Abstract
We identified a novel interactome, circ_0001018/miR-338-3p/SOX4, in papillary thyroid cancer (PTC), and we intended to confirm the regulatory relationship between the three and to study the effects of the three in PTC. The bioinformatics method was used to screen out the circular RNA and mRNA of interest. A cellular fractionation assay and fluorescence in situ hybridization (FISH) assay were conducted to prove that circ_0001018 and CCT4 (the host gene of circ_0001018) mRNA primarily localized in the cytoplasm of PTC cell lines. By qRT-PCR analysis, the expression of circ_0001018 and SOX4 mRNA was found upregulated while the expression of miR-338-3p was found downregulated in PTC tissues and cells. circ_0001018 silence significantly inhibited the tumor growth in xenografted nude mice. A series of cytological experiments such as a Cell Counting Kit-8 (CCK-8) assay, a 5-ethynyl-2′-deoxyuridine (EdU) assay, cell cycle profiling, wound healing, a transwell assay, and cell apoptosis were conducted and showed that circ_0001018 and SOX4 promoted cell proliferation, migration, and invasion, inhibited cell apoptosis, and reduced the cell cycle arrest at the G1 phase in PTC cells. Compared with circ_0001018 and SOX4, miR-338-3p held the opposite function. The regulatory relationship between circ_0001018 and miR-338-3p, and between miR-338-3p and SOX4 mRNA, was validated using a luciferase reporter gene assay and/or RNA immunoprecipitation (RIP assay). Our findings showed that circ_0001018 acted as the tumor promoter via sponging miR-338-3p to elevate SOX4 expression level in PTC. Importantly, this novel circ_0001018/miR-338-3p/SOX4 axis has the potential to be considered as a therapy target for PTC.
Keywords: circ_0001018, miR-338-3p, SOX4, papillary thyroid carcinoma
Graphical Abstract
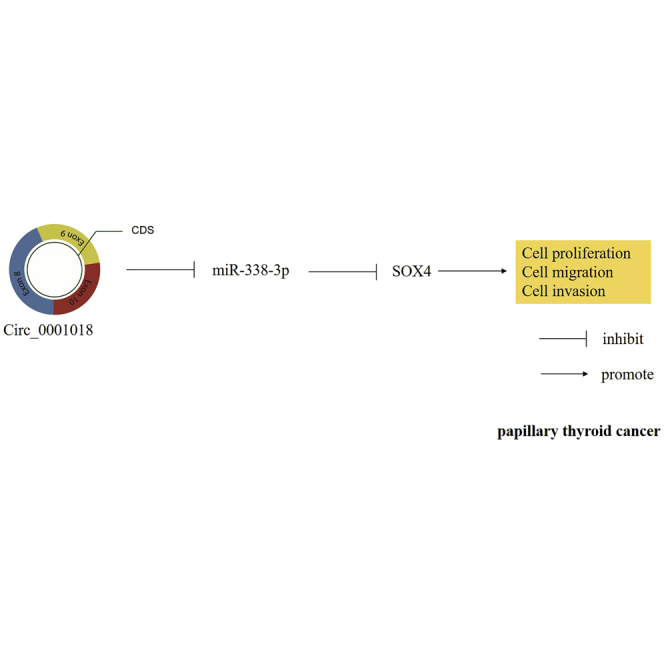
In present study, the authors’ findings showed that circ_0001018 acted as the tumor promoter via sponging miR-338-3p to elevate SOX4 expression level in papillary thyroid cancer (PTC). Importantly, this novel circ_0001018/miR-338-3p/SOX4 axis has the potential to be considered as a therapy target for PTC.
Introduction
Thyroid cancer is one of the most prevalent tumors with a gradual increase in morbidity and mortality, constantly threating the health of human beings for decades.1 The data on thyroid cancer occurrence, recurrence, and metastasis from the American Cancer Society2 show that more than 50,000 new cases of thyroid cancer are estimated to be diagnosed. Among them, about 14,260 cases are male and 37,810 are female, and, unfortunately, 2,170 of them fail to survive. In China, data from the National Cancer Center shows that about 22,000 males and 67,900 females are diagnosed with thyroid cancer each year.3 Papillary thyroid cancer (PTC), one of the subtypes of thyroid cancer, has dramatically increased in incidence since 2009 and become the most threatening subtype of thyroid cancer.4 Although traditional therapeutic methods could effectively treat PTC,5 several patients in advanced stage with metastasis still show poor prognosis outcome.6 The main reason is that the invasion and metastasis of PTC is uncontrollable.7 Therefore, it is of great significance to study the molecular mechanisms of PTC development, searching for novel therapeutic targets to conquer the disease.
Circular RNA (circRNA) is an elusive class of non-coding RNA with poor understanding about the mechanisms and functions.8 The latest research confirms that circRNAs are distributed in mammals to play important roles in multiple biological processes, such as cell growth, cell apoptosis, and cell proliferation.9, 10, 11, 12, 13 Interestingly, an increasing number of studies have found that circRNAs are abnormally expressed in tumor tissues, which is closely related to the development of cancer.13 Also, circRNA has been reported to be a sponge of microRNAs (miRNAs) to regulate the translation of downstream target mRNAs.14, 15, 16 In PTC, Cai et al.7 proved that circBACH2 was an oncogenic RNA in PTC by sponging miR-139-5p to increase the LMO4 expression. circ-ITCH impaired progression of PTC in vivo and in vitro via binding to miR-22-3p to promote CBL expression.17 miR-338-3p that is downregulated in a variety of cancer species has been reported to act as a cancer suppressor by binding to a variety of circRNAs such as circRNA MAT2B, circ_0000326, and circRNA HIPK3.18, 19, 20 In our previous study, we proved that miR-338-3p was downregulated in thyroid cancer and inhibited the progression of thyroid cancer21 so that we were eager to explore the upstream circRNAs sponging to miR-338-3p in PTC. By bioinformatics analysis, circ_0001018 (circBase: hsa_circ_0001018) is predicted to sponge miR-338-3p (miRBase: hsa-miR-338-3p), and the regulatory mechanism of circ_0001018 and the relationship between circ_0001018 and miR-338-3p are still barely investigated in PTC.
SRY-box transcription factor 4 (SOX4, GenBank: NM_003107.3) is a member of SOX (SRY-related HMG-box) family that encodes a number of transcription factors that might participate in embryonic development and cell apoptosis. SOX4 has been found to express in a large amount of tissues and to be involved in the development of the heart, B cells, and the reproductive system.22 Recently, a close relationship between SOX4 and tumorigenesis has attracted the attention of researchers in the field of oncogenesis.23,24 Min et al.25 reported that SOX4 could be a potential chemoprevention target in PTC, but no functional experiments were done to further prove their bioinformatics results.
In our research, circ_0001018 was prominently upregulated in PTC tissue samples, and its aberrant expression was found closely related with the phenotypic changes of PTC cells. Also, we found that circ_0001018 could sponge miR-338-3p to release the expression of its downstream effector SOX4, which then promoted PTC cell aggression. Hence, our research might reveal a novel clue for the clinical therapeutic strategies against PTC and may inspire creative thinking for subsequent scientific investigation.
Results
Identification, expression, and characterization of hsa-circ_0001018 in PTC
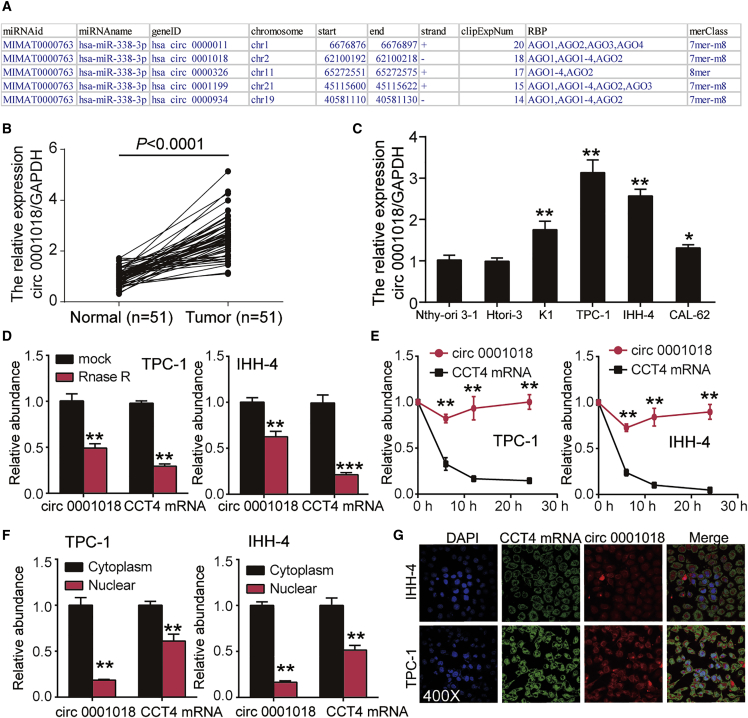
By interrogating ENCORI for the RNA interactomes (http://starbase.sysu.edu.cn/index.php) algorithm, we identified the top five most significant circRNAs based on Argonaute cross-linking and immunoprecipitation sequencing (AGO CLIP-seq) experimental evidence (Figure 1A). Among the top five circRNAs, hsa_circ_0000011 has two binding sites with miR-338-3p with an average context+ score of −0.147, and hsa_circ_0001018 has one binding site with miR-338-3p with a −0.158 context+ score. hsa_circ_0000326 and hsa_circ_0000934 have a non-exon-formed structure, and hsa_circ_0001199 has no parental gene. Then, we measured the level of expression of the top five circRNAs in PTC tissue and cells, and it was found that circ_0001018 was the most upregulated circRNA in 51 PTC tissues and cells compared with other cirRNAs (Figures 1B and 1C; Figures S1A–S1H). We then conducted a RIP assay and found that hsa_circ_0001018 was the most significantly pulled-down circRNA by miR-338-3p mimic in the presence of Argonaute 2 (Ago2) protein (Figure S2). Therefore, hsa_circ_0001018 was identified as our circRNA of interest. hsa_circ_0001018 (348 nt in length) was formed by the back-splicing of exons 8, 9, and 10 of the host gene CCT4 (Figure S3). Moreover, we divided the 51 PTC tissues samples into 26 cases of high circ_0001018 expression and 25 cases of low circ_0001018 expression according to the median of circ_0001018 expression, and we analyzed the correlation between circ_0001018 expression and clinical pathology in patients with PTC. The results revealed that high expression of circ_0001018 was found to be associated with tumor-lymph node-metastasis (TNM) staging, lymph node metastasis, and distant metastasis (Table S1). Because of the highest circ_0001018 levels in TPC-1 and IHH-4 cell lines (Figure 1C), these lines were chosen for subsequent experiments. The stability of circ_0001018 was first conformed using the RNase R method and half-life detection method: CCT4 mRNA nearly disappeared in the lysate after 20 h of incubation, while circ_0001018 stayed intact during the incubation (Figures 1D and 1E). The results confirmed that circRNA can be extremely stable whereas mRNA was particularly vulnerable. circRNAs consisting of exons have been found to be widely expressed in cytoplasm. We thus analyzed the abundance of circ_0001018 in the cytoplasm and nucleus in TPC-1 and IHH-4 cell lines using qRT-PCR assays and fluorescence in situ hybridization (FISH). CCT4 mRNA and circ_0001018 were both primarily distributed in cytoplasm (Figures 1F and 1G). In order to make clear the effect of circ_0001018 on CCT4 expression level, we designed and synthesized the vectors that could upregulate or downregulate circ_0001018 expression. Using qRT-PCR to measure transfection efficiency, circ_0001018 was found to be successfully regulated (downregulated by about 70% and upregulated by about 5-fold). Additionally, circ_0001018 short hairpin RNA (shRNA)1 did not knock down CCT4 expression whereas shRNA2 decreased CCT4 expression by 40% (Figure S1I). Similar to the results of qRT-PCR, a western blot assay showed that shRNA2 reduced the protein expression level of CCT4 by about 60% (Figure S1J). The design of shRNA should span the exon 8 and 10 junction that did not exist in the CCT4 mRNA, so that shRNA could regulate the CCT4 expression. The shRNA2 alters the expression of CCT4 mRNA and protein, and therefore we used circ_0001018 shRNA1 to perform the follow-up experiments and renamed it as sh-circRNA.
Figure 1.
hsa_circ_0001018 was highly expressed in PTC tissues and PTC cell lines
(A) An ENCORI for RNA interactomes (http://starbase.sysu.edu.cn/) algorithm was used to identify the top five most significant circular RNAs (circRNAs) based on the AGO CLIP-seq experiment support. (B) circ_0001018 levels were detected by RT-PCR in adjacent normal tissues (n = 51) compared with tumor tissue samples (n = 51); GAPDH was used as the standard reference. (C) circ_0001018 expression levels in PTC cell lines (K1, TPC-1, and IHH-4), a thyroid cell line (CAL-62), and human thyroid follicular epithelial cell lines (child Nthy-ori 3-1 cell line and parent Htori-3 cell line) were analyzed by RT-PCR; GAPDH was used as the reference. ∗p < 0.05, ∗∗p < 0.01, compared with the Nthy-ori 3-1 cell line. (D) RNase R analysis was used to determine the relative abundance of circ_0001018 and CCT4 linear mRNA in TPC-1 and IHH-4 cell lines. ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the mock group. (E) Variation of relative abundance of circ_0001018 and CCT4 mRNA in TPC-1 and IHH-4 cells during a 1-day incubation. ∗∗p < 0.01, compared with linear RNA level. (F) Distribution of circ_0001018 and CCT4 mRNA in cytoplasm and nuclear fraction in TPC-1 and IHH-4 cells. ∗∗p < 0.01, compared with cytoplasm; (G) The subcellular localization of circ_0001018 and CCT4 mRNA in cells was identified using a FISH assay. Original magnification, ×400. In (C)–(F), data indicate mean ± SD from at least three independent experiments and three repetitions where cellular experiments are indicated.
Silencing hsa_circ_0001018 inhibited tumor growth in vivo
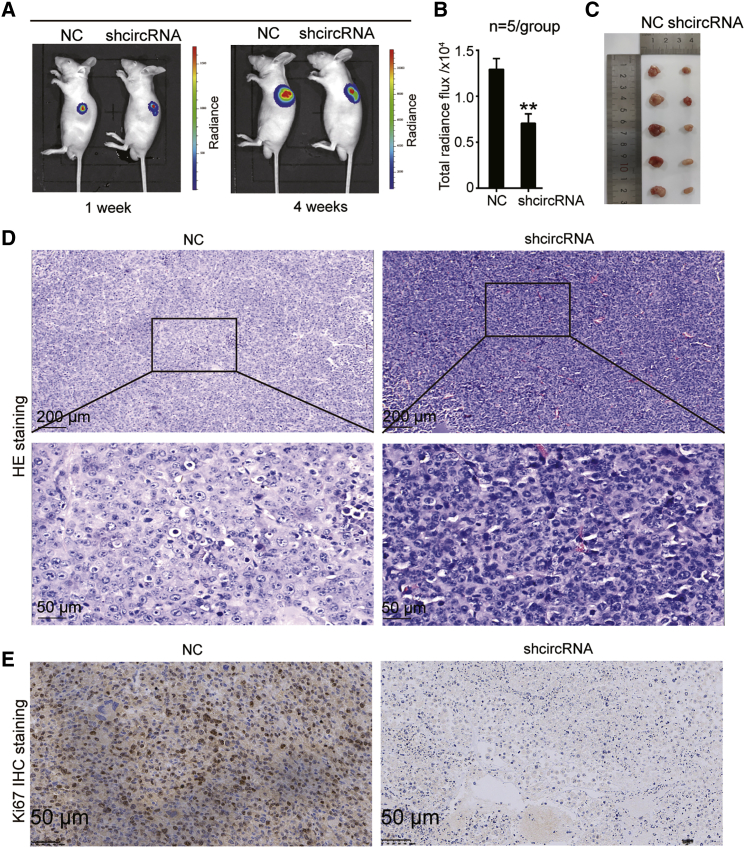
TPC-1 cells with circ_0001018 knockdown were injected into the left armpit of nude mice to assess the effect of circ_0001018 on tumor growth in vivo. The small animal live imaging showed that the total radiance flux was reduced by 45% in the circ_0001018 silencing group compared with the negative control (NC) group (Figures 2A and 2B). The xenografted tumors were also collected and their sizes were measured. The knockdown of circ0001018 resulted in smaller tumor sizes compared with the NC group (Figure 2C). Hematoxylin and eosin (H&E) staining showed that silencing circ_0001018 expression destroyed the structure of tumor tissues (Figure 2D). Ki67 immunohistochemical (IHC) assay results showed that cell proliferation was inhibited by 50% in the circ_0001018 knockdown group (Figure 2E). These data suggested that circ_0001018 knockdown inhibited tumor growth in vivo.
Figure 2.
Knockdown of circ_0001018 significantly suppressed the growth of tumors in vivo
(A) TPC-1 cells with empty vectors (NC) or circ_0001018 knockdown vectors (sh-circ_0001018) were injected into nude mice. The representative images were obtained using a small imaging system at week 1 and week 4. (B) The total radiance flux was monitored to represent tumor formation after the mice were injected (for 4 weeks). ∗p < 0.05, ∗∗p < 0.01, versus the NC group. Data indicate the mean ± SD. NC mice = 5, sh-circRNA mice = 5; repetition = 3. (C) Representative images of xenograft tumors obtained from six nude mice. (D) Representative images of H&E pathological staining. (E) The number of proliferating cells were detected by a Ki67 IHC assay. Data indicate the mean ± SD. NC mice = 5, sh-circRNA mice = 5; repetition = 3.
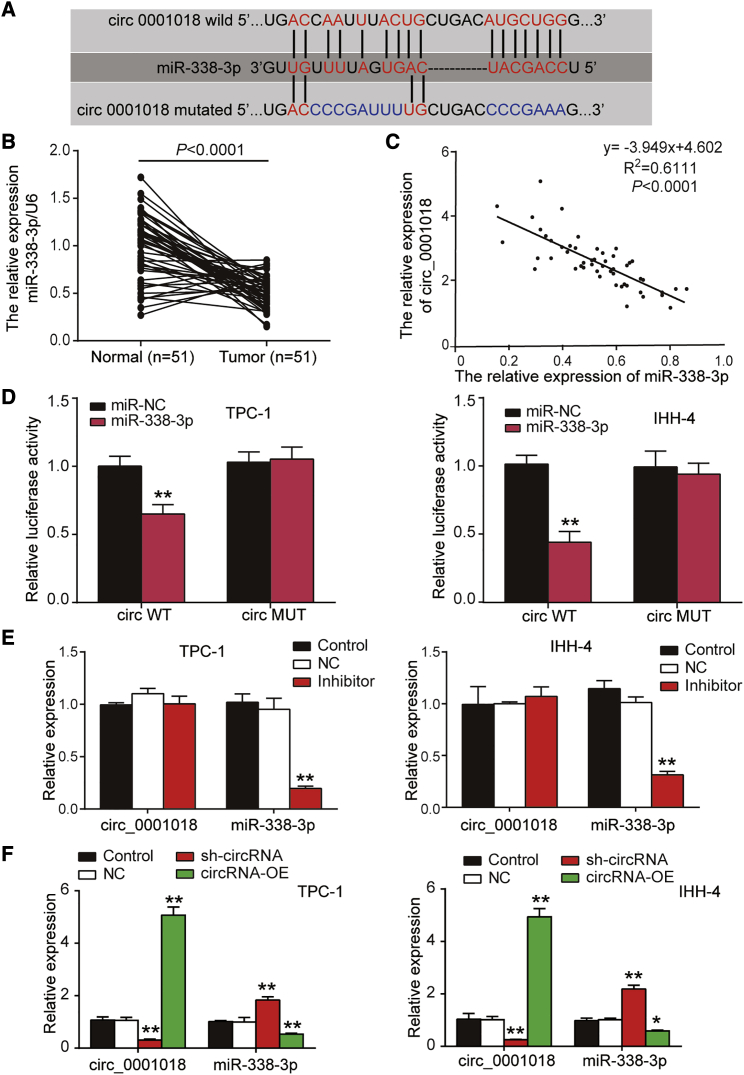
Validation of the regulatory relationship between miR-338-3p and hsa_circ_0001018
One binding site between miR-338-3p and circ_0001018 was predicted by the circRNA interactome (Figure 3A). Interestingly, the expression level of miR-338-3p was detected in healthy thyroid tissues and thyroid cancer tissues. In contrast to circ_0001018, miR-338-3p expression was lower in PTC tissues than in healthy tissues (0.52 ± 0.25 versus 1.07 ± 0.52) (Figure 3B), and the expression level of circ_0001018 showed a significant inverse correlation with the miR-338-3p expression level in PTC tissues analyzed by Pearson’s correlation analysis (Figure 3C). The luciferase activity was significantly suppressed by miR-338-3p mimics in the circ_0001018_wild-type group, but not in the circ_0001018_mut group. The luciferase activity was not affected by the miR-338-3p mimic NC (Figure 3D), suggesting that miR-338-3p directly targets wild-type circ_0001018 from sequence 286 to 292. We used anti-Ago2 RNA immunoprecipitation (RIP) and RT-PCR to analyze the enrichment of circ_0001018 in TPC-1 and IHH-4 cells. Much more enrichment of circ_0001018 was seen in the miR-338-3p mimics group with the presence of Ago2 than in the other groups (Figure S2), suggesting that circ_0001018 could sponge miR-338-3p with the presence of Ago2. A specific inhibitor for miR-338-3p was designed for downregulating the expression of miR-338-3p. RT-PCR results showed that the miR-338-3p inhibitor led to 70% downregulation of miR-338-3p, but it did not affect circ_0001018 expression (Figure 3E). Alternatively, circ_0001018 knockdown was realized by the transfection of circ0001018 shRNA: after transfection with sh-circ_0001018, the level of circ_0001018 was downregulated by 70% and that of miR-338-3p was elevated by approximately 2-fold compared with the NC group; however, circ_0001018 with an almost 5-fold increase and miR-338-3p with an almost 50% decrease were observed in the circ_0001018-overexpression (OE) group (Figure 3F).
Figure 3.
circ_0001018 targeted miR-338-3p in TPC-1 and IHH-4 cell lines
(A) Binding scheme between circ_0001018 and miR-338-3p is illustrated. The binding sites were predicted using a circRNA interactome database, which uses a TargetScan Human miRNA prediction algorithm. (B) miR-338-3p level was detected by RT-PCR in normal tissues (n = 51) compared with tumor tissues (n = 51). U6 was used as the reference. (C) The correlation between miR-338-3p and circ_0001018 expression in PTC tissues was determined by Spearman’s correlation analysis. (D) Relative luciferase activity was detected by a reporter assay in TPC-1 and IHH-4 cells. WT, wild-type of circ_0001018; MUT, mutated-type of circ_0001018. ∗∗p < 0.01, compared with the miR-NC group. (E) The expression of miR-338-3p and circ_0001018 was determined by RT-PCR in TPC-1 and IHH-4 cells transfected with miR-338-3p inhibitor. ∗∗p < 0.01, compared with the control group. (F) The expression of miR-338-3p and circ_0001018 was determined by RT-PCR in TPC-1 and IHH-4 cells transfected with sh-circRNA and circRNA overexpression vectors (circRNA-OE). ∗p < 0.05, ∗∗p < 0.01, compared with the control group. Data indicate the mean ± SD. n = 3, repetition = 3.
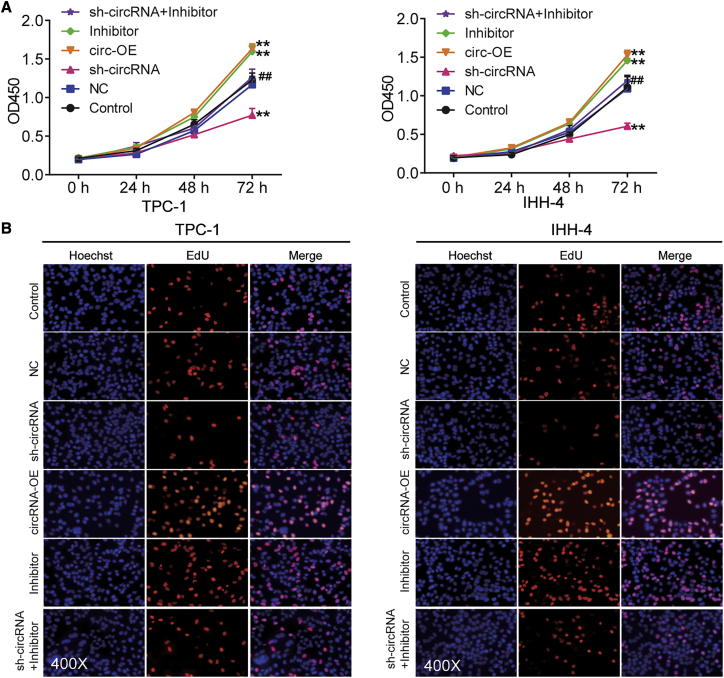
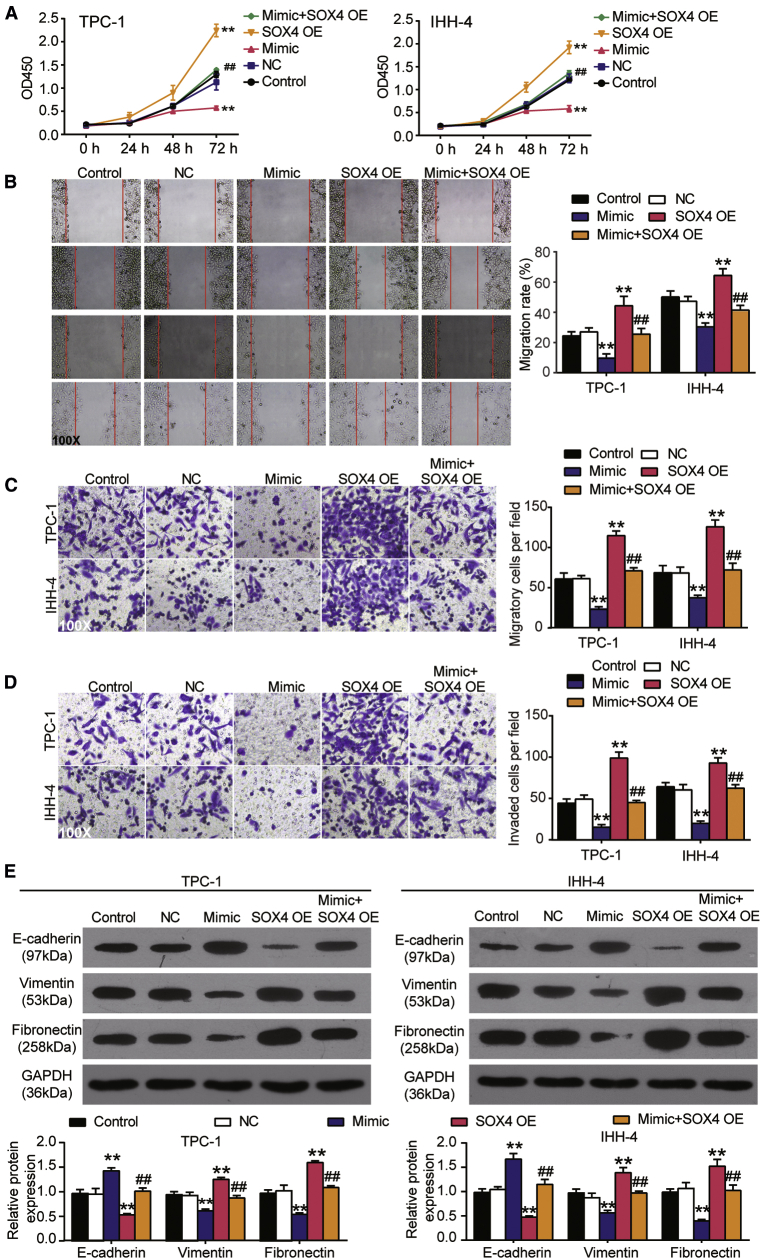
hsa_circ_0001018 promoted PTC malignancy phenotypes by sponging miR-338-3p in vitro
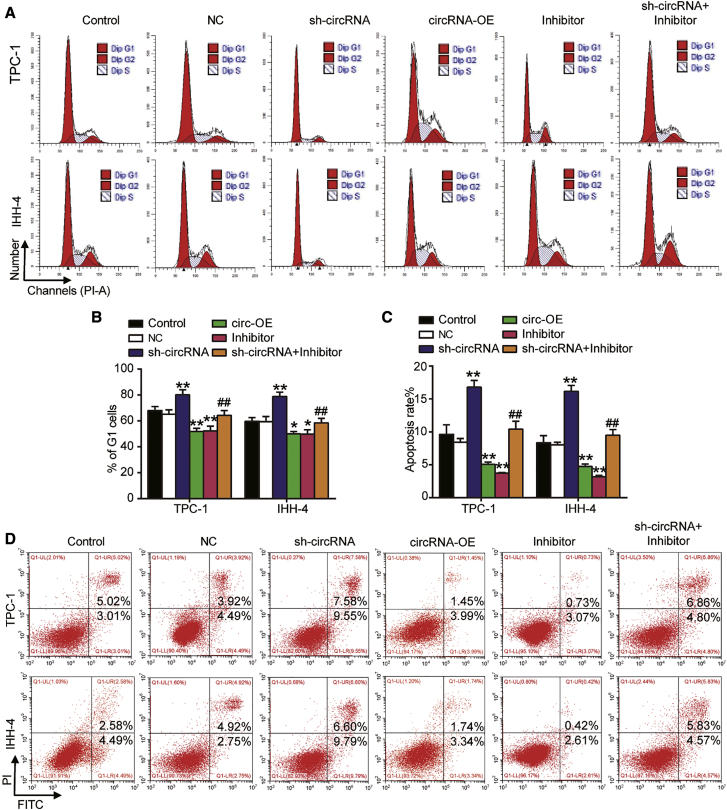
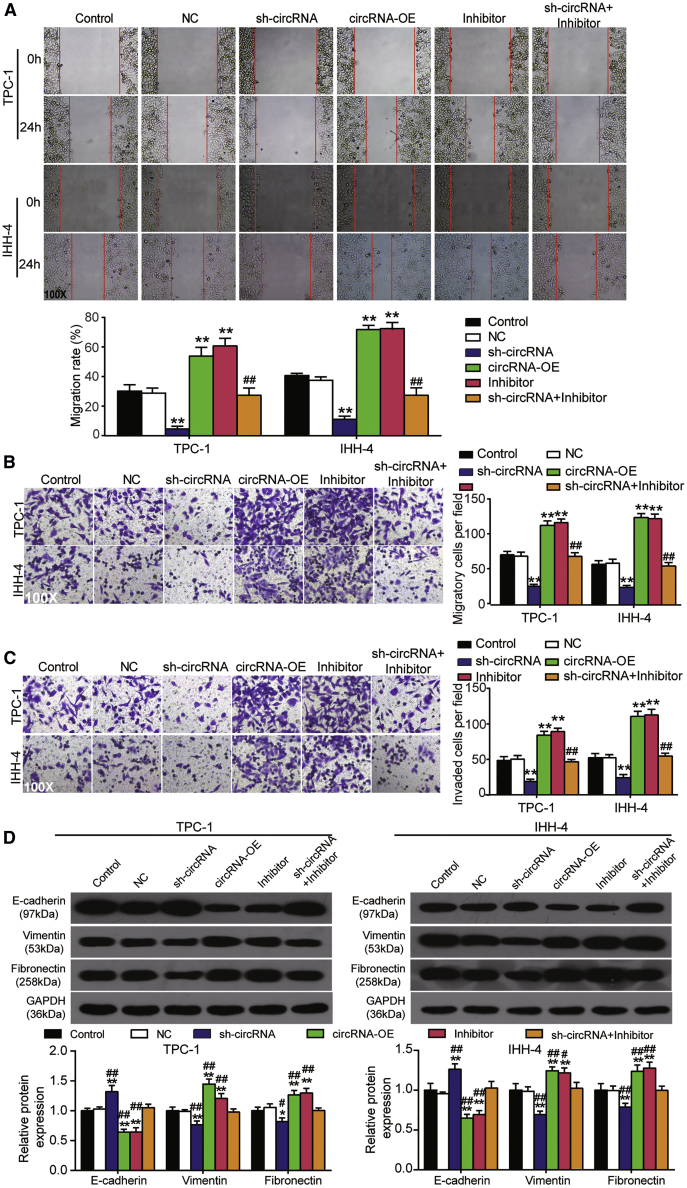
Cell survival was measured by a Cell Counting Kit-8 (CCK-8) assay in TPC-1 and IHH-4 cells transfected with circRNA overexpression vectors (circRNA-OE), sh-circRNA, miRNA inhibitor, or both: the overexpression of circ_0001018 led to enhanced cell survival at 72 h, silencing circ_0001018 weakened the survival, while miR-338-3p inhibition strengthened it in TPC-1 and IHH-4 cell lines. When both circ_0001018 and miR-338-3p were inhibited, the cell survival outcomes were similar to those of the control group (Figure 4A). The 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay proved that the upregulation of circ_0001018 enhanced DNA synthesis (reflecting proliferation), the knockdown of circ_0001018 inhibited DNA synthesis, while the inhibition of miR-338-3p promoted it in both cell lines (Figure 4B). DNA replication occurs in the G1 phase followed by cell division in nearly all eukaryotes. G1 phase arrest may suggest an impaired cell mitosis. In our experiments, cells in the G1 phase were profiled using flow cytometry analysis. We found that the upregulation of circ_0001018 led to a significantly decreased number of cells in the G1 phase, circ_0001018 knockdown led to an increased number of cells arrested in the G1 phase, whereas miR-338-3p inhibition led to an opposite result. The restore experiments showed the compromising results of knockdown of either circ_0001018 or miR-338-3p (Figures 5A and 5B). Overexpression of circ_0001018 significantly inhibited approximately 50% of cell apoptosis, silencing circ_0001018 dramatically increased apoptosis by nearly 2-fold, while miR-338-3p inhibition caused an opposite result in PTC-1 and IHH-4 cells. When the expression of circ_0001018 and miR-338-3p were both inhibited, the cells showed a similar apoptosis rate to the control group (Figure 5C and 5D). The wound-healing assay was used to evaluate cell migration ability. The result showed that the migration rate of the sh-circRNA group decreased by around 80%, while circRNA-OE or the miR-338-3p inhibitor increased the migration rate by approximately 1.8-fold compared with control group. At the same time, the miR-338-3p inhibitor partially eliminated the inhibitory effect of sh-circRNA on migration rate (Figure 6A). Furthermore, we conducted transwell migration and invasion assays to investigate the migration and invasion abilities of PTC cell lines. It was found that circ_0001018 upregulation resulted in a more than 50% higher cell migration and invasion rate, circ_0001018 knockdown resulted in an approximately 30% slower migration and invasion rate in PTC, while miR-338-3p inhibition caused an approximately 40% increase in migrated and invading cell numbers per field. The knockdown of both circ_0001018 and miR-338-3p restored the migration and invasion to a normal level (Figures 6B and 6C). Finally, the migration- and invasion-associated proteins (E-cadherin, vimentin, and fibronectin) were detected by immunoblot analysis. The results showed that overexpression of circ_0001018 and miR-338-3p inhibition reduced the protein level of E-cadherin and enhanced the protein level of vimentin and fibronectin, while circ_0001018 knockdown increased the protein level of E-cadherin and decreased the protein level of vimentin and fibronectin. Compared with control group, the levels of migration- and invasion-associated proteins in TPC-1 and IHH-4 cells with co-transfection of sh-circ_0001018 plus miR-338-3p did not show a significant difference (Figure 6D). These findings suggest that circ_0001018 could be a significant PTC promoter.
Figure 4.
circ_0001018 enhanced cell growth in TPC-1 and IHH-4 cells
(A) Cell survival was determined by a CCK-8 assay in TPC-1 and IHH-4 cells, in which circRNA-OE, sh-circRNA, miR-338-3p inhibitor, or sh-circRNA plus miR-338-3p inhibitor was transfected. OD450, optical density at 450 nm. Data indicate mean ± SD from at least three independent experiments and three repetitions. ∗∗p < 0.01, versus control; ##p < 0.01, versus sh-circRNA group. (B) An EdU assay was used to detect the cell proliferation in TPC-1 and IHH-4 cells, in which circRNA-OE, sh-circRNA, miR-338-3p inhibitor, or sh-circRNA plus miR-338-3p inhibitor was transfected.
Figure 5.
The knockdown of circ_0001018 induced cell cycle arrest and promoted cell apoptosis in TPC-1 and IHH-4 cells
(A and B) Cell cycle profiles of TPC-1 and IHH-4 cell lines were analyzed by flow cytometry. circRNA-OE, sh-circRNA, miR-338-3p inhibitor, or sh-circRNA plus miR-338-3p inhibitor was transfected. Percentage of cells in the G1 phase is visualized as a bar graph. (C and D) Cell apoptosis was detected using flow cytometry. circRNA-OE, sh-circRNA, miR-338-3p inhibitor, or sh-circRNA plus miR-338-3p inhibitor was transfected. The upper left (UL) quadrant represents necrotic cells, lower left (LL) quadrant living cells, right upper (UR) quadrant late apoptotic and necrotic cells, and the lower right (LR) quadrant represents early apoptotic cells. Data indicate mean ± SD from at least three independent experiments and three repetitions. ∗p < 0.05, ∗∗p < 0.01, versus control; ##p < 0.01, versus sh-circRNA group.
Figure 6.
hsa_circ_0001018 promoted cell migration and invasion in TPC-1 and IHH-4 cells
(A) A wound-healing assay was performed in TPC-1 and IHH-4 cells, in which circ_0001018 overexpression, sh-circ_0001018, miR-338-3p inhibitor, or sh-circ_0001018 plus miR-338-3p inhibitor was transfected. Bar graphs were drawn using the derived data. (B) A transwell migration assay was performed in TPC-1 and IHH-4 cells, in which circ_0001018 overexpression, sh-circ_0001018, miR-338-3p inhibitor, or sh-circ_0001018 plus miR-338-3p inhibitor was transfected. Bar graphs were drawn using the derived data. (C) Cell invasion was assessed using a transwell invasion assay. Bar graphs were drawn to show the number of invading cells in every group. (D) The protein levels of E-cadherin, vimentin, and fibronectin were detected by immunoblot analysis. Data are in the format of mean ± SD. n = 3, repetition = 3. ∗p < 0.05, ∗∗p < 0.01, versus control; #p < 0.05, ##p < 0.01, versus sh-circRNA group.
Identification of SOX4 as a significant downstream effector of miR-338-3p in PTC
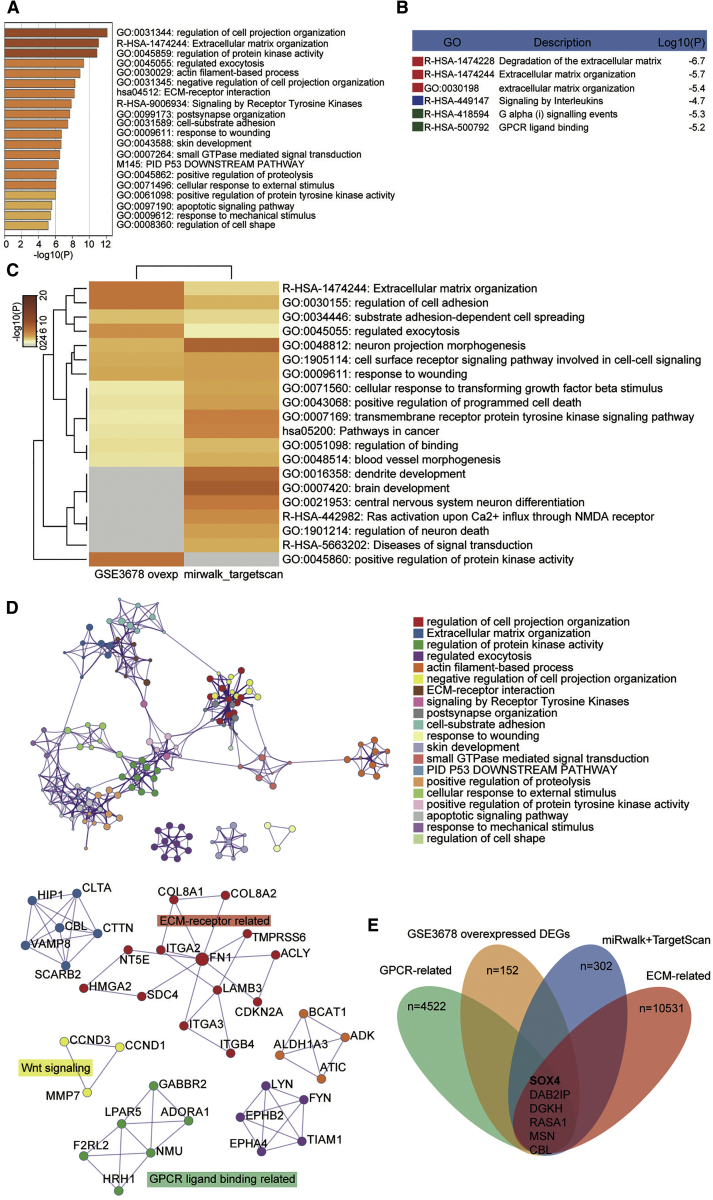
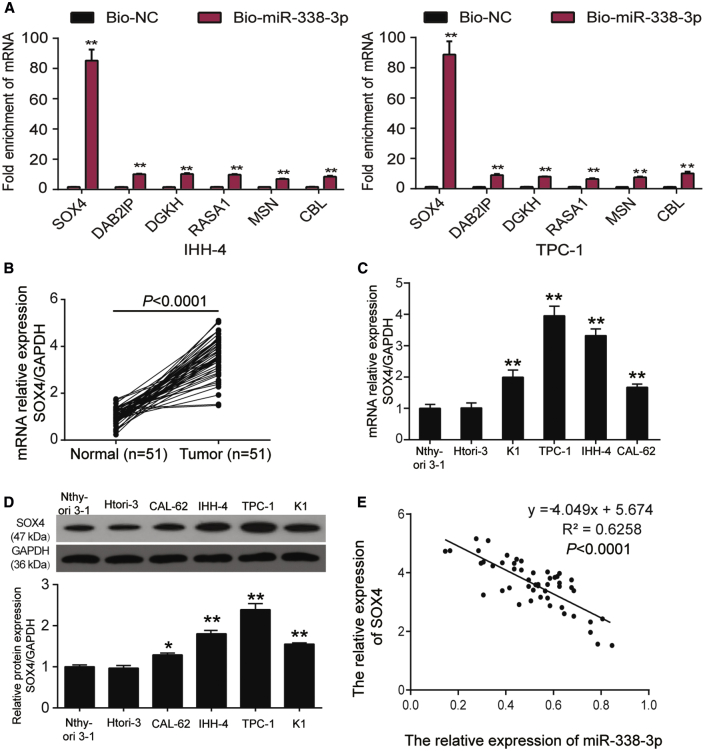
We have conducted bioinformatics analysis on GEO: GSE3678 data series to screen out the overexpressed DEGs (differentially expressed genes). We then uploaded these genes to Metascape, and we found that the significantly enriched Gene Ontology (GO) terms included extracellular matrix-related and G protein-coupled receptor (GPCR) ligand binding-related processes (Figures 7A and 7B). We then obtained the target genes of miR-338-3p from both TargetScan Human 7.2 and miRWalk databases. Subsequently, we ran a Metascape analysis of the two datasets: GEO: GSE3678 upregulated DEGs and the target genes list of miR-338-3p. We found that extracellular matrix and GPCR-related processes were again enriched (Figures 7C and 7D). We overlapped the GPCR-related gene list, the GEO: GSE3678 overexpressed DEGs list, target genes of miR-338-3p, and the extracellular matrix (ECM)-related genes list, finding out that SOX4, DAB2IP, DGKH, RASA1, MSN, and CBL could be potentially significant genes in PTC pathology (Figure 7E). We detected the enrichment of SOX4, DAB2IP, DGKH, RASA1, MSN, and CBL in TPC-1 and IHH-4 cells transfected with the biotinylated miR-338-3p (bio-miR-338-3p) by an RNA pull-down assay. The results displayed SOX4 with the highest enrichment in bio-miR-338-3p compared with other genes, which was about 85-fold higher (Figure 8A). At the same time, SOX4 was found limitedly reported in PTC in previous studies. Therefore, SOX4 was confirmed as our gene of interest to be investigated in PTC. We then acquired expression data of SOX4 in thyroid cancer from the GEPIA, GEO, and Oncomine databases. GEPIA results showed that SOX4 was significantly overexpressed in thyroid carcinomas (Figure S4A). In GEO: GSE3678 and GSE60542, the expression of SOX4 was significantly higher in tumors than in healthy tissues (Figures S4B and S4C). In GEO: GSE60542, advanced PTC (stages I, III, and IV; N1 stage) showed a significantly higher SOX4 expression level than that in healthy controls (Figures S4D and S4E). A significantly higher expression of SOX4 was observed in stage III PTC than in normal tissues; however, it was also found to be lower than in stage I PTC. Oncomine data mining also displayed that SOX4 was significantly higher in thyroid cancer than in the healthy tissues in Giordano thyroid statistics (Figure S4F). Also, in our collected samples, SOX4 mRNA was found at a higher level in thyroid tumor tissues than in the healthy thyroid tissues (Figure 8B). In selected cell lines, SOX4 was found significantly upregulated in cancer cell lines than in immortalized ones at both the mRNA and protein levels (Figures 8C and 8D). Furthermore, an inverse correlation between SOX4 and miR-338-3p expression in our collected PTC tissues was found using Pearson’s correlation analysis method (Figure 8E).
Figure 7.
SOX4 was identified to be a significant downstream effector of miR-338-3p in PTC
(A and B) GO term enrichment of GEO: GSE3678 overexpressed DEGs using a Metascape algorithm. (C and D) Comparison of GO term enrichment between GEO: GSE3678 overexpressed DEGs and target genes of miR-338-3p using a Metascape algorithm. The results are presented in a histogram (C) and an interaction network (D). (E) Overlap of GEO: GSE3678 overexpressed DEGs, GPCR-related genes, target genes of miR-338-3p, and extracellular matrix-related genes.
Figure 8.
SOX4 was detected to express more in tumor tissues than the normal
(A) The enrichment of SOX4, DAB2IP, DGKH, RASA1, MSN, and CBL using RT-PCR in the sample pulled down by biotinylated miR-338-3p. ∗∗p < 0.01, compared with biotinylated NC (Bio-NC). Data indicate the mean ± SD. n = 3, repetition = 3. (B) The relative expression of SOX4 mRNA was detected by RT-PCR in normal (n = 51) and tumor tissues (n = 51). GAPDH was used as the reference. (C) Relative expression of SOX4 mRNA in cell lines (Nthy-ori3-1, Htori-3, K1, TPC-1, CAL-62, IHH-4) was analyzed by RT-PCR. GAPDH was used as a standard. (D) Relative expression of SOX4 protein in cell lines (Nthy-ori3-1, Htori-3, K1, TPC-1, CAL-62, IHH-4) was analyzed by western blot. GAPDH was used as a standard. (E) The correlation between miR-338-3p and SOX4 mRNA expression in PTC tissues was determined by Spearman’s correlation analysis. ∗p < 0.05, ∗∗p < 0.01, compared with Nthy-ori 3-1. Data indicate the mean ± SD. n = 3, repetition = 3.
Validation of the complementary relationship between miR-338-3p and SOX4 mRNA
The putative binding scheme between the 3ʹ UTR of SOX4 and miR-338-3p was predicted using TargetScan Human 7.2 and is illustrated in Figure 9A. Wild-type and mutated-type SOX4 mRNA 3′ UTR luciferase reporter constructs, together with miR-338-3p mimic or miR-338-3p mimic NC, were transfected into TPC-1 and IHH-4 cell lines. The miR-338-3p mimic significantly inhibited approximately 50% of the luciferase activity of the wild-type but not the mutated-type SOX4 mRNA 3′ UTR luciferase reporter constructs (Figure 9B), suggesting that miR-338-3p targeted SOX4 mRNA directly. Before we further studied the effects of miR-338-3p and SOX4, we measured the transfection efficiency of SOX4 overexpression plasmids (SOX4-OE) and miR-338-3p mimic. miR-338-3p mimic transfection caused around a 4-fold increase of miR-338-3p expression (Figure 9C), around a 50% decrease of SOX4 mRNA expression (Figure 9D), and around a 50% decrease of SOX4 protein expression in the two cell lines (Figure 9E). The transfection of SOX4-OE caused an approximate 3-fold increase of SOX4 mRNA expression and a 1.5-fold increase of SOX4 protein expression. The co-transfection of miR-338-3p mimic and SOX4-OE did not show any significant differences of SOX4 mRNA and protein compared with the control group (Figures 9D and 9E). At this stage, we also detected SOX4 protein expression after cells were transfected with sh-circ_0001018. The results showed that silencing circ_0001018 downregulated the SOX4 protein expression by 50% in TPC-1 and IHH-4 cells, while miR-338-3p inhibition upregulated it by 1.25-fold in TPC-1 cells and 1.32-fold in IHH-4 cells. When miR-338-3p inhibition and circ_0001018 silencing occurred together, the expression of SOX4 protein showed a similar level with the control group (Figure 9F).
Figure 9.
miR-338-3p directly targeted SOX4 mRNA in TPC-1 and IHH-4 cell lines
(A) The complementary pairing between wild-type miR-338-3p and 3′ UTR of SOX4 is illustrated based on a TargetScan Human 7.2 algorithm. The mutated type of the SOX4 3′ UTR represents a mismatched pairing with wild-type miR-338-3p in three positions. (B) Relative luciferase activity was detected by a luciferase reporter assay in TPC-1 and IHH-4 cells. ∗∗p < 0.01, compared with miR-NC group, in which group the cells were transfected with only the transfection reagent. (C) The transfection efficiency of miR-338-3p mimics was determined by RT-PCR in TPC-1 and IHH-4 cells. U6 was used as the reference. (D) The expression of SOX4 mRNA was determined by qRT-PCR in TPC-1 and IHH-4 cells transfected with miRNA mimic, SOX4 overexpression plasmids, or co-transfected with both. GAPDH was used the reference gene. ∗∗p < 0.01, versus control; ##p < 0.01, compared with the mimic group. (E) The relative expression of SOX4 protein was detected by western blotting in TPC-1 and IHH-4 cell lines transfected with miRNA mimic, SOX4 overexpression plasmids, or co-transfected with both. GAPDH was used as the reference. ∗∗p < 0.01, versus control; ##p < 0.01, compared with the mimic group. (F) The expression of SOX4 protein in both cell lines transfected with circ_0001018 shRNA, miR-338-3p inhibitor, or both was detected using the immunoblotting method. ∗∗p < 0.01, versus control; ##p < 0.01, versus the sh-circRNA group. n = 3, repetition = 3. Data indicate mean ± SD. Statistical analysis was done using one-way ANOVA except for the luciferase assay, in which the Student’s t test was used. OE, overexpression; mimic, miR-338-3p mimic; sh-circRNA, circ_0001018 shRNA; inhibitor, miR-338-3p inhibitor.
SOX4 augmented PTC cell survival and mobility
We conducted a CCK-8 assay to measure the survival of cells with SOX4 overexpression and miR-338-3p upregulation. As shown in Figure 10A, the overexpression of SOX4 promoted cell survival dramatically in the two cell lines at 72 h, and miR-338-3p upregulation suppressed the survival to a significantly low level at 72 h. The co-upregulation of miR-338-3p and SOX4 led to significantly better survival than that in the miR-338-3p mimic group but significantly worse survival than that in the SOX4-OE group at 72 h. In the wound-healing experiment, the upregulation of miR-338-3p reduced the migration rate of PTC cells by about 60%, the upregulation of SOX4 increased the migration rate by about 1.8-fold in TPC cells and 1.3-fold in IHH-4 cells, and the upregulation of SOX4 partially reversed the inhibitory effect of miR-338-3p on the migration rate (Figure 10B). In transwell migration assays, the upregulation of miR-338-3p led to a 53% decrease and 42% decrease in the number of migratory cells in the TPC-1 and IHH-4 cell lines, respectively; the upregulation of SOX4 resulted in a 42% and 41% increase in the number of migratory cells in the TPC-1 and IHH-4 cell lines, respectively; additionally, the co-upregulation of miR-338-3p and SOX4 resulted in an approximately equal level with that in the control group (Figure 10C). In the transwell invasion assay, miR-338-3p mimic transfection weakened the invasion by approximately a third, whereas SOX4-OE transfection enhanced the invasion by around 2-fold in the two cell lines. The co-transfection of mimic and SOX4-OE resulted in an even level with the control group (Figure 10D). After detection of migration and invasion associated proteins, the protein level of E-cadherin increased by around 1.5-fold in the miR-338-3p mimic group and decreased by approximately 50% in the SOX4-OE group, while the protein levels of vimentin and fibronectin decreased by almost 50% in the miR-338-3p mimic group and increased by 1.3-fold in the SOX4-OE group. However, co-transfection of the miR-338-3p mimic and SOX4-OE did not significantly affect the protein levels of E-cadherin, vimentin, and fibronectin compared with the control (Figure 10E). From these results, we can conclude that the phenotype changes caused by the miR-338-3p mimic can be reversed by SOX4 overexpression, suggesting that miR-338-3p directly regulates SOX4, thus affecting PTC cell aggression.
Figure 10.
miR-338-3p inhibited cell migration and invasion in TPC-1 and IHH-4 cells, which was reversed by SOX4 overexpression
(A) Cell survival was determined by a CCK-8 assay (reflected by OD450 values) in TPC-1 and IHH-4 cells transfected with miRNA mimic, SOX4 overexpression plasmids, or both continuing for 3 days. (B) After transfection with a miRNA mimic, SOX4 overexpression plasmids, or both, a wound-healing assay was performed in TPC-1 and IHH-4 cells. (C) Cell migration was assessed by transwell migration assays, and miRNA mimic, SOX4 overexpression plasmids, or both were transfected into cells. Bar graphs were produced to illustrate the migratory cell numbers. (D) Cell invasion was assessed by transwell invasion assays. miRNA mimic, SOX4 overexpression plasmids, or both were transfected. (E) The protein levels of E-cadherin, vimentin, and fibronectin were detected by immunoblot analysis. n = 3, repetition = 3. Data indicate mean ± SD. Statistical analysis was conducted using a one-way ANOVA. All other groups were compared with the control group. ∗∗p < 0.01, versus control; ##p < 0.01, compared with mimic group.
Discussion
In our previous studies, we confirmed the tumor suppression effect of miR-338-3p in human PTC.21,26 In this study, we identified a novel downstream target gene, SOX4, and a novel upstream regulator, hsa_circ_0001018. We then studied the effects of circ_0001018, miR-338-3p, and SOX4 in PTC. We found that circ_0001018 enhanced tumor growth and tumor cell aggression by sponging miR-338-3p, which thereby released SOX4 mRNA. We thus concluded that the novel upstream regulator circ_0001018 and the downstream effector SOX4 could be potential oncogenes in PTC.
Aberrant miR-338-3p expression has been reported to affect the phenotypes of tumor cells in human cancers, e.g., thyroid cancer, liver cancer, gastric cancer, and neuroblastoma.27, 28, 29, 30 Specifically, Huang and colleagues27 once reported that miR-338-3p targeted SMO to suppress canceration of liver cancer cells, inspiring a novel therapy method for the treatment of liver cancer. Chen et al.28 reported that miR-338-3p suppressed cell propagation and mobility by regulating the expression of PREX2a. Together with our previous and current studies, miR-338-3p could be a tumor suppressor in PTC, and possibly a universal tumor suppressor in human cancers, which could be an interesting topic. In addition, miR-338-3p has been reported to interact with circRNAs in various cancers such as nasopharyngeal carcinoma,31 cervical cancer,20 hepatocellular carcinoma,32 colon cancer,33 lung cancer,19,34 and breast cancer.35 In our previous studies, we used TPC-1 cells stably expressing miR-338-3p to inject male BALB/mice and found that forced expression of miR-338-3p inhibited tumorigenesis.21 In all of the interactions, miR-338-3p was reported to be sponged by the circRNAs and acted as a tumor suppressor. We herein report a novel circRNA, circ_0001018, that also sponged miR-338-3p to promote PTC development. The role of circ_0001018 in human cancers has not been studied, including in PTC. In addition, we identified a novel downstream effector of miR-338-3p, SOX4. The study of the miR-338-3p-SOX4 axis has been reported in lung cancer,36 breast cancer,37 renal cell carcinoma,38 and idiopathic pulmonary fibrosis.39 To our knowledge, this is the first report of miR-338-3p and SOX4 in PTC. miR-338-3p fulfilled its tumor-suppressing function in PTC cells by suppressing SOX4. Our study contributes to the comprehension of the competing endogenous RNA (ceRNA) network that involves miR-338-3p in PTC.
An increasing number of circRNAs have been defined and reported, and many of them have been found to be important in tumor pathology; for example, some of the circRNAs have been correlated with prognosis of human PTC. For instance, Ren et al.40 reported that hsa_circRNA_047771 and hsa_circRNA_007148 might be potential prognostic biomarkers for PTC. Lan et al.41 reported that downregulation of hsa_circ_0137287 predicted aggression of papillary thyroid carcinoma. In addition, upregulation of hsa-circ_0067934 was correlated with thyroid carcinoma malignancy.42 Several other groups also reported a plethora of circRNAs to be potential PTC biomarkers.40,43, 44, 45, 46 Furthermore, there are other studies revealing that circRNAs can interfere with tumor cell activities by sponging miRNAs. A wide spectrum of circRNAs are cleaved and spliced by exons in the known protein-coding genes. Because of the back-splicing mechanism, circRNAs are rich in exposed exon sequences, allowing the complementary pairing of miRNAs, which therefore suppresses the activity of miRNAs.47,48 A previous study revealed that circ-ITCH overexpression in PTC tissues suppressed the proliferation and invasion of the cells while promoting apoptosis by sponging miR-22-3p and freed CBL mRNA.17 It was also reported that hsa_circ_0058124 knockout impaired proliferation and mobility, but induced apoptosis, in K1 and TPC-1 cell lines and in nude mice models by functioning as a ceRNA of miR-218-5p, therefore restraining NUMB mRNA in vitro and in vivo.49 hsa-circ_0025033 was also proven to facilitate cell survival, proliferation, and mobility progression, but it suppressed the apoptosis of TPC-1 and K1 cell lines by sponging miR-1231 and miR-1304.50 circ-ZFR contributes to TPC-1 and SW579 (a thyroid gland squamous cell carcinoma cell line) cell proliferation and invasion by sponging miR-1261, therefore releasing C8orf4 mRNA.51 circRNA_NEK6 was reported to promote TPC-1 cell proliferation by sponging miR-370-3p.52
These studies confirmed that circRNAs could be active facilitators of human PTC by sponging their target miRNAs. In this study, we found that circ_0001018 could act as a sponge of miR-338-3p, harboring the miRNA and thereby attenuating the suppressive function of miR-338-3p on SOX4 in TPC-1 and IHH-4 cell lines. The knockdown of circ_0001018 significantly suppressed tumor malignancy and tumor cell phenotypes.
In the last 5 years, SOX4 has been extensively studied in various human cancers, including oral cancer, breast cancer, ovarian cancer, and lung cancer, among others,53, 54, 55, 56 especially lung cancer. For instance, SOX4 was reported to be targeted by miR-363-3p to inhibit non-small cell lung cancer cell invasion.57 Zhong et al.58 reported in 2019 that SOX4 was upregulated by lncRNA OR3A4, potentially predicting poor clinical outcomes of patients with non-small cell lung cancer. However, SOX4 has not been studied in human papillary cancer. This research was thus conducted to fill this gap. We found that miR-338-3p directly targeted SOX4 mRNA and suppressed its expression. The forced overexpression of SOX4 induced cell survival, proliferation, and mobilities of TPC-1 and IHH-4 cell lines. From another perspective, when studying the SOX4-circRNA feedback loop, Ding’s team59 reported in 2019 that circ-DONSON promoted gastric cancer cell invasion by recruiting the NURF complex to the promoter region of SOX4, thus activating SOX4 transcription. In our study, SOX4 was in a ceRNA network that involves circ_0001018 and miR-338-3p: circ_0001018 promoted the neoplastic phenotypes of the PTC cells by sponging miR-338-3p, thus releasing SOX4 and resulting in an elevated SOX4 level. Taken together, we think that SOX4 can be a solid facilitator of PTC cancer genesis. Our study not only provided insights into the tumor-promoting role of SOX4 in PTC, but it also identified a novel ceRNA network involving SOX4 in PTC.
However, there are still some concerns that need to be addressed in the future. On the one hand, the study on the downstream signaling pathway of SOX4 could go deeper. On the other hand, commercial drugs are often used to treat PTC, such as cabozantinib60 and anlotinib.61 Therefore, it is important to explore the effect of the circ_0001018/miR-338-3p/SOX4 axis on the specific drugs for the treatment of PTC.
Conclusions
Our study demonstrated that circ_0001018 promoted tumor growth in vivo, and it enhanced the malignant phenotypes of PTC cells via sponging miR-338-3p to release SOX4. Our findings might provide a new target for the treatment of PTC.
Materials and methods
Clinical tissue samples
Clinical PTC and adjacent healthy tissues were collected from patients diagnosed with PTC in the China-Japan Union Hospital of Jilin University Hospital. No radiotherapy, chemotherapy, or other adjuvant therapies had been introduced. 51 pairs of tissue samples were collected followed by liquid nitrogen freezing and storage at −80°C. 51 PTC tissues and 51 adjacent healthy tissues were used for the analysis. The analysis of the tissue sample was authorized by the patients and approved by the Ethics Committee of the hospital. The baseline characteristics of all participants are presented in Table S1.
Cell lines and cell culture
Human PTC cell lines K1, IHH-4, and TPC-1, human thyroid gland undifferentiated carcinoma cell line CAL-62, as well as immortalized human thyroid follicular epithelial cell lines Nthy-ori 3-1 (child cell line) and Htori-3 (parent cell line) were used in this study. K1, IHH-4, TPC-1, Nthy-ori3-1, and Htori-3 cell lines were purchased from BNCC (Beijing, China). The CAL-62 cell line was purchased from the Chinese Academy of Sciences (Shanghai, China). The Nthy-ori3-1 cell line was cultured in RPMI 1640 medium with 10% fetal bovine serum (FBS). TPC-1, K1, CAL-62, and IHH-4 cell lines were kept in 90% DMEM-H + 10% FBS. The Htori-3 cell line was kept in 90% F-12K medium + 10% FBS. The TPC-1 cell line was derived from a differentiated thyroid gland papillary carcinoma,62 and it harbored the RET/PTC1 gene rearrangement.63 The TPC-1 cell line has been confirmed to be a thyroid cancer cell line that represents PTC.64,65 It is a highly characterized model for thyroid cancer research. The IHH-4 cell line was from a metastasis (site: left cervical lymph node) of a differentiated thyroid gland papillary carcinoma, and it is highly metastatic.66 In addition, previously published work has regularly used these two cell lines in diverse pathogenesis and progression studies.49,67, 68, 69, 70, 71, 72
Nude mice xenograft assay (a live imaging method with tumor tissue pathology analysis)
Male BALB/c nude mice (at 4 weeks of age) were purchased from Beijing Vital River Laboratory Animal Technology and randomly assigned to the sh_circRNA_0001018 group (n = 5) and NC group (n = 5). TPC-1 cells stably transfected with sh-circRNA_0001018 or empty vector were suspended in PBS. 100 μL of cell suspension (approximately 1 × 108 cells/mL) was injected into the left armpit of the nude mice. A small animal imaging system was used to monitor the tumor growth in a 4-week-long time course. Fluorescence images were obtained at week 1 and week 4. Finally, the nude mice were killed at week 4, and the tumors were resected and subjected to H&E pathologic analysis and Ki67 IHC staining. Experiments were approved by the Institutional Animal Care and Use Committee of the hospital.
Luciferase reporter gene assay
The complementary relationship between hsa_circ_0001018 and miR-338-3p was predicted using the ENCORI for RNA interactomes algorithm. TargetScan Human 7.2 and miRWalk algorithms were used to predict the complementary sequences between miR-338-3p and SOX4 mRNA. To construct luciferase reporter vectors, the sequences of circ_0001018 and the SOX4 mRNA 3′ UTR that are complementary to miR-338-3p were amplified by PCR and cloned into the pmirGLO Dual-Luciferase vector. TPC-1 and IHH-4 cells were transfected with approximately 50 ng of the corresponding vectors together with approximately 25 ng of miR-338-3p mimic or the mimic control. Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) was used for transfection. The Dual-Luciferase reporter assay system (Promega, Fitchburg, WI, USA) was used to detect the luciferase activity 2 days afterward. The ratio of firefly to Renilla luciferase was calculated.
Cell transfection
circ_0001018 shRNA, circ_0001018 overexpression vectors, SOX4 overexpression vectors, miR-338-3p mimic, and miR-338-3p inhibitor were purchased from GenePharma (Shanghai, China). For the construction of the circ_0001018 overexpression and SOX4 overexpression vectors, the full length of circ_0001018 and SOX4 was amplified by PCR, and the primers containing BamHI and XhoI sites were designed. Next, the full length of circ_0001018 and the full-length of SOX4 were inserted into pLCDH-cir (GenePharma, China) and pcDNA3.1 (GenePharma, China), respectively, at the BamHI and EcoRI sites. Finally, puromycin was used to select the transfected cells. As for circ_0001018 shRNA, two shRNAs for knockdown of circ_0001018 were designed, synthesized, and inserted between the BamHI and EcoRI sites of the pLV-CMV-puro-U6-shRNA lentiviral vector. Puromycin also used to screen out the transfected cells. The sequences of circ_0001018 shRNA (sh-1 and sh-2), circRNA-OE, miR-338-3p mimic, and miR-338-3p inhibitor are given in Table S2. The cell transfection was performed using circ_0001018 shRNA (sh-1 and sh-2), circ_0001018 overexpression vectors, SOX4 overexpression vectors, miR-338-3p mimic, and miR-338-3p inhibitor. After transfection for 48 h using Lipofectamine 3000 (Invitrogen), the transfected cells were collected to assess the transfection efficiency by qRT-PCR.
Quantitative PCR analysis
TRIzol reagent (Invitrogen) was used for RNA isolation from tissues and cell lines. The SuperScript IV first-strand synthesis system (Thermo Fisher Scientific, USA) was used for cDNA synthesis from 1 μg of total RNA. A One Step PrimeScript RT-PCR kit (TaKaRa, Tokyo, Japan) was mixed with cDNA for fluorescence quantitative PCR. The primer sequences are shown in Table S3. circ_0001018 was amplified from cDNA by divergent primers (an expected 85-bp fragment).
Half-life detection
To detect the half-life of circ_0001018 and its linear mRNA counterpart, the TPC-1 and IHH-4 cells were incubated with 5 μg/mL actinomycin D (ActD) (a transcription inhibitor) for 0, 10, 20, and 30 h. After incubation, the cells were collected, and the total RNA was isolated from the collected cells using TRIzol reagent. The half-life of circ_0001018 and its linear mRNA was detected using qRT-PCR.
Immunoblot analysis
50 μg of protein samples from lysed cells went through 10% SDS-PAGE before transfer to nitrocellulose membranes. The membranes were blocked with skim milk for 2 h. Thereafter, the primary antibodies against SOX4 (1:1,000, catalog no. ab80261, Abcam, UK), E-cadherin (1:1,000, catalog no. ab76055, Abcam, UK), vimentin (1:1,000, catalog no. ab925475, Abcam, UK), fibronectin (1:1,000, catalog no. ab24135, Abcam, UK), CCT4 (1:1,000, catalog no. 21524-1-AP, Proteintech, IL, USA), and GAPDH (1:1,000, catalog no. ab1816025, Abcam, UK) were incubated with the membranes overnight, and horseradish peroxidase (HRP)-conjugated secondary antibodies were used for subsequent incubation. Immunocomplex visualization was enhanced using enhanced chemiluminescence (ECL) reagent (Applygen, Beijing, China).
Cell fractionation
circ_0001018 and CCT4 mRNA abundance was detected in fractioned cytoplasm and nucleus. Most of the circ_0001018 and CCT4 mRNA was then localized. Nuclear and cytoplasmic RNA were extracted from cells using a cytoplasmic and nuclear RNA purification kit (Invitrogen, CA, USA). Briefly, TPC-1 and IHH-4 cells were incubated with lysis solution on ice for 10 min. Then, the cells were centrifuged at 12,000 × g for 3 min. Cytoplasmic RNA from the supernatant and nuclear RNA from the nucleus were collected and purified. GAPDH (primarily detected in the cytoplasmic fraction) and U6 (primarily detected in the nuclear fraction) were used as the endogenous controls for the cytoplasmic and nuclear fraction, respectively.
FISH assay
The FISH assay was used to confirm the subcellular localization of circ_0001018 and linear mRNA (CCT4) using a FISH kit (RiboBio, China) according to the instructions with the kit. Briefly, the FISH probes targeting the back-splicing site of circ_0001018 were designed and labeled by Cy5, and the FISH probes of linear mRNA (CCT4) were designed and labeled by fluorescein isothiocyanate (FITC). The TPC-1 and IHH-4 cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 for 10 min. Then, the cells were hybridized with Cy5-labeled FISH probes overnight. After hybridization, the cells were washed using saline sodium citrate (SSC) solution for 5 min and stained with DAPI for 10 min without light. Finally, the images were acquired by a fluorescence microscope.
RIP assay
A RIP assay was performed in TPC-1 and IHH-4 cell lines 48 h after transfection with miR-338-3p mimic or mimic control. A Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) was chosen for the experiment. Briefly, 1 × 107 cells were lysed in RNA lysis buffer, and magnetic beads were added to the lysate in RIP buffer. The magnetic beads were conjugated to human anti-Ago2 antibody or mouse immunoglobulin G (IgG) (Millipore). IgG was used as the control. Proteinase K was added to the samples. Precipitated RNA was released and collected. The enrichment of circ_0001018 and circ_0000011 was respectively examined using RT-PCR. The divergent primers were provided in Table S2.
RNA pull down
The RNA pull-down assay was carried out according to the previous study.73 bio-miR-338-3p and bio-NC (Ribobio, Guangzhou. China) were introduced into TPC-1 and IHH-4 cells. 48 h later, the cells were collected, washed with PBS, and incubated with lysis buffer (Thermo Fisher Scientific, DE, USA). The lysate was then placed in streptavidin beads (Thermo Fisher Scientific, DE, USA) and pretreated with RNase-free bovine serum albumin (BSA) and yeast tRNA at 4°C for 2 h. Next, a pull-down test was performed using a magnetic stent (Promega, Fitchburg, WI, USA). The RNA complex bound to the beads was eluted and extracted using TRIzol for the RT-PCR analysis.
Cell survival assays
CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan) was used to analyze cell survival. The two cell lines were seeded and incubated in 96-well plates and the density was about 5,000 cells per well. sh-circ_0001018 and/or miR-338-3p inhibitor, miR-338-3p mimic, and/or SOX4-OE were transfected into cells for 48 h and CCK-8 reagent (10 μL) was added at the time points of 0, 24, 48, or 72 h. An enzyme-labeling instrument (Thermo Fisher Scientific, USA) was used to measure the optical absorbance at 450 nm of every sample. The optical absorbance reflects the cell survival conditions.
EdU assay
An EdU detection kit (Ribobio, China) was used to assess cell proliferation. Briefly, TPC-1 and IHH-4 cells in the exponential phase were cultured in medium containing 50 μM EdU for 12 h at 37°C. Then, the cells were fixed in 4% paraformaldehyde and permeabilized using 0.5% Triton X-100 for 5 min. After the wash with PBS, the cells were stained with Apollo staining solution for 30 min, followed by counterstaining with Hoechst 33342 dye (Invitrogen, USA). Finally, the cells were observed under a fluorescence microscope.
Flow cytometry analysis
Cell apoptosis and cell cycle profiling were obtained using a flow cytometer. For cell apoptosis, an annexin V-FITC/propidium iodide (PI) cell apoptosis detection kit (BioLegend, USA) was used. 5 × 106 cells were collected and resuspended in 100 μL of binding buffer. Then, annexin V-FITC and PI (1:1 in volume) were added to the cells for 25 min in the dark. As for cell cycle profiling, a cell cycle assay kit (Vazyme, China) was used. Briefly, the transfected cells were resuspended in pre-cooling PBS and fixed in 75% anhydrous ethanol overnight. Next, after RNase digestion, the cells were stained with 20 μL of PI without light for 30 min. All apoptosis and cell cycle profiling data were obtained from the built-in software.
Wound-healing assay
A wound-healing assay was performed for analysis of migration rate in vitro. 5 × 105 cells/well (TPC-1 and IHH-4 cells) were cultured in a six-well plate and incubated overnight. After 90% cell confluence, the medium was replaced with the serum-free medium. After incubation for 12 h, the cells were scratched using 200-μL pipette tips. After removing the exfoliated cells with PBS, the fresh serum-free medium was added for 24 h of incubation. At 0 and 24 h after scratching, the images were obtained using an inverted microscope (Nikon, Japan), and the wound width was measured by Image-Pro Plus software (Media Cybernetics, USA).
Transwell migration and invasion assays
The migration and invasion abilities of both cell lines were assessed using a transwell assay. Initially, we seeded PTC cells in the upper chambers of every 8-μm transwell (Millipore, Billerica, MA, USA) with (invasion assays) or without (migration assays) the coating of Matrigel. For migration assays, the upper chamber was seeded with PTC cells (2 × 104 cells per well) in medium without serum while the lower chamber was added with 10% serum. Matrigel-coated chambers were used for invasion assays. A 24 h of incubation to allow the cells to migrate and 48 h to allow them to invade into lower chambers, the upper chambers were collected. PTC cells on the upper surface of the filter were removed, and the cells attached to the lower chambers were quantified under a microscope. We chose five random fields and counted the cell numbers in every field using an optical inverted microscope (Nikon, Tokyo, Japan).
Bioinformatics analysis
Datasets of GEO: GSE3678 and GSE60542 were downloaded from GEO database. The upregulated DEGs of GEO: GSE3678 were screened out using a threshold of an adjusted p <0.05 and a log fold changed (FC) >0. The upregulated DEGs as well as the overlapped target genes of miR-338-3p from TargetScan Human 7.2 and miRWalk databases were then imported into Metascape, thus showing the enriched GO terms of the two gene lists. In addition, we acquired expression data of SOX4 in thyroid cancer from GEPIA, GEO (GEO: GSE3678 and GSE60542), and Oncomine (Giordano thyroid statistics) databases.
Statistical analysis
Data are reported in the format of mean ± SD from at least three independent experiments. A Student’s two-tailed unpaired t test was used to determine differences between two groups. One-way ANOVA with Dunnett’s post hoc method was used to determine the differences among multiple groups. p < 0.05 was considered statistically significant.
Ethics approval and consent to participation
The scientific experiment with the tissue samples was certificated by the participants and approved by the Ethics Committee of China-Japan Union Hospital of Jilin University. The written informed consent was obtained from every participant.
Availability of data and materials
The datasets used in the study are available from the corresponding author upon reasonable request.
Acknowledgments
This work was supported by the Horizontal Scientific Research Project of Jilin University (no. 2018YX389).
Author contributions
Q.L. and Q.F. carried out the experiments. F.G. wrote the manuscript with support from Q.F. and Q.L. G.S. helped supervise the project. Q.L. and G.S. conceived the original idea.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.02.023.
Supplemental information
References
- 1.Xing M., Alzahrani A.S., Carson K.A., Viola D., Elisei R., Bendlova B., Yip L., Mian C., Vianello F., Tuttle R.M. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013;309:1493–1501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Haugen B.R., Alexander E.K., Bible K.C., Doherty G.M., Mandel S.J., Nikiforov Y.E., Pacini F., Randolph G.W., Sawka A.M., Schlumberger M. 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: the American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid. 2016;26:1–133. doi: 10.1089/thy.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay I.D., Thompson G.B., Grant C.S., Bergstralh E.J., Dvorak C.E., Gorman C.A., Maurer M.S., McIver B., Mullan B.P., Oberg A.L. Papillary thyroid carcinoma managed at the Mayo Clinic during six decades (1940–1999): temporal trends in initial therapy and long-term outcome in 2444 consecutively treated patients. World J. Surg. 2002;26:879–885. doi: 10.1007/s00268-002-6612-1. [DOI] [PubMed] [Google Scholar]
- 6.Fröhlich E., Wahl R. The current role of targeted therapies to induce radioiodine uptake in thyroid cancer. Cancer Treat. Rev. 2014;40:665–674. doi: 10.1016/j.ctrv.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Cai X., Zhao Z., Dong J., Lv Q., Yun B., Liu J., Shen Y., Kang J., Li J. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10:184. doi: 10.1038/s41419-019-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Alhasan A.A., Izuogu O.G., Al-Balool H.H., Steyn J.S., Evans A., Colzani M., Ghevaert C., Mountford J.C., Marenah L., Elliott D.J. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood. 2016;127:e1–e11. doi: 10.1182/blood-2015-06-649434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zlotorynski E. Non-coding RNA: circular RNAs promote transcription. Nat. Rev. Mol. Cell Biol. 2015;16:206. doi: 10.1038/nrm3967. [DOI] [PubMed] [Google Scholar]
- 11.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 12.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Yang C., Yuan W., Yang X., Li P., Wang J., Han J., Tao J., Li P., Yang H., Lv Q., Zhang W. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer. 2018;17:19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 15.Granados-Riveron J.T., Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim. Biophys. Acta. 2016;1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Nana-Sinkam S.P., Croce C.M. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15:445. doi: 10.1186/s13059-014-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M., Chen B., Ru Z., Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018;504:283–288. doi: 10.1016/j.bbrc.2018.08.175. [DOI] [PubMed] [Google Scholar]
- 18.Li Q., Pan X., Zhu D., Deng Z., Jiang R., Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70:1298–1316. doi: 10.1002/hep.30671. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y., Yu J., Huang Z., Fu B., Tao Y., Qi X., Mou Y., Hu Y., Wang Y., Cao Y. Circular RNA hsa_circ_0000326 acts as a miR-338-3p sponge to facilitate lung adenocarcinoma progression. J. Exp. Clin. Cancer Res. 2020;39:57. doi: 10.1186/s13046-020-01556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian W., Huang T., Feng W. Circular RNA HIPK3 promotes EMT of cervical cancer through sponging miR-338-3p to up-regulate HIF-1α. Cancer Manag. Res. 2020;12:177–187. doi: 10.2147/CMAR.S232235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui G.Q., Fei D., Guo F., Zhen X., Luo Q., Yin S., Wang H. MicroRNA-338-3p inhibits thyroid cancer progression through targeting AKT3. Am. J. Cancer Res. 2017;7:1177–1187. [PMC free article] [PubMed] [Google Scholar]
- 22.Dy P., Penzo-Méndez A., Wang H., Pedraza C.E., Macklin W.B., Lefebvre V. The three SoxC proteins—Sox4, Sox11 and Sox12—exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno C.S. SOX4: the unappreciated oncogene. Semin. Cancer Biol. 2020;67:57–64. doi: 10.1016/j.semcancer.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanieh H., Ahmed E.A., Vishnubalaji R., Alajez N.M. SOX4: epigenetic regulation and role in tumorigenesis. Semin. Cancer Biol. 2020;67:91–104. doi: 10.1016/j.semcancer.2019.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Min X.S., Huang P., Liu X., Dong C., Jiang X.L., Yuan Z.T., Mao L.F., Chang S. Bioinformatics analyses of significant prognostic risk markers for thyroid papillary carcinoma. Tumour Biol. 2015;36:7457–7463. doi: 10.1007/s13277-015-3410-6. [DOI] [PubMed] [Google Scholar]
- 26.Guo F., Fu Q., Wang Y., Sui G. Long non-coding RNA NR2F1-AS1 promoted proliferation and migration yet suppressed apoptosis of thyroid cancer cells through regulating miRNA-338-3p/CCND1 axis. J. Cell. Mol. Med. 2019;23:5907–5919. doi: 10.1111/jcmm.14386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X.H., Chen J.S., Wang Q., Chen X.L., Wen L., Chen L.Z., Bi J., Zhang L.J., Su Q., Zeng W.T. miR-338-3p suppresses invasion of liver cancer cell by targeting smoothened. J. Pathol. 2011;225:463–472. doi: 10.1002/path.2877. [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Pan M., Han L., Lu H., Hao X., Dong Q. miR-338-3p suppresses neuroblastoma proliferation, invasion and migration through targeting PREX2a. FEBS Lett. 2013;587:3729–3737. doi: 10.1016/j.febslet.2013.09.044. [DOI] [PubMed] [Google Scholar]
- 29.Huang N., Wu Z., Lin L., Zhou M., Wang L., Ma H., Xia J., Bin J., Liao Y., Liao W. miR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget. 2015;6:15222–15234. doi: 10.18632/oncotarget.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P., Chen X., Su L., Li C., Zhi Q., Yu B., Sheng H., Wang J., Feng R., Cai Q. Epigenetic silencing of miR-338-3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS ONE. 2013;8:e66782. doi: 10.1371/journal.pone.0066782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H.S., Zheng R.N., Guo L.B., Fu X.J. Circular RNA circ_0000615 knockdown suppresses the development of nasopharyngeal cancer through regulating the miR-338-3p/FGF2 axis. Neoplasma. 2020;67:1032–1041. doi: 10.4149/neo_2020_191019N1061. [DOI] [PubMed] [Google Scholar]
- 32.Pu J., Wang J., Li W., Lu Y., Wu X., Long X., Luo C., Wei H. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J. Cell. Mol. Med. 2020 doi: 10.1111/jcmm.15010. Published online February 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L., Peng X., Lu X., Wei Q., Chen M., Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol. Res. Pract. 2019;215:689–696. doi: 10.1016/j.prp.2018.12.032. [DOI] [PubMed] [Google Scholar]
- 34.Li S., Niu X., Li H., Liang Y., Sun Z., Yan Y. circ_0000003 promotes the proliferation and metastasis of non-small cell lung cancer cells via miR-338-3p/insulin receptor substrate 2. Cell Cycle. 2019;18:3525–3539. doi: 10.1080/15384101.2019.1690883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Ding F. hsa_circ_0008945 promoted breast cancer progression by targeting miR-338-3p. OncoTargets Ther. 2019;12:6577–6589. doi: 10.2147/OTT.S213994. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Li Y., Chen P., Zu L., Liu B., Wang M., Zhou Q. MicroRNA-338-3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am. J. Cancer Res. 2016;6:127–140. [PMC free article] [PubMed] [Google Scholar]
- 37.Jin Y., Zhao M., Xie Q., Zhang H., Wang Q., Ma Q. MicroRNA-338-3p functions as tumor suppressor in breast cancer by targeting SOX4. Int. J. Oncol. 2015;47:1594–1602. doi: 10.3892/ijo.2015.3114. [DOI] [PubMed] [Google Scholar]
- 38.Tong Z., Meng X., Wang J., Wang L. MicroRNA-338-3p targets SOX4 and inhibits cell proliferation and invasion of renal cell carcinoma. Exp. Ther. Med. 2017;14:5200–5206. doi: 10.3892/etm.2017.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J.X., Lu J., Xie H., Wang D.P., Ni H.E., Zhu Y., Ren L.H., Meng X.X., Wang R.L. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10:182. doi: 10.1038/s41419-019-1430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren H., Liu Z., Liu S., Zhou X., Wang H., Xu J., Wang D., Yuan G. Profile and clinical implication of circular RNAs in human papillary thyroid carcinoma. PeerJ. 2018;6:e5363. doi: 10.7717/peerj.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan X., Cao J., Xu J., Chen C., Zheng C., Wang J., Zhu X., Zhu X., Ge M. Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. J. Clin. Lab. Anal. 2018;32:e22573. doi: 10.1002/jcla.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Yan X., Zhang H., Zhan X. circRNA circ_0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med. Sci. Monit. 2019;25:1342–1349. doi: 10.12659/MSM.913463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lan X., Xu J., Chen C., Zheng C., Wang J., Cao J., Zhu X., Ge M. The landscape of circular RNA expression profiles in papillary thyroid carcinoma based on RNA sequencing. Cell. Physiol. Biochem. 2018;47:1122–1132. doi: 10.1159/000490188. [DOI] [PubMed] [Google Scholar]
- 44.Peng N., Shi L., Zhang Q., Hu Y., Wang N., Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS ONE. 2017;12:e0170287. doi: 10.1371/journal.pone.0170287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C., Wei Y., Yu L., Xiao Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med. Sci. Monit. 2019;25:2785–2791. doi: 10.12659/MSM.915658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q., Pan L.Z., Hu M., Ma J.Y. Molecular network-based identification of circular RNA-associated ceRNA network in papillary thyroid cancer. Pathol. Oncol. Res. 2020;26:1293–1299. doi: 10.1007/s12253-019-00697-y. [DOI] [PubMed] [Google Scholar]
- 47.Dong R., Zhang X.O., Zhang Y., Ma X.K., Chen L.L., Yang L. circRNA-derived pseudogenes. Cell Res. 2016;26:747–750. doi: 10.1038/cr.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Song Y., Shi X., Liu J., Xiong S., Chen W., Fu Q., Huang Z., Gu N., Zhang R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018;28:132–143. doi: 10.1101/gr.224386.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao Y., Chen X., Yang H., Chen W., Qian Y., Yan Z., Liao T., Yao W., Wu W., Yu T. hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J. Exp. Clin. Cancer Res. 2019;38:318. doi: 10.1186/s13046-019-1321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan Y., Xu T., Liu Y., Li W., Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem. Biophys. Res. Commun. 2019;510:334–338. doi: 10.1016/j.bbrc.2019.01.108. [DOI] [PubMed] [Google Scholar]
- 51.Wei H., Pan L., Tao D., Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem. Biophys. Res. Commun. 2018;503:56–61. doi: 10.1016/j.bbrc.2018.05.174. [DOI] [PubMed] [Google Scholar]
- 52.Chen F., Feng Z., Zhu J., Liu P., Yang C., Huang R., Deng Z. Emerging roles of circRNA_NEK6 targeting miR-370-3p in the proliferation and invasion of thyroid cancer via Wnt signaling pathway. Cancer Biol. Ther. 2018;19:1139–1152. doi: 10.1080/15384047.2018.1480888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song Z., Zhang X., Lin Y., Wei Y., Liang S., Dong C. LINC01133 inhibits breast cancer invasion and metastasis by negatively regulating SOX4 expression through EZH2. J. Cell. Mol. Med. 2019;23:7554–7565. doi: 10.1111/jcmm.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J., Lin X., Jiang W., Wu J., Lin L. lncRNA LEF1-AS1 promotes malignancy in non-small-cell lung cancer by modulating the miR-489/SOX4 axis. DNA Cell Biol. 2019;38:1013–1021. doi: 10.1089/dna.2019.4717. [DOI] [PubMed] [Google Scholar]
- 55.Li Y., Hao J., Jiang Y.M., Liu Y., Zhang S.H. Long non-coding RNA DSCAM-AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in ovarian cancer via upregulating SOX4. Eur. Rev. Med. Pharmacol. Sci. 2019;23:4143–4148. doi: 10.26355/eurrev_201905_17916. [DOI] [PubMed] [Google Scholar]
- 56.Wei D., Wang W., Shen B., Zhou Y., Yang X., Lu G., Yang J., Shao Y. MicroRNA-199a-5p suppresses migration and invasion in oral squamous cell carcinoma through inhibiting the EMT-related transcription factor SOX4. Int. J. Mol. Med. 2019;44:185–195. doi: 10.3892/ijmm.2019.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang J., Gao F., Chu H., Lou L., Wang H., Chen Y. miR-363-3p inhibits migration, invasion, and epithelial-mesenchymal transition by targeting NEDD9 and SOX4 in non-small-cell lung cancer. J. Cell. Physiol. 2020;235:1808–1820. doi: 10.1002/jcp.29099. [DOI] [PubMed] [Google Scholar]
- 58.Zhong M., Wang W.L., Yu D.J. Long non-coding RNA OR3A4 is associated with poor prognosis of human non-small cell lung cancer and regulates cell proliferation via up-regulating SOX4. Eur. Rev. Med. Pharmacol. Sci. 2019;23:6524–6530. doi: 10.26355/eurrev_201908_18537. [DOI] [PubMed] [Google Scholar]
- 59.Ding L., Zhao Y., Dang S., Wang Y., Li X., Yu X., Li Z., Wei J., Liu M., Li G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer. 2019;18:45. doi: 10.1186/s12943-019-1006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruan M., Liu M., Dong Q., Chen L. Iodide- and glucose-handling gene expression regulated by sorafenib or cabozantinib in papillary thyroid cancer. J. Clin. Endocrinol. Metab. 2015;100:1771–1779. doi: 10.1210/jc.2014-3023. [DOI] [PubMed] [Google Scholar]
- 61.Ruan X., Shi X., Dong Q., Yu Y., Hou X., Song X., Wei X., Chen L., Gao M. Antitumor effects of anlotinib in thyroid cancer. Endocr. Relat. Cancer. 2019;26:153–164. doi: 10.1530/ERC-17-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ribeiro F.R., Meireles A.M., Rocha A.S., Teixeira M.R. Conventional and molecular cytogenetics of human non-medullary thyroid carcinoma: characterization of eight cell line models and review of the literature on clinical samples. BMC Cancer. 2008;8:371. doi: 10.1186/1471-2407-8-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saiselet M., Floor S., Tarabichi M., Dom G., Hébrant A., van Staveren W.C., Maenhaut C. Thyroid cancer cell lines: an overview. Front. Endocrinol. (Lausanne) 2012;3:133. doi: 10.3389/fendo.2012.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meireles A.M., Preto A., Rocha A.S., Rebocho A.P., Máximo V., Pereira-Castro I., Moreira S., Feijão T., Botelho T., Marques R. Molecular and genotypic characterization of human thyroid follicular cell carcinoma-derived cell lines. Thyroid. 2007;17:707–715. doi: 10.1089/thy.2007.0097. [DOI] [PubMed] [Google Scholar]
- 65.Schweppe R.E., Klopper J.P., Korch C., Pugazhenthi U., Benezra M., Knauf J.A., Fagin J.A., Marlow L.A., Copland J.A., Smallridge R.C., Haugen B.R. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J. Clin. Endocrinol. Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sekiguchi M., Shiroko Y., Arai T., Kishino T., Sugawara I., Kusakabe T., Suzuki T., Yamashita T., Obara T., Ito K., Hasumi K. Biological characteristics and chemosensitivity profile of four human anaplastic thyroid carcinoma cell lines. Biomed. Pharmacother. 2001;55:466–474. doi: 10.1016/s0753-3322(01)00087-7. [DOI] [PubMed] [Google Scholar]
- 67.Song B., Li R., Zuo Z., Tan J., Liu L., Ding D., Lu Y., Hou D. lncRNA ENST00000539653 acts as an oncogenic factor via MAPK signalling in papillary thyroid cancer. BMC Cancer. 2019;19:297. doi: 10.1186/s12885-019-5533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge M.H., Jiang L.H., Wen Q.L., Tan Z., Chen C., Zheng C.M., Zhu X., Chen J.W., Zhu Z.Y., Cai X.J. Preliminary screening and analysis of metastasis-related lncRNA and co-expressed papillary thyroid carcinoma mRNA. Oncol. Lett. 2018;16:3715–3725. doi: 10.3892/ol.2018.9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z.L., Wang C., Liu W., Ai Z.L. Emerging roles of the long non-coding RNA 01296/microRNA-143-3p/MSI2 axis in development of thyroid cancer. Biosci. Rep. 2019;39 doi: 10.1042/BSR20182376. BSR20182376. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.He Y., Cao L., Wang L., Liu L., Huang Y., Gong X. Metformin inhibits proliferation of human thyroid cancer TPC-1 cells by decreasing LRP2 to suppress the JNK pathway. OncoTargets Ther. 2020;13:45–50. doi: 10.2147/OTT.S227915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Esposito T., Lucariello A., Hay E., Contieri M., Tammaro P., Varriale B., Guerra G., De Luca A., Perna A. Effects of curcumin and its adjuvant on TPC1 thyroid cell line. Chem. Biol. Interact. 2019;305:112–118. doi: 10.1016/j.cbi.2019.03.031. [DOI] [PubMed] [Google Scholar]
- 72.Yang Y., Ding L., Li Y., Xuan C. hsa_circ_0039411 promotes tumorigenesis and progression of papillary thyroid cancer by miR-1179/ABCA9 and miR-1205/MTA1 signaling pathways. J. Cell. Physiol. 2020;235:1321–1329. doi: 10.1002/jcp.29048. [DOI] [PubMed] [Google Scholar]
- 73.Sui H., Fan S., Liu W., Li Y., Zhang X., Du Y., Bao H. LINC00028 regulates the development of TGFβ1-treated human tenon capsule fibroblasts by targeting miR-204-5p. Biochem. Biophys. Res. Commun. 2020;525:197–203. doi: 10.1016/j.bbrc.2020.01.096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in the study are available from the corresponding author upon reasonable request.