Significance
Studies on bacterial and eukaryotic viruses have revealed a range of strategies used by viruses to subdue host cells for efficient virus replication. How archaeal viruses interact with their hosts remains largely unknown. We characterize a new strategy employed by a nonlytic archaeal virus STSV2 to transform its host into a giant virion-producing factory, whereby the virus infection blocks normal cell division by binary fission, leading to gradual cell growth to unprecedented sizes. The giant infected cells divide asymmetrically by budding, replenishing the pool of susceptible hosts. Thus, although tinkering with the cell cycle is a common practice among evolutionarily unrelated viruses from different domains of life, the mechanisms and manifestation of these actions can be highly diverse and unexpected.
Keywords: archaea, archaeal viruses, asymmetric cell division, ESCRT system, Saccharolobus
Abstract
Archaeal viruses represent one of the most mysterious parts of the global virosphere, with many virus groups sharing no evolutionary relationship to viruses of bacteria or eukaryotes. How these viruses interact with their hosts remains largely unexplored. Here we show that nonlytic lemon-shaped virus STSV2 interferes with the cell cycle control of its host, hyperthermophilic and acidophilic archaeon Sulfolobus islandicus, arresting the cell cycle in the S phase. STSV2 infection leads to transcriptional repression of the cell division machinery, which is homologous to the eukaryotic endosomal sorting complexes required for transport (ESCRT) system. The infected cells grow up to 20-fold larger in size, have 8,000-fold larger volume compared to noninfected cells, and accumulate massive amounts of viral and cellular DNA. Whereas noninfected Sulfolobus cells divide symmetrically by binary fission, the STSV2-infected cells undergo asymmetric division, whereby giant cells release normal-sized cells by budding, resembling the division of budding yeast. Reinfection of the normal-sized cells produces a new generation of giant cells. If the CRISPR-Cas system is present, the giant cells acquire virus-derived spacers and terminate the virus spread, whereas in its absence, the cycle continues, suggesting that CRISPR-Cas is the primary defense system in Sulfolobus against STSV2. Collectively, our results show how an archaeal virus manipulates the cell cycle, transforming the cell into a giant virion-producing factory.
Viruses and cells have likely coexisted since the emergence of the first living organisms (1). In this context, viruses have evolved a spectrum of infection strategies, with some eliciting almost no detectable impact on the physiology of the cell and others extensively reprogramming the host metabolism for maximal progeny production (2–4). Many eukaryotic viruses have been shown to be master manipulators of the cell cycle, subverting it to their advantage by tinkering with specific steps of the cycle (5, 6). For instance, some viruses induce a G1-to-S phase transition in order to replicate their genomes concomitantly with the synthesis of cellular chromosomes, whereas others arrest the progression from the G2 phase, a period of rapid cell growth and protein synthesis, to the M phase during which cells divide (5). Occasionally, virus-mediated deregulation of the cell cycle has dramatic consequences, including development of certain types of cancer (7). Whether viruses of prokaryotes, bacteria, and archaea, which represent the dominant part of the global virosphere (8–11), also actively manipulate the cell cycle of their hosts remains largely unknown. Although some bacteriophages have been shown to block cell division (12–14), the reproductive benefits of this action are not always apparent.
In archaea, the cell cycle has been most extensively investigated in hyperthermophiles of the genus Sulfolobus (phylum Crenarchaeota), which grow optimally at ∼80 °C and pH 3. Similar to eukaryotes, an exponentially growing Sulfolobus cell goes through 1) a prereplicative growth period called the G1 phase; 2) the chromosome replication stage, the S phase; 3) a second period of cellular growth, the G2 phase; and 4) rapid genome segregation and cell division periods, known as the M and D phases, respectively (15). Cell division in Sulfolobus is mediated by the eukaryotic-like ESCRT (endosomal sorting complexes required for transport) machinery, which consists of protein CdvA, four ESCRT-III proteins—ESCRT-III (CdvB), ESCRT-III-1 (CdvB1), ESCRT-III-2 (CdvB2), and ESCRT-III-3 (CdvB3)—and the AAA+ ATPase Vps4 (16–19). The ESCRT-III proteins and Vps4 are homologous to the eukaryotic counterparts, whereas CdvA is specific to archaea.
One of the remarkable features of hyperthermophilic archaea is the diversity of their viruses, most of which do not show structural or genomic relatedness to viruses of bacteria or eukaryotes (20–22). Most of the genes in these virus genomes encode unique proteins of unknown function (23). However, recent studies have uncovered that some of these genes encode diverse anti-CRISPR proteins (24, 25), which allow viruses to subvert the CRISPR-Cas systems, the primary antiviral defense mechanism in archaea (26, 27). Unlike most bacteriophages but similar to viruses of eukaryotes, many archaeal viruses are nonlytic and can be continuously released from the infected cells (21). However, how such viruses transform their hosts into virion-producing factories—sometimes referred to as the virocells (3)—and how virus replication is coordinated with the cell cycle and antiviral defense mechanisms remains largely unknown. Notably, transcriptomic studies have shown that upon infection with certain archaeal viruses, genes encoding ESCRT proteins can be either up-regulated or down-regulated (28, 29), suggesting the existence of an interplay between virus infection and cell cycle in archaea. In the case of lytic Sulfolobus turreted icosahedral virus, overexpression of escrt genes was linked to virion assembly (30), whereas down-regulation of the escrt genes during nonlytic Sulfolobus tengchongensis spindle-shaped virus 2 (STSV2) remained unexplained (28, 31).
Here we show that upon STSV2 infection, expression of all ESCRT machinery components is repressed but the growth of the infected cells continues, yielding cells with up to ∼20 times larger diameters and ∼8,000 times larger volumes compared to noninfected cells. The giant cells serve as virion factories producing infectious viral particles in the course of days, until eventual collapse. Remarkably, the gigantic infected cells underwent asymmetric cell division in an ESCRT-dependent manner, spawning normal-sized cells, which upon reinfection produced a new generation of giant cells, locking the system in a cyclic process. However, in the presence of an active CRISPR-Cas system, new spacers targeting the virus were acquired within the giant cells and the released normal-sized cells were immune to virus infection, and eventually took over the population. Collectively, our results show that an archaeal virus tinkers with the cell cycle, inducing cell gigantism and asymmetric cell division reminiscent of that occurring in budding yeast. Furthermore, we provide evidence that CRISPR adaptation takes place in productively infected cells, providing insights into CRISPR-Cas response in archaea.
Results
STSV2 Infection Induces Cell Gigantism.
To study virus–host interactions in archaea and to investigate the potential link between cell cycle and virus infection, we focused on the nonlytic virus STSV2 (31) and its host, Sulfolobus islandicus REY15A (32). The cells were infected using a multiplicity of infection (MOI) of 10 and the growth dynamics of infected and noninfected cells was followed for up to 10 d by measuring the optical density (OD600) of the corresponding cultures. Virus infection resulted in substantial growth retardation (SI Appendix, Fig. S1A), which was accompanied with continuous increase in the virus titer until 7 d postinfection (dpi) (SI Appendix, Fig. S1B). Although no cell lysis was observed, we could establish a plaque test for STSV2 (SI Appendix, Fig. S1C), which was used for virus enumeration in subsequent infection experiments. Notably, 7 dpi, there was a steep increase in the optical density of the infected culture, suggesting the emergence of a population of cells resistant to STSV2 infection (see below). Consistently, the titer of the virus in the culture started to decrease. The noninfected cell culture reached the maximal density after 3 d of incubation and entered into the death phase, characterized by gradual lysis (SI Appendix, Fig. S1A), likely due to consumption of all available nutrients.
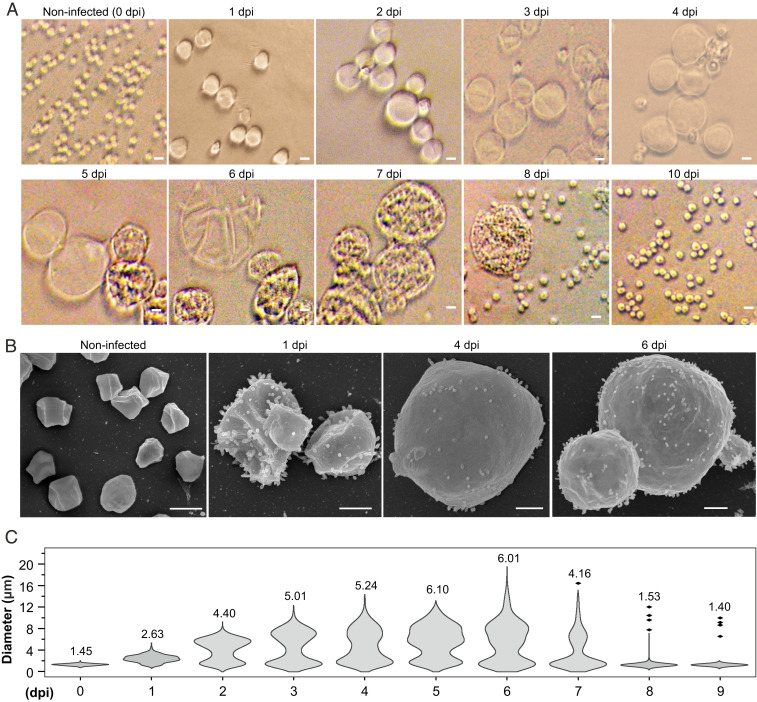
To gain further understanding on the progression of the infection, aliquots collected at different time points postinfection were observed using bright-field microscopy. Unexpectedly, we found that STSV2 infection resulted in a dramatic increase in the host cell size (Fig. 1A). After 1 dpi, the infected cells became more than twice bigger in diameter compared to the noninfected control and progressively enlarged up to 20 μm in diameter (Fig. 1A and SI Appendix, Fig. S1D). By contrast, the average diameter (1.2 ± 0.3 μm) of noninfected cells remained constant (SI Appendix, Fig. S1E). The integrity of the giant cells was further validated by scanning electron microscopy, which revealed the presence of numerous STSV2 virions on the cell surface (Fig. 1B).
Fig. 1.
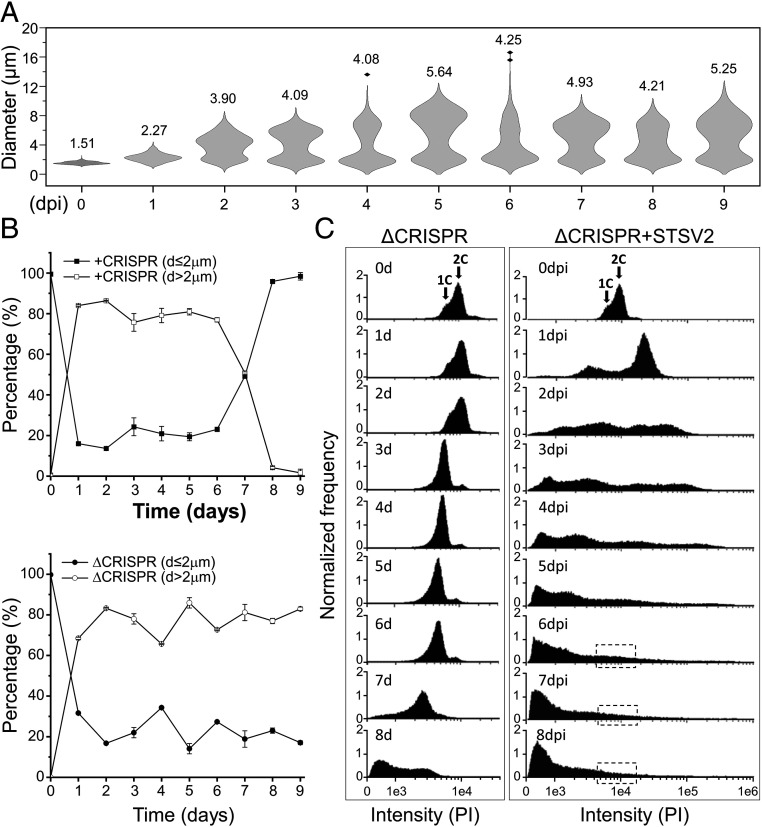
STSV2 infection induces cell gigantism. Differential interference contrast (A) and scanning electron microscopy analysis (B) of noninfected and STSV2-infected REY15A cells. (Scale bars, 2 μm in A and 1 μm in B.) (C) Size distribution of the STSV2-infected REY15A cells during different time points after infection. The numbers above the plots represent median diameters of cells for each time point. Cell cultures were sampled at the indicated time points and the diameters of 600 cells from two independent experiments were measured for each time point using ImageJ (NIH).
To quantify the changes in the infected population, we estimated the fractions of cells with different diameters at different time points postinfection. For convenience, we refer to all cells with a diameter of more than 2 μm (d > 2 μm) as “big” cells and those with a diameter of no more than 2 μm (d ≤ 2 μm) as “normal” cells. As shown in Fig. 1C, after 1 dpi, only ∼16% of cells were of normal size (d ≤ 2 μm), whereas ∼80% of the cells had a diameter ranging from 2 to 4 μm, and about 4% had a diameter between 4 and 8 μm. The overall ratio of normal and big cells was highly reproducible and remained stable (around 20% and 80%, respectively) from 1 to 6 dpi, although the number of cells with larger diameters increased in a time-dependent manner. The fraction of cells with diameters greater than 12 μm reached maximum (∼5% of all cells) at 6 dpi (Fig. 1C). However, starting with 7 dpi, the overall ratio began to change. Namely, the number of normal (d ≤ 2 μm) and big (d > 2 μm) cells became roughly equal after 7 dpi and after 8 dpi, the cell culture was dominated by normal-sized cells (96%) (Fig. 1C).
To investigate whether the ability to induce cell gigantism is unique to STSV2, we infected REY15A cells with two other nonlytic viruses, Sulfolobus spindle-shaped virus 2 (SSV2) (SI Appendix, Fig. S2A) (33) and Sulfolobus monocaudavirus 1 (SMV1) (SI Appendix, Fig. S2B) (34), both of which can efficiently replicate in REY15A cells. SMV1 is only distantly related to STSV2, although both viruses are tentative members of the family Bicaudaviridae, whereas SSV2 belongs to an unrelated virus family, the Fuselloviridae (23). SSV2 infection did not induce any changes in cell dimensions discernible by bright-field microscopy (SI Appendix, Fig. S2C). By contrast, infection with SMV1 led to increase in cell size, similar to that described above for STSV2, albeit SMV1-infected cells did not grow as large as those infected with STSV2 (SI Appendix, Fig. S2 D and E). Collectively, these results indicate that bicaudaviruses have a dramatic effect on the biology of their host, leading to an unprecedented increase in cell size. The fact that this phenomenon is not induced by SSV2 suggests that the process is virus specific and does not represent a general cell response to virus infection.
STSV2-Infected Giant Cells Contain Increased DNA Content.
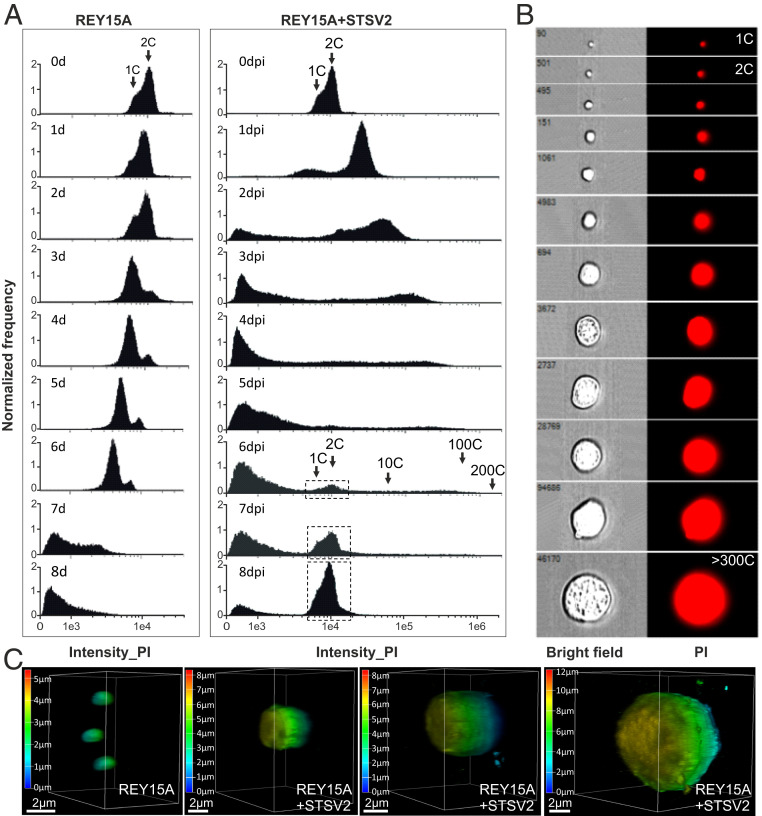
In asynchronous Sulfolobus population, most cells are in G2 phase (>50% of the cell cycle) and contain two copies (2C) of the chromosome, whereas in G1 phase, which is considerably shorter (<5% of the cell cycle), cells contain only one copy (1C) of the chromosome (35). The DNA content in the population can be readily assessed by flow cytometry, which produces characteristic profiles (15, 18). Thus, to characterize the infected population and to investigate what happens with the cellular DNA content during STSV2 infection, we performed flow cytometry analysis. As expected, during the first 2 d of active growth, the majority of noninfected cells contained two chromosomes (days 1 to 2; Fig. 2A). However, during the stationary growth stage (SI Appendix, Fig. S1A), the population became dominated by cells with 1C DNA content (day 3; Fig. 2A), signifying the arrest in G1 phase, potentially due to nutrient limitation. Finally, when the population progressed into the death phase (days 4 to 8, SI Appendix, Fig. S1A), the DNA was gradually degraded, with the peaks of the DNA content shifting from right to the left (Fig. 2A).
Fig. 2.
STSV2-infected giant cells contain increased DNA content. (A) Flow cytometry analysis of noninfected (Left) and STSV2-infected (Right) REY15A cells. Cell cultures were sampled and analyzed at the indicated time points. Arrows indicate cells with the DNA content corresponding to the equivalents of 1 copy (1C), 2C, 10C, 100C, and 200C of genomic DNA. The dotted boxes indicate the region of cells containing the DNA content corresponding to 1 and 2 copies, which reappeared in the infected cells at 6 dpi. (B) Representative images showing single cells sorted by flow cytometry with different diameters and DNA content equivalents ranging from 1 copy to more than 300 copies. PI, propidium iodide. (C) Three-dimensional reconstruction images of noninfected and STSV2-infected REY15A cells with different diameters. The cells were stained with DAPI and observed using a Leica SP8 confocal microscope. The images were analyzed by the Leica Application Suite X (LAS X) software and displayed in the volume mode. The color scale indicates the Z-depth. (Scale bars, 2 μm.)
The profiles of DNA content in STSV2-infected cultures were radically different. After 1 dpi, around 80% of the infected cells contained more than 4C equivalents of DNA, and about 10% of the cells showed the DNA content of less than 1C. As the infection progressed, there appeared cells containing an even larger number of DNA copy equivalents (Fig. 2A), with some of the giant cells harboring the DNA content corresponding to more than 300 copies (Fig. 2B). Over time, the population appeared as a continuum of cells with highly variable DNA contents. Indeed, sorting of individual cells labeled with fluorescent DNA-binding dye (propidium iodide) allowed us to visualize this continuum (Fig. 2B). Notably, starting with 2 dpi, we observed appearance of cells with DNA content lower than one chromosome copy, which could correspond to either partially degraded cellular DNA, viral DNA, or both. Starting with 6 dpi, two peaks corresponding to 1C and 2C DNA content, characteristic of noninfected cells, started to reappear in infected cells, and became dominant at 8 dpi (Fig. 2A). This result is consistent with the observation that at 8 dpi the population became dominated by normal-sized cells (Fig. 1 A and C and SI Appendix, Fig. S1A).
To get further information on the viral and cellular DNA content during the infection, we collected the infected cells at different time points (1 to 9 dpi), extracted the total (viral+cellular) DNA, and performed qPCR with chromosome- and virus-specific The ratio between the viral and cellular genome copy numbers increased gradually, peaking at 6 dpi with ∼800 viral genome copies to one cellular chromosome copy (SI Appendix, Fig. S3A). Following the emergence of resistant cells, the viral-to-host genome ratio decreased sharply. To estimate whether both the viral and cellular genomes were replicated in the big cells, we sorted the infected cells by flow cytometry and collected those with diameters larger than 5 μm (from ∼6 to ∼16 μm, median diameter 9.45 μm) for qPCR analysis. Knowing the exact cell number, we determined average numbers of viral and cellular genome copies per cell. The big cells, on average, harbored 111 ± 62 copies of the cellular chromosome and 2,426 ± 261 copies of the viral genome per cell (SI Appendix, Fig. S3B). These results clearly show that both the viral and cellular genomes are replicated during the infection.
To gain insights into the intracellular organization of the DNA, the noninfected and STSV2-infected cells were stained with DAPI and analyzed by confocal microscopy. Regardless of the cell diameter (1 to 10 μm), the DNA was evenly distributed in the cytoplasm, with no obvious condensation foci (Fig. 2C). The three-dimensional (3D) reconstruction of the infected cells also confirmed the integrity and spherical morphology of the big cells.
Expression of Cell Division Genes Is Severely Down-Regulated upon STSV2 Infection.
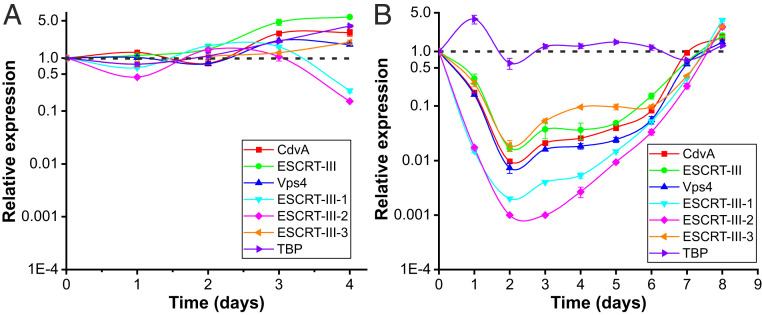
The microscopy and flow cytometry data suggest that in STSV2-infected cells, synthesis of the components of cell envelope and DNA replication continue, but the cell division is blocked. Thus, to analyze the expression of the genes involved in cell division throughout the infection, we performed qRT-PCR with primers specific to all six components of the Sulfolobus ESCRT machinery. A housekeeping gene encoding the TATA-binding protein (TBP) was used as a control. The expression level in the T0 culture (day 0) was considered as unity and expression levels at other time points were plotted relative to this level. In noninfected cells, the transcription levels of the ESCRT genes were relatively stable and fluctuated around one during exponential and stationary growth stages (days 1 to 4; Fig. 3A). However, starting with day 5, the total RNA in the noninfected cells started to be degraded (SI Appendix, Fig. S4A), consistent with cell lysis and DNA degradation (Fig. 2A and SI Appendix, Fig. S1A). By contrast, in STSV2-infected cells, the RNA remained stable throughout the experiment (SI Appendix, Fig. S4B). qRT-PCR analysis showed that the transcription levels of all ESCRT machinery components in the infected cells were down-regulated, reaching the lowest levels at 2 dpi (Fig. 3B). Expression of the gene encoding ESCRT-III-2 was most severely affected, with 57-fold decrease after 1 dpi, and 1,000-fold decrease after 2 dpi (Fig. 3B). Notably, expression level of TBP remained stable throughout the experiment, except for the temporary increase at 1 dpi. Importantly, the transcription level of all ESCRT components was stable during days 3 to 6 dpi, whereas after 7 to 8 dpi, when the culture became dominated by normal-sized cells, the expression level of the cell division genes reached the level of noninfected control cells (Fig. 3B). Consistent with the derepression of the transcription of the ESCRT machinery components, there was a rapid increase in cell division, as can be judged from the increase in optical density (SI Appendix, Fig. S1A).
Fig. 3.
Expression of cell division genes is down-regulated upon STSV2 infection. Transcriptional analysis of noninfected (A) and STSV2-infected (B) REY15A cells. Cell cultures were sampled and analyzed at the indicated time points. 16S rRNA was used as the reference and tbp, a housekeeping gene encoding TATA-binding proteins, was used as the control. The transcription levels of the target genes in the cell cultures at 0 dpi (i.e., noninfected cells prior to infection) were defined as 1 (indicated by the dashed lines). Three biological replicates were analyzed for each time point. Error bars represent SD from three independent experiments.
To further confirm the link between the cell division genes and cell diameter, we expressed in REY15A cells the C-terminally truncated ESCRT-III and CdvA proteins (ESCRT-IIIΔC and CdvAΔC, respectively), both of which have a negative effect on cell division (17, 36), and observed the cell morphology using bright-field microscopy. In both cases, the cell diameter increased from 1.2 ± 0.3 μm to 4 to 5 μm (SI Appendix, Fig. S5A). To more directly mimic the down-regulation of the expression of the cell division genes, we depleted the ESCRT-III and CdvA transcripts by 30% and 70%, respectively, using the CRISPR-based knockdown system (37). Cells with up to 4 μm in diameter were observed (SI Appendix, Fig. S5B). Notably, however, neither approach yielded cells as big as those infected with STSV2, possibly due to an additive effect of simultaneous repression of all cell division genes in the case of virus infection. These results further support the link between the repression of the cell division genes and remarkable increase in the dimensions of STSV2-infected cells.
Giant Cells Release Normal-Sized Cells through Asymmetric Cell Division.
As mentioned above, the fraction of normal-sized cells remained around 20% throughout several days of the experiment (Fig. 1C), suggesting dynamic renewal of the normal-sized cells. To gain insights into this process, we analyzed the population by bright-field microscopy and observed that some of the infected big cells displayed surface bulges (SI Appendix, Fig. S6A). Cell sorting by flow cytometry followed by fluorescence microscopy (SI Appendix, Fig. S6B) as well as confocal microscopy and 3D reconstruction (Movie S1) further suggested that the bulges are an integral part of the big cells, rather than normal-sized cells colocalizing with the big cells. Finally, the continuity between the big cells and the bulges was confirmed by electron microscopy (SI Appendix, Fig. S6C). In terms of dimensions (∼1 to 1.5 μm) and shape, these bulges resembled the normal-sized cells present in the population. Thus, we hypothesized that the bulges represent budding of normal-sized cells from the big cells, a phenomenon superficially resembling the asymmetric cell division of budding yeast (SI Appendix, Fig. S6D).
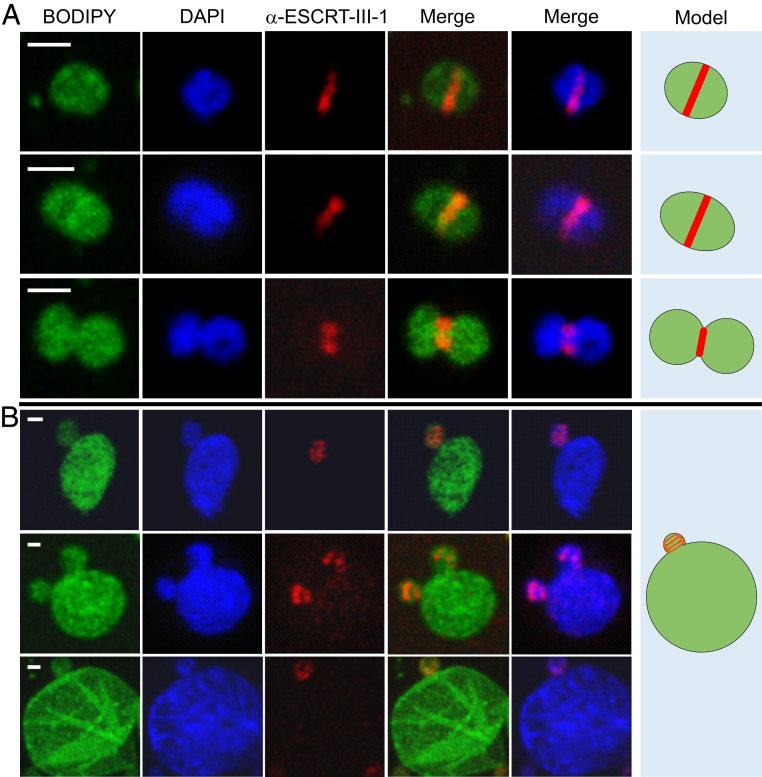
Cell division in Sulfolobus occurs by binary fission and depends on the archaeal ESCRT machinery (15, 17, 38–43). To analyze if Sulfolobus ESCRT system participates in the formation of budding-like structures in STSV2-infected cells, we performed fluorescence microscopy with antibodies against ESCRT-III-1, one of the three Sulfolobus ESCRT-III homologs previously shown to participate in S. islandicus REY15A cell division (17). In noninfected cells, ESCRT-III-1 formed ring-like structures in the midcell at different stages of cell division, including cytokinesis whereby the membrane is constricted between the two daughter cells (Fig. 4A). No such midcell ring-like structures could be detected in the STSV2-infected big cells, in which ESCRT-III-1 formed only small dot-like foci (SI Appendix, Fig. S7). However, when the STSV2-infected cells contained the “buds,” ESCRT-III-1 became organized into ring- or spiral-like structures, which localize at the budding sites (Fig. 4B). These results strongly suggest that the normal-sized cells are produced by the big cells through a budding or asymmetric cell division mechanism, thereby replenishing the subpopulation of normal-sized cells, and that ESCRT machinery participates in this process.
Fig. 4.
Immunofluorescence microscopy analysis of noninfected and STSV2-infected REY15A cells. (A) Noninfected REY15A cells at different stages of cell division. ESCRT-III-1 localizes at the midcell forming a band-like structure, which constricts with the progression of the cell division process. (B) STSV2-infected cells undergo asymmetric cell division or budding with ESCRT-III-1 localizing at the budding site forming a ring or spiral-like structures. Fixed cells were stained with BODIPY (green) to visualize the membrane, DAPI (blue) to visualize DNA, and fluorescently labeled anti-ESCRT-III-1 antibody (red) to visualize ESCRT-III-1. Hypothetical models are shown on the Right. (Scale bars, 1 μm.)
STSV2-Infected Cells Develop CRISPR-Based Resistance.
As mentioned above, after 8 dpi, the normal-sized cells outnumbered the big cells (Fig. 1), which coincided with the derepression of the cell division genes (Fig. 3) and sharp increase in the optical density of the culture (SI Appendix, Fig. S1A). We hypothesized that the observed changes in the infected population resulted from emergence of cells resistant to the STSV2 infection. The CRISPR-Cas system is the most extensively studied antiviral mechanism of Sulfolobus and has been shown to be effective against different viruses and plasmids (24, 34, 44–47). S. islandicus REY15A carries two CRISPR loci, three distinct CRISPR interference modules (one type IA and two type IIIB systems), and a single adaptation module, which integrates virus-derived spacers into both CRISPR loci (32, 45) (SI Appendix, Fig. S8A). Notably, a previous study has failed to detect spacer acquisition from STSV2, unless the cells were coinfected with SMV1 (44).
To analyze if de novo CRISPR adaptation occurred in the course of STSV2 infection, we amplified the leader-proximal regions of both CRISPR loci at different time points postinfection. PCR products corresponding to newly acquired spacers were observed in both CRISPR loci starting with 7 dpi (SI Appendix, Fig. S9A). Notably, the bands corresponding to the ancestral CRISPR loci were also visible, albeit much fainter, suggesting that at all times the population was a mixture of cells with and without spacers against STSV2. To verify that the new spacers were indeed acquired from STSV2, the infected culture after 10 dpi was plated on solid medium and three colonies of cells with a variable number of spacers (S1 to S3) in both CRISPR loci (SI Appendix, Fig. S9B) were selected for isolation. Sequencing of the leader-proximal regions of CRISPR1 and CRISPR2 loci of S1 to S3 has confirmed that the newly acquired spacers are derived from STSV2 (SI Appendix, Fig. S8B). Spot assay has shown that all three strains are resistant to STSV2 (SI Appendix, Fig. S8C). To further confirm this result, S2 strain was infected with STSV2 in liquid culture and observed by bright-field microscopy; no appreciable changes in cell morphology or size were detected (SI Appendix, Fig. S9D), consistent with the resistance to STSV2 infection. Collectively, these results demonstrate that STSV2 infection is countered by the CRISPR-Cas system and leads to de novo acquisition of multiple new spacers targeting STSV2. To study what happens with the viral genome in STSV2 spacer-containing cells, we infected spacer-lacking (REY15A) and spacer-containing (S2) strains and tracked the presence of the STSV2 genome by PCR. Whereas the STSV2 genome accumulated in the wild-type (WT) cells (SI Appendix, Fig. S9E), it was degraded in the S2 cells (SI Appendix, Fig. S9F). The STSV2 genome-specific band started to diminish at 10 h postinfection and became barely detectable after 2 dpi. These results strongly suggest that CRISPR targeting leads to degradation of the STSV2 genome.
Given that normal-sized cells are released by budding from the big cells, the STSV2-targeting CRISPR spacers could be acquired either in productively infected big cells or directly in the normal-sized cells potentially upon infection with defective viruses, as has been demonstrated for certain bacteriophages (48). To distinguish between the two possibilities, we sorted the infected cells using flow cytometry into populations of d ≤ 2 μm and d > 5 μm at different time points postinfection, and analyzed the collected populations for the presence of new CRISPR spacers by PCR, as described above. The newly acquired spacers were detected not only in the normal-sized cells (SI Appendix, Fig. S9G) but also in the big cells (SI Appendix, Fig. S9H), indicating that CRISPR adaptation could take place in cells successfully infected with STSV2. Consequently, normal-sized cells budding from the big cells carrying spacers against STSV2 would be resistant to virus infection.
CRISPR-Cas System Is Indispensable for the Emergence of Resistant Population.
To test if resistance to STSV2 infection can emerge by a mechanism independent of the CRISPR-Cas system, e.g., mutation of the receptor, we infected with STSV2 a mutant strain of S. islandicus REY15A, ΔC1C2 (hereinafter ΔCRISPR), bearing a large chromosomal deletion encompassing the only adaptation module, type IA interference module, and both CRISPR loci (SI Appendix, Fig. S8A) (49). Infected ΔCRISPR cells increased in size, similar to the wild-type REY15A cells (Fig. 5A). However, unlike in the wild-type cells, the resistant population did not emerge. Instead, even after 8 dpi, when the wild-type population was dominated by normal-sized cells (d ≤ 2 μm), the number of such cells in the ΔCRISPR culture remained stable at around 20% (Fig. 5B). Consistently, there was no increase in the optical density of the ΔCRISPR culture (SI Appendix, Fig. S1A) nor was there a reappearance of the population with 1C-2C chromosomes detectable by flow cytometry (Fig. 5C). These results show that even if cells with CRISPR-independent resistance to STSV2 did emerge in the population, they were below the detection limit during our experiment, allowing the population of big cells to be stably renewed and maintained.
Fig. 5.
CRISPR-Cas system is indispensable for the emergence of a resistant population. (A) Size distribution of the STSV2-infected CRISPR-deficient ΔC1C2 cells during different time points after infection. The width of the distribution corresponds to the frequency of occurrence. The numbers above the plots represent median diameters of cells during each time point. Cell culture was sampled at the indicated time points and the diameters of 600 cells from two independent experiments were measured at each time point. (B) Changes in the diameter of cells with (Top) and without (Bottom) the CRISPR immune system during STSV2 infection. For convenience of presentation, cells were grouped into two categories—those with d ≤ 2 µm and those with d > 2 µm. (C) Flow cytometry analysis of the DNA content in the ΔCRISPR mutant during the infection with STSV2. Cell cultures were sampled and analyzed at the indicated time points. Arrows indicate cells with the DNA content corresponding to the equivalents of one and two copies (1C and 2C, respectively) of genomic DNA. The dotted boxes indicate the region corresponding to the DNA content of one and two copies, which reappeared in the CRISPR-containing cell culture (Fig. 2A).
Discussion
Viruses are the master manipulators of their hosts at both cellular and population levels (50). Studies on virus–host interactions have greatly contributed to uncovering many fundamental aspects of cell biology, especially in eukaryotes, including mechanisms of membrane fusion, membrane scission by the ESCRT machinery, apoptosis, cytoskeleton remodeling, functioning of plasmodesmata in plants, and many more (51–54). How archaeal viruses affect the biology of their hosts remains largely unknown. Here we described a phenomenon, whereby an archaeal virus interferes with the cell cycle of its host to orchestrate the transformation of the infected cell into a gigantic virion factory. The 20-fold increase in cell diameter compared to the noninfected spherical Sulfolobus cell translates to over an 8,000-fold increase in cell volume, as can be calculated using a simple formula 4/3πr3, where r is radius. The volume of a 20-μm cell would be 4.2 × 103 fL (or 4.2 × 103 μm3), which is three to four orders of magnitude larger than the volume (∼0.4 to 3 µm3) of typical model bacteria, such as Bacillus subtilis, Staphylococcus aureus, Escherichia coli, and Caulobacter crescentus (55). Even many unicellular eukaryotes, such as budding yeast and green algae, are considerably smaller, with the diameters of 3 to 6 μm (SI Appendix, Fig. S5D) (56, 57). To the best of our knowledge, such virus-induced increase in cell dimension has not been reported for any other virus. We propose a model whereby the archaeal virus STSV2 manipulates the cell cycle of its host causing cell gigantism and asymmetric cell division (Fig. 6).
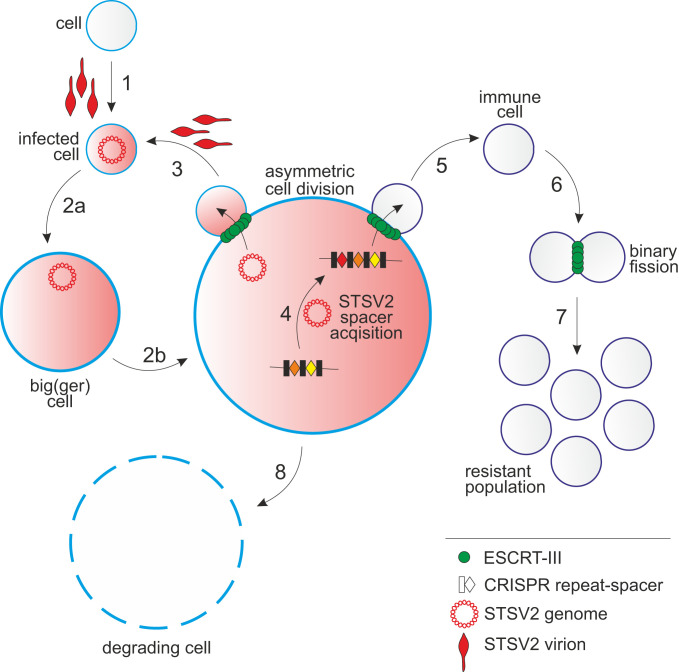
Fig. 6.
A schematic representation of the STSV2-Sulfolobus interactions. 1: infection of a normal-sized cell; 2a and 2b: gradual increase in the diameter of STSV2-infected cells; 3: asymmetric division of a STSV2-infected giant cell leading to the budding of a normal-sized cell, which can be reinfected (by exogenous virus or by virus genome vertically transmitted from the giant cell) to restart the cycle; 4: acquisition of CRISPR spacers against STSV2; 5: asymmetric division of a STSV2-infected giant cell leading to the budding of a normal-sized cell resistant to STSV2 infection due to the presence of CRISPR spacers against STSV2; 6: division of the STSV2-resistant cells by binary fission; 7: proliferation of the resistant population; and 8: gradual decay of the giant cell.
Diffusion is one of the factors believed to restrict the size of most prokaryotes (58). High surface-to-internal volume ratio of prokaryotic cells ensures efficient diffusion of nutrients, elimination of waste, and the timely movement of biomolecules, alleviating the need for dedicated transport systems found in the larger eukaryotic cells. Indeed, compartmentalization, emergence of motor protein-facilitated trafficking over a complex cytoskeletal network, and acquisition of energy-generating organelles have all been credited for the advancement of the size and complexity of eukaryotic cells (55, 58). A prevailing hypothesis posits that eukaryotes have evolved from a lineage of archaea (59, 60). However, most extant archaea, and in particular, the postulated archaeal ancestor of eukaryotes (61), have small cell size. Our results suggest that dramatic increase in cell size and volume can be readily achieved through reprogramming of the preexisting cellular machineries. We obtained similar results with two different archaeal viruses, STSV2 and SMV1, indicating that the observed increase in the cell size is not an artifact. Interestingly, it has been reported that hyperthermophilic crenarchaeon Staphylothermus marinus, which belongs to the same class (Thermoprotei) as Sulfolobus, upon growth in the presence of high concentrations of yeast extract as the sole substrate increased in diameter from the typical 0.5 to 1 μm up to 15 μm (62). These observations illuminate the plasticity of archaeal cells, possibly enabled by the absence of a rigid peptidoglycan layer found in most bacteria.
The size increase of STSV2-infected cells appears to be linked to the repression of the genes encoding ESCRT machinery components. In synchronized noninfected Sulfolobus cells, expression of some of the cell division genes is cell cycle dependent: whereas Vps4 is expressed throughout the cell cycle, ESCRT-III is nearly undetectable during G1 and S (synthesis) phases and is produced only starting with the G2 phase, when DNA replication is complete (18, 37). In STSV2-infected cells, expression of cell division genes is severely down-regulated, whereas genome replication continues to an extravagant extent, with some cells containing over 300 equivalents of chromosome copies. This state resembles arrest of the cell cycle in S phase. Indeed, a number of eukaryotic viruses, such as hepatitis B virus (63), polyomaviruses and papillomaviruses (64), and adenovirus (65), promote the transition and/or arrest of the cell cycle in the S phase. For small eukaryotic DNA viruses, which do not encode a complete DNA replication machinery, entry into S phase ensures access to the host enzymatic activities and cellular DNA precursors for virus DNA replication (6). Notably, like most crenarchaeal viruses, STSV2 does not encode its own DNA polymerase (31) and thus, in all likelihood, relies on the host machinery for genome replication.
Repression of cell division genes in STSV2-infected cells, although severe, is not total or irreversible and after 2 dpi there is a partial release of the repression, which coincides with increase in the fraction of normal-sized cells in the population to ∼20%. Remarkably, whereas noninfected Sulfolobus cells invariably divide by binary fission (15–17, 39, 40, 42), in the infected population, normal-sized cells are produced by budding from the giant cells. This mode of cell division has never been described for any archaeon, but is highly reminiscent of the asymmetric cell division characteristic of budding yeast, Saccharomyces cerevisiae (66), and that observed during self-renewal and differentiation of stem cells (67) as well as during tumorigenesis (68). The major difference between symmetric and asymmetric cell division lies in the selection of the division site, where the cytokinetic furrow including the ESCRT machinery is assembled (66). Interestingly, like in the case of symmetric cell division, the asymmetric division of STSV2-infected cells appears to be dependent on the action of the ESCRT machinery, consistent with the formation of ESCRT-III-1-containing rings and spiral-like structures in the outgoing budding cells. It is remarkable that the budding cells are of normal size, suggesting the existence of a common mechanism determining the size of the daughter cells in both infected and noninfected cells. Further research will be required to determine the full composition of the division apparatus in the infected cells and the mechanism of its asymmetric positioning. Regardless, our current results, in combination with the recent demonstration that ESCRT system is responsible for the budding of extracellular vesicles in Sulfolobus (37), implicate the archaeal ESCRT machinery in membrane remodeling processes beyond membrane abscission during normal cell division by binary fission. Whether the involvement of the ESCRT system can be extended to the budding of archaeal viruses (69), including STSV2, as has been demonstrated for diverse enveloped viruses of eukaryotes, such as HIV-1 and Ebola virus (52, 70, 71), remains to be investigated. Topologically, however, the budding of cells (as shown in this study), vesicles, and viruses are equivalent processes, whereby ESCRT proteins mediate the so-called “reverse”-topology membrane scission at the narrow membrane necks contiguous with the cytoplasm (52).
The normal-sized cells released by budding replenish the pool of susceptible hosts and can be reinfected with STSV2 produced in the previous rounds of infection or, alternatively, the infection can be propagated to the daughter cell in the form of virus genomes during the budding process (Fig. 6). The latter strategy of infection would be particularly efficient, because the virus would not have to face the harsh extracellular environment, and loss (or mutation) of the receptor would not block the virus propagation. Regardless of the mechanism, the outcome of the infection is the same—the newly produced cells again increase in size, yielding the next generation of giant cells. This cycle repeats itself, unless virus resistance develops. During the 9 d of our experiment, no such resistance had arisen, unless the CRISPR-Cas system was functional. Indeed, in the absence of the CRISPR-Cas system, the ratio of giant and normal-sized cells remained stable, with no signs of resistance development. However, when the CRISPR-Cas system was operational, resistant cells emerged 7 dpi, carrying a variable number of CRISPR spacers in both CRISPR loci, seeding a resistant population which was maintained during subsequent passages of the culture (SI Appendix, Fig. S8).
The mechanism of spacer acquisition in archaea has been studied in vitro (72–76) and has been documented during infection with different viruses in vivo (34, 44, 77). However, it remained unclear how CRISPR adaptation is coordinated with the virus infection at the cellular level. That is, are spacer acquisition and antivirus response sufficiently rapid to save a productively infected cell? In bacteria, it has been shown that replication-deficient phages are responsible for the vast majority of the acquisition of CRISPR-mediated phage immunity (48). By contrast, infection with virulent, replication-competent phages often results in abortive infection and demise of the infected cells, rather than immunity (78, 79). In the case of STSV2 infection, newly acquired spacers were identified in the giant cells, suggesting that adaptation took place despite active virus replication. Resistant cells rapidly took over the population, terminating the virus propagation.
To our knowledge, STSV2 and SMV1 are the only archaeal viruses suggested to manipulate the cell cycle of their host. We have shown that STSV2 infection blocks the normal cell division in Sulfolobus, which leads to unprecedented cell growth and asymmetric division reminiscent of that operating in budding yeast. In the presence of the CRISPR-Cas system, the population can recover and revert to division by binary fission. The plasticity of Sulfolobus cells is remarkable and similar properties could have played a key role during eukaryogenesis. The STSV2-Sulfolobus system might serve as a powerful model for addressing fundamental unanswered questions of archaeal cell biology, including transition between binary fission and asymmetric cell division, cell cycle control, determination of the optimal size of the daughter cell, de novo CRISPR adaptation in archaea, and more. As a first step in this direction, it will be important to identify the viral factor(s) responsible for repression of the cell division genes. STSV2 and SMV1 encode several putative transcriptional regulators (31, 34) which are the prime suspects involved in this process.
Materials and Methods
Strains and Growth Conditions.
Strains used in this study are listed in SI Appendix, Table S1. S. islandicus REY15A was grown aerobically at 75 °C in TSV (tryptone-sucrose-vitamins) medium, as described previously (80). TSV medium supplemented with 0.01% (wt/vol) uracil (U), TSVU, was used for culturing of the S. islandicus CRISPR deletion mutant ΔC1C2 (49). The S. cerevisiae Y2H Gold strain was grown aerobically at 30 °C with shaking (180 rpm) in YPD medium containing 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) dextrose.
Infection Assays.
For infection, REY15A and ΔC1C2 cells were collected at midlogarithmic phase and mixed with the virus. The MOI used for infection was 10. The MOI was calculated based on the plaque assays. The infected cultures were incubated at 75 °C for 1 h without shaking. Following the incubation, the cells were pelleted and washed with 7% sucrose three times (7,000 rpm for 10 min) to remove the unadsorbed virions. Finally, the infected cells were resuspended in the TSVU medium and incubated at 75 °C with shaking (140 rpm). Infections with SSV2 and SMV1 were also carried out at an MOI of 10.
Further details on microscopy and flow cytometry techniques used are provided in SI Appendix, Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (Grant 2020YFA0906800) and the National Natural Science Foundation of China (Grants 31970546 and 31670061) to Y.S.; l’Agence Nationale de la Recherche (Grant ENVIRA, ANR-17-CE15-0005-01) and the Emergence(s) project MEMREMA from Ville de Paris to M.K.; and the PRESTIGE post-doctoral program from the European Union's Seventh Framework Programme and the National Key Research and Development Program of China (Grant 2020YFA0906800) to J.L. We thank Pierre-Henri Commere from the Flow Cytometry Platform of Institut Pasteur for help with cell sorting, as well as Christine Schmitt and Olivier Gorgette from Ultrastructural Bioimaging (UTechS UBI) unit, of Institut Pasteur for their help with scanning electron microscopy. We are also grateful to the UTechS UBI unit of Institut Pasteur for access to electron microscopes. We gratefully acknowledge the UtechS Photonic BioImaging (Imagopole), C2RT, Institut Pasteur, supported by the French National Research Agency (France BioImaging; Grant ANR-10-INSB-04; Investments for the Future), for access to the confocal microscope.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.T. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022578118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Krupovic M., Dolja V. V., Koonin E. V., Origin of viruses: Primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 17, 449–458 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Howard-Varona C., et al., Phage-specific metabolic reprogramming of virocells. ISME J. 14, 881–895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forterre P., The virocell concept and environmental microbiology. ISME J. 7, 233–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moniruzzaman M., Martinez-Gutierrez C. A., Weinheimer A. R., Aylward F. O., Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat. Commun. 11, 1710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan Y., Sanyal S., Bruzzone R., Breaking bad: How viruses subvert the cell cycle. Front. Cell. Infect. Microbiol. 8, 396 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagga S., Bouchard M. J., Cell cycle regulation during viral infection. Methods Mol. Biol. 1170, 165–227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesri E. A., Feitelson M. A., Munger K., Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe 15, 266–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danovaro R., et al., Virus-mediated archaeal hecatomb in the deep seafloor. Sci. Adv. 2, e1600492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitbart M., Bonnain C., Malki K., Sawaya N. A., Phage puppet masters of the marine microbial realm. Nat. Microbiol. 3, 754–766 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Dion M. B., Oechslin F., Moineau S., Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18, 125–138 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Mushegian A. R., Are there 1031 virus particles on Earth, or more, or fewer? J. Bacteriol. 202, e00052-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haeusser D. P., et al., The Kil peptide of bacteriophage λ blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 10, e1004217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart C. R., Deery W. J., Egan E. S., Myles B., Petti A. A., The product of SPO1 gene 56 inhibits host cell division during infection of Bacillus subtilis by bacteriophage SPO1. Virology 447, 249–253 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Kiro R., et al., Gene product 0.4 increases bacteriophage T7 competitiveness by inhibiting host cell division. Proc. Natl. Acad. Sci. U.S.A. 110, 19549–19554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindås A. C., Bernander R., The cell cycle of archaea. Nat. Rev. Microbiol. 11, 627–638 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Samson R. Y., Duggin I. G., Bell S. D., Analysis of the archaeal ESCRT apparatus. Methods Mol. Biol. 1998, 1–11 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Liu J., et al., Functional assignment of multiple ESCRT-III homologs in cell division and budding in Sulfolobus islandicus. Mol. Microbiol. 105, 540–553 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Samson R. Y., Obita T., Freund S. M., Williams R. L., Bell S. D., A role for the ESCRT system in cell division in archaea. Science 322, 1710–1713 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindås A. C., Karlsson E. A., Lindgren M. T., Ettema T. J., Bernander R., A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 105, 18942–18946 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson-McGee J. H., Snyder J. C., Young M. J., Archaeal viruses from high-temperature environments. Genes (Basel) 9, E128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prangishvili D., et al., The enigmatic archaeal virosphere. Nat. Rev. Microbiol. 15, 724–739 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Krupovic M., Cvirkaite-Krupovic V., Iranzo J., Prangishvili D., Koonin E. V., Viruses of archaea: Structural, functional, environmental and evolutionary genomics. Virus Res. 244, 181–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iranzo J., Koonin E. V., Prangishvili D., Krupovic M., Bipartite network analysis of the archaeal virosphere: Evolutionary connections between viruses and capsidless mobile elements. J. Virol. 90, 11043–11055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Athukoralage J. S., et al., An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F., et al., Publisher Correction: Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat. Microbiol. 3, 1076 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Makarova K. S., et al., Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohanraju P., et al., Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016). [DOI] [PubMed] [Google Scholar]
- 28.León-Sobrino C., Kot W. P., Garrett R. A., Transcriptome changes in STSV2-infected Sulfolobus islandicus REY15A undergoing continuous CRISPR spacer acquisition. Mol. Microbiol. 99, 719–728 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Ortmann A. C., et al., Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 82, 4874–4883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder J. C., Samson R. Y., Brumfield S. K., Bell S. D., Young M. J., Functional interplay between a virus and the ESCRT machinery in archaea. Proc. Natl. Acad. Sci. U.S.A. 110, 10783–10787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erdmann S., et al., A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species. Extremophiles 18, 51–60 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Guo L., et al., Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus-host interaction studies. J. Bacteriol. 193, 1672–1680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stedman K. M., et al., Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res. Microbiol. 154, 295–302 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Erdmann S., Garrett R. A., Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol. Microbiol. 85, 1044–1056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernander R., Poplawski A., Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 179, 4963–4969 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samson R. Y., et al., Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol. Cell 41, 186–196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., et al., Archaeal extracellular vesicles are produced in an ESCRT-dependent manner and promote gene transfer and nutrient cycling in extreme environments. bioRxiv [Preprint] (2021). 10.1101/2021.02.09.430445. [DOI] [PMC free article] [PubMed]
- 38.Samson R. Y., Bell S. D., Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 17, 507–513 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Charles-Orszag A., Lord S. J., Mullins R. D., High-temperature live-cell imaging of cytokinesis, cell motility and cell-cell adhesion in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. bioRxiv [Preprint] (2020). 10.1101/2020.02.16.951772. [DOI] [PMC free article] [PubMed]
- 40.Samson R. Y., Bell S. D., Cell cycles and cell division in the archaea. Curr. Opin. Microbiol. 14, 350–356 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Samson R. Y., Dobro M. J., Jensen G. J., Bell S. D., The structure, function and roles of the archaeal ESCRT apparatus. Subcell. Biochem. 84, 357–377 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Tarrason Risa G., et al., The proteasome controls ESCRT-III-mediated cell division in an archaeon. Science 369, eaaz2532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pulschen A. A., et al., Live imaging of a hyperthermophilic archaeon reveals distinct roles for two ESCRT-III homologs in ensuring a robust and symmetric division. Curr. Biol. 30, 2852–2859.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erdmann S., Le Moine Bauer S., Garrett R. A., Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol. Microbiol. 91, 900–917 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Deng L., Garrett R. A., Shah S. A., Peng X., She Q., A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 87, 1088–1099 (2013). [DOI] [PubMed] [Google Scholar]
- 46.He F., Vestergaard G., Peng W., She Q., Peng X., CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 45, 1902–1913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manica A., Schleper C., CRISPR-mediated defense mechanisms in the hyperthermophilic archaeal genus Sulfolobus. RNA Biol. 10, 671–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hynes A. P., Villion M., Moineau S., Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nat. Commun. 5, 4399 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Gudbergsdottir S., et al., Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 79, 35–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roux S., Brum J. R., A viral reckoning: Viruses emerge as essential manipulators of global ecosystems. Environ. Microbiol. Rep. 11, 3–8 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Hay S., Kannourakis G., A time to kill: Viral manipulation of the cell death program. J. Gen. Virol. 83, 1547–1564 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Schöneberg J., Lee I. H., Iwasa J. H., Hurley J. H., Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reagan B. C., Burch-Smith T. M., Viruses reveal the secrets of plasmodesmal cell biology. Mol. Plant Microbe Interact. 33, 26–39 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Walsh D., Naghavi M. H., Exploitation of cytoskeletal networks during early viral infection. Trends Microbiol. 27, 39–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levin P. A., Angert E. R., Small but mighty: Cell size and bacteria. Cold Spring Harb. Perspect. Biol. 7, a019216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vander Wiel J. B., et al., Characterization of Chlorella vulgaris and Chlorella protothecoides using multi-pixel photon counters in a 3D focusing optofluidic system. Rsc Adv. 7, 4402–4408 (2017). [Google Scholar]
- 57.Ahmad M. R., Nakajima M., Kojima S., Homma M., Fukuda T., The effects of cell sizes, environmental conditions, and growth phases on the strength of individual W303 yeast cells inside ESEM. IEEE Trans. Nanobioscience 7, 185–193 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Schulz H. N., Jorgensen B. B., Big bacteria. Annu. Rev. Microbiol. 55, 105–137 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Zaremba-Niedzwiedzka K., et al., Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 60.López-García P., Moreira D., The Syntrophy hypothesis for the origin of eukaryotes revisited. Nat. Microbiol. 5, 655–667 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Imachi H., et al., Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiala G., Stetter K. O., Jannasch H. W., Langworthy T. A., Madon J., Staphylothermus marinus sp. nov. represents a novel genus of extremely thermophilic submarine heterotrophic archaebacteria growing up to 98 °C. Syst. Appl. Microbiol. 8, 106–113 (1986). [Google Scholar]
- 63.Benn J., Schneider R. J., Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. U.S.A. 92, 11215–11219 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shadan F. F., Cowsert L. M., Villarreal L. P., n-Butyrate, a cell cycle blocker, inhibits the replication of polyomaviruses and papillomaviruses but not that of adenoviruses and herpesviruses. J. Virol. 68, 4785–4796 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bagchi S., Raychaudhuri P., Nevins J. R., Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: A novel mechanism for E1A trans-activation. Cell 62, 659–669 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Bhavsar-Jog Y. P., Bi E., Mechanics and regulation of cytokinesis in budding yeast. Semin. Cell Dev. Biol. 66, 107–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berika M., Elgayyar M. E., El-Hashash A. H., Asymmetric cell division of stem cells in the lung and other systems. Front. Cell Dev. Biol. 2, 33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knoblich J. A., Asymmetric cell division: Recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849–860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quemin E. R., et al., Eukaryotic-like virus budding in Archaea. mBio 7, e01439-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurley J. H., Cada A. K., Inside job: How the ESCRTs release HIV-1 from infected cells. Biochem. Soc. Trans. 46, 1029–1036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vietri M., Radulovic M., Stenmark H., The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Grainy J., Garrett S., Graveley B. R., P Terns M., CRISPR repeat sequences and relative spacing specify DNA integration by Pyrococcus furiosus Cas1 and Cas2. Nucleic Acids Res. 47, 7518–7531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shiimori M., Garrett S. C., Graveley B. R., Terns M. P., Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell 70, 814–824.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rollie C., Graham S., Rouillon C., White M. F., Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res. 46, 1007–1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rollie C., Schneider S., Brinkmann A. S., Bolt E. L., White M. F., Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. eLife 4, e08716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garrett S., et al., Primed CRISPR DNA uptake in Pyrococcus furiosus. Nucleic Acids Res. 48, 6120–6135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M., Wang R., Zhao D., Xiang H., Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watson B. N. J., et al., Type I-F CRISPR-Cas resistance against virulent phages results in abortive infection and provides population-level immunity. Nat. Commun. 10, 5526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strotskaya A., et al., The action of Escherichia coli CRISPR-Cas system on lytic bacteriophages with different lifestyles and development strategies. Nucleic Acids Res. 45, 1946–1957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng L., Zhu H., Chen Z., Liang Y. X., She Q., Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13, 735–746 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.