenic gram-positive bacterium Clostridium perfringens secrete an enterotoxin (CpE) that afflicts humans and other mammals with very common foodborne and antibiotic-associated forms of gastrointestinal disease, and in some cases severe or fatal enterotoxemia (1, 2). In C. perfringens, sporulation triggers expression and release of CpE into the gastrointestinal tract of its host, where it binds to its cell-surface receptor and induces breakdown of the gastrointestinal barrier and cytotoxicity (1). CpE is 35 kDa in size, has structural homology to β-barrel pore-forming toxins (3), and recognizes and binds its receptors via its C-terminal domain (cCpE) (4, 5). The process of gastrointestinal breakdown in humans initiates with CpE binding to claudins—a 27-member family of plasma membrane proteins that assemble to fortify tight junctions (TJs) in epithelia (Fig. 1A)—and culminates with claudin-bound CpE oligomers that dissociate TJs and form transcellular ion pores that ultimately induce cell death (6).
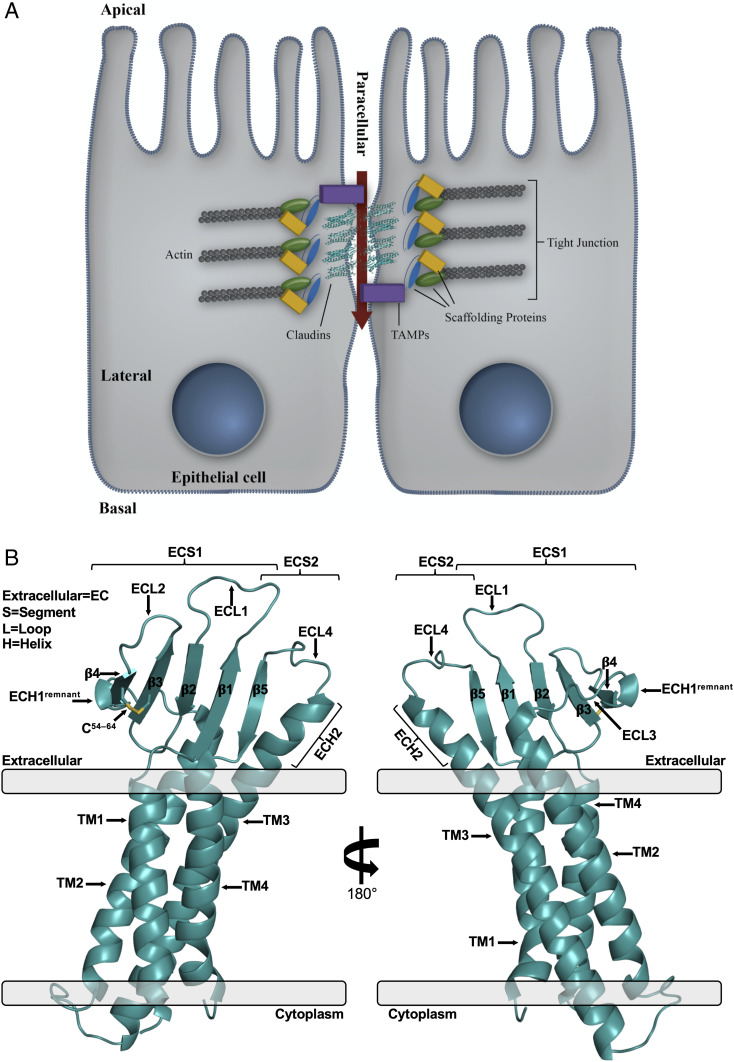
Fig. 1.
Structure of TJs and hCLDN-4. (A) Model epithelial cell–cell contact at a TJ. Claudins are shown in cartoon representation (teal) with accessory proteins called tight junction-associated Marvel proteins (TAMPs). (B) Overall structure of hCLDN-4 (teal, cartoon). Membrane borders (gray, rectangles) and membrane insertion orientation are based on calculations from the PPM server (33).
Human claudins range in size from 23 to 34 kDa and are classified by a conserved WGLWCC motif. The 13 “classic” human claudins have additional homology outside of this motif and share 30 to 71% sequence identity (SI Appendix, Fig. S1) (7). Claudins have conserved structural topologies that consist of four ⍺-helical transmembrane (TM) segments and two extracellular segments (ECSs) that form from a five-stranded antiparallel β-sheet (SI Appendix, Fig. S2A) (8–12). ECS1 contains β-strands 1 to 4 and links TM1 to TM2, while ECS2 contains β5 and links TM3 to TM4. Claudin TM and ECS domains interact laterally in cis (SI Appendix, Fig. S2B) and perpendicularly in trans, forming permselective barriers to ions while simultaneously adhering adjacent cells. Both interaction types serve as the foundation for TJ ultrastructure and function (13, 14).
CpE may disrupt gut integrity through toxin-induced dissociation of claudin cis and trans interactions, leading to breakdown of TJ barrier function, in addition to killing epithelial cells via a claudin-bound ion pore (9, 10, 12). The gastrointestinal-specific expression of a claudin is not requisite for CpE binding, however, as subtypes with no to low gut abundance bind CpE due to claudin sequence and structural conservation (9, 12, 15). Yet, sufficient sequence and structure divergence exists to impart only select subtypes the ability to bind CpE (16, 17). The recalcitrant nature of claudins to in vitro and in vivo biochemical and biophysical study has yielded contradictory findings concerning which subtypes are physiological CpE receptors, and a lack of quantitative data on claudin–CpE interactions obscures the subtle structural differences that must arise in receptor and nonreceptor claudins (16).
Claudin-3 and -4 were the first CpE receptors identified in humans and mice (4, 5). In the gut of these mammals, expression patterns and levels of claudin-3 and -4 vary (15, 18). In humans, CpE incidence in the small intestine causes morphological tissue damage and a reduction in TJ barrier integrity in vitro, while incidence in the large intestine exhibits no effect (19). Human claudin-3 and -4 overabundance in the large but not the small intestine does not coincide with CpE’s requirements for pathogenesis, making their receptor capacities unclear. In mice, claudin-3 and -4 overexpress in the small intestine, making both candidate CpE receptors. Several other claudins overexpress in subdivisions of human and mouse gut (SI Appendix, Fig. S3), rendering it increasingly challenging to categorize individual subtypes as receptors or nonreceptors for CpE using our current understanding.
The structure of human claudin-4 (hCLDN-4) in complex with cCpE allows us to define the features CpE employs for claudin subtype-specific targeting and its action on claudins in cells. Altered in vivo expression patterns of human and mouse claudins, coupled with a lack of biophysical binding data, led us to characterize and quantify CpE’s ability to recognize, bind, and destroy claudin-expressing cells to resolve the structural and functional consequences of CpE targeting. Using these findings, we identify divergence in claudin–CpE binding interactions that explains subtype-specific targeting by CpE and how this process influences cytotoxicity. Our discoveries advance categorization of claudin receptors and nonreceptors for CpE in mammalian gut, and elucidate the molecular mechanism by which CpE prompts targeted dissociation of claudins and the breakdown of TJ barriers.
Results
Structure of hCLDN-4.
The structure of hCLDN-4 in complex with cCpE was resolved at 3.37 Å (SI Appendix, Table S1). The hCLDN-4 portion includes residues 5 to 186 and contains the four–⍺-helix bundle TM domain and five-stranded antiparallel β-sheet that comprise the two ECSs, hallmarks of the claudin fold (Fig. 1B). TM3 extends outside of the membrane forming an extracellular helix (ECH), ECH2, and four extracellular loops (ECLs) connect sequentially structured subdomains. The two ECSs of hCLDN-4 resemble a left “hand” with five “fingers”—ECH2 as the “thumb” and β5, β1, β2, and disulfide-linked β3–β4 following along in order—and are presented as a “cupped hand” to create a large surface for enterotoxin binding (Fig. 1B).
The hCLDN-4 structure lacks a fully formed ECH1 like other claudins bound to cCpE (8–10, 12). ECH1 is a helix in ECL3 involved in claudin cis assembly and is found in the unbound structure of mouse claudin-15 (mCLDN-15) (11, 20, 21). In hCLDN-4, electron density in the ECH1 region appears bulbous and the loop has helical ɸ/ψ values at the N terminus of the SLLALP74 sequence that constitutes it (SI Appendix, Fig. S4A). Structural comparison of hCLDN-4’s ECH1 region with the same area in cCpE-bound hCLDN-9 shows differences despite the two subtypes having identical SLLALP74 sequences, while contrasting these subtypes with mCLDN-15 highlights the effect of cCpE binding (SI Appendix, Fig. S4B). ECH1 is unstructured in both cCpE-bound hCLDN-4 and -9 but helical in mCLDN-15. We performed sequence analysis of the ECH1 region in classic claudins to decipher if homology exists (SI Appendix, Fig. S1). We hypothesized that homology would suggest ECH1’s natural formation in claudins and that its inability to form in hCLDN-4 is a result of cCpE binding. This prediction is required because no unbound structures of hCLDN-4 or -9 are available. Sequence alignments show 33 to 83% residue identity in this range and that the consensus sequence for ECH1 in classic claudins is SLLALP74 (SI Appendix, Fig. S4C). SLLALP74 is present in 6 of 13 classic claudins, including hCLDN-4, indicating ECH1 is likely structured in cCpE’s absence. The SLLALP74 helix is hydrophobic with a predicted aliphatic index of 211.7 and grand average of hydropathicity of 1.8. With its bulbous density and helical nature, the SLLALP74 region of hCLDN-4 may represent a helical remnant of ECH1 after disruption by cCpE. Prompting structural plasticity of ECH1 could be how enterotoxin disables claudin lateral assemblies, as recognized side-chain interactions that enable cis homodimers are abrogated after cCpE binding (SI Appendix, Fig. S2B).
Structure of the hCLDN-4–cCpE Complex.
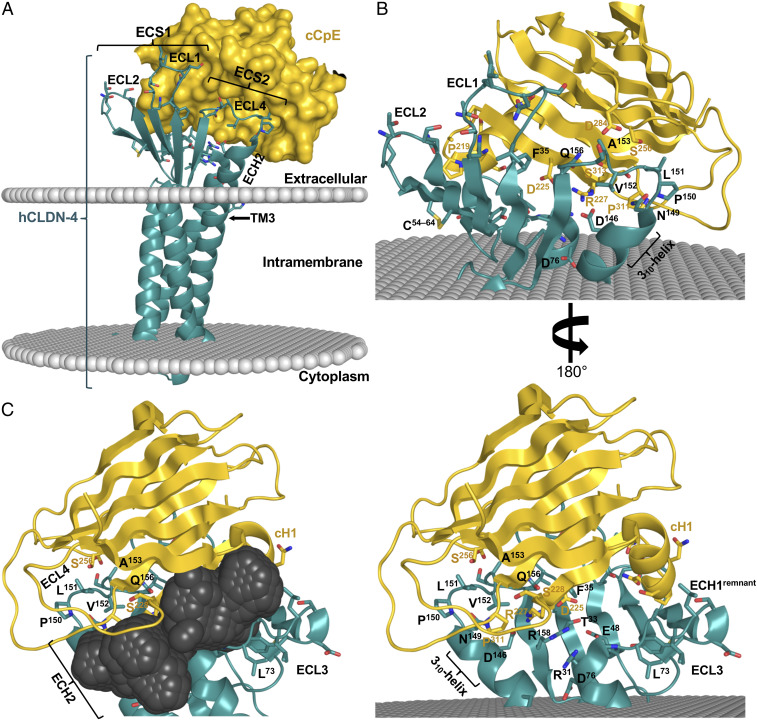
The cCpE binds target claudins while the N-terminal domain oligomerizes to form cytotoxic claudin-bound pore complexes (3, 22). Structure of the hCLDN-4–cCpE complex shows 1:1 stoichiometry in the crystal and packing of a single complex in the asymmetric unit (SI Appendix, Fig. S5). The hCLDN-4–cCpE complex is intricately associated with cCpE contacting all five fingers and resting within the extracellular cupped hand of hCLDN-4 (Fig. 2). ECS2 contains an NPLVA153 motif that penetrates a groove between β-strands of cCpE, while ECS1 contacts cCpE’s surface (Fig. 2 A and B). Most ECS interactions with cCpE occur at the periphery of the claudin fingers, which creates a solvent-accessible pocket at the intermolecular interface near the “palm” (Fig. 2C). The size and shape of this pocket are influenced by unique interactions between hCLDN-4 and cCpE.
Fig. 2.
Structure of hCLDN-4 in complex with cCpE. (A) Overall structure of cCpE (gold, surface) bound to hCLDN-4 (teal, cartoon) with model membrane borders (gray, spheres) calculated by the PPM server (33). (B) Extracellular residues used for interactions between hCLDN-4 (teal, cartoon) and cCpE (gold, cartoon) outside of the membrane (gray, spheres). (C) The solvent-accessible pocket (dark gray, spheres) at the hCLDN-4–cCpE complex interface calculated by MetaPocket 2.0 (34).
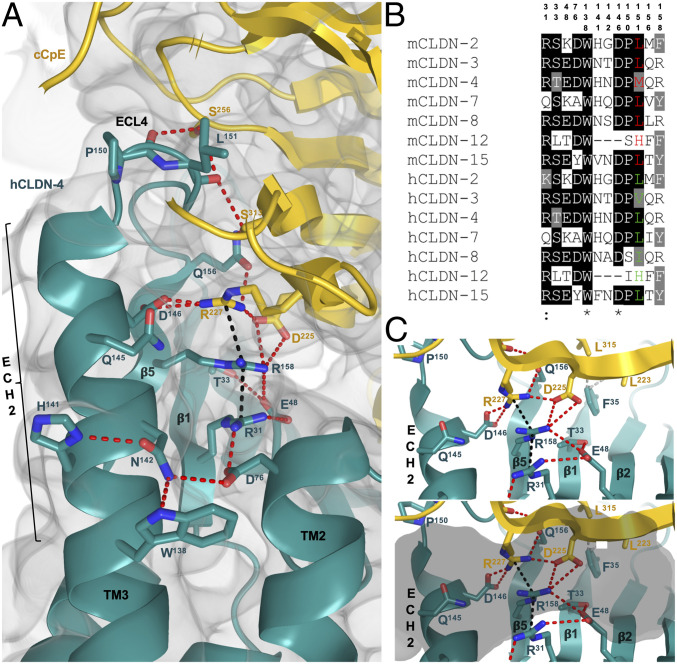
Intermolecular ion pairs and nonpolar and polar interactions coordinate the hCLDN-4–cCpE complex and define the molecular determinants for cCpE targeting of claudins. In the ECS1 and ECS2 of hCLDN-4, nine and eight residues, respectively, establish the primary interactions with cCpE (SI Appendix, Figs. S6 and S7). Structural analysis of the complex reveals an elaborate hydrogen-bonding network in hCLDN-4 that extends from the membrane boundary of TM3 and leads to the NPLVA153 motif (Fig. 3). This network utilizes residues from hCLDN-4 and cCpE, and includes many of the side chains that form the major polar interactions in the complex (SI Appendix, Fig. S7). The hCLDN-4 portion of the network, which we term the “cCpE-binding motif,” contains 12 residues—Arg31, Thr33, Glu48, Asp76, Trp138, His141, Asn142, Asp146, Pro150, Leu151, Gln156, and Arg158. The network starts at Nε1 of hCLDN-4 Trp138 and connects to Asn142, Asp76, Arg31, Glu48, Arg158, Asp225 (cCpE), Arg227 (cCpE), Asp146, Gln156, Ser313 (cCpE), Leu151, and Ser256 (cCpE), and ends at the carbonyl oxygen of Pro150 (Fig. 3A). Sequence alignment of these residues in gut-expressing claudins shows homology in mouse and human subtypes (Fig. 3B). The core of this interaction network is ionically driven and centers at Arg158 of hCLDN-4. Arg158 is sandwiched between hCLDN-4 Arg31 and cCpE Arg227, forming a string of stacked arginines with Cζ–Cζ distances of 3.5 and 3.8 Å, respectively (Fig. 3C). These Cζ distances are in agreement with those from high-resolution crystal structures with arginine stacking (23). Acidic residues Glu48, Asp76, and Asp146 of hCLDN-4 and Asp225 of cCpE surround the three arginines, forming ionic intra- and intermolecular interactions that stabilize the arginine stack conformations (Fig. 3 A and C). The three acidic residues are well-conserved in gut-expressing claudins (Fig. 3B). The ionic core residues inhabit the solvent-accessible pocket at the hCLDN-4–cCpE interface (Figs. 2C and 3C), illustrating that water, disordered in the structure, may enable its formation and maintenance. The hydrogen-bonding network ends at Phe35, which forms nonpolar interactions with two leucines of cCpE. The structure of the hCLDN-4–cCpE complex agrees with other studies showing both ECSs support cCpE binding—but provides salient details that illustrate residues inside and outside of ECS2 are co-opted by cCpE for subtype-specific targeting.
Fig. 3.
cCpE-binding motif of hCLDN-4. (A) Intra- and intermolecular interactions between hCLDN-4 (teal, cartoon) and cCpE (gold, cartoon). Side chains are shown as sticks and colored as carbon (teal/gold), oxygen (red), and nitrogen (blue). Potential hydrogen bonds (red) and arginine stacking (black) are depicted as dashed lines. The 2Fo − Fc electron density map (gray, volume) is overlaid and contoured to 1.0σ. (B) Sequence alignment of the 12-amino acid cCpE-binding motif in gut expressing claudins from mice and humans. Position 3 in the NPLVA153 motif is colored for mouse (red) and human (green) claudins. Sequences were aligned with T-Coffee (35). (C) Stacking of arginine residues around hCLDN-4 Arg158 without (Top) or with (Bottom) an overlaid solvent-accessible pocket (gray, semitransparent surface) from Fig. 2C. Bonds and side chains are shown as in A with addition of nonpolar interactions (gray, dash) around Phe35.
Sequence Analyses of cCpE-Binding Motif Residues.
We investigated whether sequence and structural conservation of this hydrogen-bonding network in claudins could explain selective targeting by cCpE. Alignment of gut-expressing mouse and human claudin sequences shows 8 to 92% identity in the 12-amino acid cCpE-binding motif, indicating divergence in certain subtypes (Fig. 3B). While it was known that not all subtypes express highly in the gut of mice and humans (15, 18), the receptive capacities of many gut claudins were unknown. Alignment analysis of the cCpE-binding motif in the characteristic CpE receptors CLDN-3 and -4 reveals 67 to 92% residue identity in the four mouse and human subtypes—signifying high conservation and receptor potential. In CLDN-3 and -4 the most diversity appears at Leu151, position 3 in the NPLVA153 motif of hCLDN-4, where side-chain hydrophobic lengths vary from valine, leucine, to methionine (Fig. 3B). We further examine the position 3 residue and the NPLVA153 motif using structural and biophysical methods to decipher their roles in enterotoxin binding and subtype-specific targeting.
Affinities and Kinetics of Claudin–Enterotoxin Complexes.
We performed comparative structural analyses between hCLDN-4–cCpE and five other claudin–cCpE complexes comprising three different subtypes (SI Appendix, Results and Discussion and Figs. S8–S11). These analyses show that cCpE poses vary widely when bound to specific subtypes (SI Appendix, Fig. S8). But, these variations are qualitative and difficult to relate to in vivo function. We therefore performed biophysical measurements on mouse and human claudin–enterotoxin complexes using biolayer interferometry (BLI) to quantitate these structural differences and determine a role for the NPLVA153 motif in binding (Table 1 and SI Appendix, Fig. S12). BLI measurements show that 1) hCLDN-4 binds cCpE with an equilibrium dissociation constant (KD) of 2.5 nM, which agrees with a reported KD of 3.4 nM measured by surface plasmon resonance (24); 2) hCLDN-4 has 100-fold higher affinity for cCpE than hCLDN-3; and 3) hCLDN-4 has a 4-fold higher cCpE affinity compared with mCLDN-3—therefore mCLDN-3 has 26-fold higher affinity for cCpE than hCLDN-3. Measurements with CpE demonstrate similar trends, owing to its N terminus playing little role in binding (Table 1). These findings establish enterotoxin binding affinities to previously undetermined mouse and human claudins and provide a quantitative approach to interpret qualitative cCpE binding poses from crystal structures of claudin–cCpE complexes (8–10, 12).
Table 1.
Claudin–enterotoxin binding affinities and kinetics
| Claudin | ECL4 motif | cCpE binding | CpE binding | |||||||
| kon, 1/Ms | koff, 1/s | KD, nM | t1/2, min | kon, 1/Ms | koff, 1/s | KD, nM | t1/2, min | |||
| hCLDN-3 | NPVVP* | 1.25E+04 | 30.74E−04 | 246.40 | 3.76 | 1.12E+04 | 35.90E−04 | 320.13 | 3.22 | |
| ± 0.01E+04 | ± 0.08E−04 | ± 1.48 | ± 0.01E+04 | ± 0.09E−04 | ± 2.16 | |||||
| hCLDN-4 | NPLVA* | 4.16E+04 | 1.02E−04 | 2.46 | 112.97 | 4.26E+04 | 1.52E−04 | 3.56 | 76.13 | |
| ± 0.01E+04 | ± 0.04E−04 | ± 0.11 | ± 0.02E+04 | ± 0.04E−04 | ± 0.17 | |||||
| mCLDN-3 | NPLVP† | 4.01E+04 | 3.77E−04 | 9.40 | 30.67 | 3.44E+04 | 4.34E−04 | 12.62 | 26.59 | |
| ± 0.01E+04 | ± 0.02E−04 | ± 0.57 | ± 0.01E+04 | ± 0.02E−04 | ± 0.77 | |||||
| hCLDN-3V150L/P152A | NPLVA* | 1.08E+04 | 2.30E−04 | 21.39 | 50.20 | 8.42E+03 | 1.84E−04 | 21.84 | 62.84 | |
| ± 0.01E+04 | ± 0.02E−04 | ± 0.13 | ± 0.11E+03 | ± 0.05E−04 | ± 1.04 | |||||
| hCLDN-4L151V/A153P | NPVVP* | 3.45E+04 | 3.82E−04 | 11.07 | 30.25 | 2.80E+04 | 2.09E−04 | 7.47 | 55.22 | |
| ± 0.01E+04 | ± 0.02E−04 | ± 0.07 | ± 0.01E+04 | ± 0.03E−04 | ± 0.11 | |||||
| hCLDN-1 | DPMTP* | 1.63E+04 | 1.53E−02 | 936.20 | 0.75 | 1.36E+04 | 1.84E−02 | 1,354.28 | 0.63 | |
| ± 0.07E+04 | ± 0.02E−02 | ± 52.68 | ± 0.07E+04 | ± 0.03E−02 | ± 95.20 | |||||
Biolayer interferometry was used to measure binding affinities and kinetics of claudin–enterotoxin interactions. Results represent averages from at least duplicate experiments. hCLDN-3 and -4 were measured in quadruplicate.
Position 3 in the NPLVA153 motif is colored for human subtypes.
Position 3 in the NPLVA153 motif is colored for a mouse subtype.
The basis for hCLDN-3–expressing cells exhibiting lower CpE binding capacities compared with hCLDN-4 was unresolved (4, 22). Comparing the affinities of enterotoxin to hCLDN-3 and hCLDN-4 shows stark differences, while similar affinities exist for mCLDN-3 and hCLDN-4. We investigated the root of these variances by assessing sequence alignments of mouse and human claudins that are overabundant in the gut (SI Appendix, Fig. S3). Position 3 of potential receptors varies—hCLDN-3 has NPVVP152, hCLDN-4 has NPLVA153, and mCLDN-3 has NPLVP152. We hypothesized changes to these residues accounted for the contrasting receptor capacities of claudins. Swapping these motifs by mutating two of the five residues in hCLDN-3 and -4 and assessing enterotoxin binding show that 1) an hCLDN-3 V150L/P152A mutant increases cCpE affinity 12-fold compared with wild-type hCLDN-3; and 2) an hCLDN-4 L151V/A153P mutant decreases cCpE affinity 5-fold compared with wild-type hCLDN-4 (Table 1). With further measurements of this motif’s influence on cCpE affinity using hCLDN-1, which has a DPMTP154 sequence and disputed CpE receptor capacity (17, 25–27), we find a KD of 936 nM—381-fold lower than hCLDN-4. As before, BLI measurements show CpE binding to these mutant and wild-type claudins results in comparable trends (Table 1). These results clarify the role of position 3 side-chain length and the NPLVA153 motif in enterotoxin binding and begin to elucidate the origin of varied CpE receptive capacities in claudin-expressing cells.
Using BLI, we established the kinetics of claudin–enterotoxin binding by obtaining the second-order association rate constants (kons), first-order dissociation rate constants (koffs), and half-lives (t1/2s) of the protein complexes (Table 1 and SI Appendix, Fig. S12). The kons for cCpE and claudins show no more than 4-fold differences, while koffs vary up to 150-fold between tested claudins, demonstrating that koff influences KD significantly (Table 1). The koff of cCpE to hCLDN-4 is 30- and 4-fold less than hCLDN-3 and mCLDN-3, respectively, accounting for hCLDN-4’s higher affinity. Mutant hCLDN-3V150L/P152A decreases koff 13-fold compared with wild-type hCLDN-3 while hCLDN-4L151V/A153P increases koff 4-fold compared with wild-type hCLDN-4, indicating that leucine at position 3 of the NPLVA153 motif improves affinity. The koff of cCpE to hCLDN-1 is 150-fold faster than hCLDN-4. The claudin–cCpE complex t1/2 (ln 2/koff) is the time at which half of the initially present complexes have dissociated. Comparing the t1/2 of cCpE with hCLDN-3 and hCLDN-4 indicates that hCLDN-4 maintains a complex with cCpE 30 times longer. mCLDN-3 binds cCpE 8 times longer than hCLDN-3. Mutant hCLDN-3V150L/P152A bound cCpE 13 times longer than wild-type hCLDN-3 while hCLDN-4L151V/A153P bound cCpE 4 times shorter than wild-type hCLDN-4, verifying that a position 3 leucine extends complex half-lives. The t1/2 between cCpE and hCLDN-1 was 45 s. Similar association and dissociation rate constants and t1/2 times were measured for claudin complexes with CpE (Table 1). These results offer a function for the position 3 leucine in enterotoxin binding, and provide a means to distinguish CpE receptors from nonreceptors in a physiological context based on the stark differences in claudin–enterotoxin kinetics.
Peripheral Expression of Claudins Renders Insect Cells Susceptible to CpE Cytotoxicity.
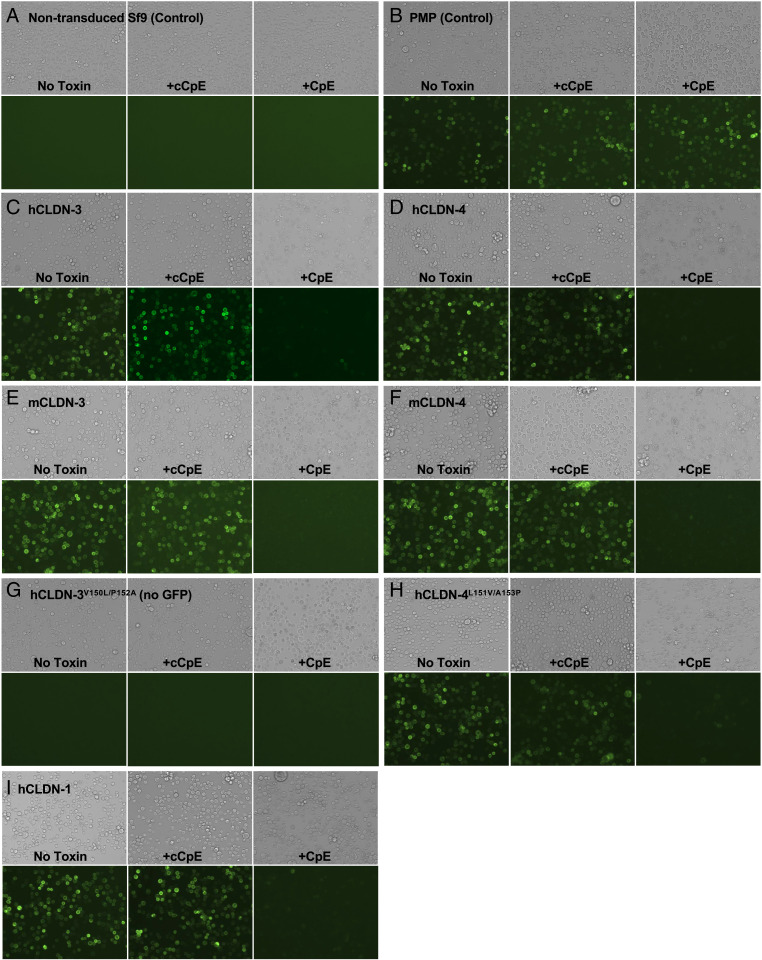
We next investigated the cellular consequences of claudin–enterotoxin complex formation ex vivo, which for CpE-induced cytotoxicity requires CpE binding to claudins and pore assembly on cellular surfaces. Sf9 cells are a model system because they lack TJs and endogenous claudins (28) but form TJ-like strands upon exogenous expression of claudins (11). We transduced Sf9 cells with baculoviruses containing green fluorescent protein (GFP)-tagged claudins from our biophysical studies, and then treated individual wells with no toxin, cCpE, or CpE (Fig. 4). Claudin-expressing cells treated with CpE and not cCpE, which lacks a cytotoxic N-terminal domain, exhibit specific morphological changes. Changes in morphology upon CpE treatment consist of more numerous large swollen and small flattened cells and loss to plasma membrane fluorescence (Fig. 4 C–I). This cellular response is not seen in nontransduced Sf9 cells, or Sf9 cells transduced with a nonclaudin membrane protein (PMP) treated with CpE, demonstrating morphological damage is both CpE- and claudin-dependent (Fig. 4 A and B). The extent of morphological damage is indistinguishable in all claudins tested, indicating claudin localization to the plasma membrane rendered Sf9 cells sensitive to CpE. Moreover, plasma membrane localization of claudins did not change upon treatment with cCpE even after 12 h, departing from results in Madin–Darby canine kidney cells (Fig. 4 C–I) (22). Injury to hCLDN-1–expressing cells treated with CpE was seen, despite 1.4 µM affinity (Fig. 4I). Our findings indicate that morphological damage to claudin-expressing cells after CpE treatment is a result of CpE-induced cytotoxicity via pore assembly on cell surfaces.
Fig. 4.
CpE-induced morphological damage to claudin-expressing cells. Sf9 cells were treated with no toxin (Left), cCpE (Middle), or CpE (Right) for 12 h and then imaged at 20× magnification using bright-field (Top) and GFP fluorescence (Bottom) after 36-h baculovirus transduction encoding (A) no virus (nontransduced) control; (B) PMP-GFP control; (C) hCLDN-3-GFP; (D) hCLDN-4-GFP; (E) mCLDN-3-GFP; (F) mCLDN-4-GFP; (G) hCLDN-3V150L/P152A (no GFP); (H) hCLDN-4L151V/A153P-GFP; and (I) hCLDN-1-GFP.
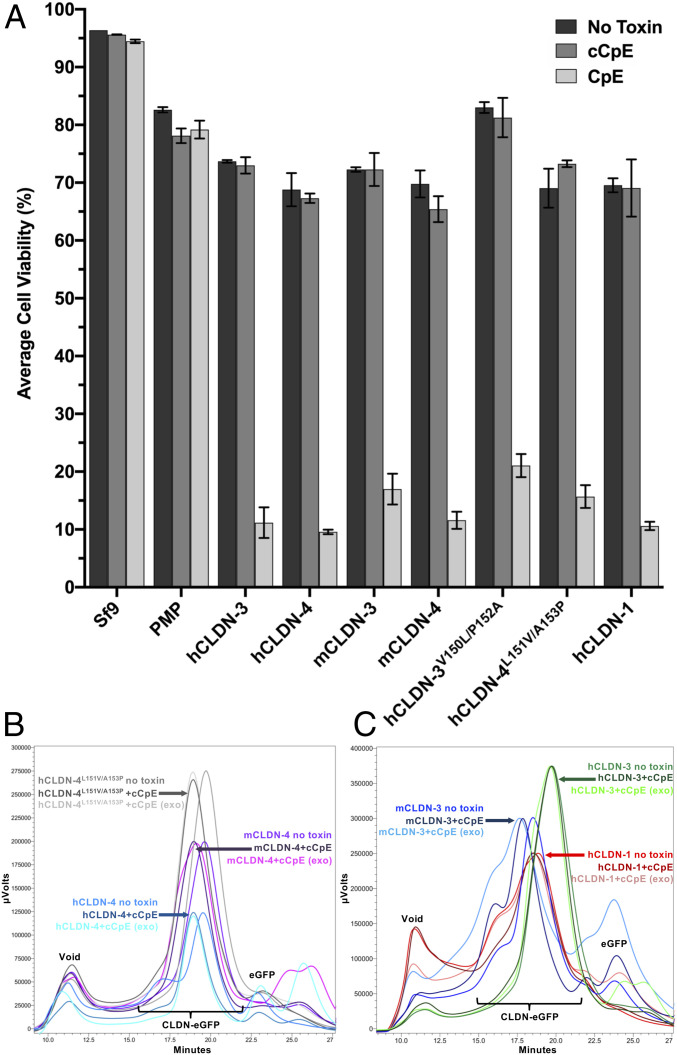
To verify that morphological damage correlates to cell death, we quantified the viability of toxin-treated cells. For cells transduced with baculovirus but not treated with toxin (Fig. 4A), we measure an average 22% decrease in cell viability, which represents normal infectivity after 48-h incubation with high-titer baculoviruses (Fig. 5A). These measurements show that only cells expressing claudins and treated with CpE undergo cell death, verifying that morphological damage signifies dead or dying cells (Fig. 4 C–I). No cell death occurs in nontransduced Sf9 cells, Sf9 cells expressing PMP treated with CpE or cCpE, or claudin-expressing cells treated with cCpE (Fig. 5A). Cytotoxicity therefore requires both CpE and peripherally expressed claudins. CpE decreases viability of claudin-expressing cells an average of 58.5% compared with cells treated with no toxin, and 57.9% compared with cells treated with cCpE. This similar decrease confirms cCpE has no cytotoxic effect. hCLDN-1, which was shown to be CpE-resistant (22, 27) or slightly CpE-sensitive (26) in other cell types, exhibits CpE-induced cell death equal to other claudins (Fig. 5A). Our cell-based cytotoxicity assays verify that CpE does not require accessory claudins or other TJ-associated proteins to form functional pore complexes on cell surfaces, and that peripheral expression of claudins renders insect cells susceptible to CpE cytotoxicity.
Fig. 5.
Cytotoxicity of CpE to claudin-expressing cells and ex vivo extracted claudin–cCpE complexes. (A) Average viability of duplicate counts with SD from Sf9 cells from Fig. 4 treated with no toxin (dark gray), cCpE (gray), or CpE (light gray). (B) FSEC traces of hCLDN-4 (blue), mCLDN-4 (purple), and hCLDN-4L151V/A153P (gray) with and without cCpE added. (C) FSEC traces of hCLDN-3 (green), mCLDN-3 (blue), and hCLDN-1 (red) with and without cCpE added. Peak heights were normalized to the highest grouped value for clarity. Note the peak shift in hCLDN-4, hCLDN-4L151V/A153P, mCLDN-4, and mCLDN-3 with cCpE added both ex vivo and in vitro whereas no shift occurs for hCLDN-1 or -3 under the same conditions.
Claudin–cCpE Complexes Are Intact on Insect Cell Surfaces.
Because BLI measurements reveal subtype-specific kinetic values and plasma membrane localization of claudins does not change upon cCpE treatment, we assessed the degree of cCpE binding to claudins exposed on insect cell surfaces. Cells expressing GFP-tagged claudins from the cytotoxic assay were harvested, and claudins were detergent-solubilized, ultracentrifuged, and then subjected to fluorescence-detection size-exclusion chromatography (FSEC) (29) within 2 h, the approximate t1/2 of the hCLDN-4–cCpE complex (Table 1). Comparison of FSEC traces from claudin-expressing cells not treated with toxin with cells treated with cCpE shows that cells expressing hCLDN-4, mCLDN-3, mCLDN-4, and hCLDN-4L151V/A153P maintain a complex with cCpE as indicated by decreases in peak elution times (Fig. 5 B and C). No change in peak elution times was seen for hCLDN-1 and hCLDN-3, signifying an inability to preserve a complex with cCpE (Fig. 5C). hCLDN-3V150L/P152A was not tested as we have no GFP-fused construct. Next, we tested if shifts in peak elution times from FSEC correspond to cCpE binding to claudins by taking supernatant from cells not treated with toxin, adding exogenous pure cCpE, and running FSEC. FSEC shows that these in vitro formed claudin–cCpE complexes elute at the same time as complexes formed ex vivo and extracted from insect cell membranes (Fig. 5 B and C). This observation verifies that cCpE remains bound to surface-exposed detergent-solubilized claudins, and that this cCpE binding accounts for the nonexogenous FSEC peak shifts. Using FSEC, we confirm that cCpE binds hCLDN-4, mCLDN-3, mCLDN-4, and hCLDN-4L151V/A153P on insect cell surfaces ex vivo and that these complexes are preserved for at least 2 h in vitro, and that cCpE complexes with hCLDN-1 and -3 are not maintained over the same period of time. Attempts to assess whether cytotoxic claudin-bound CpE pores are maintained using FSEC resulted in no fluorescence signal detected due to degradation. These biochemical results coincide with claudin–enterotoxin biophysical measurements and, when taken together with our structural findings, can be used to distinguish CpE receptors from nonreceptors in a physiological context.
Discussion
The current study uncovers salient insights into the structure–function relationship between claudin subtype-specific targeting by CpE and cytotoxic complex formation at mammalian gut TJs. The hCLDN-4 structure reveals a helical remnant in the ECH1 region and sequence alignment suggests ECH1 is present and structured in classic claudins due to the conservation of SLLALP74. It has been proposed that 1) ECH1 exists in other claudins (11); 2) CpE binding to claudins abolishes this motif (8–10, 12); and 3) some ECH1 residues function in cis assembly (12, 20, 21). Our results verify that the binding of cCpE abrogates ECH1 structure and a corresponding loss of side-chain interactions at potential cis interfaces explains how cCpE may disable lateral assembly of claudins at TJs (SI Appendix, Figs. S2B and S4). The binding of cCpE to hCLDN-4 also causes helical perturbations to ECH2, which contains additional residues known to facilitate cis assembly (SI Appendix, Fig. S11) (20). The cCpE-induced disruptions to ECH1 and ECH2 provide complementary mechanisms for cCpE to disable claudin cis assemblies in order to dissociate TJs in the gut.
The structure of the hCLDN-4–cCpE complex demonstrates that both ECSs contribute to cCpE binding and reveals a cCpE-binding motif formed from a hydrogen-bonded network that is employed by cCpE for subtype-specific targeting of claudins. At the core of this network, a three-arginine stack centered at Arg157/158 mediates important hCLDN-4–cCpE polar interactions (Fig. 3). Mutations to this arginine stack affect cCpE binding. Previously, a cCpE R227A mutant decreased cCpE binding to mCLDN-3 and hCLDN-4 in vitro and in vivo while a conservative R157Y mutant in mCLDN-3 that could preserve stacking had no influence (30). Discovery of a cCpE-binding motif connects two previously disparate theories that proposed either the NPLVA153 motif or electrostatic interactions governed cCpE binding (17, 25, 26, 31). Here we show how both classes of interactions comprise the cCpE-binding motif and cooperate in cCpE binding. Importantly, the sequence of the cCpE-binding motif is highly conserved in select gut-expressing claudins and limited in others, making it a notable structural characteristic of CpE receptors (Fig. 3).
Sequence and structure analyses pointed to a role for position 3 and NPLVA153 motif residues in enterotoxin selectivity so we determined its function in the processes of complex affinity and kinetics (Table 1). Measured affinities show a clear hierarchy for subtype binding, with cCpE preferring hCLDN-4 > mCLDN-3 > hCLDN-3 > hCLDN-1. These findings prove that a leucine at position 3 of the NPLVA153 motif—present in hCLDN-4 and mCLDN-3 but not hCLDN-1 or -3—significantly increases the binding affinities (KDs), slows the dissociation rate constants (koffs), and increases the half-lives (t1/2s) of claudin–enterotoxin complexes. The extended hydrophobic length of leucine versus valine more deeply penetrates a surface groove on cCpE, holding it in a different orientation, and provides a structural basis for the higher affinities of leucine-containing subtypes (Fig. 2A and SI Appendix, Fig. S11D). Notably, our results for hCLDN-3 contradict the presumption that it is a high-affinity CpE receptor comparable to hCLDN-4, and offers a molecular basis for the lower CpE binding capacity of hCLDN-3–expressing cells (4, 22). Our finding that a leucine at position 3 of the NPLVA153 motif differentiates high- versus low-affinity CpE binders was verified further when swapping Val150 of hCLDN-3 for Leu151 of hCLDN-4 and vice versa, and observing that a leucine improved enterotoxin affinity for hCLDN-3 while the valine diminished it for hCLDN-4 (Table 1). This experiment also showed, however, that neither mutant achieved a level of affinity of the swapped wild-type claudin. This demonstrates that the NPLVA153 motif alone is not responsible for all processes of enterotoxin binding and that residues outside of it must play accessory yet important roles. This idea is validated by our finding that hCLDN-1, with its divergent DPMTP154 motif, binds enterotoxin and that cells expressing hCLDN-1 are killed by CpE (Fig. 4I and Table 1). We hypothesize that the 12-amino acid cCpE-binding motif, which contains 10 residues outside of the NPLVA153 motif, functions in this accessory capacity by imparting recognition and association (kon) of enterotoxin to claudins through its network of hydrogen bonds. The NPLVA153 motif and position 3 would therefore primarily direct the process of complex preservation via koff, driving KD. Conservation of cCpE-binding motif residues in classic claudins explains how cCpE recognizes and binds previously thought nonreceptors like hCLDN-1 and kills cells expressing it, while the NPLVA153 motif variation explains why we find 3.8- to 380-fold variability in enterotoxin affinities for claudins. The length of time that claudin–CpE complexes are maintained thus becomes a significant factor for whether a subtype has receptive capacity, because these individual complexes are prerequisites for multimeric CpE pore formation (6).
Because hCLDN-1 and -3 have much lower affinities, faster dissociation rate constants, and shorter complex half-lives with enterotoxin compared with hCLDN-4, we suggest neither are ideal receptors for CpE. This would appear to be contradicted by our cell-death assay but confirmed by our biochemical FSEC analyses (Figs. 4 and 5). CpE was shown to induce no or slight damage to hCLDN-1–expressing cells previously (22, 27) whereas, in our assay, cells expressing hCLDN-1 and -3 were fully prone to CpE toxicity and exhibit no changes compared with cells expressing high-affinity receptors (Figs. 4I and 5A). We attribute published differences in claudin receptive capacities to the low KDs of CpE for claudins like hCLDN-1 and -3, where using CpE below 0.4 to 1.4 μM in assays would decrease binding and thus cytotoxicity. Our results indicate that at concentrations of ∼1.0 μM in the gut, CpE could cause cytotoxicity to epithelial cells expressing claudins with poor binding capabilities. However, after extraction from cell culture, we find that hCLDN-1 and -3 fail to maintain a complex with cCpE in vitro (Fig. 5C). This result coincides with affinity and half-life measurements demonstrating that complex lifetimes can distinguish CpE receptors, and validates that hCLDN-1 and -3 are nonreceptors. This in vitro discovery appears true in vivo too, as pathophysiologically relevant CpE concentrations range from 3 to 350 nM, below CpE’s KD for both hCLDN-1 and -3 (31). Our biophysical characterizations provide perspectives for distinguishing claudin receptors of CpE and uncover a role for time in CpE targeting, binding, and cytotoxicity.
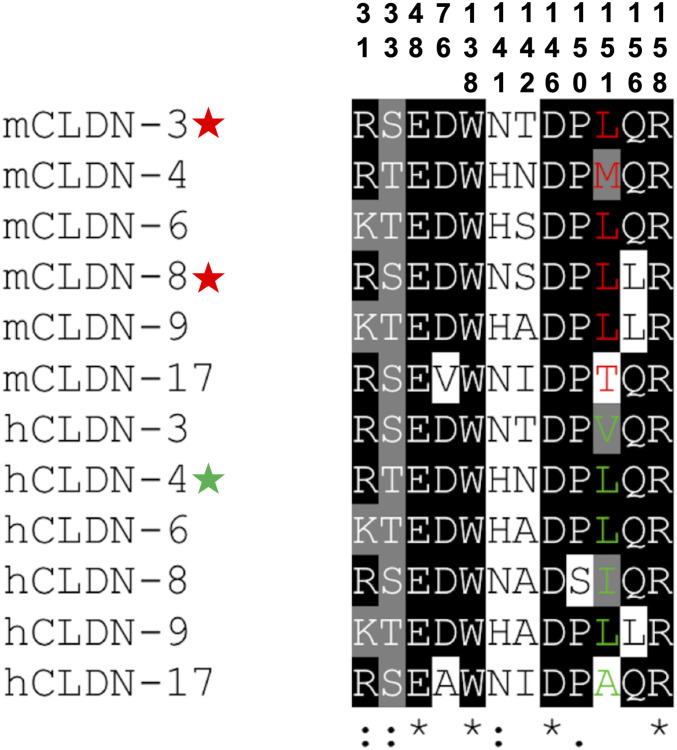
To synergize our findings and identify claudin receptors for CpE, we aligned the cCpE-binding motif of mouse and human subtypes that express highly in the gut but found considerable sequence divergence (Fig. 3B). However, performing the same alignment of classic claudins regardless of tissue overexpression and then eliminating subtypes with low cCpE-binding motif conservation reveal that this motif is predictive of high-affinity CpE receptors (Fig. 6). In CLDN-3, -4, -6, -8, -9, and -17 the motif has 42 to 92% homology. Including this study, CLDN-3–, -4–, -6–, -8–, and -9–expressing cells have been shown to be susceptible to CpE cytotoxicity (12, 17, 26, 27, 31). This verifies that the cCpE-binding motif defines receptors. Our discovery of the cCpE-binding motif in claudins coupled with the critical role of a leucine at position 3 of the NPLVA153 motif in toxin binding allows us to conclude that the high-affinity (<10 nM) CpE receptors are CLDN-3, -6, -8, and -9 in mice and CLDN-4, -6, and -9 in humans. We have shown that CpE binds hCLDN-9 with a KD of 5.1 nM and that hCLDN-9 is homologous to hCLDN-6 previously (12). By correlating these seven high-affinity subtypes to subtype expression patterns in the gut, we uncover that the primary CpE receptors for mice are CLDN-3 and -8 and for humans is CLDN-4 (Figs. 3B and 6). Because CpE demonstrates hierarchical binding preferences for individual claudins, subtypes with slight changes to position 3, like mCLDN-4 (methionine) and hCLDN-8 (isoleucine), may act as secondary receptors. Indeed, we show mCLDN-4 maintains a complex with cCpE for periods approaching hCLDN-4 and that CpE kills cells expressing it (Figs. 4F and 5A and Table 1). hCLDN-8 has been shown previously to impart CpE-mediated death to cells at concentrations below 300 nM, albeit at lower cytotoxic efficiency than hCLDN-4 (31). These findings explain the varied receptive capacities of claudin subtypes for CpE and can be applied to aid design of new cCpE- and CpE-based therapeutics to treat claudin-specific diseases in a variety of mammalian cells and tissues.
Fig. 6.
Prediction of mammalian claudin receptors for CpE using the cCpE-binding motif. Sequence alignments from T-Coffee (35) of the 12-amino acid cCpE-binding motif from classic claudins. Position 3 in the NPLVA153 motif is shown for mouse (red) and human (green) subtypes. Note the higher conservation of the motif in these claudins compared with a similar alignment in gut expressing claudins (Fig. 3B). All 12 claudins likely bind CpE, and if expressed on cell surfaces would be killed by CpE. But only subtypes with a leucine at position 3 of the NPLVA153 motif and expressed highly in gut would constitute the CpE receptors in mice (red, star) and humans (green, star).
Summary
As the major structural elements of TJs, claudins simultaneously direct lateral cell adhesion and paracellular transport of molecules between epithelial sheets. CpE binding to claudins dissociates TJs and disrupts these dual functionalities, altering gastrointestinal homeostasis. The mechanism of claudin-specific targeting by CpE was unresolved. Deciphering this process is vital to remedy CpE-induced foodborne disease and for structure-guided design of cCpE-based therapeutics aimed at modulating TJ barriers. This work elucidates the structural basis for CpE subtype-specific targeting of claudins by uncovering a toxin-binding motif that CpE employs to recognize, bind, and maintain a complex with claudins. Although CpE recognizes and binds claudins broadly, only select subtypes bind with sufficient affinity to preserve a complex with CpE long enough to impart cytotoxicity. These subtypes possess high homology in toxin-binding motif residues, are experimentally verified high-affinity CpE binders, and are thus probable receptors for CpE. Using our characterization of these subtypes in conjunction with claudin expression levels in the gut, we provide evidence that the primary CpE receptors—CLDN-3 in mice and CLDN-4 in humans—have diverged. The CpE concentrations known to induce pathophysiological damage to gut agree with our structural- and biophysical-based classification of receptors. During various physiological processes, remodeling of TJs in gut or its subtissues may display TJs composed of different subtypes in the apical and subapical layers (32). For CpE to be effective in these scenarios for targeted disruption of TJs, it would exploit the conserved toxin-binding motifs of multiple subtypes with ranges of receptive capacities. CpE has likely coevolved with this select group of classic claudins in mammals as a way to combat their dynamic and heterogeneous reintegration into TJs. Our study provides support that 1) CpE exploits subtype-specific features of claudins but also generally recognizes the conserved claudin fold; 2) hierarchical binding preferences allow some subtypes at low concentrations (<10 nM) and others at higher concentrations (>300 nM) to produce gastrointestinal disease; and 3) unaltered cCpE- or CpE-based therapeutics aimed to target specific claudins and pathologies will likely lack intended specificity. These molecules may trigger unintended side effects in normal tissues, so thorough and off-target analyses are warranted in efforts to design and test them. Further elucidation of the claudin-bound and pore-forming processes that occur during CpE cytotoxicity of epithelial tissues will help inform these endeavors.
Materials and Methods
Protein Expression and Purification.
Plasmid pFastBac1 encoding wild-type and mutant claudins, cCpE, CpE, and PMP was expressed in insect cells. For claudins, membranes were solubilized in 1% (weight/volume) n-undecyl-β-d-maltopyranoside (UDM). All proteins were purified via immobilized metal-affinity chromatography (IMAC) with NiNTA and resin was captured, washed, and treated with thrombin. Cleaved proteins were eluted and then used for binding, cytotoxicity, and crystallization assays. IMAC-purified hCLDN-4 and cCpE were mixed, incubated at 4 °C, concentrated, and loaded onto a SEC column. Peak fractions from the complex were pooled, concentrated to 15 mg/mL, and used for crystallization. More detailed procedures are described in SI Appendix, Methods.
Crystallization and Structure Determination.
Crystals grew from a mother liquor containing 100 mM dl-malic acid, 2-(N-morpholino)ethanesulfonic acid, Tris base (1:2:2, pH 6.0), and 25% polyethylene glycol 1500. Crystals were cryoprotected with oil and then flash-frozen in liquid nitrogen. Diffraction data from a single crystal were collected at Advanced Light Source beamline 8.3.1 and processed in space group P212121, and the structure was determined by molecular replacement using a poly-Ala model of hCLDN-9 bound to cCpE (12) as search model. SI Appendix, Table S1 shows data and refinement statistics with further details given in SI Appendix, Methods.
Claudin–Enterotoxin Affinity and Kinetics Using BLI.
IMAC-pure CpE- and cCpE-His10 were immobilized on NiNTA or anti-His (HIS2) biosensors and BLI was performed using an Octet RED384 System (FortéBIO). Kinetics experiments included a wash, toxin loading, baseline, and association and dissociation steps. During association, toxins were incubated with 0 to 250 or 0 to 2,000 nM free claudins depending on claudin–toxin affinity. Data from at least duplicate trials were analyzed, modeled, and fit. More detailed procedures are described in SI Appendix, Methods.
Cytotoxicity and Microscopy.
Baculoviruses containing GFP-tagged or untagged claudins and PMP were added to adherent Sf9 cells in a 24-well plate and then placed at 27 °C. After 36 h, no toxin (untreated) or IMAC-pure cCpE and CpE were each added to the medium of two individual wells and placed back at 27 °C for 12 h. Morphological damage of Sf9 cells was assessed with a combination of light and fluorescence microscopy. Postimaging, cells were gently removed from the wells and stained with trypan blue. Cell viability (total live divided by total cells) was quantified in duplicate manually using a hemacytometer. Further detail is provided in SI Appendix, Methods.
Postinfection Binding Assay.
Untreated and cCpE-treated Sf9 cells expressing GFP-tagged claudins from Cytotoxicity Assay were harvested, pelleted, and resuspended in 1% UDM-supplemented buffer. Proteins were solubilized for 1 h and ultracentrifuged, and then the supernatant was subjected to FSEC (29). Control samples for verifying complex formation came from untreated cells expressing GFP-claudins that were then incubated with IMAC-pure cCpE (exogenous) and monitored via FSEC. SI Appendix, Methods describes the assay in greater detail.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants R35GM138368 (to A.J.V.) and R01GM024485 (to R.M.S.). A.J.V. also acknowledges funding provided by the Nebraska Center for Integrated Biomolecular Communication (NCIBC) and for access to the NCIBC Systems Biology Microscopy Core (NIH National Institutes of General Medical Sciences Grant P20 GM113126). Beamline 8.3.1 at the Advanced Light Source was supported by the University of California Office of the President, Multi-Campus Research Programs and Initiatives Grant MR-15-338599 and by the Program for Breakthrough Biomedical Research and is partially funded by the Sandler Foundation. The Advanced Light Source (Berkeley, CA) is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under Contract DE-AC02-05CH11231, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by the US Department of Energy Office of Biological and Environmental Research.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024651118/-/DCSupplemental.
Data Availability
The atomic coordinates and structure factors reported in this article have been deposited in the Protein Data Bank (PDB ID code 7KP4). All study data are included in the article and/or SI Appendix.
References
- 1.McClane B. A., The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon 39, 1781–1791 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Rood J. I., et al., Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 53, 5–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitadokoro K., et al., Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J. Biol. Chem. 286, 19549–19555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katahira J., et al., Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J. Biol. Chem. 272, 26652–26658 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Katahira J., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N., Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J. Cell Biol. 136, 1239–1247 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell L. A., Koval M., Specificity of interaction between Clostridium perfringens enterotoxin and claudin-family tight junction proteins. Toxins (Basel) 2, 1595–1611 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lal-Nag M., Morin P. J., The claudins. Genome Biol. 10, 235 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura S., et al., Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat. Commun. 10, 816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitoh Y., et al., Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347, 775–778 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Shinoda T., et al., Structural basis for disruption of claudin assembly in tight junctions by an enterotoxin. Sci. Rep. 6, 33632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H., et al., Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344, 304–307 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Vecchio A. J., Stroud R. M., Claudin-9 structures reveal mechanism for toxin-induced gut barrier breakdown. Proc. Natl. Acad. Sci. U.S.A. 116, 17817–17824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krause G., et al., Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Van Itallie C. M., Anderson J. M., Claudin interactions in and out of the tight junction. Tissue Barriers 1, e25247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes J. L., Van Itallie C. M., Rasmussen J. E., Anderson J. M., Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr. Patterns 6, 581–588 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Veshnyakova A., et al., On the interaction of Clostridium perfringens enterotoxin with claudins. Toxins (Basel) 2, 1336–1356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winkler L., et al., Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 284, 18863–18872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lameris A. L., et al., Expression profiling of claudins in the human gastrointestinal tract in health and during inflammatory bowel disease. Scand. J. Gastroenterol. 48, 58–69 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Fernández Miyakawa M. E., Pistone Creydt V., Uzal F. A., McClane B. A., Ibarra C., Clostridium perfringens enterotoxin damages the human intestine in vitro. Infect. Immun. 73, 8407–8410 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J., et al., Multiple claudin-claudin cis interfaces are required for tight junction strand formation and inherent flexibility. Commun. Biol. 1, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hempel C., et al., Assembly of tight junction strands: Claudin-10b and claudin-3 form homo-tetrameric building blocks that polymerise in a channel-independent manner. J. Mol. Biol. 432, 2405–2427 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Sonoda N., et al., Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 147, 195–204 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neves M. A., Yeager M., Abagyan R., Unusual arginine formations in protein function and assembly: Rings, strings, and stacks. J. Phys. Chem. B 116, 7006–7013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinoda T., et al., Cell-free methods to produce structurally intact mammalian membrane proteins. Sci. Rep. 6, 30442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita K., et al., Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 476, 258–261 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Kimura J., et al., Clostridium perfringens enterotoxin interacts with claudins via electrostatic attraction. J. Biol. Chem. 285, 401–408 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson S. L., Smedley J. G. III, McClane B. A., Identification of a claudin-4 residue important for mediating the host cell binding and action of Clostridium perfringens enterotoxin. Infect. Immun. 78, 505–517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitic L. L., Unger V. M., Anderson J. M., Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci. 12, 218–227 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawate T., Gouaux E., Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Veshnyakova A., et al., Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 protein suggests structural modifications of the toxin to target specific claudins. J. Biol. Chem. 287, 1698–1708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrestha A., McClane B. A., Human claudin-8 and -14 are receptors capable of conveying the cytotoxic effects of Clostridium perfringens enterotoxin. MBio 4, e00594-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Hernandez V., Quiros M., Nusrat A., Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann. N. Y. Acad. Sci. 1397, 66–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B., MetaPocket: A meta approach to improve protein ligand binding site prediction. OMICS 13, 325–330 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Notredame C., Higgins D. G., Heringa J., T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors reported in this article have been deposited in the Protein Data Bank (PDB ID code 7KP4). All study data are included in the article and/or SI Appendix.