Significance
Targeting energy metabolism in Mycobacterium tuberculosis has emerged as a new paradigm in antituberculosis drug discovery. Succinate dehydrogenase is considered the regulator of respiration in M. tuberculosis. Mycobacteria contains two different succinate dehydrogenase enzymes designated Sdh1 and Sdh2. Sdh1 has recently been identified as a new class of succinate dehydrogenase. In this study, we have determined M. smegmatis Sdh1 structures alone and in the presence of ubiquinone-1, revealing that Sdh1 has a novel electron transfer pathway and a unique substrate-binding site. These data show that the structure of M. tuberculosis Sdh1 is significantly different by comparison with the human counterpart making a good antituberculosis drug target.
Keywords: succinate dehydrogenase, Mycobacterium smegmatis, electron transport chain, cryoelectron microscopy
Abstract
Complex II, also known as succinate dehydrogenase (SQR) or fumarate reductase (QFR), is an enzyme involved in both the Krebs cycle and oxidative phosphorylation. Mycobacterial Sdh1 has recently been identified as a new class of respiratory complex II (type F) but with an unknown electron transfer mechanism. Here, using cryoelectron microscopy, we have determined the structure of Mycobacterium smegmatis Sdh1 in the presence and absence of the substrate, ubiquinone-1, at 2.53-Å and 2.88-Å resolution, respectively. Sdh1 comprises three subunits, two that are water soluble, SdhA and SdhB, and one that is membrane spanning, SdhC. Within these subunits we identified a quinone-binding site and a rarely observed Rieske-type [2Fe-2S] cluster, the latter being embedded in the transmembrane region. A mutant, where two His ligands of the Rieske-type [2Fe-2S] were changed to alanine, abolished the quinone reduction activity of the Sdh1. Our structures allow the proposal of an electron transfer pathway that connects the substrate-binding and quinone-binding sites. Given the unique features of Sdh1 and its essential role in Mycobacteria, these structures will facilitate antituberculosis drug discovery efforts that specifically target this complex.
During respiration, cells harvest energy from their environment via redox reactions. The harvested energy is converted into adenosine triphosphate (ATP) by ATP synthase (also called as complex V). This process requires the formation of a transmembrane electrochemical gradient that is produced by the electron flux catalyzed by four integral-membrane respiratory complexes (designated complexes I to IV) of the oxidative phosphorylation system (1).
Respiratory complex II, also named succinate dehydrogenase (succinate:quinone reductase or SQR) or fumarate reductase (quinol:fumarate reductase or QFR), depending on the preferred direction of the reaction in vivo, performs the reversible oxidation of succinate to fumarate, a function that is coupled to the presence of mobile quinone electron carriers (2). This process is central to cellular metabolism and energy conversion, bridging the Krebs cycle and oxidative phosphorylation (3). All complex II enzymes across the different kingdoms of life share a common overall architecture with a smaller membrane-bound domain and a large soluble domain (4). The soluble domain consists of two hydrophilic subunits, A and B. The structure of subunit A is composed of four domains (5, 6): an FAD-binding domain, a capping domain, a helical domain, and a C-terminal domain. The active site for succinate-fumarate interconversion is located between the flavin-binding domain and the capping domain. Subunit B is a small iron-sulfur protein harboring a [2Fe-2S], a [4Fe-4S], and a [3Fe-4S] cluster, that facilitate electron movement. The two soluble subunits, A and B, are highly conserved across bacteria and mammals. In contrast, the structure and components of the integral-membrane domain, SdhC, can vary containing 0, 1, or 2 membrane-bound subunits with 5 or 6 transmembrane helices and possess varying numbers of heme b groups (0, 1, or 2). Variations also occur according to the type of quinone they use (ubiquinone or menaquinone) and the number of quinone-binding sites (7, 8). As a result, the membrane-spanning regions have distinct evolutionary origins. Thus, the complex II superfamily has been further divided into five subfamilies (types A through E) depending on their biophysical properties (8). Several structures of complex II superfamily enzymes have been determined: type A (exemplified by the Mycobacterium smegmatis (Msm) Sdh2) (9), type B (exemplified by the Wolinella succinogenes QFR) (6), type C [exemplified by the Escherichia coli (10) and porcine (5) SQRs], and type D (exemplified by the E. coli QFR) (11). However, Sdh1 has only recently been identified in mycobacteria and is suggested to represent a new class of respiratory complex II referred to as type F (12). It has been shown that Sdh1 activity is essential for Mycobacterium tuberculosis to adapt to optimal growth and survival under aerobic conditions (13–15). It may accomplish the oxidation of succinate by using higher potential quinones (12, 16). It is suggested that there are enough structural differences between mycobacterial Sdh1 and mitochondrial SQR for anti-TB drug discovery and design (17).
In the present study, cryoelectron microscopy (cryo-EM) structures of the M. smegmatis Sdh1 in the presence and absence of substrate, ubiquinone-1, have been determined. These allow the visualization of the electron transport path of a type F complex II system.
Results and Discussion
Overall Fold of Msm Sdh1.
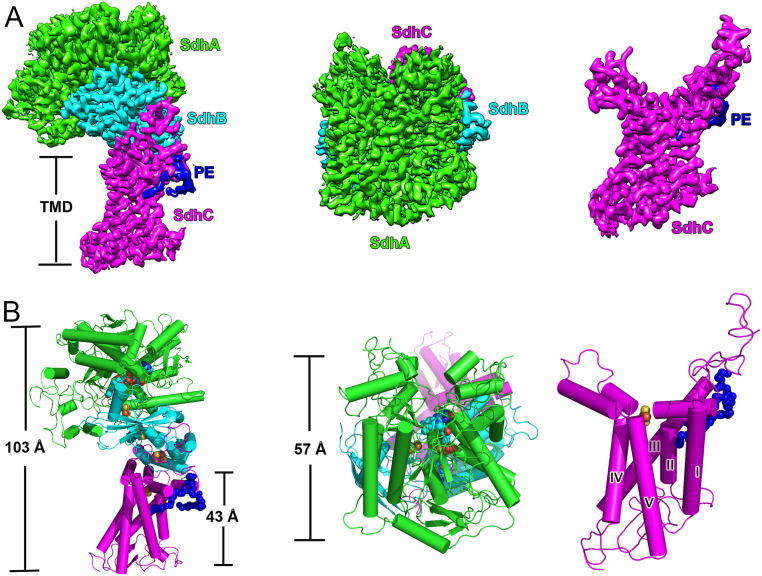
Msm Sdh1 was expressed recombinantly and purified to homogeneity (SI Appendix, Fig. S1 A and B). Folding was confirmed by a succinate-quinone oxidoreductase activity which gave a kcat 0.87 ± 0.02 s−1 (SI Appendix, Fig. S1C and Table S4). The three-dimensional structure of Msm Sdh1 was determined to 2.88-Å resolution using cryo-EM (SI Appendix, Figs. S2 and S4 and Tables S1 and S2). It is monomeric with dimensions of 57 Å by 103 Å (Fig. 1) and has the shape of the letter “q” with a hydrophilic head and a hydrophobic multipass transmembrane-anchored tail. The monomeric assembly is similar to that observed in porcine SQR (5), a protomer of E. coli SQR/QFR (10, 11), and W. succinogenes QFR (6). The hydrophilic head of Sdh1 consists of the FAD (flavin adenine dinucleotide)-binding protein (SdhA) and the iron-sulfur protein (SdhB). The entire hydrophobic domain contains one membrane-anchored subunit (SdhC), which has five transmembrane helices designated I to V (Fig. 1).
Fig. 1.
Overall structure of Msm succinate dehydrogenase 1. (A) Cryo-EM density map at 2.88-Å resolution. (Left) Front view; (Middle) top view (from cytoplasm); (Right) transmembrane region of Sdh1. SdhA, SdhB, and SdhC are colored in green, cyan, and magenta, respectively. PE is in blue. (B) Same views as a cartoon representation. The five transmembrane helices are labeled and PE is shown as blue spheres.
Density for a phosphatidylethanolamine (PE) unit was also observed (Fig. 1 and SI Appendix, Fig. S4), and such a unit was subsequently detected by mass spectrometry (SI Appendix, Fig. S1D). According to the structure, it is located between transmembrane helices II and III on the cytoplasmic side. However, we did not observe cardiolipin (CL) in the map density, despite it being also detected by mass spectrometry (SI Appendix, Fig. S1D), suggesting it adopts a highly flexible structure in the complex.
Unique Topology of the Sdh1 Transmembrane Domain.
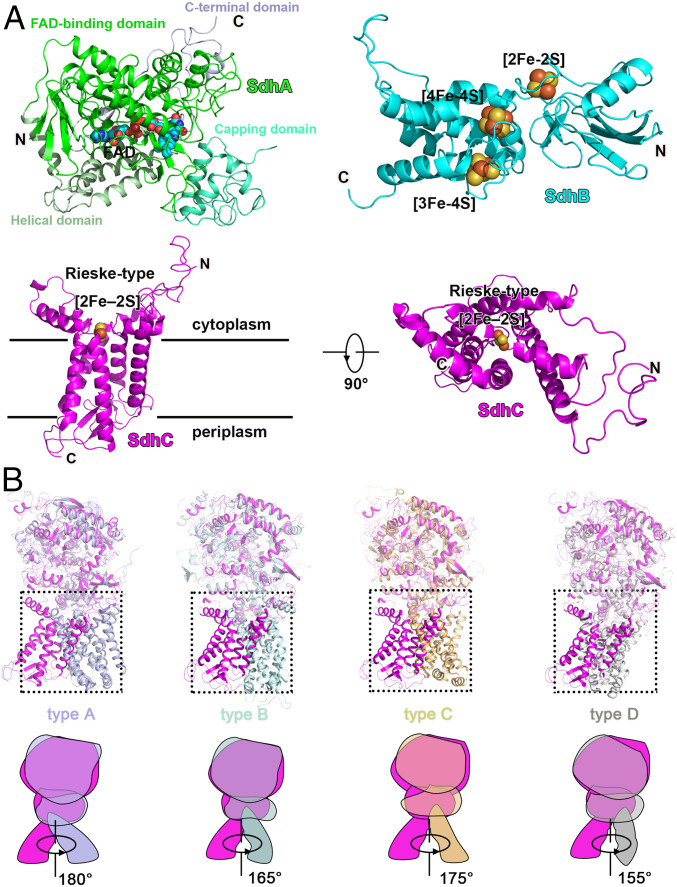
The SdhA subunit can be observed as four subdomains (5, 6), termed the FAD-binding domain, capping domain (significantly disordered), helical domain, and C-terminal domain (Fig. 2A). In contrast, the SdhB subunit is shaped like a butterfly and organized into only two domains (Fig. 2A). The N-terminal domain is composed of a three-stranded β-sheet and a small α-helix, while the C-terminal α-helical domain contains six α-helices linked by short loop regions. The capping domain is disordered, which may be due to the fact no ligand is bound. Previous studies on other complex II systems have shown that ligand (18) or substrate (19) binding can result in a change of structural order for this domain. Overall, the soluble domains (subunits A and B) including that of Sdh1 are similar in fold to the previously reported bacterial and mitochondrial respiratory complex IIs (5, 6, 10, 11) (Fig. 2B and SI Appendix, Fig. S5).
Fig. 2.
The composition of Msm Sdh1 and comparison with types A–D complex IIs. (A) SdhA, SdhB, and SdhC with prosthetic groups are presented in cartoon mode. FAD and FeS clusters are shown as spheres. (B) Structural superimposition of Msm Sdh1 with Msm Sdh2 (type A, PDB code 6LUM), W. succinogenes QFR (type B, PDB code 1E7P), Sus scrofa SQR, (type C, PDB code 1ZOY), and E. coli QFR (type D, PDB code 1L0V). Corresponding cartoon models and rotation angles are shown. Msm Sdh1, Sdh2, and W. succinogenes QFR, S. scrofa SQR, and E. coli QFR are in magenta, light-blue, pale cyan, orange, and gray, respectively.
The transmembrane domain of Sdh1 resembles a flower, in which a Rieske-type [2Fe-2S] cluster (described below) and transmembrane helices appear as pistil and leaf shapes, respectively (Fig. 2A). The N and C termini of the SdhC subunit are on opposite sides of the membrane-spanning region, corresponding to the cytoplasm and periplasm, respectively (Fig. 2A). The relative orientation between the soluble domain and the transmembrane domain is different in Sdh1 by comparison with other complex II classes (Fig. 2B). Also, the membrane-embedded domain cannot be aligned to any other complex II (Fig. 2B). The Rieske-type [2Fe-2S] cluster that replaces the equivalent b-type heme group (found in all other complex IIs) in the membrane region mediates electron transfer (described below). The structural differences in the membrane-anchored domain suggest that Sdh1 has a different quinone-binding site by comparison with other types of complex II (described below). Collectively, these features, especially the unique membrane-spanning domain, highlight that Sdh1 is a respiratory complex II that is different from those previously characterized to date (12).
Electron Transfer Pathway in Msm Sdh1.
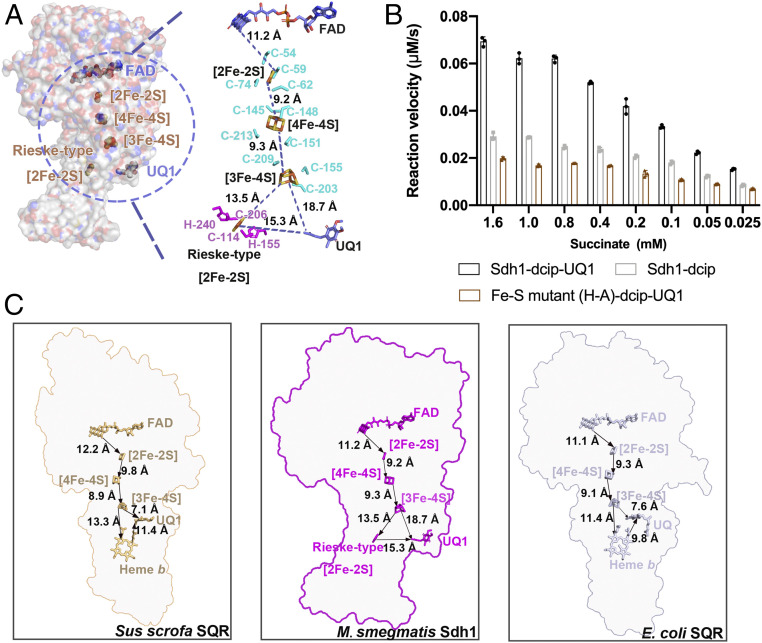
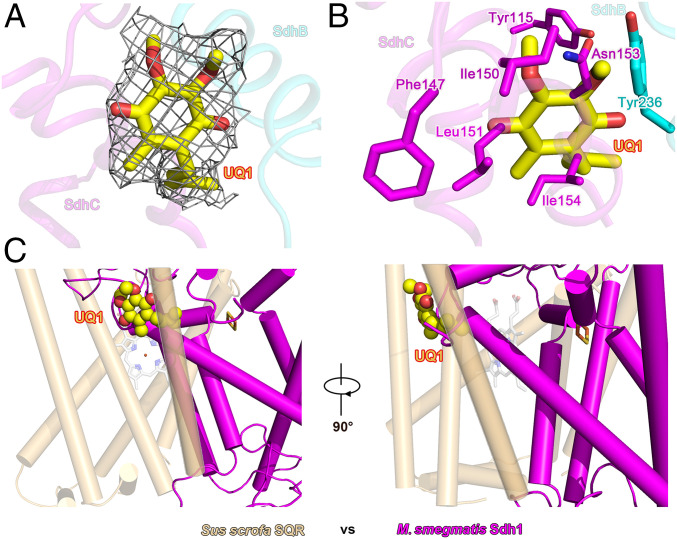
All of prosthetic groups (FAD, [2Fe-2S], [4Fe-4S], [3Fe-4S], and a Rieske-type [2Fe-2S]) required for electron transfer were unambiguously assigned into the cryo-EM density map (SI Appendix, Fig. S4). The edge-to-edge distance between these four prosthetic groups is less than 14 Å (Fig. 3A), a distance range that can efficiently allow the delivery of electrons between these sites (20). The FAD is in Sdh1 at the center of the catalytic site for fumarate reduction and succinate oxidation. The [2Fe-2S] cluster is coordinated by a loop located in the N-terminal domain, and the C-terminal domain is responsible for binding the [4Fe-4S] and [3Fe-4S] clusters (Fig. 2A and SI Appendix, Fig. S4B). The three different iron-sulfur clusters of the SdhB subunit are coordinated by conserved cysteine residues previously observed in SQRs and QFRs (5, 6, 10, 11, 21). The Rieske-type [2Fe-2S] cluster (22) is coordinated by two histidine and cysteine residues of the SdhC subunit and embedded in the transmembrane domain. In E. coli and porcine SQRs, the position equivalent to the Rieske-type [2Fe-2S] cluster site is occupied by a heme b (5, 10) (Fig. 3C). In order to identify the location for quinone binding, we determined the structure in complex with ubiquinone-1 (UQ1) at 2.53-Å resolution (SI Appendix, Figs. S3 and S4 and Tables S1 and S3). Ubiquinone-1 is bound at the entrance of the pocket formed by the cytoplasmic N termini of transmembrane III and the C-terminal segment of SdhB and interacts with Tyr-B236, Phe-C147, Ile-C150, Leu-C151, and Ile-C154 (Fig. 4 A and B). The edge-to-edge distances between UQ1 and Rieske-type [2Fe-2S] and [3Fe-4S] are about 15.3 and 18.7 Å, respectively, which are too far for efficient electron transfer (20). Thus, we speculate the endogenous electron acceptor quinone would bind closer to these two redox centers to facilitate electron transfer, and what we observe here might be a representation of a leaving state of the UQ1.
Fig. 3.
Prosthetic groups in Msm Sdh1 and the path of electron transfer. (A) Msm Sdh1. (Left) The overall structure is shown as surface representation with the prosthetic groups shown in spheres. (Right) Prosthetic groups labeled with edge-to-edge distances. The prosthetic groups and amino acids around the Fe-S clusters are shown as sticks and colored corresponding to that of the subunits. (B) Comparison of wild-type and mutant succinate dehydrogenase activities. Mean values result from technical triplicates and error bars represent the SD. (C) Comparison of prosthetic groups from S. scrofa SQR, Msm Sdh1, and E. coli SQR, along with their edge-to-edge distances. Prosthetic groups are shown as stick models. Arrows indicate the likely direction of electron transport.
Fig. 4.
Quinone-binding site bound with ubiquinone-1. (A) The quinone-binding site and the map density for ubiquinone-1 (UQ1). SdhB and SdhC are shown as cartoons. UQ1 is shown as a sticks model and colored in yellow. Map density of UQ1 is shown as mesh with a threshold of 0.6. (B) Amino acids in SdhB and SdhC that form the quinone-binding site. (C) Quinone-binding site in Msm Sdh1 superimposed onto the S. scrofa SQR. Msm Sdh1 and S. scrofa SQR are colored magenta and orange, respectively. Atoms for UQ1 are shown as spheres.
In E. coli and porcine SQRs, the electrons released from succinate oxidation are transferred to the [3Fe-4S] by a chain of redox centers including FAD, [2Fe-2S] and [4Fe-4S], and then to either the heme b and/or the ubiquinone (ubiquinone is preferable) (Fig. 3C) (5, 10). Once heme b is reduced, it can in turn reduce ubiquinone (5, 10). The heme b is proposed to serve as an electron sink in the electron transfer pathway (23, 24). In this process, quinone likely receives the first electron from the [3Fe-4S] cluster to form the semiquinone that can be detected by electron paramagnetic resonance (EPR) spectroscopy. Here, rapid electron equilibration occurs between the heme b and quinone to stabilize the semiquinone radical species (23, 25). Next the semiquinone accepts a second electron from either the heme b or the [3Fe-4S] cluster to complete full reduction. Given the overall arrangement of redox centers between Sdh1 and SQRs (Fig. 3C), they very likely have a similar electron transfer pathway. Therefore, in Sdh1 the substrate-binding site and quinone-binding site are also proposed to be connected by a chain of redox centers consisting of FAD, [2Fe-2S], [4Fe-4S], [3Fe-4S], and Rieske [2Fe-2S]. It is noteworthy that electron tunneling rates can be affected by distances between, as well as midpoint potentials for each prosthetic group (20). It is well known that the presence of the His and Cys ligands can increase the reduction potential of the [2Fe-2S] cluster (which attract electrons) (26). Although the distance between the [3Fe-4S] cluster and the quinone-binding site is long (18.7 Å) in the present structure, its native distance is believed to be within the limit for effective electron tunneling according to the enzyme activity of the wild-type Sdh1 (Fig. 3B) and the quinone-bound SQRs from E. coli and porcine (5, 10). Moreover, [3Fe-4S] clusters generally have a lower potential in enzymes operating with low potential quinones, regardless of whether they are SQRs or QFRs (27). Hence, overall, although electrons can be transferred either to UQ1 and/or to Rieske [2Fe-2S] from the [3Fe-4S] cluster, transfer to Rieske [2Fe-2S] is very likely preferable here (Fig. 3A). Additionally, using EPR spectroscopy, no semiquinone signal was observed during catalytic succinate oxidation (SI Appendix, Fig. S1E), a feature which is observed in the E. coli SQR (23). Taken together, these results suggest that the Rieske-type [2Fe-2S] also serves as an electron sink accepting electrons from the [3Fe-4S] first, and when both the [3Fe-4S] and the Rieske-type [2Fe-2S] are reduced, this would allow for the reduction of bound quinone by the concomitant two electrons. In this mechanism, the Rieske-type [2Fe-2S] cluster would hold an electron until the [3Fe-4S] cluster is reduced, which can also potentially prevent the formation of reactive oxygen species that have been shown to be harmful to cells (28).
The heme b is not an absolute structural requirement and not essential for quinone reduction (29, 30). In the present study, we replaced the two [2Fe-2S] cluster His-ligands (His-C155 and His-C240) with two alanine residues. Sdh1 completely lost quinone reduction activity (Fig. 3B and SI Appendix, Table S4). According to the description above, compared to the heme b in the E. coli and porcine SQRs, the mutant of Rieske-type [2Fe-2S] ligand may strongly affect the start of normal electron transfer mediated by the Rieske-type [2Fe-2S], which is an essential stepping stone of electron transfer in the current structure. The gel filtration and cryo-EM data on the His-Ala mutant also suggest that the mutation destabilizes the protein, which could have affected the binding or the bound orientation of the Rieske [2Fe-2S] cluster (SI Appendix, Fig. S6). In addition, the residue His-C155 is not only the ligand of the Rieske-type [2Fe-2S] cluster, but also potentially plays a role in stabilizing the quinone binding. The alteration of the histidyl ligand may result in the disruption of the quinone-binding site and affect its ability to interact the quinone. Overall, these structural features are in reasonable agreement with the inactivation of the Sdh1 mutant to the quinone reduction activity. The emergence of a Rieske-type [2Fe-2S] cluster and potential electron transport path in Sdh1 may be a crucial adaptive feature of mycobacteria. The physiological roles of these need to be investigated in the future.
It has been reported that M. smegmatis Sdh1 does not generate or consume electrochemical gradients (12). Moreover, Sdh1 is suggested to use higher potential quinones such as polyketides in vivo (12, 16). Therefore, these observations suggest that Sdh1 does not contain the canonical E pathway (transmembrane electron transfer via the heme groups is strictly coupled to cotransfer of protons involving a prominent component residue, glutamate) used to produce the proton motive force to promote succinate oxidation and quinone reduction, as identified in W. succinogenes QFR (31). Correspondingly, the canonical E pathway is not observed in the Sdh1 structure.
Quinone-Binding Site Contributes to the Rational Structure-Assisted Drug Discovery.
Respiratory complex IIs are found in all realms of life including bacterial pathogens. However, they have been largely neglected for drug development (17). The quinone-binding sites in different respiratory complex IIs have different structural features. Thus, like other quinone-binding sites (1, 32, 33), the Sdh1 quinone-binding site is also of great interest as a potential drug target.
Mammalian complex II is coupled to ubiquinone whereas Sdh1 enzymes use the polyketide quinones (12, 16). This difference provides opportunities for developing inhibitors that selectively target Sdh1. M. smegmatis and M. tuberculosis (Mtb) have a similar quinone-binding site according to the sequence alignments of the membrane-spanning subunit involved with quinone binding (SI Appendix, Fig. S7A). Similarly, there is also a quinone-binding site in mammals (e.g., human and porcine) (SI Appendix, Fig. S7B). However, the quinone-binding sites in Msm Sdh1 and porcine SQR poorly align together (Fig. 4C). Moreover, after 3D superimposition of these two structures, the quinone-binding site of Msm Sdh1 is blocked by a transmembrane helix in the porcine SQR (Fig. 4C). These observations show that the quinone-binding site of Msm and Mtb significantly differ in comparison with that found in the mammalian counterparts, and therefore is a target site for drug development.
Conclusions
We have determined structures of Msm Sdh1 in the presence and absence of ubiquinone-1. Sdh1 comprises three subunits, two water-soluble, SdhA and SdhB, and SdhC which is membrane anchored. A Rieske-type [2Fe-2S] cluster embedded in the membrane has been identified. It appears this cluster is required for quinone reduction. The essential role that Mtb Sdh1 plays in electron transport and its different structural features compared to the mammalian counterparts suggest it is a good antimycobacterial drug target.
Materials and Methods
Cloning and Expression of Wild-Type and Mutant Msm Sdh1.
The Msm Sdh1 complex is encoded by five putative genes (MSMEG_0416-0420). Genes were amplified from M. smegmatis strain mc251 genomic DNA by PCR using Phanta Max DNA polymerase (Vazyme), and two-step PCR was used to inset a 10× His tag at the C terminus of the SdhC (MSMEG_0419). Genes encoding the entire Sdh1 operon were then cloned into the vector pMV261 which harbors an acetamide promoter. The resultant plasmid was transformed into M. smegmatis mc251 cells by electroporation. The cells were cultivated in Luria broth (LB) liquid media supplemented with 50 μg/mL kanamycin, 20 μg/mL carbenicillin, and 0.1% Tween-80. When the cells were grown to an optical density (OD600 of 1.0 to 1.2) at 37 °C, overexpression of the recombinant protein was induced by 0.2% (wt/vol) acetamide at 16 °C. After 4 d, cell pellets were harvested by centrifugation and frozen at −20 °C until use. The cloning and expression of the mutant Sdh1 were performed as for the wild-type Sdh1 as described above. The primers are listed in SI Appendix, Table S5.
Purification and Characterization of Wild-Type and Mutant Msm Sdh1.
Cell pellets were thawed and resuspended in buffer A containing 20 mM Hepes, pH 7.5, 150 mM NaCl, and then lysed by passing through a French press at 1,200 bar. Cell debris was removed by centrifugation at 14,000 rpm for 10 min at 4 °C. The supernatant was collected and ultracentrifuged at 150,000 × g, 4 °C for 1.5 h. The membrane fraction was solubilized by addition of 1% (wt/vol) LMNG (lauryl maltose neopentyl glycol) in buffer A and incubated for 2 h at 4 °C. The suspension was ultracentrifuged and the supernatant was applied to Ni-NTA agarose beads (GE Healthcare) in 10 mM imidazole at 4 °C. The beads were rinsed in buffer A with 50 mM imidazole and 0.004% (wt/vol) LMNG. The buffer was exchanged to buffer B with 20 mM Hepes, pH 7.5, 150 mM NaCl, and 0.02% (wt/vol) GDN (glyco-diosgenin) and then washed in resin in batch mode. The protein was eluted from the beads with buffer B containing 500 mM imidazole. Protein was then concentrated and loaded onto a Superose 6 increase (10/300 GL, GE Healthcare) with buffer B. Peak fractions were concentrated to ∼5 mg/mL for electron microscopy studies.
In order to determine the composition of the wild-type Sdh1 complex, the purified protein sample was mixed with 2× loading buffer and analyzed by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Only three bands were observed and they were subjected to mass spectrometry analysis. Phospholipids were detected by using normal-phase liquid chromatography-mass spectrometry (LCMS) (34) and samples were processed as described previously (1).
EPR spectra were acquired as previously described (1, 9) using a Bruker X-band (9.4 GHz) EMX plus 10/12 spectrometer with ESR-910 flowing helium cryostat, at a temperature of 10K, and a 5-Gauss modulation amplitude at 100 kHz under nonsaturating microwave power conditions (100 μW). To reveal the iron-sulfur clusters in the complex, samples were in the “air-oxidized” state (as isolated) or in the “reduced” state (by addition of 200 μM succinate or 1 mM dithionite) (35–37).
Activity Measurement.
The succinate:quinone oxidoreductase activity of wild-type and mutant Msm Sdh1 was determined using a succinate-2,6-dichlorophenolindophenol (DCIP) assay according to the method previously described (12). In the present study, we used ubiquinone-1 as the intermediate electron acceptor to test succinate dehydrogenase activity. The reaction mixture contained buffer B, 0.025 to 1.6 mM sodium succinate, 0.5 mM ubiquinone-1, and 100 μM DCIP. The reaction was initiated by addition of DCIP. All assays were performed at 37 °C, using the Molecular Devices iD3 Reader. Data and figures were processed with GraphPad Prism 6.
Electron Microscopy Sample Preparation and Imaging.
A total of 3 μL of 5 mg/mL Sdh1 complex with 1 mM dithiothreitol was applied to H2/O2 glow-discharged 200-mesh Quantifoil R 0.6/1 grids (Quantifoil, Micro Tools GmbH) and subsequently blotted using a FEI Vitrobot and then frozen in liquid ethane. The grids were imaged in a 300-keV Titan Krios (Thermo Fisher Scientific) with a Gatan K2 Summit direct electron detector (Gatan) and a Gatan Quantum energy filter. Data were collected in superresolution mode at a pixel size of 0.82 Å with a dose rate of 8e−/(pixel·s). Images were recorded for 5 s in 40 subframes to give a total dose of 60 electrons per Å2. To obtain Msm Sdh1 in complex with ubiquinone-1, they were mixed in a mole ratio of 1:20 and incubated for 30 min at 4 °C. The cryo-EM sample preparation and image processing were performed as for the Sdh1 sample as described above.
Imaging Processing and 3D Reconstruction.
A total of 5,508 dose-fractionated image stacks were subjected to beam-induced motion correction using MotionCor2 (38) and the contrast transfer functions were estimated by Gctf (39). Particles were picked automatically from 5,230 micrographs and 429,485 particles were extracted with a box size of 384 pixels in cryoSPARC (40). The 2D and 3D classifications and refinements were all performed in cryoSPARC. A total of 50,000 particles were used to perform Ab initio reconstruction. Three classes including 316,482 particles as the reference model were used to perform hetero refinement. The best class contained 254,341 particles. After nonuniform refinement and local refinement with mask, a 2.88-Å resolution map was obtained. A total of 6,004 image stacks for the samples of Msm Sdh1 in complex with ubiquinone-1 were collected, resulting in a 2.53-Å map.
Model Building and Refinement.
The atomic model of apo-Msm Sdh1 was manually built ab initio using Coot 0.8.9.1 (41). The density quality of the interior region was higher, so model building commenced here, followed by iterative manual fitting adjustment in Coot (41) and real space refinement in PHENIX (42). The atomic model of Msm Sdh1 in complex with ubiquinone-1 was built using the apo-Sdh1 model. To start, the cryo-EM model was docked into the EM density map using Chimera, this was then followed by manual adjustment and refinement as for apo-Sdh1. All reported resolutions were based on the gold-standard FSC 0.143 criteria (43). All figures were made using University of California San Francisco Chimera (44) and PyMOL (45).
Supplementary Material
Acknowledgments
We thank Dr. Chao Peng of the Mass Spectrometry System at the National Facility for Protein Science in Shanghai, Zhangjiang Lab, SARI, China, for data collection and analysis and Prof. Kaixia Mi (CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, CAS) for sharing the strain M. smegmatis mc251. We thank the Bio-Electron Microscopy Facility of ShanghaiTech University, and we are grateful to Dr. Qianqian Sun for her help with cryo-EM technical support. This work was supported by grants from the National Key Research and Development Program of China (Grant 2017YFC0840300), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grants XDB37030201, XDB37040000, and XDB37020203), the National Natural Science Foundation of China (Grants 81520108019 and 813300237), and the Tianjin Natural Science Foundation (Grant 20JCQNJC01430).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022308118/-/DCSupplemental.
Data Availability
All data presented in this study are available within the figures and the supplementary information. EM maps and atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb) and Protein Data Bank (PDB; http://www.rcsb.org), respectively, under the following ID codes: EMD-30595 and PDB 7D6X for the apo-Sdh1, and EMD-30594 and PDB 7D6V for the Sdh1-UQ1.
References
- 1.Gong H., et al., An electron transfer path connects subunits of a mycobacterial respiratory supercomplex. Science 362, eaat8923 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Sharma P., Maklashina E., Cecchini G., Iverson T. M., Crystal structure of an assembly intermediate of respiratory Complex II. Nat. Commun. 9, 274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher M. J., Herath A. S., Udagedara S. R., Dougan D. A., Truscott K. N., Crystal structure of bacterial succinate:quinone oxidoreductase flavoprotein SdhA in complex with its assembly factor SdhE. Proc. Natl. Acad. Sci. U.S.A. 115, 2982–2987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P., Maklashina E., Cecchini G., Iverson T. M., Maturation of the respiratory complex II flavoprotein. Curr. Opin. Struct. Biol. 59, 38–46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun F., et al., Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121, 1043–1057 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Lancaster C. R., Kröger A., Auer M., Michel H., Structure of fumarate reductase from Wolinella succinogenes at 2.2 A resolution. Nature 402, 377–385 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Moosavi B., Berry E. A., Zhu X. L., Yang W. C., Yang G. F., The assembly of succinate dehydrogenase: A key enzyme in bioenergetics. Cell. Mol. Life Sci. 76, 4023–4042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancaster C. R., The di-heme family of respiratory complex II enzymes. Biochim. Biophys. Acta 1827, 679–687 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Gong H., et al., Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF. Nat. Commun. 11, 4245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yankovskaya V., et al., Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299, 700–704 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Iverson T. M., Luna-Chavez C., Cecchini G., Rees D. C., Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284, 1961–1966 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Hards K., Rodriguez S. M., Cairns C., Cook G. M., Alternate quinone coupling in a new class of succinate dehydrogenase may potentiate mycobacterial respiratory control. FEBS Lett. 593, 475–486 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Griffin J. E., et al., High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7, e1002251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman T., et al., Succinate dehydrogenase is the regulator of respiration in Mycobacterium tuberculosis. PLoS Pathog. 10, e1004510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rittershaus E. S. C., et al., A lysine acetyltransferase contributes to the metabolic adaptation to hypoxia in Mycobacterium tuberculosis. Cell Chem. Biol. 25, 1495–1505.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand A., et al., Polyketide quinones are alternate intermediate electron carriers during mycobacterial respiration in oxygen-deficient niches. Mol. Cell 60, 637–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hards K., et al., Two for the price of one: Attacking the energetic-metabolic hub of mycobacteria to produce new chemotherapeutic agents. Prog. Biophys. Mol. Biol. 152, 35–44 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Starbird C. A., et al., New crystal forms of the integral membrane Escherichia coli quinol:fumarate reductase suggest that ligands control domain movement. J. Struct. Biol. 202, 100–104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomasiak T. M., Maklashina E., Cecchini G., Iverson T. M., A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II. J. Biol. Chem. 283, 15460–15468 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page C. C., Moser C. C., Chen X., Dutton P. L., Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Guan H. H., et al., Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas. Sci. Rep. 8, 14935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y., Assembly and transfer of iron–sulfur clusters in the plastid. Front. Plant Sci. 9, 336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran Q. M., Rothery R. A., Maklashina E., Cecchini G., Weiner J. H., The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J. Biol. Chem. 281, 32310–32317 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Horsefield R., et al., Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): A mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem. 281, 7309–7316 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Anderson R. F., Hille R., Shinde S. S., Cecchini G., Electron transfer within complex II. Succinate:ubiquinone oxidoreductase of Escherichia coli. J. Biol. Chem. 280, 33331–33337 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Bak D. W., Elliott S. J., Alternative FeS cluster ligands: Tuning redox potentials and chemistry. Curr. Opin. Chem. Biol. 19, 50–58 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Hägerhäll C., Succinate: Quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320, 107–141 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Finkel T., Holbrook N. J., Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Maklashina E., Rothery R. A., Weiner J. H., Cecchini G., Retention of heme in axial ligand mutants of succinate-ubiquinone xxidoreductase (complex II) from Escherichia coli. J. Biol. Chem. 276, 18968–18976 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Vibat C. R., Cecchini G., Nakamura K., Kita K., Gennis R. B., Localization of histidine residues responsible for heme axial ligation in cytochrome b556 of complex II (succinate:ubiquinone oxidoreductase) in Escherichia coli. Biochemistry 37, 4148–4159 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Lancaster C. R., et al., Experimental support for the “E pathway hypothesis” of coupled transmembrane e- and H+ transfer in dihemic quinol:fumarate reductase. Proc. Natl. Acad. Sci. U.S.A. 102, 18860–18865 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pethe K., et al., Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 19, 1157–1160 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., et al., The compound TB47 is highly bactericidal against Mycobacterium ulcerans in a Buruli ulcer mouse model. Nat. Commun. 10, 524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam S. M., et al., Extensive characterization of human tear fluid collected using different techniques unravels the presence of novel lipid amphiphiles. J. Lipid Res. 55, 289–298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearce L. L., et al., The resistance of electron-transport chain Fe-S clusters to oxidative damage during the reaction of peroxynitrite with mitochondrial complex II and rat-heart pericardium. Nitric Oxide 20, 135–142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolaj-Robin O., et al., Biochemical and biophysical characterization of succinate: Quinone reductase from Thermus thermophilus. Biochim. Biophys. Acta 1807, 68–79 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Beinert H., Ackrell B. A., Kearney E. B., Singer T. P., Iron-sulfur components of succinate dehydrogenase: Stoichiometry and kinetic behavior in activated preparations. Eur. J. Biochem. 54, 185–194 (1975). [DOI] [PubMed] [Google Scholar]
- 38.Zheng S. Q., et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al., Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems. Annu. Rev. Biophys. 42, 265–287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenthal P. B., Henderson R., Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Schrodinger, The PyMOL Molecular Graphics System (Schrodinger LLC, 2010), Version 1.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this study are available within the figures and the supplementary information. EM maps and atomic coordinates have been deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb) and Protein Data Bank (PDB; http://www.rcsb.org), respectively, under the following ID codes: EMD-30595 and PDB 7D6X for the apo-Sdh1, and EMD-30594 and PDB 7D6V for the Sdh1-UQ1.