Significance
In this work, we show that the last step of cytokinesis, termed abscission, is delayed in early zebrafish embryos. As a result, sibling cells remain connected to one another by a thin membrane bridge for several cycles, forming clusters of interconnected cells. Bridge severing (i.e., abscission) commences at the 10th cell cycle when embryos enter the midblastula transition switch, in which embryonic cells become individualized and exhibit the characteristics of mature cells. Cells connected by intercellular bridges shared similar cellular behaviors, such as transcription onset and cell shape. Our data suggest that cell–cell connectivity is maintained in early embryos through persistent bridge connections that allow cells to coordinate their behavior during embryonic development.
Keywords: cell division, microtubules, development, MBT, abscission
Abstract
Animal cytokinesis ends with the formation of a thin intercellular membrane bridge that connects the two newly formed sibling cells, which is ultimately resolved by abscission. While mitosis is completed within 15 min, the intercellular bridge can persist for hours, maintaining a physical connection between sibling cells and allowing exchange of cytosolic components. Although cell–cell communication is fundamental for development, the role of intercellular bridges during embryogenesis has not been fully elucidated. In this work, we characterized the spatiotemporal characteristics of the intercellular bridge during early zebrafish development. We found that abscission is delayed during the rapid division cycles that occur in the early embryo, giving rise to the formation of interconnected cell clusters. Abscission was accelerated when the embryo entered the midblastula transition (MBT) phase. Components of the ESCRT machinery, which drives abscission, were enriched at intercellular bridges post-MBT and, interfering with ESCRT function, extended abscission beyond MBT. Hallmark features of MBT, including transcription onset and cell shape modulations, were more similar in interconnected sibling cells compared to other neighboring cells. Collectively, our findings suggest that delayed abscission in the early embryo allows clusters of cells to coordinate their behavior during embryonic development.
Cytokinesis is the part of cell division during which the cytoplasm of a single eukaryotic cell divides into two sibling cells. Animal cytokinesis begins with furrow ingression and is followed by the formation of a thin (∼1 μm) and long (∼10 μm) intercellular bridge that is packed with dense microtubule stalks and is surrounded by membrane (1–3). The intercellular bridge is ultimately severed in a process termed abscission that leads to the physical separation of the two daughter cells (1, 4). Abscission is a highly coordinated process that is executed by the Endosomal Sorting Complex Required for Transport (ESCRT) membrane fission machine and the microtubule-severing enzyme, Spastin, and is temporally regulated by cytokinetic proteins such as Aurora B (4–7).
The intercellular bridge persists for different durations, ranging from minutes to hours, depending on cell type and cellular context (2, 8–11). During this period, the cells are connected to one another and are able to exchange cytosolic components until abscission commences and the two cells become physically separated (10–13). Although chronologically, abscission is the last step of cell division, it was shown that cells can continue to advance in cell cycle without resolving their intercellular bridges and that in mammalian cells, abscission commences at the gap 1 (G1) phase of the following cell cycle (7, 8, 14, 15). The ability to maintain cell connectivity while progressing through cell cycle, which can be mediated by regulating abscission timing, may have implications during early stages of development. Yet, although mitosis in general and cytokinesis in particular have been extensively studied during development, the role of abscission itself in a developmental setup has not been fully elucidated (3, 16–19).
Although the molecular components that drive and regulate abscission were shown to be similar in various organisms (i.e., tissues and cellular contexts), the spatiotemporal characteristics of abscission and its regulation vary between systems and, in some cases, is associated with specific developmental phenotypes (7, 9, 10, 20). For example, in Drosophila germ cells, delayed abscission leads to the formation of stem-cysts, where all cells share the same cytoplasm but each cell remained individualized (10). Furthermore, abscission timing is differentially regulated in Drosophila germ cells, and interfering with abscission timing caused developmental abnormalities, such as encapsulation defects (10, 20). In early mice embryos, zebrafish (Danio rerio) oocytes, Caenorhabditis elegans embryos, and Madin-Darby canine kidney (MDCK) cysts, the position of the intercellular bridge and the timing of abscission were directly related to cell polarization (7, 15, 21–23). Finally, the duration of abscission was recently shown to regulate pluripotency exit in mouse embryonic stem cells (14). Together, these findings suggest that abscission may play a role in regulating developmental processes.
Zebrafish embryogenesis begins with 10 cycles of rapid (∼15 min), synchronized divisions composed of mitotic and synthesis phases while missing the G1 and G2 phases (24, 25). At the end of the fourth cell cycle, complete plasma membrane furrowing occurs for the first time and intercellular bridges can be formed (25). During the fast cell cycles, transcription and protein synthesis are inhibited. As a consequence, cell volume decreases in each cycle and cellular behavior is mediated by maternally deposited messenger RNAs (mRNAs) and proteins, which are diluted from one cycle to the other (24, 26). At the 10th cell cycle, midblastula transition (MBT) occurs. The MBT is characterized by cell cycle lengthening (including gap phases), loss of cell synchrony, apoptosis, appearance of cell motility, and onset of zygotic transcription (called zygotic genome activation [ZGA]) (27). These changes are prerequisite for acquiring different cell fates and specific morphological forms during development (24, 26).
The dramatic changes that occur at the onset of MBT in aquatic animal species are precisely timed in the embryo. The nuclear to cytoplasmic ratio (N/C), which gradually increases due to the reductive nature of early divisions, plays a critical role in triggering MBT (26, 28, 29). In parallel, gradual dilution of maternally expressed transcription repressors and reduction of replication and transcription factors, resulting from the lack of transcription, were shown to control ZGA (19, 26, 30–33). Whether and how these factors are coordinated between single cells in the embryo is largely unknown. Previous work by Kane and Kimmel describing the changes that occur during MBT in zebrafish embryos suggested that the timing of MBT onset is more similar in immediate linear ancestor cells (e.g., sibling cells) than in cells residing at physical proximity (27). Yet, how cells distinguish their siblings from other neighboring cells has not been defined.
Regulating abscission timing can potentially affect coordination between sibling cells versus nonsibling cells by maintaining connectivity between cells through intercellular bridges. Here, we characterized abscission timing during the development of zebrafish embryos and found that abscission is delayed until the 10th cell cycle (e.g., MBT). Consistent with the abscission delay, expression levels of abscission-related genes, including the core ESCRT component Chmp4bb, were found to rise at MBT. Prolonged inhibition of Chmp4bb expression, via morpholino, delayed abscission beyond the 10th cell cycle. Prior to MBT, sibling cells continued to divide rapidly without resolving their intercellular bridges, giving rise to the formation of interconnected sibling cell clusters. Notably, interconnected sibling cells exhibited a similar pattern of MBT-associated transcription onset and cell shape changes compared to nonconnected neighboring cells in the embryo. Collectively, our data indicate that abscission is part of the MBT switch and suggest that cells can temporally coordinate specific cellular behaviors by maintaining cell connectivity through intercellular bridges, which may ultimately influence their developmental program.
Results
Characterizing Cytokinetic Abscission in Blastula Stage.
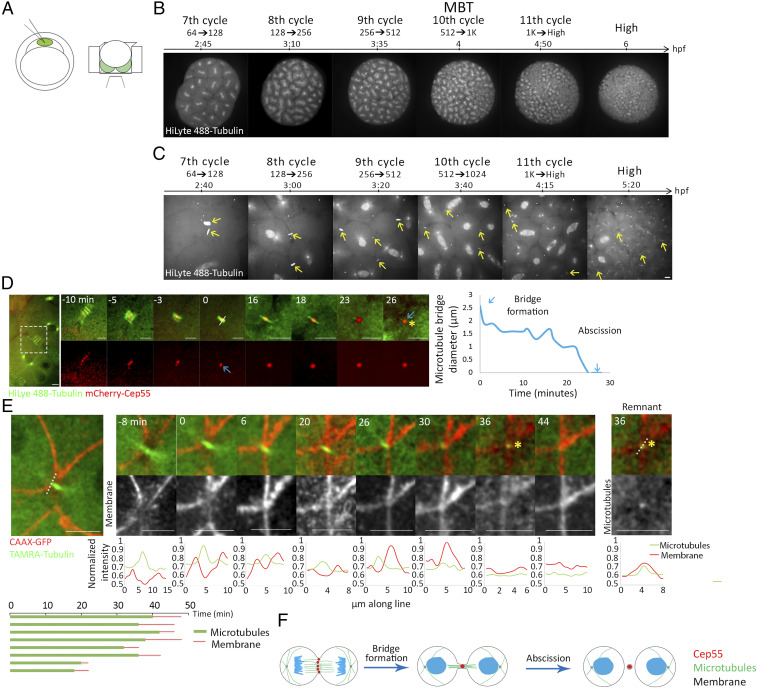
Formation of the intercellular bridge is characterized by compaction of the mitotic spindle microtubules. Proteins that accumulated at the cell equator during anaphase, such as CEP55, condense during this process, giving rise to the formation of a dense structure called the midbody (3, 34). Cleavage of the intercellular bridge is characterized by narrowing of the microtubule stalk at either side of the midbody followed by the release of a midbody remnant (3, 4, 35). To visualize these features in developing embryos, single-cell embryos were injected with fluorescently labeled tubulin and with an mRNA encoding for the midbody protein Cep55 tagged with mCherry, and imaged on a confocal spinning disk using custom-designed agar chambers [previously described by Mitchison and colleagues (36)] (Fig. 1A and SI Appendix, Fig. S1A). This injection procedure did not affect the ability of the embryo to develop into a fully differentiated fish, and embryos imaged under these conditions divided normally under the microscope and followed the previously described timeline for zebrafish at the blastula stage (Fig. 1B and SI Appendix, Fig. S1B) (27). Importantly, using this setup, mitotic divisions and intercellular bridges could be readily recorded over time (Fig. 1 B and C and Movies S1 and S2).
Fig. 1.
Cytokinetic abscission in zebrafish embryos at blastula stage. (A) Schematic illustration of the experimental setup used to visualize abscission in live zebrafish embryos during blastula. One-cell-stage embryos were injected with mRNA encoding for specific proteins and with HiLyte 488-Tubulin (Left), dechorionized and mounted in custom-made agar chambers (described in Methods and SI Appendix, Fig. S1B) with the animal pole facing the coverslip, and imaged using an inverted spinning disk (Right). (B, C) Whole embryos (B) and intercellular bridges (C) visualized throughout blastula stage in live embryos. Shown are selected frames from time lapse movies demonstrating mitosis at 7th to 11th cell cycle stages using tubulin labeling. Maximum intensity projection (30 Z slices at 1 µm) is shown (Movies S1 and S2). (D) Representative time frames from live imaging recordings of an embryo at the 8th to 10th cell cycles injected with HiLyte488-tubulin (green) and mRNA encoding to mCherry-cep55 (red) (Movie S3). A zoomed-out image of the two dividing cells is shown to the left. Time 0, bridge formation, was set as the first time point at which a single microtubule stalk with a packed Cep55 puncta was observed. Formation of a midbody remnant (asterisk) was defined as the completion of abscission. Plot on the right: the diameter of the microtubule stalk was measured at the rim of the midbody from bridge formation (white line, time 0) and plotted through time. Blue arrows represent bridge formation and abscission events. This analysis was repeated for each of the intercellular bridges measured in this study. (Scale bar, 10 µm.) n = 18 bridges, five embryos. (E) Representative time frames from live imaging recordings of an embryo at the 10th cell cycle injected with TAMRA-tubulin (green) and mRNA encoding CAAX-GFP (red) (Movie S4). A zoomed-out image of the two dividing cells is shown to the left. Time 0, bridge formation; asterisk, midbody remnant. Line intensities along the line drawn in the zoomed-out image are plotted for each channel at the bottom of each image. Note that the microtubule stalk (single peak, red) is surrounded by a membrane tube (two peaks, green). Midbody remnant formation (asterisk) is accompanied by decreased tubulin signal at the location of the bridge. A continuous membrane staining is observed several minutes later, representing closure of the membrane tube. (Right) An image and a line intensity plot of the midbody remnant showing that the tubulin foci is surrounded by a membrane staining. (Bottom) Measurements of intercellular bridge durations based on microtubule signal (green) and membrane signal (red) as determined for individual bridges. (Scale bar, 10 µm.) n = 8 bridges, two embryos. (F) Schematic illustration of the major steps in cytokinesis in zebrafish embryos.
Using this experimental setup, intercellular bridges were not detected in embryos prior to the 5th cell cycle. At the end of the 5th cell cycle (32 cells), few bridges were detected, and from the 6th cell cycle (64 cells) onward, bridges could be readily detected and followed through time. To test whether the absence of intercellular bridges reflects the incomplete membrane furrowing previously reported for zebrafish embryos at early stages (25), we examined the ability of proteins to diffuse between cells using a photoactivation assay. Activating PA-mCherry in a single cell of an embryo at the beginning of the 5th cell cycle (16 cells) quickly resulted in mCherry fluorescence in the entire embryo, indicating that all cells are indeed connected to one another at this developmental stage (SI Appendix, Fig. S2A). The data suggest that intercellular bridges do not form prior to complete furrowing and first occur in the embryo at the 5th cell cycle.
Intercellular bridges imaged in embryos at the 6th cell cycle or later exhibited the typical bridge formation and abscission, previously characterized in other systems (2, 34, 35, 37, 38). Contraction of the spindle microtubules was accompanied by CEP55 condensation, giving rise to the formation of an intercellular bridge. The diameter of the microtubule stalk decreased over time until a single punctum of CEP55 and microtubules was observed, suggesting the formation of a midbody remnant (Fig. 1D and Movie S3). Membrane labeling, using a CAAX-GFP marker, showed a gap in the membrane where the microtubules condensed to form the intercellular bridge, which became narrower over time (Fig. 1E and Movie S4). Complete gap closure was observed several minutes after the disappearance of the bridge microtubules, indicating that microtubule severing precedes membrane closure, as previously described (Fig. 1 E, Bottom) (39). The midbody remnant was surrounded by membrane staining, indicating that microtubule severing was accompanied by membrane severing (Fig. 1 E, Right). Based on these observations, we concluded that bridge formation and abscission exhibit the typical characteristics observed in mammalian cells and that tracking microtubule diameter and midbody remnant formation can be used as a readout of abscission timing (Fig. 1F).
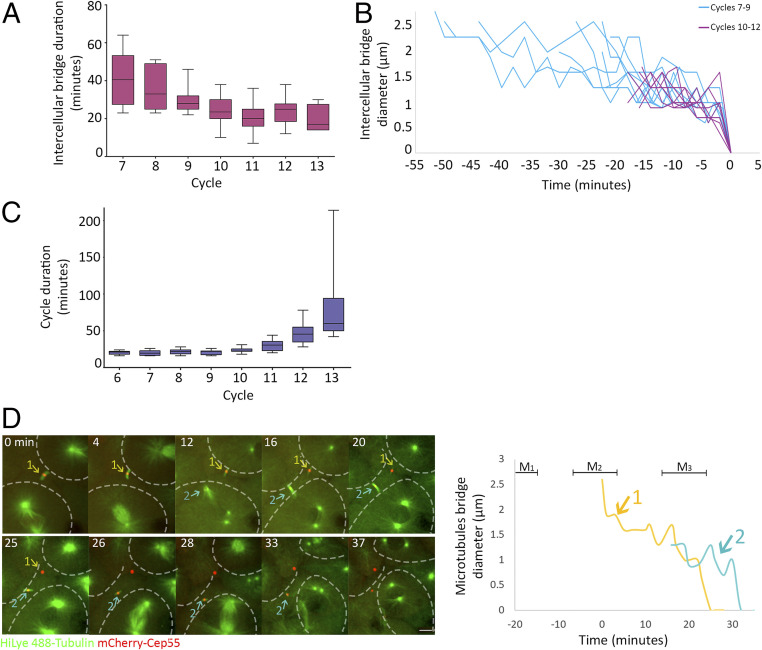
Measurements of intercellular bridge durations at different division cycles revealed that intercellular bridges that were formed at subsequent cycles persisted for shorter times, ranging from over 40 min in 7th cell cycle to ∼20 min in the 10th to 13th cell cycles (Fig. 2A). The analysis of intercellular bridge diameter, used to determine the duration of abscission, revealed that the long-lasting intercellular bridges formed during the 7th to 9th cell cycles were wider (>2 μm) than the short-lived bridges, formed at later cycles, which exhibited the characteristic 1 μm diameter (Fig. 2B). The observed shortening in intercellular bridge duration was in contrast to the overall cell cycle duration, which exhibited the previously characterized fast divisions until the 10th cell cycle (every ∼20 min) and was followed by gradual elongation of the cell cycle duration to over an hour (Fig. 2C) (25, 27). The time duration of intercellular bridges (∼40 min) relative to the short division times (∼20 min) in 7th and 8th cell cycles indicated that intercellular bridges formed in these cycles persists through the following cycles. Indeed, live imaging of embryos at these stages revealed that cells connected via intercellular bridges continued to the next mitotic cycle, ultimately forming new intercellular bridges before severing the previous ones (Fig. 2D and Movie S5). Thus, prior to the 10th cell cycle (MBT), individual cells progress through the cell cycle without resolving their intercellular bridges and can be simultaneously connected by intercellular bridges to more than one sibling cell.
Fig. 2.
Intercellular bridges persist for longer times in early cell cycles. (A) Averaged durations of intercellular bridges (measured based on Cep55 and tubulin signal, as described in Fig. 1D) formed at different cell cycles. n = 49 bridges, five embryos. Error bars, SD. (B) Intercellular bridge diameters measured in bridges that formed before (blue) or after (purple) the 10th cell cycle. Time 0, time of abscission. (C) Averaged cell cycle durations measured in embryos at different division cycles. Values were measured for individual cells, and averaged values obtained for each embryo were plotted according to the division cycle. n = 9 embryos; error bars, SD. (D) Live imaging of dividing cells in zebrafish embryos at the 8th to 10th cell cycles. Maximum intensity projections (30 Z slices at 1 µm) taken from selected time points of a representative embryo are shown (Movie S5). Microtubules, green; Cep55, red; intercellular bridges, arrows. Cells outlines are marked with dashed lines. The durations of two intercellular bridges observed in the movie sequence are plotted to the right. Colors in plot correspond to arrows in the movie sequence. Mitosis (M); numbers refer to bridge numbers (M1, bridge 1). n = 8 cells, five embryos. (Scale bar, 10 µm.)
Abscission Is Delayed Prior to MBT.
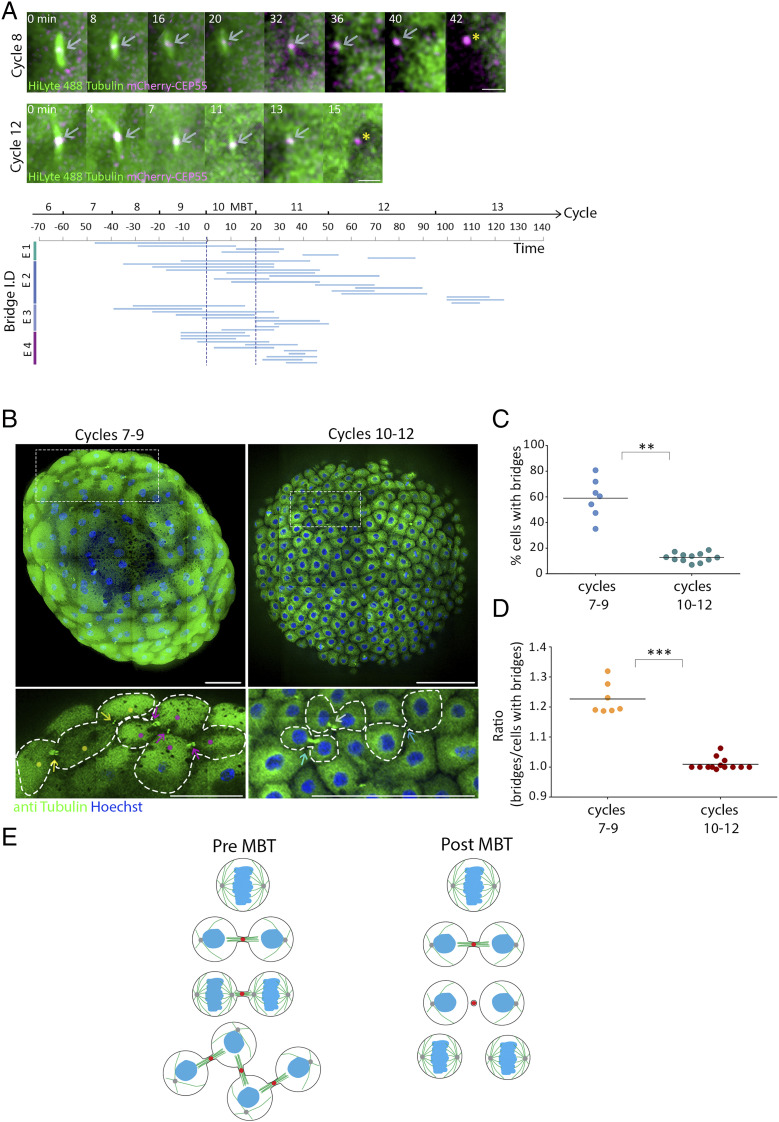
As we found that cells can be connected by multiple intercellular bridges before MBT, we hypothesized that intercellular bridge duration and abscission timing are related to the MBT switch. To test this possibility, we traced individual bridges over time in single embryos and measured the precise timing of abscission in the developing embryo. Interestingly, abscission events were rarely documented in embryos before entering the 10th cell cycle of MBT (Fig. 3A). After MBT, abscission of bridges that formed either before or after the 10th cell cycle was observed. Therefore, the prolonged durations observed for intercellular bridges that formed in early cell cycles (6th to 9th) may arise from inhibition of abscission prior to MBT.
Fig. 3.
Abscission is stalled until MBT in zebrafish embryos. (A) Embryos injected with mRNA encoding to mCherry-Cep55 (magenta) and HiLyte488-tubulin (green) were imaged live, and individual intercellular bridges were tracked and measured over time (as described in Fig. 1D). (Upper) Maximum intensity projection images (10 Z slices of 0.7 µm) of representative intercellular bridges formed pre-MBT (8th cell cycle) or post-MBT (12th cell cycle), as indicated. Gray arrows, intercellular bridges; asterisks, midbody remnants. (Scale bar, 5 µm.) (Bottom) The timeline of intercellular bridges obtained from single embryos in relation to MBT. Time 0, entrance to the 10th cell cycle. Each line represents an individual intercellular bridge (from formation to abscission). (B) Tile images of whole embryos fixed pre- (7th to 9th cycles) or post-MBT (10th to 12th cycles) and stained with anti-tubulin and Hoechst. Enlarged images of the regions in dashed rectangles in upper panels are shown on the bottom. Dots, cells; arrows, intercellular bridges; dashed lines, cells outline. Clusters of connected cells are labeled with similar colors. (Scale bar, 100 µm.) (C) Percentage of cells with intercellular bridges visualized in embryos fixed at the 7th to 9th cell cycles (average = 59% ± 12.21%, n = 7 embryos) and in the 10th through 12th cell cycles (average = 12.8% ± 3.43%, n = 12 embryos). The percentage of cells in late cytokinesis in pre-MBT embryos (7th to 9th cell cycles) is significantly higher than in post-MBT embryos (10th to 12th cell cycles) according to t test (t0.05,6 = 7.921, P = 0.002). (D) The ratio of intercellular bridges per cell at late cytokinesis was measured in pre- and post-MBT embryos and plotted on the graph. Averaged ratios for pre-MBT embryos = 1.009 ± 0.019 (n = 7 embryos); post-MBT embryos = 1.22 ± 0.048 (n = 12). Ratios in pre-MBT embryos are significantly higher than in post-MBT embryos (Mann–Whitney U test, U7,9 = 0, P = 0.0002). (E) Schematic model of intercellular bridge connection in pre- and post- MBT embryos. **P = 0.001 to 0.01, ***P = 0.0001 to 0.001.
The findings that abscission is delayed during the rapid mitotic cycles prior to MBT suggests that cells at early blastula accumulate intercellular bridges over time. To estimate the extent of this phenomenon, intercellular bridges were visualized in whole embryos that were fixed either before MBT (pre-MBT, 7th to 9th cell cycles) or after MBT (post-MBT, 10th and 11th cell cycles), and the number of bridges was compared across stages (Fig. 3B). As expected by the high rate of synchronized divisions prior to MBT, the percentage of cells connected by intercellular bridges was considerably higher in pre-MBT compared to post-MBT embryos (60% versus 10%, respectively) (Fig. 3C). Interestingly, while in post-MBT embryos, cytokinetic cells exhibited the typical morphology of a pair of sibling cells connected to each other with an intercellular bridge, in pre-MBT embryos, cytokinetic cells often obtained intercellular bridge connections to more than one cell, forming local clusters of interconnected sibling cells (Fig. 3B). Quantification of the number of bridges connected per cell in post-MBT embryos (10th to 12th cell cycles) resulted the typical 1:1 ratio, indicating that each cell is connected by a single bridge. However, in pre-MBT embryos (7th to 9th cell cycles), we observed a significantly higher ratio of 1:1.24, indicating that, on average, each cell is connected by more than one intercellular bridge (Fig. 3D).
Next, we aimed to validate that the persisted intercellular bridges observed in pre-MBT embryos allow the exchange of cytosolic components between cells, as previously demonstrated in other cellular contexts (10–13). To this end, we injected fluorescently labeled dextran into a single cell at a 128-cell (7th cycle) live embryo and tracked its diffusion over time. In contrast to the complete cellular interconnectivity observed in cells at the 16-cell stage (SI Appendix, Fig. S2A), dextran injected at 128 cells quickly diffused between interconnected cells in the same cluster but was excluded from other neighboring cells (SI Appendix, Fig. S2B and Movie S6). Collectively, these data suggest that prior to MBT, cells continue to divide rapidly without resolving their intercellular bridges, which allows for the formation of interconnected sibling cells that can exchange cytosolic material (Fig. 3E).
The Spatiotemporal Expression of ESCRTs Is Involved in Cytokinetic Abscission at Blastula Stage.
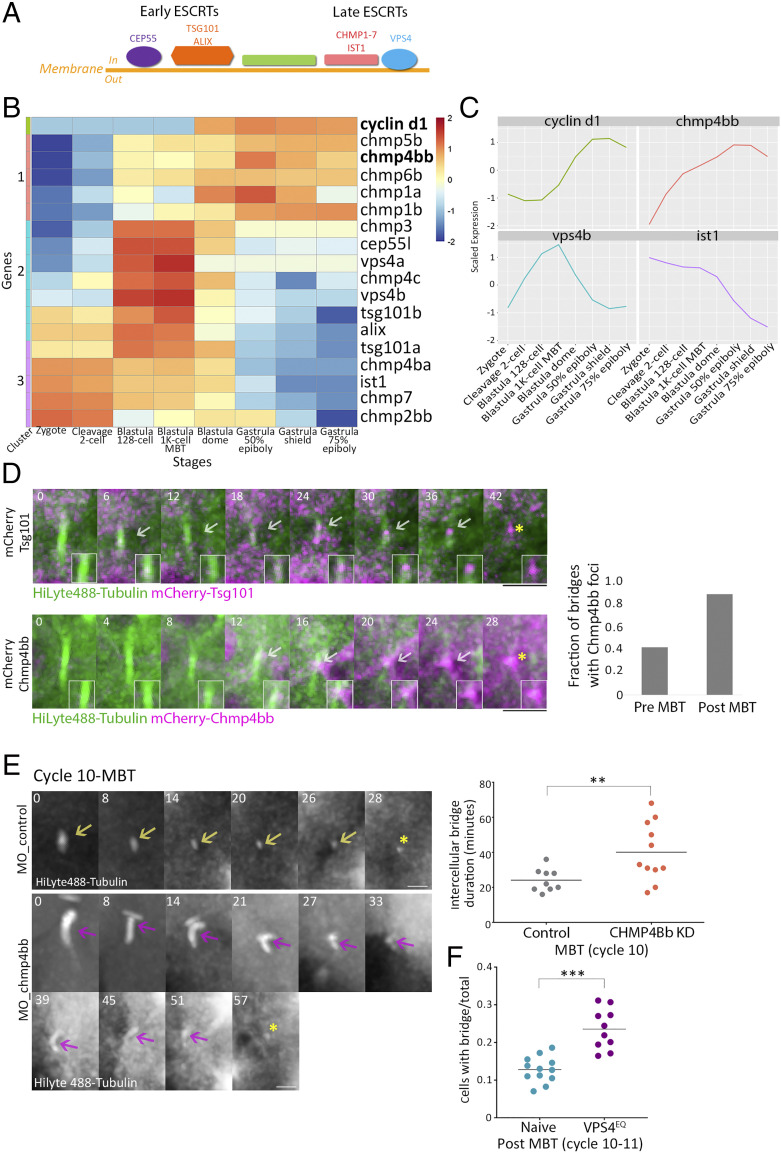
MBT is manifested by the onset of massive transcription and global gene expression. While proteins that are essential for the first division cycles are maternally expressed, other proteins begin to transcribe from maternally deposited mRNA at early blastula stage (minor wave), and most proteins are expressed after MBT onset (major wave) (26, 40). The temporal inhibition of gene expression prior to MBT raises the possibility that expression absence of abscission-related genes is associated with the observed abscission delay. To test this notion, we analyzed the temporal transcription of 48 abscission-related genes across eight developmental stages, pre- and post-MBT, using the gene expression atlas of zebrafish embryogenesis (41) (SI Appendix, Fig. S3A). Among the 48 abscission genes, we focused our analysis on the 17 genes in the zebrafish genome that belonged to the ESCRT machinery, a multiprotein complex that actively executes scission of the intercellular bridge membrane and is composed of early components (e.g., CEP55, TSG101, and ALIX) and late components (CHMP1-7, IST1, and VPS4) (5, 42) (Fig. 4A). Therefore, the availability of ESCRT components, and particularly of late ESCRT components, can directly affect abscission timing.
Fig. 4.
ESCRT proteins are involved in abscission during zebrafish embryogenesis. (A) Schematic representation of the canonical ESCRT cascade. Early ESCRT proteins, such as CEP55, ALIX, and TSG101, recruit late ESCRTs (CHMP1-7 and IST1) and VPS4 to the inner side of the plasma membrane where scission occurs. (B) Hierarchical clustering of the expression levels of the zygotically activated gene cyclin d1 and 17 ESCRT genes taken from RNA sequencing data from eight developmental stages [downloaded from White et al. (41)]. Rows, individual genes; columns, developmental stages. Colors in the heatmap indicate the expression levels of each gene across the samples relative to its mean expression and trimmed to range [−2, 2] (SI Appendix, Fig. S3B for raw data). Clusters 1–3, color coded to the left of the heatmap, are based on expression patterns. (C) Normalized mRNA levels obtained for Cyclin d1 and representative ESCRT genes plotted through time. x-axis, developmental stages; y-axis, normalized expression levels. (D) Embryos were injected with mRNA that encodes for Tsg101 (Upper) or Chmp4bb (Bottom), fused to mCherry (magenta) and with HiLyte 488-Tubulin (green). Shown are maximum intensity projection images of subvolumes of the cells that include the intercellular bridge (10 through 25 Z slices of 0.7 µm intervals) taken from time lapse movies of representative cells imaged in embryos at the 12th cell cycle. Both proteins localized to the intercellular bridge (gray arrow) and were detected in the midbody remnants after abscission (asterisks). Time 0, bridge formation. (Insets) Zoomed-in images of the midbody. (Scale bar, 10 µm.) (Right) A fraction of intercellular bridges exhibiting chmp4bb localization in pre- and post-MBT of embryos. Tsg101, n = 8 bridges, two embryos; chmp4bb, n = 46 bridges, three embryos. (E) Embryos were injected with morpholino oligos (control or Chmp4bb specific), and intercellular bridges were recorded through time in embryos at the 10th cell cycle. Shown are maximum intensity projection images (6 through 10 Z slices of 1 µm interval) taken from movies of representative intercellular bridges (Movie S7). Asterisks, midbody remnant. (Scale bar, 5 µm.) (Right) Intercellular bridge durations measured for individual intercellular bridges (as described in Fig. 1D) in control and knockdown (KD) embryos. Averaged bridge duration in Chmp4bb KD embryos = 40.09 ± 16.82 (n = 11 bridges, five embryos); in control embryos, averaged bridge duration = 24.11 ± 6.47 min (n = 9 bridges, three embryos). Intercellular bridge duration was significantly longer in Chmp4bb KD embryos compared to control embryos (unpaired t test with Welch's correction for unequal variances, t0.05,13 = 2.9, P = 0.012). (F) Percentage of cells with intercellular bridges in embryos at the 10th to 12th cell cycles (post-MBT). VPS4EQ-injected embryos had a significantly higher ratio than naive embryos (unpaired t test with Welch's correction for unequal variances, t0.05,14 = 2.9, 5.48, P < 0.0001). n = 22 embryos. **P = 0.001 to 0.01, ***P = 0.0001 to 0.001.
Using cluster analysis of the temporal transcription of different genes, three different expression patterns were found for ESCRT genes (clusters 1–3). The expression pattern of genes in cluster 1 was characterized by low expression prior to MBT that elevated afterward, resembling the expression pattern of cyclin d1, which was used as a reference for zygotic-expressed genes (43, 44). The expression pattern of genes in cluster 2 was characterized by elevated expression at the 128-cell stage, correlating with the expression levels previously documented for minor wave genes (40). Genes in cluster 3 showed relatively high expression levels from one cell stage (zygote) and can therefore be considered as maternally expressed genes. Among the 17 ESCRT genes analyzed, five genes, including tsg101a and ist1, were found in cluster 3, suggesting that these genes are maternally expressed. A total of seven ESCRT genes, including cep55, vps4a, and vps4b, were in cluster 2, and another five genes, including chmp4bb, were in cluster 1, indicating that more than half of the ESCRT genes are likely not being deposited maternally (Fig. 4 B and C and SI Appendix, Fig. S3B).
As CHMP4B and VPS4 manifest the minimal evolutionary conserved complex required for membrane scission (45), we further analyzed their protein expression levels. Using western blot analysis, we found that Vps4 could be detected at embryos of 256 cells, while Chmp4bb could only be detected at MBT or later, supporting the spatiotemporal expression patterns of these genes and indicating, in particular, that chmp4bb is not expressed prior to MBT (SI Appendix, Fig. S3C). Live imaging of fluorescently tagged versions of early ESCRTs (Tsg101) and late ESCRTs (Chmp4bb and Vps4) showed that they all localize to the intercellular bridge prior to abscission in post-MBT embryos, confirming that ESCRTs are involved in cytokinetic abscission of cells in early zebrafish embryos (Fig. 4D and SI Appendix, Fig. S4A). The additional Chmp4B homolog in the zebrafish genome, Chmp4ba, which has a similar sequence homology to the mammalian CHMP4B, could not be detected at intercellular bridges in the embryo. We therefore concluded that, at least during embryogenesis, Chmp4bb is the functional CHMP4B homolog in zebrafish cytokinetic abscission (SI Appendix, Fig. S4 B and C). Notably, the fraction of intercellular bridges in which Chmp4bb could be detected was significantly higher in post-MBT than pre-MBT embryos, validating the observed abscission delay and supporting a role for Chmp4bb in timing abscission in the embryo (Fig. 4 D, Right). Altogether, these results suggest that ESCRTs are involved in abscission of intercellular bridges of embryos at blastula stage and that the availability of specific ESCRT components, including Chmp4bb, can potentially be involved in the abscission delay observed prior to MBT.
To test the effect of ESCRT expression on abscission timing in cells at the blastula stage, we used a chmp4bb morpholino, which reduced Chmp4bb levels by 70% (compared to embryos injected with a control morpholino sequence) (SI Appendix, Fig. S4D). Live imaging of cytokinetic cells in embryos injected with chmp4bb morpholino revealed a significant abscission delay at the 10th cell cycle (40.1 ± 16.8 min versus 24.1 ± 6.47 min) (Fig. 4E and SI Appendix, Fig. S4E). No major difference in intercellular bridge durations were recorded in cells at the 9th cell cycle, in accordance with the lack of Chmp4bb expression at these stages (SI Appendix, Fig. S4E). A noticeable but milder delay was observed at the 11th cell cycle, and no delay was observed at later cycles. Therefore, reducing Chmp4bb levels in early embryos stall abscission at MBT. Consistently, injection of mRNA encoding a dominant negative version of VPS4 (VPS4EQ) resulted in a significant increase in the number of intercellular bridges observed post-MBT (Fig. 4F). The milder phenotype observed upon depletion of Chmp4bb at later cell cycles may reflect a critical need for high levels of Chmp4bb at MBT to allow the severing of the intercellular bridges that accumulated in the embryo. Alternatively, the transient reduction in Chmp4bb levels caused by the morpholino approach may not have been sufficient for inducing a more severe phenotype. That said, embryos injected with Chmp4bb morpholino exhibited a significantly reduced survival rate (<30% in embryos injected with Chmp4bb morpholino versus >60% in embryos injected with a control morpholino sequence, measured 24 h postfertilization), suggesting that these phenotypes are sufficient to interfere with proper development of the embryo (SI Appendix, Fig. S4F). Together with the zygotic expression pattern of Chmp4bb, these results suggest that the levels of Chmp4bb may regulate the timing of abscission onset in the embryo and are crucial for the overall survival of the embryo.
Cells Connected by Intercellular Bridges Exhibit a Similar Transcription Pattern and Cell Shape Changes at MBT.
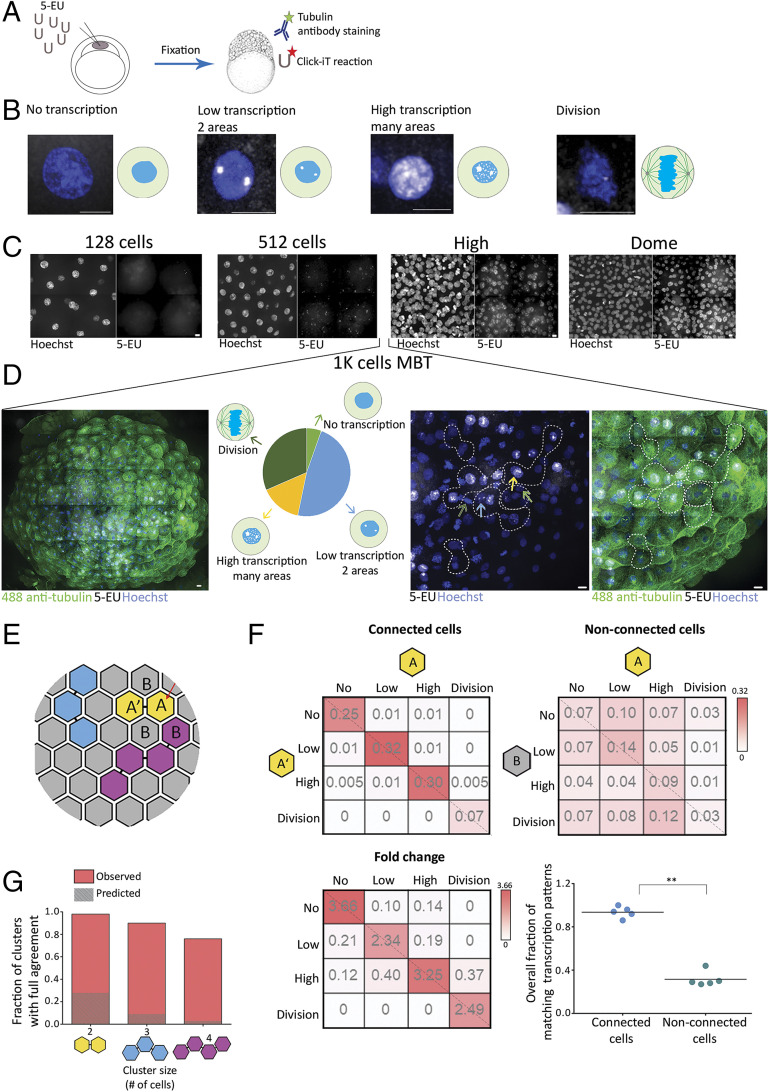
Next, we asked, what could be the developmental role of stalling abscission prior to MBT? As the global transcription of the zygote genome mainly occurs at the MBT, we set to examine the onset of transcription in relation to cell–cell connectivity in zebrafish embryogenesis. Initially, we characterized the overall transcription levels in single cells of pre- and post-MBT embryos. To this end, one-cell-stage embryos were injected with a modified uridine that is capable of binding a fluorescent dye via a click reaction (Fig. 5A). Upon transcription, the modified uridine is incorporated into the newly synthesized transcripts and can serve as a reporter for the overall transcription level in the nucleus, as previously described (46). Using this analysis, we identified the following four transcription patterns in embryos at blastula stage: 1) no transcription, only background uridine labeling; 2) low transcription, uridine labeling at two foci in the nucleolus; 3) high transcription, the entire nucleus is labeled with uridine; and 4) dividing cells, no uridine labeling accompanied by condensed chromatin staining at the cell equator (Fig. 5B). These transcription patterns were observed in different embryonic stages: In early stages (7th cell cycle, 128 cells), no transcription was observed; in the 9th cell cycle (512 cells), cells exhibited the characteristic staining of low transcription; and at later stages (12th to 15th cycles, high and dome), transcription levels were high (Fig. 5C and SI Appendix, Fig. S5A). Interestingly, while in embryos at early and late blastula stages (corresponds to 128 to 512 cells and high dome, respectively) a uniform transcription pattern was observed throughout the embryo, considerable variations in transcription patterns were observed between individual cells in embryos at the 10th cell cycle (Fig. 5D). In fact, all four transcription patterns could be observed in a single embryo at this stage. This heterogeneity in transcription levels during MBT indicates that cells autonomously regulate the timing of their transcriptional onset, as previously suggested (27).
Fig. 5.
Cells connected by intercellular bridges exhibit a similar transcription pattern. (A) A schematic illustration of the experimental setup. One-cell-stage embryos were injected with 5-ethynyl uridine (5-EU) and fixed at the indicated stages. Embryos were stained with anti-tubulin (for intercellular bridge detection) and Hoechst (for nuclear detection) and click-labeled using Alexa594-azide for detection of mRNA-incorporated 5-EU. (B) Single cells labeled with 5-EU (white) and Hoechst (blue), exhibiting the 5-EU labeling patterns observed in the embryos. Transcription patterns were classified into four different stages based on 5-EU labeling: no, low, high, and dividing cells (46). (Scale bar, 10 µm.) (C) Tile images of maximum intensity projection images (15 through 30 Z slices of 1 to 1.5 µm interval) of representative regions in embryos at the indicated developmental stages. n = 4–5 embryos from each stage. (Scale bar, 10 µm.) (D) Heterogeneous transcription patterns observed in embryos at MBT. (Far Left) A tile image of a representative embryo labeled with tubulin (green), Hoechst (blue), and 5-EU (white). (Middle Left) The relative distribution of the four transcription patterns (classified in B) observed in a representative embryo (no transcription, 5%; low transcription, 48%; high transcription, 15%; and dividing cells, 32%). (Middle Right and Far Right) Enlarged images of subset regions in the embryo with high levels of cell–cell variations in transcription patterns. Different transcription patterns are marked in arrows at different colors (corresponding to arrows in the Middle Left). Clusters of interconnected cells (marked in dashed lines) were identified based on tubulin staining and were used in the analysis described in E–G. (Scale bar, 10 µm.) (E–G) Quantitative analysis of the similarity in transcription patterns observed between connected and nonconnected cells. (E) Schematic representation of an embryo at MBT. Connected cells are filled with similar colors; letters correspond to the matrixes presented in F. (F) Resemblance matrix of transcription patterns of embryos at MBT. (Top) Pairs of connected (Left) and nonconnected (Right) cells (A, A′, and A, B, respectively) were scored according to their transcription pattern, and the fraction of specific combinations of transcription patterns between cell pairs was plotted in the matrix. Color indicates probability. Data were pooled across five embryos (206 connected and 768 nonconnected cell pairs). (Bottom Left) The fold change increase in fraction of connected versus nonconnected cell pairs. (Bottom Right) The accumulated probability of agreement in transcriptional patterns in connected versus nonconnected cell pairs (values were calculated independently for each embryo, n = 5). Connected cell pairs obtained a significantly higher transcriptional agreement compared to nonconnected pairs (0.94 ± 0.05 versus 0.32 ± 0.07, respectively; Mann–Whitney U test, U5,5 = 0.8, P = 0.003). (G) Clusters of two, three, and four connected cells were scored for matched transcription patterns such that only clusters in which all cells exhibited a similar transcription pattern were considered as clusters with intracluster agreement (red bars). Values were compared to simulated data, generated as described in SI Appendix, Supplementary Methods (dashed bars). Two-cell clusters (n = 150), 98.6% versus 28%; three-cell clusters (n = 30), 90% versus 9.1%; four-cell clusters (n = 12), 76.9% versus 3.1%. **P = 0.001 to 0.01.
Given that the MBT switch is thought to be driven by the ratios of cellular components and that diffusion occurs through intercellular bridges of interconnected sibling cells, we asked whether interconnected sibling cells exhibit similar transcription patterns. For that, embryos at the 10th cycle were labeled both for transcription (using modified uridine) and for intercellular bridges (using anti-tubulin antibodies). The transcription patterns of pairs of cells connected by bridges (connected cells) and adjacent pairs of neighboring cells that are not connected by a bridge (nonconnected cells) were determined (Fig. 5 E and F, A-A′ versus A-B, correspondingly). We then pooled all connected and nonconnected cell pairs and calculated the transcription agreement probability matrix, in which the bin at row r and column c holds the probability for a pair (connected or nonconnected) to have one cell in the transcriptional pattern corresponding to r and the other in the pattern corresponding to c (Fig. 4F). The accumulated probabilities at the diagonal of the agreement matrix is the probability of two cells within a pair to have the same transcriptional pattern. This analysis revealed that connected cells exhibit a higher degree of agreement in their transcription pattern compared to nonconnected neighboring cells (Fig. 5F, 0.94 versus 0.33 correspondingly). This observation of matched transcription patterns was not limited to cell pairs. In fact, the intracluster transcriptional agreement was even more striking in clusters of three and four connected cells in respect to a null model that assumed no spatial relations (Fig. 5G and SI Appendix, Supplementary Methods). These data show that the transcription pattern is strongly correlated in interconnected cells, suggesting that these cells are subjected to similar transcription regulation.
Last, we asked whether cellular properties downstream of transcription are coupled in interconnected cells at MBT. We focused on changes in cell polarization and shape that were previously described to occur during MBT (27). We thus compared the deviation in the cell’s aspect ratio (i.e., the major axis of each cells divided by its minor axis) in pairs of adjacent connected cells versus nonconnected neighboring cells (SI Appendix, Fig. S5B and SI Appendix, Supplementary Methods). More specifically, we considered a triplet of neighboring cells that included two connected cells and one nonconnected cell and measured their aspect ratio (SI Appendix, Fig. S5B and Movie S8). We calculated, for each triplet, the absolute difference in the aspect ratio between two connected and nonconnected cells as a measure of cell shape similarity. The difference in the aspect ratio of connected pairs was significantly smaller compared to neighboring nonconnected pairs, as verified by unmatched (0.23 ± 0.05 compared to 0.35 ± 0.19, respectively) or matched analysis, directly comparing cell pairs from the same triplet (SI Appendix, Fig. S5 C and D). These results establish a stronger agreement of cell shape in connected cells. Taken together with the similarity in the transcription pattern, these results suggest that abscission may be an integral part of MBT that allows coordinating cellular behavior during the MBT period.
Discussion
In this work, we showed that cells do not resolve their intercellular bridges prior to MBT and that abscission commences at the 10th cell cycle. This phenomenon was associated with the formation of clusters of cells that are connected by intercellular bridges and exhibit matching transcriptional and morphological features during MBT, raising the possibility that cells can communicate through their intercellular bridges. Indeed, intercellular bridges were previously shown to enable the exchange of molecules between daughter cells in both cultured mammalian cells and in whole organisms (Drosophila and C. elegans). Additionally, protein exchange was documented between cells in zebrafish embryos at gastrula stage that maintained long ranged intercellular bridges (13). Consistently, we showed that clusters of interconnected sibling cells exchange cytoplasmic dextran. Together, these findings suggest that delayed abscission prior to MBT plays a role in the formation of interconnected cell clusters in the embryos that may allow cells to coordinate their behavior at early stages of embryogenesis.
In 1995, Kimmel and colleagues provided a detailed description of the stages of embryonic development in zebrafish (25). In this seminal work, they report that in the first four cell divisions of zebrafish embryos, furrowing is not complete, and the cells remain partly connected to one another and to the yolk (as also confirmed by our photoactivation experiments). Once the embryo reaches the 32-cell stage, furrowing is complete, and the cells are considered as fully separated. Yet, several lines of evidence suggest that cells at these stages (32 to 1,000 cells, 5th to 10th cell cycles) sustain memory of their lineage. For example, in 1995, Kane and Kimmel reported that although cells autonomously set on the MBT switch, sibling cells exhibit more similar behaviors compared to other nonconnected neighboring cells (27, 47). Additionally, in cell lineage reconstruction experiments, cells in embryos at the 128- to 512-cell stage appeared to reside in clusters based on their 8-cell blastomere lineage (47). More recently, the levels of zygotically activated genes were shown to be more similar in daughter cells compared to other neighboring cells in embryos at blastula stage (31). All these observations indicated that sibling cells in the embryo maintain a cellular memory of their lineage, but how this memory is transmitted between the cells was not addressed. Given our observations of persistent intercellular bridges prior to MBT, we propose that the memory documented between cells at these stages originates from their cell–cell connectivity through intercellular bridges. Thus, we would like to suggest the following scenario for zebrafish development: Early embryogenesis begins by incomplete furrowing during mitosis until the embryo reaches the 32-cell stage. Then, complete furrowing occurs but abscission is delayed, and cells remain connected to one another until they reach 1,000-cell stage (MBT). After MBT, complete furrowing followed by abscission gives rise to independent cells. This scenario adds zebrafish embryogenesis to the long list of developmental processes mediated by incomplete cytokinesis (48) and suggests that abscission timing and cell–cell connectivity should be considered when addressing cellular behaviors in early zebrafish embryos.
How cells connected by intercellular bridges coordinate their transcription onset is still elusive. One possibility is that cytosolic diffusion through bridges leads to similar N/C ratios or to equilibration transcription repressor levels that directly affect ZGA. Alternatively, intercellular bridge connections may affect transcription onset indirectly through synchronizing other cellular behaviors, such as cell cycle duration and division time, or by maintaining a physical connection between the cells that affect their packing (49, 50). Further studies that combine quantitative analysis of these parameters with lineage tracking and intercellular bridge tracing in the early embryo will allow for this fundamental question to be resolved (50).
What governs the completion of abscission at MBT is still not fully understood. Our data suggests that the ESCRT-III component Chmp4bb contributes to this process. Notably, levels of the CHMP4B homolog in Drosophila, Shrub, were also shown to regulate abscission timing in the Drosophila germline, and loss of Shrub induced the formation of egg chambers with 32 connected cells (10). Given the fundamental role of CHMP4B in executing ESCRT-mediated fission in vitro and in vivo, it is possible that the cellular levels of Chmp4bb are tightly regulated during development and are served to control cell–cell connectivity in the embryo by regulating abscission timing (10, 35, 38, 51, 52). Expression levels of other abscission-related genes, such as rhoab, rab11a, and arf6b, and additional late ESCRT genes, such as CHMP3 and CHMP1B, were found to increase toward MBT, implying that the availability of additional genes may affect abscission timing in embryos to ensure a precise developmental program (SI Appendix, Figs. S3A and S4B). Additionally, known abscission timing regulators, such as Aurora B and Chmp4C, were found to be expressed prior to MBT, raising the possibility that the Aurora B–mediated abscission checkpoint also plays a role in delaying abscission prior to MBT.
The role of abscission and intercellular bridge connections in cellular regulations remained elusive over the years. Recently, Paluch and colleagues demonstrated that abscission timing is directly related to pluripotency exit with stem cells displaying slow abscission that is significantly accelerated upon pluripotency exit (14). Interestingly, several of the observations made in stems cells were highly similar to our observations in the developing embryo. First, similar to our coordinated transcription pattern, sibling cells displayed correlated pluripotency exit timing. Second, the slow abscission phase was accompanied with wider bridges. Third, the number of bridges exhibiting CHMP4B localization was significantly increased during pluripotency exit (i.e., abscission shortening). It is therefore tempting to speculate that the physiological role of abscission is to link cell division to cell fate decisions at various levels and cellular contexts.
Methods
Microinjection and Mounting for Live Imaging.
mRNA (0.8 to 1 ng) or morpholino oligos (5 ng) (SI Appendix, Table S1) were prepared and injected into one-cell embryos as described in SI Appendix, Supplementary Methods. Under these conditions, embryos were viable and showed normal cell division and physiological morphology at blastula stage (Fig. 1B and SI Appendix, Fig. S1B). Injected embryos were dechorionized and placed in specialized agar chambers for imaging according to Whur et al. (36) (SI Appendix, Supplementary Methods and Fig. S1A and Fig. 1A). Two morpholino oligos were injected and analyzed by immunoblot analysis for Chmp4bb knockdown (MO_1chmp4bb and MO_2chmp4bb) (SI Appendix, Fig. S4D). MO_1chmp4bb was found to be more efficient by western blot analysis and was therefore used in all the presented experiments.
Whole-Mount Immunostaining.
Naïve or injected embryos were fixed and labeled with monoclonal anti–α-Tubulin (DM1A; Sigma-Aldrich) followed by the secondary antibody Alexa Flour 488 anti-mouse (A21202, Invitrogen) and Hoechst (H-3570 Invitrogen). Embryos were dissected to remove the yolk and mounted in fluoromount-G (SouthernBiotech) on a coverslip with the animal pole facing the coverslip. Samples were imaged by confocal microscopy as described below. Detailed protocols are provided in SI Appendix, Supplementary Methods.
Click-iT Labeling of Transcription Pattern.
One-cell-stage embryos were microinjected with 50 pmols of 5 ethynyl uridine (5 EU, E10345), fixed at the indicated stages and labeled via Click-iT reaction kit [as previously described by Chan et al. (46)] and α-tubulin and Hoechst as described above. A detailed protocol is provided in SI Appendix, Supplementary Methods.
Imaging and Image Processing.
Live embryos were imaged from the 8th-cell cycle to dome stage (13th to 14th cell cycles) on a fully incubated, confocal spinning disk microscope at 26 °C (Marianas; Intelligent Imaging) using a ×63 oil objective (numerical aperture 1.4), ×40 oil objective (numerical aperture 1.3), or ×20 air objective (numerical aperture 0.8). Embryos were recorded on an electron-multiplying charge-coupled device camera (Evolve; Photometrics). A total of 25 to 32 confocal slices of 0.7 to 1.0 µm were captured for each time point at 1.5 to 2 min time intervals. Only the two cell layers close to the animal pole of the embryo were imaged and analyzed.
Fixed embryos were imaged with a ×40 oil objective (numerical aperture 1.4) or ×63 oil objective (numerical aperture 1.4). A total of 30 to 100 slices at 0.6 to 1 μm intervals were acquired, providing a Z range of up to 150 μm. Tile acquisition was applied for capturing the entire embryo, and three-dimensional volumes were merged to a montage using SlideBook (version 6, Intelligent Imaging Innovations). To improve visualization, images were subjected to Gaussian filtering in SlideBook.
Data Analysis.
Cell cycle length was defined by microtubules morphology as follows: two centrosomes, prophase; spindle, metaphase. Cell cycle length was defined as the time interval between one prophase to the next in the same cell.
Abscission duration was measured by annotating and tracking intercellular bridges through time. Intercellular bridge formation was defined based on the formation of condensed microtubule fibers between the cells (Tubulin channel) with a dense Cep55 puncta at the center (Fig. 1D). Completion of abscission was determined based on the diameter of the intercellular bridge and the formation of a midbody remnant, as previously described (2). In the absence of Cep55 expression, bridge duration was measured according to tubulin diameter and midbody remnant formation.
For measuring bridge to cell ratios, intercellular bridges and cells that carry intercellular bridges were manually annotated, and the ratio between them was calculated for each embryo. For calculations, intercellular bridges were counted twice (once for each cell it is connected to), resulting in a 1:1 ratio between intercellular bridges and cells under typical conditions.
Information on the analysis use for transcription pattern similarities in connected and nonconnected cells and on RNA-sequencing data visualization is provided in SI Appendix, Supplementary Methods.
Statistics.
Data were analyzed for column statistics in GraphPad Prism version 5.00 for Windows. Statistical significance was determined by t test, Mann–Whitney U test, or Wilcoxon matched-pairs signed rank test (Z) as specified in the figure legends.
Supplementary Material
Acknowledgments
We thank all members of the Elia and Birnbaum laboratories for their help and feedback throughout the project. We also thank members of the Peyrieras laboratory for hosting S.A.-L. and sharing with her their knowledge and expertise. The Elia laboratory is funded by the Israeli Science Foundation (ISF) Grant No. 1323/18. The Birnbaum laboratory is funded by the ISF (960/2016) and Research without Borders–St. Boniface Hospital Foundation–Ben-Gurion University of the Negev. A.Z. was supported by the Israeli Council for Higher Education via Data Science Research Center, Ben-Gurion University of the Negev, Israel. S.T.G.-O. was supported by Hi-Tech, Bio-Tech, and Chemo-tech fellowship and the Negev fellowship of Ben-Gurion University of the Negev.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021210118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.D’Avino P. P., Giansanti M. G., Petronczki M., Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 7, a015834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gershony O., et al., Measuring abscission spatiotemporal dynamics using quantitative high-resolution microscopy. Methods Cell Biol. 137, 205–224 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Green R. A., Paluch E., Oegema K., Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Elia N., Ott C., Lippincott-Schwartz J., Incisive imaging and computation for cellular mysteries: Lessons from abscission. Cell 155, 1220–1231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Addi C., Bai J., Echard A., Actin, microtubule, septin and ESCRT filament remodeling during late steps of cytokinesis. Curr. Opin. Cell Biol. 50, 27–34 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Nähse V., Christ L., Stenmark H., Campsteijn C., The abscission checkpoint: Making it to the final cut. Trends Cell Biol. 27, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Mathieu J., et al., Aurora B and cyclin B have opposite effects on the timing of cytokinesis abscission in Drosophila germ cells and in vertebrate somatic cells. Dev. Cell 26, 250–265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gershony O., Pe’er T., Noach-Hirsh M., Elia N., Tzur A., Cytokinetic abscission is an acute G1 event. Cell Cycle 13, 3436–3441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.König J., Frankel E. B., Audhya A., Müller-Reichert T., Membrane remodeling during embryonic abscission in Caenorhabditis elegans. J. Cell Biol. 216, 1277–1286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matias N. R., Mathieu J., Huynh J. R., Abscission is regulated by the ESCRT-III protein shrub in Drosophila germline stem cells. PLoS Genet. 11, e1004653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green R. A., et al., The midbody ring scaffolds the abscission machinery in the absence of midbody microtubules. J. Cell Biol. 203, 505–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steigemann P., et al., Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Caneparo L., Pantazis P., Dempsey W., Fraser S. E., Intercellular bridges in vertebrate gastrulation. PLoS One 6, e20230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaigne A., et al., Abscission couples cell division to embryonic stem cell fate. Dev. Cell 55, 195–208.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zenker J., et al., A microtubule-organizing center directing intracellular transport in the early mouse embryo. Science 357, 925–928 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Eno C., Gomez T., Slusarski D. C., Pelegri F., Slow calcium waves mediate furrow microtubule reorganization and germ plasm compaction in the early zebrafish embryo. Development 145, dev156604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yabe T., et al., The maternal-effect gene cellular island encodes aurora B kinase and is essential for furrow formation in the early zebrafish embryo. PLoS Genet. 5, e1000518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng B., Schwarz H., Jesuthasan S., Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp. Cell Res. 279, 14–20 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Lee M. T., et al., Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eikenes A. H., et al., ALIX and ESCRT-III coordinately control cytokinetic abscission during germline stem cell division in vivo. PLoS Genet. 11, e1004904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elkouby Y. M., Jamieson-Lucy A., Mullins M. C., Oocyte polarization is coupled to the chromosomal bouquet, a conserved polarized nuclear configuration in meiosis. PLoS Biol. 14, e1002335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai X., et al., Aurora B functions at the apical surface after specialized cytokinesis during morphogenesis in C. elegans. Development 147, dev181099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luján P., et al., PRL-3 disrupts epithelial architecture by altering the post-mitotic midbody position. J. Cell Sci. 129, 4130–4142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siefert J. C., Clowdus E. A., Sansam C. L., Cell cycle control in the early embryonic development of aquatic animal species. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 178, 8–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Langley A. R., Smith J. C., Stemple D. L., Harvey S. A., New insights into the maternal to zygotic transition. Development 141, 3834–3841 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Kane D. A., Kimmel C. B., The zebrafish midblastula transition. Development 119, 447–456 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Newport J., Kirschner M., A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 (1982). [DOI] [PubMed] [Google Scholar]
- 29.Zhang M., Kothari P., Lampson M. A., Spindle assembly checkpoint acquisition at the mid-blastula transition. PLoS One 10, e0119285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collart C., Allen G. E., Bradshaw C. R., Smith J. C., Zegerman P., Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341, 893–896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadzhiev Y., et al., A cell cycle-coordinated Polymerase II transcription compartment encompasses gene expression before global genome activation. Nat. Commun. 10, 691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M., Kothari P., Mullins M., Lampson M. A., Regulation of zygotic genome activation and DNA damage checkpoint acquisition at the mid-blastula transition. Cell Cycle 13, 3828–3838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier M., et al., Cohesin facilitates zygotic genome activation in zebrafish. Development 145, dev156521 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Zhao W. M., Seki A., Fang G., Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell 17, 3881–3896 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elia N., Sougrat R., Spurlin T. A., Hurley J. H., Lippincott-Schwartz J., Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U.S.A. 108, 4846–4851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wühr M., Obholzer N. D., Megason S. G., Detrich H. W. III, Mitchison T. J., Live imaging of the cytoskeleton in early cleavage-stage zebrafish embryos. Methods Cell Biol. 101, 1–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee H. H., Elia N., Ghirlando R., Lippincott-Schwartz J., Hurley J. H., Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 322, 576–580 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H., Membrane scission by the ESCRT-III complex. Nature 458, 172–177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guizetti J., et al., Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Aanes H., et al., Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res. 21, 1328–1338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White R. J., et al., A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 6, e30860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guizetti J., Gerlich D. W., Cytokinetic abscission in animal cells. Semin. Cell Dev. Biol. 21, 909–916 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Duffy K. T., et al., Coordinate control of cell cycle regulatory genes in zebrafish development tested by cyclin D1 knockdown with morpholino phosphorodiamidates and hydroxyprolyl-phosphono peptide nucleic acids. Nucleic Acids Res. 33, 4914–4921 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamir E., Kam Z., Yarden A., Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol. Cell. Biol. 17, 529–536 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurley J. H., ESCRTs are everywhere. EMBO J. 34, 2398–2407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan S. H., et al., Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev. Cell 49, 867–881.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olivier N., et al., Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329, 967–971 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Haglund K., Nezis I. P., Stenmark H., Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 4, 1–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alsous J. I., Villoutreix P., Stoop N., Shvartsman S. Y., Dunkel J., Entropic effects in cell lineage tree packings. Nat. Phys. 14, 1016–1021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villoutreix P., et al., An integrated modelling framework from cells to organism based on a cohort of digital embryos. Sci. Rep. 6, 37438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caballe A., Martin-Serrano J., ESCRT machinery and cytokinesis: The road to daughter cell separation. Traffic 12, 1318–1326 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Chiaruttini N., et al., Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866–879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.