Significance
Respiration-coupled ATP production yields most of the energy in eukaryotic cells and relies on the function of mitochondrial oxidative phosphorylation complexes. Biogenesis of these complexes involves both nuclear and mitochondrial genes, which must be coordinately expressed to assure stoichiometric assembly. The regulatory system that provides this coordination remains poorly understood. Here, we report on the function of a nuclear gene product Rmd9, a feasible component of such a system from yeast. We show that Rmd9 is the factor that binds to the dodecamer element present in mitochondrial mRNAs and provide structural insights into Rmd9-RNA interaction. Combined with previous genetic analyses, our results indicate that Rmd9 controls yeast mitochondrial gene expression by stabilizing and enabling the processing of the mitochondrial mRNAs.
Keywords: mitochondria, gene expression, PPR proteins, protein-RNA complex, PAR-CLIP
Abstract
Stabilization of messenger RNA is an important step in posttranscriptional gene regulation. In the nucleus and cytoplasm of eukaryotic cells it is generally achieved by 5′ capping and 3′ polyadenylation, whereas additional mechanisms exist in bacteria and organelles. The mitochondrial mRNAs in the yeast Saccharomyces cerevisiae comprise a dodecamer sequence element that confers RNA stability and 3′-end processing via an unknown mechanism. Here, we isolated the protein that binds the dodecamer and identified it as Rmd9, a factor that is known to stabilize yeast mitochondrial RNA. We show that Rmd9 associates with mRNA around dodecamer elements in vivo and that recombinant Rmd9 specifically binds the element in vitro. The crystal structure of Rmd9 bound to its dodecamer target reveals that Rmd9 belongs to the family of pentatricopeptide (PPR) proteins and uses a previously unobserved mode of specific RNA recognition. Rmd9 protects RNA from degradation by the mitochondrial 3′-exoribonuclease complex mtEXO in vitro, indicating that recognition and binding of the dodecamer element by Rmd9 confers stability to yeast mitochondrial mRNAs.
RNA 3′ polyadenylation is ubiquitous in cells. Polyadenylation can stabilize RNA or aid in its decay, it enables nuclear mRNA export, and facilitates translation (1–7). In organelles, poly(A) tails are generally known to alter the stability of mRNA; however, the mitochondria of the yeast Saccharomyces cerevisiae lack polyadenylation machinery (8, 9). Thus, similar to bacteria and most organelles, yeast mitochondria utilize an alternative mechanism of mRNA stabilization. Specifically, the 3′-untranslated regions (UTRs) of all mitochondrial mRNAs in the yeast contain a conserved sequence that was identified as the processing site of the polycistronic ATP9/tRNAS/VAR1 (10–12) and COX1/ATP8/ATP6 (13, 14) primary transcripts. This sequence, AAUAA(U/C)AUUCUU, termed the “conserved dodecameric motif” (called here “dodecamer”), has been associated with 3′-end processing and stabilization of mitochondrial RNA (15). However, the mechanism by which the dodecamer mediates these functions has remained unclear. It has been postulated that a dodecamer binding protein may associate with mature mRNAs species, thus preventing their exonucleolytic degradation (16). Yet, the identity of this factor is not known.
In this work, we demonstrate that Rmd9 is the factor that targets the dodecameric sequence. We confirm the specificity of this interaction in a defined in vitro system and show that Rmd9 promotes RNA stability in the presence of ribonucleases. In addition, we demonstrate that Rmd9 contacts the 3′ ends of the mitochondrial mRNAs in vivo. Finally, we present a crystal structure of Rmd9 in association with the dodecamer RNA target. Our results clarify the mechanisms by which yeast mitochondrial mRNAs undergo 3′-end maturation and stabilization and provide insights into the structural basis for single-stranded RNA recognition by a fungal PPR protein.
Results
Identification of Rmd9 as the Dodecamer-Binding Factor.
Two decades ago the H. P. Zassenhaus laboratory isolated a dodecamer-binding activity by affinity separation of yeast mitochondrial proteins on a column derivatized with a dodecamer-carrying chimeric DNA/RNA oligonucleotide (17). This activity was associated with a 55-kDa protein, termed DBP (Dodecamer-Binding Protein), which was then further characterized in vitro (17, 18). However, the identity of the protein remained unknown.
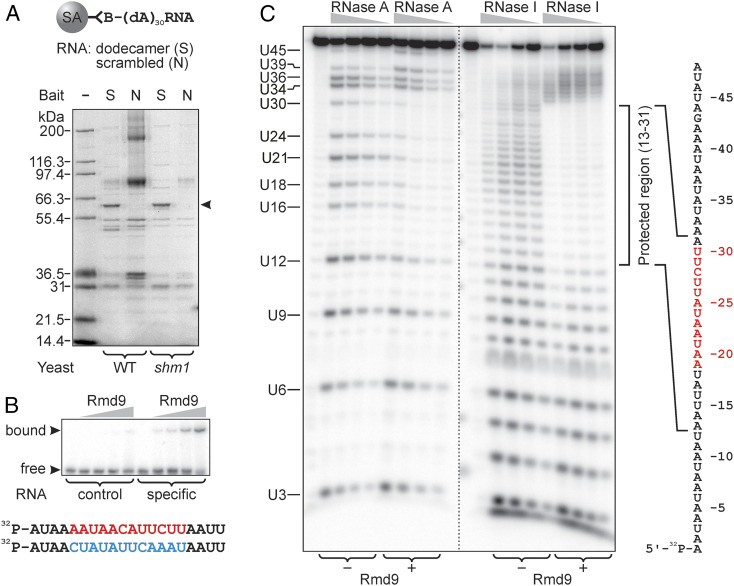
Here, we used RNA-protein affinity pull-down to partially purify and identify the factor that binds the dodecameric signal. We used a similar chimeric bait with the 5′ end attached to biotin to allow immobilization via streptavidin (Fig. 1A). As a control, a nonspecific bait was used, in which the sequence of the dodecamer was scrambled. Streptavidin beads charged with the RNA baits were incubated with lysates of purified yeast mitochondria and the proteins associated with the beads were examined by polyacrylamide gel electrophoresis (PAGE). Compared to the nonspecific control, the pull-down revealed a single unique protein band with an apparent electrophoretic mobility of 62 kDa (Fig. 1A). The protein was identified by peptide mass fingerprinting as Rmd9. Thus, we conclude that Rmd9 is the factor that associates with the dodecamer element in yeast mitochondrial mRNAs. Consistently, Rmd9 was previously implicated in processing and stabilization of mitochondrial transcripts and it was assigned to the family of PPR proteins, the representatives of which are known to function in organellar RNA metabolism (see Discussion for details).
Fig. 1.
Rmd9 specifically associates with the dodecamer element in vitro. (A) A Coomassie-stained polyacrylamide gel showing the proteins pulled from mitochondrial extracts by streptavidin beads carrying either specific (S) or nonspecific (N) bait RNA molecules. The composition of the baits is explained at the Top. The protein in the band pointed to by the arrowhead was identified as Rmd9. The same pull-down experiment was also performed using Δshm1 BY4743 yeast because Shm1 showed high background in several repeats. (B) An EMSA experiment demonstrating selective binding of Rmd9 to an RNA probe that contained the dodecamer (shown in red at the Bottom), which was scrambled in the control probe (blue). (C) A 5′-[32P]-labeled RNA probe (shown on the Right with the dodecamer indicated in red) was subjected to RNase A or RNase I digestion, as indicated. Where specified, Rmd9 was present in the reactions. The digestion products were resolved by denaturing PAGE. The region in the probe where the cleavage efficiency decreased due to the presence of Rmd9 is indicated by the bracket on the Right.
Recombinant Rmd9 Specifically Binds the RNA Dodecamer In Vitro.
To determine if Rmd9 can bind to the dodecamer element independently of other factors present in the lysate, we expressed and purified an N-terminally histidine-tagged form corresponding to the mature protein Δ51-Rmd9 (SI Appendix, Fig. S1). We first performed an electrophoretic mobility shift assay (EMSA), which showed that Rmd9 binds a synthetic RNA probe containing the dodecamer, but not a scrambled sequence (Fig. 1B). Based on this observation and the fact that the specific probe is not expected to form stable secondary structures, we conclude that Rmd9 is a sequence-specific RNA-binding protein that recognizes its target in the single-stranded context without requiring additional factors.
To further confirm that the protein specifically binds the dodecameric element, we formed an Rmd9-RNA complex and subjected it to RNase footprinting. A 5′-[32P]-labeled RNA probe, which represented a dodecamer-containing sequence downstream of the COX2 gene, was subjected to limited endonucleolytic cleavage in the absence or presence of Rmd9. As shown in Fig. 1C, treatment of the probe with RNase A produced a pattern of products resulting from cleavage after pyrimidine residues that are followed by purines, as expected. In the presence of Rmd9, formation of the labeled products due to cleavage in the region from U16 to U30 was greatly diminished, indicating protection of this part of the probe by the protein. The protected region contained the entire sequence of the dodecamer.
We then determined the footprint of Rmd9 with greater precision by utilizing RNase I in the same assay. In the absence of Rmd9, RNase I produced a uniform pattern of products by cleaving the probe at every nucleotide. Protection by Rmd9 almost completely blocked the cleavage of RNA in the region between positions 13 and 31. These results demonstrate that Rmd9 binds directly to the dodecamer element and in addition protects one nucleotide downstream and six nucleotides upstream of the dodecamer as indicated by the schematic in Fig. 1C. The nuclease protection assay was also used to assess the affinity of Rmd9 toward its target, which we found to be in the low nanomolar range (SI Appendix, Fig. S2).
Earlier genetic work has identified a mutation near the end of the SCEI open reading frame (ORF) that disrupted processing of the SCEI mRNA at the dodecamer element (15). The observed phenotype was linked to a 3U>A,4A>U substitution inside the dodecameric sequence. Here we examined the effect of this substitution on Rmd9-RNA binding. We performed an EMSA, in which two RNA probes carrying the wild-type and mutated dodecamer elements were allowed to compete for binding to Rmd9 (SI Appendix, Fig. S3). The experiment showed that the wild-type dodecamer-containing probe could efficiently outcompete the mutant probe. Thus, we concluded that the previously observed loss-of-function phenotype can be correlated to poor binding of the mutant dodecamer to Rmd9.
Rmd9 Is Associated with the Mitochondrial mRNA at the Dodecamer Element In Vivo.
To assess whether Rmd9 contacts RNA in vivo, we applied photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) analysis (19). Cellular RNA was photosensitized by incorporation of 4-thiouracil (4-thioU) and photo cross-linked to the associated proteins. The cross-linked RNA was coimmunoprecipitated with Rmd9 and deep sequenced. The sequencing reads were analyzed for the presence of U>C substitutions, which mark the points of covalent attachment of Rmd9 due to photo cross-linking (20). We found a high frequency of substitutions at U positions just upstream of the dodecameric sequences of the mitochondrial mRNAs (SI Appendix, Fig. S4), indicating that Rmd9 is associated with these regions in vivo. In addition, we detected a number of potential binding sites within highly structured RNA species such as introns, tRNAs, and rRNAs (SI Appendix, Fig. S4A). The latter is consistent with previous reports that found Rmd9 in association with the mitochondrial ribosome (21, 22). The 4-thioU residues situated within the dodecamer element showed less efficient cross-linking, although they are expected to contact the protein. Such variation in photoreactivity of the 4-thioU residues inside a protein complex has been noted before and was attributed to a particular local structural environment that may be suboptimal for the photo reaction (23). Alternatively, the introduction of 4-thioU substitutions within the dodecameric sequence may have inhibited Rmd9 binding, thus causing this effect.
We also examined how the Rmd9 PAR-CLIP sequencing reads were distributed within the 3′-end regions of mRNA species (SI Appendix, Fig. S5). The majority of the reads were concentrated close to the dodecamer element in the AI5β, ORF1, and SCEI mRNAs, as expected. In other mRNAs, unexpectedly, regions of very high abundance of the sequencing reads were observed inside the ORFs and away from the dodecamer element, which did not fit the pattern of distribution of high U>C transition regions (SI Appendix, Fig. S4A). However, the true sites of cross-linking should be associated with the regions of high frequency of U>C substitutions rather than with clusters showing high abundance of the sequencing reads (20).
Rmd9 Protects RNA from Exonucleolytic Cleavage In Vitro.
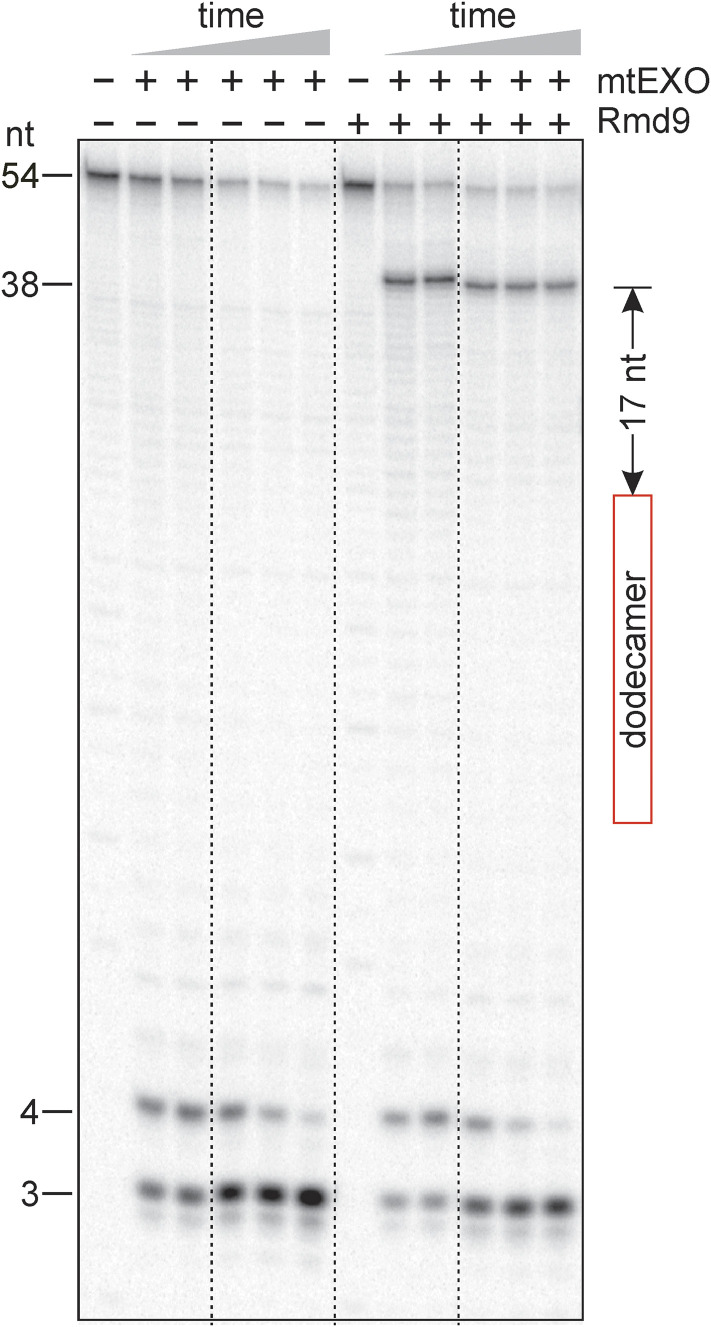
To probe the hypothesis that binding of Rmd9 can stabilize mRNAs, we tested whether Rmd9 can inhibit RNA digestion in the 3′ to 5′ direction. We employed the processive adenosine triphosphate (ATP)-dependent 3′-exoribonuclease mtEXO, which is the primary degradosome in yeast mitochondria (24). As shown in Fig. 2, mtEXO readily degraded a 5′-[32P]-labeled dodecamer-containing RNA probe in the absence of Rmd9. However, when Rmd9 was bound to the probe, the digestion exhibited a prominent pause at position 38 of the RNA. These results indicate that Rmd9 provides a roadblock to mtEXO, thereby protecting RNA from further degradation. The pause occurred 17 nucleotides downstream from the dodecameric sequence, consistent with the observation that the RNA-binding channel of the homologous mtEXO from Candida glabrata can accommodate 16 to 17 RNA nucleotides (25).
Fig. 2.
Rmd9 protects RNA from the action of mtEXO in vitro. A phosphor image showing the products of digestion of a 5′-[32P]-labeled RNA probe by mtEXO. The position of the dodecamer in the RNA is indicated by the red rectangle. The distance between the 3′ nucleotide of the dodecamer and the position of the Rmd9-induced pause by mtEXO is indicated.
Structure of the Rmd9-Dodecamer RNA Complex.
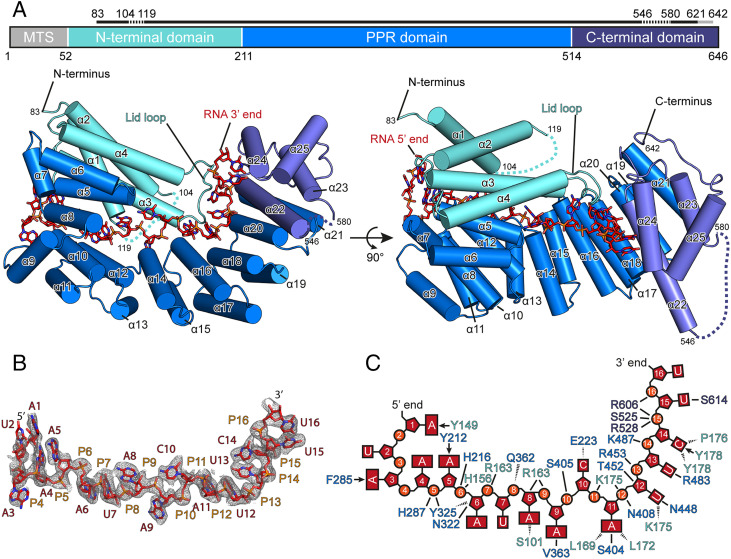
To investigate how Rmd9 recognizes the RNA dodecamer, we cocrystallized Rmd9 with its target RNA (SI Appendix, Fig. S6) and determined the structure of the complex at a resolution of 2.5 Å (Fig. 3 and SI Appendix, Fig. S7). The structure shows that Rmd9 belongs to the PPR protein family, as predicted (26). The central part of Rmd9 (residues 211 to 514, “PPR domain”) consists of eight helix-turn-helix PPR motifs, which form a superhelical structure. The PPR domain is flanked by an N-terminal “lid” domain (residues 80 to 210) that consists of four bundled helices, and a C-terminal domain (residues 514 to 642). The C-terminal domain also comprises four α-helices, of which one (α22) protrudes markedly from the protein body and contains a pronounced kink at the point where its length exceeds that of the PPR helices. This helix marks the boundary with the PPR domain (Fig. 3A). Of the eight structural repeats in the PPR domain, two (labeled PPR3 and PPR7 in SI Appendix, Fig. S7B) conform to the previously observed PPR motifs of plant proteins, featuring a pair of four-turn helices separated by a two-amino acid turn and followed by a five-amino acid unstructured tail (27, 28).
Fig. 3.
Structure of Rmd9 bound to RNA. (A) A cartoon representation of the Rmd9-RNA20 complex with α-helices shown as cylinders and the bound RNA shown as sticks. A schematic of the protein domain architecture is given at the Top. A solid black line above the schematic indicates the regions with interpretable electron density in the crystal structure, with missing regions indicated by a dashed line and regions of weak density by a gray line. The structure is colored according to domains (N-terminal lid domain: aquamarine; PPR domain: marine; C-terminal domain: slate; and RNA: red). Helices are numbered from the N to C terminus. MTS: mitochondrial targeting sequence. (B) Stick representation of the final, refined model of the RNA nucleotides visible in the Rmd9-RNA20 structure. RNA nucleotides are colored in red and blue, and the backbone phosphates are in orange. The unbiased experimental electron density is shown as gray mesh at 1.0σ with 1.7-Å carve. (C) A schematic representation of the RNA in the Rmd9-RNA20 complex. Interactions of the amino acid residues with the RNA are shown. Arrows indicate stacking interactions, solid lines show hydrogen bonding or electrostatic interactions with side chains, and dashed lines show hydrogen bonding to the peptide backbone.
RNA Dodecamer Recognition by Rmd9.
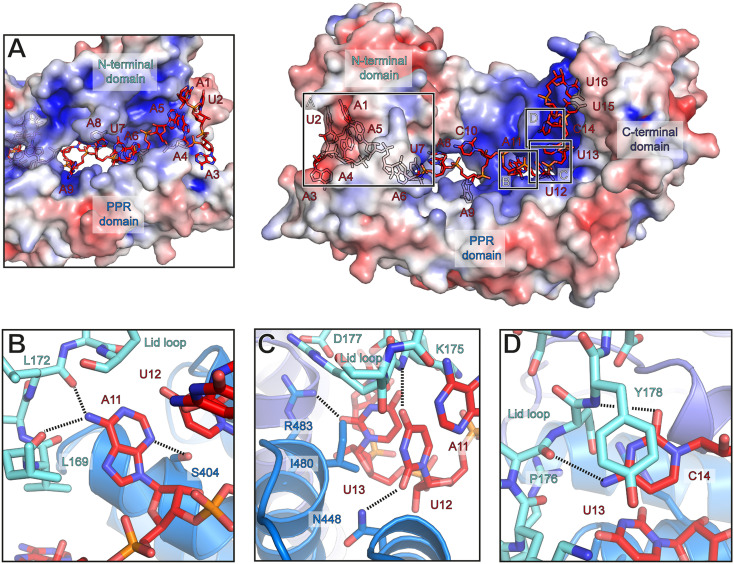
Similar to other PPR proteins, the inner side surface of the superhelical PPR domain of Rmd9 provides a positively charged RNA-binding groove (Fig. 4). The RNA runs along the entire length of the PPR domain with its 5′ end bound to the N-terminal region of the protein and the 3′ end positioned at the C-terminal domain (Figs. 3A and 4). This orientation is similar to that in previously described RNA-PPR protein complexes (27, 29). The observed position of the RNA relative to Rmd9 appears to be specific, as identical binding was found in two complexes containing different RNA molecules, RNA16 and RNA20 (Materials and Methods), and because all RNA bases and backbone moieties are in highly defined positions and orientations. The protein-RNA contacts stabilizing the complex are summarized in Fig. 3C. The central part of the RNA runs along a portion of the PPR domain in a fashion resembling that observed in the plant PPR proteins (Fig. 4).
Fig. 4.
Protein-RNA interactions in the Rmd9-RNA complex. A surface representation of Rmd9 colored according to electrostatic potential (blue: positive; red: negative; +5 kT/e to −5 kT/e). Boxes indicate regions shown as close-up in indents A–D. In indents B–D, protein residues that may interact with the RNA and are discussed in the text are shown as sticks. (A) A view into the RNA-binding tunnel formed by the N-terminal lid domain and the PPR domain. (B) Close-up view of the interactions between base A11 and Rmd9. (C) Close-up view of the interactions of bases U12 and U13 with Rmd9. (D) Close-up view of the interactions between base C14 and Rmd9.
The 5′-end proximal nucleotides are tightly enclosed in a tunnel formed by the PPR domain and the N-terminal lid domain (Fig. 4A). This region does not appear to contribute strongly to the sequence specificity of Rmd9, as the interactions here are primarily backbone interactions. At the entrance to the tunnel, base A5 stacks against Y212 and the trajectory of the following base (A6) is rotated by 180°, with backbone phosphate P6 stabilized by H216 and H156. The primary amine group of base A8 is positioned within 3.2 Å of S101, enabling potential interactions. The trajectory of base A9 is blocked by helix α4 of the lid domain, and this base is flipped by 90° into a hydrophobic pocket (Fig. 4A). Nucleotide position 10 is the only position not strictly conserved in the dodecamer motif (30). This base, which is C in the oligonucleotides RNA20 and RNA16, is flipped to face in the opposite direction as A9 and appears in hydrogen bonding distance to the backbone carbonyl of E223 in motif PPR1 (SI Appendix, Fig. S7B), which may favor C instead of U at this position (Fig. 3C).
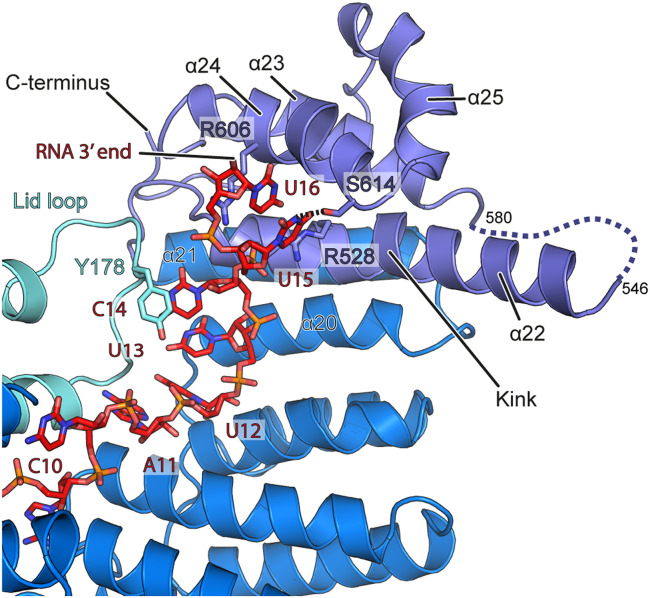
Further toward the 3′ end, the RNA contacts the C-terminal domain and the loop of the lid domain of Rmd9 and appears to be involved in more numerous protein-base contacts. In particular, A11 is positioned in hydrogen bonding distance to S404 in the PPR domain and L169 and L172 in the lid loop (Fig. 4B). U12 may contact N448 in the PPR domain and is wedged apart from the following base U13 by I480 (Fig. 4C). The latter is in close vicinity to R483 and may make additional water-mediated contacts to K175 and D177 in the lid loop. Nucleotide C14 stacks against Y178 of the lid loop and may form hydrogen bonds to its backbone carbonyl or that of P176 (Fig. 4D). The trajectory of the following RNA nucleotides is directed by α22, which protrudes from the protein body. The C-terminal domain of Rmd9 engages primarily in electrostatic interactions with the RNA backbone, with only few interactions mediated by the bases of U15 and U16 (Fig. 5). The structural elements that coordinate the middle and 3′-proximal regions of the RNA (the lid loop, parts of the PPR domain inner surface, and the N-terminal portion of α22) show strong sequence conservation (SI Appendix, Figs. S8 and S9).
Fig. 5.
Rmd9-RNA interactions at the C-terminal domain. Cartoon representation of Rmd9 and stick representation of the RNA. Coloring as in Fig. 3. Nucleotides 11 to 14 run along the PPR domain and nucleotides 15 and 16 face toward helix α24. Helix α22 contains a marked kink and protrudes from the protein body into the solvent. Base U15 may hydrogen bond to S614.
Discussion
Here we show that Rmd9 is the long-sought-after factor that binds to the dodecamer element in the 3′-UTRs of mitochondrial mRNAs in yeast. We present the structural basis of Rmd9-RNA recognition and provide evidence that Rmd9 can impair RNA exonucleolytic cleavage by the major mitochondrial 3′ exonuclease.
Previous bioinformatic analysis had placed Rmd9 into the PPR protein family (26). PPR proteins are primarily involved in the metabolism of organellar RNA and are especially numerous in plants (31, 32). Each of these proteins contains a series of degenerately conserved sequence motifs that are typically 35 amino acids in length. These PPR motifs are folded into helix-turn-helix structures, which in turn assemble into superhelices when present as head-to-tail arrays within the protein (27, 28, 33, 34). Many PPR proteins can recognize sequences in single-stranded RNA. The recognition by plant PPR proteins is thought to occur in a modular fashion, with one PPR motif recognizing a single RNA base. Certain combinations of the amino acids present at positions 5 and 35 of each motif determine the base specificity in accordance with a set of recognition rules termed the “PPR code” (27, 35–37).
While far fewer in number, proteins that fit the “plant” PPR consensus have also been identified in organisms other than plants. In Saccharomyces cerevisiae, the mitochondrial factors Aep3, Dmr1, and Pet309 are the only three plant-like PPR proteins (33). Yet, profile hidden Markov model analyses (26) revealed 13 additional potential yeast PPR proteins, including Rmd9. This group defined the yeast-specific signature of the PPR motifs, which differs from that in plants (38). Due to the lack of structural data, it remained unknown whether the fungal PPR proteins have the same structural organization as those found in plants. In particular it was unclear whether they were capable of modular RNA base recognition, or if the plant PPR code extended into nonplant eukaryotes.
Our high-resolution structure shows that Rmd9 is indeed a PPR protein. However, its RNA interactions differ from those observed in plant PPR proteins. In particular, none of the PPR motifs observed in the structure are engaged in typical plant PPR-like interactions with the RNA. Still, Rmd9 contains two canonical structural PPR motifs, marked PPR3 and PPR7 in SI Appendix, Fig. S7B. PPR3 (S in position 5 and N in position 35) would be expected to recognize an adenine and PPR7 (N in position 5 and T in position 35) has undefined specificity per the plant PPR code. PPR motifs 2, 4, 5, and 8 are not canonical and show small variations in the length of their helices, turns, and/or tails. However, such minor structural discrepancies are also seen in some plant PPR motifs (27, 28). Two repeats in Rmd9 appear to be unusual: PPR1 is followed more closely by helix α7 and PPR6 has helices that are one turn longer compared to canonical plant PPR motifs. Obviously, the eight PPR motifs of Rmd9 are not enough to enable modular recognition of all 12 bases of the dodecameric sequence.
Given the high specificity and affinity for the dodecamer sequence we observed in our biochemical assays, surprisingly few base-specific contacts are apparent in the Rmd9-RNA structure. Instead of binding in a strictly modular one base per one PPR motif fashion, nucleotides 5 to 9 are enclosed by the tunnel formed between the PPR domain and the N-terminal lid, suggesting that the latter may be able to open and close to allow initial binding of the RNA. Nucleotides 11 through 13 run along PPR motifs 6 to 8 in a fashion reminiscent of that observed in plant PPR proteins, with A11, U12, and U13 recognized by residues in position 5 of the repeats. However, they are situated too far to interact with the amino acids at positions 35 in the respective motifs. Instead, the lid loop of the N-terminal domain, which connects helix α3 to α4 and extends to be in proximity to the 3′ end of the RNA, forms extensive interactions with each of these bases. It is not obvious from the structure how this RNA binding is initially achieved, and we therefore speculate that the lid may be in an “open” position prior to the RNA binding at the N-terminal tunnel region. It is thus possible that the “closed lid” configuration captured in the structure corresponds to a “postrecognition” state of the complex.
Our analysis showed that swapping nucleotides U3 and A4 in the dodecamer significantly inhibits its binding to Rmd9 (SI Appendix, Fig. S3). In the Rmd9-RNA structure, these two nucleotides correspond to RNA positions U7 and A8, which are located in a cleft formed by the lid and PPR domains. While it is not readily apparent from the structure why the mutation should cause the observed drop in binding affinity, it may partially be due to a loss of the S101 contact with A8 and possible steric clashes that could occur upon substituting U7 with the bulkier adenine.
Results of previous genetic analyses are consistent with Rmd9 being the factor that acts through the dodecamer element and stabilizes mRNA. Rmd9 is essential for respiration and stability of the mitochondrial genome (21, 39). It is also essential for the efficient production of at least three gene products, Cox2, Cytb, and Atp6, which are all subunits of the oxidative phosphorylation system (39). Deletion of the RMD9 gene leads to a global depletion of all but one mature mitochondrial mRNA species, consistent with its proposed role in mRNA stabilization (39). Interestingly, the small subunit rRNA (15S) was also considerably destabilized in the absence of Rmd9; however, it is unclear whether this effect was direct or indirect because the 15S rRNA neither contains nor is in the vicinity of a canonical dodecamer.
Rmd9 was implicated in RNA maturation. The processing of two polycistronic precursors, COX1/ATP8/ATP6 and ATP9/tRNAS/VAR1, was disrupted in a RMD9 deletion mutant (39). Combined with our data, this suggests that the dodecamer-dependent RNA processing may rely on an endonuclease that Rmd9 could recruit to the site of processing.
Rmd9 has also been proposed to play a role in mRNA recruitment to specific sites where mitochondrially encoded subunits of the oxidative phosphorylation complexes are translated and simultaneously inserted into the membrane (21, 39). This suggestion appears to be plausible, as Rmd9 demonstrated a tendency to weakly associate with the monosomal fraction of the mitoribosome (21) and also because the protein was found to be a part of the mitochondrial organization of gene expression (MIOREX) complex (40).
Together with the results of previous genetic analyses, our data establish Rmd9 as a central player in maintaining the steady-state levels of the mitochondrial mRNAs that encode the components of the yeast oxidative phosphorylation system. By forming tight, nanomolar-affinity complexes with the dodecamer elements in the 3′-UTRs, Rmd9 promotes maturation of the corresponding primary transcripts, possibly by recruiting a yet unknown processing endonuclease, and also protects the mRNAs from 3′-exonucleolytic degradation, particularly by mtEXO. The exoribonuclease-blocking function of Rmd9 described here is paralleled by a group of proteins (including PPR proteins) that bind specifically to certain RNA species in chloroplasts (32, 41). However, a distinctive feature of the yeast mitochondrial system is that this function appears to be carried out by a single protein that relies on a target sequence common in all mRNAs.
Materials and Methods
Oligonucleotides.
Sequences of the DNA and RNA oligonucleotides (Integrated DNA Technology and GE Healthcare Dharmacon) used in the study are listed in SI Appendix, Table S1.
RNA Affinity Pull-Down of the Dodecamer-Binding Protein.
The chimeric DNA/RNA bait used in the pull-down (RNA-bait-DD) was based on the design previously described by Li and Zassenhaus (17). It carried the dodecameric sequence within a 20-nucleotide RNA segment, which was attached at the 5′ end to a (dA)30 spacer. Here, we added a biotin moiety to the 5′ end of the construct in order to utilize an immobilized avidin stationary phase in place of the previously used poly(dT)-cellulose support (17). To prepare the affinity beads, 10 μL of Streptavidin Plus UltraLink resin (Pierce) was equilibrated with phosphate-buffered saline (PBS) buffer (100 mM Na-phosphate, pH 7.2, 150 mM NaCl) and incubated with 1 nmol of the dodecamer-containing bait (RNA-bait-DD; see SI Appendix, Table S1) for 2 min at room temperature. The beads were then washed with PBS and stored at 4 °C. Control beads were prepared in the same way by loading the resin with the nonspecific bait (RNA-bait-NS), in which the dodecameric sequence was scrambled. Mitochondria were isolated from the BY4743 S. cerevisiae strain by ultracentrifugation in a discontinuous sucrose density gradient as previously described (42). Settled mitochondria (3 mL) were resuspended in 15 mL of 20 mM Hepes·KOH (pH 7.4), 250 mM sucrose, and pelleted by centrifugation (12,000 × g, 10 min, 4 °C). The pellet was resuspended in 27 mL of lysis buffer (20 mM Hepes·KOH, pH 7.4, 250 mM sucrose, 250 mM KCl, 10 mM ethylenediaminetetraacetic acid [EDTA], 10% glycerol, 2 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with Complete Mini EDTA-free protease inhibitor mixture (Roche). Triton X-100 was mixed in dropwise to a final concentration of 0.5% and the mixture was left on ice for 15 min. The lysate was treated on a Sonic Dismembrator 500 (Fisher Scientific) using 30-s pulses intermitted with 30-s pauses over a total time of 5 min at 4 °C and then clarified by centrifugation (20,000 × g, 1 h, 4 °C). KCl was added to the supernatant to a final concentration of 1 M and the resulting mixture was dialyzed against 2 L of binding buffer (10 mM Hepes·KOH, pH 7.4, 100 mM KCl; 1 mM EDTA, 5% glycerol, 2 mM DTT, 0.1 mM PMSF) overnight. The lysate (47 mL total volume containing ∼2 mg/mL of protein according to a Bradford assay) was then cleared by centrifugation (20,000 × g, 1 h, 4 °C), distributed in 1.5-mL aliquots and kept frozen at −20 °C until used. Streptavidin beads carrying RNA-bait-DD (10 μL) were combined with 3 mL of the lysate and slowly rotated for 1 h at 4 °C. The control beads were incubated with the lysate in the same manner. After the incubation, the lysate was removed, the beads were washed with binding buffer (5 × 1.5 mL), and heated at 95 °C in 30 μL of 1× NuPAGE LDS sample buffer (Invitrogen) for 2 min. The eluted proteins were separated by electrophoresis in 4 to 12% Bis-Tris NuPAGE gels run in 3-morpholinopropane-1-sulfonate (MOPS) buffer (Invitrogen) and stained with Coomassie Blue R-250 (Bio-Rad). The bands of interest were cut out of the gel and submitted for liquid chromatography-tandem mass spectrometry protein identification.
PAR-CLIP Analysis.
The PAR-CLIP experiment and data preprocessing were performed as previously described (43) using yeast expressing C-terminally TAP-tagged Rmd9 (SI Appendix, Fig. S4). Further, P values for true cross-linking sites were calculated using a previously described statistical model (44). For the analysis shown in SI Appendix, Fig. S4A we selected high-rate U>C transition sites by considering genomic positions with P value <0.001 and read coverage ≥250. At the selected sites we show the fraction of reads with U>C substitutions.
Preparation of Plasmids for Expression of Recombinant Proteins.
Cloning procedures and construction of plasmids for the expression of Rmd9 and mtEXO in Escherichia coli are described in SI Appendix.
Expression and Purification of Recombinant Rmd9.
Plasmid pMA27O2THP was transformed into E. coli BL21-CodonPlus (DE3)-RIPL cells (Agilent). The cells were grown at 37 °C in 6 L Luria–Bertani (LB) medium supplemented with 0.1 mg/mL ampicillin and 0.8% glucose to an optical density (OD)600 of 0.6 AU and the culture was then cooled to 16 °C. Isopropyl β-D-1-thiogalactopyranoside (IPTG, 0.2 mM) was added and the cells were grown for an additional 12 h at 16 °C. The cells were collected by centrifugation, resuspended in 200 mL of lysis buffer (40 mM Tris·HCl, pH 7.9, 0.3 M NaCl, 15 mM imidazole, 5% glycerol, 5 mM β-mercaptoethanol), and supplemented with 1 mM PMSF, 50 μM benzamidine, 1 μM pepstatin, and 10 μM leupeptin. The suspension was placed on ice and treated with ultrasound on a Sonic Dismembrator 500 (Fisher Scientific) using 30-s pulses intermitted with 30-s pauses for a total pulse time of 20 min. The lysate was cleared by centrifugation (37,000 × g, 4 °C, 30 min) and the supernatant was filtered through a column packed with 1 mL of Ni-IDA agarose beads (Gold Biotechnology). The beads were washed by passing through the column 15 mL of the lysis buffer supplemented with NaCl to 1.5 M and followed by 15 mL of the lysis buffer. The protein was eluted with 5 mL of the lysis buffer supplemented with imidazole to 0.2 M and loaded on a 5 mL HiTrap Heparin HP column (GE Healthcare). The column was eluted by a linear salt concentration gradient (0.15 to 0.8 M NaCl) in 15 mM Tris·HCl (pH 7.9), 5% glycerol, and 5 mM β-mercaptoethanol. The fractions containing purified Rmd9 were combined, concentrated on an Ultracel-50K centrifugal filter (Merck Millipore), flash-frozen in small aliquots with liquid N2, and stored at −80 °C.
Electrophoretic Mobility Shift Assay.
Rmd9 (67.5, 125, 250, or 500 nM) was combined with 5′-[32P]-labeled probe RNA20 or RNA20c (200 nM) and yeast tRNA (800 nM) in 10 μL of 20 mM Tris·HCl (pH 7.9), 25 mM NaCl, 0.1 mM EDTA, and 5 mM MgCl2. The mixtures were incubated for 10 min at room temperature and supplemented with 2 μL of gel loading solution (50% glycerol spiked with bromophenol blue). Control mixtures were set up in the same way except Rmd9 was not added. The mixtures were loaded on a 10% native polyacrylamide gel (37.5:1, acrylamide:bis-acrylamide) cast in 1× Tris-borate-EDTA (TBE) buffer and containing 2% glycerol and 0.02% Noniodet P-40 (Sigma), and the gel was run in 0.5× TBE at 4 °C (200 V, 30 min). The radioactive bands in the gel were visualized with a Typhoon 9200 PhosphorImager (GE Healthcare).
RNase Footprinting.
The probe RNA48 was 5′-[32P]-labeled and gel purified as described (43). The probe (50 nM) was incubated with Rmd9 (100 nM) and yeast tRNA (800 nM) for 20 min at 30 °C in 10 μL of 50 mM Tris·HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT. RNase A (Sigma-Aldrich) or RNase I (New England Biolabs) was added and the reaction was incubated for 15 min at 30 °C. Multiple reactions were performed to vary the concentration of RNase A (0.08, 0.04, 0.02, and 0.01 pg/µL) and RNase I (0.02, 0.01, 0.005, and 0.0025 U/µL). An additional set of similar reactions was carried out, in which Rmd9 was omitted. After the incubation, the reactions were mixed with preheated 2× gel loading solution (90% formamide and 50 mM EDTA spiked with bromophenol blue and xylene cyanol FF) and treated for 2 min at 95 °C. The reaction products were resolved by denaturing 20% PAGE (7 M urea, 19:1, acrylamide:bisacrylamide) and visualized by phosphor imaging on a Typhoon 9200 scanner (GE Healthcare).
Expression and Purification of Recombinant mtEXO.
To prepare mtEXO, we took the approach previously described by Malecki et al. (45). E. coli BL21-CodonPlus (DE3)-RIPL cells (Agilent) were transformed with plasmids pMA31 or pMA35 expressing Dss1 and Suv3, respectively (SI Appendix). Each culture was grown at 37 °C in 6 L LB supplemented with 0.8% glucose and 0.1 mg/mL ampicillin to an OD600 between 0.6 and 0.8 AU. The cultures were then rapidly cooled to 16 °C, IPTG (0.2 mM) was added, and the cells were incubated at 16 °C for an additional 12 h. The cells were harvested and mixed so that the cells producing Dss1 were in a threefold excess by weight. The cells were lysed and mtEXO was purified by Ni-affinity chromatography as described (45). The complex was loaded on a 5-mL HiTrap Heparin HP column (GE Healthcare) and eluted by a linear salt concentration gradient (0.2 to 1.2 M NaCl) in 20 mM Hepes·KOH (pH 7.0), 5% glycerol, and 5 mM β-mercaptoethanol. The fractions containing mtEXO were combined, concentrated on an Ultracel-50K centrifugal filter (Merck Millipore), and loaded on a Superdex 200 10/300 GL column (GE Healthcare). The column was eluted with 20 mM Hepes·KOH (pH 7.0), 0.2 M NaCl, 5% glycerol, 5 mM β-mercaptoethanol, the purified mtEXO was concentrated, distributed into small aliquots, flash-frozen in liquid N2, and stored at −80 °C.
Assaying 3′-Exonucleolytic Cleavage by mtEXO.
Rmd9 (100 nM) was incubated with 5′-[32P]-labeled probe RNA54 (50 nM) and yeast tRNA (800 nM) for 20 min at 30 °C in 10 μL of 10 mM Tris·HCl, pH 7.9, 25 mM KCl, 10 mM MgCl2, 1 mM DTT, and 1 mM ATP. The formed Rmd9-RNA complex was treated with mtEXO (200 nM) for 0.5, 1, 4, 8, and 16 min. Control digestions were performed in the same way except that Rmd9 was not present. The products of the digestions were separated in a 20% polyacrylamide gel (7 M urea, 19:1, acrylamide:bisacrylamide) and visualized by phosphor imaging.
X-Ray Data Collection and Structure Determination.
The conditions used to prepare Rmd9-RNA crystals (SI Appendix, Fig. S6) are described in SI Appendix. Diffraction data for the native Rmd9-RNA20 crystals were collected at beamline X06SA at the Swiss Light Source synchrotron using an EIGER16M detector. The crystals belonged to space group P3121 and diffracted to 2.55-Å resolution (SI Appendix, Table S2). Initial phases were determined by single-wavelength anomalous dispersion (SAD) using the native sulfur and phosphorous atoms occurring in protein and RNA as anomalous scatterers (SI Appendix, Fig. S7A). For this, two isomorphous crystals were chosen and diffraction data were collected at beamline X06DA at the Swiss Light Source synchrotron, which is equipped with a multiaxis PRIGo goniometer. A total of 27 full-rotation low-dose datasets were collected at varying chi-angles and different positions of the rod-shaped crystals and merged with XDS, resulting in ∼277-fold redundancy (SI Appendix, Table S2). The anomalous substructure was determined using the shelx suite with the hkl2map interface (46, 47). Shelxc reported anomalous signal to a resolution of ∼3.8 Å, and the substructure search using shelxd identified 36 potential heavy atom sites, a fraction of which were arranged in a periodical fashion resembling a phosphate backbone. Phasing and density modification was performed using phenix.autosol (48) with the manually edited heavy atom sites file from shelxd as input, from which we removed low-occupancy and unlikely sites. The resulting experimental electron density was of high quality and, together with the heavy atom locations and anomalous difference map, allowed for manual building of both the protein and RNA using Coot (49). Following this, the initial model was refined using phenix.refine (48) against the higher-resolution native dataset, followed by iterative cycles of manual rebuilding, refinement, and placement of solvent molecules. As the native dataset also appeared to contain some anomalous scattering signal, Friedel pairs were kept separate during refinement. The resulting anomalous difference map showed peaks close to some cysteine residues (Cys192 and Cys315), which coincided with positive difference density peaks around the sulfhydryl groups of these residues. It has been reported previously that the sulfhydryl groups of cysteine residues can form covalent adducts with dimethylarsinic (cacodylic) acid in the presence of reducing agents (50–53). As our crystallization conditions contained both cacodylate and reducing agent, we modeled these residues as S-(dimethylarsenic)-cysteine. The final model of Rmd9-RNA20 comprises residues 80 to 104, 119 to 546, and 580 to 642 of Rmd9 as well as 16 nucleotides of continuous RNA and was refined to a free R-factor of 23.47% with excellent stereochemistry (SI Appendix, Table S2). The structure of Rmd9 in complex with the RNA16 was solved by molecular replacement using the refined structure of Rmd9-RNA20 lacking the RNA as search model. Following initial refinement, the RNA was built into the clear difference density and the structure was iteratively rebuilt and refined to a final free R-factor of 22.95% with excellent stereochemistry (SI Appendix, Table S2). In the Rmd9-RNA16 complex, the 5′-most nucleotide (A3) is not visible, most likely because in the RNA20 complex it is stabilized by A1, which is lacking in the RNA16 construct. Analysis of an anomalous difference map calculated using phases derived from the final model revealed that in the Rmd9-RNA16 crystals, an additional cysteine residue (Cys93) is converted to S-(dimethylarsenic)-cysteine. The protein portions in the Rmd9-RNA20 and Rmd9-RNA16 structures are essentially identical, with a rmsd of 0.172 Å over all atoms.
To analyze the conservation of structural elements that interact with the RNA, we generated a manually curated sequence alignment of eight Rmd9 homologs using ClustalΩ within the MPI bioinformatics toolkit (54). The eight yeast species included in the alignment showed the presence of sets of dodecamer elements in their mitochondrial RNA. Conservation scores and alignment visualizations were calculated with ConSurf (55).
Supplementary Material
Acknowledgments
We thank William T. McAllister for helpful comments on the manuscript. Parts of this work were performed at Beamline X06SA and X06DA at the Swiss Light Source at the Paul-Scherrer-Institut, Villigen, Switzerland. We thank the staff at the Swiss Light Source for their support, in particular Vincent Olieric and Justyna Woydyla for help with anomalous data collection. This work was supported by New Jersey Health Foundation (Grant PC127-13 to M.A.) and the NIH (Grant R35 GM131832 to D.T.). H.S.H. was supported by the Deutsche Forschungsgemeinschaft (Grants FOR2848, SFB1190, EXC 2067/1 39072994). P.C. was supported by the Deutsche Forschungsgemeinschaft (Grants EXC 2067/1 39072994, SFB860, SPP2191) and the ERC Advanced Investigator Grant CHROMATRANS (Grant no. 882357). We also acknowledge the support from the Rowan University Graduate School of Biomedical Studies.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009329118/-/DCSupplemental.
Data Availability
The PAR-CLIP RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE139514). Atomic coordinates and structure factors of Rmd9-RNA16 and Rmd9-RNA20 complexes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 7A9X and 7A9W, respectively).
References
- 1.Wickens M., Anderson P., Jackson R. J., Life and death in the cytoplasm: Messages from the 3′ end. Curr. Opin. Genet. Dev. 7, 220–232 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Sachs A. B., Sarnow P., Hentze M. W., Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell 89, 831–838 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Gagliardi D., Dziembowski A., 5′ and 3′ modifications controlling RNA degradation: from safeguards to executioners. Philos. Trans. R. Soc. London B Biol. Sci. 373, 20180160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proudfoot N. J., Ending the message: Poly(A) signals then and now. Genes Dev. 25, 1770–1782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart M., Polyadenylation and nuclear export of mRNAs. J. Biol. Chem. 294, 2977–2987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slomovic S., Portnoy V., Liveanu V., Schuster G., RNA polyadenylation in prokaryotes and organelles; Different tails tell different tales. Crit. Rev. Plant Sci. 25, 65–77 (2006). [Google Scholar]
- 7.Hajnsdorf E., Kaberdin V. R., RNA polyadenylation and its consequences in prokaryotes. Philos. Trans. R. Soc. London B Biol. Sci. 373, 20180166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster G., Stern D., RNA polyadenylation and decay in mitochondria and chloroplasts. Prog. Mol. Biol. Transl. Sci. 85, 393–422 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Chang J. H., Tong L., Mitochondrial poly(A) polymerase and polyadenylation. Biochim. Biophys. Acta 1819, 992–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thalenfeld B. E., Bonitz S. G., Nobrega F. G., Macino G., Tzagoloff A., oli1 Transcripts in wild type and in a cytoplasmic “petite” mutant of yeast. J. Biol. Chem. 258, 14065–14068 (1983). [PubMed] [Google Scholar]
- 11.Zassenhaus H. P., Martin N. C., Butow R. A., Origins of transcripts of the yeast mitochondrial var 1 gene. J. Biol. Chem. 259, 6019–6027 (1984). [PubMed] [Google Scholar]
- 12.Hofmann T. J., Min J., Zassenhaus H. P., Formation of the 3′ end of yeast mitochondrial mRNAs occurs by site-specific cleavage two bases downstream of a conserved dodecamer sequence. Yeast 9, 1319–1330 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Osinga K. A., De Vries E., Van der Horst G., Tabak H. F., Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 3, 829–834 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon M., Faye G., Organization and processing of the mitochondrial oxi3/oli2 multigenic transcript in yeast. Mol. Gen. Genet. 196, 266–274 (1984). [DOI] [PubMed] [Google Scholar]
- 15.Butow R. A., Zhu H., Perlman P., Conrad-Webb H., The role of a conserved dodecamer sequence in yeast mitochondrial gene expression. Genome 31, 757–760 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Min J., Zassenhaus H. P., Identification of a protein complex that binds to a dodecamer sequence found at the 3′ ends of yeast mitochondrial mRNAs. Mol. Cell. Biol. 13, 4167–4173 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Zassenhaus H. P., Purification and characterization of an RNA dodecamer sequence binding protein from mitochondria of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 261, 740–745 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Li H., Zassenhaus H. P., Phosphorylation is required for high-affinity binding of DBP, a yeast mitochondrial site-specific RNA binding protein. Curr. Genet. 37, 356–363 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Schulz D., et al., Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155, 1075–1087 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Hafner M., et al., Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams E. H., Butler C. A., Bonnefoy N., Fox T. D., Translation initiation in Saccharomyces cerevisiae mitochondria: Functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics 175, 1117–1126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A. P., et al., Molecular connectivity of mitochondrial gene expression and OXPHOS biogenesis. Mol. Cell 79, 1051–1065.e10 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ascano M., Hafner M., Cekan P., Gerstberger S., Tuschl T., Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev. RNA 3, 159–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szczesny R. J., et al., RNA degradation in yeast and human mitochondria. Biochim. Biophys. Acta 1819, 1027–1034 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Razew M., et al., Structural analysis of mtEXO mitochondrial RNA degradosome reveals tight coupling of nuclease and helicase components. Nat. Commun. 9, 97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipinski K. A., Puchta O., Surendranath V., Kudla M., Golik P., Revisiting the yeast PPR proteins—application of an Iterative Hidden Markov Model algorithm reveals new members of the rapidly evolving family. Mol. Biol. Evol. 28, 2935–2948 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Yin P., et al., Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Ban T., et al., Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J. Biol. Chem. 288, 31540–31548 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C., et al., Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7, 11285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turk E. M., Das V., Seibert R. D., Andrulis E. D., The mitochondrial RNA landscape of Saccharomyces cerevisiae. PLoS One 8, e78105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz-Linneweber C., Small I., Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13, 663–670 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Barkan A., Small I., Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Small I. D., Peeters N., The PPR motif–A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Manna S., An overview of pentatricopeptide repeat proteins and their applications. Biochimie 113, 93–99 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Barkan A., et al., A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke J., et al., Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 20, 1377–1382 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Yan J., et al., Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 47, 3728–3738 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbert C. J., Golik P., Bonnefoy N., Yeast PPR proteins, watchdogs of mitochondrial gene expression. RNA Biol. 10, 1477–1494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nouet C., et al., Rmd9p controls the processing/stability of mitochondrial mRNAs and its overexpression compensates for a partial deficiency of oxa1p in Saccharomyces cerevisiae. Genetics 175, 1105–1115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kehrein K., et al., Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 10, 843–853 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Manavski N., Schmid L.-M., Meurer J., RNA-stabilization factors in chloroplasts of vascular plants. Essays Biochem. 62, 51–64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markov D. A., et al., Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast 26, 423–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones J. L., et al., Yeast mitochondrial protein Pet111p binds directly to two distinct targets in COX2 mRNA, suggesting a mechanism of translational activation. J. Biol. Chem. 294, 7528–7536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baejen C., et al., Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell 55, 745–757 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Malecki M., Jedrzejczak R., Stepien P. P., Golik P., In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J. Mol. Biol. 372, 23–36 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick G. M., A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Pape T., Schneider T. R., HKL2MAP: A graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Cryst. 37, 843–844 (2004). [Google Scholar]
- 48.Adams P. D., et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maignan S., Guilloteau J. P., Zhou-Liu Q., Clément-Mella C., Mikol V., Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: High level of similarity of the active site with other viral integrases. J. Mol. Biol. 282, 359–368 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Greenwald J., Le V., Butler S. L., Bushman F. D., Choe S., The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry 38, 8892–8898 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Brooks M. A., Ravelli R. B., McCarthy A. A., Strub K., Cusack S., Structure of SRP14 from the Schizosaccharomyces pombe signal recognition particle. Acta Crystallogr. D Biol. Crystallogr. 65, 421–433 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Liu X., Zhang H., Wang X. J., Li L. F., Su X. D., Get phases from arsenic anomalous scattering: De novo SAD phasing of two protein structures crystallized in cacodylate buffer. PLoS One 6, e24227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmermann L., et al., A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Ashkenazy H., et al., ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PAR-CLIP RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE139514). Atomic coordinates and structure factors of Rmd9-RNA16 and Rmd9-RNA20 complexes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 7A9X and 7A9W, respectively).