Significance
Endocytosis controls the proteome and lipidome of the plasma membrane and therefore the communication between the cell and the outside world. Because of its importance, many attempts have been made to develop tools to interfere with this process. These include genetic approaches and small chemicals. Thus far, however, the tools developed to modulate plant endocytosis either lack specificity, are irreversible, or require several days to produce an effect. Here, we generated a conditional and reversible tool to specifically interfere with endocytosis. Short-term heat treatment destabilizes an engineered adaptor complex subunit, which abolishes the function of the endocytic TPLATE complex. This causes accumulation of endocytic cargoes at the plasma membrane and provides a genetic background for future research.
Keywords: Arabidopsis, clathrin-mediated endocytosis, TPLATE complex, autophagy, temperature-sensitive mutant
Abstract
In plants, endocytosis is essential for many developmental and physiological processes, including regulation of growth and development, hormone perception, nutrient uptake, and defense against pathogens. Our toolbox to modulate this process is, however, rather limited. Here, we report a conditional tool to impair endocytosis. We generated a partially functional TPLATE allele by substituting the most conserved domain of the TPLATE subunit of the endocytic TPLATE complex (TPC). This substitution destabilizes TPC and dampens the efficiency of endocytosis. Short-term heat treatment increases TPC destabilization and reversibly delocalizes TPLATE from the plasma membrane to aggregates in the cytoplasm. This blocks FM uptake and causes accumulation of various known endocytic cargoes at the plasma membrane. Short-term heat treatment therefore transforms the partially functional TPLATE allele into an effective conditional tool to impair endocytosis. Next to their role in endocytosis, several TPC subunits are also implicated in actin-regulated autophagosomal degradation. Inactivating TPC via the WDX mutation, however, does not impair autophagy, thus enabling specific and reversible modulation of endocytosis in planta.
Endocytosis is an evolutionarily conserved eukaryotic pathway by which extracellular material and plasma membrane (PM) components are internalized via vesicles (1, 2). Clathrin-mediated endocytosis (CME), relying on the scaffolding protein clathrin, is the most prominent and the most studied endocytic pathway (3–5). As clathrin does not interact directly with the PM, nor does it recognize cargoes, adaptor proteins are required to act as essential links between the clathrin coat and the PM (6). In plant cells, material selected for CME is recognized by two adaptor complexes, the adaptor complex 2 (AP-2) and the TPLATE complex (TPC) (7–9). In contrast to TPC, single subunit mutants of AP-2 are viable (7, 8, 10–13) and AP-2 recruitment and dynamics appear to rely on TPC function (8, 14).
TPC represents an ancestral adaptor complex, which is however absent in present-day metazoans and yeasts. It was experimentally identified as an octameric complex in Arabidopsis and as a hexametric complex in Dictyostelium (8, 15). Plants, however, are the only eukaryotic supergroup identified so far where TPC is essential for life (8, 15), as knockout or severe knockdown of single subunits of TPC in Arabidopsis leads to pollen or seedling lethality, respectively (8, 13). Two TPC subunits, AtEH1/Pan1 and AtEH2/Pan1, were not associated with the other TPC core components when the complex was forced into the cytoplasm by truncating the TML subunit and did not copurify with the other TSET components in Dictyostelium. This indicates that they may be auxiliary components to the core TPC (8, 15). These AtEH/Pan1 proteins were recently identified as important players in actin-regulated autophagy in plants. AtEH/Pan1 proteins recruit several components of the endocytic machinery to the autophagosomes, and are degraded together with them under stress conditions (16). However, whether this pathway serves to degrade specific cargoes or to regulate the endocytic machinery itself (17), and whether the whole TPC is required for this degradation pathway, remains unclear.
Genetic and chemical tools to manipulate endocytosis have been extensively investigated via interfering with the functions of endocytic players, such as clathrin (18–22), adaptor proteins (7, 10–12, 14, 23–25), and dynamin-related proteins (26–30). The chemical inhibitors originally used to affect CME in plants have recently been described to possess undesirable side effects (31) or to affect proteins that are not only specific for endocytosis: for example, clathrin itself, as it is also involved in TGN trafficking (19, 22). The same is true for several genetic tools currently available to affect CME in plants (18, 21, 22, 30). Manipulation of TPC, functioning exclusively at the PM, represents a very good candidate to affect CME more specifically. So far however, there are no chemical tools to target TPC functions or dominant-negative mutants available. Inducible silencing works, but causes seedling lethality and takes several days to become effective (8). The only tools to manipulate TPC function in viable plants consist of knock-down mutants with very mild reduction of expression and consequently similar mild effects on CME (8, 14, 16, 32).
Results and Discussion
Mutations in the Evolutionarily Conserved WDX Domain of TPLATE Destabilize TPC.
In this study, we employed a targeted mutagenesis strategy to obtain stronger partial loss-of-function alleles, which would however still allow us to overcome the genetic barriers caused by TPC single subunit mutations. We mutated selected evolutionary conserved motifs in the trunk domain of TPLATE, a fundamental subunit of TPC, and assessed the functionality of these isoforms by their capacity to complement the tplate loss-of-function mutant (SI Appendix, Fig. S1 A and B). We selected the motifs to be mutated either because of an earlier indication of their functionality (EF-loop) (8, 33–35), because of amino acid conservation across plant species (Linker subdomain) or across all studied species (WDX domain, after WenDingXing, which means “stability” in Chinese). These conserved amino acid residues were mutated to alanine, glycine, or serine based on the original amino acid composition (Fig. 1A and SI Appendix, Fig. S1A). Our substitutions aimed to maintain similar flexibility and hydrophobicity, not to completely abolish TPLATE function.
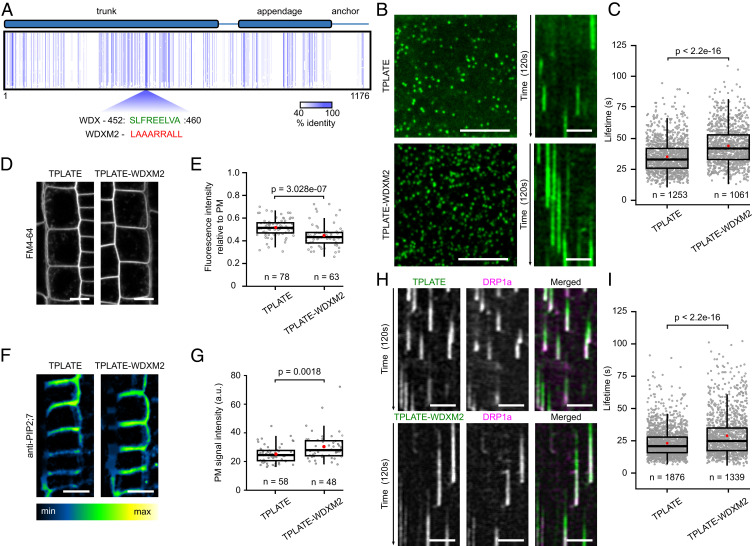
Fig. 1.
Substituting the WDX domain of TPLATE delays endocytosis. (A) Schematic representation of TPLATE domain organization and sequence conservation. Arabidopsis TPLATE was aligned against various eukaryotic species and the schematic view of the amino acid alignment data normalized on the Arabidopsis TPLATE sequence shows highly conserved amino acids as vertical blue lines. The conserved sequence of the WDX motif is shown in green and the corresponding substituted amino acids in WDXM2 are below in red. Numbers represent the amino acid positions of the WDX motif within Arabidopsis TPLATE. (B) Representative spinning disk images and related kymographs showing endocytic foci and lifetimes of TPLATE and TPLATE-WDXM2 in etiolated Arabidopsis epidermal hypocotyl cells. (Scale bars, 7 μm for spinning disk images and 25 µm for kymographs.) (C) Lifetime quantification of TPLATE- and TPLATE-WDXM2–marked foci in etiolated epidermal hypocotyl cells. The number of events analyzed is indicated. At least 12 movies from 4 seedlings for each transgenic line were analyzed. (D and E) Representative confocal images and quantification of FM4-64 uptake in TPLATE and TPLATE-WDXM2 in root epidermal cells. Numbers represent the amount of cells analyzed. At least eight individual seedlings for each transgenic line were imaged. (Scale bars, 10 μm.) (F and G) Representative immunolocalization images and quantification of fluorescence intensity at the PM of endogenous PIP2;7 in roots from TPLATE- and TPLATE-WDXM2–complemented lines. (Scale bars, 10 µm.) Numbers of measured cells are indicated at the bottom. At least nine independent roots for each genotype were imaged and measured. (H and I) Kymograph analysis and lifetime quantification of DRP1a in TPLATE and TPLATE-WDXM2 epidermal hypocotyl cells. (Scale bars, 25 μm.) The number of events analyzed is indicated. At least 10 movies from 3 seedlings for each transgenic line were analyzed. Red circles represent the mean in C, E, G, and I. The P values were calculated by the Mann–Whitney U test.
The LAT52p-driven TPLATE complements the male-sterility phenotype of the tplate T-DNA insertion line (13). We evaluated the functionality of our TPLATE isoforms, taking advantage of this strategy. Unlike substitutions of the sandwich and platform subdomains located in the appendage domain, which abolish assembly of TPC (36), our complementation assays confirmed that mutations in these selected motifs of TPLATE did not abolish its functionality (SI Appendix, Fig. S1C and Table S1).
The dynamic behavior of endocytic proteins is correlated with the efficiency of endocytosis as reduced or prolonged lifetimes correlate with aborted or deficient CME, respectively (12, 14, 18). All TPC subunits are recruited together to highly dynamic endocytic spots at the PM (8, 35, 37). We evaluated whether our TPLATE isoforms showed affected dynamic behavior at the PM and, therefore, would have a potential effect on CME. Lifetime analyses showed that all isoforms were dynamically recruited to the PM and exhibited various resident lifetimes (SI Appendix, Fig. S2 A and B). Of all isoforms tested, those where the most conserved WDX domain was substituted (independently in TPLATE-WDXM1 and in TPLATE-WDXM2) prolonged the resident lifetime most severely and were chosen for further analysis. TPLATE-WDXM1 and M2 showed increased densities of endocytic dots at the PM as well as reduced FM4-64 uptake compared to control lines (Fig. 1 A–E and SI Appendix, Fig. S2). TPLATE-WDXM2, which is an extension of the TPLATE-WDXM1 substitution (SI Appendix, Fig. S1A), reduced FM uptake more severely compared to TPLATE-WDXM1 and had protein expression levels comparable to the original complemented line (SI Appendix, Fig. S1C). We therefore largely focused on that particular isoform. Along with the reduced FM4-64 uptake, we also observed increased PM accumulation of the aquaporin PIP2;7 in TPLATE-WDXM2–complemented plants compared to TPLATE-GFP–complemented plants (Fig. 1 F and G). The above results suggest a minor impairment of endocytic capacity in TPLATE-WDXM2–complemented plants.
To further correlate the delay in dwell-time at the PM with the other CME machinery, we examined the dynamics of DRP1a-mRFP in TPLATE- and TPLATE-WDXM2–complemented lines. Consistent with the prolonged lifetimes of the mutated TPLATE isoform, DRP1a also exhibited prolonged lifetimes in TPLATE-WDXM2–complemented lines (Fig. 1 H and I).
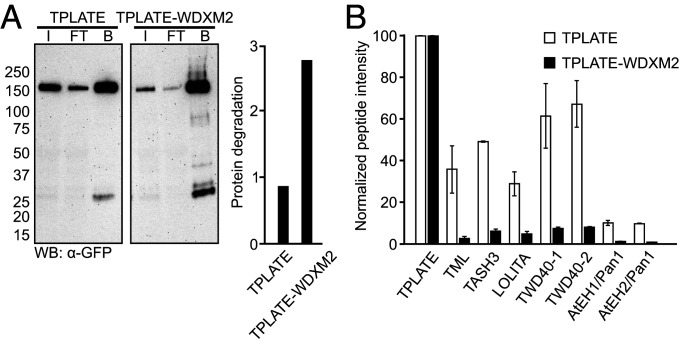
Using an integrative structural approach, we recently revealed TPC architecture and showed the central location of the TPLATE subunit inside TPC and its crucial role for complex assembly (36). Given the central position of the TPLATE subunit in TPC, we therefore evaluated whether substitutions in the WDX domain affected complex assembly and stability. Coimmunoprecipitation (co-IP) analysis revealed an increased degradation of the bait proteins TPLATE-WDXM1 and TPLATE-WDXM2 compared to TPLATE-GFP (Fig. 2A and SI Appendix, Fig. S3A). Modifications in the WDX domain therefore destabilize TPLATE. The results are consistent with the integrative TPC structure, revealing that the WDX domain is a part of the region exhibiting a substantial amount of intramolecular interactions (36). Affinity purification–mass spectrometry analysis further confirmed reduced levels of the whole TPC in the WDX domain substituted complemented lines compared to TPLATE-GFP–complemented lines. Relative to the bait protein, the average intensity of commonly detected peptides for all other TPC subunits was reduced in TPLATE-WDXM–complemented lines compared to the original TPLATE-complemented line (Fig. 2B and SI Appendix, Fig. S3B). Together, these results imply that substitutions in the WDX domain of TPLATE delay endocytosis by lowering the amount of TPC.
Fig. 2.
Substitutions in its WDX domain destabilize TPLATE and affect TPC levels. (A) GFP-Trap of complemented TPLATE-GFP and WDXM2-GFP substitution lines. Input (I), flow through (FT), and bound (B) fractions were analyzed using an anti-GFP antibody. Protein degradation was quantified as the ratio between full-length protein and all smaller bands within each lane. (B) MS analysis following co-IP on complemented TPLATE-GFP and TPLATE-WDXM2-GFP lines. For each TPC subunit, the intensities of only those peptides that were present in all experiments (both baits and all in all replicas) were averaged and normalized to the values of the corresponding bait protein. SDs are based on three technical repeats.
Conditional Destabilization of WDX Does Not Impair Autophagy.
These partially functional WDXM-complemented lines provide us with an opportunity to study the involvement of TPC in endocytosis, as well as to evaluate whether the whole TPC is required for actin cytoskeleton-regulated autophagy from the endoplasmic reticulum–PM contact sites. Autophagy-defective mutants are typically characterized by their hypersensitivity to nutrient deprivation (38–47). Down-regulation of AtEH/Pan1 leads to susceptibility to nutrient deficiency, especially fixed carbon depletion stress, and the seedlings phenocopy atg5 mutants (16). To evaluate whether destabilizing TPC results in susceptibility to nutrient deficiency, we compared TPLATE-, TPLATE-WDXM2–complemented lines, and atg mutants under fixed carbon starvation conditions. Long-term constant dark exposure leads to significant reduction of root growth as well as yellowing of the cotyledons in atg5 and atg7 mutants. This was not the case for the TPLATE- and TPLATE-WDXM2–complemented lines (SI Appendix, Fig. S3 C–E). This indicates that the AtEH/Pan1-dependent autophagy pathway is not impaired in TPLATE-WDXM–complemented lines under fixed carbon starvation.
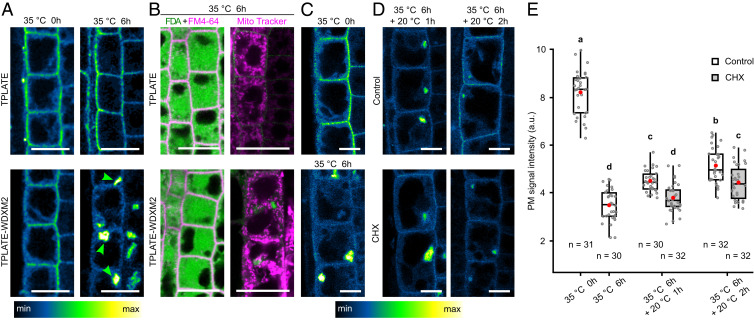
In vivo, functional protein yields, solubility, and proper cellular or extracellular localization are often correlated with protein stability (48). Protein misfolding and aggregation can be accelerated under stress conditions, like high temperature, denaturing conditions, or altered pH (48). To enhance the destabilization of WDXM-containing TPC, we used a short-term heat treatment. Time-course monitoring of TPLATE-GFP localization in complemented lines revealed that, in contrast to TPLATE, short-term heat treatment (35 °C, up to 6 h) incrementally abolished its PM localization and caused a complete loss of the PM signal after 6 h (SI Appendix, Fig. S4). The loss of PM signal correlated with induced aggregation of TPLATE-WDXM2 in the cytoplasm (Fig. 3A and SI Appendix, Fig. S4). FM4-64, fluorescein diacetate (FDA), as well as Mitotracker red staining confirmed the cell viability following the treatment (49, 50). After 35 °C treatment for 6 h, both TPLATE- and TPLATE-WDXM2–complemented plants were able to convert FDA to its fluorescent form, maintained PM integrity, and showed similar mitochondria staining, confirming that cell viability and mitochondrial function was not affected by the treatment (Fig. 3B). Moreover, the heat-treated plants recovered at room temperature, whereby the aggregates disappeared and PM recruitment of TPLATE-WDXM2 returned (Fig. 3 C and D). The recovery was delayed when translation was inhibited using cycloheximide (Fig. 3 C and D), suggesting a substantial contribution of the de novo protein synthesis in the recovery process.
Fig. 3.
Short-term heat treatment induces an incremental and reversible aggregation of TPLATE-WDXM2 in the cytoplasm. (A) Confocal images of root cells from TPLATE-GFP– and TPLATE-WDXM2-GFP–complemented lines subjected to normal conditions (20 °C, indicated as 35 °C 0 h) or heat treatment (35 °C 6 h). Arrowheads indicate the aggregated TPLATE-WDXM2 in the cytoplasm. (Scale bars, 20 µm.) (B) Confocal images of FDA and MitoTracker red (CM-H2XRos) staining in TPLATE- and TPLATE-WDXM2–complemented lines after 6 h heat treatment. (Scale bars, 25 µm.) (C–E) Confocal images (C and D) and quantification (E) of PM signal intensity in root cells from TPLATE-WDXM2-GFP–complemented lines before and after heat treatment. Seedlings after heat treatment were recovered in liquid 1/2 MS medium supplemented with or without 50 µM cycloheximide (CHX) at 20 °C. (Scale bars, 10 µm.) Numbers at the bottom represent the number of cells analyzed from at least six individual roots for each time point. Red circles represent the mean and letters represent significantly different groups (P < 0.001) evaluated by the Tukey multiple pairwise-comparisons test.
Short-term heat shock was recently employed as a strategy to study autophagy in plants (51–53). In our hands, time-course experiments monitoring autophagic body formation visually indicated autophagosome formation in roots expressing the autophagic vesicle marker YFP-ATG8a within 1 h of heat treatment (SI Appendix, Fig. S5A). Moreover, Concanamycin A (ConcA) treatment, which suppresses autophagic body breakdown and stabilizes their content in vacuoles (16, 51, 54), confirmed that autophagic flux was maintained under our conditions (SI Appendix, Fig. S5B).
To test if destabilizing TPC would impair autophagy, we phenotypically compared TPLATE- and TPLATE-WDXM–complemented lines with atg mutant lines under long-term heat stress. TPLATE-WDXM inactivation under these stress conditions affected Arabidopsis seedling development more severely than the atg mutations did. Root growth was reduced and the cotyledons showed extensive whitening, which was not observed in any of the other lines tested (SI Appendix, Fig. S6). The effects of blocking endocytosis therefore outweigh those of blocking autophagy under the applied stress conditions. The question whether autophagy was affected upon TPLATE-WDXM inactivation thus remained unanswered.
We therefore compared autophagosome formation directly in TPLATE, TPLATE-WDXM2–complemented lines, and atg mutants expressing the autophagic marker mCherry-ATG8e. Although the TPLATE protein was removed from the PM and aggregated in the cytoplasm, TPLATE-WDXM2–complemented plants exhibited comparable amounts of mCherry-ATG8e–labeled autophagosomes. Conversely, mCherry-ATG8e–labeled autophagosomes were not observed in both atg mutants (SI Appendix, Fig. S5 C and D). The detection of autophagosomes in TPLATE-WDXM2–complemented lines suggested that destabilizing TPC does not affect autophagy under heat stress. Besides, in TPLATE-WDXM2 plants, partial colocalization between mCherry-ATG8e–labeled autophagosomes and the aggregated TPLATE spots indicated that autophagy likely plays a role in clearing the aggregated TPLATE proteins destabilized by heat stress (SI Appendix, Fig. S5C).
AtEH/Pan1 proteins are important for autophagosome formation as overexpression of AtEH/Pan1 boosts autophagosome formation while down-regulation impairs this (16). AtEH/Pan1 proteins recruit other endocytic proteins, including several TPC subunits, AP-2 subunits, and clathrin to AtEH/Pan1-labeled autophagosomes (16). The observation that TPLATE-WDXM2 aggregation by heat did not impair autophagosome formation would imply that AtEH/Pan1 function is not affected under these conditions. We therefore examined whether AtEH/Pan1 proteins would be coaggregated along with TPLATE-WDXM2. We monitored AtEH2/Pan1 localization in the TPLATE-WDXM2–complemented lines under the short heat treatment. Aggregated TPLATE-WDXM2 failed to recruit AtEH2/Pan1, which remained at the PM (SI Appendix, Fig. S5E). The observations that AtEH/Pan1 proteins were not aggregated together with TPLATE-WDXM2, their role in autophagy, and the fact that autophagy was not inhibited upon functional inactivation of TPLATE favors the hypothesis that AtEH/Pan1 proteins might drive autophagosome formation independently of the other TPLATE complex subunits.
Conditional Destabilization of WDX Impairs Endocytosis.
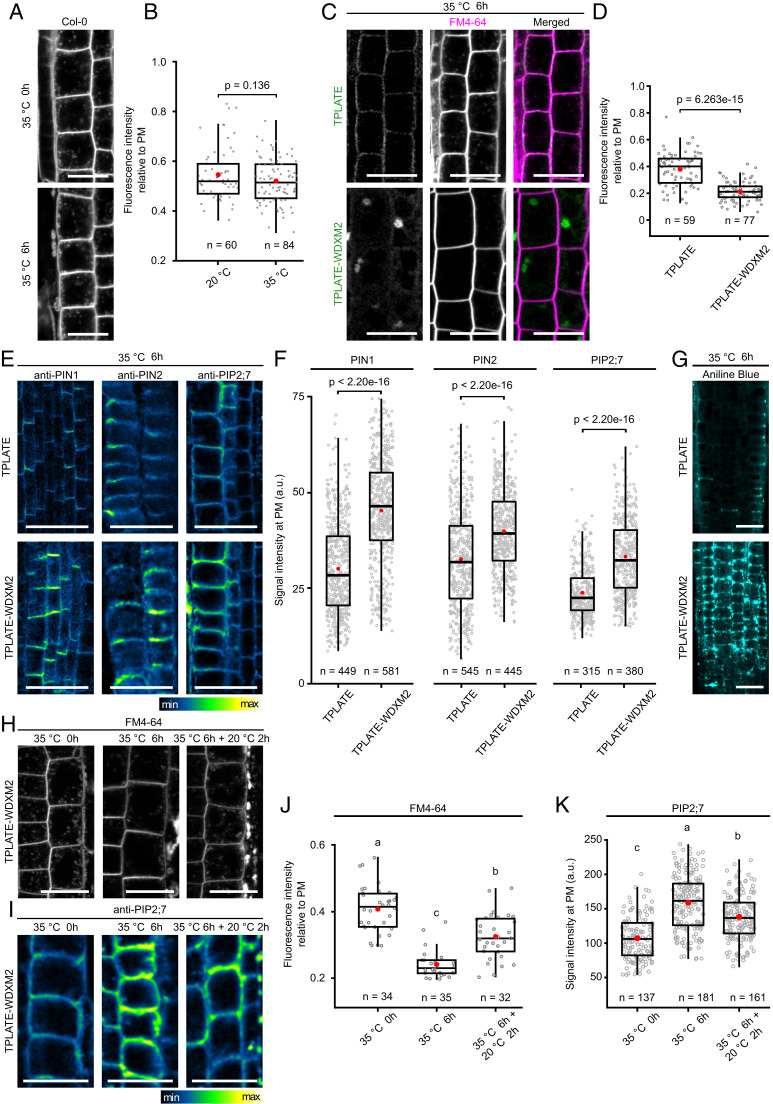
TPC is hypothesized to function as an early adaptor protein complex that internalizes PM cargoes during CME in plants (8, 32). Our data already showed that destabilizing TPC partially affected the efficiency of endocytosis under normal conditions (Fig. 1 D and E). Given that the destabilized TPLATE protein was removed from the PM and aggregated in the cytoplasm, we further addressed the endocytic capacity of WDXM2-complemented lines under short-term heat stress. As an initial control, we evaluated whether short-term heat treatment would affect endocytosis. We compared FM4-64 uptake in Col-0 seedlings before and after heat treatment. Quantification of internalization showed that CME was not significantly affected by increasing the temperature (Fig. 4 A and B). In contrast and in line with the aggregation of TPLATE-WDXM2 in the cytoplasm, FM4-64 uptake after short-term heat stress was strongly impaired (Fig. 4 C and D). The cytoplasm appeared devoid of FM4-64–labeled endosomes, similarly to what was observed upon strong down-regulation of TPC single subunits (8).
Fig. 4.
Destabilizing TPLATE-WDXM2 by short-term heat stress impairs endocytosis. (A and B) Representative confocal images (A) and quantification (B) of FM4-64 uptake in WT Col-0 seedlings before and after 6 h heat treatment. (Scale bars, 20 µm.) Numbers at the bottom represent the number of cells analyzed from nine individual roots for each temperature. (C and D) Representative confocal images (C) and quantification (D) of FM4-64 uptake in TPLATE- and TPLATE-WDXM2–complemented lines after 6-h heat treatment. (Scale bars, 25 µm.) Numbers at the bottom represent the number of cells analyzed from 10 roots for each transgenic line respectively. (E and F) Representative immunolocalizations (E) and quantification (F) of fluorescence intensity on the PM of endogenous PIN1, PIN2, and PIP2;7 proteins after 6 h heat treatment in roots from TPLATE- and TPLATE-WDXM2–complemented lines. (Scale bars, 25 µm.) Numbers of measured PM are indicated at the bottom. At least 18 transgenic plants for each protein were measured. (G) Confocal images of Aniline blue staining after 6-h heat treatment in roots from TPLATE- and TPLATE-WDXM2–complemented lines. (Scale bars, 25 µm.) (H and J) Representative confocal images (H) and quantification (J) of FM4-64 uptake in TPLATE-WDXM2–complemented seedlings before and after heat treatment as well as following a 2-h recovery period. (Scale bars, 20 µm.) Numbers at the bottom indicate measured cells from at least five seedlings. (I and K) Representative immunolocalization images (I) and quantification (K) of PM fluorescence intensity of endogenous PIP2;7 proteins before and after heat treatment as well as following a 2-h recovery period in roots from TPLATE-WDXM2–complemented lines. (Scale bar, 25 µm.) Numbers at the bottom indicate measured cells from at least nine seedlings. Red circles represent the mean in B, D, F, J, and K. The P values in B, D, and F were calculated by the Mann–Whitney U test. Letters in J and K represent significantly different groups (P < 0.001) evaluated by the Tukey multiple pairwise-comparisons test.
To further evaluate the specific internalization of PM cargoes, we examined the PM levels of several reported cargoes that undergo CME via immunolocalization (8, 20, 55). Consistently with the reduction of the FM4-64 internalization, we observed increased PM accumulation of the auxin efflux carriers PIN1 and PIN2 (56, 57) and the aquaporin PIP2;7 (58) in the TPLATE-WDXM2–complemented plants compared to control lines (Fig. 4 E and F). Loss-of-function of TPLATE correlates with ectopic callose deposition in mature pollen as well as in Arabidopsis roots (13). Aniline blue staining confirmed an ectopic callose deposition in TPLATE-WDXM2– compared with TPLATE-complemented lines after short-term heat treatment (Fig. 4G). To evaluate the reversibility of endocytic capacity of TPLATE-WDXM2 plants upon short-term heat treatment, we compared FM4-64 uptake as well as aquaporin PIP2;7 PM accumulation after a short recovery at 20 °C. In line with the reversibility of the TPLATE-WDXM2 localization, we observed a partial recovery of FM4-64 uptake as well a partial recovery of the internalization of PIP2;7 (Fig. 4 H–K). Together, these results support that the heat-inducible cytoplasmic aggregation of TPLATE in our TPLATE-WDXM2–complemented line inactivates TPC, and represents a tool to selectively and reversibly impair endocytosis in Arabidopsis.
In conclusion, we generated a partially functional TPLATE allele via substituting the WDX domain of TPLATE, which allows to conditionally destabilize TPC. By generating the TPLATE-WDXM2–complemented tplate(−/−) line, we developed a highly specific and reversible tool to inhibit plant CME. Our tool works directly on the protein level and therefore is faster than the previously reported artificial microRNA approaches (8). We demonstrate that this tool is efficient in disrupting CME without impairing autophagosome formation under short-term heat treatment.
Methods
Construction of Multiple Sequence Alignment.
To identify TPLATE homologs, the predicted proteins of each genome were searched using BLASTP (59) with Arabidopsis TPLATE as an input sequence. Used databases were GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and the Joint Genome Institute (https://genome.jgi.doe.gov/portal/). Multiple sequence alignment (Dataset S1) was constructed with the MAFFT algorithm in einsi mode (60) and visualized by the Jalview program (61)
Molecular Cloning.
Primers used to generate TPLATE domain substitution fragments are listed in SI Appendix, Table S2. Constructs carrying various mutations in conserved regions of TPLATE were generated by mutagenesis PCR. Two-step PCR reactions were performed to introduce the mutations. The entry clone pDONR207-TPLATE without stop codon (62) was used as a template with the combinations of sewing primers and mutation primers to obtain the mutated fragments, respectively. Then sewing PCR reactions were performed with the mixture of mutation fragments as template and combinations of sewing primers. To generate the TPLATE-EFM2 construct, two-step PCR reactions were performed with a TPLATE–EF1 construct as a template. To generate domain substituted TPLATE entry clones, all sewing PCR products were introduced into pDONR221 via Gateway BP reactions (Invitrogen) and confirmed by sequencing. To yield the expression constructs, the TPLATE domains substituted entry clones were combined with pB7m34GW (63), pDONRP4-P1r-Lat52 (13), and pDONRP2R-P3-EGFP (13) in triple gateway LR reactions (Invitrogen).
Plant Materials and Growth Conditions.
All the plant materials used in this research are in the Columbia-0 (Col-0) ecotype and listed in SI Appendix, Table S3. To generate transgenic lines expressing isoforms of TPLATE, tplate heterozygous mutant plants that were confirmed by genotyping PCR were transformed by floral dip with various expression constructs of TPLATE substitution motifs fused to GFP under the control of the pLAT52 promoter, similar to the original complementation approach (13). To identify transgenic plants possessing the tplate T-DNA insertion, the primary transformants (T1 plants) expressing TPLATE isoforms were selected with 10 mg/L Basta and further identified by genotyping PCR. The T2 plants expressing TPLATE isoforms were genotyped again to identify homozygous tplate mutants. Genotyping PCR was performed with genomic DNA extracted from rosette leaves. Genotyping LP and RP primers for tplate have been described previously (13), and the T-DNA–specific primer (LBb 1.3) is described on the SIGnAL website (http://signal.salk.edu/tdnaprimers.2.html).
The UBQ10p::YFP-ATG8a and UBQ10p::mCherry-ATG8e reporter lines, atg5-1 and atg7-3 mutants have been described previously (16). The same male parent expressing mCherry-ATG8e was used to pollinate atg5-, atg7-, TPLATE-, and TPLATE-WDXM2–complemented lines. F2 plants were identified by genotyping PCR to obtain homozygous mutant background (atg5-1, atg7-3, tplate for TPLATE and TPLATE-WDXM2). The progeny expressing mCherry-ATG8e were selected based on fluorescence. The dual-color line expressing LAT52p::TPLATE-GFP and H3.3p::AtEH2-mRuby3 in the double-complemented mutant (tplate/tplate ateh2/ateh2) was described in Wang et al. (16). Similarly, the pLAT52p::TPLATE-WDXM2-GFP–complemented line was crossed with the H3.3p::AtEH2/Pan1-mRuby3–complemented line (16). F2 plants were identified by genotyping PCR to obtain double homozygous mutant (tplate/tplate ateh2/ateh2) backgrounds.
Seeds were sterilized by chlorine gas sterilization and sown on 1/2 MS medium plates. After a 3-d vernalization period, seedlings were grown 1/2 MS media (without sucrose) under continuous light conditions (68 μE m−2 s−1 photosynthetically active radiation) at 21 °C in a growth chamber. When required, seedlings grown under continuous light conditions were kept in constant darkness or moved to Lovibond incubators with continuous light (75 μE m−2 s−1 photosynthetically active radiation) at 35 °C for specific time periods.
Staining for Live-Cell Imaging.
The 5-d-old seedlings treated without or with short-term heat treatment (35 °C for 6 h) or following with recovery in 1/2 MS liquid medium or on 1/2 MS medium plates at 20 °C were processed with specific staining and imaged with a Leica SP8X or Zeiss 710 confocal microscope. During the staining, the staining solution was protected from light.
For FM4-64 uptake, seedlings were incubated with 1/2 MS liquid medium containing 2 μM FM4-64 (Invitrogen) at 20 °C for 15 min prior to confocal imaging. For FM4-64 and FDA costaining, whole Arabidopsis seedlings were incubated with 1/2 MS liquid medium supplemented with 5 μg/mL FDA (ThermoFisher) and 2 μM FM4-64 at room temperature for 15 min prior to confocal imaging. For the heat-recovery experiment, seedlings after short-term heat treatment were recovered in 1/2 MS with 0.1% DMSO or 50 µM cycloheximide (Sigma-Aldrich) at 20 °C for certain hours and counterstained with 10 µg/mL propidium iodide (Invitrogen) for 2 min. The MitoTracker red CMXRos (ThermoFisher) was used to stain the mitochondria. Whole Arabidopsis seedlings were incubated with 1/2 MS liquid medium supplemented with 100 nM MitoTracker red at room temperature for 15 min. Callose staining was performed as described previously (64). After fixation, seedlings were washed thoroughly with KH2PO4 (pH = 9.5) solution and incubated with the same solution supplemented with 0.01% (wt/vol) Aniline blue for 2 h under vacuum.
Immunofluorescence.
The Arabidopsis roots of short-term heat-treated seedlings were analyzed by immunofluorescence as described previously (65). The anti-PIN1 antibody, anti-PIN2 antibody, anti-PIP2 antibody, as well as the fluorochrome-conjugated secondary anti-sheep antibodies Alexa488 for PIN1 and the anti-rabbit Alexa555 for PIN2, and PIP2 were used in this study and listed in SI Appendix, Table S4. All presented immunofluorescence experiments were performed at least two times with a similar outcome.
Live-Cell Imaging and Analysis.
Dynamic imaging of TPLATE and TPLATE domain substitution isoforms at the PM was performed on 4-d-old etiolated hypocotyl epidermal cells using a Nikon Ti microscope equipped with an Ultraview spinning-disk system and the Volocity software package (PerkinElmer), as described previously (8, 16, 37). Images were acquired with a 100× oil-immersion objective (Plan Apo, NA = 1.45). Seedlings expressing GFP-fused proteins were imaged with 488-nm excitation light and an emission window between 500 nm and 530 nm in single camera mode, or 500 to 550 nm in dual camera mode. Seedlings expressing mRFP- and tagRFP-labeled proteins were imaged with 561-nm excitation light and an emission window between 570 nm and 625 nm in single camera mode or 580 to 630 nm in dual camera mode. Single-marker line movies were acquired with an exposure time of 500 ms per frame for 2 min. Dual-color lines were acquired simultaneously (dual camera mode) with an exposure time of 500 ms per frame for 2 min. The CherryTemp system (Cherry Biotech) was used to maintain the temperature of samples constant at 20 °C during imaging, as performed previously (37).
Arabidopsis seedlings were imaged between slide and cover glass. The FM4-64 uptake of TPLATE-, TPLATE-WDXM1–, and TPLATE-WDXM2–complemented lines at normal condition were imaged with a Zeiss 710 inverted confocal microscope equipped with the ZEN 2009 software package and using a C-Apochromat 40× water Korr M27 objective (NA 1.2). FM4-64 was visualized using 561-nm laser excitation and a 650- to 750-nm detection window. The stained seedlings after short-term heat treatment were imaged using a Leica SP8X microscope equipped with a WhiteLight laser. Images were taken using a 40× water-corrected objective (40× HC APO CS2, NA = 1.10). Fluorescence for FM4-64 (excitation 561 nm, emission 650 to 750 nm), FDA (excitation 488 nm, emission 500 to 550 nm), MitoTracker red (excitation 579 nm, emission 600 to 650 nm), aniline blue (excitation 405 nm, emission 490 to 510 nm) were collected with Hybrid detectors (HyDTM) with or without (Aniline blue) time gating between 0.3 and 6 ns. Fluorescence for GFP (excitation 488 nm, emission 500 to 540 nm), YFP (excitation 514 nm, emission 520 to 550 nm), mCherry/mRuby3 (excitation 561 nm, emission 590 to 650 nm) were imaged with the WhiteLight laser and collected with hybrid detector (HyDTM) using a time-gating window between 0.3 and 6.0 ns. Images of the dual-color lines were acquired in line sequential mode. For the immunofluorescence imaging, seedlings were imaged with a 20× objective (HC PL APO CS2 20×/0.75 DRY). Fluorophores were excited using a WhiteLight laser with 488-nm (Alexa488) or 555-nm (Alexa555) excitation wavelength detected with a Hybrid detector with a gain of 100, (HyD 495 nm to 549 nm or HyD 561 nm to 621 nm) and time gating between 1 and 8 ns.
To measure the lifetimes or TPLATE isoforms at the PM, kymographs were generated using the Volocity software package and the lifetimes of individual endocytosis events were measured manually from the generated kymographs. Only endocytic events with a clear start or end present during the duration of the time lapse were retained for the measurements. Density measurements of endocytic dots were performed with movies exported from the Volocity software package using the ImageJ software package. Movies were processed with the “Walking Average” tool (number of frames to average, 4), followed by a t-projection of the first 10 frames (with Max Intensity). The number of endocytic foci in a certain region were counted using the “Find Maxima” tool. To determine the threshold of “Prominence” when do the “Find maxima”, the t-projected images were processed with the “MorphoLibJ” plugin (Plugins >>> MorphoLibJ >>> Morphological filters >>> Operation “White TOP Hat”; Element “disk”; Radius “2”) to get rid of background. Then the images were processed with “Gaussian Blur” filter (σ = 10) and ran “Histogram” to get the “max value” for the “Prominence” threshold. After being processed with the “MorphoLibJ” plugin, the images were used to quantify the density of the endocytic foci.
To measure the number of autophagosomes, single confocal slices of short-term heat-treated roots expressing mCherry-ATG8e were analyzed with ImageJ using the “Find Maxima” tool. The number of ATG8e-labeled autophagosomes in the regions of interest were selected and counted using the “Find Maxima” tool. Prior to counting, images were processed with a “smooth” and subtract background function (rolling ball radius with 1.0 pixels).
Phenotypic Analysis.
For fixed-carbon starvation phenotypical analysis, seedlings were grown on 1/2 MS medium vertically or horizontally in the growth chamber under continuous light conditions at 20 °C for 5 d. After that, root growth was marked, and the plates were covered by aluminum foil and kept in constant darkness for an extra 1 wk (root growth) or 9 d (cotyledons observations). Root growth of seedlings was measured with ImageJ equipped with the NeuronJ plugin. The percentage of yellowing seedlings was visually determined. For heat treatment phenotypical analysis, 5-d-old seedlings were grown vertically on 1/2 MS medium under continuous light conditions at 20 °C were moved to Lovibond incubators with continuous light (75 μE m−2 s−1 photosynthetically active radiation) at 35 °C for 3 or 4 additional days. Prior to moving plants to the incubator, root growth was marked. Root growth of seedlings was measured with ImageJ equipped with the NeuronJ plugin. After 4-d heat treatment, seedlings exhibited whitening cotyledons were visually counted.
SDS/PAGE and Western Blot.
Samples were analyzed by loading on 4 to 20% gradient gels (Bio-Rad), after addition of 4× Laemmli sample buffer (Bio-Rad) and 10× NuPage sample reducing agent (Invitrogen). Gels were transferred to PVDF or Nitrocellulose membranes using the Trans-Blot Turbo system (Bio-Rad). Blots were imaged on a ChemiDoc Imaging System (Bio-Rad) and the ImageJ program was used for a quantitative analysis.
Arabidopsis Seedling Protein Extraction.
Arabidopsis seedlings were grown for 7 d on 1/2 MS medium under constant light. Seedlings were harvested, flash-frozen, and ground in liquid nitrogen. Proteins were extracted in a 1:1 ratio, buffer (mL): seedlings (g), in HB+ buffer, as described in Van Leene et al. (66). Protein extracts were incubated for 30 min at 4 °C on a rotating wheel before spinning down twice at 20,000 × g for 20 min. The protein content of the supernatant was measured using Qubit (ThermoFisher) and equal amounts of proteins were loaded for analysis.
Coimmunoprecipitation.
Arabidopsis seedlings were grown for 7 d on 1/2 MS medium at 20 °C under constant light. Seedlings were harvested, flash-frozen, and ground in liquid nitrogen. Proteins were extracted in a 1:1 ratio, buffer (mL): seedlings (g), in 50 mM Tris⋅HCl, 150 mM NaCl, 5mM DTT supplemented with 1× cOmplete ULTRA Tablets, Mini, EDTA-free, EASYpack Protease Inhibitor Mixture (Roche). Protein extracts were incubated for 30 min at 4 °C on a rotating wheel and cleared by subsequent centrifugation at 20,000 × g for 20 min. The protein content of the supernatant was measured using Qubit (ThermoFisher) and an equal amount of total proteins were incubated with 20 µL GFP-Trap, magnetic agarose beads (Chromotek, gtma-20, lot 90122001MA). Beads were washed three times with the lysis buffer and eluted by boiling at 70 °C for 10 min. Protein degradation was analyzed by ImageJ by comparing pixel intensities (gray values) of full-length protein versus all smaller molecular weight products.
Identification of Interacting Proteins Using IP/MS-MS.
IP experiments were performed for three biological replicates as described previously (67), using 3 g of 4-d-old seedlings. Interacting proteins were isolated by applying total protein extracts to αGFP-coupled magnetic beads (Milteny Biotech). Three replicates of TPLATE motif substitution mutants (WDXM1 and WDXM2) were compared to three replicates of Col-0 and TPLATE-GFP [in tplate(−/−)] as controls. Tandem mass spectrometry (MS-MS) and statistical analysis using MaxQuant and the Perseus software was performed as described previously (68).
Relative amounts of TPC subunits detected compared to the bait protein (TPLATE-GFP, WDXM1-GFP, or WDXM2-GFP) were calculated based on peptide intensity levels. Peptides of the three bait proteins (TPLATE, WDXM1, and WDXM2) as well as of each specific TPC subunit that were identified in all experiments (i.e., in all three biological repeats by MS-MS or by matching) and for which an intensity value was assigned were selected for the quantification. Those peptides are highlighted in color in Dataset S2. The total number of common peptides that were taken into account for the measurement are as follows: TPLATE, 29; TML, 4; TASH3, 11; LOLITA, 1; TWD40-1, 13; TWD40-2, 20; AtEH1, 2; and AtEH2, 2. Peptides used for the calculations of the respective proteins are color coded in Dataset S2. The intensities of the common peptides for each subunit were normalized to the averaged intensity of the bait proteins. The normalized intensity values of each prey peptide were then averaged for each prey protein, the SD was calculated, and the values are plotted in Fig. 2B and SI Appendix, Fig. S3.
Supplementary Material
Acknowledgments
We thank the Vlaams Instituut voor Biotechnologie proteomics core facility of LC/MS-MS analysis, and to the developers of the open-source programs used in this study, particularly Gimp, Inkscape, ImageJ, Jalview, R, and Rstudio. This work was supported by the European Research Council Grants 682436 (to D.V.D.) and 714055 (to B.D.R.); Research Foundation–Flanders (FWO) Odysseus II Grants G0D0515N (to B.D.R.) and 1226420N (to P.G.); China Scholarship Council Grants 201508440249 (to J.W.) and 201906760018 (to Q.J.); and by a Ghent University Special Research cofunding Grant ST01511051 (to J.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023456118/-/DCSupplemental.
Data Availability
All other study data are included in the article and supporting information.
References
- 1.Kaksonen M., Roux A., Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Mettlen M., Chen P. H., Srinivasan S., Danuser G., Schmid S. L., Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robert S., et al., ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143, 111–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitsikas V., Corrêa I. R. Jr, Nichols B. J., Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife 3, e03970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paez Valencia J., Goodman K., Otegui M. S., Endocytosis and endosomal trafficking in plants. Annu. Rev. Plant Biol. 67, 309–335 (2016). [DOI] [PubMed] [Google Scholar]
- 6.McMahon H. T., Boucrot E., Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Di Rubbo S., et al., The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 25, 2986–2997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadeyne A., et al., The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 156, 691–704 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., et al., Change your TPLATE, change your fate: Plant CME and beyond. Trends Plant Sci. 20, 41–48 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Kim S. Y., et al., Adaptor protein complex 2-mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25, 2970–2985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaoka S., et al., Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25, 2958–2969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan L., et al., Dynamic analysis of Arabidopsis AP2 σ subunit reveals a key role in clathrin-mediated endocytosis and plant development. Development 140, 3826–3837 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Van Damme D., et al., Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell 18, 3502–3518 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bashline L., Li S., Zhu X., Gu Y., The TWD40-2 protein and the AP2 complex cooperate in the clathrin-mediated endocytosis of cellulose synthase to regulate cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 12870–12875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirst J., et al., Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife 3, e02866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P., et al., Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 10, 5132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C., Shen W., Yang C., Zeng L., Gao C., Knowns and unknowns of plasma membrane protein degradation in plants. Plant Sci. 272, 55–61 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Adamowski M., et al., A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 30, 700–716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejonghe W., et al., Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 15, 641–649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhonukshe P., et al., Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17, 520–527 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Kitakura S., et al., Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23, 1920–1931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C., et al., Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis. Plant Cell 25, 499–516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bashline L., Li S., Anderson C. T., Lei L., Gu Y., The endocytosis of cellulose synthase in Arabidopsis is dependent on μ2, a clathrin-mediated endocytosis adaptin. Plant Physiol. 163, 150–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C., et al., Differential regulation of clathrin and its adaptor proteins during membrane recruitment for endocytosis. Plant Physiol. 171, 215–229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinari A., et al., Polar localization of the borate exporter BOR1 requires AP2-dependent endocytosis. Plant Physiol. 179, 1569–1580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backues S. K., Korasick D. A., Heese A., Bednarek S. Y., The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22, 3218–3231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collings D. A., et al., Arabidopsis dynamin-like protein DRP1A: A null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J. Exp. Bot. 59, 361–376 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Konopka C. A., Bednarek S. Y., Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 147, 1590–1602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor N. G., A role for Arabidopsis dynamin related proteins DRP2A/B in endocytosis; DRP2 function is essential for plant growth. Plant Mol. Biol. 76, 117–129 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Yoshinari A., et al., DRP1-Dependent endocytosis is essential for polar localization and boron-induced degradation of the borate transporter BOR1 in Arabidopsis thaliana. Plant Cell Physiol. 57, 1985–2000 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Dejonghe W., et al., Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat. Commun. 7, 11710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez-Rodríguez C., et al., The cellulose synthases are cargo of the TPLATE adaptor complex. Mol. Plant 11, 346–349 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Buevich A. V., Lundberg S., Sethson I., Edlund U., Backman L., NMR studies of calcium-binding to mutant alpha-spectrin EF-hands. Cell. Mol. Biol. Lett. 9, 167–186 (2004). [PubMed] [Google Scholar]
- 34.Janssens V., et al., Identification and functional analysis of two Ca2+-binding EF-hand motifs in the B"/PR72 subunit of protein phosphatase 2A. J. Biol. Chem. 278, 10697–10706 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Van Damme D., et al., Adaptin-like protein TPLATE and clathrin recruitment during plant somatic cytokinesis occurs via two distinct pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 615–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yperman K., et al., Molecular architecture of the endocytic TPLATE complex. Sci. Adv., 10.1126/sciadv.abe7999 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., et al., High temporal resolution reveals simultaneous plasma membrane recruitment of TPLATE complex subunits. Plant Physiol. 183, 986–997 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung T., Phillips A. R., Vierstra R. D., ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62, 483–493 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Doelling J. H., Walker J. M., Friedman E. M., Thompson A. R., Vierstra R. D., The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Hanaoka H., et al., Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F., Chung T., Vierstra R. D., AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26, 788–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phillips A. R., Suttangkakul A., Vierstra R. D., The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi H., et al., TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 29, 890–911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suttangkakul A., Li F., Chung T., Vierstra R. D., The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson A. R., Doelling J. H., Suttangkakul A., Vierstra R. D., Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiong Y., Contento A. L., Bassham D. C., AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Yoshimoto K., et al., Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldenzweig A., Fleishman S. J., Principles of protein stability and their application in computational design. Annu. Rev. Biochem. 87, 105–129 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Greco M., Chiappetta A., Bruno L., Bitonti M. B., In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 63, 695–709 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones K., Kim D. W., Park J. S., Khang C. H., Live-cell fluorescence imaging to investigate the dynamics of plant cell death during infection by the rice blast fungus Magnaporthe oryzae. BMC Plant Biol. 16, 69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jung H., et al., Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 71, 73–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X., Srivastava R., Howell S. H., Bassham D. C., Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 85, 83–95 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Zhou J., et al., NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9, e1003196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall R. S., Vierstra R. D., Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Li R., et al., A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24, 2105–2122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gälweiler L., et al., Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Müller A., et al., AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johanson U., et al., The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126, 1358–1369 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altschul S. F., et al., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K., Rozewicki J., Yamada K. D., MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Damme D., Bouget F. Y., Van Poucke K., Inzé D., Geelen D., Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40, 386–398 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Karimi M., Depicker A., Hilson P., Recombinational cloning with plant gateway vectors. Plant Physiol. 145, 1144–1154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulich I., et al., Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 168, 120–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sauer M., et al., Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20, 2902–2911 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Leene J., et al., A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6, 1226–1238 (2007). [DOI] [PubMed] [Google Scholar]
- 67.De Rybel B., et al., A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Wendrich J. R., Boeren S., Möller B. K., Weijers D., De Rybel B., In vivo identification of plant protein complexes using IP-MS/MS. Methods Mol. Biol. 1497, 147–158 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other study data are included in the article and supporting information.