Abstract
Objective
We present the Berlin-Tübingen-Oncology corpus (BRONCO), a large and freely available corpus of shuffled sentences from German oncological discharge summaries annotated with diagnosis, treatments, medications, and further attributes including negation and speculation. The aim of BRONCO is to foster reproducible and openly available research on Information Extraction from German medical texts.
Materials and Methods
BRONCO consists of 200 manually deidentified discharge summaries of cancer patients. Annotation followed a structured and quality-controlled process involving 2 groups of medical experts to ensure consistency, comprehensiveness, and high quality of annotations. We present results of several state-of-the-art techniques for different IE tasks as baselines for subsequent research.
Results
The annotated corpus consists of 11 434 sentences and 89 942 tokens, annotated with 11 124 annotations for medical entities and 3118 annotations of related attributes. We publish 75% of the corpus as a set of shuffled sentences, and keep 25% as held-out data set for unbiased evaluation of future IE tools. On this held-out dataset, our baselines reach depending on the specific entity types F1-scores of 0.72–0.90 for named entity recognition, 0.10–0.68 for entity normalization, 0.55 for negation detection, and 0.33 for speculation detection.
Discussion
Medical corpus annotation is a complex and time-consuming task. This makes sharing of such resources even more important.
Conclusion
To our knowledge, BRONCO is the first sizable and freely available German medical corpus. Our baseline results show that more research efforts are necessary to lift the quality of information extraction in German medical texts to the level already possible for English.
Keywords: medical information extraction, German language, corpus annotation
LAY SUMMARY
In this work we present the Berlin-Tübingen-Oncology (BRONCO) corpus, a German medical text corpus of 200 discharge summaries from the oncology departments of two hospitals. In the corpus mentions of diagnoses, treatments and medications are annotated together with a number of attributes as laterality, negation, and speculation. The corpus will be freely available for the research community for training and evaluating models for information extraction. To our knowledge, BRONCO will be the first freely available German medical corpus. To obey data protection law, we anonymized all documents and shuffled sentences in the publicly available version of the corpus. Consequently, applications are limited to the sentence level. We also provide baselines for named entity recognition, named entity normalization, and negation and speculation detection using state-of-the-art techniques.
INTRODUCTION
Clinical documentation contains a vast amount of patient-specific information, including disease etiology, family background, symptoms, examination results, and treatments. A systematic analysis of large quantities of documents can help to improve clinical care, to support clinical decision making, and to quality-control clinical pathways.1 However, documentation is mostly available in free text format at least in Germany, and its retrospective analysis for a given research hypothesis requires reading and understanding often hundreds or thousands of long and complex texts. Clinical natural language processing (NLP) investigates methods for automated information extraction (IE) specifically designed to process clinical text containing incomplete sentences, complex syntax, medical vocabulary, and idiosyncratic abbreviations. The quality of clinical NLP tools depends on the availability of annotated medical corpora for training and evaluation. Thus, the sharing of annotated corpora is indispensable:2 (1) The performance of different tools can be evaluated and compared, (2) reproducibility of previous results can be checked, and (3) machine-learning based NLP tools can be developed by groups world-wide without the time-consuming effort of corpus annotation, which furthermore requires high levels of medical knowledge.
To secure patient privacy, sharing of medical reports is only allowed either with explicit patient consent or when texts are fully anonymized. In the United States, HIPAA (Health Insurance Portability and Accountability Act of 1996; https://www.hipaajournal.com/de-identification-protected-health-information/) defines anonymization of medical data as the removal of 18 distinct Protected Health Information (PHI) identifiers. Based on these regulations, the NLP community developed tools for automatic deidentification of medical narratives,3–5 which greatly eased the development and publication of annotated corpora, such as MIMIC6 or corpora provided through shared tasks such as i2b2/n2c2 (https://portal.dbmi.hms.harvard.edu/projects/n2c2-nlp/), SemEval (http://alt.qcri.org/semeval2014/), and the CLEF ehealth lab series (http://clef-ehealth.org/). The access to corpora, in turn, enabled the development of freely available high-quality IE tools, among them MetaMap and cTakes.7,8
Compared to English, development of clinical NLP tools processing German medical text is still in its infancy.9–11 Similar to the United States, anonymized patient data can be shared in principle. However, there exists no clear definition of the PHI identifiers that need to be removed to obtain a fully anonymized medical document. Instead, the decision whether a certain approach achieves anonymization rests with the data protection officers at each institution. It is therefore extremely difficult to (1) obtain medical documents for NLP research outside of hospitals and (2) to share those data with other research groups. Consequently, although several annotation studies on German clinical corpora have been carried out previously, all those corpora are kept closed.12–17 The most recent corpus is 3000PA containing 3000 documents from 3 clinical sites that has been annotated with medication parameters.15 For German clinical texts, there are also no freely available IE or anonymization tools, and the reported quality of IE methods on closed corpora can neither be evaluated independently nor reproduced externally. Supplementary TableS1 gives key characteristics of selected clinical corpora used for IE for different languages, showing that several corpora for languages other than German are freely available for years, especially for English.
In this work, we present the freely available Berlin-Tübingen-Oncology corpus (BRONCO). It consists of shuffled sentences from 200 German discharge summaries from cancer patients annotated with medical entities (BRONCO was created by the nationally funded project “Personalizing Oncology via Semantic Integration of Data” (PersOnS), see https://persons-project.informatik.uni-tuebingen.de/). The ultimate aim of BRONCO is to foster the development of high-quality NLP tools for extracting the precise disease history of cancer patients. As a first step toward this goal, we manually anonymized documents and annotated diagnoses, treatments, and medication. Additionally, medical entities were annotated with attributes (laterality, negation, speculation, and possible in the future). We also created baselines for a set of IE tasks using state-of-the-art technologies.
To allow unbiased evaluation of IE tools, we randomly split the corpus in 2 parts: The larger subset, BRONCO150, contains 8976 sentences and 8760 annotations and is available under a liberal license for training and evaluation of IE tools (https://www2.informatik.hu-berlin.de/∼leser/bronco/index.html). The second subset, BRONCO50, with 2458 sentences and 2364 annotations, is kept closed as held-out data. As a further mean to prevent deanonymization sentences in both corpora are randomly shuffled. In the future, we will offer the service to evaluate new IE tools on BRONCO50 in our lab.
MATERIALS AND METHODS
Corpus design and preprocessing
We randomly selected 200 discharge summaries of patients suffering from hepatocellular carcinoma or melanoma treated between 2013 and 2016 at the university hospitals in Berlin or Tübingen. After careful anonymization the study on this data and publication of BRONCO was approved by the Data Protection Officers of both hospitals and the ethics committee of Charité (EA1/322/20). Documents were extracted from electronic patient records, converted to plain text, and manually anonymized by 1 or 2 clinicians at each hospital. Anonymization included removal of direct identifiers as names, age, contact details, IDs, and locations. Dates, persons, and hospital names were preannotated using regular expressions with the annotation tool Ellogon (http://www.ellogon.org/index.php/annotation-tool). All dates within each document were automatically modified by a fixed number of days to keep chronological order of events. The number of days was chosen randomly for each document.
Annotation scope
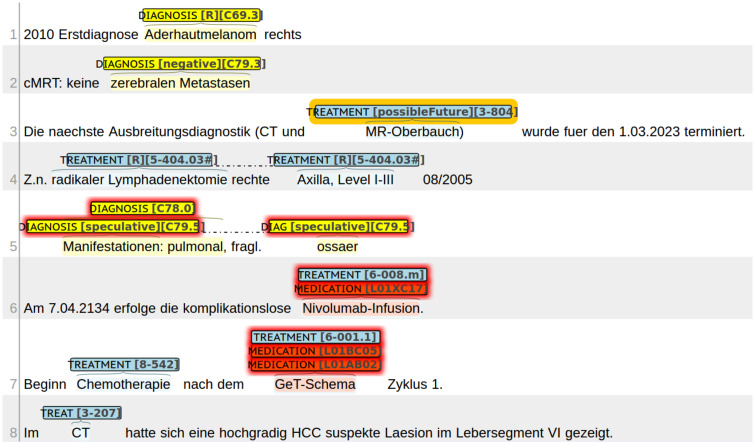
At first, we annotated section headings in all discharge summaries (see Supplementary Material for details). In specific sections, we annotated medical entities that are particularly important for the disease history of cancer patients, namely: diagnosis, treatment, and medication. As a common practice in NLP research, by “medical entities,” we mean linguistic entities, that is, word or phrases that designate objects or processes relevant in health, including expressions clinicians use to describe patient-related matters. For terminology grounding (normalization) of medical entities, we utilized terminologies commonly used in clinical practice in Germany. A diagnosis is a disease, a symptom or a medical observation that can be matched with the German Modification of the International Classification of Diseases (ICD10; www.dimdi.de/dynamic/de/klassifikationen/icd/icd-10-gm/). A treatment is a diagnostic procedure, an operation or a systemic cancer treatment that can be found in the Operationen und Prozedurenschlüssel (OPS; www.dimdi.de/dynamic/de/klassifikationen/ops/). A medication names a pharmaceutical substance or a drug that can be related to the Anatomical Therapeutic Chemical Classification System (ATC; www.dimdi.de/dynamic/de/arzneimittel/atc-klassifikation/). Examples for each entity type are shown in Figure 1. Whenever applicable, medical entities were annotated with laterality (right, left, and both sided), negation (e.g., a diagnosis is ruled out or a medication is paused), speculation (e.g., a diagnosis is unclear), or whether it is expressed as a possible future event (e.g., a procedure is planned for the future). Examples for each attribute are shown in lines 1–5 in Figure 1. We defined a number of rules in our annotation guideline (available on the BRONCO website) to clarify any ambiguous situation we encountered in our corpus. These rules are shown in Supplementary Appendix A.
Figure 1.
Exemplary excerpts from original discharge summaries and annotations, shown in BRAT visualization. Attributes in brackets have the following meaning: laterality right (R), negated entity (negative), speculative entity (speculative), and entity possible in the future (possibleFuture). Additionally, codes resulting from entity normalization are given in brackets.
Annotation process
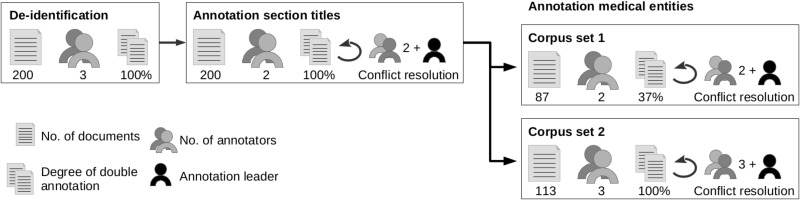
The annotation process was conducted by an annotation leader who prepared documents for annotation, developed annotation guidelines together with the medical experts, and organized conflict resolution but did not perform any annotations. For organizational reasons, annotations were performed by 2 groups of annotators, group A (2 medical experts) and group B (3 medical experts and 3 medical students). Annotation guidelines were developed by the annotation leader and group A using 9 documents (see Supplementary Appendix B); adaptations required by situations encountered only later were possible. An overview of the complete annotation process is illustrated in Figure 2. Technically, we used the Brat Rapid Annotation Tool (BRAT).18
Figure 2.
Annotation procedure including deidentification, annotation of section titles, and annotation of medical entities with attributes. Altogether, 1 annotation leader and 9 medical annotators were involved in different parts of the process.
Group A annotated 87 documents, of which 32 documents were double annotated for quality control. Differences in annotations were discussed with the annotation leader and resolved based on the guidelines and mutual agreement. To speed up annotation, we preannotated 59 documents with frequently annotated phrases, such as “CT Thorax/Abdomen/Becken” (computed tomography of thorax, abdomen, and pelvis), using exact matching. Annotators had to check and correct preannotations.
Group B annotated 113 documents. Here, we used a different procedure because medical students performed differently well during training. First, 3 medical students double annotated all documents without preannotations. Then the 3 medical experts of group B resolved conflicts using BRAT as shown in Figure 3. Training of annotators and considerations that lead to this procedure are described in Supplementary Appendix B.
Figure 3.
Visualization of mismatches between annotations of 2 annotators, shown in BRAT visualization. (A) One of the annotations misses Laterality R and (B) “Oberbauchsonographie” (sonography of the upper abdomen) is annotated only by 1 annotator and “Ausschluss von Leberfiliae” (exclusion of liver metastasis) is annotated with different text spans and only once with attribute possibleFuture.
Interannotator agreement (IAA) is calculated as microaveraged phrase-level F1-score19 before conflict resolution. We used phrase-level IAA, because most diagnosis and treatment annotations comprise of multiple tokens like “hepatozelluläres Karzinom” (hepatocellular carcinoma). In such cases, phrase-level IAA is more suitable than token-level IAA as phrases with different boundaries are detected as disagreement and can be resolved during conflict resolution. We used average F1-score instead of Cohen’s κ as the number of negative (not marked) phrases is poorly defined.20
Corpus creation
After annotation, the corpus was split in 2 parts containing only annotated sections of 150 and 50 documents, referred to as BRONCO150 and BRONCO50, respectively. In each part, sentences were split based on punctuation. To avoid splitting sentence after abbreviations like “Z.n.” (condition after), we used a list of common German medical abbreviations retrieved from Wikipedia (https://de.wikipedia.org/wiki/Medizinische_Abk%C3%BCrzungen) as exceptions. As an additional measure against potential deanonymization, we randomly shuffled sentences within each part of BRONCO. Finally, we further split BRONCO150 into 5 sets for allowing reproducible cross validation.
We performed 2 analyses on BRONCO150 to evaluate the effect of shuffling sentences. First, we calculated similarity scores between sentences originating from the same document and those coming from different documents. Secondly, we tried to reconstruct the original documents through clustering of sentences. We performed hierarchical clustering to segment the sentences into 150 groups, that is, 1 group per original document. For each group, we measure from how many documents the sentences originate. For both analyses, we used cosine similarity over TF-IDF representations of the documents.
Baseline methods for information extraction
We developed baseline tools using state-of-the-art techniques for named entity recognition (NER), named entity normalization (NEN), and detection of negated and uncertain entities. Performances of all baselines were measured as microaveraged precision, recall and F1. To this end, both BRONCO corpora were tokenized and tagged with part-of-speech using JCORE models that have been trained on a closed German clinical corpus (FRAMED).21,22 Gold standard annotations were used to convert the corpora to IOB format.
Named entity recognition
We applied the conditional random fields (CRF) implementation CRFsuite23 and a bidirectional long short-term memory network with a final CRF layer (LSTM-CRF),24 respectively. For both, CRF and LSTM-CRF, we used default feature sets plus a number of further lexical and orthographic features. We also tested the impact of FastText embeddings trained on German Wikipedia articles.25 CRF and LSTM-CRF models were evaluated with 5-fold cross validation on BRONCO150 and trained on the full BRONCO150 corpus for evaluation on the held-out corpus.
Named entity normalization
We implemented a simple approach using a dictionary lookup with Apache Solr 7.5.0 (https://lucene.apache.org/) followed by a reranking of candidates using the inference method from.26 Additionally to the dictionaries used for annotation, we applied Rote Liste (Rote Liste, Service GmbH, 2/2019) for mentions of branded drug names. To evaluate NEN, gold standard entity annotations were extracted from the BRONCO corpora and subjected to normalization.
Negation and speculation detection
We applied NegEx,27 which detects negated and uncertain (speculated) entities using a list of trigger terms and rules for defining their scope. We applied the original list of German trigger terms from28 as well as an updated list from.29 For evaluation, all sentences and gold standard entity annotations were fed into NegEx.
More details on all applied methods are shown in Supplementary Appendix C.
RESULTS
We first report on the estimated quality and frequency of annotated entities in BRONCO and both subsets. Next, we study whether shuffling of sentences in BRONCO150 actually prevents reconstruction of documents. Finally, we report, separately for both parts of BRONCO, on baseline results for information extraction using state-of-the-art techniques.
Quality of annotations
The quality of annotation is measured separately for annotation and normalization of medical entities and attributes for both groups of annotators (A and B). Table 1 shows the IAA for all double annotated documents for group A (32 documents) and group B (113 documents). Annotation of text spans reaches high agreement for all medical entities in group A (IAA 0.81–0.94). Agreement for normalization is also high with IAA 0.73–0.90. For both levels of annotation quality, text span and normalization, agreement increases from treatment to diagnosis to medication. Also, the agreement between annotations of attributes is high, especially for negation and laterality with IAA of 0.81 and 0.75, respectively. Agreement is generally lower for group B: 0.66–0.87 for text spans, 0.47–0.75 for normalization, and 0.37–0.53 for attributes. Note that all conflicting annotations were manually resolved in the final BRONCO.
Table 1.
Interannotator agreement (IAA) calculated as microaveraged phrase-level F1 for 2 corpus sets annotated by 2 groups of annotators (A, B)
| Group A |
Group B |
|||||
|---|---|---|---|---|---|---|
| Annotation type | No. of instances | Text span | Code/attribute | No. of instances | Text span | Code/attribute |
| Diagnosis | 734 | 0.88 (0.94) | 0.84 | 2860 | 0.69 (0.79) | 0.54 |
| Treatment | 522 | 0.81 (0.93) | 0.73 | 1730 | 0.66 (0.77) | 0.47 |
| Medication | 300 | 0.94 (0.96) | 0.90 | 927 | 0.87 (0.92) | 0.75 |
| Laterality | 104 | – | 0.75 | 452 | – | 0.53 |
| Negation | 76 | – | 0.81 | 319 | – | 0.50 |
| Speculation | 81 | – | 0.69 | 288 | – | 0.44 |
| Possible Future | 37 | – | 0.68 | 244 | – | 0.37 |
Note: IAA was calculated before conflict resolution. For text spans, IAA is also given as (token level) Cohen’s κ in paratheses. Number of double annotated documents: group A (32) and group B (113).
Frequency of entities
Corpus statistics and frequencies of annotated entities for both parts of BRONCO as well as for the complete corpus are shown in Table 2. Overall, BRONCO contains 11 124 annotations of medical entities and 3118 annotations of attributes. Most frequent annotations are diagnosis (5245), followed by treatment (3866) and medication (2013). Judged by the number of unique instances (26–45% of all annotations), the vocabulary is quite versatile for each type of entity. Overall, 1256 medical entities (10%) are related to a specific laterality, and about 15% are either negated (630 entities), speculated (613 entities), or may possibly occur in the future (619 entities). Overall, 796 medical entities (7%) are noncontinuous.
Table 2.
Frequency of annotated medical entities and attributes in BRONCO and its 2 subsets, together with general statistics
| Annotation type | BRONCO150 | BRONCO50 | BRONCO complete | Unique instances |
|---|---|---|---|---|
| Diagnosis | 4080 | 1165 | 5245 | 2394 |
| Treatment | 3050 | 816 | 3866 | 1101 |
| Medication | 1630 | 383 | 2013 | 532 |
| Total medical entities | 8760 | 2364 | 11 124 | – |
| Laterality | 1033 | 223 | 1256 | |
| Negation | 503 | 127 | 630 | |
| Speculation | 474 | 139 | 613 | |
| Possible future | 479 | 140 | 619 | |
| Total attributes | 2489 | 629 | 3118 | |
| #Documents | 150 | 50 | 200 | |
| #Sentences | 8976 | 2458 | 11 434 | |
| #Tokens | 70 572 | 19 370 | 89 942 |
Note: Unique instances are the number of unique mentions within the complete corpus.
No reconstruction of documents
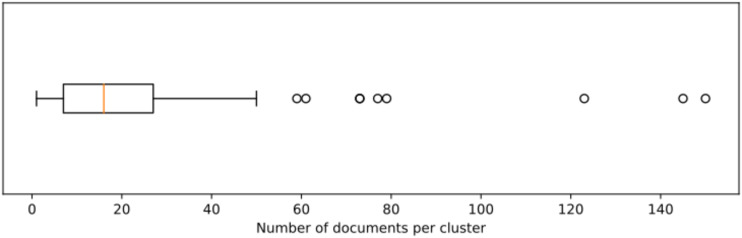
First, we compared the similarity of sentences in BRONCO150. Supplementary Figure S2 shows the distributions of pairwise similarities for sentences of the same (left) and different original documents (right) having at least 1 word in common. There is almost no difference regarding in- and cross-document sentence pairs. Furthermore, about 90% of all sentence pairs do not share a single word and therefore have zero similarity in the 1-hot encoding we applied here (note that these pairs were excluded to create Supplementary Figure S2, since otherwise the boxplots degenerate to flat lines). Furthermore, we studied how much a hierarchical clustering of sentences (with cutoffs to create 150 clusters) reconstructs the original documents. Figure 4 shows the distribution of numbers of documents per cluster. On average, clusters consist of 60 sentences originating from 22 different documents. There are only 3 clusters having sentences just from a single original document. These clusters contain only 10–13 sentences which cover 6–21% of a complete document. As there are also 18 clusters of similar sizes and similar average pairwise similarity between cluster members, we see no way of identifying pure (yet still very incomplete) clusters without knowledge of the original document.
Figure 4.
Distribution of documents per cluster after hierarchical clustering of sentences in BRONCO150.
Performance of IE baselines
Named entity recognition
We compared the performance of a CRF and a LSTM-CRF with and without using German (nonbiomedical) word embeddings. Results are listed in Table 3. On the BRONCO150 subset, the CRF approach outperforms LSTM-CRF by ∼3pp F1 on diagnosis, by ∼1pp on treatment, and by ∼2pp on medication; differences are very similar on the BRONCO50 subset. Word embeddings have only marginal impact on the CRF, but considerably improve performance of the LSTM-CRF approach (+5pp, +3pp, and +3pp for diagnosis, treatment, and medication, respectively). A notable difference exists between the results for diagnosis and treatment on BRONCO150 versus BRONCO50 for both approaches, where F1 scores are lower for the held-out part. We attribute this drop to the fact that results for BRONCO150 are obtained using cross-validation over randomly shuffled sentences, which means that sentences from the same document often are contained in the training and the test data. This increases the chances that individual entities of the test split already have been seen in the training data. Though this might be considered as a form of information leakage, we decided against creating the folds in BRONCO150 at the level of documents, as this would make document reconstruction easier and reidentification of individuals possible. Clearly, results for the BRONCO50 subset should be considered as more realistic.
Table 3.
Performance for baseline methods for NEN and NER (CRF and LSTM-CRF) with and without pretrained word embeddings (WE)
| Annotation type | Task | Method | BRONCO150 |
BRONCO50 |
||||
|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |||
| Diagnosis | NER | CRF | 0.80(0.01) | 0.71(0.02) | 0.75(0.02) | 0.79 | 0.67 | 0.72 |
| CRF+WE | 0.782(0.006) | 0.70(0.02) | 0.74(0.01) | 0.77 | 0.66 | 0.71 | ||
| LSTM | 0.75(0.03) | 0.69(0.03) | 0.72(0.01) | 0.78 | 0.65 | 0.71 | ||
| LSTM+WE | 0.81(0.08) | 0.74(0.08) | 0.77(0.08) | 0.79 | 0.65 | 0.72 | ||
| NEN | Dictionary lookup | 0.58 | 0.54 | 0.56 | 0.54 | 0.50 | 0.52 | |
| Treatment | NER | CRF | 0.86(0.02) | 0.78(0.01) | 0.82(0.01) | 0.83 | 0.73 | 0.78 |
| CRF+WE | 0.85(0.02) | 0.78(0.01) | 0.81(0.01) | 0.81 | 0.73 | 0.76 | ||
| LSTM | 0.83(0.04) | 0.79(0.03) | 0.81(0.02) | 0.85 | 0.69 | 0.76 | ||
| LSTM+WE | 0.85(0.06) | 0.82(0.07) | 0.84(0.06) | 0.76 | 0.74 | 0.75 | ||
| NEN | Dictionary lookup | 0.18 | 0.13 | 0.15 | 0.15 | 0.12 | 0.13 | |
| Medication | NER | CRF | 0.96(0.008) | 0.85(0.02) | 0.90(0.009) | 0.94 | 0.87 | 0.90 |
| CRF+WE | 0.96(0.004) | 0.84(0.009) | 0.90(0.006) | 0.95 | 0.85 | 0.90 | ||
| LSTM | 0.91(0.05) | 0.86(0.03) | 0.88(0.02) | 0.95 | 0.85 | 0.89 | ||
| LSTM+WE | 0.96(0.02) | 0.87(0.06) | 0.91(0.04) | 0.91 | 0.89 | 0.90 | ||
| NEN | Dictionary lookup | 0.66 | 0.68 | 0.67 | 0.64 | 0.69 | 0.66 | |
Note: Results for BRONCO150 are averaged over 5-fold with standard deviation in brackets. Best (highest) values per entity type, corpus, and w/o WE are bold.
Named entity normalization
We applied a dictionary lookup approach combined with a candidate reranking. Results are listed in Table 3. We find the best performance in terms of F1 for medication (0.67 and 0.66) followed by diagnosis (0.56 and 0.52) for BRONCO150 and BRONCO50, respectively. For treatment, performance only reaches F1 0.15 (BRONCO150) and F1 0.13 (BRONCO50).
Negation and speculation detection
We applied NegEx using 2 available lists of trigger terms, Chapman et al28 and Cotik et al.29 Using the Chapman list, negation detection reaches F1 0.44 (BRONCO150) and F1 0.37 (BRONCO50), as shown in Table 4. Speculation detection is worse. F1 only reaches 0.02 and 0.09 on BRONCO150 and BRONCO50, respectively. The recently published Cotik list improves results, but F1 scores nevertheless do not exceed 0.55 for negation and 0.33 for speculation detection in both corpora.
Table 4.
Negation and speculation detection of entities using NegEx with 2 lists of German trigger terms: Chapman et al28 and Cotik et al29
| BRONCO150 |
BRONCO50 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Annotation type | Trigger list | #GSC | P | R | F1 | #GSC | P | R | F1 |
| Negation | Chapman | 503 | 0.57 | 0.35 | 0.44 | 127 | 0.45 | 0.31 | 0.37 |
| Cotik | 503 | 0.62 | 0.50 | 0.55 | 127 | 0.52 | 0.55 | 0.54 | |
| Speculation | Chapman | 474 | 0.13 | 0.01 | 0.02 | 139 | 0.26 | 0.06 | 0.09 |
| Cotik | 474 | 0.54 | 0.24 | 0.33 | 139 | 0.71 | 0.22 | 0.33 | |
DISCUSSION
We present the BRONCO, a large and freely available corpus of German oncological discharge summaries. The corpus consists of shuffled sentences and is annotated with medical entities (diagnosis, treatment, medication) and their attributes (laterality, negation, speculation, possible in the future). Additionally, we developed baselines for NER, NEN, and negation and speculation detection and evaluated them on 2 subsets of the corpus. BRONCO150 will be published openly. Application of BRONCO is limited to sentence-level IE tasks. Nevertheless, we believe BRONCO can have a positive impact on German clinical NLP because it is the first sizable corpus that will become freely available.
Some previously built German medical corpora for IE exceed the size of BRONCO. For instance, 3000PA, created by large German research consortium (https://www.medizininformatik-initiative.de/), contains 3000 documents annotated with medication and related parameters.15 A subset of 3000PA annotated with diagnosis, findings, and symptoms contains 1.5M tokens.17 However, none of these is publicly available. The main reason for this situation undoubtedly is the uncertainty among researchers and data protection officers when a corpus can be given the status of being “fully anonymized,” as required by German and European regulations. We reacted in 3 ways to this issue: first, the corpus was completely manually deidentified. This process was confirmed by Charité and UKT data protection officers. Second, we only annotate and publish certain sections of the discharge summaries, avoiding all sections containing mostly biographic information. Third, we shuffled all sentences in the 2 subcorpora to blur their order and relationships. We performed an attempt to break this shuffling using sentence clustering and showed that it failed. Our method for this attempt has, however, limitations. The most important one probably is that in our 1-hot encoding words must appear syntactically identical to be matched between sentences, ignoring their semantics. One could try to overcome the limitation by using precomputed language models.25,30,31 However, extremely large and domain-specific corpora necessary to train good language models are either not available or kept closed. The same is true for any potentially existing language models.
BRONCO provides high-quality annotations (1) because in all double annotated documents (145 out of 200) conflicts were dissolved in a controlled process and (2) because all single annotated documents were annotated by persons that achieved high IAA with their peers for all levels of annotation. The IAA for entity annotation (0.69–0.88 for diagnosis, 0.66–0.81 for treatment, and 0.87–0.94 for medication) is comparable to previous annotations studies:15 achieved 0.88–0.99 for medication and17 reached 0.637 for diagnosis. Annotation studies on English clinical corpora are in the range of 0.7–0.88 (F1) or 0.73 (Cohen’s κ) for entities like disorder, procedures, or chemicals and drugs.8,32,33 As expected, normalization of entities was more difficult than merely finding entity mentions, especially for treatment concepts. Annotators were much less familiar with OPS (especially medical students) than with the other terminologies. Documentation officers, who create ICD10, OPS, and DRG coding as part of their professional activities, probably would have been a better fit for this task.
The 2-step annotation process we used for group B achieved the best balance between work time/cost and annotation quality. For comparable medical annotation projects, we therefore recommend the following procedure: First, annotations should be performed by persons, preferably more senior medical students, specifically hired for the annotation task. Every document should be annotated at least twice, and annotators are asked to highlight phrases where they are not sure how to proceed. In a second step, trained staff members only correct such phrases and conflicting annotations which significantly reduces the time they have to invest. The first step may include preannotations of frequently annotated terms to further speed up the process. However, this procedure certainly is a challenge for building truly large corpora containing thousands of documents.
Generally, recent years have shown that neural network based NER taggers outperform all other methods for biomedical texts, at least for English.34 In recent NER studies on German clinical corpora, CRF and LSTM-CRF methods have been used. A CRF and a character-level Bi-LSTM-CRF was trained for several types of medical entities, including medical condition, treatments and medications on 627 clinical notes from nephrology annotated with UMLS.35 Their F1-scores for treatment and medication are ∼5pp and ∼11pp worse for the CRF and ∼3pp and ∼2pp worse for the Bi-LSTM-CRF, when compared to BRONCO150 results (the precise setup for evaluation is not clear in the paper, but it certainly used a form of cross-validation. Therefore, we compare to BRONCO150 and not BRONCO50), though in their case the Bi-LSTM-CRF always outperformed the CRF. Their F1-scores for medical condition are ∼9pp and 13pp better for CRF and the Bi-LSTM-CRF, respectively. The rule-based system JUMEx extracted among other entities medication names from 3000PA reaching F1-scores of 0.65 compared to 0.90 for BRONCO50.15 On the Jena subset of 3000PA the CRF-based JCORE pipeline extracted among other entities diagnosis mentions with F1-score of 0.48 compared to 0.72 for BRONCO50.17 The latter 2 studies work on much larger corpora (more than 1.5M tokens) using 10-fold cross validation while for BRONCO150 we could only apply 5-fold. Additionally, 3000PA covers documents from a broader domain than BRONCO. Further progress in NER may be achieved by adding more fine-grained language models. For German medical texts, such models are not available, yet. However, it would be worth testing the German instance of the multilingual BERT language model.31
For NEN, we applied dictionary matching followed by a reranking of candidate terms. Results are mixed; whereas F1-scores for diagnosis and medication are somewhat encouraging (52% vs 66% on BRONCO50), the performance for treatments is very low (13%). These poor results can be related to the well-known vocabulary mismatch between the language of controlled vocabularies and the clinical jargon. Especially, OPS contains very complex concepts. Building interface terminologies may help to overcome this issue36,37 as well as making German translations of rich terminologies such as SNOMED-CT augmented with proper synonym sets accessible to the research community. Abbreviations are often specific within organizations and thus notoriously difficult to include in general terminologies. Additionally, tools for abbreviation resolution, like,38–40 might be worthwhile here to improve terminology grounding.
Negation and speculation detection using NegEx showed diverse results. We achieved the best performance using trigger terms from Cotik et al.29 F1-scores of 0.55 for negation can be considered as a promising basis for future improvements, yet a score of 0.33 for speculation is clearly not satisfying. Note that Cotik et al report F1-scores of 0.91 for negation and 0.55 for speculation on their corpus,29 indicating the still highly corpus-specific nature of the trigger term lists. To improve negation and speculation detection, one could either largely extend the list of trigger terms and their scopes, or adapt other tools like ConText. a more advanced version of NegEx currently available only for English.41 Also training of polarity models, as in,42 could be tested.
CONCLUSION
We provide the BRONCO, the first annotated German medical corpus freely available to the research community. This corpus offers the possibility to compare, evaluate, and train basic NLP tasks for the medical domain such as NER, NEN, and detection of different attributes of named entities.
FUNDING
This work was funded by the German Bundesministerium für Bildung und Forschung (BMBF), grants 031L0030B and 031L0023B, and the Deutsche Forschungsgemeinschaft, grant LE1428/1-1. D.T.R. is a participant in the BIH-Charité Clinician Scientist Program funded by the Charité – Universitätsmedizin Berlin and the Berlin Institute of Health.
AUTHOR CONTRIBUTIONS
U.L. and O.K. conceived the idea of the project. U.L., N.P.M., and U.K. supervised the work. M.K. organized the annotation process and performed data analysis, supported by M.S., H.H., J.Š., J.S., and M.H. M.L., D.T.R., I.J., G.R., M.Z., T.B., J.G., B.B., and L.O. anonymized and annotated the corpus. M.K. and U.L. wrote the manuscript with input from all authors.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this article (BRONCO150) will be shared on reasonable request and based on a data usage agreement. Please visit https://www2.informatik.hu-berlin.de/∼leser/bronco/index.html.
Supplementary Material
REFERENCES
- 1. Jensen PB, Jensen LJ, Brunak S.. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012; 13 (6): 395–405. [DOI] [PubMed] [Google Scholar]
- 2. Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc 2011; 18 (5): 540–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dernoncourt F, Lee JY, Uzuner O, et al. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 24 (3): 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu Z, Tang B, Wang X, et al. De-identification of clinical notes via recurrent neural network and conditional random field. J Biomed Inform 2017; 75: S34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stubbs A, Filannino M, Uzuner Ö.. De-identification of psychiatric intake records: overview of 2016 CEGS N-GRID shared tasks Track 1. J Biomed Inform 2017; 75: S4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 17 (3): 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hellrich J, Matthies F, Faessler E, et al. Sharing models and tools for processing German clinical texts. Stud Health Technol Inform 2015; 210: 734–8. [PubMed] [Google Scholar]
- 10. Starlinger J, Kittner M, Blankenstein O, et al. How to improve information extraction from German medical records. IT Inform Technol 2017; 59: 171–9. [Google Scholar]
- 11. Lohr C, Buechel S, Hahn U. Sharing copies of synthetic clinical corpora without physical distribution—a case study to get around IPRs and Privacy Constraints Featuring the German JSYNCC Corpus. In: proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018); May 7–12, 2018; Miyazaki, Japan.
- 12. Fette G, Ertl M, Wörner A, et al. Information extraction from unstructured electronic health records and integration into a data warehouse. In: INFORMATIK 2012 – Proceedings 42 Jahrestagung Der Gesellschaft Für Informatik (GI); September 16–21, 2012: 1237–51; Braunschweig, Germany.
- 13. Toepfer M, Corovic H, Fette G, et al. Fine-grained information extraction from German transthoracic echocardiography reports. BMC Med Inform Decis Mak 2015; 15: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roller R, Uszkoreit H, Xu F, et al. A fine-grained corpus annotation schema of German nephrology records. In: proceedings of the Clinical Natural Language Processing Workshop (ClinicalNLP). Osaka, Japan: The COLING 2016 Organizing Committee; 2016: 69–77.
- 15. Hahn U, Matthies F, Lohr C, et al. 3000PA-towards a national reference corpus of German clinical language. Stud Health Technol Inform 2018; 247: 26–30. [PubMed] [Google Scholar]
- 16. Lohr C, Luther S, Matthies F, et al. CDA-compliant section annotation of German-language discharge summaries: guideline development, annotation campaign, section classification. In: proceedings of the 2018 Annual Symposium of the American Medical Informatics Association. Data, Technology, and Innovation for Better Health; San Francisco, CA, USA; 2018. [PMC free article] [PubMed]
- 17. Lohr C, Modersohn L, Hellrich J, Kolditz T, Hahn U. An evolutionary approach to the annotation of discharge summaries. Stud Health Technol Inform 2020; 270: 28–32. [DOI] [PubMed] [Google Scholar]
- 18. Stenetorp P, Pyysalo S, Topić G, et al. BRAT: a web-based tool for NLP-assisted text annotation. In: proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics. San Diego, CA: Association for Computational Linguistics; 2012: 102–7.
- 19. Uzuner Ö, Solti I, Xia F, et al. Community annotation experiment for ground truth generation for the i2b2 medication challenge. J Am Med Inform Assoc 2010; 17 (5): 519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hripcsak G, Rothschild AS.. Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005; 12 (3): 296–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn U, Buyko E, Landefeld R, et al. An overview of JCoRe, the JULIE lab UIMA component repository. In: proceedings of the Workshop “Towards Enhanced Interoperability for Large HLT Systems: UIMA for NLP,” Marrakech, Morocco, May 31, 2008: 1–7.
- 22. Wermter J, Hahn U. An annotated German-language medical text corpus as language resource. In: proceedings 4th International Conference on Language Resources and Evaluation; May 24–30, 2004: 473–6; Lisbon, Portugal.
- 23. Okazaki N. CRFsuite: a fast implementation of conditional random fields (CRFs); 2007. http://www.chokkan.org/software/crfsuite/ Accessed April 3, 2018.
- 24. Lample G, Ballesteros M, Subramanian S, et al. Neural architectures for named entity recognition. In: proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. San Diego, CA: Association for Computational Linguistics; 2016: 260–70.
- 25. Mikolov T, Grave E, Bojanowski P, et al. Advances in pre-training distributed word representations. In: proceedings of the International Conference on Language Resources and Evaluation (LREC 2018); May 7–12, 2018: 52–55; Miyazaki, Japan.
- 26. Dogan R, Lu Z. An inference method for disease name normalization. In: AAAI Fall Symposium – Technical Report; November 2–4, 2012: 8–13; Arlington, Virginia.
- 27. Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 34 (5): 301–10. [DOI] [PubMed] [Google Scholar]
- 28. Chapman WW, Hillert D, Velupillai S, et al. Extending the NegEx lexicon for multiple languages. Stud Health Technol Inform 2013; 192: 677–81. [PMC free article] [PubMed] [Google Scholar]
- 29. Cotik V, Roller R, Xu F, et al. Negation detection in clinical reports written in German. In: proceedings of the Fifth Workshop on Building and Evaluating Resources for Biomedical Text Mining (BioTxtM2016); December 13–16, 2016: 115–24; Osaka, Japan.
- 30. Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. In: Workshop Proceedings of the 1st International Conference on Learning Representations; May 2–4, 2013; Scottsdale, AZ, USA.
- 31. Devlin J, Chang M-W, Lee K, et al. Bert: pre-training of deep bidirectional transformers for language understanding. In: proceedings of the 2019 Conference of the North American Chapter of the Association for Computer Linguistics: Human Language Technologies. Minneapolis, MN: Association for Computational Linguistics; 2019: 4171–86.
- 32. Wang Y. Annotating and recognising named entities in clinical notes. In: proceedings of the ACL-IJCNLP 2009 Student Research Workshop;August 2–7, 2009: 18–26; Suntec, Singapore.
- 33. Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013; 20 (5): 922–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Habibi M, Weber L, Neves M, et al. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 2017; 33 (14): i37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roller R, Rethmeier N, Thomas P, et al. Detecting named entities and relations in German clinical reports. In: International Conference of the German Society for Computational Linguistics and Language Technology; September 13–14, 2017: 146–54; Berlin, Germany: Springer.
- 36. Schulz S, Rodrigues J-M, Rector A, et al. Interface terminologies, reference terminologies and aggregation terminologies: a strategy for better integration. Stud Health Technol Inform 2017; 245: 940–4. [PubMed] [Google Scholar]
- 37. Schulz S, Hammer L, Hashemian-Nik D, et al. Localising the clinical terminology SNOMED CT by semi-automated creation of a German interface vocabulary. In: proceedings of the LREC 2020 Workshop on Multilingual Biomedical Text Processing (MultilingualBIO 2020). Marseille, France: European Language Resources Association; 2020: 15–20.
- 38. Kreuzthaler M, Oleynik M, Avian A, et al. Unsupervised abbreviation detection in clinical narratives. In: proceedings of the clinical natural language processing workshop (ClinicalNLP); December 13–16, 2016: 91–8; Osaka, Japan. [Google Scholar]
- 39. Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. In: Altman RB, Dunker AK, Hunter L, Jung TA, Klein TE, eds. Biocomputing 2003. Singapore: World Scientific; 2002: 451–62. [PubMed] [Google Scholar]
- 40. Wu Y, Denny JC, Trent Rosenbloom S, et al. A long journey to short abbreviations: developing an open-source framework for clinical abbreviation recognition and disambiguation (CARD). J Am Med Inform Assoc 2017; 24 (e1): e79–86–e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009; 42 (5): 839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu S, Miller T, Masanz J, et al. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLOS One 2014; 9 (11): e112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article (BRONCO150) will be shared on reasonable request and based on a data usage agreement. Please visit https://www2.informatik.hu-berlin.de/∼leser/bronco/index.html.