Significance
Mendelian susceptibility to mycobacterial disease (MSMD) is defined by selective vulnerability to weakly virulent mycobacteria. The 32 known inborn errors of IFN-γ immunity account for MSMD in about half of the patients, and for a smaller proportion of cases of tuberculosis (TB). We report homozygous ZNFX1 variants in two families in which the index cases had MSMD or TB with intermittent monocytosis. Upon overexpression, the two variants encode truncated proteins. We show that human ZNFX1 is localized in ribonucleoprotein granules called stress granules. The patients’ production of and response to IFN-γ are apparently intact, and the patients have not experienced severe viral illnesses. Inherited deficiency of stress granule-associated ZNFX1 is a genetic etiology of MSMD or TB with intermittent monocytosis.
Keywords: mycobacteria, monocytosis, inflammation, inborn error of immunity, ZNFX1
Abstract
Human inborn errors of IFN-γ underlie mycobacterial disease, due to insufficient IFN-γ production by lymphoid cells, impaired myeloid cell responses to this cytokine, or both. We report four patients from two unrelated kindreds with intermittent monocytosis and mycobacterial disease, including bacillus Calmette–Guérin-osis and disseminated tuberculosis, and without any known inborn error of IFN-γ. The patients are homozygous for ZNFX1 variants (p.S959* and p.E1606Rfs*10) predicted to be loss of function (pLOF). There are no subjects homozygous for pLOF variants in public databases. ZNFX1 is a conserved and broadly expressed helicase, but its biology remains largely unknown. It is thought to act as a viral double-stranded RNA sensor in mice, but these patients do not suffer from severe viral illnesses. We analyze its subcellular localization upon overexpression in A549 and HeLa cell lines and upon stimulation of THP1 and fibroblastic cell lines. We find that this cytoplasmic protein can be recruited to or even induce stress granules. The endogenous ZNFX1 protein in cell lines of the patient homozygous for the p.E1606Rfs*10 variant is truncated, whereas ZNFX1 expression is abolished in cell lines from the patients with the p.S959* variant. Lymphocyte subsets are present at normal frequencies in these patients and produce IFN-γ normally. The hematopoietic and nonhematopoietic cells of the patients tested respond normally to IFN-γ. Our results indicate that human ZNFX1 is associated with stress granules and essential for both monocyte homeostasis and protective immunity to mycobacteria.
Mycobacteria cause three major endemic illnesses—tuberculosis (TB), leprosy, and Buruli ulcer—and rarer illnesses caused by less virulent live bacillus Calmette−Guérin vaccines or “atypical” environmental mycobacteria (EM). Rare patients with a selective predisposition to clinical disease caused by bacillus Calmette–Guérin vaccine and EM have Mendelian susceptibility to mycobacterial disease (MSMD) (1, 2). These patients are also prone to disease caused by the more virulent Mycobacterium tuberculosis, and intramacrophagic pathogens, such as Salmonella species. Their mycobacterial infections range from acute to recurrent or chronic, with localized to regional or disseminated clinical manifestations. Germline mutations of 17 genes (CYBB, IFNG, IFNGR1, IFNGR2, IL12B, IL12RB1, IL12RB2, IL23R, IRF8, ISG15, JAK1, NEMO, RORC, SPPL2A, STAT1, TBX21, and TYK2) have been reported, defining 32 genetic etiologies due to the allelic heterogeneity at multiple loci (1, 3, 4). The products of these genes are involved in the production of, or response to, interferon-γ (IFN-γ), which is therefore more a macrophage-activating factor than an anti-viral IFN (5). Most genetic etiologies underlie “isolated MSMD,” with few clinical features other than mycobacterial and related infections. Fewer etiologies underlie “syndromic MSMD,” in which additional pathways other than that governed by IFN-γ (e.g., IFN-α/β, IL-17A/F) are also disrupted, resulting in one or a few additional clinical phenotypes, infectious or otherwise (e.g., viral or fungal infections) (6–10). Outside of the “syndromic MSMD” umbrella, which has somewhat arbitrary boundaries, other inborn errors of immunity (IEI) can underlie mycobacterial disease with a broader range of phenotypes, such as MonoMAC due to inherited GATA2 deficiency, which is invariably associated with monocytopenia and a lack of dendritic cells (DCs) (11–13). The study of MSMD led to the discovery of both the first rare and common monogenic etiologies of TB (14–17). No genetic explanation has yet been found for about half of the cases of MSMD. In this context, we were intrigued by the discovery of two unrelated patients with MSMD or TB displaying intermittent monocytosis.
Results
Patients from Two Unrelated Kindreds Are Homozygous for ZNFX1 Variants.
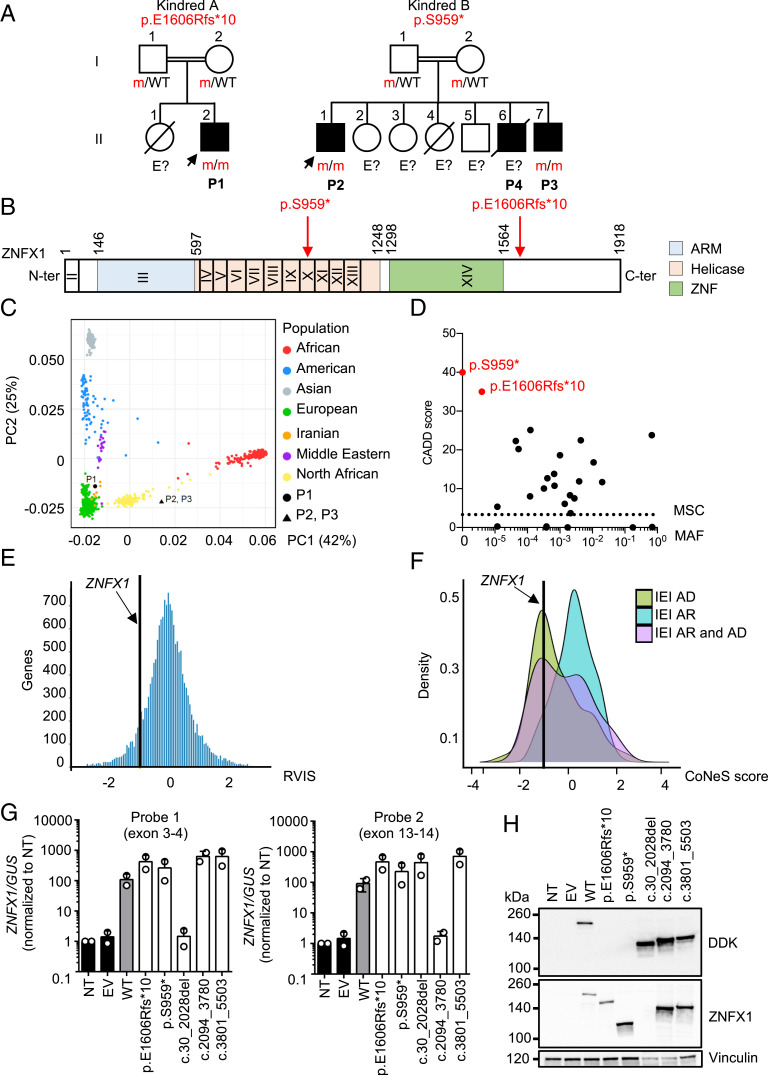
We studied the exomes of two index cases (P1 and P2)—one with MSMD (P1, kindred A, II.2) and the other with severe TB (P2, kindred B, II.1)—and a sibling of P2 (P3, kindred B, II.7), who also displayed the distinctive feature of intermittent monocytosis (P1, P2, and P3), which has never before been reported in any of the patients with known genetic etiologies of MSMD or TB (Fig. 1A and SI Appendix, Fig. S1). The patients’ clinical features are detailed in SI Appendix (case reports, SI Appendix, Table S1). Two of the patients (P2 and P3) also had unexplained episodes of organomegaly, fever, and thrombocytopenia. However, they were never hospitalized for severe viral disease despite seropositivity for multiple viruses, attesting to previous infections, as detected by phage immunoprecipitation sequencing (PhIP-Seq) (SI Appendix, Fig. S2). P1 is from Iran (kindred A) and is 13 y old, whereas P2 and P3 originate from Morocco and are living in Belgium (kindred B) and are 22 and 11 y old, respectively. The two families were unrelated to each other, but each family was consanguineous. We therefore searched for autosomal recessive (AR) genetic etiologies by testing a hypothesis of genetic homogeneity (i.e., the three patients being homozygous for variants of the same gene), using whole-exome sequencing (WES) data for the three patients. We considered only nonsynonymous coding variants that were rare in public databases (minor allele frequency, MAF < 0.01) and absent from our in-house cohort of > 10,000 patients with various infectious diseases. This approach identified ZNFX1 (zinc finger NFX1-type containing 1, NM_021035.2, GRCh37) as the only candidate gene in the three patients (P1, P2, and P3) (Fig. 1B and SI Appendix, Fig. S3A). No other genes, including known IEI and MSMD/TB genes (1, 3, 18, 19), presented homozygous rare nonsynonymous coding variants (single-nucleotide variant or copy number variant) common to the three patients. A brother of P2 and P3 (kindred B, II.6, P4) died at 14 mo of age from M. tuberculosis meningo-encephalitis, suggesting that he shared this recessive phenotype. He was not genotyped, because no material was available. The ancestries of these patients were confirmed by principal component analysis (PCA) (Fig. 1C). Their high homozygosity rates (4.9%, 2.2%, and 1.7%), as also shown by WES (20), confirmed parental consanguinity (SI Appendix, Fig. S3A). P1 carried a homozygous small deletion of four nucleotides (c.4815_4818del) in ZNFX1, which was predicted to create a premature stop codon 10 amino acids (aa) downstream (p.E1606Rfs*10) (Fig. 1B). P2 and P3 carried a homozygous single-nucleotide substitution (c.2876C > G) resulting in a premature stop codon (p.S959*) (Fig. 1B).
Fig. 1.
AR ZNFX1 deficiency in four patients from two unrelated kindreds. (A) Pedigrees of the two unrelated kindreds. Generations are indicated by Roman numerals (I and II), and each individual is indicated by an Arabic numeral (1 to 7). Affected patients are represented by closed black symbols, and index cases are indicated by an arrow. “E?” indicates individuals with an unknown genotype. Symbols crossed with a black diagonal line correspond to deceased individuals. (B) Schematic representation of the ZNFX1 gene. Coding exons are indicated by Roman numerals (II to XIV). The ZNFX1 protein is represented, with three predicted domains: the ARM, helicase, and ZNF domains. The reported mutations are indicated in red with an arrow. (C) PCA showing the origins of P1, P2, and P3 plotted on main ethnic origins extracted from the 1000 Genomes database and our own WES database. (D) MAF and CADD score of the homozygous ZNFX1 variants found in P1 to P3 (red symbols) and of all homozygous variants in gnomAD v.2.1.1 and BRAVO/TOPmed (black symbols). The dotted line corresponds to the MSC, with its 99% CI. The MSC for ZNFX1 is 3.3. (E) According to its RVIS, ZNFX1 is highly intolerant of genetic variation. (F) CoNeS for ZNFX1 and its distribution for genes causing IEI, according to disease mode of inheritance (AD). (G) RT-qPCR on cDNA from HEK293T cells nontransfected (NT) or transfected with an empty plasmid (EV), WT-ZNXF1, mutated-ZNXF1 (E1606Rfs*10 and S959*), or constructs with in-frame deletions of each ZNFX1 domain, for the ARM (c.30_2028del), helicase (c.2094_3780del), and ZNF (c.3801_5503del) domains, with a probe spanning the junction between exons 3 and 4 (probe 1, Left) or exons 13 and 14 (probe 2, Right). GUSB was used for normalization. Values are expressed as means ± SEM. (H) Western blot of total protein extracts from HEK293T cells either left NT or transfected with EV, WT, or mutated ZNXF1, or constructs with in-frame deletions of the ARM (c.30_2028del), helicase (c.2094_3780del), or ZNF (c.3801_5503del) domain, all inserted into pCMV6 with a C-terminal Flag-tag. ZNXF1 was detected with an mAb directed against the N terminus of ZNXF1 or an antibody directed against the C-terminal DDK (Flag) tag. An antibody against vinculin was used as a loading control. The results shown are representative of two independent experiments.
The Two ZNFX1 Variants Are Very Rare and Predicted to Be Damaging.
The c.4815_4818del variant (p.E1606Rfs*10) is exceedingly rare, and has been reported only once before, in the heterozygous state, in South Asia (allele count n = 1 in the Genome Aggregation Database [gnomAD] v2.1.1 database; rs775817125; MAF = 3.98 × 10−6). The p.S959* variant is not reported in public databases (gnomAD v2.1.1, BRowse All Variants Online [BRAVO]/Trans-Omics Precision Medicine [TOPMed]) or our in-house database consisting of >10,000 exomes, including >1,000 exomes from patients from northern Africa or the Middle East (Fig. 1D) (21). Moreover, both variants were predicted to be deleterious, with combined annotation-dependent depletion (CADD) scores of 35 and 40, respectively, well above the corresponding mutation significance cutoff (MSC) for ZNFX1 (99% CI) (22) (SI Appendix, Fig. S3B). The p.E1606Rfs*10 variant is located downstream from the ZNF domain, whereas the p.S959* variant is located within the helicase domain (Fig. 1B). Finally, Sanger sequencing of ZNFX1 from genomic DNA extracted from whole blood (P1, P2, and P3), simian virus 40 (SV40)-immortalized fibroblasts (P1 and P2), and Epstein−Barr virus-immortalized B lymphocytes (EBV-B cells) (P1) confirmed the patients’ homozygosity for the corresponding variant (Fig. 1A and SI Appendix, Fig. S3C). The genotypes of the other individuals are consistent with an AR pattern of inheritance, with incomplete penetrance for MSMD or TB but complete penetrance for monocytosis. Overall, these findings suggest that homozygosity for very rare or private ZNFX1 variants predicted to be loss of function (pLOF) underlies monocytosis and/or mycobacterial diseases in the four patients.
The ZNFX1 Gene Is under Strong Negative Selection.
ZNFX1 is a highly conserved helicase protein from the Up-frameshift 1 (UPF1)-like superfamily 2 (SF2) (23). Very little is known about its biological function. It plays a crucial role in RNA interference-mediated gene silencing in Caenorhabditis elegans (24, 25). In mouse and human cell lines, ZNFX1 has been reported to act as a cytosolic double-stranded RNA (dsRNA) sensor with antiviral activity through interaction with mitochondrial antiviral signaling protein (26). There are no homozygotes for pLOF variants in any public database (BRAVO/gnomAD/TOPmed), and there are no other homozygotes for any nonsynonymous rare variants of this gene in our in-house database of >10,000 patients with diverse severe infectious diseases, further suggesting that the variants carried by the patients are disease causing. The cumulative frequency of heterozygous pLOF variants in public databases is no higher than 1.70 × 10−4 after filtering out variants with low coverage (covered in 50% of individuals) and noncanonical transcripts. There is no major ethnic group in which pLOF variants are less rare. The ZNFX1 gene has a residual variation intolerance score (RVIS) of −0.96 (placing it in the top 14% of human genes most intolerant to genetic variation), a gene damage index of 5.5 (top 29%), and a consensus negative selection (CoNeS) score of −1.31 (top 11%), suggesting that ZNFX1 is a target of strong negative selection (Fig. 1 E and F) (20, 27, 28). Comparisons with known genes for which variants have been shown to underlie IEI showed that ZNFX1 falls at the intersection of the groups of genes underlying AR and autosomal dominant (AD) disorders (20). Overall, these findings suggested that homozygosity for pLOF variants of the negatively selected ZNFX1 gene would be exceedingly rare in the general population and that the very rare ZNFX1 pLOF variants observed in these two kindreds would be deleterious, causing monocytosis and/or mycobacterial disease when present in homozygosity, in the patients.
Overexpression of the E1606Rfs*10 and S959* ZNFX1 Mutants Leads to the Production of Truncated Proteins.
The canonical ZNFX1 messenger RNA (mRNA) (NM_021035.2, GRCh37) consists of 14 exons. It encodes a protein of 1,918 aa with a predicted molecular weight (MW) of 220 kDa. We studied the production of the mutant ZNFX1 proteins by plasmid-mediated overexpression of the corresponding complementary DNAs (cDNAs) in human embryonic kidney (HEK) 293T cells. We generated C-terminal DDK-tagged cDNA constructs for the expression of the two variants, p.E1606Rfs*10 and p.S959*. In transient transfection experiments, we evaluated the impact of the two ZNFX1 variants relative to the wild-type (WT) cDNA. Transfection with p.E1606Rfs*10 and p.S959* resulted in the production of abundant ZNFX1 mRNA, as shown by RT-qPCR with a probe binding the region spanning the junction between exons 3 and 4 (Fig. 1 G, Left) or exons 13 and 14 (Fig. 1 G, Right). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (WB) of cell extracts, followed by immunodetection with a specific monoclonal antibody (mAb) against the N-terminal region of anti-ZNFX1, showed that the WT proteins were produced at the expected MW of about 220 kDa. By contrast, overexpression of the p.E1606Rfs*10 (predicted MW of ∼179 kDa) and p.S959* (∼106 kDa) alleles resulted in truncated proteins (Fig. 1H). Using a mAb directed against a C-terminal DDK tag, we were unable to detect the p.E1606Rfs*10 and p.S959* mutant proteins on WB, implying that there was no reinitiation of translation. Analyses of the nuclear and cytosolic protein fractions showed that the WT protein and mutant proteins lacking one of the three domains (Armadillo [ARM], Helicase, and ZNF) of the ZNFX1 protein were located in the cytoplasm (SI Appendix, Fig. S3D). We also tested 25 nonsynonymous variants of this gene present in the homozygous state in public databases (gnomAD, n = 23 and BRAVO/TOPmed, n = 2). All were missense, and all presented normal protein expression on WB (SI Appendix, Fig. S3E). These results suggest that the patients’ nonsense and ultrarare private and frameshift variants encode truncated proteins, whereas all the nonsynonymous variants found in the homozygous state in the general population are normally expressed. These findings show that the patients’ ZNFX1 variants strongly alter the nature of the ZNFX1 proteins produced, consistent with the notion that they are deleterious and pathogenic.
ZNFX1 Colocalizes with Cytoplasmic Ribonucleoprotein Stress Granules.
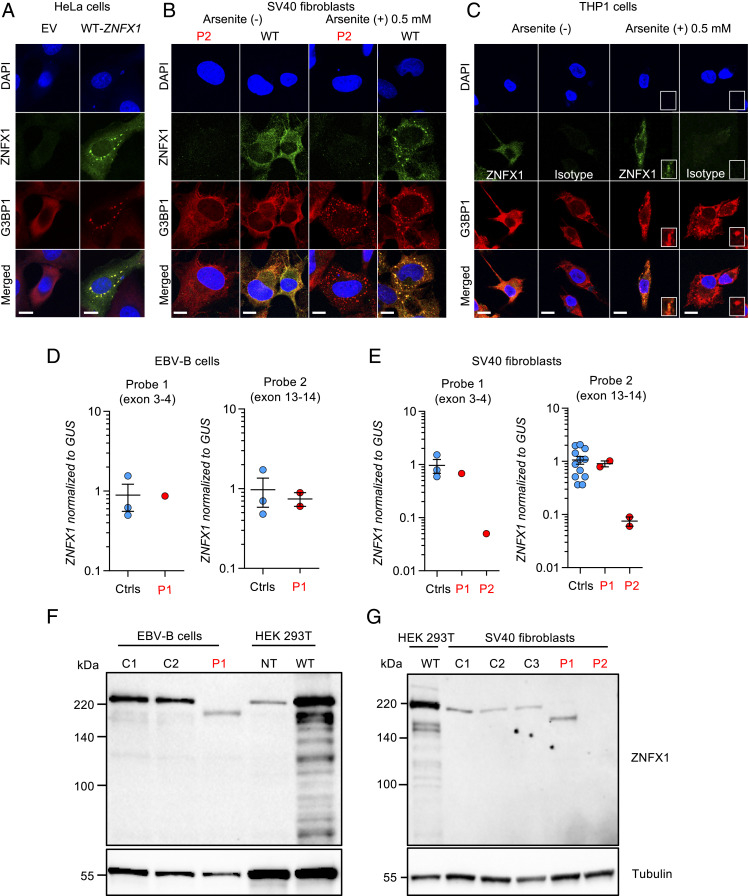
Human ZNFX1 mRNA is broadly detected in adult tissues, but predominantly in hematopoietic cells (Human Protein Atlas, GTEx, and BioGPS) (29). The pattern of expression of ZNFX1 protein in human tissues is not well delineated. Its subcellular distribution is also unclear. In silico analyses of its primary structure with various software did not detect known targeting signals for specific subcellular organelles (30). Its helicase and ZNF domains suggest that it is capable of interacting with nucleic acids (26, 31). Human ZNFX1 has been reported to be present at the outer mitochondrial membrane following transient transfection of A549 epithelial cell lines, whereas, in C. elegans, worm WT-ZNFX1 was located in germ granules (P granules) (25, 26). We performed confocal microscopy with an mAb against ZNFX1 in A549 and HeLa epithelial cells transiently transfected with human WT-ZNFX1. We found that ZNFX1 was located within the cytoplasm in HeLa and A549 cells, with two different nonexclusive fluorescence patterns, a diffuse and a punctuated distribution (Fig. 2A and SI Appendix, Fig. S4). ZNFX1 did not colocalize with the endoplasmic reticulum, peroxisomes, endosomes, lysosomes, membrane-associated Golgi apparatus, or mitochondria (SI Appendix, Fig. S4A). A similar localization was confirmed in SV40 fibroblasts stimulated with poly(I:C) from a healthy control, whereas no ZNFX1 protein was detected in SV40 fibroblasts of P2 using a mAb recognizing an epitope in the N-terminal part of ZNFX1, upstream of the mutation site (SI Appendix, Fig. S4B). We then tested the colocalization of WT-ZNFX1 with membraneless ribonucleoprotein (RNP) granules in transiently transfected A549 and HeLa cells. We found that ZNFX1 localized to stress granules formed upon transfection, which is typical of RNA-binding proteins, as shown using antibodies against two stress granule markers, G3BP1 and TIA1 (Fig. 2A and SI Appendix, Fig. S4C) (32–35). Moreover, transient transfection of WT-ZNFX1 into A549 and HeLa cells induced stress granule formation in about half of the transfected cells. By comparison, stress granules were observed in less than 1% of the cells transfected with an empty vector (EV) (Fig. 2A and SI Appendix, Fig. S4 C and D). Similarly, endogenous ZNFX1 localized in arsenite-induced stress granules in SV40 fibroblasts from healthy control and activated THP1 cells (Fig. 2 B and C). However, human ZNFX1 did not localize in P bodies, as shown using antibodies against DCP1a. The striking juxtaposition of DCP1a-containing P bodies and ZNFX1-containing stress granules has been observed in many studies (SI Appendix, Fig. S4E) (36). These findings suggest that ZNFX1 is an RNA-binding protein that localizes to stress granules.
Fig. 2.
Expression of ZNFX1 in cell lines. (A) Confocal microscopy of HeLa cells transiently transfected with an EV or the WT-ZNFX1, stained with the anti-G3BP1 antibody, DAPI, or an antibody directed against the C terminus of ZNFX1. (Scale bar, 10 µm.) (B) Colocalization of ZNFX1 and G3BP1 in SV40 fibroblasts stimulated for 24 h with 25 µg/mL of poly(I:C) from a healthy control and a ZNFX1-deficient patient (P2), treated or not with 0.5 mM arsenite for 30 min. (Scale bar, 10 µm.) (C) Colocalization of ZNFX1 and G3BP1 in adherent THP1 cells treated or not with 0.5 mM arsenite for 30 min. An IgG isotype is used as a negative control. The results shown are representative of at least two independent experiments. (Scale bar, 10 µm.) (D) RT-qPCR for ZNFX1 with a probe spanning the junction between exons 3 and 4 (Left) or spanning the junction between exons 13 and 14 (Right) in EBV-B cells from healthy controls (Ctrls, n = 3) and a patient (P1). GUSB was used for normalization. Values are expressed as means ± SEM. (E) RT-qPCR for ZNFX1 with a probe spanning the junction between exons 3 and 4 (Left) and a probe spanning the junction between exons 13 and 14 (Right) in SV40 fibroblasts from healthy controls (n = 3) and patients (P1 and P2). GUSB was used for normalization. Values are expressed as means ± SEM. (F) Western blot of total protein extracts from EBV-B cells from healthy controls (C1 and C2) and a patient (P1). ZNFX1 was detected with an mAb directed against the N terminus. An antibody against tubulin was used as a loading control. The results shown are representative of two independent experiments. (G) Western blot of total protein extracts from SV40 fibroblasts from healthy controls (C1, C2, and C3) and patients (P1 and P2). ZNFX1 was detected with an mAb directed against the N terminus. An antibody against tubulin was used as a loading control. The results shown are representative of two independent experiments.
ZNFX1 Expression in the Patients’ Cell Lines.
We assessed the impact of the homozygous ZNFX1 mutations on both mRNA and protein levels in the patients’ cells, using both immortalized nonhematopoietic (SV40 fibroblasts, P1 and P2) and hematopoietic (EBV-B cells, P1, not available for P2) cell lines. No cell line was available for P3 and P4. We first amplified the WT and mutant full-length cDNAs. We performed RT-qPCR with two sets of probes for the ZNFX1 mRNA. Comparison with cells from healthy controls showed that SV40 fibroblasts and EBV-B cells from P1 displayed normal levels of ZNFX1 mRNA production (Fig. 2 A and B). By contrast, the SV40 fibroblasts of P2 contained about 5% of the level of ZNFX1 mRNA found in the WT (Fig. 2C), suggesting that nonsense-mediated mRNA decay was occurring. We then used SDS-PAGE, followed by WB with an mAb against the N terminus of ZNFX1, to assess the levels of endogenous ZNFX1. The WT protein was produced at the expected MW. By contrast, SV40 fibroblasts and EBV-B cells from P1 produced a mutant protein with an MW lower than that of the WT protein, consistent with truncation. Moreover, SV40 fibroblasts from P2 contained no detectable ZNFX1 protein (Fig. 2 C and D). However, protein production was rescued by introducing the WT ZNFX1 cDNA by lentiviral transduction, confirming that the p.S959* variant leads to a loss of expression (SI Appendix, Fig. S3F). Overall, these results show that homozygosity for the p.E1606Rfs*10 and p.S959* ZNFX1 variants results in the production of truncated proteins in SV40 fibroblasts and EBV-B cells from P1 and a lack of detectable protein in SV40 fibroblasts from P2, respectively. Together with our previous findings, these data suggest that at least three of these patients, and possibly all four, have AR complete ZNFX1 deficiency.
Frequency of Lymphoid and Myeloid Cells among the Cells of ZNFX1-Deficient Patients.
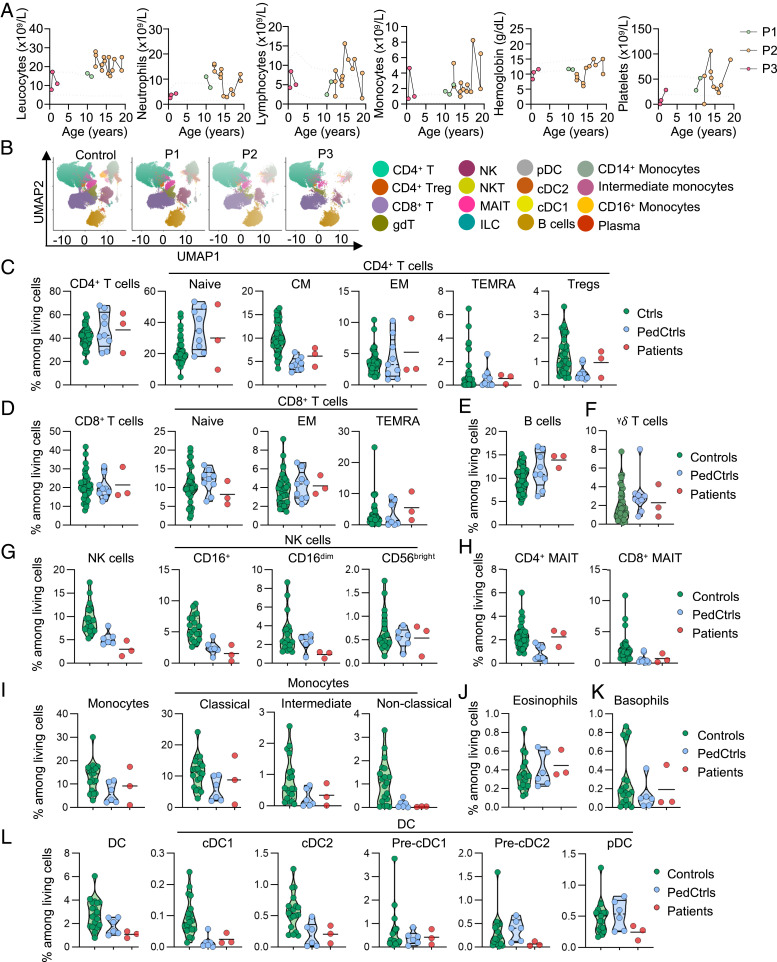
We analyzed the impact of ZNFX1 deficiency on leukocyte development. Complete blood counts on fresh samples from the three patients tested (P1, P2, and P3) revealed peripheral leukocytosis with intermittent monocytosis and variable neutrophilia and lymphocytosis, even in the absence of clinical infection (Fig. 3A). No leukocyte counts were available for P4 (already deceased). Bone marrow aspiration in P2 at the age of 12 y showed that all hematopoietic lineages were present, with multiple cells of the monocyte lineage (42%) bearing lobular nuclei without chromosomal abnormalities (SI Appendix, Fig. S1). Abundant monocytes were detected on concomitant blood smears (SI Appendix, Fig. S1). We then studied cryopreserved peripheral blood mononuclear cells (PBMCs) from P1, P2, and P3 by conventional flow cytometry and mass cytometry (cytometry by time of flight [CyTOF]) (Fig. 3B). At the time of sampling, P1 and P2 (aged 12 and 15 y, respectively) had high relative and absolute numbers of monocytes in complete blood counts (Fig. 3A). Flow cytometry and CyTOF showed that the proportions of CD4+ and CD8+ T cells and B cells among PBMCs were normal, relative to age-matched controls (Fig. 3 B–E). The proportions of naive CD4+ T cells and memory CD4+ and CD8+ subsets, regulatory T cells (Tregs), and γδ T cells were also normal (Fig. 3 C, D, and F). The proportion of NK cells was low among the PBMCs of the three patients tested, with a low frequency of CD56dim NK cells and a normal frequency of CD56bright NK cells (Fig. 3G). The proportions of CD4+ and CD8+ mucosal associated invariant T (MAIT) cells were conserved in the three patients (Fig. 3H). At the time of sampling, the proportion of total monocytes had not increased in any of the patients tested, as detected by CyTOF. Two of the three monocyte subpopulations, classical (CD14+CD16−) and intermediate (CD14+CD16+) monocytes, were in the range of healthy controls, whereas the proportion of nonclassical (CD14dimCD16+) monocytes was low (Fig. 3I). The frequency of basophils and eosinophils was normal (Fig. 3 J and K). The proportions of total DCs and of conventional (cDC1 and cDC2) and plasmacytoid DCs were normal on CyTOF, in all three patients tested (Fig. 3L). Overall, the development of other myeloid and lymphoid subsets was largely normal at the time of sampling among the patients with inherited ZNFX1 deficiency tested.
Fig. 3.
Hematological and immunological profile of patients with inherited ZNFX1 deficiency. (A) Absolute numbers of peripheral total leukocytes, neutrophils, lymphocytes, monocytes and platelets, and hemoglobin from P1, P2, and P3, determined from complete blood counts. (B) UMAP analysis of CyTOF showing the distribution of the main peripheral leukocyte populations identified in a healthy control, P1, P2, and P3. (C) Frequency of living CD4+ T cells and their subsets in adult controls (green circles), age-matched controls (PedCtrls, blue circles), and P1, P2, and P3 (red circles), as determined by CyTOF. Frequency of living (D) CD8+ T cells and their subsets, (E) B cells, (F) γδ T cells, (G) NK cells and their subsets, and (H) MAIT cells, and frequency of (I) monocytes and their subsets, (J) eosinophils, (K) basophils, and (L) dendritic cells (DCs) and their subsets, as determined by CyTOF. Results are expressed as a proportion of living single PBMCs.
Normal Production of IFN-γ by ZNFX1-Deficient Lymphocytes.
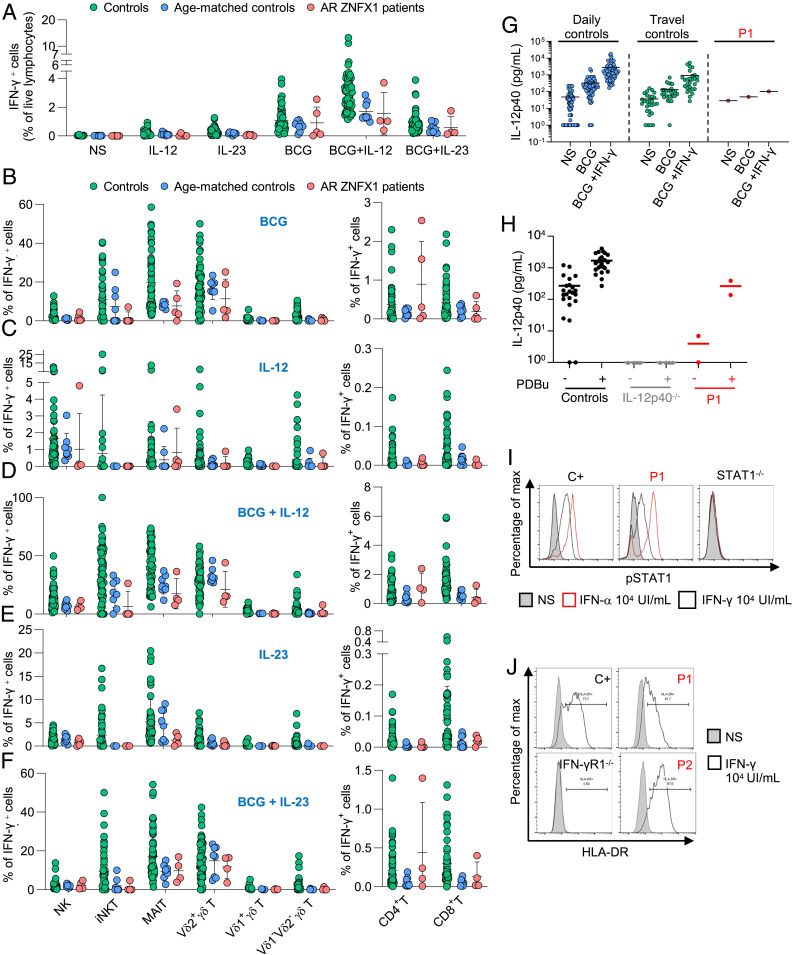
IFN-γ is essential for protective immunity to mycobacteria in humans (4, 37, 38). The related IL-12 and IL-23 cytokines, and secreted ISG15, are required for human antimycobacterial immunity and are induced by IFN-γ (6, 15, 39). An impaired production of, or response to, IFN-γ is characteristic of MSMD patients (1, 3). Among leukocyte subsets, human ZNFX1 is most abundantly expressed in neutrophils, monocytes, and macrophages (ProteinAtlas and BioGPS databases). Mouse ZNFX1 is also more abundant in macrophages, but the antimycobacterial immunity of ZNFX1-deficient mice has not been evaluated (26). We found that ZNFX1 transcription was induced after bacillus Calmette–Guérin infection, more so than after stimulation with phorbol 12-myristate 13-acetate (PMA) in the THP1 monocyte cell line (SI Appendix, Fig. S5A). We also assessed the impact of ZNFX1 deficiency on IFN-γ production by fresh lymphocytes. Whole-blood IFN-γ production after stimulation with bacillus Calmette–Guérin alone or bacillus Calmette–Guérin plus IL-12 was normal in P1 (data not available for the other patients) (SI Appendix, Fig. S5B). The production of IFN-γ and TNF-α after stimulation with bacillus Calmette–Guérin alone, bacillus Calmette–Guérin plus IL-12, bacillus Calmette–Guérin plus IL-23, or PMA−ionomycin was also normal for cryopreserved PBMCs (Fig. 4A and SI Appendix, Fig. S5C). In addition, the proportion of innate (NK cells), “innate-like” (iNKT, MAIT, and γδ T cells), and adaptive (CD4+ and CD8+ T cells) lymphocytes producing IFN-γ was normal following stimulation with bacillus Calmette–Guérin alone, bacillus Calmette–Guérin plus IL-12, or bacillus Calmette–Guérin plus IL-23, as measured by flow cytometry (Fig. 4 B–F). Overall, these findings indicate that the development of the major lymphocyte subsets and their IFN-γ production were normal in ZNFX1-deficient patients.
Fig. 4.
Conserved IFN-γ immunity in ZNFX1-deficient patients. PBMCs from controls (green dots), age-matched controls (blue dots), and patients with AR ZNFX1 deficiency (red dots) were left unstimulated or were stimulated with IL-12, or IL-23, with or without bacillus Calmette–Guérin activation. (A) IFN-γ levels in the supernatant of PBMCs with and without stimulation with IL-12, IL-23, bacillus Calmette–Guérin, bacillus Calmette–Guérin+IL-12, and bacillus Calmette–Guérin+IL-23, as assessed by intracellular flow cytometry. Proportion of IFN-γ−producing lymphocytes of various subsets involved in innate (NK, iNKT, and MAIT cells), adaptive (CD4+ and CD8+ T cells), and both adaptive and innate (γδ T cells including Vδ1+, Vδ2+, and Vδ1−Vδ2− subsets) immunity after stimulation with (B) bacillus Calmette–Guérin, (C) IL-12, (D) bacillus Calmette–Guérin+ IL-12, (E) IL-23, or (F) bacillus Calmette–Guérin+IL-23. (G) Secretion of IL-12p40 by whole blood from daily controls (n = 69), travel controls (n = 14), P1, with and without stimulation with bacillus Calmette–Guérin alone, or bacillus Calmette–Guérin and IFN-γ. ELISA was used to determine the levels of this cytokine. (H) Secretion of IL-12p40 by EBV-B cells from controls, patients with AR complete IL-12p40 deficiency (IL-12p40−/−), or P1, after stimulation with 10−7 M PDBu for 24 h. ELISA was used to determine the levels of these cytokines. (I) Phosphorylation of STAT1 after 20 min of stimulation with IFN-γ (104 IU/mL) or IFN-α (104 IU/mL) in EBV-B cells from a healthy control (C+), P1, and a patient with AR complete STAT1 deficiency (STAT1−/−), as determined by flow cytometry. (J) Induction of HLA-DR expression in SV40 fibroblasts from controls (C+), P1, P2, and a patient with AR complete IFN-γR1 deficiency (IFN-γR1−/−) after 48 h of stimulation with IFN-γ (104 IU/mL), as determined by flow cytometry. The results of I and J are representative of two independent experiments.
Normal Response to IFN-γ in the Cells of ZNFX1-Deficient Patients.
We then tested cellular responses to IFN-γ. ZNFX1 transcription is increased by IFN-γ in human monocyte-derived macrophages (40). We investigated whether ZNFX1 deficiency led to an impaired IFN-γ response in primary cells and cell lines from the patients. IL-12p40 secretion by whole blood following IFN-γ stimulation was normal in P1, although its induction by bacillus Calmette–Guérin or bacillus Calmette–Guérin+IFN-γ was in the lower range of controls (Fig. 4G). Moreover, the production of IL-12p40 and IL-12p70, and of TNF-α, was normal after stimulation with phorbol 12, 13-dibutyrate (PDBu) in EBV-B cells from P1 (Fig. 4H and SI Appendix, Fig. S5 E and F). Early responses to IFN-γ, as evaluated by STAT1 phosphorylation in EBV-B cells (P1), and late responses to IFN-γ, as evaluated by human leukocyte antigen-DR (HLA-DR) induction in SV40 fibroblasts from P1 and P2, were normal (Fig. 4 I and J). The production of reactive oxygen species by neutrophils from P1 and P2 in response to PMA was normal (SI Appendix, Table S2). Overall, leukocytes from ZNFX1-deficient patients responded normally to IFN-γ, and immortalized hematopoietic and nonhematopoietic cell lines from patients displayed apparently normal responses to IFN-γ.
Discussion
We report here four patients from two unrelated kindreds, originating from Iran and Morocco, with a novel form of “syndromic” MSMD or TB, associated with intermittent but marked monocytosis. The patients have AR ZNFX1 deficiency. Two of these patients also had recurrent and unexplained episodes of fever, thrombocytopenia, and organomegaly. ZNFX1 is ubiquitously expressed, but its function remains elusive. It would not, therefore, be surprising for these and, perhaps, other manifestations to be documented as clinical phenotypes when more ZNFX1-deficient patients are characterized. Penetrance for MSMD or TB is difficult to calculate. Only one of the four patients was vaccinated with bacillus Calmette–Guérin, and he had clinical disease. It is unknown whether the patients are prone to clinical disease caused by EM, another defining feature of MSMD (1, 2, 41). Two patients had severe TB, and the fourth did not suffer from any mycobacterial disease. Severe mycobacterial disease occurred at the ages of 3 mo (P1, bacillus Calmette–Guérin-osis), 11 y (P2, TB), and 14 mo (P4, TB). The three surviving patients are now 13 y (P1), 22 y (P2), and 11 y (P3) old. P3 had not developed mycobacterial disease by the age of 11 y and had not received the bacillus Calmette–Guérin vaccine. His exposure to M. tuberculosis is unknown. The penetrance of ZNFX1 deficiency for TB is probably greater than that for MSMD, if only because M. tuberculosis is about 1,000 times more virulent than bacillus Calmette–Guérin (15–17).
The vast majority of MSMD patients have normal counts of myeloid cells, except patients with AD partial IRF8 and AR complete SPPL2a deficiencies, which present defects of both cDC1s and cDC2s and of cDC2s alone, respectively (42, 43). Patients with AR complete IRF8 deficiency have monocytopenia associated with a complete loss of DCs (42). Monocytopenia is also seen in MonoMAC caused by germline heterozygous GATA2 mutations, which is characterized by a wide range of clinical manifestations, including mycobacterial disease (13, 44, 45). Monocytosis can be found during mycobacterial disease, in the course of autoimmune conditions, such as Ras-associated autoimmune leukoproliferative disorder, or associated with various myeloid neoplasms (46–48). The monocytosis of the ZNFX1-deficient patients is unlikely to be due to their mycobacterial disease, because their monocyte counts remained high up to 5 y after the clinical remission of mycobacterial disease. In P1, blood monocytosis was associated with bone marrow monocytosis, as reported twice in studies performed during acute episodes of fever, which resolved. By contrast to clonal disorders, such as juvenile myelomonocytic leukemia (JMML, which frequently present with monocytosis, cytopenia, and organomegaly), the bone marrow of P2 presented no cellular dysplastic features or chromosomal abnormalities (49, 50). Furthermore, mycobacterial infection is uncommon in patients with JMML (51). AR ZNFX1 deficiency is, therefore, a genetic etiology of MSMD or TB associated with intermittent monocytosis. The mechanisms underlying monocytopoiesis in these patients remain to be studied.
Given that impaired production of, or response to, IFN-γ is the defining characteristic of the known genetic etiologies of MSMD, we checked for these cellular phenotypes in primary cells and cell lines from the patients. We found no evidence of any major defect of IFN-γ production by the various subsets of lymphoid cells. We also found no evidence of any major disruption of the IFN-γ response pathway in whole blood and cell lines. However, ZNFX1 is expressed predominantly in myeloid cells, particularly in neutrophils and monocytes, and its transcription is induced following the infection of THP1 cells with bacillus Calmette–Guérin. Both the Mycobacterium-responsive and IFN-γ−responsive pathways should be studied in the various subsets of myeloid cells ex vivo or with induced pluripotent stem cell-derived myeloid cells in vitro (52). All the genes involved in MSMD affect the response and/or production of IFN-γ. It is tempting to speculate that ZNFX1 may affect the production of IFN-γ−inducing cytokines, such as IL-12p70, IL-23, and ISG15, by certain subsets of myeloid cells. It would also be of interest to test ZNFX1-deficient mice for antimycobacterial immunity. ZNFX1 is the first product of an MSMD gene shown to have RNA-binding activity. If an IFN-γ−related cellular phenotype were identified, it would be essential to try to understand the causality of ZNFX1 defects. This will involve deciphering the mechanism by which a protein bound to RNAs can disrupt IFN-γ−related immunity, or other mechanisms of antimycobacterial immunity (53, 54).
Our findings suggest that ZNFX1 is recruited in stress granules, at least in A549, HeLa, THP1 cells, and SV40 fibroblasts. Stress granules can associate with P bodies, but the components, structures, and functions of stress granules and P bodies are different (55, 56). While ZNFX1 was not detected in P bodies, we found that it induced stress granule formation in the absence of (other) stress, which is typical of RNA-binding proteins, such as CPEB, TIA1, G3BP, and FMRP (32–35). Interestingly, T. Ishidate et al. (24) previously reported the localization of ZNFX1 in germline-specific membraneless RNP organelles called P granules in C. elegans. Some cellular environment conditions, such as oxidative stress, heat shock, hyperosmolarity, viral infection, and ultraviolet irradiation, can trigger the assembly of stress granule components (56–58). Further studies will be required to determine whether the localization of ZNFX1 in stress granules is involved in protective immunity to mycobacteria. Finally, it is intriguing that our patients did not suffer from severe viral illnesses, whereas mouse ZNFX1 has been reported to be important for antiviral immunity via the sensing of dsRNA (26). More patients should be identified, their clinical histories analyzed, and their cells tested, before any firm conclusions are drawn. Overall, our study indicates that human ZNFX1 is a protein recruited in stress granules. Human ZNFX1 is essential for monocyte homeostasis and protective immunity to mycobacteria, via mechanisms potentially involving myeloid cells, as the patients described here displayed monocytosis, and ZNFX1 levels are highest in monocytes. Further studies are required to delineate more clearly the essential and redundant functions of human ZNFX1 at the cellular, immunological, and clinical levels.
Materials and Methods
Patient Recruitment and Ethics.
Clinical history and biological specimens were obtained from the referring clinicians, with the signed consent of the patients, parents, and siblings participating in the study. All the experiments involving human subjects conducted in this study were performed in accordance with institutional, local, and national ethical guidelines. Approval was obtained from the French Ethics Committee “Comité de Protection des Personnes,” the French National Agency for Medicine and Health Product Safety, the “Institut National de la Santé et de la Recherche Médicale,” in France and the Rockefeller University Institutional Review Board in New York.
Genomic DNA, WES, and Sanger Sequencing.
Genomic DNA (gDNA) was extracted from PMBCs after Ficoll-Paque Plus gradient purification, or from whole blood with the iPrep PureLink gDNA Blood kit and iPrep Instruments (Life Technologies, Thermo Fisher Scientific). WES was performed on gDNA from patients (P1 to P3), as previously described (4). Briefly, an adapter-ligated library was prepared with the TruSeq DNA Sample Prep Kit (Illumina). Exome capture was performed with the SureSelect Human All Exon 50 Mb kit (Agilent Technologies). Paired-end sequencing was performed on an Illumina HiSeq 2000 (Illumina), generating 100 base reads. The sequences were aligned with the human genome reference sequence (hg19 build), with the Burrows-Wheeler Aligner (59). Downstream processing was performed with the Genome Analysis Toolkit (GATK), SAMtools (60), and Picard Tools (http://broadinstitute.github.io/picard). Substitution and indel calls were made with GATK Unified Genotyper and GATK IndelGenotyperV2, respectively. All calls with a Phred-scaled single-nucleotide polymorphism quality of ≤20 and a read coverage of ≤2 were filtered out. All mutations found in ZNFX1 were confirmed by Sanger sequencing on genomic DNA. Familial segregation was analyzed if DNA samples from relatives were available. Sanger sequencing was performed on amplified PCR products purified by ultracentrifugation through Sephadex G-50 Superfine resin (Amersham-Pharmacia-Biotech) and sequenced with the Big Dye Terminator 3.1 Cycle Sequencing Kit on an ABI Prism 3700 apparatus (Applied Biosystems). Sequence files and chromatograms were analyzed with Genalys 2.8 software (CNG, Centre National de Génotypage, https://www.cnrgh.fr/index.html).
Cell Culture, Stimulation, and Flow Cytometry.
EBV-B cells were cultured in Roswell Park Memorial Institute medium (RPMI) medium 1640 (GIBCO BRL) supplemented with 10% heat-inactivated pooled fetal bovine serum (FBS) (GIBCO BRL), referred to as complete RPMI 1640. HEK293T cells and SV40 fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (GIBCO BRL) supplemented with 10% FBS. Cells were grown at 37 °C, under an atmosphere containing 5% CO2. For the study of IL-12p40/IL-12p70/TNF-α production by EBV-B cells, 2 × 106 cells in 1 mL of RPMI-2% fetal calf serum were stimulated by incubation for 24 h with 10−7 M PDBu (Sigma-Aldrich) (61). EBV-B cells or SV40 fibroblasts were washed in phosphate-buffered saline (PBS) and dispensed into a 24-well plate for labeling. Aqua (Live/Dead Fixable Dead Cell Stain kit from Invitrogen) staining was performed to exclude dead cells. The cells were then left unstimulated, or were stimulated by incubation with IFN-γ (105 IU/mL, Imukin), or IFN-α2b (105 IU/mL, IntronA) for 20 min. The reaction was then stopped by adding cold PBS, and the cells were harvested with trypsin and stained directly. Intracellular staining was performed with the Cytofix/Cytoperm Plus Fixation/Permeabilization kit (BD) with anti-STAT1-PE (BD) or anti-Y701-pSTAT1-Alexa468 (BD) antibodies or an equivalent concentration of isotype-matched control mAb (Becton Dickinson) in 2% FBS in PBS, for 1 h at 4 °C. Compensation was performed on single-stained nonstimulated samples. Staining was assessed on a Gallios Flow cytometer (Beckman Coulter), and the results were analyzed with FlowJo v10 (Tree Star).
SV40 fibroblasts were plated at a density of 105 cells in 1 mL of DMEM-10% FBS in 12-well plates. The following day, SV40 fibroblasts were left unstimulated or were stimulated for 48 h with recombinant IFN-α2b (Introna, MSD) or recombinant IFN-γ (Imukin). After stimulation, cells were harvested with trypsin, and stained with fluorescein isothiocyanate (FITC)-HLA-DR antibody (clone L243, #307604, Biolegend) and the Aqua Dead Cell Stain kit (#L34957 Thermo Fisher Scientific). Cells were acquired on a Beckman Coulter Gallios flow cytometer and analyzed with FlowJo Software.
Site-Directed Mutagenesis, and Transient and Stable Transfection.
A DDK-tagged ZNFX1 cDNA plasmid was obtained from Origene (#RC214589). Constructs carrying single-nucleotide mutant alleles were generated from this plasmid by mutagenesis with appropriate primers, with the Pfu Ultra II Fusion HS DNA (#600674, Agilent) polymerase. They were then digested with DpnI (#R0176L, New England Biolab). Site-directed deletions were created with CloneAmp HiFi (Takara, #639298), and the blunt ends were 5′-phosphorylated with the T4 DNA polymerase (#E1201S, New England Biolab), and ligated with the Quick ligation kit (#M2200S, New England Biolab). Plasmids were amplified in competent cells of E. coli (#C3019H, New England Biolab) and purified with the Maxiprep kit (#12162, Qiagen). HEK293T cells were transiently transfected with the various constructs, with X-tremeGENE9 (#XTG9-RO, Roche), used in accordance with the manufacturer’s instructions.
The WT coding sequence of ZNFX1 was inserted into pTRIP-SSFV-deltaNGFR-2A. For lentivirus production, HEK293T cells were transfected with 1.6 µg of pTRIP-SSFV-DNGFR-2A-ZNFX1-WT, 0.2 µg of pCMV-VSV-G (Addgene #8454), 0.2 µg of pHXB2 (NIH-AIDS Reagent Program #1069), and 1 µg of psPAX2 (Addgene #12260), with X-treme gene 9 (Roche), according to the manufacturer’s instructions. Supernatants were harvested after 24 h, and 8 µg/mL protamine sulfate was added. The lentiviral suspension obtained was used to transduce 2 × 105 SV40 fibroblasts by spinoculation at 1,200 × g for 2 h. The transduced cells were subjected to magnetic-activated cell sorting (MACS) purification with anti-CD271 microbeads (Miltenyi Biotec), and the purified cells were used for further experiments. Transduction was confirmed by flow cytometry with an anti-CD271-PE antibody (BD Bioscience).
Western Blot.
Total protein extracts were prepared by mixing cells with lysis buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 0.5% Triton X-100, and 2 mM (ethylenedinitrilo)tetraacetic acid) supplemented with protease inhibitors (Complete, Roche), phosphatase inhibitor mixture (PhosphoStop, Roche), 0.1 mM dithiothreitol (DTT, Life Technologies), phenylmethylsulfonyl fluoride and incubating for 30 min on ice. The cytoplasmic and nuclear fractions of the cells were separated with NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific, #78835). Protein fractions were separated by SDS-PAGE, and the resulting bands were electrotransferred onto poly(vinylidene difluoride) membranes. The membrane was blocked by incubation in TBS supplemented with 0.1% Tween 20 and 5% skimmed milk powder for 60 min at room temperature and incubated with antibodies against ZNFX1 (Abcam, ab179452), HRP-DDK (Sigma-Aldrich, #A8592), tubulin (Cell Signaling, sc-239448), and HRP-lamin A/C (Santa Cruz, sc-376248). Membranes were washed several times in 0.05% Tween-20 in PBS, and antibody binding was detected by incubation with horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (GE Healthcare), by enhanced chemiluminescence (GE Healthcare).
RNA Extraction, RT-qPCR, and cDNA.
Total RNA was extracted with the RNeasy Mini Kit (#74104, Qiagen), according to the manufacturer’s instructions. Reverse transcription was performed on 1 μg of RNA with random primers and the High-Capacity RNA-to-cDNA Kit (Applied Biosystems), according to the manufacturer’s protocol. Quantitative PCR was then performed with the TaqMan Fast Universal PCR Master Mix (2×), No AmpErase UNG (Thermo Fisher Scientific, #4352042), and the following probes, all from Thermo Fisher Scientific: ZNFX1 exons 13 and 14 (Hs01105231_m1), ZNFX1 exons 3 and 4 (Hs01105222_m1), GUSB (#1702016), in an ABI PRISM 7700 Sequence Detection System. The relative expression levels of the genes were calculated with the formula 2−ΔCt, and the results were normalized against the levels of mRNA for the housekeeping gene encoding β-glucuronidase (GUS).
Whole-Blood Activation Enzyme-Linked Immunosorbent Assay for Cytokines.
Whole-blood samples were collected from healthy controls (fresh blood and travel blood) and patients, in heparin-containing collection tubes (61, 62). These samples were diluted 1:2 in RPMI 1640 (GibcoBRL). Diluted blood samples were then dispensed into five wells (1 mL per well) of a 48-well plate (Nunc). The samples were incubated for 48 h at 37 °C, under an atmosphere containing 5% CO2, and under three different sets of activation conditions: with medium alone, with bacillus Calmette–Guérin (M. bovis-bacillus Calmette–Guérin, Pasteur substrain), or with bacillus Calmette–Guérin plus recombinant (rh) IL-12 (20 ng/mL; R&D Systems), or bacillus Calmette–Guérin plus IFN-γ (Imukin). The cell culture supernatant or whole blood was collected for determinations of IL-12p40 (#DP400, R&D Systems) or IFN-γ (Sanquin) by enzyme-linked immunosorbent assay (ELISA), according to the kit manufacturer’s protocol.
Confocal Microscopy.
HeLa or A549 cells were plated on chambered coverslips (#80826, iBidi) and were left untransfected or were transiently transfected with the ZNFX1 cDNA plasmid or an empty pCMV6 vector for 24 h. SV40 fibroblasts or THP1 cells were plated on chamber coverslips and left stimulated or not with bacillus Calmette–Guérin or poly(I:C), respectively, for 24 h. The cells were fixed in 4% formaldehyde in PBS, pH 7.4. Cells were incubated overnight at 4 °C with primary antibody: mouse anti-G3BP (Abcam #ab56574), anti-DCP1A (Abnova #H00055802-M06), anti-RAB7 (Abcam #ab50533), anti-PMP70 (Sigma #SAB4200181), anti-LAMP-2 (Santa Cruz # sc-18822), anti-EEA1 (BD # 610457), anti-TIA1 (Santa Cruz #sc-166247), and rabbit anti-ZNFX1 (Abcam, ab179452) antibodies. The cells were washed three times with PBS 1× and stained by incubation with secondary antibodies for 1 h at room temperature (goat anti-rabbit IgG Alexa Fluor 488 [#A-11034]; goat anti-mouse IgG Alexa Fluor 633 [#A-21052]) before mounting in Prolong-gold and visualization by confocal microscopy (×63 oil immersion lens).
Immunophenotyping of Leukocytes by Mass Cytometry.
We performed mass cytometry (CyTOF) with two different antibody panels (termed as regular surface and myeloid panels hereafter), as previously described by Yang and coworkers (3, 63). For the regular surface panel, we analyzed PBMCs from 37 healthy adult controls and nine healthy age-matched controls (aged from 1 y to 17 y) from diverse ethnic backgrounds, and three patients (P1, P2, and P3) with homozygous ZNFX1 mutations in seven batches of experiments performed on different dates. For the myeloid panel, we analyzed PBMCs from 19 healthy adult controls, six healthy age-matched controls (aged from 1 y to 17 y) from diverse ethnic backgrounds, P1, P2, and P3 in five batches of experiments performed on different dates. In particular, the three ZNFX1-deficient patients were studied with eight and three healthy adult controls in a single batch of regular surface and myeloid panel experiments, respectively. Freshly thawed PBMCs (1.0 × 106 to 1.5 × 106 cells per panel) were incubated with Fc block and stained with a panel of metal-conjugated antibodies obtained from Fluidigm or through customized conjugation. The regular surface panel consisted of the following antibodies: anti-CD45-89Y, anti-CD57-113In, anti-CD11c-115In, anti-CD33-141Pr, anti-CD19-142Nd, anti-CD45RA-143Nd, anti-CD141-144Nd, anti-CD4-145Nd, anti-CD8-146Nd, anti-CD20-147Sm, anti-CD16-148Nd, anti-CD127-149Sm, anti-CD1c-150Nd, anti-CD123-151Eu, anti-CD66b-152Sm, anti-PD-1-153Eu, anti-CD86-154Sm, anti-CD27-155Gd, anti-CCR5-156Gd, anti-CD117-158Gd, anti-CD24-159Tb, anti-CD14-160Gd, anti-CD56-161Dy, anti-γδTCR-162Dy, anti-CRTH2-163Dy, anti-CLEC12A-164Dy, anti-CCR6-165Ho, anti-CD25-166Er, anti-CCR7-167Er, anti-CD3-168Er, anti-CX3CR1-169Tm, anti-CD38-170Er, anti-CD161-171Yb, anti-CD209-172Yb, anti-CXCR3-173Yb, anti-HLA-DR-174Yb, anti-CCR4-176Yb, and anti-CD11b-209Bi antibodies. The myeloid panel consisted of the following antibodies: anti-CD45-89Y, anti-HLA-ABC-113In, anti-CD11c-115In, anti-CD33-141Pr, anti-CD19-142Nd, anti-CD45RA-143Nd, anti-CD141-144Nd, anti-CD4-145Nd, anti-CD8-146Nd, anti-CLEC9A-147Sm, anti-CD16-148Nd, anti-FcεRIα-149Sm, anti-CD1c-150Nd, anti-CD123-151Eu, anti-CD66b-152Sm, anti-CD83-153Eu, anti-CD86-154Sm, anti-CD27-155Gd, anti-PD-L1-156Gd, anti-CD163-158Gd, anti-CD103-159Tb, anti-CD14-160Gd, anti-CD56-161Dy, anti-CD64-162Dy, anti-CD40-164Dy, anti-CD169-166Er, anti-CD117-167Er, anti-CD3-168Er, anti-CX3CR1-169Tm, anti-CD38-170Er, anti-CD207-171Yb, anti-CD206-172Yb, anti-HLA-DR-174Yb, anti-Axl-175Lu, anti-CD209-176Yb, and anti-CD11b-209Bi antibodies. The stained cells were washed, fixed, and permeabilized with barcode permeabilization buffer (Fluidigm), and barcoded with Fluidigm’s Cell-ID 20-Plex Pd Barcoding Kit. Samples were then washed and pooled into a single tube. Dead cells and doublets were excluded by staining the cells with a rhodium-based dead cell exclusion intercalator (Rh103) before fixation and with cationic iridium nucleic acid intercalators (Ir191 and Ir193) after fixation. After surface staining, the samples were fixed and stored until acquisition on a Helios mass cytometer (Fluidigm).
We investigated the immunophenotype of the ZNFX1-deficient patients, by combining data from seven batches of experiments, as performed by Yang et al. (3). We manually inspected the data from each batch separately, and computationally compared immunophenotypes across batches in an objective and unbiased manner, with Integration of Multi-Batch Cytometry Datasets (iMUBAC) (63). Briefly, this method is a combination of high-dimensional batch correction, unsupervised clustering, and batch-specific cell type classification through machine learning. Data from each of the samples were first preprocessed for the automatic exclusion of dead cells and doublets. Cells from healthy controls (but not the ZNFX1-deficient patients) were then used for high-dimensional batch correction with Harmony (64). Data were down-sampled to 200,000 cells per batch before batch correction. Unsupervised clustering was then performed on the batch-corrected marker expression values, with the FlowSOM and ConsensusClusterPlus packages in Bioconductor (65, 66). Sixty clusters were retained after metaclustering. A batch-specific cell type classifier was trained on the uncorrected expression values and cell type annotations. We randomly sampled a maximum of 100 cells per cluster to improve computation efficiency and to alleviate class imbalance issues, as some of the cell types are much rarer than others. An extremely randomized tree algorithm was employed, with mtry and numRandomCuts hyperparameters tuned for each batch (67). Classifiers were trained through five rounds of 10-fold cross-validation with internal upsampling. Internal accuracy during cross-validation systematically exceeded ∼90%. The trained classifier was then applied to all cells, including the patients’ cells, for probabilistic cell type determination. Finally, each cluster was inspected manually, identified, and merged on the basis of marker expression patterns, if necessary, to “translate” the assigned clusters into cell types. For visualization purposes, cells from one healthy adult control and three ZNFX1-deficient patients were used to generate Uniform Manifold Approximation and Projection (UMAP) plots. Data were down-sampled to 100,000 cells per individual. All available markers were used for dimension reduction. Figures are color-coded according to the cell types identified through the unsupervised clustering approach described above. All analyses were performed with R v.4.0.
Stimulation of PBMCs with Live Bacillus Calmette–Guérin Infection for Cytokine Production.
This experiment was performed in four batches, on four dates. The data from the four batches were compiled for the final analysis, as already performed by Yang et al. (3). PBMCs from adult healthy donors, age-matched healthy donors (1 y to 17 y old) and P1, P2, and P3 were thawed, resuspended in RPMI 10% FBS, and counted. Cells (300,000 per well in 96-well U-bottomed plates) were plated at a density of 1.5 × 106 cells/mL. PBMCs from P1 and P2 were plated for all conditions but bacillus Calmette–Guérin+IL-12 and bacillus Calmette–Guérin+IL-23 conditions were PBMCs from P1 and PBMCs from P3 were plated in simplicate. Cells were stimulated in the presence and absence of M. bovis-bacillus Calmette–Guérin at a multiplicity of infection (MOI) = 1, and in the presence and absence of recombinant IL-12 (5 ng/mL, R&D) or recombinant IL-23 (10 ng/mL, R&D). Golgiplug (BD Biosciences) was added to each well after 40 h of stimulation. Eight hours later, supernatants were harvested for cytokine determinations in a 13-plex Legendplex assay (BioLegend), and cells were collected by centrifugation for fluorescence-activated cell sorter staining. In brief, cells were first stained with the Zombie NIR Viability kit (BioLegend) for 15 min. They were then stained with FcBlock (Miltenyi Biotec), anti-γδTCR-alexa 647 (BioLegend), anti-CD3-V450 (BD Biosciences), anti-CD56-BV605 (BioLegend), anti-CD4-BUV563 (BD Biosciences), anti-Vδ1TCR-FITC (Miltenyi Biotec), anti-CD8-BUV737 (BD Biosciences), anti-Vδ2TCR-APC/Fire750 (BioLegend), anti-CD20-BV785 (BioLegend), anti-Vα7.2-Alexa 700 (BioLegend), MR1-5-OP-RU-tetramer (NIH tetramer core facility), anti-Vβ11-APC (Miltenyi Biotec), and anti-iNKT-BV480 (BD Biosciences) antibodies for 30 min. The stained cells were fixed with the FOXP3/Transcription Factor Buffer set (Thermo Fisher Scientific) and intracellularly stained with anti-IFN-γ-BV711 (BioLegend) and anti-TNF-α-BV510 (BioLegend) antibodies in Perm/Wash buffer. Cells were acquired on a CyTek Aurora spectral flow cytometer, and the data were analyzed with Cytobank.
PhIP-Seq.
For antibody profiling by PhIP-Seq, plasma samples were obtained from two patients (P1 and P2). For comparison, and as additional controls, we also tested 10% liquid IVIg from pooled human plasma (Privigen CSL Behring AG) and human IgG-depleted serum (Supplier No. HPLASERGFA5ML, Molecular Innovations, Inc.). The total IgG levels in the plasma samples were determined with a Human IgG total ELISA Ready-SET-Go kit (Thermo Fisher Scientific). Diluted plasma samples containing ∼4 μg of total IgG were incubated at 4 °C overnight with 2 × 1010 plaque-forming units of a modified version of the original PhIP-Seq phage library. This modified T7 phage library was used to display a total of 115,753 peptides, each 56 aa long, including the same viral peptides as the original PhIP-Seq phage library and additional peptides derived from the protein sequences of various microbial B-cell antigens and allergens made available by the Immune Epitope Database and Analysis Resource (http://www.iedb.org). Subsequent steps were performed as previously described (4).
Statistical Methods.
Statistical analysis was performed with GraphPad Prism, version 7.02 (GraphPad Software). A P value of less than 0.05 was considered statistically significant.
Web Resources.
Web resources are CADD (https://cadd.gs.washington.edu/) and gnomAD Browser, (https://gnomad.broadinstitute.org/).
Supplementary Material
Acknowledgments
We warmly thank to Jie Chen and David Langlais for helpful discussions, and Yelena Nemirovskaya, Christine Rivalain, Dominick Papandrea, and Lazaro Lorenzo for administrative support. We also thank Stephen Elledge (Brigham and Women’s Hospital, Harvard University Medical School) for kindly providing the VirScan phage library used in this study. This research was supported, in part, by a grant from the National Institute of Allergy and Infectious Diseases (Grant R37AI095983), the National Center for Research Resources and the National Center for Advancing Sciences of the National Institutes of Health (Grant UL1TR001866), the Yale Center for Mendelian Genomics funded by the National Human Genome Research Institute (Grant UM1HG006504), the Genome Sequencing Project (GSP) Coordinating Center (Grant U24HG008956), the High Performance Computing Center (Research Infrastructure Program S10OD018521), The Rockefeller University, the St. Giles Foundation, National Institute of Health and Medical Research (INSERM), University of Paris, the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (Grant ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (Grant EQU201903007798), the SCOR Corporate Foundation for Science, the French National Research Agency under the “Investments for the future” (Program ANR-10-IAHU-01) and GENMSMD (Grant ANR-16-CE17.0005-01, to J.B.), Sidra Medicine (Program SDR400048), a Strategic Positioning Fund for Genetic Orphan Diseases (Grant SPF2012/005), and an inaugural Use Inspired Basic Research (UIBR) grant from the Agency for Science, Technology and Research in Singapore (to B.R.). M.O. was supported by the David Rockefeller Graduate Program, the Funai Foundation for Information Technology, the Honjo International Scholarship Foundation, the New York Hideyo Noguchi Memorial Society, and the Cooperative Center on Human Immunology at The Rockefeller University. T.L.V, J.R., and P.B. are supported by the MD-PhD program of the Imagine Institute with the support of the Bettancourt-Schueller Foundation. A.-L.N. is supported by the international PhD program of the Imagine Institute with the support of the Bettancourt-Schueller Foundation. J.R. was also supported by a “poste d’Accueil INSERM.”
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102804118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Bustamante J., Mendelian susceptibility to mycobacterial disease: Recent discoveries. Hum. Genet. 139, 993–1000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustamante J., Boisson-Dupuis S., Abel L., Casanova J. L., Mendelian susceptibility to mycobacterial disease: Genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin. Immunol. 26, 454–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang R., et al., Human T-bet governs innate and innate-like adaptive IFN-gamma immunity against mycobacteria. Cell 183, 1826–1847.e31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerner G., et al., Inherited human IFN-γ deficiency underlies mycobacterial disease. J. Clin. Invest. 130, 3158–3171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathan C. F., Murray H. W., Wiebe M. E., Rubin B. Y., Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 158, 670–689 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogunovic D., et al., Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreins A. Y., et al., Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212, 1641–1662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada S., et al., IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science 349, 606–613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eletto D., et al., Biallelic JAK1 mutations in immunodeficient patient with mycobacterial infection. Nat. Commun. 7, 13992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuis S., et al., Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33, 388–391 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Hsu A. P., et al., Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood 118, 2653–2655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickinson R. E., et al., Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood 118, 2656–2658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oleaga-Quintas C., et al., Inherited GATA2 deficiency is dominant by haploinsufficiency and displays incomplete clinical penetrance. J. Clin. Immunol. 41, 639–657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boisson-Dupuis S., et al., Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol. Rev. 264, 103–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boisson-Dupuis S., et al., Tuberculosis and impaired IL-23-dependent IFN-γ immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol. 3, eaau8714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerner G., et al., Human ancient DNA analyses reveal the high burden of tuberculosis in Europeans over the last 2,000 years. Am. J. Hum. Genet. 108, 517–524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerner G., et al., Homozygosity for TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc. Natl. Acad. Sci. U.S.A. 116, 10430–10434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tangye S. G., et al., Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J. Clin. Immunol. 40, 24–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boisson-Dupuis S., The monogenic basis of human tuberculosis. Hum. Genet. 139, 1001–1009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rapaport F., et al., Negative selection on human genes underlying inborn errors depends on disease outcome and both the mode and mechanism of inheritance. Proc. Natl. Acad. Sci. U.S.A. 118, e2001248118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott E. M.et al.; Greater Middle East Variome Consortium , Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 48, 1071–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itan Y., et al., The mutation significance cutoff: Gene-level thresholds for variant predictions. Nat. Methods 13, 109–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairman-Williams M. E., Guenther U. P., Jankowsky E., SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 20, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishidate T., et al., ZNFX-1 functions within perinuclear nuage to balance epigenetic signals. Mol. Cell 70, 639–649.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan G., et al., Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., et al., Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat. Cell Biol. 21, 1346–1356 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Itan Y., et al., The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U.S.A. 112, 13615–13620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrovski S., et al., The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet. 11, e1005492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim M. S., et al., A draft map of the human proteome. Nature 509, 575–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satori C. P., et al., Bioanalysis of eukaryotic organelles. Chem. Rev. 113, 2733–2811 (2013).Correction in: Chem. Rev.113, 5699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Font J., Mackay J. P., Beyond DNA: Zinc finger domains as RNA-binding modules. Methods Mol. Biol. 649, 479–491 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Mazroui R., et al., Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007–3017 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Tourrière H., et al., The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Gilks N., et al., Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D., The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118, 981–992 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Hubstenberger A., et al., P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell 68, 144–157.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Devos R., et al., Molecular cloning of human immune interferon cDNA and its expression in eukaryotic cells. Nucleic Acids Res. 10, 2487–2501 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray P. W., Goeddel D. V., Structure of the human immune interferon gene. Nature 298, 859–863 (1982). [DOI] [PubMed] [Google Scholar]
- 39.Martínez-Barricarte R., et al., Human IFN-γ immunity to mycobacteria is governed by both IL-12 and IL-23. Sci. Immunol. 3, eaau6759 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takata K., et al., Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity 47, 183–198.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Rosain J., et al., Mendelian susceptibility to mycobacterial disease: 2014-2018 update. Immunol. Cell Biol. 97, 360–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hambleton S., et al., IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 365, 127–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong X. F., et al., Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat. Immunol. 19, 973–985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bigley V., Cytlak U., Collin M., Human dendritic cell immunodeficiencies. Semin. Cell Dev. Biol. 86, 50–61 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Collin M., Dickinson R., Bigley V., Haematopoietic and immune defects associated with GATA2 mutation. Br. J. Haematol. 169, 173–187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi C., Pamer E. G., Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutta P., Nahrendorf M., Regulation and consequences of monocytosis. Immunol. Rev. 262, 167–178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calvo K. R., et al., JMML and RALD (Ras-associated autoimmune leukoproliferative disorder): Common genetic etiology yet clinically distinct entities. Blood 125, 2753–2758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niemeyer C. M., Flotho C., Juvenile myelomonocytic leukemia: Who’s the driver at the wheel? Blood 133, 1060–1070 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Hasle H., Myelodysplastic and myeloproliferative disorders of childhood. Hematology (Am. Soc. Hematol. Educ. Program) 2016, 598–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donadieu J.et al.; French GATA2 study group , Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica 103, 1278–1287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lachmann N., et al., Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports 4, 282–296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdel-Nour M., et al., The heme-regulated inhibitor is a cytosolic sensor of protein misfolding that controls innate immune signaling. Science 365, eaaw4144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomes E., Shorter J., The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kedersha N., et al., Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson P., Kedersha N., Stress granules. Curr. Biol. 19, R397–R398 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Burgess H. M., Mohr I., Defining the role of stress granules in innate immune suppression by the herpes simplex virus 1 endoribonuclease VHS. J. Virol. 92, e00829-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCormick C., Khaperskyy D. A., Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 17, 647–660 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esteve-Solé A., et al., Laboratory evaluation of the IFN-γ circuit for the molecular diagnosis of Mendelian susceptibility to mycobacterial disease. Crit. Rev. Clin. Lab. Sci. 55, 184–204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feinberg J., et al., Bacillus Calmette Guerin triggers the IL-12/IFN-gamma axis by an IRAK-4- and NEMO-dependent, non-cognate interaction between monocytes, NK, and T lymphocytes. Eur. J. Immunol. 34, 3276–3284 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Ogishi M., et al., Multibatch cytometry data integration for optimal immunophenotyping. J. Immunol. 206, 206–213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korsunsky I., et al., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Gassen S., et al., FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Wilkerson M. D., Hayes D. N., ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meuwis M. A., et al., Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem. Pharmacol. 73, 1422–1433 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.