Significance
Telomeres play an important role in aging, and having relatively short telomeres is associated with an increased risk of death in humans and other animals. Telomere length is influenced by both genetic and environmental factors, both of which could potentially drive the positive association with survival. We used lifelong telomere length measurements from a population of wild sheep to disentangle this relationship. Our analyses reveal a genetic correlation between telomere length and longevity but no association between telomere shortening and mortality risk. These findings have important implications for our understanding of telomere dynamics and their role in health and lifespan.
Keywords: survival, longevity, aging, senescence, biomarker
Abstract
Telomere length (TL) is considered an important biomarker of whole-organism health and aging. Across humans and other vertebrates, short telomeres are associated with increased subsequent mortality risk, but the processes responsible for this correlation remain uncertain. A key unanswered question is whether TL–mortality associations arise due to positive effects of genes or early-life environment on both an individual’s average lifetime TL and their longevity, or due to more immediate effects of environmental stressors on within-individual TL loss and increased mortality risk. Addressing this question requires longitudinal TL and life history data across the entire lifetimes of many individuals, which are difficult to obtain for long-lived species like humans. Using longitudinal data and samples collected over nearly two decades, as part of a long-term study of wild Soay sheep, we dissected an observed positive association between TL and subsequent survival using multivariate quantitative genetic models. We found no evidence that telomere attrition was associated with increased mortality risk, suggesting that TL is not an important marker of biological aging or exposure to environmental stress in our study system. Instead, we find that among-individual differences in average TL are associated with increased lifespan. Our analyses suggest that this correlation between an individual’s average TL and lifespan has a genetic basis. This demonstrates that TL has the potential to evolve under natural conditions, and suggests an important role of genetics underlying the widespread observation that short telomeres predict mortality.
Telomeres are repetitive sequences of noncoding DNA found at the terminal ends of linear chromosomes, and they play an important role in maintaining DNA stability and integrity (1–3). Telomeres shorten during cell replication and in response to oxidative stress (4, 5), and cellular senescence and apoptosis is triggered once telomeres reach a critically short threshold (2). The important role of telomeres in cellular senescence has led to telomere shortening being considered as one of nine “hallmarks of aging,” and average telomere length (TL) as an important biomarker of whole-organism health and biological aging (6). In humans, relatively short leukocyte telomeres have been linked to a range of age-related diseases such as diabetes, cancer, and cardiovascular disease (7–9) and increased subsequent mortality risk (10–12). A recent metaanalysis suggests this pattern may generalize beyond humans: Across studies from 20 nonmodel vertebrate species (predominantly birds), there was an overall positive association between TL and subsequent survival (13). Although evidence for a causal role for telomeres in whole-organism aging and longevity remains weak (14), these findings highlight the potential significance of TL as a biomarker of human and animal health (15, 16) and for our understanding of life history evolution (17, 18).
Studies in humans and other vertebrates have found evidence for consistent differences in TL among individuals over multiple measurements (19, 20). Such repeatable among-individual differences in any trait may result from the trait being under genetic influence, from long-term effects of the early-life environment, and/or environmental conditions that persist across the lifetime. There is good evidence that variation in average TL in blood cells has a genetic basis in humans and other vertebrates, although estimates of the heritability (the proportion of variation attributed to additive genetic effects) of TL are variable (21, 22). Recent studies of wild vertebrates have also revealed considerable variation in adult TL among birth cohorts, suggesting persistent impacts of early-life environment (23, 24). At the same time, there is growing evidence that TL is highly dynamic across an individual’s lifetime, and metaanalyses of human and nonhuman animal studies show that experience of diverse forms of environmental stress are predictive of shorter TL (25–27). Indeed, some studies using longitudinal TL data have found that telomere shortening over successive measurements rather than TL per se is predictive of mortality (28–30). Thus, the emerging picture from studies in humans and other vertebrates is that shorter TL generally predicts increased risk of subsequent mortality, and that variation in TL is under the influence of both genetics and environmental stressors.
The observation that shorter TL measurements predict increased mortality risk could be underpinned by two nonmutually exclusive processes operating across the lifetimes of individuals. Firstly, individuals may differ in their average TL across life, and individuals with shorter TL may be shorter lived. This pattern is referred to as the “selective disappearance” of individuals with shorter telomeres, and it implies that TL reflects constitutive differences among individuals (for example, due to genetics or differences in early-life environment) which shape their longevity (31, 32). Secondly, individuals may differ in their pattern of TL change over time, and individuals showing the greatest telomere loss across successive measurements are more likely to die subsequently. This pattern is consistent with the idea that within-individual telomere dynamics reflect recent and cumulative experiences of environmental stress and physiological deterioration that also predict mortality. Neither pattern necessarily implies a causal role for telomeres in driving the mortality risk of an organism, because associations between TL and survival could result from both traits being correlated with underlying, unmeasured variables which causally impact survival (14, 18). Nevertheless, unraveling the contribution of genetics, early-life environment, and more immediate telomere shortening to the observed association between TL and survival is essential for our understanding of TL as a biomarker of health and aging (19).
To our knowledge, no study to date has assessed the relative importance of the different processes underlying the relationship between TL and mortality risk across the entire lifespan. To do so demands repeated measurements from across life to characterize among- and within-individual variation in TL, a population pedigree or genomic information to separate genetic and environmental sources of variation, and detailed information on individual health and fitness outcomes over the lifetime. Here, we use a multivariate mixed-effects modeling approach to analyze extensive, longitudinal data from a long-term study of wild Soay sheep living on St Kilda, Scotland, to distinguish between possible models of why shorter TL predicts increased mortality risk. We find that the observed positive association between TL and mortality in this system is underpinned by selective disappearance of individuals with shorter average TL. Importantly, our results suggest this is largely driven by genetically based differences in both TL and longevity.
Results
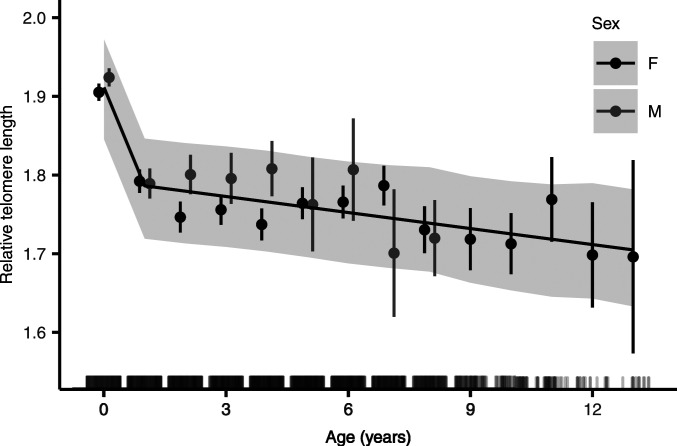
Soay sheep resident to our study area on St Kilda have been individually marked at birth and closely monitored and repeatedly blood sampled across their lifetimes. Here, we measured relative TL (RTL) in 3,641 samples collected from 1,586 individual sheep over a 19-y period (see Materials and Methods). We found that RTL declined with age in Soay sheep, with a more rapid initial decline between measurements at around 4 and 16 mo, followed by a slower linear decline thereafter (Fig. 1). The best fitting age function in our models of RTL included a two-level factor for age class (lambs and adults aged ≥1 y) and a linear term for age in years, which is equivalent to a segmented regression with a threshold at 1 y of age (SI Appendix, Tables S1 and S2). There was considerable variation in RTL within age groups (SI Appendix, Fig. S1). There was limited evidence for a sex difference in either average RTL or the rate of change in RTL with age, although the significance of sex in our model depended on model structure (SI Appendix). The individual repeatability of RTL over the lifespan was 0.214 [95% credible intervals (CI) 0.169 to 0.252; SI Appendix, Table S2]. Excluding the variance attributed to qPCR plate and qPCR row (which represents measurement error) from the denominator, the repeatability was 0.241 (95% CI 0.204 to 0.282; SI Appendix, Table S2). Although RTL declined with age on average, we found evidence for consistent differences in RTL among individuals.
Fig. 1.
RTL varied with age over the lifespan in Soay sheep (n = 3,641 observations of 1,586 individuals). The points show raw data medians and SEs for each age, with females in black and males in gray (for clarity, n = 3 observations of females aged > 13 y and n = 6 observations of males aged > 8 y are grouped with ages 13 and 8 y, respectively). The black lines show predictions from the best model (SI Appendix, Table S2); gray shading represents 95% credible intervals around those predictions. The rug plot on the inside of the x axis shows the distribution of observations across the age range. Note that the aging pattern in the raw data and best-fitting age functions from mixed-effects models are not expected to align perfectly when selective disappearance effects are present.
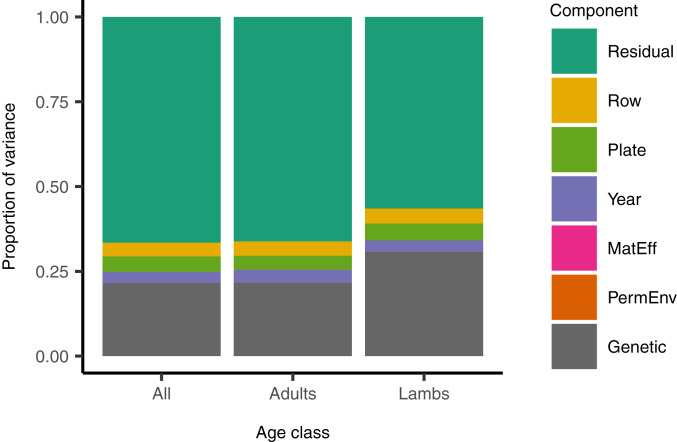
There was detectable additive genetic variance for RTL across all ages in the population (Fig. 2 and SI Appendix, Table S3). The heritability of RTL (the proportion of variance explained by additive genetic effects) was 0.204 (95% CI: 0.158 to 0.252). The permanent environment effect was bound at zero (<0.001, 95% CI: <0.001 to 0.017), indicating that individual repeatability in RTL could almost entirely be attributed to genetic rather than environmental effects. Maternal effects also explained a very small proportion of the variance (<0.001, 95% CI <0.001 to 0.034, estimate bound at zero). The year the sample was collected, qPCR plate and qPCR row each explained 3 to 4% of the variance in RTL (year: 0.031, 95% CI 0.014 to 0.084; qPCR plate: 0.043, 0.027 to 0.066; qPCR row: 0.038, 0.014 to 0.172). Excluding the measurement error terms of qPCR plate and row from the total phenotypic variance, the heritability of TL was 0.233 (95% CI 0.189 to 0.279). This shows that variation in RTL has a genetic basis in wild Soay sheep.
Fig. 2.
The proportion of variance in RTL in Soay sheep explained by different variance components. Estimates are based on the mode of the posterior distribution from Bayesian quantitative genetic animal models. Estimates for age class “all” came from a univariate model incorporating all observations (n = 3,632 of 1,582 individuals) that included sex, age class, and age in years as fixed effects (SI Appendix, Table S3). The separate estimates for adults (age ≥ 1 y) and lambs (4 mo old) came from a bivariate model that accounted for the covariance between lamb and adult TL at the genetic, year, and residual levels (SI Appendix, Table S4). MatEff, maternal effect; PermEnv, permanent environment.
We went on to estimate the genetic correlation between RTL expressed in lambs, which are still developing at the time of measurement (aged 4 mo), and in older individuals, which are sexually mature and have largely completed growth. When estimated with a bivariate model of lamb and adult RTL, the heritability of RTL in lambs was estimated to be 0.285 (95% CI 0.206 to 0.369), and in adults 0.210 (95% CI: 0.156 to 0.255) (Fig. 2 and SI Appendix, Table S4). The genetic correlation between lambs and adults was close to 1 (0.916, 95% CI: 0.806 to 0.996; SI Appendix, Table S4), implying that largely the same or linked genes influenced RTL across age groups. The residual correlation between lamb and adult RTL was close to zero (0.036, 95% CI: −0.196 to 0.008; SI Appendix, Table S4). The negligible residual correlation was expected given the lack of permanent environment effect underlying repeatable differences in individual RTL across all ages (SI Appendix, Table S3). Repeatable among-individual differences across ages in RTL were therefore driven predominantly by genetic rather than environmental effects, and a similar set of genes influenced RTL across ages.
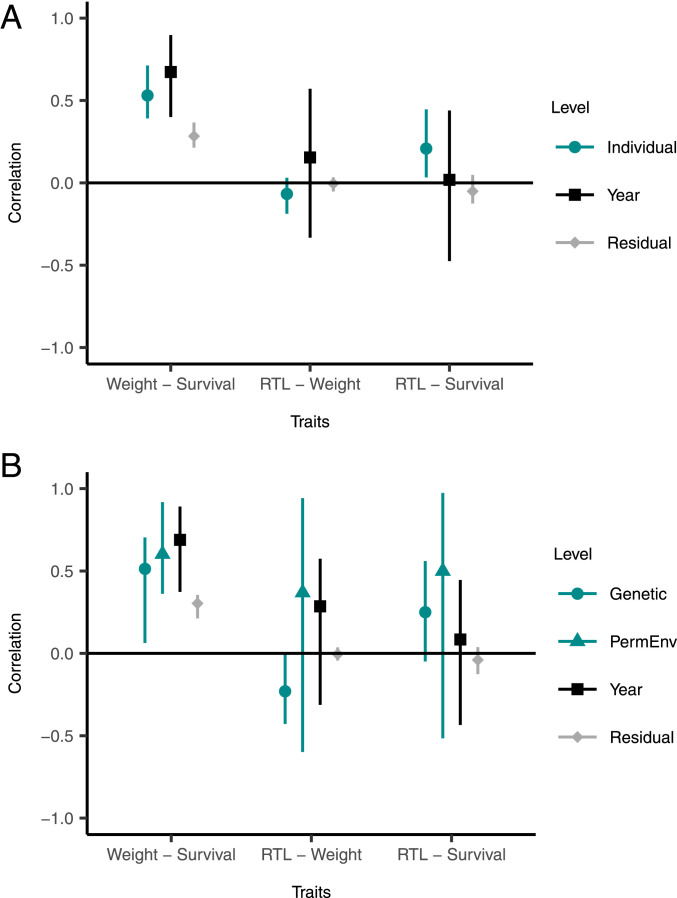
Using multivariate mixed-effects models, we next tested the strength and direction of correlations among RTL, body weight (both measured in August), and subsequent overwinter survival at different hierarchical levels (among-individual, genetic, among-year, within-individual). Although we were primarily interested in the RTL–survival association, the inclusion of weight in our models allowed us to contrast the magnitude of the RTL–survival association with a well-studied trait which is known to be linked to condition and fitness in our study system (33). As previously documented, August body weight was strongly predictive of improved winter survival prospects (33–35) (Fig. 3). Our multivariate models revealed that this association was present at all hierarchical levels (Fig. 3 and SI Appendix, Tables S5 and S6). Our initial phenotypic model indicated that heavier individuals, on average, tended to have longer lifespans (Fig. 3A and SI Appendix, Table S5). Developing this model into a pedigree-based quantitative genetic model revealed that this among-individual effect was driven by similar sized contributions from genes and environment (Fig. 3B and SI Appendix, Table S6). Overwinter survival probabilities were higher in years where the average body weight was higher (among-year effect), and individuals with relatively low weight compared to their average weight were less likely to survive (residual effect; Fig. 3). In our phenotypic model, there was little evidence for an association between RTL and body weight at any hierarchical level (Fig. 3A and SI Appendix, Table S5). However, the quantitative genetic model revealed a negative genetic correlation (Fig. 3B and SI Appendix, Table S6). This was consistent in separate models of lambs and adults, although, here, credible intervals overlapped zero more widely (SI Appendix, Tables S7–S10).
Fig. 3.
The correlation among RTL, August body weight, and overwinter survival probability at different hierarchical levels in Soay sheep (n = 3,569 observations of 1,574 individuals). Correlations were estimated as the mode of the posterior distribution with 95% higher probability density intervals from multivariate Bayesian mixed-effects models. (A) Estimates for the among-individual (teal), among-year (black), and residual correlations (gray) from a phenotypic model (SI Appendix, Table S5). (B) Estimates for the genetic (teal circles), permanent environment (teal triangles), among-year (black), and residual correlations (gray) from a quantitative genetic animal model (SI Appendix, Table S6).
We found a positive association between RTL and overwinter survival probability at the among-individual level (Fig. 3A and SI Appendix, Table S5). This suggests that individuals with longer telomeres, on average, across their lives, tended to have longer lifespans. The 95% credible intervals overlapped zero for both the among-year and the residual covariance between RTL and overwinter survival (Fig. 3A and SI Appendix, Table S5). The absence of an association at the within-individual (or residual) level indicates that, if an individual had a relatively short RTL measurement compared to their average in a particular year, this was not associated with an increased risk of mortality. Overall, these findings imply that average RTL after weaning, rather than more immediate changes relative to an individual’s average RTL, predicted overwinter survival.
The quantitative genetic model revealed a positive association between RTL and overwinter survival probability at the genetic level, although the credible intervals narrowly overlapped zero (Fig. 3B and SI Appendix, Table S6). The covariance between RTL and survival was very small (<0.001) at the permanent environment level (SI Appendix, Table S6), and the correlation at this level had wide credible intervals which extensively crossed zero (Fig. 3B). Our results were broadly consistent for lambs and adults across separate models, although the credible intervals of the among-individual and genetic effects did narrowly cross zero within some age groups (SI Appendix, Figs. S2 and S3, Supplementary Methods and Results, and Tables S7–S10). Overall, our results suggest that the among-individual positive association between RTL and survival identified in the phenotypic model was driven largely by genetic effects (Fig. 3B and SI Appendix, Table S6). In other words, genes conferring longer average telomeres after weaning also tended to be associated with longer lifespan.
Discussion
A growing number of studies across vertebrate species have shown that TL is heritable (21) and that relatively short TL is predictive of increased mortality risk (13). We have dissected the TL–mortality relationship across the natural lifespan of wild Soay sheep to identify the processes that drive this association. We show that selective disappearance of individuals with shorter average TL is the key process, rather than an association between the pattern of TL attrition and mortality. If TL was a marker of biological aging, or it reflected variation in condition resulting from environmental stress, we would expect telomere shortening to predict mortality, resulting in a positive residual correlation between TL and survival in our models. We found no support for a correlation at the residual level, and only the among-individual correlations between TL and survival were consistently different from zero. This suggests that TL is not a useful marker of biological aging in our system. Furthermore, it is inconsistent with the hypothesis that TL reflects variation in physiological state or condition resulting from recent or accumulating experience of environmental stress during adulthood, which also influences mortality risk. However, our first TL measurement was taken at 4 mo of age—around the time of weaning for most lambs in our study population—and it is possible that associations between average TL and survival are driven by some combination of initial TL and the rate of telomere attrition prior to first measurement. An individual’s average TL could therefore reflect its ability to maintain homeostasis and resist environmental stress during very early development, which, in turn, could predict its subsequent lifespan. Nonetheless, our results demonstrate that variation in TL is heritable and under directional selection, meaning it has the potential to evolve under natural conditions.
Our moderate estimate of the heritability of TL (around 20%; Fig. 2) represents evidence for an appreciable genetic contribution to variation in TL in a wild mammal. Recent studies in wild badgers and bats have used similar pedigree-based “animal models” but found both the repeatability and heritability of TL to be negligible (36, 37). Our estimate was lower than in a recent longitudinal study of farmed dairy cattle [32–38% (38)], which is unsurprising given the expectation of reduced environmental variation in livestock compared to wild systems. It is notable that both studies found negligible permanent environment effects, implying that consistent differences in TL across life were largely the result of genetic rather than early environmental effects in both systems (38) (Fig. 2). More broadly, while human studies have tended to find moderate to high heritability of TL, the growing literature in birds presents a much more variable picture (21). Two laboratory studies of birds have estimated the heritability of TL to be >100% (39, 40), while some studies in the wild suggest little genetic influence on TL [heritability <5% (41, 42)]. The reasons for the variation in heritability estimates across studies remain an important area for future research. As well as variation in the way that genes influence telomere dynamics, differences in the environmental variation experienced by different populations or species, and the degree of error associated with different telomere measurement methodologies, are also likely to play a role (21). Our study used a qPCR method to measure TL, which is often described as having greater measurement error than the “gold standard” terminal restriction fragment (TRF) approach (43). However, we note that the technical repeatability of our qPCR assay is high and compares favorably to those reported for TRF studies (see Materials and Methods). Furthermore, most previous studies in humans and birds have focused on specific age classes (e.g., elderly humans or prefledging birds). Our data span the entire lifetimes of individuals, and we were able to demonstrate a very high genetic correlation between TL measured in lambs and adults. This provides clear evidence that the same or linked genes influence TL in early and later life, an assumption that has rarely been tested in any study system (although see ref. 44).
In a previous study, we identified an association between TL and mortality in a much smaller sample of female Soay sheep (45). In that study, we reported a considerably lower repeatability for TL (13%) than in the present study and were only able to detect a TL–mortality relationship among younger females experiencing a high-mortality winter. This previous study was focused on a subset of females from four birth cohorts, and thus had relatively few samples from later adulthood (45). Our present study is distinguished by the much larger dataset and more complete population coverage, which has allowed us to assess the repeatability of TL and determine that it is the among-individual differences in TL across life that are predictive of mortality. We note that, when splitting our multivariate models by age groups, the among-individual covariance between TL and survival was positive and of a similar magnitude in adults and lambs, although the credible intervals overlapped zero (SI Appendix, Figs. S2 and S3 and Table S4). This indicates that the association was not simply due to effects on lamb survival. Although some longitudinal vertebrate studies have found that telomere attrition predicted survival better than average or recent TL (28, 29, 46), others have identified extremely high consistency in individual TL across measurements (19, 20) and long-term associations between early-life TL and adult lifespan (24, 47, 48). Most of these studies involved only one or two measurements of TL per individual, and none used a multivariate mixed-effects modeling approach capable of fully dissecting the contributions of genetic, individual, annual, and residual sources to observed TL–survival covariance.
Recent studies of elderly human cohorts have identified candidate single-nucleotide polymorphism (SNP) loci associated with telomerase genes, which are involved in maintenance of telomeres and genomic integrity, that not only predict average leukocyte TL but also subsequent morbidity and mortality (49–51). However, human twin studies also suggest that relatively short TL, independent of genetic factors, predicts mortality (52). In support of a causal role for telomerase genetics in mortality and aging, studies of laboratory mice without telomerase appear to show early onset of aging phenotypes, while mice with genetically enlarged telomeres are longer lived (53, 54). Our findings provide support for a role for genetics in observed TL–mortality relationships from a nonhuman system outside the laboratory. Further work is required to determine whether specific genes, such as those involved in telomere maintenance, are implicated in this relationship or whether the observed genetic correlations result from minor effects of many different genes.
Our results imply that the observed relationship between TL and mortality is not causal. A causal effect of short telomeres on survival would lead to the expectation that both individuals with low average TL (among-individual covariance) and those with short TL at measurement relative to their average (residual covariance) should be positively related to survival. But this is not what we observed. Instead, it seems likely that the genes influencing some yet to be determined aspect of an individual’s overall frailty (or robustness) have a correlated influence on TL. These genes could be influencing TL determined during early embryonic development and/or the rate of telomere attrition during gestation and neonatal life, as our first TL measurement was not taken until around 4 mo of age. However, comparing the magnitude of the TL–survival correlations with the correlations between body weight and survival highlights that the association between TL and mortality is modest compared to a measure that is more directly relatable to condition and health. It remains to be determined whether our findings in Soay sheep will generalize to other species and systems, but our work highlights the importance of using large datasets that span the entire lifetimes of many individuals, in order to fully understand the drivers of associations among TL, health, and mortality risk. Overall, our results provide important insights into the genetics of lifespan in the wild and highlight the importance of long-term, longitudinal studies across different species for our understanding of TL as a biomarker of health and fitness.
Materials and Methods
Study System and Sample Collection.
The Soay sheep (Ovis aries) is a primitive breed of domestic sheep that has been living on the remote St Kilda archipelago with minimal human management for the last few millennia [57°49′N, 8°34′W (33)]. Since 1985, the sheep resident within the Village Bay area of the main island in the archipelago, Hirta, have been the subject of an individual-based study (33). Individuals are caught and tagged within a few days of birth in the spring. Ten censuses are conducted on each of three annual field seasons, during spring (March through April), summer (July through August) and autumn (October through November), meaning the timing of individual’s disappearance is known with a high degree of accuracy (33). The vast majority of sheep mortality occurs in late winter (85% of adult deaths occur in January through April), and daily carcass searches during this period mean that death dates are known to the nearest month for most individuals. Each August, 50 to 60% of the resident population are caught in temporary corral traps and blood sampled. Blood is collected from each individual into 9-mL lithium heparin Vacuettes and kept in a cool box or fridge from the point of sampling. The blood is processed within 24 h to separate the plasma and buffy coat fractions. The Vacuette is then spun at 1,008 × g for 10 min, and the plasma layer is drawn off and replaced by the same quantity of 0.9% NaCl solution, gently mixed, and spun again at 1,008 × g for 10 min. The intermediate buffy coat layer, comprising mainly white blood cells, is then drawn off into a 1.5-mL Eppendorf tube and stored at −20 °C until used to assay leukocyte TL. All data collection was approved by the UK Home Office and carried out in accordance with the relevant guidelines.
Pedigree Reconstruction.
Parentage was inferred by genetic methods, except for some maternal links inferred by observation (55). Multigenerational pedigree reconstruction was performed in the R package Sequoia (56), using 431 unlinked SNP markers. This likelihood-based approach infers not only parent−offspring relationships but also siblings and second-degree relatives. In the resulting pedigree, a mother and/or father was assigned to 6,082 individuals. After pruning to only those individuals informative to the current analyses [using the R package MasterBayes (57)], the pedigree had a maximum depth of 13 generations and consisted of 2,411 individuals, of which 2,273 were nonfounders, and a total of 2,050 maternities and 2,172 paternities were assigned.
Sample Selection and Randomization.
A total of 3,891 August buffy coat samples from 1,647 animals of known age collected between 1998 and 2016 were selected for TL measurement. These samples were selected from the total available buffy coat freezer archive (n = 6,775 from 3,315 sheep) based on the following exclusion criteria: samples collected before 1998 (n = 1,924); samples from individuals of unknown age (n = 97); samples collected between 2013 and 2016 from lambs or yearlings that were caught only once (n = 454); and samples collected from individuals that were only captured once but survived, and were therefore available for sampling in subsequent years (n = 409). These criteria were designed to maximize the longitudinality of the dataset while avoiding biasing the dataset against short-lived individuals (i.e., individuals only sampled once because they died). Sample years were then randomly allocated to one of four batches, each comprising 4 y to 5 y (to reduce the number of samples that needed to be removed from the freezer at any one time). Samples were then fully randomized within each batch, assigned a unique identifier from the start of batch 1 to the end of batch 4, and processed from DNA extraction through to qPCR in this order.
DNA Extraction.
Genomic DNA was extracted from buffy coat on 96-well plates using the Macherey−Nagel Nucleospin 96 Blood kit (Cat# 740665). The samples were extracted on a liquid handling robot (Freedom Evo-2 150; Tecan) using a vacuum manifold. In order to facilitate passage of the sample through the DNA binding plate, the following step was included prior to automated extraction on the robot: 50 µL of buffy coat was mixed with 300 µL of red blood cell lysis solution (Qiagen; Cat# 158902) and incubated at room temperature for 5 min, before centrifugation at 12,200 rpm for 30 s; 250 µL of the supernatant was discarded, and the cell pellet was resuspended in the residual supernatant, before it was transferred to a 96-well MN lysis block (Macherey−Nagel Nucleospin 96 Blood kit; Cat# 740665) and sealed. The liquid handling robot was loaded with the MN lysis block with the seal removed, the silica DNA binding plate, 100% ethanol, lysis buffer BQ1, wash buffers B5 and BW, and, finally, a master mix containing phosphate-buffered saline (PBS), proteinase K, and RNase A (binding plate, buffers, and proteinase K supplied with Macherey−Nagel Nucleospin 96 Blood kit, Cat# 740665; RNase A, Qiagen Cat# 158924; PBS, Sigma Cat# D1408). The extraction protocol followed manufacturer’s guidelines for use with a vacuum manifold with the following amendments. The robot added 96 µL of 1× PBS, 25 µL of proteinase K, and 4 µL of RNase A to each sample. Lysis was performed on a shaker for 10 min. The vacuum steps for binding and the first two washes were increased to 5 min, and the final wash was increased to 10 min, with an additional 10-min vacuum at the end to dry the membrane. If any lysate/wash failed to pass through the silica membrane after a vacuum step, the plate was removed from the robot and centrifuged for 3 min at 4,000 rpm. For the few samples that failed to pass through after centrifugation, the wash/lysate was removed by hand with a pipette. After dry centrifugation (3 min, 4,000 rpm), DNA was eluted in a total of 150 µL of elution buffer BE (Macherey−Nagel Nucleospin 96 Blood kit; Cat# 740665) which was warmed to 60 °C prior to adding it onto the silica membrane. Elution was performed in two steps: first, 100 µL of buffer BE followed by centrifugation (3 min, 4,000 rpm), then 50 µL of buffer BE followed by a final centrifugation (3 min, 4,000 rpm).
DNA extraction quality control.
Following DNA extraction, a strict quality control protocol was implemented to measure DNA concentration, integrity, and purity (SI Appendix, Fig. S4). Of the 3,891 samples selected for analysis, 42 samples were missing or accidentally omitted through human error. The available samples were measured on a Nanodrop ND-1000 9 spectrophotometer (Thermo Scientific). Samples yielding <20 ng/µL were rejected, and reextraction was attempted for 491 of these samples. Overall, 87 samples failed to yield sufficient DNA and were excluded from the analyses. Samples yielding ≥20 ng/µL were checked for DNA purity. The acceptable range for absorption for 260/280 nm ratio (a measure of DNA vs. protein and RNA contamination) was 1.7 to 2.0. Samples falling outside of this range were excluded from subsequent analyses (n = 66, representing a failure rate of 1.76%). Two-thirds of samples were assayed for 260/230 nm ratio (a measure of salt and other impurities), and samples with a ratio of <1.8 were excluded from subsequent analyses (n = 41, representing a failure rate of 1.73%). Samples of sufficient yield and purity were standardized to 10 ng/µL, and DNA integrity was assessed by running 200 ng of DNA on a 0.5% agarose gel. Samples were scored for integrity on a scale of one to five by visual examination of their DNA crowns, and samples scoring three to five were excluded from further analyses (see ref. 58). Integrity was initially assayed for all of the first 1,667 samples run. Only 16 of these samples were given a DNA integrity score of two, and 7 failed (failure rate of 0.42%). We randomly tested a quarter of all subsequent samples for DNA integrity, with only four failing (representing a failure rate of 0.76%). In total, 11 samples were excluded due to poor DNA integrity. Overall, DNA was successfully extracted and passed quality control requirements from 3,644 buffy coat samples (SI Appendix, Fig. S4).
TL Measurement.
qPCR.
RTL was measured using real-time qPCR (59), using protocols we have previously developed and validated using blood samples from sheep and cattle (45, 58). The qPCR method estimates the total amount of telomeric sequence present in a sample relative to the amount of a nonvariable copy number reference gene. In this study, we used the beta-2-microglobulin (B2M) as our reference gene (45, 58), with primers supplied by Primer Design (Cat# HK-SY-Sh-900). For telomeric amplification, tel1b (5′-CGG TTT GTT TGG GTT TGG GTT TGG GTT TGG GTT TGG GTT-3′) and tel 2b (5′-GGC TTG CCT TAC CCT TAC CCT TAC CCT TAC CCT TAC CCT-3′) primers were used. Telomere primers were manufactured, purified by high performance liquid chromotography (HPLC), and supplied by Integrated DNA Technologies (IDT). Telomere and reference gene reactions were run in separate wells of the same qPCR plate at a concentration of 300 nM and 900 nM, respectively. Samples were diluted to 0.5 ng/µL with buffer BE (Macherey−Nagel Nucleospin 96 Blood kit; Cat# 740665) immediately prior to qPCR analysis. Each reaction was prepared using 5 µL of LightCycler 480SYBR Green I Master Mix (Cat # 04887352001, Roche) and 1 ng of sample DNA in a total reaction volume of 10 µL. We used 384-well plates which were loaded with sample DNA and master mix using an automated liquid handling robot (Freedom Evo-2 150; Tecan).
Each plate included eight calibrator samples (1 ng/µl) to account for plate-to-plate variation and two nontemplate controls (NTC) consisting of nuclease free water. The calibrator sample was extracted from a large quantity of buffy coat prepared from blood supplied from a single domestic sheep (Cat# SHP-BUFCT-LIHP, Sera Laboratories International LTD). We carried out a large number of extractions from this sample, applied the same quality control as above, and then pooled the extracts and aliquoted them for subsequent use. A five-point standard curve, consisting of a fourfold serial dilution of the calibrator sample (at concentrations: 20 ng/µL, 5 ng/µL, 1.25 ng/µL, 0.3125 ng/µL, and 0.078125 ng/µL) was included on the plate to provide a visual check that the samples amplified at the correct cycle. Samples, calibrators, standard curve, and NTCs were all run in triplicate. All qPCRs were performed using a Roche LC480 instrument using the following reaction protocol: 10 min at 95 °C (enzyme activation), followed by 50 cycles of 15 s at 95 °C (denaturation) and 30 s at 58 °C (primer annealing), then 30 s at 72 °C (signal acquisition). Melting curve protocol was 1 min at 95 °C, followed by 30 s at 58 °C, then 0.11 °C/s increase to 95 °C followed by 10 s at 40 °C (cool down).
Calculation of RTL.
We used the LinRegPCR software package [version 2016.0; (60)] to correct our amplification curves for baseline fluorescence, and to calculate well-specific reaction efficiencies and Cq values. A constant fluorescent threshold was set within the window of linearity for each amplicon group, calculated using the average Cq across all plates. The threshold values used were 0.2 and 0.25, and the average PCR efficiency across all plates was 1.876 and 1.881 for the B2M and telomere amplicon groups, respectively. Samples were excluded from further analysis if the coefficient of variation (CV) across triplicate Cq values and triplicate PCR efficiency values for either amplicon was >5% (n = 1; note that, for >95% of our samples, the triplicate CV was <2%), or if at least one of their triplicate reactions had an efficiency that was 5% higher or lower than the mean efficiency across all wells on that plate for the respective amplicon (n = 2; > 95% of samples were within 2% of mean plate efficiencies; SI Appendix, Fig. S4).
RTL for each sample was calculated, following Pfaffl (61), using average reaction efficiencies for each plate and Cq for each sample determined by LinRegPCR as follows:
where and are the mean reaction efficiencies for the respective amplicon group across all samples on a given plate; and are the average Cqs for the relevant amplicon across all calibrator samples on the plate; and and are the average of the triplicate Cqs for the sample for each amplicon.
Repeatability of TL.
To assess the repeatability of our qPCR assay, the first qPCR plate (n = 48 samples) was run eight times over four consecutive days: four times over 2 d with samples in the same position and then four times over 2 d with samples in an alternative row within the qPCR plate. We calculated the overall repeatability of RTL as the proportion of variance explained by sample identity over the total variance, in a linear mixed-effects model including only sample identity as a random intercept term [using restricted maximum likelihood estimation in glmmTMB v.0.2.3 (62)]. The overall repeatability of RTL was 0.866 (95% confidence intervals 0.807 to 0.908). The repeatability of RTL measured in the same location across the first four qPCR plates was 0.948 (95% CI 0.922 to 0.965), illustrating how repeatability can be inflated if samples are consistently run in the same location. In a second model which included sample identity, qPCR plate, and qPCR row as random intercept terms, the proportion of variance explained by qPCR plate and row was 0.005 and 0.063, respectively. The repeatability of RTL in this model was 0.824 (95% CI 0.748 to 0.880), or 0.884 (95% CI 0.829 to 0.923) if the plate and row terms were excluded from the total variance (since they represent measurement error). These repeatability estimates compare favorably with other studies of TL measured by both qPCR [interassay repeatability: 0.85 (48), 0.82 (24)] and TRF [TRF repeatability: 0.86 (63)].
Data Analysis.
All analyses were conducted in the program R version 3.6.1 (64) using the package MCMCglmm v.2.29 (65) unless otherwise specified.
Relationship among TL, age, and sex.
RTL was approximately normally distributed in lambs, adults, and overall (SI Appendix, Fig. S5). We ran a series of linear mixed-effects models to determine the function that best described variation in TL with age. We included a two-level factor for age class (lamb: ∼4 mo of age; adult: ≥1 y). To account for age-related variation within the adult age class, we included age in years as a fixed covariate. We tested linear, quadratic, and cubic age terms, as well as threshold age functions with a range of breakpoints (2 y to 11 y). We also tested a four-level factor for age class (lamb: ∼4 mo of age; yearling: ∼16 mo; adult: 2 y to 6 y; geriatric: >6 y. All models included individual identity and sample year as random intercept terms to account for nonindependence among observations. The qPCR plate and row for each sample were also included as crossed random intercept terms to account for variation associated with measurement error. The models were run using maximum likelihood estimation in glmmTMB v.0.2.3 (62), and Akaike information criterion (AIC) model selection was used to determine the best age function. We selected the model with the fewest parameters within two ΔAIC of the model with the lowest AIC value. Once we determined the best-fitting age function, we tested for a difference in average TL between the sexes by including a two-level factor for sex in our model. To test whether the aging patterns differed between the sexes, we also tested for interactions between sex and the selected age terms. The significance of additive and interactive effects of sex was assessed using likelihood ratio tests (SI Appendix). The repeatability of TL over the lifespan was estimated as the variance explained by the random effect of individual over the total phenotypic variance (the sum of the random effects variance components plus the residual variance). There were 3,641 observations of TL from 1,586 sheep available for this analysis. Of these individuals, 836 had one RTL measurement available, 271 had two, 281 had three or four, and 198 had five or more measurements.
Heritability of TL.
A quantitative genetic animal model was used to estimate the additive genetic variance for TL in Soay sheep. The model contained a two-level fixed factor for age class (lamb: ∼4 mo; adult: ≥1 y) and a linear covariate for age in years. We also included sex as a two-level fixed factor to account for differences in average TL between the sexes. The additive genetic effect was estimated using information on individual relatedness from the population pedigree. We included maternal identity in addition to the additive genetic component to capture similarity among maternal siblings that is not explained by the additive genetic effect (known as the maternal effect). We also included individual identity as a random effect to capture consistent differences in measures from the same individual that are not attributed to genetic effects, influenced, for example, by where the individual lives or aspects of their early-life environment (the permanent environment effect). Year of sample collection, qPCR plate, and qPCR row were included as random intercept terms. The heritability of TL was calculated as the variance explained by the additive genetic effect over the total phenotypic variance. This model was run for 1.1 × 105 iterations, with 1 × 104 burn-in and thinning interval of 50, resulting in 2,000 stored samples of the Markov chain Monte Carlo (MCMC) chain with minimal autocorrelation (<0.2). Parameter estimates are presented as the posterior mode with 95% highest posterior density (HPD) intervals. Parameter expanded priors were used for all variance components, and inverse-Wishart priors were used for the residual variance. There were 3,632 observations of TL from 1,582 sheep available for this analysis (n = 9 observations from four sheep were excluded because maternal identity was unknown).
We also ran a bivariate model to estimate the genetic correlation between TL in lambs (∼4 mo) and adults (≥1 y) which treated TL in lambs and adults as separate response variables. We included sex as a two-level fixed factor for both lambs and adults, with age in years included as a fixed covariate for adults only. The qPCR plate and row were included as random intercept terms across both models. Maternal identity in the lamb model and individual identity in the adult model were included as random effects to estimate maternal and permanent environment effects, respectively. We estimated the unstructured variance−covariance matrix for the genetic, year, and residual effects, which enabled us to estimate the correlations across age classes at these different hierarchical levels. There is a possibility that the lamb residual variance could covary with the adult permanent environment effect, but we did not attempt to model this covariance because we detected no permanent environment effect in adults (see Results). The model was run 2.1 × 105 iterations, with 1 × 104 burn-in and thinning interval of 200, resulting in 1,000 stored samples of the MCMC chain (autocorrelation < 0.2). The genetic correlation was taken from the posterior correlation of the stored samples with 95% HPD intervals. The bivariate model was run using 3,632 measurements from 1,582 individuals. Both lamb and adult TL measurements were available for 424 individuals.
Associations among TL, August weight, and overwinter survival.
Phenotypic model.
We used a multivariate mixed-modeling approach to examine the association among TL, weight, and overwinter survival at different hierarchical levels. RTL, body weight in kilograms (both measured August year t), and annual survival (to 1 May year t + 1) were our response variables, modeled as Gaussian, Gaussian, and threshold distributions, respectively (corresponding to identity and probit link functions). Unstructured variance–covariance matrices were estimated for some of the random effects, allowing us to estimate the covariance among the three traits at different hierarchical levels. For each of these random effects, we obtained a posterior distribution for the variance and covariance among TL (“RTL”), weight (“Wt”), and overwinter survival (“Surv”),
We estimated the covariance at the among-individual, among-year, and residual levels. The among-individual level captures consistent differences between individuals (e.g., if individuals that are consistently heavier tended to have longer lifespans). The covariance at the year level captures associations between average trait values among years (e.g., whether the survival rate is higher in years when the average body weight is higher). The residual level reflects covariance that is not captured by the among-individual and among-year levels (i.e., if an individual has a relatively low body weight compared to its average has lower chances of surviving).
The model was run for 5.3 × 105 iterations, with 3 × 104 burn-in and thinning interval of 500, resulting in 1,000 stored samples of the MCMC chain (autocorrelation < 0.1). The residual variance was fixed at one for survival (as is the standard recommendation for threshold models), and the latent variables were constrained to be between ±7 to avoid numerical difficulties as the probabilities approached zero and one. In addition to the random effects of identity and year, the TL model included random intercept terms for qPCR plate and row, and the weight and survival models included random intercept terms for maternal identity and birth year. We fitted age and sex in slightly different ways across the three models, to reflect our understanding of differences in age-related variation in these three traits. In the telomere model, we included age class (lamb vs. adult) as a two-level fixed factor. No other fixed effects were included in the telomere model, since we were interested in how TL covaried with the other traits across the lifespan. In the weight model, we included age class (lamb vs. adult) plus linear, quadratic, and cubic age terms (capturing variation in weight with age during adulthood) and their interactions with sex. In a previous study, we detected a decline in weight across the year prior to death in this system, using univariate models of weight (35). We did not include a term of final year alive here, as this effect is captured in the residual covariance between weight and survival in our multivariate model. In the survival model, we included age class (lamb vs. adult) plus sex and its interaction with age class, as well as linear and quadratic age terms (capturing variation in survival with age during adulthood). This model was run using 3,569 measurements from 1,574 individuals for which TL, August weight, and overwinter survival measurements were available. TL and body weight were z-transformed prior to inclusion in the model (mean = 0, SD = 1). To check the consistency of our results, we additionally ran separate models for lambs aged 4 mo and adults aged ≥1 y (SI Appendix, Supplementary Methods and Results).
Genetic model.
We extended our multivariate analysis to examine the association among TL, August body weight, and overwinter survival probability at the genetic level (the G matrix). As with the univariate heritability analysis, we used information on individual relatedness from the population pedigree to estimate the additive genetic variances and covariances. In addition to the additive genetic (co)variance, we estimated the covariance at the residual, among-year, and among-individual levels (the permanent environment covariance: the covariance among traits at the individual level that is not explained by genetic effects). The model was run for 5.3 × 105 iterations, with 3 × 104 burn-in and thinning interval of 500, resulting in 1,000 stored samples of the MCMC chain (autocorrelation < 0.1). Parameter expanded priors were used for all variance components, and inverse-Wishart priors were used for the residual variances. The residual variance was fixed at one for survival (as is the standard recommendation for threshold models), and the latent variables were constrained to be between ±7 to avoid numerical difficulties as the probabilities approached zero and one. Other fixed and random effects were as stated above for the phenotypic models. We additionally ran separate animal models for lambs aged 4 mo and adults aged ≥1 y to check the consistency of our results (SI Appendix, Supplementary Methods and Results).
Supplementary Material
Acknowledgments
We thank the many project members and volunteers who have contributed to the long-term study of Soay sheep on St Kilda over many years; the National Trust for Scotland for permission to work on St Kilda; and QinetiQ and Kilda Cruises for logistical support in the field. We thank Camillo Bérénos and Jisca Huisman for reconstruction of the population pedigree, Lorraine Kerr and Eliane Salvo‐Chirnside for laboratory support, Jarrod Hadfield and Craig Walling for statistical advice, and Jon Slate for constructive feedback on the manuscript. The long-term field study has been largely funded by the UK Natural Environment Research Council, SNP genotyping was mostly funded by the European Research Council, and the telomere project was funded by the UK Biotechnology and Biological Sciences Research Council (Grant BB/L020769/1).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020563118/-/DCSupplemental.
Data Availability
Data used in this paper are available in the Supporting Information. The R code used in our analyses is available at: https://github.com/hfroy/PNAS_2021.
References
- 1.Blackburn E. H., Structure and function of telomeres. Nature 350, 569–573 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Aubert G., Lansdorp P. M., Telomeres and aging. Physiol. Rev. 88, 557–579 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Shay J. W., Wright W. E., Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 20, 299–309 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Levy M. Z., Allsopp R. C., Futcher A. B., Greider C. W., Harley C. B., Telomere end-replication problem and cell aging. J. Mol. Biol. 225, 951–960 (1992). [DOI] [PubMed] [Google Scholar]
- 5.von Zglinicki T., Oxidative stress shortens telomeres. Trends Biochem. Sci. 27, 339–344 (2002). [DOI] [PubMed] [Google Scholar]
- 6.López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benetos A., et al., Short telomeres are associated with increased carotid atherosclerosis in hypertensive subjects. Hypertension 43, 182–185 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Blasco M. A., Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Zhao J., Miao K., Wang H., Ding H., Wang D. W., Association between telomere length and type 2 diabetes mellitus: A meta-analysis. PLoS One 8, e79993 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cawthon R. M., Smith K. R., O’Brien E., Sivatchenko A., Kerber R. A., Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361, 393–395 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Kimura M., et al., Telomere length and mortality: A study of leukocytes in elderly Danish twins. Am. J. Epidemiol. 167, 799–806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boonekamp J. J., Simons M. J., Hemerik L., Verhulst S., Telomere length behaves as biomarker of somatic redundancy rather than biological age. Aging Cell 12, 330–332 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Wilbourn R. V., et al., The relationship between telomere length and mortality risk in non-model vertebrate systems: A meta-analysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simons M. J., Questioning causal involvement of telomeres in aging. Ageing Res. Rev. 24, 191–196 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Aviv A., Telomeres and human somatic fitness. J. Gerontol. A Biol. Sci. Med. Sci. 61, 871–873 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Bateson M., Cumulative stress in research animals: Telomere attrition as a biomarker in a welfare context? BioEssays 38, 201–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monaghan P., Ozanne S. E., Somatic growth and telomere dynamics in vertebrates: Relationships, mechanisms and consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young A. J., The role of telomeres in the mechanisms and evolution of life-history trade-offs and ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benetos A., et al., Tracking and fixed ranking of leukocyte telomere length across the adult life course. Aging Cell 12, 615–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bichet C., et al., Telomere length is repeatable, shortens with age and reproductive success, and predicts remaining lifespan in a long-lived seabird. Mol. Ecol. 29, 429–441 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Dugdale H. L., Richardson D. S., Heritability of telomere variation: It is all about the environment! Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broer L., et al., Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. 21, 1163–1168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spurgin L. G., et al., Spatio-temporal variation in lifelong telomere dynamics in a long-term ecological study. J. Anim. Ecol. 87, 187–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Lieshout S. H. J., et al., Individual variation in early-life telomere length and survival in a wild mammal. Mol. Ecol. 28, 4152–4165 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Oliveira B. S., et al., Systematic review of the association between chronic social stress and telomere length: A life course perspective. Ageing Res. Rev. 26, 37–52 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Pepper G. V., Bateson M., Nettle D., Telomeres as integrative markers of exposure to stress and adversity: A systematic review and meta-analysis. R. Soc. Open Sci. 5, 180744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chatelain M., Drobniak S. M., Szulkin M., The association between stressors and telomeres in non-human vertebrates: A meta-analysis. Ecol. Lett. 23, 381–398 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Boonekamp J. J., Mulder G. A., Salomons H. M., Dijkstra C., Verhulst S., Nestling telomere shortening, but not telomere length, reflects developmental stress and predicts survival in wild birds. Proc. R. Soc. B 281, 20133287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood E. M., Young A. J., Telomere attrition predicts reduced survival in a wild social bird, but short telomeres do not. Mol. Ecol. 28, 3669–3680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett E. L., Burke T. A., Hammers M., Komdeur J., Richardson D. S., Telomere length and dynamics predict mortality in a wild longitudinal study. Mol. Ecol. 22, 249–259 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Vaupel J. W., Manton K. G., Stallard E., The impact of heterogeneity in individual frailty on the dynamics of mortality. Demography 16, 439–454 (1979). [PubMed] [Google Scholar]
- 32.Nussey D. H., Coulson T., Festa-Bianchet M., Gaillard J. M., Measuring senescence in wild animal populations: Towards a longitudinal approach. Funct. Ecol. 22, 393–406 (2008). [Google Scholar]
- 33.Clutton-Brock T. H., Pemberton J. M., Soay Sheep: Dynamics and Selection in an Island Population (Cambridge University Press, 2004). [Google Scholar]
- 34.Milner J., Albon S., Illius A., Pemberton J., Clutton‐Brock T., Repeated selection of morphometric traits in the Soay sheep on St Kilda. J. Anim. Ecol. 68, 472–488 (1999). [Google Scholar]
- 35.Nussey D. H., et al., Patterns of body mass senescence and selective disappearance differ among three species of free-living ungulates. Ecology 92, 1936–1947 (2011). [DOI] [PubMed] [Google Scholar]
- 36.van Lieshout S., et al., Estimation of environmental and genetic contributions to telomere length variation in a wild mammal. J. Evol. Biol. 34, 296–308 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Foley N. M., et al., Drivers of longitudinal telomere dynamics in a long-lived bat species, Myotis myotis. Mol. Ecol. 29, 2963–2977 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Seeker L. A., et al., Bovine telomere dynamics and the association between telomere length and productive lifespan. Sci. Rep. 8, 12748 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asghar M., Bensch S., Tarka M., Hansson B., Hasselquist D., Maternal and genetic factors determine early life telomere length. Proc. Biol. Sci. 282, 20142263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atema E., et al., Heritability of telomere length in the Zebra Finch. J. Ornithol. 156, 1113–1123 (2015). [Google Scholar]
- 41.Becker P. J. J., et al., Mother–offspring and nest-mate resemblance but no heritability in early-life telomere length in white-throated dippers. Proc. Biol. Sci. 282, 20142924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sparks A. M., et al., Telomere heritability and parental age at conception effects in a wild avian population. Mol. Ecol., 10.1111/mec.15804 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Nussey D. H., et al., Measuring telomere length and telomere dynamics in evolutionary biology and ecology. Methods Ecol. Evol. 5, 299–310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seeker L. A., et al., Longitudinal changes in telomere length and associated genetic parameters in dairy cattle analysed using random regression models. PLoS One 13, e0192864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fairlie J., et al., Lifelong leukocyte telomere dynamics and survival in a free-living mammal. Aging Cell 15, 140–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telomere attrition rates are associated with weather conditions and predict productive lifespan in dairy cattle. Sci. Rep. 11, 5589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heidinger B. J., et al., Telomere length in early life predicts lifespan. Proc. Natl. Acad. Sci. U.S.A. 109, 1743–1748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eastwood J. R., et al., Early-life telomere length predicts lifespan and lifetime reproductive success in a wild bird. Mol. Ecol. 28, 1127–1137 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Atzmon G., et al., Genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc. Natl. Acad. Sci. U.S.A. 107, 1710–1717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soerensen M., et al., Genetic variation in TERT and TERC and human leukocyte telomere length and longevity: A cross-sectional and longitudinal analysis. Aging Cell 11, 223–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarabino D., Peconi M., Pelliccia F., Corbo R. M., Analysis of the association between TERC and TERT genetic variation and leukocyte telomere length and human lifespan—A follow-up study. Genes (Basel) 10, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakaysa S. L., et al., Telomere length predicts survival independent of genetic influences. Aging Cell 6, 769–774 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Armanios M., et al., Short telomeres are sufficient to cause the degenerative defects associated with aging. Am. J. Hum. Genet. 85, 823–832 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muñoz-Lorente M. A., Cano-Martin A. C., Blasco M. A., Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nat. Commun. 10, 4723 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bérénos C., Ellis P. A., Pilkington J. G., Pemberton J. M., Estimating quantitative genetic parameters in wild populations: A comparison of pedigree and genomic approaches. Mol. Ecol. 23, 3434–3451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huisman J., Pedigree reconstruction from SNP data: Parentage assignment, sibship clustering and beyond. Mol. Ecol. Resour. 17, 1009–1024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hadfield J. D., Richardson D. S., Burke T., Towards unbiased parentage assignment: Combining genetic, behavioural and spatial data in a Bayesian framework. Mol. Ecol. 15, 3715–3730 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Seeker L. A., et al., Method specific calibration corrects for DNA extraction method effects on relative telomere length measurements by quantitative PCR. PLoS One 11, e0164046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cawthon R. M., Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruijter J. M., et al., Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfaffl M. W., A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks M. E., et al., glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9, 378–400 (2017). [Google Scholar]
- 63.Vedder O., Verhulst S., Zuidersma E., Bouwhuis S., Embryonic growth rate affects telomere attrition: An experiment in a wild bird. J. Exp. Biol. 221, jeb181586 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Team R. C., R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019), Version 3.6.1.
- 65.Hadfield J. D., MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. J. Stat. Softw. 33, 1–22 (2010).20808728 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this paper are available in the Supporting Information. The R code used in our analyses is available at: https://github.com/hfroy/PNAS_2021.