Significance
Myristoylation is a major type of cotranslational lipid modification to many proteins that play important roles in numerous cell biology processes. Myristoylation serves diverse roles in protein–membrane association, protein structure, protein stability, and protein–protein interaction. With regard to protein–membrane association, it is believed that myristoylation alone does not confer sufficient energy to associate a protein to the membrane unless assisted by a second membrane-affinity motif. We find that the myristoylation on the PKA catalytic subunit (PKA-C) is sufficient to associate a fraction of PKA-C molecules to the plasma membrane in neuronal dendrites in a functionally significant manner without a canonical second motif. Our results remove a conceptual constraint for understanding myristoylated proteins and their function.
Keywords: myristoylation, cAMP-dependent kinase, PKA, fractional membrane association
Abstract
Myristoylation is a posttranslational modification that plays diverse functional roles in many protein species. The myristate moiety is considered insufficient for protein–membrane associations unless additional membrane-affinity motifs, such as a stretch of positively charged residues, are present. Here, we report that the electrically neutral N-terminal fragment of the protein kinase A catalytic subunit (PKA-C), in which myristoylation is the only functional motif, is sufficient for membrane association. This myristoylation can associate a fraction of PKA-C molecules or fluorescent proteins (FPs) to the plasma membrane in neuronal dendrites. The net neutral charge of the PKA-C N terminus is evolutionally conserved, even though its membrane affinity can be readily tuned by changing charges near the myristoylation site. The observed membrane association, while moderate, is sufficient to concentrate PKA activity at the membrane by nearly 20-fold and is required for PKA regulation of AMPA receptors at neuronal synapses. Our results indicate that myristoylation may be sufficient to drive functionally significant membrane association in the absence of canonical assisting motifs. This provides a revised conceptual base for the understanding of how myristoylation regulates protein functions.
Myristoylation is a major type of posttranslational modification that occurs at the N terminus of a myriad of proteins (1–4). Depending on the target, myristoylation can contribute to the structure, stability, protein–protein interactions, and subcellular localization of the modified proteins (2–4). In particular, myristoylation often facilitates protein association with the membrane. However, it is thought that, with an acyl chain of only 14 carbons, myristate confers insufficient energy for stable association of a protein with the membrane (5, 6). Subsequent studies have shown that a second membrane-affinity motif, such as a stretch of basic residues or a second lipid modification, is required for the membrane association of several myristoylated proteins (reviewed in refs. 2–4). When the second membrane-affinity motif is removed or neutralized, either physiologically or via mutagenesis, the membrane localization of the protein is disrupted. Thus, the canonical view is that myristoylation alone is not sufficient to provide a functionally significant association of a protein with the plasma membrane, even though myristoylation has been observed to be associated with reconstituted lipid bilayers (7).
Myristoylation was first discovered in the catalytic subunit of protein kinase A (PKA) (1, 8), which is a primary mediator of the second messenger cAMP that plays diverse essential roles in nearly all organisms, from bacteria to humans. At rest, PKA is a tetrameric protein that consists of two regulatory subunits (PKA-Rs) and two catalytic subunits (PKA-Cs). PKA holoenzymes are anchored to specific subcellular locations via the binding of PKA-R with A-Kinase anchoring proteins (9–12). In the presence of cAMP, PKA-C is released from PKA-R and becomes an active kinase (8, 13–16).
Despite being myristoylated, PKA-C is thought to function as a cytosolic protein because of its high solubility (14, 15). Consistently, a PKA-C mutant with disrupted myristoylation has been shown to support the phosphorylation of certain substrates and to maintain several PKA functions in heterologous cells (17). Structural studies found that the PKA-C myristoylation is folded into a hydrophobic pocket, and it was proposed that this myristoylation serves a structural role (18–20). This view has started to shift based on recent reports showing that activated PKA-C can associate with the membrane in a myristoylation-dependent manner (16, 21, 22), including in neurons. However, the extent to which PKA-C associates with the plasma membrane in living cells and its functional significance are not known. Furthermore, as discussed above, myristolyation-mediated membrane association is thought to require a second-membrane motif. The identity of this second membrane-affinity motif has not been determined. Therefore, we set out to address these questions.
Results
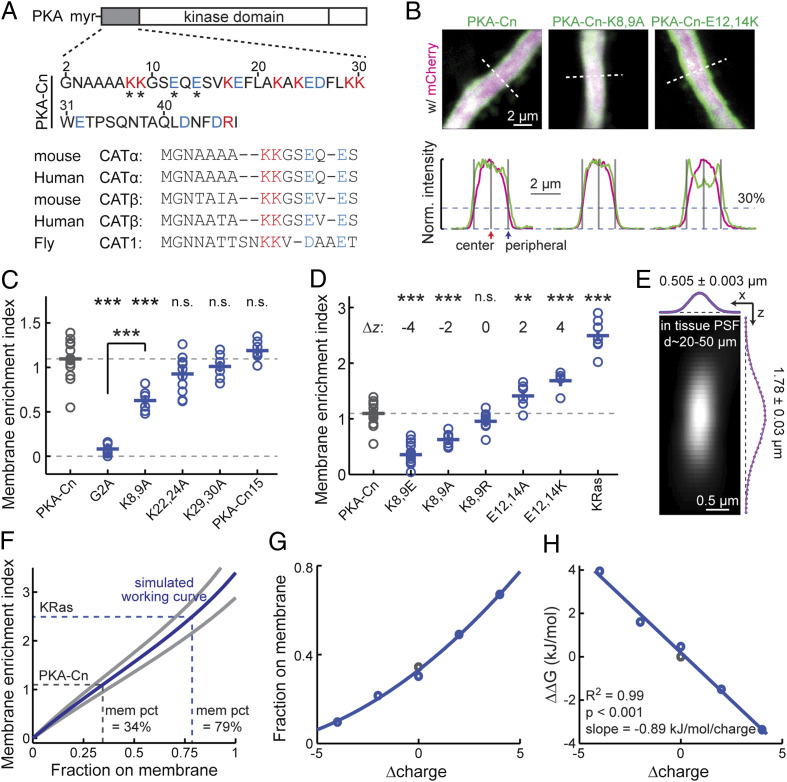
First, our previous results (16) suggested that the N-terminal 47 residues of PKA-C (called PKA-Cn) (Fig. 1 A, Upper), which lack the kinase domain and the C-terminal domains, and cannot bind to PKA-R, were sufficient to target a fluorescent protein (FP) to the plasma membrane in neuronal dendrites. In order to quantitatively assay the membrane affinity, cultured hippocampal slices were transfected with PKA-Cn fused to the N terminus of the yellow FP mVenus (PKA-Cn-mVenus) together with a red cytosolic FP (mCherry). The thick (ϕ ≥ 1.5 µm) apical dendrites of CA1 neurons were imaged live at 2 to 4 d posttransfection under a two-photon microscope. An optical section through the center of the dendrite (Fig. 1 B, Upper Left and SI Appendix, Fig. S1) was used to compare the localizations of PKA-Cn-mVenus and mCherry. The PKA-C-mVenus fluorescence (shown as green) was more enriched at the periphery of the dendrite compared to mCherry fluorescence (red, shown as magenta). To quantify the membrane association, a membrane-enrichment index (MEI) was computed:
where F is the fluorescence intensity. The peripheral fluorescence intensities were measured at the edge of the dendrite, as determined by the location where the red was 30% of the peak fluorescence (Fig. 1 B, Lower) (16). MEI values greater than 0 would indicate membrane enrichment, whereas MEI values less than 0 would indicate membrane exclusion. PKA-Cn-mVenus exhibited an MEI value significantly greater than 0 (MEI = 1.09 ± 0.05; P < 0.001, cf. 0; Fig. 1C). These values were independent of the expression level of PKA-Cn-mVenus (SI Appendix, Fig. S2), with a linear fit extrapolating to the zero expression level at an MEI value of 1.17. These results further support an affinity of PKA-Cn for the membrane.
Fig. 1.
PKA-Cn is sufficient to localize a fraction of FPs to the plasma membrane. (A) Sequence of PKA-C N terminus showing balanced positive (magenta) and negative (blue) charges. (Upper) The 47 residues of mouse PKA-C used in this study. The residues affecting membrane affinity are indicated (asterisks). (Lower) Alignment of the first 15 residues of the indicated PKA-C subtypes and species. (B) Representative images and traces along the indicated dashed white lines of thick neuronal apical dendrites transfected with the indicated constructs for quantifying MEIs. The red fluorescence of mCherry is shown as magenta (to accommodate color-blind readers). Gray bars mark the locations where the fluorescence intensities are used to calculate MEIs: at the center and at the periphery where the red signal is at 30% of peak (corresponding to the plasma membrane) (16). (C) Collective MEI quantifications for the indicated PKA-Cn-mVenus constructs. From left to right, n = 16, 12, 6, 11, 7, and 7 for the indicated constructs. (D) Collective MEI quantifications for the indicated PKA-Cn-mVenus constructs and KRas-mEGFP. The charge relative to PKA-Cn (Δz) is indicated for each construct. The data for PKA-Cn is the same as in C. For other constructs, from left to right, n = 18, 6, 11, 6, 5, and 7. (E) Measured PSF in cultured hippocampal slices at imaging depths of 20 to 50 μm, n = 36. (F) MEI versus membrane-fraction working curves computed based on simulation using the measured PSF size and the experimental dendritic width of PKA-Cn at mean (blue) and one SD (gray). As indicated, the percentages of PKA-Cn and KRas on the membrane (mem pct) were estimated based on the mean-size curve. (G) Charge versus membrane-fraction relationships for the PKA-Cn constructs in D. Membrane fractions were estimated based on the mean MEI values and the corresponding dendrite sizes. (H) The relationship between the charge versus Gibbs free energy relative to wild-type PKA-Cn for the constructs in G and its linear fit. *P ≤ 0.05 and is statistically significant after Bonferroni correction for multiple tests, **P ≤ 0.01, and ***P ≤ 0.001.
We then asked how strong the membrane affinity of PKA-Cn was. Mutating the glycine residue at position 2 to alanine (PKA-Cn-G2A-mVenus), which prevented myristoylation (17, 23, 24), nearly completely abolished the membrane localization (MEI = 0.08 ± 0.02, P < 0.001 cf. PKA-Cn-mVenus) (Fig. 1C), indicating that the membrane affinity is myristoylation dependent (see also ref. 16). To evaluate the degree of membrane affinity of PKA-Cn-mVenus, its MEI was compared with that of mEGFP-tagged KRas (25), a known peripheral membrane protein. The MEI of PKA-Cn-mVenus was ∼45% of that of KRas (Fig. 1D), indicating that its membrane affinity is moderate. To estimate the fraction of PKA-Cn on the membrane, simulation was performed based on experimentally measured parameters. The point spread function (PSF) of our two-photon microscope was measured under the same conditions as the MEI experiments (20 to 50 µm deep in cultured hippocampal slices) (Fig. 1E). The lateral and axial sizes of the PSF and the diameters of PKA-Cn-mVenus dendrites (ϕ = 2.18 ± 0.32; mean ± SD) were used to simulate how different fractions of membrane localization affected the image and MEI (Fig. 1F and SI Appendix, Fig. S3; see Materials and Methods for details). Using the measured MEI of PKA-Cn-mVenus on the working curve derived from the simulation, ∼34% of the protein was estimated to be localized to the plasma membrane (Fig. 1F). This fraction on the membrane likely represented the lower bound, since PKA-Cn may also have affinity for intracellular membranes, which was not taken into account here. Overall, PKA-Cn was sufficient to confer a fractional membrane affinity in a myristoylation-dependent manner.
Because PKA-Cn alone was sufficient to confer association with the plasma membrane, we reasoned that if there was a second membrane-affinity motif, it must exist within PKA-Cn. Because PKA-Cn did not contain any additional lipid modification site, the presence of positively charged residues (e.g., see refs. 26 and 27) would be the only mechanism that could enhance the affinity of PKA-Cn to the typically negatively charged plasma membrane. There were eight basic residues (seven lysines and one arginine) and eight acidic residues (glutamate and aspartate) within PKA-Cn, resulting in an overall net-neutral charge (Fig. 1A). Nevertheless, it is possible that the positive charges could not be functionally counteracted by negative charges because of their unique positions or orientations in the three-dimensional space. To address this possibility, we mutated the lysine residues to alanine residues two at a time and assayed their MEIs. Mutating the two lysine residues closest to the N terminus (K8,9A), but not those further away (K22,24A and K29,30A, respectively), to alanine resulted in decreased MEIs (MEI = 0.63 ± 0.05 for PKA-Cn-K8,9A-mVenus, P < 0.001 cf. PKA-Cn-mVenus) (Fig. 1 B and C). Therefore, it appeared that only charges close to the N terminus modulated the membrane affinity of PKA-Cn. Indeed, another PKA-C N-terminal construct containing only the first 15 residues (named PKA-Cn15-mVenus) was localized to the membrane to a degree comparable to that of PKA-Cn (MEI = 1.19 ± 0.04 for PKA-Cn15-mVenus, P = 0.27, cf. PKA-Cn-mVenus) (Fig. 1C). The charge, rather than the residue identity, is likely the primary factor for the MEI decrease because when these lysine residues were mutated to similarly charged arginine residues (K8,9R), the MEI was not altered significantly (Fig. 1D). Notably, even for PKA-Cn-K8,9A-mVenus, significant plasma membrane affinity remained (P < 0.001, cf. PKA-Cn-G2A-mVenus), suggesting that these residues quantitatively, rather than qualitatively, perturbed the membrane affinity of PKA-Cn.
The above results indicate that the two aforementioned lysine residues (K8 and K9) are the only candidates for the second membrane-affinity motif. However, within the first 15 residues of PKA-C, the net charge is also neutral, with these positively charged lysine residues counterbalanced by two negatively charged glutamate residues (E12 and E14) (Fig. 1A). This net-zero charge at the N terminus of PKA-C appears to be evolutionarily conserved from flies to mammals and across different PKA-C isoforms (Fig. 1 A, Lower), which were also sufficient to direct mVenus to the membrane (SI Appendix, Fig. S4). We asked whether the two glutamates could effectively counter the effects of the K8 and K9 lysine residues with regard to plasma membrane affinity. If they could not, mutating these glutamates would have little or smaller effects on the MEI compared to the lysine mutations. The opposite was found: mutating the glutamate residues to alanine or lysine (E12,14A and E12,14K, respectively) resulted in significantly increased MEIs (Fig. 1D). When combining the mutations at the lysine and glutamate positions, the membrane affinity is monotonically dependent on the summed charge from the four positions (Fig. 1D). The membrane fractions (Fig. 1G) and the Gibbs free energy relative to the wild-type PKA-Cn (Fig. 1H) were determined based on working curves computed using the corresponding dendrite sizes. A linear relationship was found between the charge and the binding energy regardless of whether the lysine or glutamate residues were mutated (R2 = 0.99, P < 0.001). Each positive charge contributed an energy of −0.89 kJ ⋅ mol−1. A simple explanation from these results is that charges at the lysine and glutamate positions have equal weights in affecting the membrane affinity of PKA-Cn, although unique roles of individual residues cannot be fully ruled out. Overall, although charges can modulate the membrane affinity, the observed affinity is intrinsic to myristoylation because PKA-Cn has a net-zero charge.
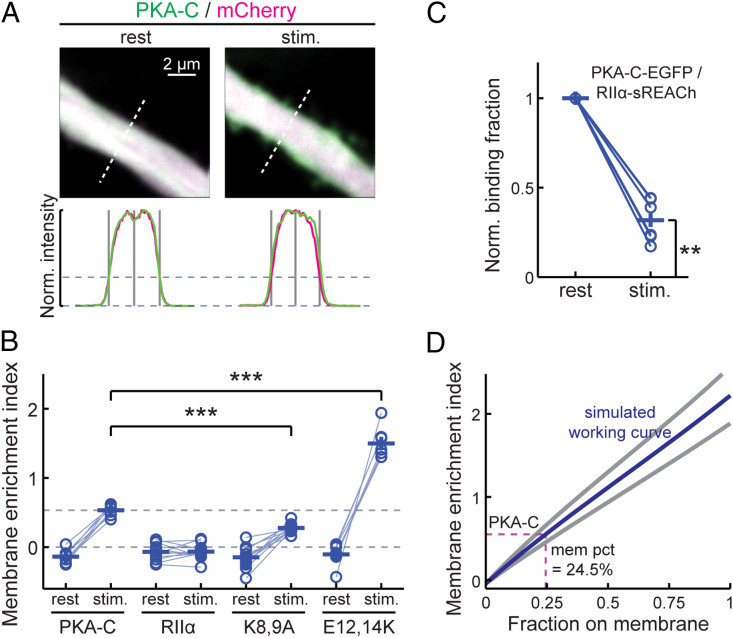
We next examined whether full-length PKA-C is associated with the plasma membrane in a manner similar to that of PKA-Cn. PKA-C-mEGFP was coexpressed with untagged PKA-RII-α and the cytosolic marker mCherry. Consistent with our previous results using a different PKA regulatory subunit (PKA-RII-β) (16), full-length PKA-C was slightly excluded from the plasma membrane at rest as reflected by a negative MEI (MEI = −0.14 ± 0.03; P = 0.004, cf. 0) (Fig. 2 A and B). This is likely because most PKA-C molecules at rest are bound to PKA-RII-α, the majority of which are in turn anchored to the microtubule cytoskeleton via MAP2 (9, 28). By increasing intracellular cAMP concentrations using the adenylyl-cyclase activator forskolin along with the phosphodiesterase inhibitor IBMX, PKA-C translocated to become associated with the plasma membrane as reflected by a shift in the MEI to a positive value (MEI = 0.53 ± 0.03 upon stimulation; P < 0.001, both cf. rest and 0). In contrast, in separate control experiments, mEGFP-tagged PKA-RII-α did not translocate under the same conditions (Fig. 2B), indicating that the fraction of PKA-C that translocated had dissociated from PKA-RII-α. This dissociation was consistent with the canonical understanding of PKA activation. The tendency to translocate to the plasma membrane upon stimulation was attenuated but not abolished when the K8 and K9 lysine residues were mutated to alanine (K8,9A) and were increased when E12 and E14 were mutated to lysines (E12,14K; Fig. 2B). The fraction of liberated PKA-C on the plasma membrane was estimated using an approach similar to that for the PKA-Cn mutants with two additional considerations: 1) PKA-C is slightly excluded from the membrane at rest (Fig. 2B) and 2) not all PKA-C dissociated from the PKA-R under our stimulation conditions. To quantify the latter, we used two-photon fluorescence lifetime imaging microscopy (2pFLIM) (29, 30) to measure the binding ratio between PKA-C-mEGFP and PKA-RII-α-sREACh (sREACh is a low-emitting yellow FP optimized for being a Föster resonance–energy transfer acceptor to mEGFP for 2pFLIM) (31). When normalized to the resting state, less than 70% (68 ± 5%) of PKA-C-mEGFP dissociated from PKA-RII-α-sREACh under our stimulation conditions (Fig. 2C). Using this dissociation fraction and the corresponding dendrite sizes (2.25 ± 0.39 µm for PKA-C-mEGFP, mean ± SD), ∼24.5% of liberated PKA-C was estimated to become associated with the plasma membrane. Since the membrane affinity of full-length PKA-C was not higher than that of PKA-Cn (compare Fig. 2D with Fig. 1F), it is unlikely that there is an additional membrane-affinity motif present in the remainder of PKA-C.
Fig. 2.
Association of full-length PKA-C with the plasma membrane. (A) Representative images of thick neuronal apical dendrites, and traces along the dashed white lines for quantifying MEIs for wild-type full-length PKA-C before (rest) and after stimulation (stim.) of PKA activity by forskolin (25 μM) and IBMX (50 μM). (B) Collective MEI quantifications for the indicated full-length PKA-C constructs (coexpressed with untagged PKA-RII-α) and PKA-RII-α-mEGFP before and after forskolin and IBMX stimulation. From left to right, n = 8, 12, 12, and 7 for the indicated constructs. (C) The binding ratio normalized to the resting state of PKA-C-mEGFP with RII-α-sREACh, as measured and quantified using 2pFLIM. (D) MEI versus membrane-fraction working curves using the experimental dendritic widths of PKA-C and the degree of PKA-C liberation, as measured in C. The estimated membrane fraction of liberated PKA-C is indicated (dash lines). *P ≤ 0.05 and is statistically significant after Bonferroni correction for multiple tests, **P ≤ 0.01, and ***P ≤ 0.001.
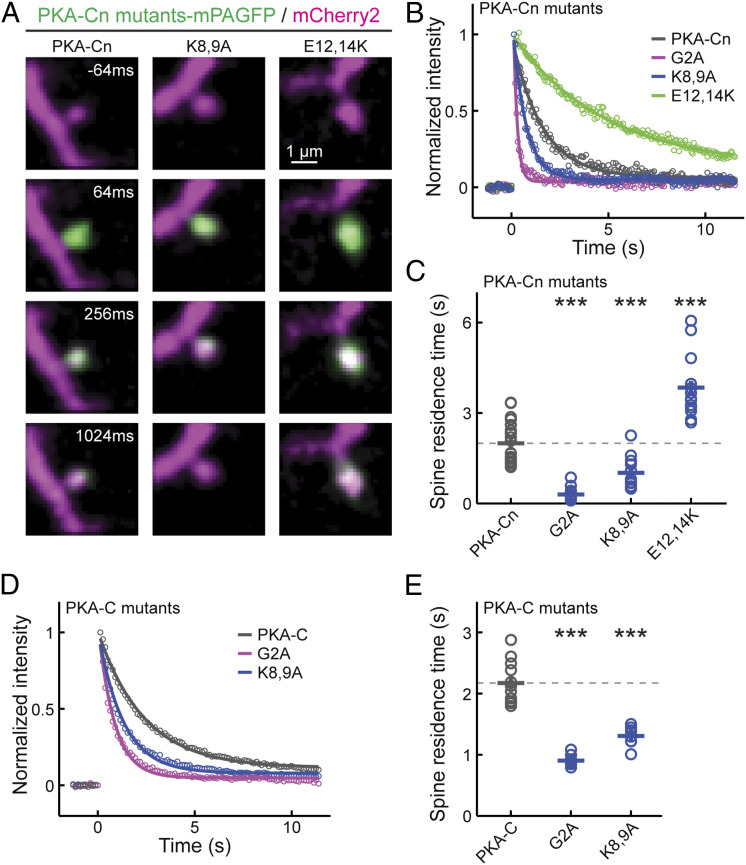
We used a second approach to confirm the tendency of PKA-C and its N-terminal mutants to associate with the membrane. Given that peripheral membrane-associated proteins diffuse much more slowly than cytosolic proteins (16, 32), we assayed the protein motility of PKA-C, PKA-Cn, and their respective mutants. PKA-Cn and its mutants were fused to monomeric photoactivatable GFP (mPAGFP) (33) and were coexpressed with the cytosolic marker mCherry or mCherry2. The mobility of these constructs was assayed by using focal two-photon photoactivation of mPAGFP at individual spines and monitoring the resulting fluorescence decay over time (Fig. 3 A and B) (34, 35). We focused on the spine because the time constant of the fluorescence decay at the spine (i.e., the spine-residence time) is linearly inversely proportional to the diffusion coefficient of a protein (34). The G2A mutant of PKA-Cn exhibited a spine-residence time (τ = 0.30 ± 0.05) (Fig. 3C) comparable to previously reported cytosolic protein diffusion (16, 34). The mobility of PKA-Cn was much slower (τ = 2.00 ± 0.15 s, P < 0.001, cf. G2A mutant) (Fig. 3C), which is consistent with its association with the membrane. The mobility of PKA-Cn was further decreased by the E12,14K mutations, which is consistent with a higher membrane affinity. While the mobility of PKA-Cn was increased by the K8,9A mutations, it was still much lower than that of the G2A mutant (Fig. 3C), corroborating the earlier conclusions based on MEIs that myristoylation is sufficient for membrane affinity in the absence of positively charged residues. Similarly, when full-length PKA-C-mEGFP was coexpressed with PKA-RII-α and activated by forskolin and IBMX, the K8,9A mutation increased the mobility of full-length PKA-C, but the degree of increase was less than that observed for the G2A mutants (Fig. 3 D and E). These results support the conclusions based on MEI measurements that the N-terminal fragment of PKA alone is sufficient for membrane association and that the moderate membrane affinity can be modulated bidirectionally by changing charges near the myristoylation site.
Fig. 3.
Mobility measurements of PKA-Cn and full-length PKA-C constructs. (A) Representative time-lapse images of the indicated constructs. Two-photon focal activation of mPAGFP was carried out at time 0 on a single spine. (B and C) Representative traces (B) and collective results (C) of the spine residence time of the indicated PKA-Cn constructs. From left to right, n = 21, 15, 12, and 13. (D and E) Representative traces (D) and collective results (E) of the spine residence time of indicated full-length PKA-C constructs coexpressed with untagged PKA-RII-α. Experiments were done in the presence of forskolin and IBMX to allow PKA-Cs to be liberated from PKA-Rs and translocate to the spine and the membrane. From left to right, n = 12, 9, and 8. *P ≤ 0.05 and is statistically significant after Bonferroni correction for multiple tests, **P ≤ 0.01, and ***P ≤ 0.001.
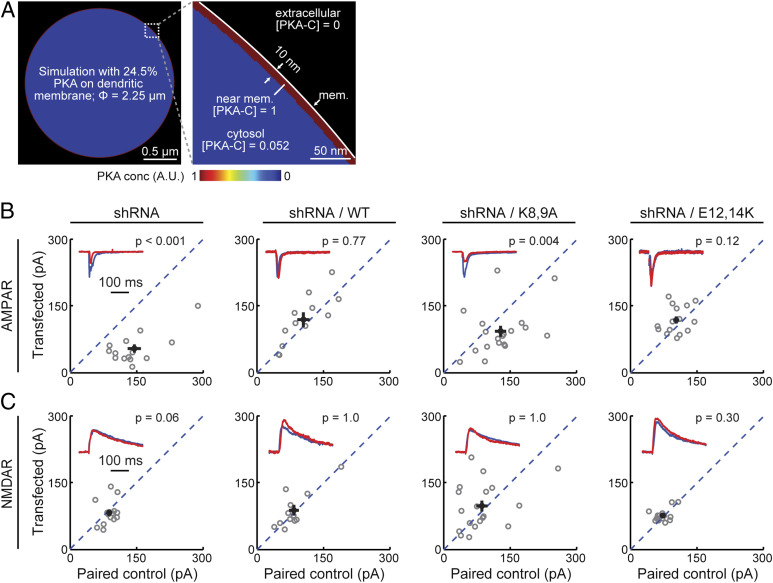
If only approximately one-fourth of liberated PKA-C is associated with the plasma membrane (Fig. 2D), can it explain the previous finding that PKA-C is more efficient in phosphorylating the same substrate on the membrane than in the cytosol (16)? To address this question, we performed a computational analysis. Assuming that membrane-bound PKA-C can phosphorylate substrates up to 10 nm away from the membrane (∼size of PKA-C) (18) and distribute the remaining liberated PKA-C evenly throughout the dendritic cytosol, with a dendrite size of 2.25 µm (the dendrite size used for Fig. 2D) and a membrane fraction of 24.5%, the PKA-C concentration at the membrane is more than 19-folds as high as its concentration in the cytosol (Fig. 4A). This analysis explains the results of previous biochemical experiments that a PKA substrate is much more efficiently phosphorylated on the membrane than in the cytosol (16). Overall, this analysis indicates that moderate membrane affinity of PKA effectively concentrates its protein activity at the plasma membrane.
Fig. 4.
Fractional-membrane association of PKA-C is important for its physiological function. (A) Simulation experiment showing that the observed moderate membrane affinity of full-length PKA-C is sufficient to enrich its activity at the membrane to over 19 times higher than its activity in the cytosol. (B and C) Representative traces (red) normalized to the paired control (blue) (insets) and scatter plots of paired AMPA (B) and NMDA (C) receptor currents from neighboring untransfected CA1 neurons paired with those transfected with shRNA against PKA-C and the indicated shRNA-resistant rescue constructs. Statistical P values were obtained using a sign test (MATLAB). From left to right, n = 12, 14, 19, and 15. *P ≤ 0.05 and is statistically significant after Bonferroni correction for multiple tests, **P ≤ 0.01, and ***P ≤ 0.001.
Finally, is the moderate membrane affinity of PKA-C important for its regulation of synaptic functions? To address this, a previously established short hairpin RNA (shRNA) construct (16) was used to selectively knock down PKA-C-α in CA1 neurons in cultured hippocampal slices. PKA has been implicated to regulate synaptic transmissions (16, 36). We therefore assayed evoked AMPA and NMDA receptor (AMPAR and NMDAR, respectively) currents in paired, transfected, and adjacent untransfected CA1 neurons in cultured hippocampal slices. As shown previously (16), neurons expressing the shRNA construct exhibited significantly lower AMPAR currents (Fig. 4B, leftmost panel) but not NMDAR currents (Fig. 4C, leftmost panel), compared to the paired controls. As a result, the AMPAR/NMDAR current ratio was also reduced (SI Appendix, Fig. S5, leftmost panel). The reduced AMPAR currents and reduced AMAPR/NMDAR current ratios were rescued by coexpression of shRNA-resistant wild-type PKA-C-EGFP (Fig. 4 B and C and SI Appendix, Fig. S5). However, with decreased, but not abolished, membrane affinity, PKA-C bearing the K8,9A mutations failed to rescue the phenotype. In contrast, the E12,14K mutant, which exhibited an enhanced membrane affinity, did rescue the phenotype. In addition, we have previously shown that PKA-C bearing the G2A mutation also cannot rescue knockdown of wild-type PKA-C in neuronal dendrites (16). Taken together, these results indicate that the membrane affinity resulting from myristoylation alone, while moderate, is important for normal synaptic function.
Discussion
In summary, we found that the PKA-C N-terminal fragment is sufficient to associate liberated PKA-C, as well as functionally irrelevant FPs, to the plasma membrane in living neuronal dendrites. Taken together with the observation that the PKA-C N-terminal fragment is both electrically neutral and lacks other membrane-affinity motifs, we concluded that myristoylation is sufficient to associate a protein to the plasma membrane in neuronal dendrites in the absence of a canonical assisting motif. Furthermore, while the myristoylation only confers a moderate membrane affinity and only associates a fraction of PKA-C molecules with the plasma membrane, this fraction is sufficient to enrich PKA activity at the membrane over the cytosol by over an order of magnitude and is indispensable for physiological function. Overall, these results suggest that the previous thought that myristoylation can only support functional membrane association in the presence of a second membrane-affinity motif must be revised. The revised view removes a conceptual constraint in understanding the wide range of proteins that are myristoylated (2–4, 37).
Historically, PKA has not been considered as a membrane-associated protein in part because of the following: 1) it lacks a second membrane-affinity motif, which is traditionally deemed necessary for membrane association; 2) PKA-C appears to be highly soluble; and 3) the myristoylation of PKA-C does not appear to be essential for its catalytic function. Although this view has been modified recently (16, 21, 22), the above results remain to be reconciled. Here, we show that the membrane association PKA-C does not require a second membrane-affinity motif. At the same time, the membrane affinity of PKA-C is moderate, which explains its solubility. In addition, while the PKA catalytic function does not depend on the myristoylation, our results, together with previous results (16), show that the fractional membrane affinity is still required for its successful regulation of synaptic function, presumably by concentrating its activity toward membrane substrates. It is also worth noting that small cellular compartments, such as neuronal dendrites, have a high membrane-to-cytosol ratio, which may have contributed to the detection of PKA-C at the membrane. Interestingly, although the membrane affinity of PKA-C appears to be tunable (e.g., by changing the negatively charged glutamate residues near the N terminus to alanine or lysine residues), its net-zero charge is evolutionally conserved (Fig. 1A). Moderate membrane affinity may have advantages, such as maintaining certain levels of kinase activity in the cytosol, allowing the kinase to travel between discontinuous membranes, allowing rapid inactivation of the kinase by regulatory subunits that are anchored away from the membrane, and potentially allowing bidirectional regulation of enzyme concentration on the membrane.
Materials and Methods
Plasmid Constructs.
Constructs were made using standard mutagenesis and subcloning methods. All previously unpublished constructs and their sequences have been submitted to Addgene (https://www.addgene.org/Haining_Zhong/; ID 168482 to 168499).
Organotypic Hippocampal Slice Cultures and Transfections.
Cultured rat hippocampal slices were prepared from P6 to P8 (typically P7) pups, as described previously (28, 38). Animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (39) of the NIH and were approved by the Institutional Animal Care and Use Committee of the Oregon Health and Science University (no. IS00002274). Plasmid constructs were transfected after 1.5 to 3 wk in vitro via the biolistic gene-transfer method using the Helios gene gun and 1.6 μm gold beads or with single-cell electroporation (electroporation was used for Fig. 4 and SI Appendix, Fig. S5) (40), where long-term expression (∼1 wk) was required.
Two-Photon Imaging and Photoactivation.
A custom built two-photon microscope based on an Olympus BW51WI microscope body was used. Laser beams from two different Ti:Sapphire lasers (Mai Tai, Newport) were aligned to allow for simultaneous two-photon excitation and photoactivation. Laser intensities were controlled by Pockels cells (Conoptics). Imaging and photoactivation were controlled by ScanImage (Vidrio Technologies) (41). Slices were perfused in gassed artificial cerebral spinal fluid containing 4 mM Ca, 4 mM Mg, and 0.5 µM tetrodotoxin during imaging. MEI imaging experiments were carried out at 960 nm at the center z position of thick (> 1.5 µm, mean ∼2.2 µm) apical dendrites of CA1 neurons. mEGFP and mVenus fluorescence (green) were unmixed with that of the cytosolic marker (mCherry or mCherry2) using a dichroic (Chroma 565DCXR) and band-pass filters (Chroma HQ510/70 for green and Semrock FF01-630/92 for red). For in-tissue PSF measurements, 0.1 µm green fluorescent beads were puffed into tissue using a Picospritzer.
Photoactivation experiments were carried out using an 810 nm laser for focal photoactivation of mPAGFP and a separate imaging laser set at 990 nm to simultaneously image the activated mPAGFP (green) and the cytosolic marker (red). Spines expressing PKA-Cn constructs were imaged at 16 Hz at a 32 × 32 image size, and full-length constructs were imaged at 8 Hz with a 64 × 64 image size. Power for photoactivation was determined empirically at the time of individual experiments.
Image Analysis.
Image analysis was performed using custom software written in MATLAB. For MEI measurements, line profiles of five-pixel width were manually drawn across the apical dendrites that were smooth on both sides (i.e., void of spines). The profiles were obtained from a single optical section centered along the middle z-plane of the dendrite. The background subtracted fluorescence intensity in the green (PKA constructs) and red (cytosol marker) channels were then used to calculate the MEI using the equation described in the main text.
Photoactivation experiments were analyzed by manually drawing regions of interest (ROIs) over the x-y projection of the spines of interest. The green fluorescence emission from the ROI was then averaged and determined over each image frame of the experiment. Background values calculated from a manually drawn background ROI at the same frames were subtracted from the spine ROI values. Only spines that showed an activation signaling more than three times the SD of the baseline fluorescence fluctuation (before photoactivation) were included in data analysis. The green fluorescence intensity decay postphotoactivation was then averaged across three separate trials and subsequently fit with a single exponential decay. The spine residence time was defined as the time constant of the exponential fit.
Computation and Simulation.
Computation and simulation were carried out in MATLAB. Simulation was used to generate the membrane fraction and MEI working curve. The full-width half maximum of averaged experimental PSFs was used to generate a Gaussian PSF with 10 nm pixel sizes in the x-z dimensions. The cytosolic distribution in the x-z dimensions was simulated as an evenly distributed round disk, and membrane distribution was simulated as signals being localized only at the edge of the disk. The diameter of the dendrite was specified by experimental measurements. Different percentages of signals were distributed to the membrane and the cytosol, and the resulting spatial distribution was convoluted with the PSF to produce the simulated images. MEI was measured at the z-center of the simulated image using the same algorithms as for experiments. For different constructs in Fig. 1G, the membrane fraction estimations were based on the corresponding average dendrite sizes. Gibbs free energy (Fig. 1H) was further calculated as follows:
where Fm is the fraction on membrane, R is the gas constant, and T is the absolute temperature.
For Fig. 2D, the above simulation was modified to include one additional component of full-length PKA distribution: a fraction (31.8% of total) of immobile PKA-C was restricted from a zone that was 20 nm to the membrane. The restricted distribution by itself mimicked the observed slightly negative MEI of PKA-C at rest. The fraction of mobile PKA-C on the membrane (with all mobile PKA-C summed to the 100%) was then plotted against MEIs to yield Fig. 2D.
For Fig. 4A, the pixel density was set to 2 nm and the membrane PKA activity was evenly distributed between r and r-0.01 µm of the circle (assuming that membrane-bound PKA-C can phosphorylate up to 10 nm [i.e., ∼1 molecular size of PKA-C] away from the membrane), where r is the radius of the dendrite and was specified using the experimental data for PKA-C MEI measurements. The membrane distribution was then added to the cytosolic distribution where the remaining fraction of PKA-C is evenly distributed throughout the entire dendrite. The activity ratio between the membrane and cytosolic PKA concentrations can also be analytically derived as follows:
where Fm is the fraction of PKA on the membrane, and d is the effective depth of membrane-PKA phosphorylation and is set to 10 nm. Computation and simulation gave essentially identical results.
Two-Photon Fluorescence Lifetime Imaging and Analyses.
2pFLIM was carried out in the time domain as described recently (29, 30). Briefly, a two-photon microscope was modified by adding hardware components to compare the timing of each photon with the pulse timing of the Ti:Sapphire laser. Data acquisition was controlled by custom software (called FLIMimage) written in MATLAB provided by Dr. Ryohei Yasuda with modifications. Excitation and imaging conditions are similar to conventional two-photon imaging for MEIs with the exception that a Chroma ET500/40× barrier filter was used for green to allow better suppression of residual sREACh fluorescence in the green channel. Data analyses were performed using custom software written in MATLAB named FLIMview (30). Binding ratios were the result of fitting the fluorescence lifetime curve in FLIMview with two exponentials, with one reflecting the unbound state and the other reflecting the bound state.
Electrophysiology.
Whole-cell voltage-clamp recordings were performed using a MultiClamp 700B amplifier (Molecular Devices). Electrophysiological signals were filtered at 2 kHz and digitized and acquired at 20 kHz using custom software written in MATLAB. Slices were perfused with artificial cerebrospinal fluid containing 4 mM Ca and 4 mM Mg. The internal solution contained (in millimolar) 132 Cs-gluconate, 10 Hepes, 10 Na-phosphocreatine, 4 MgCl2, 4 Na2-ATP, 0.4 Na-GTP, 3 Na-ascorbate, 3 QX314, and 0.2 EGTA with an osmolarity of 295 mOsmol/kg. The junction potential was calculated to be −17 mV using a built-in function in the Clampfit software (Molecular Devices). Several less-abundant anions (phosphocreatine, ATP, GTP, and ascorbate) were omitted in the calculation due to lack of data in the program. The Cl reversal potential was −75 mV.
To reduce recurrent activities, cultured hippocampal slices were cut on both sides of CA1, and 4 µM 2-chloroadenosine (Sigma) was present in all recording experiments. A total of 10 µM GABAzine (SR 95531, Tocris) was also included to suppress GABA currents. For electrical stimulation, a bipolar, θ-glass stimulating electrode (Warner Instruments) was positioned in the stratum radiatum 100 to 150 μm lateral to the recorded neuron. For all recordings, a transfected neuron and an untransfected neuron located within 50 µm of each other were sequentially recorded without repositioning the stimulation electrode. Measurements were carried out on averaged traces from ∼20 trials under each condition. For AMPAR currents, the cells were held at −60 mV (before correcting for the junction potential), and the current was measured as the baseline-subtracted peak current within a window of 2 to 50 ms after electric stimulation. For NMDAR currents, the average currents at 140 to 160 ms after stimulation were used when the cells were held at +55 mV (before correcting for the junction potential).
Data Analysis, Presentation, and Statistics.
Quantification and statistical tests were performed using custom software written in MATLAB. Averaged data are presented as mean ± SEM, unless noted otherwise. Throughout the paper, “n” indicates the number of neurons for MEI analysis and the number of spines for photoactivation experiments, unless noted otherwise. For photoactivation experiments, no more than four spines were from a single neuron. P values were obtained from one-way ANOVA tests, unless noted otherwise.
Supplementary Material
Acknowledgments
We thank all members of the Mao and Zhong laboratories at the Vollum Institute for constructive discussions. We thank Drs. Tianyi Mao, John Williams, Wolfhard Almers, and Bart Jongbloets for critical comments on the manuscript and Ms. Teresa Newton for proofing the manuscript. This work was supported by two NIH Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative awards to H.Z. (U01NS094247 and R01NS104944).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021658118/-/DCSupplemental.
Data Availability
Plasmid sequence data have been deposited in Addgene (https://www.addgene.org/Haining_Zhong/; ID 168482–168499).
Change History
January 17, 2022: Figure S5 has been updated.
References
- 1.Carr S. A., Biemann K., Shoji S., Parmelee D. C., Titani K., n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc. Natl. Acad. Sci. U.S.A. 79, 6128–6131 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Resh M. D., Covalent lipid modifications of proteins. Curr. Biol. 23, R431–R435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright M. H., Heal W. P., Mann D. J., Tate E. W., Protein myristoylation in health and disease. J. Chem. Biol. 3, 19–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farazi T. A., Waksman G., Gordon J. I., The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276, 39501–39504 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Peitzsch R. M., McLaughlin S., Binding of acylated peptides and fatty acids to phospholipid vesicles: Pertinence to myristoylated proteins. Biochemistry 32, 10436–10443 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Murray D., Ben-Tal N., Honig B., McLaughlin S., Electrostatic interaction of myristoylated proteins with membranes: Simple physics, complicated biology. Structure 5, 985–989 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Struppe J., Komives E. A., Taylor S. S., Vold R. R., 2H NMR studies of a myristoylated peptide in neutral and acidic phospholipid bicelles. Biochemistry 37, 15523–15527 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Francis S. H., Corbin J. D., Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 56, 237–272 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Theurkauf W. E., Vallee R. B., Molecular characterization of the cAMP-dependent protein kinase bound to microtubule-associated protein 2. J. Biol. Chem. 257, 3284–3290 (1982). [PubMed] [Google Scholar]
- 10.Lohmann S. M., DeCamilli P., Einig I., Walter U., High-affinity binding of the regulatory subunit (RII) of cAMP-dependent protein kinase to microtubule-associated and other cellular proteins. Proc. Natl. Acad. Sci. U.S.A. 81, 6723–6727 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong W., Scott J. D., AKAP signalling complexes: Focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Scott J. D., Pawson T., Cell signaling in space and time: Where proteins come together and when they’re apart. Science 326, 1220–1224 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beavo J. A., Bechtel P. J., Krebs E. G., Activation of protein kinase by physiological concentrations of cyclic AMP. Proc. Natl. Acad. Sci. U.S.A. 71, 3580–3583 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nairn A. C., Hemmings H. C. Jr, Greengard P., Protein kinases in the brain. Annu. Rev. Biochem. 54, 931–976 (1985). [DOI] [PubMed] [Google Scholar]
- 15.Johnson D. A., Akamine P., Radzio-Andzelm E., Madhusudan M., Taylor S. S., Dynamics of cAMP-dependent protein kinase. Chem. Rev. 101, 2243–2270 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Tillo S. E., et al., Liberated PKA catalytic subunits associate with the membrane via myristoylation to preferentially phosphorylate membrane substrates. Cell Rep. 19, 617–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clegg C. H., Ran W., Uhler M. D., McKnight G. S., A mutation in the catalytic subunit of protein kinase A prevents myristylation but does not inhibit biological activity. J. Biol. Chem. 264, 20140–20146 (1989). [PubMed] [Google Scholar]
- 18.Zheng J., et al., Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci. 2, 1559–1573 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yonemoto W., McGlone M. L., Taylor S. S., N-myristylation of the catalytic subunit of cAMP-dependent protein kinase conveys structural stability. J. Biol. Chem. 268, 2348–2352 (1993). [PubMed] [Google Scholar]
- 20.Bastidas A. C., et al., Role of N-terminal myristylation in the structure and regulation of cAMP-dependent protein kinase. J. Mol. Biol. 422, 215–229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaffarogullari E. C., et al., A myristoyl/phosphoserine switch controls cAMP-dependent protein kinase association to membranes. J. Mol. Biol. 411, 823–836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P., et al., An isoform-specific myristylation switch targets type II PKA holoenzymes to membranes. Structure 23, 1563–1572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamps M. P., Buss J. E., Sefton B. M., Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc. Natl. Acad. Sci. U.S.A. 82, 4625–4628 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Towler D. A., et al., Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263, 1784–1790 (1988). [PubMed] [Google Scholar]
- 25.Hancock J. F., Magee A. I., Childs J. E., Marshall C. J., All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–1177 (1989). [DOI] [PubMed] [Google Scholar]
- 26.Sigal C. T., Zhou W., Buser C. A., McLaughlin S., Resh M. D., Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc. Natl. Acad. Sci. U.S.A. 91, 12253–12257 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross F. R., Garber E. A., Pellman D., Hanafusa H., A short sequence in the p60src N terminus is required for p60src myristylation and membrane association and for cell transformation. Mol. Cell. Biol. 4, 1834–1842 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong H., et al., Subcellular dynamics of type II PKA in neurons. Neuron 62, 363–374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda R., et al., Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 9, 283–291 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Ma L., et al., A highly sensitive A-kinase activity reporter for imaging neuromodulatory events in awake mice. Neuron 99, 665–679.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakoshi H., Lee S. J., Yasuda R., Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 36, 31–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvey C. D., Yasuda R., Zhong H., Svoboda K., The spread of Ras activity triggered by activation of a single dendritic spine. Science 321, 136–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson G. H., Lippincott-Schwartz J., A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Bloodgood B. L., Sabatini B. L., Neuronal activity regulates diffusion across the neck of dendritic spines. Science 310, 866–869 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Gray N. W., Weimer R. M., Bureau I., Svoboda K., Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 4, e370 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteban J. A., et al., PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat. Neurosci. 6, 136–143 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Udenwobele D. I., et al., Myristoylation: An important protein modification in the immune response. Front. Immunol. 8, 751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoppini L., Buchs P. A., Muller D., A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37, 173–182 (1991). [DOI] [PubMed] [Google Scholar]
- 39.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011).
- 40.Otmakhov N., Lisman J., Measuring CaMKII concentration in dendritic spines. J. Neurosci. Methods 203, 106–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pologruto T. A., Sabatini B. L., Svoboda K., ScanImage: Flexible software for operating laser scanning microscopes. Biomed. Eng. Online 2, 13 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmid sequence data have been deposited in Addgene (https://www.addgene.org/Haining_Zhong/; ID 168482–168499).