Significance
The ability to robustly isolate intraorganoid fluid in a high-throughput capacity, using human endometrial epithelia as a model, circumvents several constraints typically associated with obtaining biological fluids, both in vivo and in vitro. This approach also allows for direct tandem genomics, transcriptomics, proteomics, and functional metabolomics. In addition to enhancing our understanding of uterine fluid composition regulation, this innovation is directly and readily transferrable to wider fields of cell and organoid biology, with implications for personalized, or precision, medicine approach development.
Keywords: endometrium, organoid, uterine fluid, human, reproduction
Abstract
Suboptimal uterine fluid (UF) composition can lead to pregnancy loss and likely contributes to offspring susceptibility to chronic adult-onset disorders. However, our understanding of the biochemical composition and mechanisms underpinning UF formation and regulation remain elusive, particularly in humans. To address this challenge, we developed a high-throughput method for intraorganoid fluid (IOF) isolation from human endometrial epithelial organoids. The IOF is biochemically distinct to the extraorganoid fluid (EOF) and cell culture medium as evidenced by the exclusive presence of 17 metabolites in IOF. Similarly, 69 metabolites were unique to EOF, showing asymmetrical apical and basolateral secretion by the in vitro endometrial epithelium, in a manner resembling that observed in vivo. Contrasting the quantitative metabolomic profiles of IOF and EOF revealed donor-specific biochemical signatures of organoids. Subsequent RNA sequencing of these organoids from which IOF and EOF were derived established the capacity to readily perform organoid multiomics in tandem, and suggests that transcriptomic regulation underpins the observed secretory asymmetry. In summary, these data provided by modeling uterine luminal and basolateral fluid formation in vitro offer scope to better understand UF composition and regulation with potential impacts on female fertility and offspring well-being.
Uterine glands and their product, uterine lumen fluid (UF), are central to mammalian pregnancy establishment (1–5). Despite UF being the site of critical reproductive events, including sperm migration, embryo implantation, and conceptus development, our understanding of the mechanisms governing UF formation and composition regulation is poor. This is largely attributable to ethical (6) and technical (7) limitations surrounding human UF interrogation in vivo, in addition to inadequate cell culture techniques recapitulating UF formation in vitro (8). Regarding the former, several available methods for sampling UF from animals, such as in situ cannulation, perfusion, and excision and rapid sampling (7), are not transferable to women. However, UF sampling, by aspiration or lavage, remains one of the least invasive methods for collecting material indicative of a woman’s uterine health; further emphasizing the importance of, and need for, understanding UF composition.
Regarding UF investigation in vitro, previous cell culture-based efforts to capture reproductive fluids include the culture of primary bovine, porcine, and murine oviduct epithelial monolayers on semipermeable supports in an apical-basal air-liquid interface. Oviduct epithelial cells in this configuration regain ciliation (9–12), are hormonally responsive (11, 13–16), and mediate physiological-like interactions with sperm (11, 17, 18). Moreover, their apically secreted oviduct fluid surrogate differs in biochemical composition to the culture medium provided basally (16, 19), contains oviduct-specific glycoproteins (12, 16), responds to basal stromal cell coculture (16, 19), and supports early embryo development (12). However, major limitations of this culture approach include a loss of physiological attributes and monolayer integrity after ∼4 wk in culture (20). Furthermore, attempts to similarly maintain primary endometrial, as opposed to oviductal, epithelia have been less successful, as hormonal responsiveness is inconsistent, and cellular senescence is observed by the third culture passage (21–23).
Endometrial epithelial cell organoids (EEOs) derived from primary endometrial tissue represent a paradigm shift in this regard (24). EEOs are heterogeneous and comprise both luminal and glandular epithelia (25, 26) and more faithfully mimic in vivo phenotypes and genetics while retaining appropriate hormonal responsiveness over multiple passages (27). EEOs are, therefore, amenable to long-term expansion and can be cryopreserved (25, 28, 29). Interestingly, EEOs exhibit polarity, whereby apical cell membranes face the center of the organoid (inwards) while the basolateral side faces outwards (25, 30, 31), suggesting that EEOs could be a useful model for investigating UF formation and function in vitro. As such, our overarching aim was to test the hypothesis that EEO intraorganoid fluid (IOF) may represent an in vitro UF surrogate. To achieve this, EEOs from three different donors were cultured prior to the capture of their IOF by two complementary methods (micromanipulation [MMN] or high-throughput centrifugation [HTC]) and metabolomic analysis. In parallel, EEO extraorganoid fluid (EOF) was collected and similarly analyzed. Finally, bulk RNA-sequencing (RNA-seq) was performed on the EEOs from which IOF and EOF were derived to understand their transcriptome (Fig. 1A).
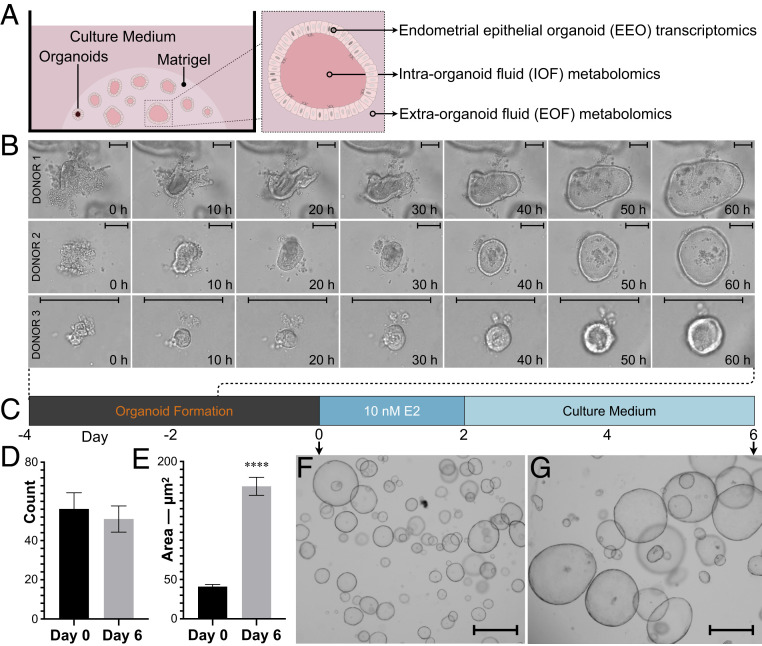
Fig. 1.
Study design and EEO monitoring. (A) Schematic depiction of study-dependent variables. (B) Time-lapse live imaging over 60 h of representative EEO formation from each donor. (Scale bars, 100 μm.) (C) Schematic depiction of the EEO preparation regimen including a 48-h 17β-estradiol (E2) exposure. Mean (±SEM) EEO (D) count and (E) areas on days 0 and 6 (n = 12); P ≤ 0.0001 (****). Representative EEO on days 0 (F) and 6 (G) at ×40 magnification. (Scale bars, 500 µm.)
Results
Isolation of EEO IOF.
The first approach to isolating IOF was aspiration by MMN, a technique commonly used for injecting material into oocytes and embryos. The primary advantage of MMN is clean IOF extraction, i.e., with minimal, if any, EOF or cell debris contamination. Disadvantages, however, include high user competency and time requirements, in addition to considerable equipment costs. Furthermore, IOF extraction by MMN is limited to a very small number of organoids, between 8 and 10 EEOs per donor in this study. This limitation results in a low IOF volume yield, translating to high IOF dilution for subsequent analysis, thereby compromising chromatographic resolution. Additionally, MMN does not allow for tandem multiomic (e.g., transcriptomic) analyses on the EEOs from which IOF was derived as easily as an HTC approach, and the requirement to overlay EEOs with mineral oil for MMN (Fig. 2A) may interfere with EOF biochemistry.
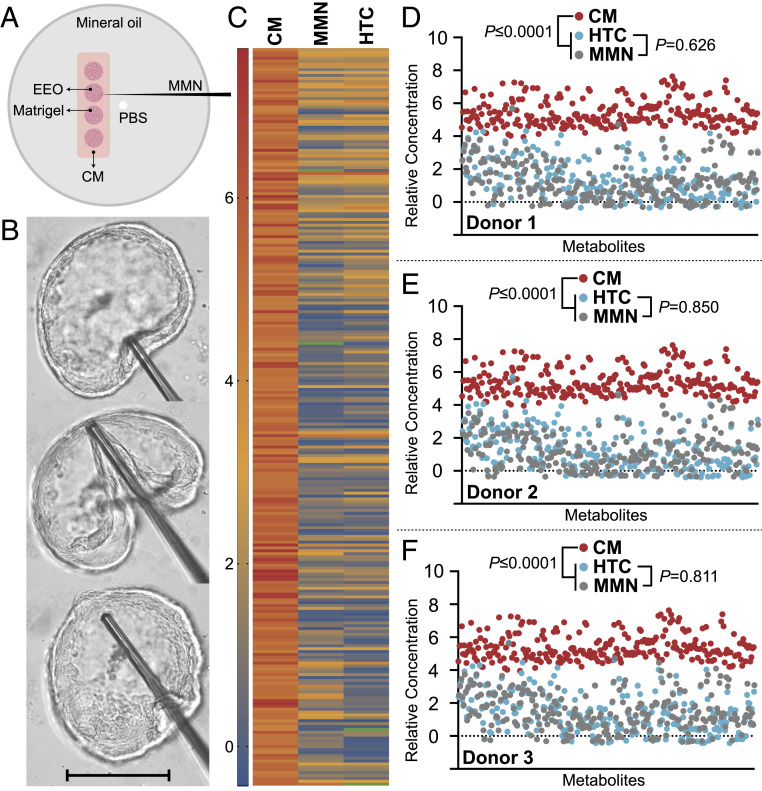
Fig. 2.
EEO IOF isolation validation. (A) Schematic depiction of the preparation for extracting IOF by MMN and transferring it to PBS. (B) Sequential images of IOF retrieval by MMN. (Scale bar, 250 μm.) (C) Mean normalized metabolomic profiles of IOF obtained by MMN vs. HTC from EEO vs. CM. Green depicts undetected metabolites. IOF profiles obtained by MMN and HTC were statistically identical (P = 0.735), whereas the CM metabolome differed from MMN and HTC counterparts (P ≤ 0.0001). (D–F) Metabolomic profiles of IOF obtained by MMN vs. HTC by donor vs. CM profiling. Legends and statistical comparisons between groups are provided above each plot.
To alleviate these constraints, a method for IOF extraction in a high-throughput capacity was developed. A centrifugal force of 3,750 × g for 15 min at 4 °C was determined sufficient for EEO disruption without compromising individual cell viability. This is in line with other human cell types, which can withstand centrifugal forces of up to 10,000 × g for up to 30 min without impacting their integrity (32). To compare the semiquantitative metabolomic profiles of IOF obtained by MMN vs. HTC, the following normalization parameters were applied. For MMN: 1) raw peak intensities were multiplied to account for gas chromatography-mass spectrometry (GC-MS) sample loading (technical normalization), to yield “adjusted peak intensities” before 2) the application of the following normalization formula: [(adjusted peak intensity) × (fold dilution)]/[(aspirated organoid count) × (mean organoid area)], wherein “fold dilution” refers to the dilution factor incurred by IOF transfer to phosphate buffered saline (PBS) (Fig. 2A), and “mean organoid area” serves as a biological material normalization factor. Thereafter, 3) normalized values were logarithmically transformed (Dataset S1). For HTC: 1) raw peak intensities were multiplied to account for PBS dilution during the IOF extraction process, before 2) dividing this value by the RNA concentration recovered in the residual cell pellet from which the IOF was derived, as a biological material normalization. Then, 3) normalized values were logarithmically transformed (Dataset S2). N.B.: For EOF metabolomic data, raw peak intensities were divided by the RNA concentration recovered in the residual cell pellet from which IOF and EOF were sampled, prior to logarithmic transformation (Dataset S2).
Comparing mean normalized semiquantitative metabolomic profiles of IOF obtained by MMN (Fig. 2B) compared to HTC found that both methods yield statistically identical (P = 0.735) results (Fig. 2C) from each donor (Fig. 2 D–F). As such, MMN and HTC are analogous approaches to isolating IOF. Moreover, the IOF metabolome differed (P ≤ 0.0001) to that of the culture medium (CM) provided basally at both the mean (Fig. 2C) and individual donor (Fig. 2 D–F) levels. Thus, EEO consistently form a biochemically selective barrier.
Qualitative Metabolomics of EEO Fluids.
Upon confirmation that HTC is a comparable approach to MMN for IOF isolation, the following were subjected to more thorough high-throughput untargeted ultrahigh-performance liquid chromatography (LC)-MS: HTC-derived IOF (n = 3); EOF (n = 3); unconditioned CM (n = 1); and Matrigel-conditioned medium (MCM; n = 1). IOF is representative of the uterine lumen (25, 30), whereas EOF is quasirepresentative of the uterine epithelial-stromal cell interface and underlying vasculature. The unconditioned or blank CM, in addition to MCM, served as negative controls. The latter was performed to determine whether biochemical leaching from Matrigel, a solubilized heterogeneous complex basement membrane protein (33), affects the biochemical composition of CM and, therefore, EEO fluid metabolomic profiles (34). Indeed, 40 metabolites were identified in MCM that were absent from CM (Fig. 3A and Dataset S2). It is worth noting that the same Matrigel batch was used throughout this study, to avoid potential lot-to-lot variation (8).
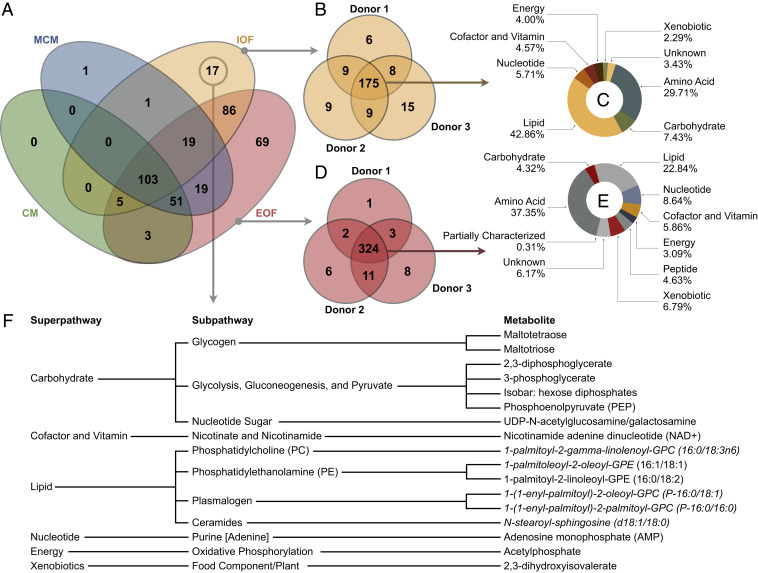
Fig. 3.
Qualitative organoid fluid metabolomics. (A) Venn diagram depicting the number of metabolites identified in organoid CM, MCM, IOF, and EOF across donors. (B) IOF metabolite presence by donor, in addition to (C) the mean superpathway breakdown of the IOF metabolites common to all donors. (D) EOF metabolite presence by donor, in addition to (E) the mean superpathway breakdown of the EOF metabolites common to all donors. (F) The 17 IOF-exclusive metabolites, arranged by pathway, wherein italicized metabolites are predicted.
In total, 374 metabolites were identified, of which 172 were EEO-derived, i.e., absent from both CM and MCM (Fig. 3A). Intriguingly, 231 metabolites were detected in IOF, of which 175 (75.8%) were common to IOF from all donors, showing a high degree of homogeneity in the qualitative (i.e., biochemical presence) metabolome of EEO IOF (Fig. 3B). Moreover, the IOF landscape was dominated by lipids (42.9%) and amino acids (29.7%), followed by carbohydrates (7.4%), among others (Fig. 3C). Regarding EOF, 355 metabolites were identified, of which 324 (91.3%) were common to EOF from all donors, showing even greater homogeneity in the EEO EOF metabolome, from a qualitative perspective (Fig. 3D). Unlike IOF, however, these EOF common metabolites primarily clustered under amino acids (37.4%) followed by lipids (22.8%), among others (Fig. 3E). Moreover, of the 172 EEO-derived metabolites, 86 (50%) were identified in both IOF and EOF, 69 (40%) were exclusive to EOF, and 17 (10%) were unique to IOF (Fig. 3A). Thus, in addition to EEO being a biochemically selective barrier, EEO metabolite turnover and secretion is asymmetrical about the apical-basolateral axis.
Quantitative Metabolomics of EEO Fluids.
Comparing the normalized semiquantitative metabolomic profiles of IOF vs. EOF (Dataset S2) by principal component analysis (PCA) highlights their clear separation (Fig. 4A), further demonstrating that IOF and EOF are biochemically distinct. In addition to differential qualitative metabolite presence (Fig. 3A), this is attributable to the increase (P ≤ 0.05) of 296 metabolites in EOF compared to IOF (Fig. 4A). Further contrasting EOF quantitative metabolic profiles by donor reveals metabolic signatures unique to EEO from each donor (Fig. 4 B and C), despite the high degree of homogeneity in respective qualitative metabolomes (Fig. 3). Thus, while qualitative metabolite presence in EOF is broadly comparable across donors, their relative concentrations are not. Total EOF metabolite levels also differed by donor (Fig. 4D).
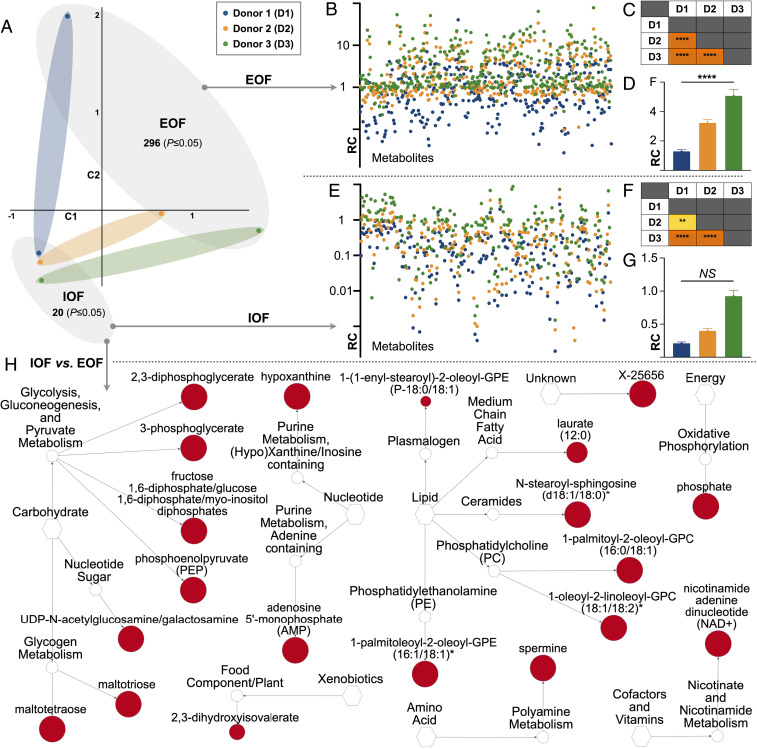
Fig. 4.
Quantitative intraorganoid fluid metabolomics. (A) Principal component analysis of IOF and EOF quantitative metabolic profiles from each donor in addition to the number of metabolites elevated (P ≤ 0.05) in each. (B) EOF metabolomic signatures by donor (relative concentration [RC]). (C) Corresponding EOF donor vs. donor statistical comparisons [P ≤ 0.0001 (****)] are also provided. (D) Mean total metabolite levels in EOF by donor. (E) IOF metabolomic signatures by donor. (F) Corresponding IOF donor vs. donor statistical comparisons [P ≤ 0.01 (**)] are also provided. (G) Mean total metabolite levels in IOF by donor (not significant [NS]). (H) Network view by pathway of the metabolites elevated (P ≤ 0.05) in IOF vs. EOF wherein node diameter is proportional to fold-change magnitude.
Regarding IOF, 20 biochemicals were elevated in IOF compared to EOF (Fig. 4A) alongside 8 additional metabolites which exhibited a trend (0.05 < P ≤ 0.10) toward an increase (Dataset S2). Interestingly, of the 20 metabolites elevated in IOF, just 12 correspond to the 17 IOF-exclusive metabolites (Fig. 3A), as concentrations of the remaining 5 IOF-exclusive metabolites did not differ. The 7 metabolites identified in both IOF and EOF but elevated (P ≤ 0.05) in IOF were: hypoxanthine, 1-(1-enyl-stearoyl)-2-oleoyl-GPE; 1-palmitoyl-2-oleoyl-GPC; 1-oleoyl-2-linoleoyl-GPC; laurate; spermine; and phosphate. In contrast, phenol red, a minimally metabolizable dye (35) and convenient CM internal standard, was 3,758-fold higher in EOF than IOF (Dataset S2), further exemplifying endometrial organoid epithelial biochemical selectivity. Lastly, comparing quantitative IOF profiles by donor revealed similarly unique EEO IOF metabolic signatures, as observed in EOF (Fig. 4 E and F); however, total metabolite levels in IOF were independent of EEO origin (Fig. 4G). These data combined demonstrate the ability to distinguish between EEO by donor based on IOF or EOF quantitative biochemical profiles alone.
EEO Transcriptome.
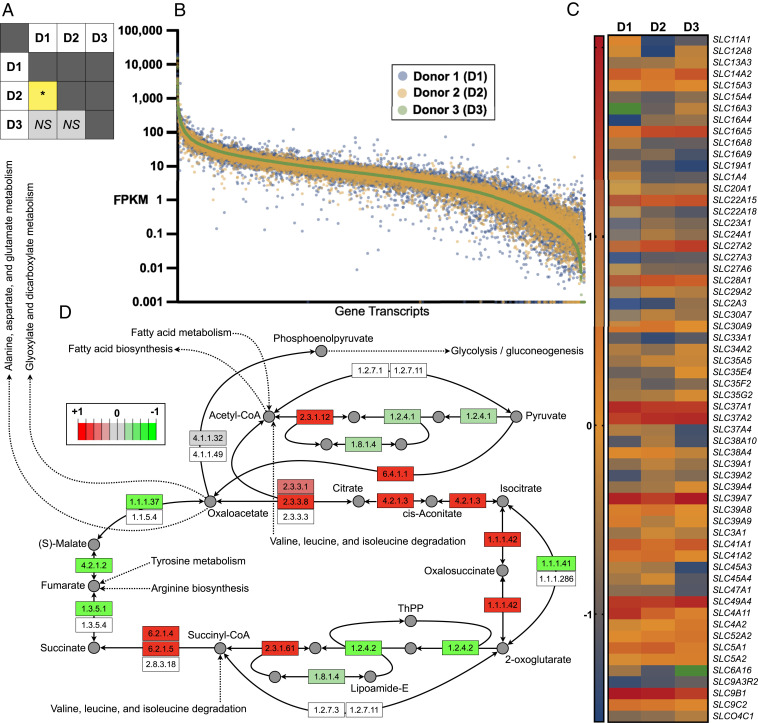
Bulk RNA-seq was performed on the EEO pellets from which IOF was extracted by HTC (i.e., in tandem). In total, 14,597 genes were expressed. Contrasting fragments per kilobase of transcript per million mapped reads (FPKM) values by donor revealed a different transcriptome of EEO from donors 1 and 2, but not donors 1 and 3 and donors 2 and 3 (Fig. 5A). Nonetheless, plotting FPKM values for each gene revealed donor-dependent expression variation (Fig. 5B). Focusing on the 301 solute carrier transporter (SLC) transcripts revealed no difference in their overall expression across donors, although expression of 60 differed by twofold or more between donors (Fig. 5C). Of these, 10% correspond to zinc transporters (SLC39), followed by monocarboxylate (SLC16; 8.3%) and nucleotide sugar (SLC35) transporters (6.7%). Gene ontology analysis of all transcripts revealed 14 (70%) of the top 20 directly related to metabolic processes (Dataset S3). Further interrogation of these transcriptomic data, specifically, comparing data from donor 1 vs. 2 using Pathview rendering, showed altered gene expression of several enzymes involved in the citric acid cycle (Fig. 5D), among others. Moreover, EEOs from all donors also expressed tight junction proteins ZO1-3 (TJ1-3), further corroborating EEO formation of a biochemically robust barrier. Complete RNA-seq data sets have been deposited at the Gene Expression Omnibus (GSE166289) (36).
Fig. 5.
Tandem organoid transcriptomics. (A) Statistical comparison [P ≤ 0.05 (*); not significant (NS)] of transcriptomic profiles of endometrial epithelial organoids obtained from all three donors. (B) Logarithmically transformed FPKM values per donor per gene (transcriptomic signatures) presented in descending order. (C) Relative transcript abundance of SLC protein genes, specifically, those exhibiting a ≥2.0-fold change between two or more donors. Green depicts undetected transcripts. (D) Pathview and Kyoto Ecyclopedia of Genes and Genomes (KEGG) database adapted extrapolation and rendering of transcriptomic profiles of donors 1 vs. 2 within the context of the citric acid cycle, wherein green denotes enzyme transcript-relative down-regulation and red depicts enzyme transcript-relative up-regulation.
Discussion
This study utilized existing technologies for the application of performing tandem multiomics experiments in organoids in a high-throughput capacity. From a technical perspective, we found that an optimized HTC approach to isolating EEO IOF was analogous to MMN. Thereafter, the HTC approach was used to determine that human EEO IOF and EOF are biochemically distinct and exhibit donor-specific biochemical signatures. Furthermore, IOF bears numerous key similarities to in vivo UF. This is a report of uterine epithelial apical fluid formation in vitro, in addition to basolateral fluid composition regulation, findings and methods directly and readily transferrable to other organoid systems.
Isolation of IOF has only been achieved by: 1) choroid plexus organoid aspiration, similarly to the MMN method presented here, to harvest in vitro cerebrospinal fluid (37); and 2) sonication for toxin extraction from snake venom gland organoids (38). Advantages of the former method include IOF aspiration with minimal, if any, EOF or cell debris contamination. However, IOF recovery from such few organoids may not be representative of the broad organoid population, particularly regarding endometrial organoids, in lieu of observed constituent cell heterogeneity (26). In contrast, the latter method is highly effective for select protein production and isolation in a high-throughput capacity; however, sonication lyses both organoids and constituent cells, rendering detailed interrogation of neat IOF challenging.
The endometrial secretome is complex and composed of proteins, cytokine peptides, ions, and metabolites, such as carbohydrates, amino acids, and lipids (2, 39–44). Perhaps the most notable metabolic trait of UF is relative hypoglycemia (44); Gardner et al. (39) measured glucose in UF from 15 women, reporting a mean of 3.2 ± 0.3 mM across all stages of the menstrual cycle. This value is in contrast to physiological systemic glucose concentrations in the region of 5.5 mM. Moreover, UF glucose levels remained unchanged in cattle intravenously infused with glucose to induce systemic hyperglycemia (45). While EOF is not directly comparable to serum, glucose was greatly reduced in IOF vs. EOF (Dataset S2), demonstrating that endometrial epithelia cultured as organoids in vitro form a selective barrier to glucose, similarly to in vivo. Furthermore, glucose transporters SLC2A1, SLC2A3, and SLC2A10-13, among others, in addition to SLC2A4RG (encoding the nuclear transcription factor involved in regulating SLC2A4 expression) (46), were expressed in EEOs. Glucose flux regulation by the uterine epithelium is particularly important as glucose, in turn, regulates human trophoblast proliferation (47). Specifically, glucose activates MTOR signaling, independently of PI3K, via intermediary metabolite activation, notably uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) (47). In addition to the central role of UDP-GlcNAc to trophoblast proliferation, it is the sole source of GlcNAc for O-linked protein glycosylation in the trophoblast (48). Intriguingly, UDP-GlcNAc was one of the 17 metabolites exclusively identified in IOF and was absent from EOF, CM, and MCM (Fig. 3F). It is tempting to suggest that UDP-GlcNAc, an integral substrate to embryo development, whose transporter (SLC35A3) (49) was expressed in EEO, may not be endogenously produced by the conceptus, but rather maternally derived. Given the established centrality of glucose and glycogen homeostasis to successful pregnancy establishment (44), it is perhaps unsurprising that 35% of IOF-exclusive metabolites pertained to glucose and glycogen metabolism (Fig. 3F).
Another metabolite exclusive to IOF was oxidized nicotinamide adenine dinucleotide (NAD+). The significance of NAD+ and its precursor, nicotinamide mononucleotide (NMN), to pregnancy has been recently elucidated. Genome sequencing of families with histories of congenital malformations revealed variants in two genes encoding enzymes involved in de novo NAD+ synthesis (50). Cuny et al. (51) subsequently found that maternal NAD+ deficiency—whether environmental or genetic in etiology—elevated congenital malformations and miscarriages in mice. Corroborating these data, NMN supplementation to murine embryos, derived from oocytes from aged mothers, restored embryo competence and development in vitro, and improved subsequent live birth rates (52). The fact that NAD+ is present in the in vitro human endometrial epithelial lumen, but not basolaterally (Fig. 3F), and is found extracellularly in other systems, rendering it unlikely to be artifactual (53), further supports this emerging notion that NAD+ plays a key role in human maternal-embryo communication and gestation. Indirectly linked to NAD+ and, therefore, conceptus redox homeostasis, are lactate and pyruvate, as their extracellular ratio (lactate-to-pyruvate ratio [LPR]) influences intracellular NAD+ levels (54). The LPR of serum is ∼15, whereas it is almost 60 in UF, on account of high lactate (5.9 ± 1.2 mM) as compared to pyruvate that is present in UF at levels comparable to those in circulation (0.1 ± 0.05 mM) (39). Intriguingly, lactate was present in IOF and EOF but not CM or MCM, whereas pyruvate was present in CM, MCM, and EOF, but absent from donor 1 and 3 IOF. Thus, the mean (±SD) EOF LPR was 3.0 ± 0.8, whereas the IOF LPR for donor 2 was 132. Although these in vitro LPR values are incomplete, and LPR values differ to in vivo counterparts, they do show that EEOs create an LPR gradient, the directionality of which is the same as that observed in vivo. Inconsistent pyruvate secretion by EEOs from different donors may be partially reflected in differential EEO monocarboxylate transporter (SLC16A) family expression. For instance, the expression of SLC16A3, which exhibits a particularly high affinity for pyruvate and is presumed to have evolved to retain cellular pyruvate for glycolytic NADH production prevention (55), was considerably variable among donor EEOs. SLC16A4, SLC16A5, SLC16A8, and SLC16A9 FPMK values were similarly inconsistent across EEOs from different donors (Fig. 5C).
Also closely related to glucose metabolism and NAD+ potentials is the adenosine monophosphate (AMP)-activated protein kinase (AMPK), which senses intracellular energy [ADP/ATP] status and glucose availability (56), and is expressed (PRKAB2) by the human morula (57). As AMPK activation by elevated AMP (another of the solely identified metabolites in IOF; Fig. 3F) leads to increased intracellular NAD+ (58), it may be that the endometrium has evolved two functionally redundant strategies for equipping the conceptus with NAD+—one direct (i.e., NAD+ secretion), the other indirect (i.e., AMP provision). Proper AMPK function is paramount to early embryo development, as perturbed activity is correlated to retarded murine blastocyst trophectoderm differentiation and poor tight junction formation (59, 60). Further, there are compelling data to suggest that pharmacological and environmental AMPK agonists contribute to miscarriages and assisted reproductive technology failures (61). Moreover, bovine early conceptus development has been recently hypothesized to revolve around a glucose-AMPK-PPARG (proliferator-activated receptor gamma) axis, modulated by uterine secretions of glucose, adenosine phosphates, and lipids (62–65). Whether such metabolic mechanisms are conserved in human embryos—in addition to the relative contribution of maternally derived lipids to human embryo development—remain open questions. While lipids dominated (42.9%) the IOF landscape, followed by amino acids (29.7%) (Fig. 3C), only a few studies have interrogated the lipid (41) and amino acid (43) composition of human UF. The former identified just 9 lipids, whereas the latter were limited to 18 free amino acids, in contrast to the 102 lipid and 125 amino acid moieties identified in the present study of EEOs. As such, the scarcity of comprehensive in vivo data (66, 67) render robust comparisons a challenge. Rectifying this is by thoroughly metabolically profiling uterine fluid from women, similarly to what has been conducted in cattle (63, 68–70), sheep (71), pigs (72), and mice (73) will shed much needed light on the biochemistry surrounding maternal-embryo communication in our species, and allow us to better evaluate the merits of IOF as a UF surrogate.
Perhaps the most interesting of the 69 EOF-exclusive metabolites identified (Fig. 3A) was adenosine 3′-5′-cyclic monophosphate (cAMP)—whose supplementation to culture media is required to induce stromal cell decidualization in vitro (74). While the mechanisms of extracellular cAMP secretion, via ATP-binding cassette C (ABCC) transporters, are well characterized in other systems (75, 76), the potential paracrine function of cAMP in endometrial epithelial-stromal cross-talk has yet to be established. Intriguingly, however, EEOs from all three donors expressed eight ABCCs, including ABCC4, whose dysregulation in vivo is associated with endometriosis (77). As such, we hypothesize that basolateral cAMP secretion by EEOs is not artifactual, rather a physiological phenotype previously unobservable in vitro and still so in vivo. Further intriguing EOF-exclusive substrates were dipeptides Cys-Gly, Tyr-Gly, and Pro-Gly. Given that glycine comprises approximately one-third of all collagen (78)—extracellular matrix protein synthesized and deposited by stromal fibroblasts (79)—it is tempting to suggest that EEOs basolaterally secrete glycine conjugates as stromal substrate for collagen production. Moreover, unconjugated glycine levels were 15-fold higher in EOF vs. IOF (Dataset S2). Teasing-out these hypotheses using EEO-stromal coculture systems, similarly to Abbas et al. (31), represent important areas for further research, as discussed below.
Despite the identification of 14,597 gene transcripts and profiling of 374 metabolites in the EEOs, greater EOF and IOF (Fig. 4) metabolomic variance was observed than transcriptomic variance (Fig. 5). Thus, metabolomics offers greater resolution in distinguishing between EEO donors. This, in addition to the fact that metabolomics offers several advantages over genomics, transcriptomics, and proteomics, as it represents the final result of interactions between genes, RNAs, and proteins (80), is an important consideration for personalized, or precision, medical applications, whose value to reproductive medicine is gaining traction (81, 82). Although the EEO sample size in this study is too small to determine whether IOF or EOF biochemistry correlates to donor attributes (e.g., age), metabolically profiling IOF, and EOF, from aged or infertile women, in a similar vein to Boretto et al. (30), may help us build an atlas of secreted substrates important to—and therefore predictors of—pregnancy success. Additional areas for future work include investigating the IOF and EOF proteome and determining the extent to which hormonal supplementation alters IOF and EOF composition. Hormonal supplementation is lacking from the current experimental design as existing data indicate that the stage of the menstrual cycle does not impact UF carbohydrate (39) or amino acid (43) composition, barring possibly taurine (83). However, investigating whether EOF composition is hormonally responsive and functionally relevant, e.g., capable of inducing stromal cell decidualization in vitro (84, 85), represents an interesting line for pursual. Furthermore, probing the influence of a pathophysiological endocrine milieu—mimicking aspects of subfertility-associated disorders such as polycystic ovary syndrome (i.e., hyperandrogenism) and diabetes mellitus (i.e., hyperglycemia)—on the EEO IOF and EOF secretome, may shed light on several metabolomic etiologies of pregnancy loss. Additional important areas for future work include investigating the extent to which Matrigel batch variation and EEO passage number influence EEO IOF and EOF dynamics. Moreover, the innovations presented here, coupled to gene editing technologies such as CRISPR-Cas9-mediated homology-independent organoid transgenesis (CRISPR-HOT) (86), represent an opportunity to conduct functional metabolomics for transporter function elucidation. Finally, the methods presented herein are directly and readily transferrable to organoids from other secretory cell types.
Materials and Methods
EEO Culture.
Primary human endometrial epithelial cells were derived from three donors. Donor 1 was a 30 y old undergoing a cystectomy 18 d postmenstruation. Donor 2 was a 25 y old admitted for bilateral tubal ligation 6 d postmenstruation. Donor 3 was a 37 y old with a body mass index (BMI) of 38, without history of irregular bleeding or endometriosis, and of proven fertility, also undergoing a bilateral tubal ligation. Current literature suggests that BMI does not alter the UF metabolome (43). Written informed consent was obtained from each donor and all experiments were approved by the University of Missouri Institutional Review Board.
EEO IOF Extraction by MMN.
To isolate IOF by MMN, EEOs from donors 1, 2, and 3 at their passage 12, 9, and 7, respectively, were plated to 100-mm Petri dishes (Fisher Scientific, FB0875712). Specifically, four 20-μL Matrigel domes per dish, in addition to a single 1.5-μL 1× PBS drop were positioned according to the schematic depiction in Fig. 2A. EEO cultures were then covered with organoid expansion culture medium (OEM), before overlaying the entire dish with mineral oil (Sigma, M8410). Mineral oil was required to enable the accurate deposition of aspirated IOF into the adjacent PBS drop (Fig. 2A). EEOs were maintained as such at 37 °C under 5% CO2 in air for 10 d without hormonal supplementation. IOF was subsequently manually extracted using a Leica DMi8 microscope coupled to an Eppendorf TransferMan 4r micromanipulator. EEOs were held in place by the surrounding Matrigel (Fig. 2B) and EEO IOF aspiration was achieved using an Eppendorf CellTram 4r Oil and Biopsy Tip I (Eppendorf, 5195000052). Eight, 10, and 10 EEOs, selected at random, were aspirated from donors 1, 2, and 3, respectively. Thereafter, the IOF was dispensed into the adjacent 1.5-μL PBS droplet (Fig. 2A). Each PBS drop—now also comprising IOF—was then transferred to a 500-μL conical tube, brought to a total volume of 25 μL with 1× PBS, snap frozen in N2(l), and stored at −80 °C until being sent on dry ice for analysis to the University of Missouri Metabolomics Core. To isolate IOF by HTC to compare against the MMN method, EEOs from donors 1, 2, and 3 at the same passage were plated to 4 wells of a 12-well plate each by seeding four 20 μL Matrigel domes per well and maintained in OEM at 37 °C under 5% CO2 in air for 10 d also without hormonal supplementation. Thereafter, IOF was extracted by HTC as described below.
EEO IOF Extraction by HTC.
Culture medium was aspirated from EEO culture wells and replaced with chilled 1× PBS twice. EEOs were then incubated with chilled Cell Recovery Solution (Corning, 354253) at 4 °C for 30 min. Using a 1-mL micropipette coupled to a wide-bore tip, EEOs were gently transferred to a 15-mL tube and centrifuged at 270 × g for 10 min at 4 °C. The supernatant was aspirated, and the pellet was gently resuspended in 3 mL chilled 1× PBS prior to recentrifugation and pellet resuspension in 250 μL chilled 1× PBS. EEOs were then transferred to a 1.5-mL tube and lightly vortexed for 5 min before centrifugation at 3,750 × g for 15 m at 4 °C. The resulting diluted IOF (supernatant) was transferred to a 1.5-mL tube, snap frozen in N2(l), and stored at −80 °C until analysis. The residual endometrial epithelial cell pellet was suspended in 1 mL TRIzol reagent (Invitrogen, 15596026) and stored at −80 °C until further processing.
EEO IOF vs. EOF Experimental Design.
EEO from donors 1, 2, and 3 were each transferred to 4 wells of a 12-well plate at a density of three 20-μL Matrigel domes per well and 10,000 cells per dome. Following 4 d of postpassage culture in OEM, the EEO culture medium was replaced by OEM lacking N2 supplement, instead comprising 10 nM 17β-estradiol (E2; Sigma E1024) for 48 h. EEOs were subsequently maintained in conventional CM—i.e., OEM without N2 supplement—for a further 96 h, with media replenishment every 48 h. Thereafter, 250 μL of 750 µL total EEO-conditioned CM (EOF) was aspirated from each well, pooled according to donor, snap frozen in N2(l), and stored at −80 °C until analysis. IOF was subsequently isolated by HTC as described above. In parallel, one well comprising four 20-μL Matrigel domes and CM alone was similarly maintained at 37 °C under 5% CO2 in air for 10 d. This MCM control, in addition to a 500-μL aliquot of the same CM, was also snap frozen in N2(l) and stored at −80 °C.
Full details pertinent to EEO culture, time-lapse imaging of EEO formation, metabolomics (MMN vs. HTC), metabolomics (IOF vs. EOF), metabolomic data normalization and presentation, EEO population and size analyses, and RNA isolation and sequencing, can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the T.E.S. laboratory and University of Missouri Metabolomics Core for helpful discussions. This work was supported by NIH Grant R01 HD096266 from the Eunice Kennedy Shriver National Institute of Child Health and Development and a Tier 2 award from the University of Missouri System Research and Creative Works Strategic Investment Program.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026804118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (accession no. GSE166289) (36). All study data are included in the article and/or supporting information.
References
- 1.Salamonsen L. A., Evans J., Nguyen H. P. T., Edgell T. A., The microenvironment of human implantation: Determinant of reproductive success. Am. J. Reprod. Immunol. 75, 218–225 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Spencer T. E., Biological roles of uterine glands in pregnancy. Semin. Reprod. Med. 32, 346–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton G. J., Watson A. L., Hempstock J., Skepper J. N., Jauniaux E., Uterine glands provide histiotrophic nutrition for the human fetus during the first trimester of pregnancy. J. Clin. Endocrinol. Metab. 87, 2954–2959 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Kelleher A. M., DeMayo F. J., Spencer T. E., Uterine glands: Developmental biology and functional roles in pregnancy. Endocr. Rev. 40, 1424–1445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer T. E., Kelleher A. M., Bartol F. F., Development and function of uterine glands in domestic animals. Annu. Rev. Anim. Biosci. 7, 125–147 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Pennings G., Uterine lavage: Ethics of research and clinical applications. Hum. Reprod. 35, 1949–1953 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Leese H. J., et al., Female reproductive tract fluids: Composition, mechanism of formation and potential role in the developmental origins of health and disease. Reprod. Fertil. Dev. 20, 1–8 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald H. C., Schust D. J., Spencer T. E., In vitro models of the human endometrium: Evolution and application for women’s health. Biol. Reprod. 104, 282–293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopal M., Tollner T. L., Finkbeiner W. E., Cherr G. N., Widdicombe J. H., Differentiated structure and function of primary cultures of monkey oviductal epithelium. In Vitro Cell. Dev. Biol. Anim. 42, 248–254 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Levanon K., et al., Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene 29, 1103–1113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S., Einspanier R., Schoen J., In vitro mimicking of estrous cycle stages in porcine oviduct epithelium cells: Estradiol and progesterone regulate differentiation, gene expression, and cellular function. Biol. Reprod. 89, 54 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Chen S., et al., An air-liquid interphase approach for modeling the early embryo-maternal contact zone. Sci. Rep. 7, 42298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miessen K., Sharbati S., Einspanier R., Schoen J., Modelling the porcine oviduct epithelium: A polarized in vitro system suitable for long-term cultivation. Theriogenology 76, 900–910 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Chen S., Einspanier R., Schoen J., Long-term culture of primary porcine oviduct epithelial cells: Validation of a comprehensive in vitro model for reproductive science. Theriogenology 80, 862–869 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Chen S., et al., In vitro mimicking of estrous cycle stages: Dissecting the impact of estradiol and progesterone on oviduct epithelium. Endocrinology 159, 3421–3432 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Simintiras C. A., et al., Modelling aspects of oviduct fluid formation in vitro. Reproduction 153, 23–33 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Gualtieri R., et al., Long-term viability and differentiation of bovine oviductal monolayers: Bidimensional versus three-dimensional culture. Theriogenology 78, 1456–1464 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Gualtieri R., et al., Bovine oviductal monolayers cultured under three-dimension conditions secrete factors able to release spermatozoa adhering to the tubal reservoir in vitro. Theriogenology 79, 429–435 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Simintiras C. A., Sturmey R. G., Genistein crosses the bioartificial oviduct and alters secretion composition. Reprod. Toxicol. 71, 63–70 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Chen S., Schoen J., Air-liquid interface cell culture: From airway epithelium to the female reproductive tract. Reprod. Domest. Anim. 54, 38–45 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Arnold J. T., Kaufman D. G., Seppälä M., Lessey B. A., Endometrial stromal cells regulate epithelial cell growth in vitro: A new co-culture model. Hum. Reprod. 16, 836–845 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Wang H., et al., A novel model of human implantation: 3D endometrium-like culture system to study attachment of human trophoblast (Jar) cell spheroids. Mol. Hum. Reprod. 18, 33–43 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Li D., et al., Development and characterization of a polarized human endometrial cell epithelia in an air-liquid interface state. Stem Cell Res. Ther. 9, 209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibaoui Y., Feki A., Organoid models of human endometrial development and disease. Front. Cell Dev. Biol. 8, 84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boretto M., et al., Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775–1786 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald H. C., Dhakal P., Behura S. K., Schust D. J., Spencer T. E., Self-renewing endometrial epithelial organoids of the human uterus. Proc. Natl. Acad. Sci. U.S.A. 116, 23132–23142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alzamil L., Nikolakopoulou K., Turco M. Y., Organoid systems to study the human female reproductive tract and pregnancy. Cell Death Differ. 28, 35–51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turco M. Y., et al., Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568–577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Z.-Y., Jia S.-Z., Liu S., Leng J.-H., Endometrial organoids: A new model for the research of endometrial-related diseases†. Biol. Reprod. 103, 918–926 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boretto M., et al., Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 21, 1041–1051 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Abbas Y., et al., Generation of a three-dimensional collagen scaffold-based model of the human endometrium. Interface Focus 10, 20190079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delahaye M., Lawrence K., Ward S. J., Hoare M., An ultra scale-down analysis of the recovery by dead-end centrifugation of human cells for therapy. Biotechnol. Bioeng. 112, 997–1011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes C. S., Postovit L. M., Lajoie G. A., Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886–1890 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Aisenbrey E. A., Murphy W. L., Synthetic alternatives to Matrigel. Nat. Rev. Mater. 5, 539–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Driscoll J. L., et al., Phenolsulfonphthalein (phenol red) metabolism in primary monolayer cultures of adult rat hepatocytes. In Vitro 18, 835–842 (1982). [DOI] [PubMed] [Google Scholar]
- 36.Simintiras C. A., Spencer T. E., Capture and metabolomic analysis of the human endometrial epithelial organoid secretome. National Center for Biotechnology Information (NCBI) Gene Expression Omnibus - Series GSE166289. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166289. Deposited on 5 February 2021. [DOI] [PMC free article] [PubMed]
- 37.Pellegrini L., et al., Human CNS barrier-forming organoids with cerebrospinal fluid production. Science 369, eaaz5626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Post Y., et al., Snake venom gland organoids. Cell 180, 233–247.e21 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Gardner D. K., Lane M., Calderon I., Leeton J., Environment of the preimplantation human embryo in vivo: Metabolite analysis of oviduct and uterine fluids and metabolism of cumulus cells. Fertil. Steril. 65, 349–353 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Cheong Y., Boomsma C., Heijnen C., Macklon N., Uterine secretomics: A window on the maternal-embryo interface. Fertil. Steril. 99, 1093–1099 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Vilella F., Ramirez L. B., Simón C., Lipidomics as an emerging tool to predict endometrial receptivity. Fertil. Steril. 99, 1100–1106 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Salamonsen L. A., et al., Proteomics of the human endometrium and uterine fluid: A pathway to biomarker discovery. Fertil. Steril. 99, 1086–1092 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Kermack A. J., et al., Amino acid composition of human uterine fluid: Association with age, lifestyle and gynaecological pathology. Hum. Reprod. 30, 917–924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dean M., Glycogen in the uterus and fallopian tubes is an important source of glucose during early pregnancy. Biol. Reprod. 101, 297–305 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Leane S., et al., The effect of exogenous glucose infusion on early embryonic development in lactating dairy cows. J. Dairy Sci. 101, 11285–11296 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Knight J. B., Eyster C. A., Griesel B. A., Olson A. L., Regulation of the human GLUT4 gene promoter: Interaction between a transcriptional activator and myocyte enhancer factor 2A. Proc. Natl. Acad. Sci. U.S.A. 100, 14725–14730 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J., Song G., Wu G., Bazer F. W., Functional roles of fructose. Proc. Natl. Acad. Sci. U.S.A. 109, E1619–E1628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen H. Y., Abbasi S., Kellems R. E., Xia Y., mTOR: A placental growth signaling sensor. Placenta 26, S63–S69 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Maszczak-Seneczko D., et al., UDP-N-acetylglucosamine transporter (SLC35A3) regulates biosynthesis of highly branched N-glycans and keratan sulfate. J. Biol. Chem. 288, 21850–21860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H., et al., NAD deficiency, congenital malformations, and niacin supplementation. N. Engl. J. Med. 377, 544–552 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Cuny H., et al., NAD deficiency due to environmental factors or gene-environment interactions causes congenital malformations and miscarriage in mice. Proc. Natl. Acad. Sci. U.S.A. 117, 3738–3747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertoldo M. J., et al., NAD+ repletion rescues female fertility during reproductive aging. Cell Rep. 30, 1670–1681.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cantó C., Menzies K. J., Auwerx J., NAD(+) metabolism and the control of energy homeostasis: A balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houtkooper R. H., Cantó C., Wanders R. J., Auwerx J., The secret life of NAD+: An old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 31, 194–223 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halestrap A. P., Meredith D., The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 447, 619–628 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Lin S. C., Hardie D. G., AMPK: Sensing glucose as well as cellular energy status. Cell Metab. 27, 299–313 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Hu B., et al., EmExplorer: A database for exploring time activation of gene expression in mammalian embryos. Open Biol. 9, 190054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cantó C., et al., AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young N. P., et al., AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 30, 535–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calder M. D., Edwards N. A., Betts D. H., Watson A. J., Treatment with AICAR inhibits blastocyst development, trophectoderm differentiation and tight junction formation and function in mice. Mol. Hum. Reprod. 23, 771–785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puscheck E. E., et al., Why AMPK agonists not known to be stressors may surprisingly contribute to miscarriage or hinder IVF/ART. J. Assist. Reprod. Genet. 35, 1359–1366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro E. S., Symposium review: Lipids as regulators of conceptus development: Implications for metabolic regulation of reproduction in dairy cattle. J. Dairy Sci. 101, 3630–3641 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Simintiras C. A., Sánchez J. M., McDonald M., Lonergan P., Progesterone alters the bovine uterine fluid lipidome during the period of elongation. Reproduction 157, 399–411 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Simintiras C. A., Sánchez J. M., McDonald M., Lonergan P., The biochemistry surrounding bovine conceptus elongation. Biol. Reprod. 101, 328–337 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Simintiras C. A., et al., Conceptus metabolomic profiling reveals stage-specific phenotypes leading up to pregnancy recognition in cattle. Biol. Reprod., 10.1093/biolre/ioab021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Wang Q., Wang H., Duan E., Uterine fluid in pregnancy: A biological and clinical outlook. Trends Mol. Med. 23, 604–614 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Bracewell-Milnes T., et al., Metabolomics as a tool to identify biomarkers to predict and improve outcomes in reproductive medicine: A systematic review. Hum. Reprod. Update 23, 723–736 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Simintiras C. A., et al., Biochemical characterization of progesterone-induced alterations in bovine uterine fluid amino acid and carbohydrate composition during the conceptus elongation window. Biol. Reprod. 100, 672–685 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Simintiras C. A., Sánchez J. M., McDonald M., Lonergan P., The influence of progesterone on bovine uterine fluid energy, nucleotide, vitamin, cofactor, peptide, and xenobiotic composition during the conceptus elongation-initiation window. Sci. Rep. 9, 7716 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moraes J. G. N., et al., Analysis of the uterine lumen in fertility-classified heifers: II. Proteins and metabolites. Biol. Reprod. 102, 571–587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J., et al., Select nutrients in the ovine uterine lumen. IX. Differential effects of arginine, leucine, glutamine, and glucose on interferon tau, ornithine decarboxylase, and nitric oxide synthase in the ovine conceptus. Biol. Reprod. 84, 1139–1147 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Walsh S. C., et al., Metabolic compounds within the porcine uterine environment are unique to the type of conceptus present during the early stages of blastocyst elongation. Mol. Reprod. Dev. 87, 174–190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Y., et al., Metabolic changes of maternal uterine fluid, uterus, and plasma during the peri-implantation period of early pregnancy in mice. Reprod. Sci. 27, 488–502 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Lucas E. S., et al., Recurrent pregnancy loss is associated with a pro-senescent decidual response during the peri-implantation window. Commun. Biol. 3, 37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jackson E. K., Mi Z., Dubey R. K., The extracellular cAMP-adenosine pathway significantly contributes to the in vivo production of adenosine. J. Pharmacol. Exp. Ther. 320, 117–123 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Hofer A. M., Lefkimmiatis K., Extracellular calcium and cAMP: Second messengers as “third messengers”? Physiology (Bethesda) 22, 320–327 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Gori I., et al., Augmented epithelial multidrug resistance-associated protein 4 expression in peritoneal endometriosis: Regulation by lipoxin A(4). Fertil. Steril. 99, 1965–73.e2 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Fidler A. L., Boudko S. P., Rokas A., Hudson B. G., The triple helix of collagens–an ancient protein structure that enabled animal multicellularity and tissue evolution. J. Cell Sci. 131, jcs203950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyazaki K., et al., Generation of progesterone-responsive endometrial stromal fibroblasts from human induced pluripotent stem cells: Role of the WNT/CTNNB1 pathway. Stem Cell Reports 11, 1136–1155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Souza R. T.et al.; Preterm SAMBA Study Group , Metabolomics applied to maternal and perinatal health: A review of new frontiers with a translation potential. Clinics (São Paulo) 74, e894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goetz L. H., Schork N. J., Personalized medicine: Motivation, challenges, and progress. Fertil. Steril. 109, 952–963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simon C., Sakkas D., Gardner D., Critchley H., Precision medicine in human reproduction. Special Issue. Biol. Reprod. 101, 1075 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Casslén B. G., Free amino acids in human uterine fluid. Possible role of high taurine concentration. J. Reprod. Med. 32, 181–184 (1987). [PubMed] [Google Scholar]
- 84.Haller M., Yin Y., Ma L., Development and utilization of human decidualization reporter cell line uncovers new modulators of female fertility. Proc. Natl. Acad. Sci. U.S.A. 116, 19541–19551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valdez J., et al., On-demand dissolution of modular, synthetic extracellular matrix reveals local epithelial-stromal communication networks. Biomaterials 130, 90–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Artegiani B., et al., Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat. Cell Biol. 22, 321–331 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (accession no. GSE166289) (36). All study data are included in the article and/or supporting information.