Abstract
Background
high recurrence rates of up to 75% within 2 years in pancreatic ductal adenocarcinoma (PDAC) patients resected for cure indicate a high medical need for clinical prediction tools and patient specific treatment approaches. Addition of the EGFR inhibitor erlotinib to adjuvant chemotherapy failed to improve outcome but its efficacy in some patients warrants predictors of responsiveness.
Patients and Methods
we analysed tumour samples from 293 R0-resected patients from the randomized, multicentre phase III CONKO-005 trial (gemcitabine ± erlotinib) with targeted sequencing, copy number, and RNA expression analyses.
Findings
a total of 1086 mutations and 4157 copy-number aberrations (CNAs) with a mean of 17.9 /tumour were identified. Main pathways affected by genetic aberrations were the MAPK-pathway (99%), cell cycle control (92%), TGFβ signalling (77%), chromatin remodelling (71%), and the PI3K/AKT pathway (65%). Based on genetic signatures extracted with non-negative matrix factorization we could define five patient clusters, which differed in mutation patterns, gene expression profiles, and survival. In multivariable Cox regression analysis, SMAD4 aberrations were identified as a negative prognostic marker in the gemcitabine arm, an effect that was counteracted when treated with erlotinib (DFS: HR=1.59, p = 0.016, and OS: HR = 1.67, p = 0.014). Integration of differential gene expression analysis established SMAD4 alterations with low MAPK9 expression (n = 91) as a predictive biomarker for longer DFS (HR=0.49; test for interaction, p = 0.02) and OS (HR = 0.32; test for interaction, p = 0.001).
Interpretation
this study identified five biologically distinct patient clusters with different actionable lesions and unravelled a previously unappreciated association of SMAD4 alteration status with erlotinib effectiveness. Confirmatory studies and mechanistic experiments are warranted to challenge the hypothesis that SMAD4 status might guide addition of erlotinib treatment in early-stage PDAC patients.
Keywords: Pancreatic cancer, Precision medicine, Erlotinib, SMAD4, MAPK9
Research in context.
Evidence before this study
The EGFR inhibitor Erlotinib is currently the only FDA-approved kinase inhibitor for patients with advanced pancreatic cancer showing some improvement in OS and DFS. When added to gemcitabine in the adjuvant setting, erlotinib failed to improve outcome for patients with early-disease stages. Molecular predictors of response are largely unknown for EGFR inhibition in PDAC. Searching PubMed using the terms “precision medicine” or “personalized medicine” or “integrative genomic profiling” together with “pancreatic cancer”, we identified and reviewed pertinent articles published in English before May 1st 2020. We were able to identify mainly review and perspective articles. In addition, some articles applying real-time genomic characterization to guide subsequent treatment strategies by actionable lesions were found. However, these latter studies were exclusively performed in advanced disease stages and only a subset of patients received molecularly matched therapy. Therefore, to the best of our knowledge there has been no large study published yet applying integrative genomics to a R0-resected PDAC patient cohort treated within a large multicentre phase III trial.
Added value of this study
We demonstrate that integrative genomic profiling of PDAC tumour samples resected for cure is capable to define distinct genetic subgroups harbouring different actionable lesions. This approach led to the discovery of a frequent molecular phenotype – defined by genetic aberrations in SMAD4 coupled with low mRNA expression of MAPK9 – as a predictor of erlotinib response. We show that such datasets can be used as a powerful tool for precision medicine approaches, especially in light of growing multimodal treatment possibilities.
Implications of all the available evidence
Mounting evidence underscores the potential of precision medicine approaches in metastatic PDAC patients. Our study underlines the rationale that tumour-based molecular profiling for patients with pancreatic cancer should also be routinely performed in early disease stages.
Alt-text: Unlabelled box
1. Introduction
With advances in next generation sequencing technologies, our knowledge of the molecular background of most cancer types has increased tremendously. This has not only led to a better understanding of the mutational processes in cancer, but has paved the way for more patient specific treatment approaches. Because driver mutations are causative, drugs that target the function of resulting proteins can be therapeutic. For example, the treatment of EGFR mutated non-small-cell lung cancers with EGFR inhibitors [1,2] like erlotinib improved patients outcome significantly and is nowadays standard of care.
However, patients with pancreatic ductal adenocarcinoma (PDAC) have so far not benefited from recent improvements made by precision medicine approaches in other malignancies. Clinical outcome of PDAC remains dismal, with a 5-year survival rate below 10% across all stages and a median survival of <11 months in advanced disease. PDAC is the fourth leading cause of cancer-related death in Western societies [3]. Several large-scale sequencing studies revealed a complex mutational landscape underlying PDAC carcinogenesis with recurrent oncogenic events in four well-known cancer genes (KRAS, TP53, SMAD4, and CDKN2A), as well as a long list of rather infrequent alterations, and established RNA-based PDAC subtyping [4], [5], [6]. However, due to the lack of molecular targets and clear predictive factors, this current knowledge could not be transferred into relevant clinical decision making so far. Limited sample size, and heterogeneity within clinical trial cohorts might have masked the impact of genetic alterations.
To address this knowledge gap, we performed a molecular in-depth characterization of 293 R0-resected PDAC patients treated within the CONKO-005 trial, a randomized, multicentre phase III trial which compared adjuvant chemotherapy of gemcitabine with or without the tyrosine kinase inhibitor erlotinib [7]. While no significant difference in disease-free survival (DFS) and overall survival (OS) could be observed between the two treatment arms, our aim of this analysis was to identify patient subgroups with a potential benefit from additional erlotinib by integrative genomics using a combination of mutation, copy number, and gene expression analyses.
2. Methods
2.1. Patients
All patients were enrolled in the CONKO-005 study, an open-label, multicentre, randomized phase III trial investigating the addition of erlotinib to gemcitabine compared to gemcitabine only as adjuvant therapy [7]. Treatment details have been published and are summarized in the Supplemental Appendix. No differences with respect to baseline characteristics and patients’ outcome between the 293 patients from our study population and the entire CONKO-005 cohort was observed (Table 1 and Supplemental Fig. S1).
Table 1.
Clinical baseline characteristics of 293 PDAC patients from the CONKO-005 trial.
| Characteristics | PDAC |
|---|---|
| (n = 293) | |
| Age, years | |
| Median | 64 |
| Range | 24–82 |
| Sex | |
| male - no. (%) | 163 (56%) |
| female - no. (%) | 130 (44%) |
| Arm | |
| Gemcitabine - no. (%) | 149 (51%) |
| Gemcitabine + Erlotnib - no. (%) | 144 (49%) |
| Karnofsky | |
| 60 - no. (%) | 1 (>1%) |
| 70 - no. (%) | 10 (3%) |
| 80 - no. (%) | 75 (26%) |
| 90 - no. (%) | 112 (38%) |
| 100 - no. (%) | 95 (32%) |
| Grading | |
| G1 - no. (%) | 7 (2%) |
| G2 - no. (%) | 180 (61%) |
| G3 - no. (%) | 96 (33%) |
| unknown - no. (%) | 10 (4%) |
| T-Stage | |
| T1 - no. (%) | 9 (3%) |
| T2 - no. (%) | 30 (10%) |
| T3 - no. (%) | 251 (86%) |
| T4 - no. (%) | 3 (1%) |
| N-Stage | |
| N0 - no. (%) | 106 (36%) |
| N1 - no. (%) | 187 (64%) |
| Postoperative CA 19–9, kU/L | |
| Median (range) | 19,5 (1–5816) |
| ≤ 100 - no. (%) | 223 (76%) |
| 101–500 - no. (%) | 29 (10%) |
| > 500 - no. (%) | 12 (4%) |
| unknown - no. (%) | 29 (10%) |
2.2. Ethics approval
Written consent was obtained from every individual in accordance with the Declaration of Helsinki and with ethical approval obtained from the local ethics committee from the Charité – Universitätsmedizin Berlin, Germany (EAl/139/05, amendment 12.08.2012).
2.3. Mutation analysis by targeted sequencing
DNA and RNA were extracted from 331 FFPE samples, with a tumour content of at least 10% (Supplemental Table S1). The sequencing panel covered full-length coding regions of 67 genes described as significantly mutated [5,6], shown as clinically relevant [8], [9], [10], included in previous panels [9,11], and/or representing major players in the EGFR pathway (Supplemental Table S2). A custom Agilent SureSelectXT Target Enrichment System for Illumina Paired-End Multiplexed Sequencing was used and libraries were paired-end sequenced with a mean sequencing depth of ~600x and a minimal reading depth of 200x. Further information on filtering criteria are outlined in the Supplemental Appendix.
To reduce the likelihood of false single nucleotide variant (SNV) calling, we established a validation pipeline using a combination of SNV frequency, EBcall p-value, DNA quality, and sequencing duplication rate. A total of 219 potential SNVs representing 17% of all detected variants were investigated in a second independent experiment either by amplicon-based targeted deep sequencing (n = 195) or ddPCR (n = 24) as previously described [12], [13], [14]. With a mean coverage of 88102x, we could validate 210 variants, which led to a high validation rate of 96%.
2.4. Copy-number alterations (CNA) detection by targeted sequencing
Copy-number analysis was performed as previously reported using an in-house pipeline CNACS (https://github.com/papaemmelab/toil_cnacs) (Y. Shiozawa and S.Ogawa, manuscript in preparation) [15], in which total number of reads covering each bait region and allele frequency of heterozygous single-nucleotide polymorphisms (SNP) were used as input data (see also Supplemental Appendix and Table S3). For 171 patients we could detect CNAs based on targeted sequencing data. For validation and remaining 112 samples, we generated information about local CNA in 11 genes based solely on a Multiplex Ligation-dependant Probe Amplification (MLPA) assay. For 10 patients we were not able to obtain CNA data with either method (Supplemental Fig. S2).
2.5. Expression profiling with nCounter technology
The NanoString nCounter Flex system was used to run a customized version of the PanCancer Pathways Panel (770 genes representing 13 canonical pathways in cancer; Supplemental Appendix and Table S4). After RNA quality controls, 230 samples were analysed. Genes were tested for differential expression in response to each selected covariate. For each gene, a single linear regression was fit using all selected covariates to predict expression and false discovery rate (FDR) was estimated according to the Benjamini-Hochberg procedure [16].
2.6. Statistical analysis
Statistical analyses were performed using IBM SPSS statistics (version 24) and R (version 3.6.1) and are detailed in the Supplemental Appendix. Co-occurrence and mutational exclusivity was calculated with Fisher's exact test and subsequently corrected for multiple testing [16]. To model clonal composition we used a modified version of the SciClone Bioconductor package as previously described [17,18]. Signatures extracted from SNV and CNA patterns with non-negative matrix factorization (NMF) were used to group patients into five distinct clusters [19]. Step-wise subsampling of the patient cohort (minus five patients per step) was performed to ensure a high stability of the identified signatures and, therefore a high cluster stability (Supplemental Fig. S3). Cox models were used for time to-event variables (OS and DFS), and p-values were calculated using the Wald test. Multivariable cox proportional hazards models were used to investigate variables associated with survival endpoints. To select input for the multivariable cox proportional hazards models, univariate cox regression analysis of clinical and genetic variables were carried out. Primary analysis endpoint was OS, followed by exploratory analysis for DFS. Kaplan-Meier analysis was performed to construct survival curves and log-rank test was applied to evaluate differences between subgroups.
2.6.1. Role of funding source
The funders of this study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. KH, RH, IY, KY, MS, and FD had access to the raw data. FD had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Genetic landscape of PDAC
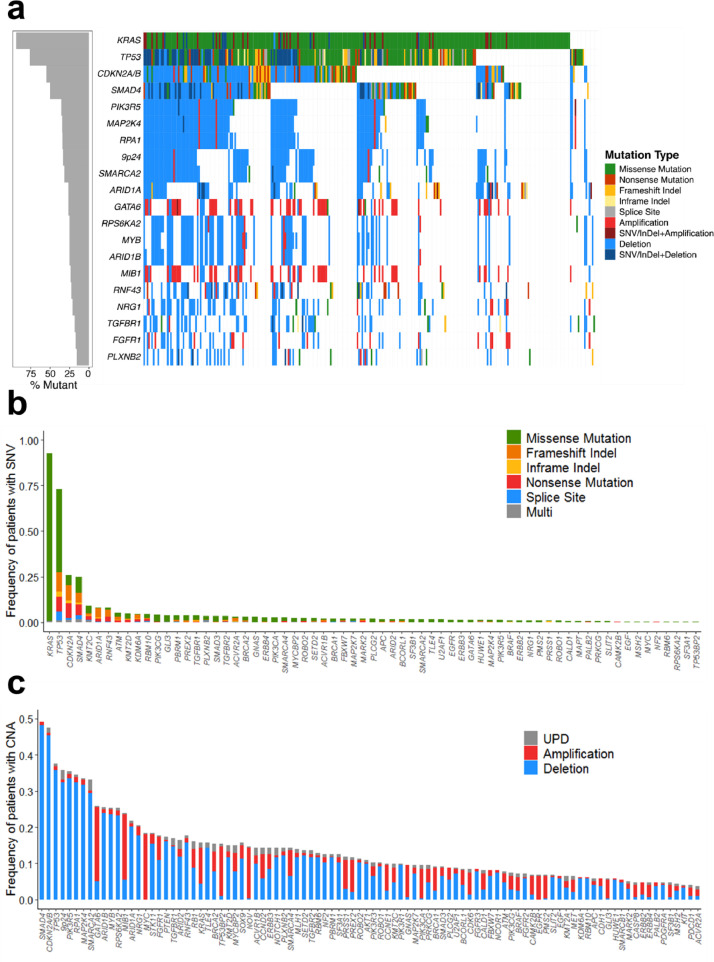
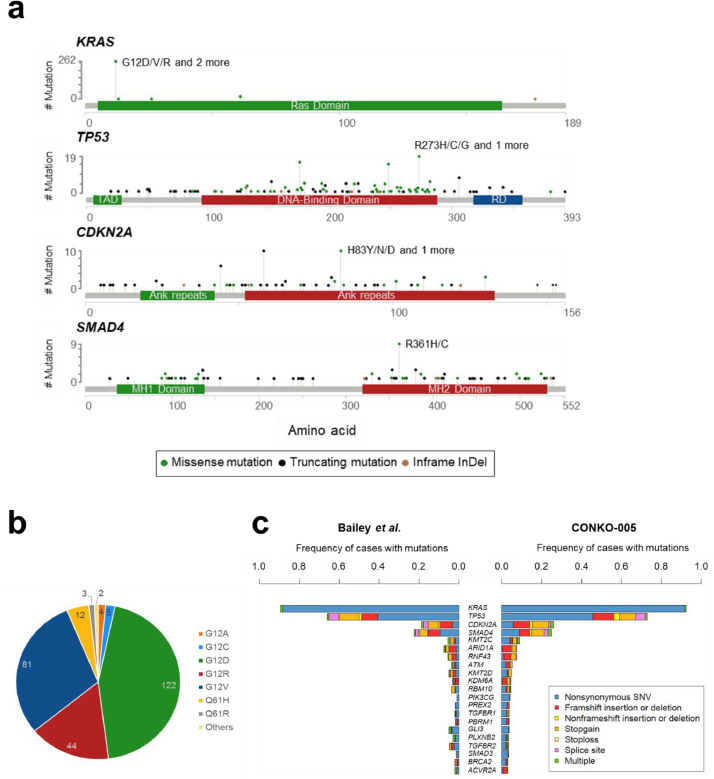
A total of 1086 SNVs and 4157 CNAs were identified in 293 patients (Fig. 1, Supplemental Tables S5 and S6). In 99% of all patients (n = 290), at least one genetic alteration was found, with a median of 4 (range: 0–23) SNVs and 11 (range: 0–66) CNAs per patient (Supplemental Figs. S4 and S5). Within the 67 genes analysed for SNVs, 58 were found to be recurrently mutated. The four most commonly mutated genes were KRAS (93%), TP53 (74%), CDKN2A (27%), and SMAD4 (27%), with mutations targeting known hot spots (e.g. KRAS G12) (Fig. 2a and b). Mutation types as well as mutation frequencies were comparable to previous PDAC sequencing studies (Fig. 2c) [5,6].
Fig. 1.
Overview of genetic alterations in R0-resected PDAC patients. (a) Landscape plot of the 20 most frequently altered genes in 293 PDAC patients. (b) Frequency and type of all SNVs within the 67 genes investigated for mutations and small insertions/ deletions (InDel). (c) Frequency and type of all CNAs within the 100 genes investigated for copy number alterations.
Fig. 2.
Detailed SNV profile of R0-resected PDAC patients. (a) Lollipop plot of four most commonly mutated genes. The most common SNVs are annotated for each gene. Modified from cbioportal [38,39]. (b) Overview of the different KRAS mutations with respect to their affected codon position and resulting amino acid change. The number of mutations for each variant is annotated. (c) Comparison of mutation frequencies and type with findings from Bailey et al. [6]. The 20 most frequently altered genes are shown in order of mutation frequency, based on the CONKO-005 cohort results. Mutations types are indicated by color. Abbreviations: TAD = trans-activation domain, RD = regulatory domain.
The major CNA loci were identified in genomic regions encoding for SMAD4 (49%), CDKN2A (47%), and TP53 (38%) as well as the 9p24 locus, containing the immune checkpoint regulators CD274, PDCD1LG2, and JAK2 (35%). About 70% of all CNAs were deletions, nevertheless several well-known proto-oncogenes were found to be almost exclusively amplified (KRAS, GATA6, MYC). Gene specific focal CNAs, such as deletion of 9p21 with CDKN2A/B and gain of 18q11 with GATA6, as well as recurrent CNAs of entire chromosomes / chromosome arms, like loss of chromosome 6 and gain of 1q, were identified (Supplemental Fig. S6). The main pathways affected by SNVs and CNAs were the MAPK-pathway (99%), cell cycle control (92%) and TGFβ signalling (77%). Genes encoding for members of the PI3K/AKT pathway (65%) and genes involved in chromatin remodelling (71%) were also frequently affected in this cohort (Supplemental Fig. S7, Supplemental Table S7). The EGFR gene was found to be mutated in four patients (1.4%) and amplified in 19 patients (6.5%).
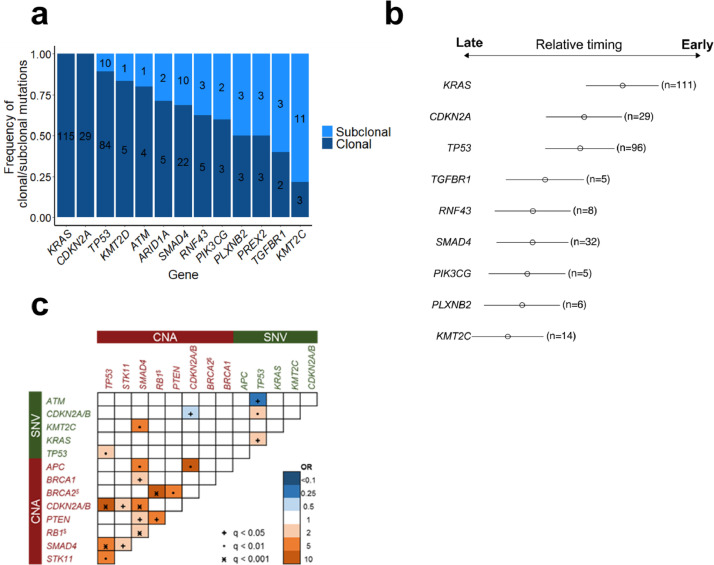
To study preferred trajectories of disease evolution, we used variant allele frequencies (VAFs) to estimate the proportion of tumour cells carrying a given mutation and to identify clonal (in all cells) or subclonal mutations (in a fraction of cells). We applied this approach to 124 copy number neutral KRAS mutated patients with 2 or more oncogenic mutations from which we were able to calculate CNA adjusted VAFs to infer clonal architecture [17]. This approach identified genetic alterations in KRAS, CDKN2A, and TP53 to be mostly clonal and thus likely to represent disease-initiating events (89 – 100% clonal), while mutations in SMAD4 were more often subclonal. Mutations affecting TGFBR1 and KMT2C (Fig. 3a and b) were likely to be later evolutionary events.
Fig. 3.
Overview of clonal hierarchies and genetic interaction pattern. (a) Clonality status of genes mutated in 124 patients without CNA in KRAS. (b) Bradley-Terry Model of all genes with sufficient mutational overlap with concomitant genetic events for model construction. 124 copy number neutral KRAS mutated patients with 2 or more additional mutations were used to calculate CNA adjusted VAFs. (c) Co-mutations and mutual exclusivity plot for all 293 patients. Genes mutated in at least 4% of patients and CNAs in all MLPA genes were included in the analysis. Significance levels of q-values (multiple testing corrected) are shown with symbols, odds ratio with colors (blue show different levels of mutual exclusivity, orange show different levels of co-mutation). $ = genes are located within same chromosomal region on chromosome 13q14.
Next, we searched for pairwise gene associations, recognizing that pairs of genes could show a tendency to either co-occurrence or mutually exclusivity. In addition, we were interested to dissect the interplay of SNV and CNA acquisition. A total of 14 pairs were significantly associated with a false discovery rate <5%. The three major CNAs affecting the genes SMAD4, CDKN2A, and TP53 showed a strong co-occurrence, which means that many patients have two or more of these genes concomitantly deleted. In addition, TP53 mutations were often accompanied by mutations in SMAD4, KRAS, and CDKN2A/B. While mutually exclusive gene pairs often imply functional redundancy, such a pattern was rarely observed. Only ATM mutations hardly co-occurred with TP53 mutations, especially with TP53 activating mutations (Figs. 3c, S8, Supplemental Table S8).
3.2. Non-negative matrix factorization (NMF) reveals molecular subgroups with distinct clinical outcomes
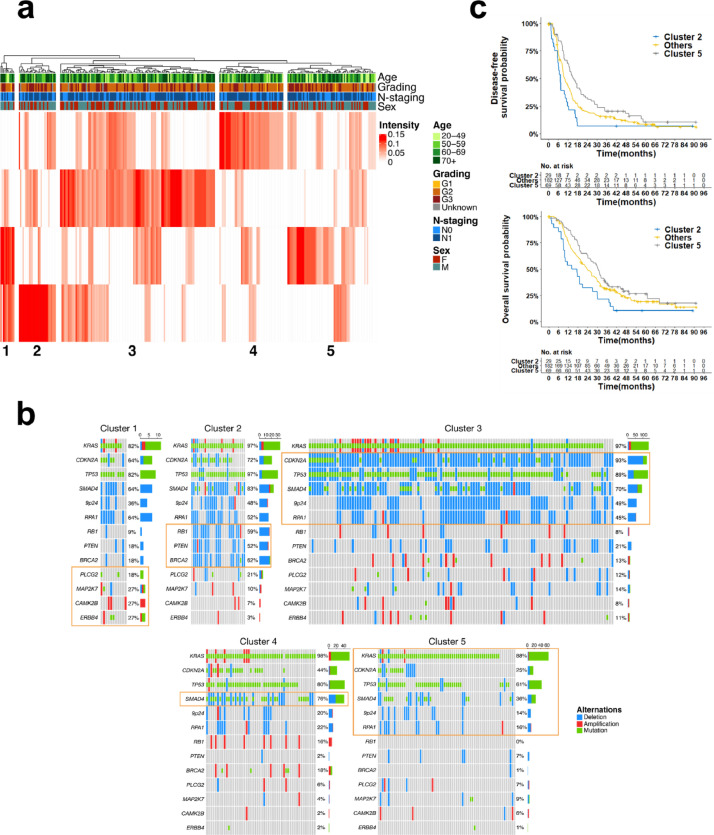
The complex genetic landscape in PDAC prompted additional analyses to better capture multiparametric patterns of mutational co-occurrence. NMF was performed to extract signatures in 78 genetic alterations (67 SNVs and 11 CNAs). The four extracted signatures were used to group the patients with hierarchical clustering into five robust patient clusters (Fig. 4a). To define the robustness of patient clusters, the stability of extracted signatures was tested with random step-wise subsampling of the patient cohort (Supplemental Fig. S3). While baseline characteristics such as age, gender, N-stage, and grading were distributed similarly between all clusters (Fig. 4a, Supplemental Table S9), a wide range of distinct genetic and clinical features could be attributed to each cluster (Supplemental Table S10). For example, tumours from patients in cluster 1 (n = 11) had significantly more mutations (8.6 vs 3.5 in all other groups) and a significant enrichment of mutations and amplifications in genes from the ERBB signalling pathway (PLCG2, MAP2K7, ERBB4, and CAMK2B). Cluster 2 (n = 29) was enriched for copy-number deletions in well-known tumour suppressor genes (RB1, BRCA2, and PTEN), while cluster 3 (n = 121) contained numerous deletions affecting major PDAC gene loci (e.g. CDKN2A/B, TP53, and 9p24). In contrast, clusters 4 (n = 50) and 5 (n = 69) both had much fewer alterations per tumour (12.2 and 9.2 vs 25.9, 24.4, and 22.4 in clusters 1, 2, and 3). In cluster 4, this was mainly based on the absence of CDKN2A/B deletions, whereas in cluster 5 frequently altered genes, such as SMAD4, CDKN2A/B, TP53, and KRAS, were all less often affected. Of note, cluster 4 had a significant enrichment of SMAD4 mutations and showed an overrepresentation of the transition base change C>A/G>T (p = 0.002) (Figs. 4b, S9).
Fig. 4.
Unsupervised patient clustering using non-negative matrix factorization. (a) Heat map showing to which extent the four signatures extracted from 78 genetic alterations with NMF match for each patient of the cohort (intensity) (n = 283). Hierarchical clustering was used to group the patients into five distinct clusters. Additional annotations show their respective clinical baseline characteristics. (b) Exemplary genes showing the molecular biological differences between clusters. Significantly enriched/depleted genes are circled in orange. (c) Kaplan-Meier curves depicting DFS and OS from patients of clusters 2 and 5 compared to all other patients.
Additionally, clusters 2 and 5 showed specific mRNA gene expression patterns. Cluster 2 associated significantly with overexpression of genes encoding receptors and effectors of the PI3K/AKT pathway (MYB, MDM2). In cluster 5, we saw overexpression of MAPK pathway activating genes (RASGRP1, PDGFA, and PRKACB) as well as the PI3K-AKT inhibitor PTEN, while several cell cycle control genes were decreased (PKMYT1, SFN, CHEK2, and SKP2) (Supplemental Fig. S10).
Notably, these genetic and transcriptomic differences were associated with clinical outcome. Patients in cluster 2 had significantly shorter DFS and OS times in multivariable cox regression analyses (HR=1.96, p = 0.002, and HR=2.06, p = 0.001). In contrast, cluster 5 patients showed longer DFS and OS (HR= 0.6, p = 0.002 and HR= 0.65, p = 0.015) (Figs. 4c, S11).
Furthermore, we could identify novel potential points of action for targeted therapies in respective clusters: in cluster 1 the prevalent mutations in ERBB4, GNAS, and KMT2D have been described as potential biomarkers [20], [21], [22], e.g. loss-of-function mutations in KMT2D were associated with sensitivity to BET inhibition in PDAC cells [23]. In cluster 2 with deletions in RB1, BRCA2 and PTEN, for example, the oral pan-AKT Inhibitor MK-2206 has been shown to decrease tumour size and CA19–9 levels in PDAC patients with PTEN loss [24] and the PARP inhibitor olaparib prolonged progression-free survival in metastatic PDAC patients with BRCA1/2 mutations [25]. Interestingly, patients in cluster 4 displayed a trend towards longer DFS and OS, when treated with erlotinib in combination with gemcitabine (GemErlo) as compared to the gemcitabine only treatment arm (DFS: p = 0.098; OS: p = 0.089) (Supplemental Fig. S12).
The largest cluster 3 showed no unique biological and/or clinical associations, likely due to persisting heterogeneity within this cluster requiring further subgrouping based on even larger patient cohorts. Nevertheless, our clustering based on the signatures extracted with NMF identified two well-defined patient clusters (clusters 2 and 5) with distinctive clinical outcome and potentially actionable genetic lesions as well one cluster with a trend for increased erlotinib sensitivity (cluster 4).
3.3. Increased responsiveness to erlotinib counteracts negative prognostic effect of SMAD4 alterations especially in patients with low MAPK9 expression
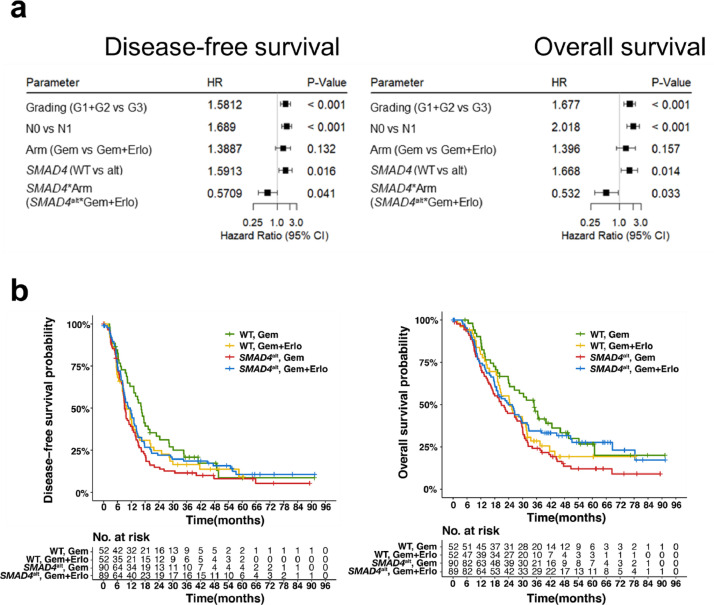
As previously shown for advanced PDAC [26], no predictive or prognostic effect on DFS or OS was observed with respect to the EGFR mutation or CNA status in this cohort (Supplemental Figure S13). The longer OS in the erlotinib treatment arm of cluster 4, which is enriched for SMAD4 mutations, provided a first hint for a genetic alteration to be predictive for erlotinib sensitivity. Since most SMAD4 SNVs were truncating mutations and almost all SMAD4 CNAs were deletions suggesting similar loss of function consequences, we grouped patients harbouring at least one SMAD4 alteration in a SMAD4 altered subgroup (SMAD4alt; n = 179). While both SMAD4alt status and treatment arm alone were not prognostic for OS in a multivariable cox hazard analysis, the interaction test of both parameters correlated with a significant longer OS and DFS (HR= 0.53, p = 0.033 and HR= 0.57, p = 0.041, respectively) (Figs. 5a, S14). After correcting for the effect of the treatment arms, SMAD4 alteration status itself became a negative prognostic marker for OS and DFS (OS: HR=1.67, p = 0.014; DFS: HR=1.59, p = 0.016, respectively) (Fig. 5b). In the gemcitabine treatment arm (Gem), SMAD4alt patients had a significantly shorter DFS and OS compared to SMAD4 wild-type (SMAD4WT) patients (log rank: DFS: p = 0.018, OS: p = 0.0078). At the same time, SMAD4alt patients showed a trend towards longer OS when treated with GemErlo compared to gemcitabine alone without reaching significance (log rank: DFS: p = 0.209, OS: p = 0.056).
Fig. 5.
Erlotinib sensitivity in SMAD4 altered PDAC patients. (a) Forrest plot of multivariable cox hazard model for the interaction of SMAD4 status and treatment arm, containing all clinical baseline characteristics. (b) Kaplan-Meier curve comparing OS between both treatment arms in SMAD4 altered (SMAD4alt) and SMAD4 wild-type (WT) patients (2-sided log-rank test).
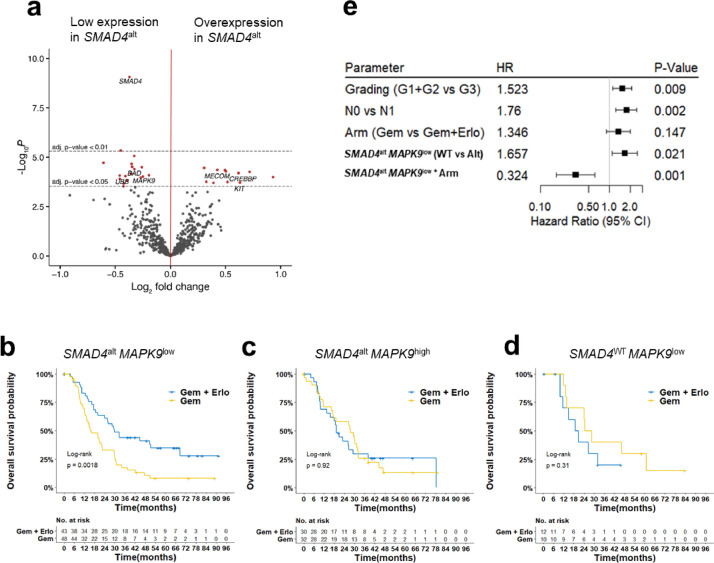
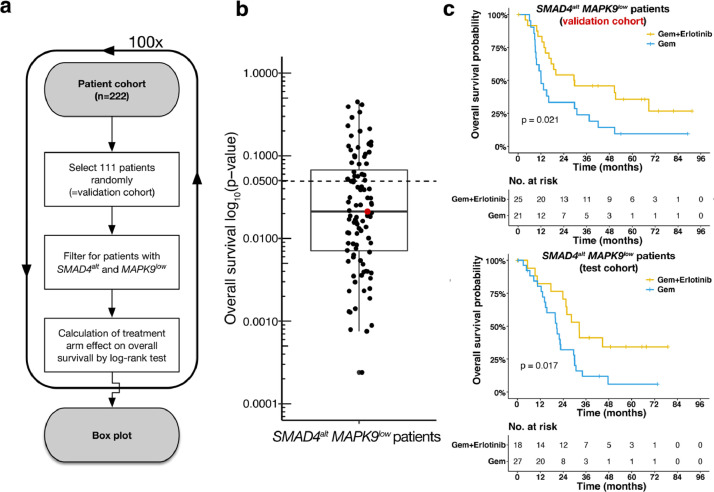
In order to get insights into differentially activated pathways with respect to the underlying SMAD4alt status, we compared gene expression profiles from 153 SMAD4alt and 69 SMAD4WT PDAC patients. After correcting for multiple testing, a total of 11 and 19 genes were significantly up- or down-regulated in SMAD4alt patients, respectively (Fig. 6a, Supplemental Table S11). A careful literature search for the relation with carcinogenesis and/or EGFR pathway inhibition of these 30 differentially expressed genes, pointed us to the down-regulated Jun-kinase MAPK9, a known transcriptional activator of the JNK pathway. As increased JNK activation has been shown to mediate acquired resistance to EGFR inhibition by bypass signalling [27], we hypothesized that effectiveness of EGFR inhibition might be altered according MAPK9 expression levels in SMAD4alt patients. Integration of SMAD4 genetic aberration status with MAPK9 gene expression levels grouped SMAD4alt patients into patients with low (91/222= 40.9%) or high (62/222=27.9%) MAPK9 expression (Supplemental Table S12). Strikingly, the beneficial effect of erlotinib was restricted to SMAD4alt patients with low MAPK9 expression (DFS: HR=0.49; test for interaction, p = 0.02) and OS (HR= 0.32; test for interaction, p = 0.001). No differences were observed neither in SMAD4alt patients with high MAPK9 expression nor in SMAD4WT patients with low or high MAPK9 expression levels (Fig. 6c and d). To further validate this finding, we randomly divided our cohort in a test and a validation cohort of equal size (n = 111, respectively). Next, we compared OS of SMAD4alt MAPK9low patients within the test cohort according to both treatment arms (Fig. 7a). After 100 random assignments a median p-value of 0.021 was observed, confirming the beneficial effect of additive erlotinib over gemcitabine alone in this specific genotype (upper quartile: 0.068, lower quartile: 0.007) (Fig. 7b). For the randomly selected cohorts with a p-value closest to the median, both the test and the validation cohort resulted in a significantly increased OS in SMAD4alt MAPK9low patients treated with additive erlotinib (Fig. 7c).
Fig. 6.
Erlotinib sensitivity in SMAD4 altered PDAC patients is mediated by MAPK9 expression levels. (a) Volcano plot of differential expression analysis between SMAD4WT (baseline) patients and SMAD4alt patients. Horizontal lines show significance levels of p-values (multiple testing adjusted with Benjamini-Hochberg). The 50 variants with the lowest p-value are labelled. Kaplan-Meier curves comparing OS between both treatment arms in patents according to their (b) SMAD4altMAPK9low, (c) SMAD4altMAPK9low, (d) SMAD4WTMAPK9low status (2-sided log-rank test). (e) Forrest plot of multivariablecox hazard model for OS showing the interaction of SMAD4altMAPK9low status and treatment arm, containing all clinical baseline characteristics.
Fig. 7.
Validation of additive erlotinib in patients with a SMAD4altMAPK9low status. (a) A validation cohort and respectively a test cohort consisting of 111 patients each were picked randomly from the patient cohort (n = 222). This step was repeated 100 times. (b) Each dot represents one out of 100 randomly assigned validation cohort. The box plot (median: 0.021, upper quartile: 0.068, lower quartile: 0.007) was generated based on overall survival differences according to the two treatment arms (i.e., Gem + Erlotinib and Gem) in patients with a SMAD4altMAPK9low status. P-values derive from log-rank tests for all 100 randomly selected validation cohorts. (c) The validation cohort with a p-value closest to the median of all p-values, as visualised by the red dot, is shown exemplary. Kaplan-Meier curves comparing OS between both treatment arms for validation and test cohorts are shown for these specific validation and test cohorts.
In summary, SMAD4alt patients had a significantly shorter OS and DFS when treated with gemcitabine alone, an effect that was negated when erlotinib was added to the adjuvant chemotherapy regimen, especially in SMAD4alt patients with low MAPK9 expression levels.
4. Discussion
Several seminal studies have defined the genetic landscape of PDAC over the last decade [4], [5], [6], and paved the ground for our panel design. As molecular data obtained from prospective and randomised clinical trials are rare to date [28], we speculated that signatures extracted from comprehensive SNV and CNA data through NMF factorization might identify patient subgroups with different response to gemcitabine ± erlotinib. We defined five tumour subgroups with important differences with respect to affected gene frequencies, underlying base change signatures, associated deregulated gene expression profiles and different survival outcome. Tumours from patients in cluster 5 had only few alterations in addition to the initiating KRAS mutations with increased MAPK pathway expression as well as high levels of PTEN. This implies that these patients’ tumours were driven by early PDAC events of hyperactive KRAS. In contrast, cluster 2 showed an enrichment of deletions in three major tumour suppressors (PTEN, RB1, and BRAC2). This lead to an uncontrolled activation of the PI3K-/AKT signalling pathway. Therefore, tumours in cluster 2 did not seem to solely rely on the initial MAPK pathway deregulation but have evolved to deregulate a second cell proliferation pathway with the effect of a more aggressive tumour biology leading to decreased DFS and OS.
Although these findings cannot directly be translated into clinical practice, they open new avenues for combined treatment approaches that should target different deregulated pathways based on respective patient clusters at the same time. This approach also helped to identify a genetically defined subgroup of PDAC patients that showed longer survival when treated with GemErlo in this phase 3 trial. Since Moore et al. showed some improvement of OS and DFS in erlotinib treated patients with advanced PDAC [29], several predictive biomarkers have been proposed. For example, KRAS mutation status was shown to be associated with improved OS after erlotinib treatment in advanced PDAC in smaller patient series [26,30]. In preclinical models, cell lines with the so-called “classical” Collison subtype, defined by GATA6 overexpression, were shown to exhibit increased erlotinib sensitivity [31]. In our study, neither KRAS mutations status nor GATA6 amplifications were associated with erlotinib sensitivity, which might be due to the fact that our patients were treated at an earlier disease stage. However, we found SMAD4 alterations to be significantly associated with OS and DFS in a treatment dependant manner. While SMAD4 is widely accepted as a poor prognostic biomarker [9], it has not been postulated in the context of predicting response to EGFR inhibition. Erlotinib has been shown to have similar or even higher affinities to other protein kinases than EGFR [32]. As neither EGFR expression levels nor genetic EGFR alterations were predictors of response in our study, contribution of erlotinib off-targets are conceivable. In line with this hypothesis, SMAD4alt tumours showed downregulation of the Jun-kinase MAPK9. Increased JNK activation has been shown to mediate acquired resistance to EGFR inhibition by bypass signalling [27]. In fact, we identified SMAD4 alterations with decreased MAPK9 expression, identified in 91 patients of this study cohort, as a predictive biomarker for erlotinib response in R0-resectable PDAC patients. Since the CONKO-005 study was the first major clinical trial comparing erlotinib treatment with standard of care in R0-resected patients, our cohort is unique in its size and homogeneity. While this gave us the opportunity to analyse previously overseen genetic interactions, it made it impossible to validate our findings in a suitable second patient cohort. Even though we performed extensive self-validation to further strengthen the reliability of our analysis, the missing independent validation is a limitation of our study. Of note, a recent study showed a previously unappreciated function for tumour cell-intrinsic SMAD4 by promoting anti-tumour immunity in PDAC. This function could be attributed to a direct link between SMAD4 and EGFR through a transcriptional axis involving KDM3A, KLF5, and SMAD4 converging on EGFR [33]. As erlotinib sensitized tumors to combination immunotherapy in vivo [33], it will be of interest to study the combination of EGFR inhibition with immunotherapies in PDAC patients with SMAD4 aberrations. Another promising combinatorial approach was recently shown using simultaneous EGFR/c-RAF inhibition [34], further indicating that the full therapeutic potential of erlotinib in PDAC is yet to be defined.
One might question the role of Gemcitabine in curable PDAC as mFOLFIRNOX showed superiority in terms of longer DFS and OS as compared to gemcitabine alone in R0-resected PDAC patients recently [35]. However, real-world data about the adjuvant use of mFOLFIRINOX are not yet available. Median age in PDAC patients is 70 to 75 years and elderly patients are often not eligible for an intensified adjuvant chemotherapy. For these patients, Gemcitabine remains a well-documented and recommended option in the postoperative situation [36]. While a transcriptomic signature has recently been identified to predict Gemcitabine sensitivity [37], our data extend current precision medicine approaches with respect to the use of EGFR inhibitors in PDAC. Further analysis are now warranted to dissect the precise mechanisms of EGFR inhibition in PDAC.
Funding
Else Kröner-Fresenius Foundation, Wegener Foundation, German Cancer Aid.
Declaration of Competing Interest
Dr. Damm reports personal fees from Roche, Novartis, AbbVie, Astra Zeneca outside the submitted work. Dr. Sinn reports personal fees from Sanofi, Astra Zeneca, Amgen, Servier, MSD, Incyte, Pfizer outside the submitted work. Dr. Bullinger reports personal fees from Abbvie, Amgen, Astellas, Bristol-Myers Squibb, Celgene, Daiichi Sankyo, Gilead, Hexal, Janssen, Jazz Pharmaceuticals, Menarini, Novartis, Pfizer, Sanofi, and Seattle Genetics outside the submitted work. Dr. Keilholz reports personal fees from Bristol-Myers Squibb, MSD, Merck Serono, Pfizer, Astra Zeneca outside the submitted work. All other authors have declared no conflicts of interest.
Acknowledgments
Contributors
KH and FD designed the study. JKS, SB, PL, UK, UP, MH, HR, and MS provided cases, data and/or clinical annotation. KH, RH, YI, KY, FB, FC, NK, TY, YShio, YShir, HP, OB, LB, SO, and FD carried out experiments and data analysis. KH, MS, and FD wrote the manuscript. All authors reviewed and approved the final manuscript.
Acknowledgements
We thank all participating patients and their families, the CONKO study group and all participating centers. This study was supported by grants from the Brigitte und Dr. Konstanze Wegener-Stiftung (#38), the Else Kröner-Fresenius-Stiftung (#2017_EKES.33), Deutsche Krebshilfe (#70113643), and the DKTK all awarded to FD. MS had financial support by the Charite-Forschungsfoerderung and the Rahel-Hirsch-Habilitationsstipendium. This work was partially sponsored by MEXT/JSPS KAKENHI (JP26221308, JP26253060), Scientific Research on Innovative Areas (15H05909, 15H05912), JSPS Core-to-Core Program, MEXT as "Priority Issue on Post-K computer", Grants-in-Aid from the Japan Agency for Medical Research and Development, AMED (JP15cm0106056, JP19cm0106501, JP16ck0106073, JP19ck0106250) all awarded to SO. We thank Dr. Alexandra Stege and Hedwig Lammert for technical assistance, Agilent Technologies for help developing the gene panel and acknowledge the assistance of the Central Biobank Charité (ZeBanC).
Data sharing statement
All DNA sequencing data have been deposited in the NCBI Sequence Read Archive (SRA): BioProject accession number is PRJNA703316. Any data not published within the article or supplementary materials will be shared in anonymized format upon reasonable request. If desired, please contact the corresponding author of this article.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103327.
Appendix. Supplementary materials
References
- 1.Shepherd F.A., Rodrigues Pereira J., Ciuleanu T. Erlotinib in previously treated non–small-cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 2.Thatcher N., Chang A., Parikh P. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicenter study (Iressa survival evaluation in lung cancer) Lancet. 2005;366(9496):1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Biankin A.V., Waddell N., Kassahn K.S. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waddell N., Pajic M., Patch A.M. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey P., Chang D.K., Nones K. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 7.Sinn M., Bahra M., Liersch T. CONKO-005: adjuvant chemotherapy with gemcitabine plus erlotinib versus gemcitabine alone in patients after R0 resection of pancreatic cancer: a multicenter randomized phase III trial. J Clin Oncol. 2017;35(29):3330–3337. doi: 10.1200/JCO.2017.72.6463. [DOI] [PubMed] [Google Scholar]
- 8.Vincent A., Herman J., Schulick R., Hruban R.H., Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sausen M., Phallen J., Adleff V. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosmidis C., Sapalidis K., Kotidis E. Pancreatic cancer from bench to bedside: molecular pathways and treatment options. Ann Transl Med. 2016;4(9):165. doi: 10.21037/atm.2016.05.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GeneDx. Pancreatic cancer panel. 2016 (accessed 12.07.2016).
- 12.Christen F., Hoyer K., Yoshida K. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): an international study on 331 patients. Blood. 2019;133(10):1140–1151. doi: 10.1182/blood-2018-05-852822. [DOI] [PubMed] [Google Scholar]
- 13.Arends C.M., Galan-Sousa J., Hoyer K. Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia. 2018;32(9):1908–1919. doi: 10.1038/s41375-018-0047-7. [DOI] [PubMed] [Google Scholar]
- 14.Young E., Noerenberg D., Mansouri L. EGR2 mutations define a new clinically aggressive subgroup of chronic lymphocytic leukemia. Leukemia. 2017;31(7):1547–1554. doi: 10.1038/leu.2016.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshizato T., Nannya Y., Atsuta Y. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129(17):2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57(1):289–300. (Methodological) [Google Scholar]
- 17.Teixeira V.H., Pipinikas C.P., Pennycuick A. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat. Med. 2019;25(3):517–525. doi: 10.1038/s41591-018-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mylonas E., Yoshida K., Frick M. Single-cell analysis based dissection of clonality in myelofibrosis. Nat Commun. 2020;11(1):73. doi: 10.1038/s41467-019-13892-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hüebschmann D., Kurzawa N., Steinhauser S. Deciphering programs of transcriptional regulation by combined deconvolution of multiple omics layers. bioRxiv. 2017 [Google Scholar]
- 20.Prickett T.D., Agrawal N.S., Wei X. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41(10):1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iorio F., Knijnenburg T.A., Vis D.J. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nault J.C., Fabre M., Couchy G. GNAS-activating mutations define a rare subgroup of inflammatory liver tumors characterized by STAT3 activation. J Hepatol. 2012;56(1):184–191. doi: 10.1016/j.jhep.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Andricovich J., Perkail S., Kai Y., Casasanta N., Peng W., Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33(3):512–526. doi: 10.1016/j.ccell.2018.02.003. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yap T.A., Yan L., Patnaik A. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol Off J Am Soc Clin Oncol. 2011;29(35):4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- 25.Golan T., Hammel P., Reni M. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeck S., Jung A., Laubender R.P. EGFR pathway biomarkers in erlotinib-treated patients with advanced pancreatic cancer: translational results from the randomised, crossover phase 3 trial AIO-PK0104. Br J Cancer. 2013;108(2):469–476. doi: 10.1038/bjc.2012.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manole S., Richards E.J., Meyer A.S. JNK pathway activation modulates acquired resistance to EGFR/HER2-targeted therapies. Cancer Res. 2016;76(18):5219–5228. doi: 10.1158/0008-5472.CAN-16-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinn M., Sinn B.V., Treue D. TP53 Mutations predict sensitivity to adjuvant gemcitabine in patients with pancreatic ductal adenocarcinoma: next-generation sequencing results from the CONKO-001 trial. Clin Cancer Res. 2020;26(14):3732–3739. doi: 10.1158/1078-0432.CCR-19-3034. [DOI] [PubMed] [Google Scholar]
- 29.Moore M.J., Goldstein D., Hamm J. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 30.Kim S.T., Lim D.H., Jang K.T. Impact of KRAS mutations on clinical outcomes in pancreatic cancer patients treated with first-line gemcitabine-based chemotherapy. Mol Cancer Ther. 2011;10(10):1993–1999. doi: 10.1158/1535-7163.MCT-11-0269. [DOI] [PubMed] [Google Scholar]
- 31.Collisson E.A., Sadanandam A., Olson P. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conradt L., Godl K., Schaab C. Disclosure of erlotinib as a multikinase inhibitor in pancreatic ductal adenocarcinoma. Neoplasia. 2011;13(11):1026–1034. doi: 10.1593/neo.111016. (New York, NY) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Yuan S., Norgard R.J. Epigenetic and transcriptional control of the epidermal growth factor receptor (EGFR) regulates the tumor immune microenvironment in pancreatic cancer. Cancer Discov. 2021;11(3):736–753. doi: 10.1158/2159-8290.CD-20-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blasco M.T., Navas C., Martin-Serrano G. Complete regression of advanced pancreatic ductal adenocarcinomas upon combined inhibition of EGFR and C-RAF. Cancer Cell. 2019;35(4):573–587. doi: 10.1016/j.ccell.2019.03.002. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conroy T., Hammel P., Hebbar M. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379(25):2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 36.Khorana A.A., McKernin S.E., Berlin J. Potentially curable pancreatic adenocarcinoma: ASCO clinical practice guideline update. J Clin Oncol. 2019;37(23):2082–2088. doi: 10.1200/JCO.19.00946. [DOI] [PubMed] [Google Scholar]
- 37.Nicolle R., Gayet O., Duconseil P. A transcriptomic signature to predict adjuvant gemcitabine sensitivity in pancreatic adenocarcinoma. Ann Oncol. 2021;32(2):250–260. doi: 10.1016/j.annonc.2020.10.601. [DOI] [PubMed] [Google Scholar]
- 38.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerami E., Gao J., Dogrusoz U. The cBio Cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.