Abstract
In the U.S. in early 2020, heterogenous and incomplete county-scale data on COVID-19 hindered effective interventions in the pandemic. While numbers of deaths can be used to estimate actual number of infections after a time lag, counties with low death counts early on have considerable uncertainty about true numbers of cases in the future. Here we show that supplementing county-scale mortality statistics with socioeconomic data helps estimate true numbers of COVID-19 infections in low-data counties, and hence provide an early warning of future concern. We fit a LASSO negative binomial regression to select a parsimonious set of five predictive variables from thirty-one county-level covariates. Of these, population density, public transportation use, voting patterns and % African-American population are most predictive of higher COVID-19 death rates. To test the model, we show that counties identified as under-estimating COVID-19 on an early date (April 17) have relatively higher deaths later (July 1) in the pandemic.
Keywords: COVID-19, Socioeconomic covariates, Pandemics, County-scale estimation, Social networks, Negative binomial regression, LASSO
1. Introduction
Coronavirus disease 2019 (COVID-19), the pandemic that emerged in Wuhan, China in 2019 (World Health Organization, 2020), increased rapidly across the United States in early 2020, challenging the capacity for a coordinated response. In the absence of a vaccine, two strategies for containing the virus have been social distancing and widespread testing (Bedford et al., 2020, Maharaj and Kleczkowski, 2012). Widespread testing reduces selection bias in estimating the numbers of undocumented infections, a crucial variable in the dynamics of spread (Munster et al., 2020, Li et al., 2020). In early 2020, however, there were not enough COVID-19 testing data in the U.S. to predict infections and health care demand (Munster et al., 2020), given substantial heterogeneity in testing rates in both geographic and socio-economic terms (Chowell and Mizumoto, 2020). Under-reporting of COVID-19 infections was likely substantial, perhaps by orders of magnitude in the U.S., both overall and at the county level (Bendavid et al., 2020, Lau et al., 2020).
During the urgent early phase of such a pandemic, decisions at the level of both individual behavior and public health response are not only crucial (e.g., Dehning et al., 2020), but “have to be made using the scarce data available” (Zhang and Qian, 2020). Rapid, approximate estimates of infection rates, using online data, are valuable in this phase (Bentley and Ormerod, 2010, McIver and Brownstein, 2014, Bancroft and Lee, 2014, Chunara et al., 2013). Here we use U.S. COVID-19 data, from early (17 April 2020) and later (1 July 2020) in the progression of the pandemic, to test a means of county-scale estimation of pandemic virus infections when testing data are still incomplete and heterogeneous. This then offers a means of identifying the most vulnerable counties that have not yet reported significant statistics.
At the scale of U.S. counties, we assume that the numbers of recorded deaths by a given day are the most complete data on the extent of the virus (Baud et al., 2020, Flaxman et al., 2020, Marchant et al., 2020). Even though there will naturally be a distribution of times between infection and death, for purposes of statistical prediction, we assume the number of infections will be proportional to the number of deaths a certain number of days afterward. With the clinical literature as a guide (Huang et al., 2020), we follow Diebner and Timmesfeld (2020) in using regressions to determine the most predictive number of days lag between cases and deaths. We expect the optimal lag, in terms of predictive value to be between one and two weeks. In two clinical studies of patients with confirmed COVID-19 cases in Wuhan, China, in early 2020, the median time from onset of symptoms to ICU admission was 10.5 days ( patients) in one study (Huang et al., 2020) and 10 days ( patients) in another study (Wilson et al., 2020). Comparing the Pearson correlations between the logarithm of cumulative COVID-19 cases versus the logarithm of cumulative COVID-19 deaths in Germany at different time lags, Diebner and Timmesfeld (2020) find that 13 days was the optimal lag. The same 13-day optimal lag was found when comparing log-transformed new (daily) deaths versus cases (Diebner and Timmesfeld, 2020). Other studies determine or use a time delay of 13 days from illness onset to death (Linton et al., 2019, Wang et al., 2020, Yang et al., 2020), so while our regression estimate of ten days (below) delay for the U.S. seems reasonable, we find only limited effect on our regression results by using a 14-day delay. Similarly, an epidemiological model found little impact between using lags of 13, 15 or 18 days (Flaxman et al., 2020).
Although COVID-19 mortality rates are age-dependent (Kucharski et al., 2020, Verity et al., 2020), we use a generalized fatality rate in our regressions, as this may be the only variable available from early county-aggregated data, as well as include two age variables in our regressions (mean age and % over 65 years old).
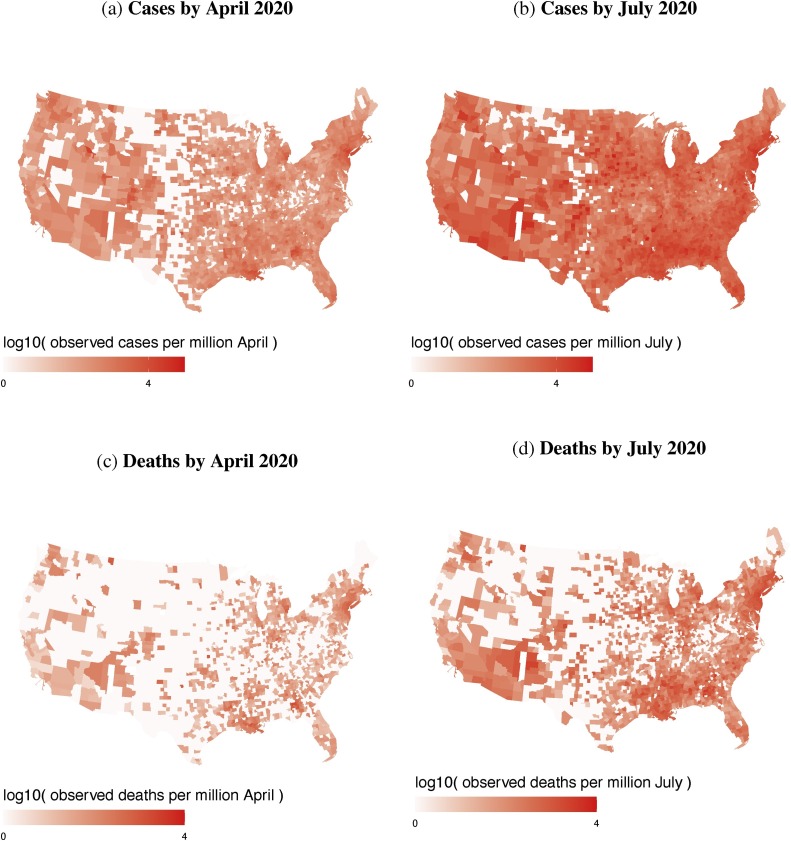
In using county-scale death statistics, a source of statistical uncertainty is the relatively low numbers of deaths early in the pandemic. On April 17, 2020, for example, a large portion of U.S. counties were not reporting any deaths, whereas by 1 July 2020 many more counties were reporting nonzero deaths (Fig. 1a and b). Since the early increase of infections was exponential (Fig. S2), as expected in compartmental models (Wu et al., 2020b, Kucharski et al., 2020), the level of under-reporting can outpace increases in testing rates. As the early numbers of COVID-19 cases in states can differ by orders of magnitude (e.g., New York vs. Great Plains states; Figs. 1a and S2), their differences in under-reporting could be substantial.
Fig. 1.
Maps of the numbers, per million in each county population, of confirmed COVID-19 cases by April (upper left) versus July 2020 (upper right) and deaths by April (lower left) versus July 2020 (lower right). Gray counties designate zero reported cases/deaths.
This raises the concern as to which counties reporting few deaths and cases early in the pandemic might be most vulnerable to relatively higher cases and deaths as the pandemic spreads. To address this, we can make use of the rich co-variate data available at the U.S. county level to improve estimates of under-reporting. Although these county-level estimates are our main objective, the multivariate regression also identifies factors most predictive of COVID-19 deaths. As our objective is prediction, we note that the most predictive variables may or may not be the most causal. Given, for example, the importance of age, physical clustering and pre-existing conditions to COVID-19 (Kucharski et al., 2020, Verity et al., 2020, Centers for Disease Control, 2020, Thompson, 2020, Lu et al., 2020, Yusef et al., 2020), certain county-level variables may have direct correlation; others will have indirect correlation. For example, a likely direct effect on the COVID-19 death to case ratio is the number of hospital ICU beds in the county (Schulte et al., 2020, King, 2020). An indirect effect may be median household income: high income counties may have more jobs that can be performed remotely (del Rio-Chanona et al., 2020). Some variables likely subsume the effects of other variables. Nonwhite populations may have higher rates of COVID-19 infections and/or deaths, for reasons that could include distrust in healthcare (Armstrong et al., 2008), reliance on public transportation (Anderson, 2016), exposure to greater air pollution (Ard and Bullock, 2020) or higher incidence of chronic health conditions (Fang et al., 2020, Lighter et al., 2020).
For our ‘early warning’ estimates, we do not use data on protective behaviors such as social distancing and mask wearing. While these behaviors affect (reduce) the number of infections, they are delayed, dynamic responses to the number of cases (see Supplementary Fig. S1) and unlikely to be available early in the pandemic. In the case of COVID-19, for example, survey data on mask wearing were collected and reported in mid-July 2020 (Katz et al., 2020), months after an early warning system could be implemented.
In essence, we use the county-level death statistics to estimate how many people in the county were infected by COVID-19 on a given date. Even if the death counts were 100% accurate, however, due to all the counties with low numbers, we need to impute the underlying likelihood of death in those counties. The county-scale likelihood estimates provide non-zero values for predictive purposes on all counties, including sparsely-populated counties or counties that the epidemic has not yet fully reached. Comparing observations of cases and deaths from April and July 2020, we can test whether the method identified in April the most vulnerable counties to subsequent infections in July.
Due to the exponential growth in numbers plus the sparseness of count data from many counties, here we will be using a negative binomial regression with the Least Absolute Shrinkage and Selection Operator (LASSO) method to estimate deaths across the country at a given date (a new regression must be carried for each selected date). When these death estimates are divided by a generalized fatality rate, the result is an estimation of the true numbers of infected people in each county. Comparing the estimated numbers of infected people to the observed number of confirmed cases gives us a measure of case under-estimation in each county.
2. Methods
To improve estimations from heterogeneous COVID-19 death data at the U.S. county level, here we introduce thirty-one covariates (Table 1 ) into a negative-binomial regression. These covariates cover nine broad categories: clustering of population, voting behavior, health-care access, preexisting health conditions, age, ethnicity, links to COVID-19 hotspots, employment and education. Included among these covariates are three scalar variables to capture some of the international spread of COVID-19 from early hotspots (Callaway et al., 2020), specifically China, Italy, and Iran (Table 1). These fixed scalar effects for each U.S. county are derived from the Facebook social connectedness index (Bailey et al., 2018).
Table 1.
Covariates for predicting cumulative COVID-19 deaths and under-reporting rates, at the county level of aggregation. The portal for U.S. Census data (U.S. Census Bureau, 2020a, U.S. Census Bureau, 2020b) is www.census.gov.
| Covariates in matrix | Description | Source |
|---|---|---|
| Clustering | ||
| Population size | U.S. Census | |
| Population density | U.S. Census | |
| Persons per house | U.S. Census | |
| Persons per bedroom | U.S. Census | |
| Persons per room | U.S. Census | |
| Public transport | U.S. Census | |
| Voteshare | ||
| % Democratic – Republican | MIT Election Lab | |
| Health care | ||
| ICU beds per capita | Schulte et al. (2020) | |
| Hospitals | Schulte et al. (2020) | |
| % no health insurance | U.S. Census | |
| Health | ||
| % Obese (BMI 30) | CDC | |
| Diabetes | CDC | |
| Hypertension | Olives et al. (2013) | |
| Air pollution ( | Wu et al. (2020a) | |
| Age | ||
| Mean age | U.S. Census | |
| % over 65 years old | U.S. Census | |
| Ethnicity | ||
| Black | Black | U.S. Census |
| Hispanic | Hispanic | U.S. Census |
| Native | Native American | U.S. Census |
| Facebook connections to: | ||
| China (per capita) | Facebook SCI | |
| Italy (per capita) | Facebook SCI | |
| Iran (per capita) | Facebook SCI | |
| Employment | ||
| Median household income | U.S. Census | |
| , , | Jobs: professional, service | U.S. Census |
| , , | Jobs: office, trade, transport | U.S. Census |
| Incarcerated | Vera Inst. (2020) | |
| Education | ||
| College educated | U.S. Census | |
| No high school | U.S. Census | |
All variables in Table 1 represent fixed, county-level effects. While dynamic effects of social networks and inter-county travel were surely a factor in coronavirus spread (Maier and Brockmann, 2020), we do not employ such dynamic data here for two reasons. The first is that we aim for early estimation using fixed county-level covariates that would be already available at the onset of a future pandemic. Secondly, there is not currently an established method for adding network covariates to the method we employ here, a LASSO Negative Binomial regression (Hays et al., 2010, Silk et al., 2017, Leifeld and Cranmer, 2019). We reserve the challenge of adding dynamic network data for future work.
Since case numbers grow exponentially, our regressions use the logarithm of county-level death counts on day , , where the vector contains entries for each of 3088 counties. Even when log-transformed, however, the count data are likely to be overdispersed and subsequent standard errors underestimated. For this reason, we use a negative binomial distribution of errors in the regressions, which allows us to use number of COVID-19 deaths as count data for predictions. To estimate the numbers of deaths, , in all counties on day , the regression relationship is:
| (1) |
where is the vector of weights for the covariates (see Table 1). The errors follow a negative binomial distribution and have a variance for a given mean, , of , where is a dispersion parameter. In the regression, we use sandwich corrected estimates of standard errors (Luque-Fern et al., 2016).
Using thirty-one related covariates, while comprehensive, will likely result in both overfitting and colinearity in the Negative Binomial regression. To mitigate these risks, we employ a LASSO penalization in the loss function of the Negative Binomial regression. LASSO selects the most predictive variables by regularization, forcing many of the estimated effect sizes to zero; the most predictive covariates are those left with non-zero effects. Importantly, as LASSO is a method of dimension reduction that focuses on prediction, covariates that are set to zero may actually be causal in the real world. Conversely, highly predictive covariates may actually not be causal, they may have just subsumed the variance of many other truly causal covariates.
In the LASSO process, the choice of the regularization parameter, , is important, as higher results in fewer non-zero parameters. We optimize using 2-fold cross-validation over a range of possible values. The LASSO regularization minimizes the following function, which plays-off the log likelihood, versus the sum of the individual coefficients in multiplied by (Lehman and Archer, 2019):
| (2) |
where in this case, indexes the 3088 counties in the sample. For a given likelihood , the higher is, the smaller must be, and the fewer non-zero parameters are allowed. We apply this negative binomial regression to COVID-19 deaths data on both 17 April and 1 July, 2020. To maximize the number of non-zero predictors, we chose the largest that did not markedly reduce the out-of-sample predictive power of the LASSO regression. This turned out to be ; higher values reduced the cross-validated log likelihood of the LASSO regression (see Fig. S3).
Next, as our estimate of the number actually infected in county on date , , we divide by the fatality rate, . We then define the under-estimation in the county, , as the ratio of confirmed cases on record, , to . Measuring under-estimation as a ratio—the difference of the log-transformed values ()—rather than a simple difference helps account for the highly skewed distribution of cases across counties. Using vector notation to represent all 3088 counties in the sample, we determine :
| (3) |
Using data from the two dates allows us to determine whether determined for low-data counties in April is predictive of unexpectedly high numbers of COVID-19 deaths in July.
3. Results
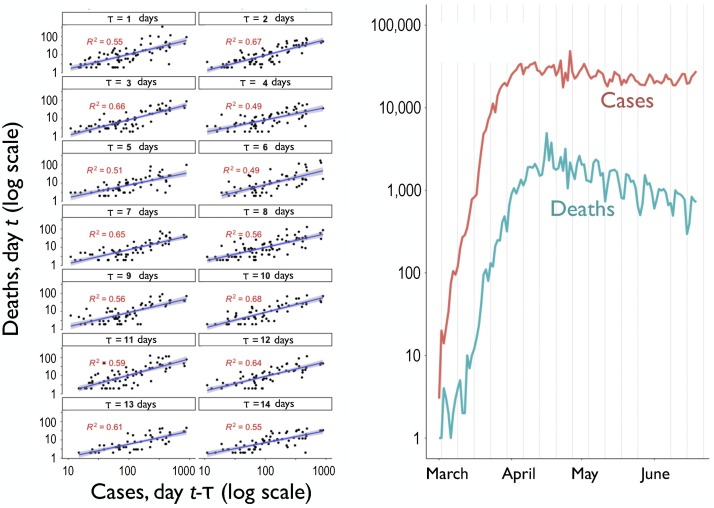
For estimating COVID-19 infection rates from death rates, we first determined an optimum lag between COVID-19 infection and death statistics (log-transformed) in 3088 counties. We ran forty regressions of the logged number of new deaths on April 13 against the logged number of new cases, at lag times ranging between 1 and 40 days. The largest value occurred with a lag of (see Fig. 2 ), which is in line with findings from other studies (Backer et al., 2020, Huang et al., 2020, Russell et al., 2020, Wilson et al., 2020).
Fig. 2.
Left: COVID-19 cases on day versus deaths on day (both log scale) for all U.S. counties with more than 1,000,000 people, from to days’ delay. All are comparing versus a case date of 04/13/2020. Right: Daily deaths versus daily cases in the U.S., with logarithmic y-axis.
We confirm that our results are robust to variation in lag time by showing our under-reporting statistic still predicts more future deaths, using mean lag times between contracting COVID-19 and death, , of 0, 7, 10 and 14 days (Supplementary Figs. S4–S6). We find positive slopes in all cases (Supplementary Tables S3–S6), indicating that our under-reporting statistic is predictive of a higher number of future COVID-19 cases. We stress that the value of is strictly a parameter for use in prediction, not a definitive statement about duration of infection for individuals. Indeed the parameter —which we estimated for COVID-19 in the U.S. in 2020—would ideally be estimated independently for each case study. The lag would likely differ, for example, in a future pandemic and/or within health/hospital systems of the world (e.g., Wood, 2020).
3.1. Five predictive covariates
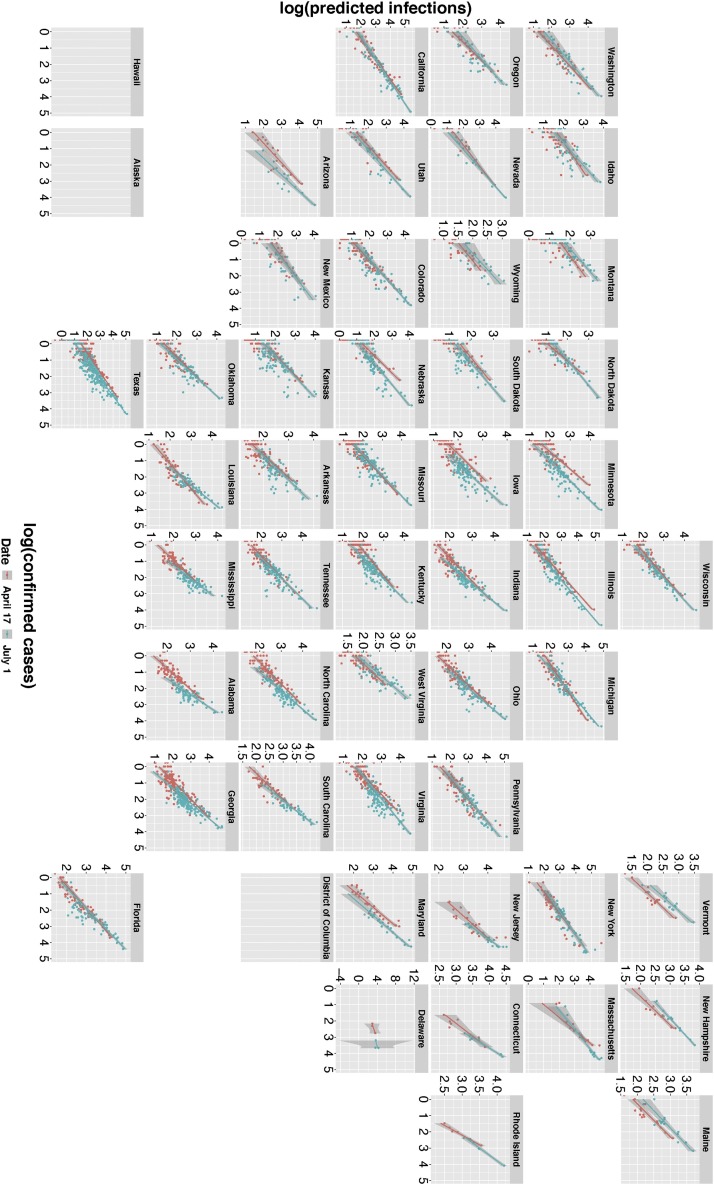
Of the thirty-one variables (Table 1) entered into the LASSO negative binomial regression, five were retained by the LASSO analysis at the optimal level of as most predictive of COVID-19 deaths by county (Table 2 ). Consistent with existing literature (discussed below), the five predictive variables are: population size, population density, public transport, voteshare, and % African-American population. Table 2 shows their regression coefficients via Eq. (1). These five variables explain a significant proportion of the variance in county death data: comparing predicted versus confirmed death counts (both log-transformed) yields for 17 April, and for 1 July (Table 2). Using these coefficients, Fig. 3 compares vs in all counties for two dates in 2020, April 17 and July 1. Using a similar LASSO regression (see Supplement), we confirm that deaths are a much better predictor of than the full 31 covariates of Table 1. By themselves, these fixed socioeconomic variables cannot predict which counties are likely to under-report COVID-19 cases.
Table 2.
Coefficients for the five variables retained by the LASSO regression () for prediction of COVID-19 deaths and cases, on 17 April 2020 and 1 July 2020. Variables from Table 1 with zero effect at are not shown. Population variables were log-transformed. There were 3088 observations. For a full list of effects at different levels of , see Supplementary Tables.
| Deaths | Deaths | |
|---|---|---|
| Co-variate | 17 April | 1 July |
| Population size () | 0.87 | 0.92 |
| Population density () | 0.14 | 0.13 |
| Public transport () | 5.36 | 1.36 |
| Voteshare () | 0.18 | 0.05 |
| % African American | 1.07 | 1.96 |
| vs. observed deaths | 0.47 | 0.60 |
Fig. 3.
Predicted log-transformed COVID-19 infections, () versus log-transformed confirmed cases (). Each plot represents a different state, and datum points are counties within each state. Colors show determinations for two dates, April 17, 2020 (red) and July 1, 2020 (blue). Regression lines use points weighted by the county populations. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
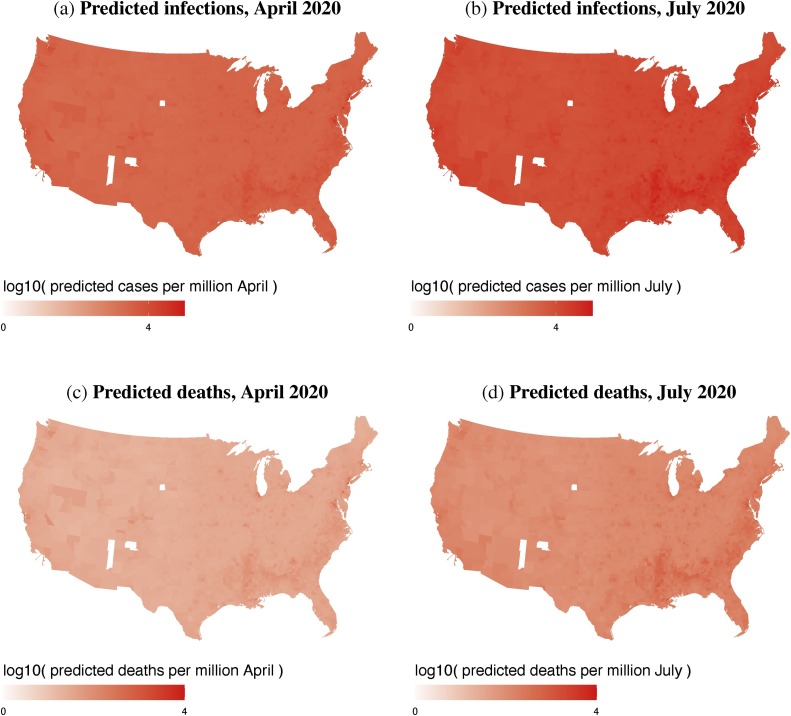
Since LASSO is a factor-reduction technique, rather than a definitive statement of causality, we discuss first the predictive value of these five covariates as an ‘early warning system’ to identify US counties of particular concern. The predictions are mapped at county level in Fig. 4 , which shows county-level predicted COVID-19 cases and deaths for April 17, and July 1, 2020. The maps fill in the data gaps across the less populated counties in the middle of the country, including the Great Plains (cf. Fig. 1). Similar to the raw numbers (Fig. 1), the predicted numbers are highest on the coasts, the northeast, Florida, and major urban areas.
Fig. 4.
County-level predicted COVID-19 infections, (top row) and deaths, (bottom row) for April 17, and July 1, 2020. Data are log-transformed.
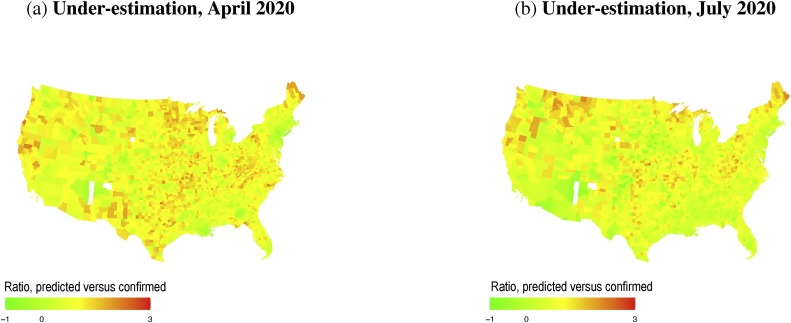
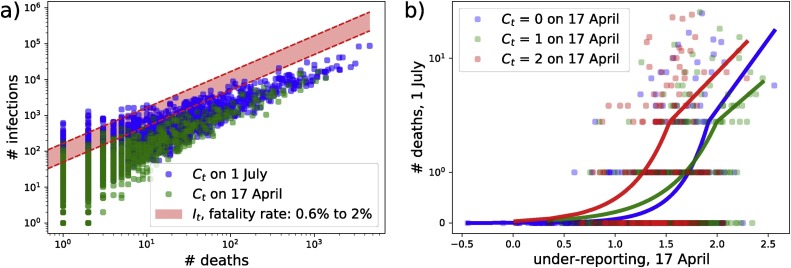
Fig. 5 shows county-level maps of the under-estimation measure, , for April 17 and July 1. These maps of have less obvious patterning than maps of observed infections or deaths (Fig. 1, Fig. 4). Many of the highest values of in July are found in counties in the northern states (Maine, Idaho, Montana and Michigan), as well as parts of northern California and Oregon. Using a fatality rate of 1%, our estimations of actual infections, , are one to two orders of magnitude larger than confirmed case numbers, , as shown in Fig. 6 a, which shows both and on April 17 and July 1. This is broadly consistent with localized estimates, such as a California county where the COVID-19 infection rate was “50–85-fold more than the number of confirmed cases” in early April (Bendavid et al., 2020). In China, by comparison, 6 out of 7 COVID-19 infections (before 23 January) were potentially not reported (Li et al., 2020).
Fig. 5.
COVID-19 under-estimation, , for April 17, and July 1, 2020.
Fig. 6.
(a) Deaths versus COVID-19 infections in each county. (Green dots: confirmed cases vs deaths on 17 April, 2020. Blue dots: confirmed cases vs deaths on 1 July, 2020. Red band: Predicted infections for a range of fatality rates between 0.6 and 2%.) (b) Under-reporting on 17 April, 2020 vs deaths on July 1, 2020; where green circles are counties where on 17 April (), blue circles where () and red circles where (). Solid lines are best fits from a negative binomial regression. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6a shows that the gap between the confirmed cases and the predicted infections (red band) increases with number of deaths (i.e., population size). We test how well estimated for low-information counties on 17 April identifies those counties with high death counts by July. For low-data counties, our estimate of can offer an indicator of risk at the county level, as shown in Fig. 6b. Determining for 17 April, we predict which counties were at most risk for relatively high levels of COVID-19 by July 1. Fig. 6b shows that, for counties with little data in April 2020, a high under-reporting estimate was an effective early warning signal of excess deaths by July 2020. Fig. 6b shows this for the 1171 counties with 0 (), 1 () or 2 () confirmed cases in April. The fits show that counties with larger under report scores had higher COVID-19 deaths, ranging between just over 0 and 5 on average. This may seem like a small number, but with our assumed fatality rate of 1%, this represents an outbreak stretching into the hundreds in counties with fewer than 3 reported cases in April. There is also a substantial range on these predictions, with 13 out of the 1171 counties reporting 15 or more deaths, indicating an outbreak of more than 1500 cases.
4. Discussion and conclusion
In this prediction exercise, certain variables identified by LASSO explain more of the variation in the outcome than others. We should avoid assigning actual causation with the value these variables have in predicting true number of infections. Nevertheless, the five covariates in Table 2 are among the prominent risk factors listed by the CDC, and we may speculate on how they relate to the twenty-two variables not retained by the LASSO at the optimized value (see Supplement for results using other values of ).
Equally notable are the twenty-two variables from Table 1 not returned in the LASSO results (Table 2). This does not mean these variables are not important in the real world, but that for the purposes of predicting case numbers, the information in the five are sufficient to supersede, or encompass, the information residing in the other twenty-six. Notably, none of the four pre-existing health conditions—obesity, diabetes, hypertension or pollution—was selected by LASSO as predictive of COVID-19 deaths or cases. Given the studies showing these to be genuine risk factors for individuals (Lighter et al., 2020), one of the five variables in Table 2 must be subsuming their predictive effects. The importance of African-American proportion of the population is consistent with the literature on socioeconomic correlates with COVID-19. In different ZIP codes of New York City, for example, Lieberman-Cribbin et al. (2020) found that, as the proportion of white residents increased, the number of COVID-19 tests increased and fraction of those testing positive decreased. Again, as LASSO is a factor-reduction method, we suspect that the proportion African American has such predictive significance that it subsumed the predictive effects of other covariates such as diabetes, hypertension, pollution and income. This may explain why income was not an important predictive variable in the LASSO results, despite the relative lack of testing resources in poorer U.S. counties (Schulte et al., 2020, van Dorn et al., 2020) and the overall disparity in COVID-19 testing efforts and resources attributable to socioeconomic and racial disparities in healthcare access (Karaye and Horney, 2020, Lieberman-Cribbin et al., 2020).
As COVID-19 mortality rates depend on an individual's age (Kucharski et al., 2020, Verity et al., 2020), it may seem surprising to see both % over 65 years old and mean adult age not among the most important factors emerging from the LASSO results (Table 2). Connections with Italy, China or Iran were also not among the five most predictive factors, in April or in July. This despite evidence that the U.S. received confirmed “index” cases from Europe and from China (Spiteri et al., 2020).
Voteshare in the last election (% Democratic % Republican) was an important variable in predicting deaths, both in April 17 and on July 1 (Table 2). The use of two dates helps us rule out the effect of counties in New York state acting as outliers. In April 2020, New York state had the highest number of reported COVID-19 cases, and was also among the most Democratic-leaning. The voteshare effect remains, however, by July 1, when both cases and deaths were higher in many other parts of the country, including counties in the U.S. South, where voteshare is much different.
It could be that democratic voteshare captures aspects left unrepresented by the other crowding variables. Various surveys in the U.S. in 2020 (FiveThirtyEight, 2020) have shown Democratic voters to have higher levels of concern about COVID-19 than Republican voters. Given that Democratic voteshare predicts more deaths, this might be due to higher levels of liberal behavior among states from the West coast and North-East driven by differences in culture (Harrington and Gelfand, 2014). Democratic voteshare may act as a proxy for more openness, tolerance (Ruck et al., 2020) and looser norms (Harrington and Gelfand, 2014). By contrast, collectivist cultures may be better equipped to mitigate a pandemic through a tendency to obey authority (Kemmelmeier et al., 2003), conform with peers (Murray et al., 2011) and engage in less physical contact (Wu et al., 2019). Further consideration of this hypothesis would need to account for the demographics of strongly Democratic counties.
Overall, we find that the use of socioeconomic determinants allows—as a supplement to existing SIR models—for rapid, early warning estimations for vulnerable counties at times when county-scale reporting data is heterogeneous and incomplete. In using death counts with a ten-day time lag, we were able to predict which counties were under-reporting COVID-19 cases in April and validate these predictions against fatality rates in July.
In future pandemics this ‘early warning system’ could be used to identify vulnerable counties where disease outbreaks have not yet occurred. False positives will be produced but this may be improved by expanding our set of 31 covariates. While the results present a parsimonious set of socioeconomic risk factors for COVID-19 prevalence, additional covariate data will inevitably become available for early warning tools in future events. With further research, the methodology we have laid out here can be adapted to incorporate dynamic and/or network data, such as seasonality and ultraviolet light exposure (Carleton et al., 2020, Merow and Urban, 2020), inter-county migration or the Facebook connectedness Index. The incorporation of the more complex data into these early warning tools is a goal for future work.
Author contributions
Conception and design of study: R.A. Bentley, D.J. Ruck, J. Borycz
Acquisition of data: D.J. Ruck, J. Borycz, R.A. Bentley
Analysis and/or interpretation of data: D.J. Ruck, R.A. Bentley, J. Borycz
Drafting the manuscript: R.A. Bentley
Revising the manuscript: R.A. Bentley, D.J. Ruck
Approval of the submitted manuscript: R.A. Bentley, D.J. Ruck, J. Borycz
Data accessibility
Data and relevant code for this research work are stored in GitHub repository: https://github.com/damianruck/EWS-pandemic-socioeconomic-variables.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgments
We acknowledge support by NSF grant 2028710. We thank Facebook for access to aggregated Facebook friendship data. DJR was supported by College of Arts and Sciences, U. of Tennessee. We thank Theodore Papamarkou, Luke Matthews, Tamara Chavez-Lindell and anonymous reviewers for comments.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ehb.2021.100988.
Appendix B. Supplementary data
The following are the supplementary data to this article:
References
- Anderson M. 2016. Who Relies on Public Transit in the U.S. Pew Research.https://www.pewresearch.org/fact-tank/2016/04/07/who-relies-on-public-transit-in-the-u-s/ [Google Scholar]
- Ard K., Bullock C. In: Spatiotemporal Analysis of Air Pollution and Its Application in Public Health. Li L., Zhou X., Tong W., editors. Elsevier; Washington, DC: 2020. Concentrating risk? The geographic concentration of health risk from industrial air toxics across America; pp. 277–292. [Google Scholar]
- Armstrong K., McMurphy S., Dean L.T., Micco E., Putt M., Halbert C.H., Schwartz J.S., Sankar P., Pyeritz R.E., Bernhardt B., Shea J.A. Differences in the patterns of health care system distrust between blacks and whites. J. Gen. Internal Med. 2008;23(6):827–833. doi: 10.1007/s11606-008-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eurosurveillance. 2020;25:2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M., Cao R., Kuchler T., Stroebel J., Wong A. Social connectedness: measurement, determinants, and effects. J. Econ. Perspect. 2018;32(3):259–280. [PubMed] [Google Scholar]
- Bancroft E.A., Lee S. Use of electronic death certificates for influenza death surveillance. Emerg. Infect. Dis. 2014;20(1):78–82. doi: 10.3201/eid2001.130471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D., Qi X., Nielsen-Saines K., Musso D., Pomar L., Favre G. Real estimates of mortality following COVID-19 infection. Lancet Infect. Dis. 2020;20(7):773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M., Sall A.A., Schuchat A., Ungchusak K., Wieler L.H. COVID-19: towards controlling of a pandemic. Lancet. 2020;395(10229):1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid E., Mulaney B., Sood N., Shah S., Ling E., Bromley-Dulfano R., Lai C., Weissberg Z., Saavedra R., Tedrow J., Tversky D., Bogan A., Kupiec T., Eichner D., Gupta R., Ioannidis J., Bhattacharya J. 2020. COVID-19 Antibody Seroprevalence in Santa Clara County, California. medRxiv 2020.04.14.20062463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R.A., Ormerod P. A rapid method for assessing social versus independent interest in health issues. Soc. Sci. Med. 2010;71(3):482–485. doi: 10.1016/j.socscimed.2010.03.042. [DOI] [PubMed] [Google Scholar]
- Callaway E., Cyranoski D., Mallapaty S., Stoye E., Tollefson J. Coronavirus by the numbers. Nature. 2020;579:482–483. doi: 10.1038/d41586-020-00758-2. [DOI] [PubMed] [Google Scholar]
- Carleton T., Cornetet J., Huybers P., Meng K., Proctor J. 2020. Evidence for Ultraviolet Radiation Decreasing COVID-19 Growth Rates: Global Estimates and Seasonal Implications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2020. COVID-19 Hospitalization and Death by Race/Ethnicity. [Google Scholar]
- Chowell G., Mizumoto K. The COVID-19 pandemic in the USA: what might we expect? Lancet. 2020;395(10230):1093–1094. doi: 10.1016/S0140-6736(20)30743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunara R., Smolinski M.S., Brownstein J.S. Why we need crowdsourced data in infectious disease surveillance. Curr. Infect. Dis. Rep. 2013;15(4):316–319. doi: 10.1007/s11908-013-0341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehning J., Zierenberg J., Spitzner P., Wibral M., Neto J.P., Wilczek M., Priesemann V. Inferring change points in the spread of COVID-19 reveals the effectiveness of interventions. Science. 2020;369(6500):160. doi: 10.1126/science.abb9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio-Chanona R.M., Mealy P., Pichler A., Lafond F., Farmer D. Supply and demand shocks in the COVID-19 pandemic: an industry and occupation perspective. COVID Econ. 2020;6:65–104. [Google Scholar]
- Diebner H.H., Timmesfeld N. Preprints 2020090628; 2020. Exploring COVID-19 Daily Records of Diagnosed Cases and Fatalities Based on Simple Non-Parametric Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Karakiulakis G., Rotha M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8(4):e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FiveThirtyEight. https://fivethirtyeight.com/coronavirus-polls. (Accessed 28 April 2020).
- Flaxman S., Mishra S., Gandy A., Unwin H.J.T., Mellan T.A., Coupland H., Whittaker C., Zhu H., Berah T., Eaton J.W., Monod M., Imperial College COVID-19 Response Team, Ghani A.C., Donnelly C.A., Riley S., Vollmer M.A.C., Ferguson N.M., Okell L.C., Bhatt S. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020;584(7820):257–261. doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- Harrington J.R., Gelfand M.J. Tightness-looseness across the 50 United States. Proc. Natl. Acad. Sci. U. S. A. 2014;111(22):7990–7995. doi: 10.1073/pnas.1317937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays J.C., Kachi A., Franzese R.J., Jr. A spatial model incorporating dynamic, endogenous network interdependence: a political science application. Stat. Methodol. 2010;7(3):406–428. [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaye I.M., Horney J.A. The impact of social vulnerability on COVID-19 in the U.S.: an analysis of spatially varying relationships. Am. J. Prev. Med. 2020;59(3):317–325. doi: 10.1016/j.amepre.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J., Sanger-Katz M., Quealy K. A detailed map of who is wearing masks in the U.S. The New York Times. 2020;(July 17) [Google Scholar]
- Kemmelmeier M., Burnstein E., Krumov K., Genkova P., Kanagawa C., Hirshberg M.S., Erb H.-P., Wieczorkowska G., Noels K.A. Individualism, collectivism, and authoritarianism in seven societies. J. Cross-Cult. Psychol. 2003;34(3):304–322. [Google Scholar]
- King J.S. Covid-19 and the need for health care reform. N. Engl. J. Med. 2020 doi: 10.1056/NEJMp2000821. [DOI] [PubMed] [Google Scholar]
- Kucharski A.J., Russell T.W., Diamond C., Liu Y., Edmunds W.J., Funk S., Eggo R.M. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect. Dis. 2020;20:553–558. doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H., Khosrawipour T., Kocbach P., Ichii H., Bania J., Khosrawipour V. Evaluating the massive underreporting and undertesting of COVID-19 cases in multiple global epicenters. Pulmonology. 2020 doi: 10.1016/j.pulmoe.2020.05.015. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman R.R., Archer K.J. Penalized negative binomial models for modeling an overdispersed count outcome with a high-dimensional predictor space. PLoS ONE. 2019;14(1):e0209923. doi: 10.1371/journal.pone.0209923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifeld P., Cranmer S.J. A theoretical and empirical comparison of the temporal exponential random graph model and the stochastic actor-oriented model. Netw. Sci. 2019;7(1):20–51. [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020;368(6490):489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Cribbin W., Tuminello S., Flores R.M., Taioli E. Disparities in COVID-19 testing and positivity in New York City. Am. J. Prev. Med. 2020;59(3):326–332. doi: 10.1016/j.amepre.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighter J., Phillips M., Hochman S., Sterling S., Johnson D., Francois F., Stachel A. Obesity in patients younger than 60 years is a risk factor for Covid-19 hospital admission. Clin. Infect. Dis. 2020;71(15):896–897. doi: 10.1093/cid/ciaa415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton N.M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A.R., Jung S.M., Yuan B., Kinoshita R., Nishiura H. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation. J. Clin. Med. 2019;9(2):538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Gu J., Li K., Xu C., Su W., Lai Z., Zhou D., Yu C., Xu B., Yang Z. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg. Infect. Dis. 2020;26(7):1628–1631. doi: 10.3201/eid2607.200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque-Fernandez M.A., Belot A., Quaresma M., Maringe C., Coleman M.P., Rachet B. Adjusting for overdispersion in piecewise exponential regression models to estimate excess mortality rate in population-based research. BMC Med. Res. Methodol. 2016;16(1) doi: 10.1186/s12874-016-0234-z. Article 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharaj S., Kleczkowski A. Controlling epidemic spread by social distancing: do it well or not at all. BMC Public Health. 2012:12. doi: 10.1186/1471-2458-12-679. Article 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B.F., Brockmann D. Effective containment explains subexponential growth in recent confirmed COVID-19 cases in China. Science. 2020;368(6492):742–746. doi: 10.1126/science.abb4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant R., Samia N.I., Rosen O., Tanner M.A., Cripps S. arXiv:2004.04734; 2020. Learning as We Go: An Examination of the Statistical Accuracy of COVID19 Daily Death Count Predictions. [Google Scholar]
- McIver D.J., Brownstein J.S. Wikipedia usage estimates prevalence of influenza-like illness in the United States in near real-time. PLoS Comput. Biol. 2014;10(4):e1003581. doi: 10.1371/journal.pcbi.1003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merow C., Urban M.C. Seasonality and uncertainty in global COVID-19 growth rates. Proc. Natl. Acad. Sci. U. S. A. 2020;117(44):27456–27464. doi: 10.1073/pnas.2008590117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China: key questions for impact assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Murray D.R., Trudeau R., Schaller M. On the origins of cultural differences in conformity: four tests of the pathogen prevalence hypothesis. Pers. Soc. Psychol. Bull. 2011;37(3):318–329. doi: 10.1177/0146167210394451. [DOI] [PubMed] [Google Scholar]
- Olives C., Myerson R., Mokdad A.H., Murray C.J., Lim S.S. Prevalence, awareness, treatment, and control of hypertension in United States counties, 2001–2009. PLOS ONE. 2013;8(4):e60308. doi: 10.1371/journal.pone.0060308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck D.J., Bentley R.A., Lawson D.J. Cultural prerequisites of socioeconomic development. R. Soc. Open Sci. 2020;7(2):190725. doi: 10.1098/rsos.190725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell T.W., Hellewell J., Jarvis C.I., van Zandvoort K., Abbott S., Ratnayake R., Cmmid Covid-Working Group, Flasche S., Eggo R.M., Edmunds W.J., Kucharski A.J. Estimating the infection and case fatality ratio for coronavirus disease (COVID-19) using age-adjusted data from the outbreak on the Diamond Princess cruise ship, February 2020. Eurosurveillance. 2020;25(12):2000256. doi: 10.2807/1560-7917.ES.2020.25.12.2000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte F., Lucas E., Rau J., Szabo L., Hancock J. Millions of older Americans Live in counties with no ICU beds as pandemic intensifies. Kaiser Health News. 2020;(March 20) Data: https://khn.org/wp-content/uploads/sites/2/2020/03/KHN-ICU-bed-county-analysis_2.zip. [Google Scholar]
- Silk M.J., Croft D.P., Delahay R.J., Hodgson D.J., Weber N., Boots M., McDonald R.A. The application of statistical network models in disease research. Methods Ecol. Evol. 2017;8:1026–1041. [Google Scholar]
- Spiteri G., Fielding J., Diercke M., Campese C., Enouf V., Gaymard A., Bella A., Sognamiglio P., Sierra Moros M.J., Riutort A.N., Demina Y.V., Mahieu R., Broas M., Bengnér M., Buda S., Schilling J., Filleul L., Lepoutre A., Saura C., Mailles A., Levy-Bruhl D., Coignard B., Bernard-Stoecklin S., Behillil S., van der Werf S., Valette M., Lina B., Riccardo F., Nicastri E., Casas I., Larrauri A., Salom Castell M., Pozo F., Maksyutov R.A., Martin C., Van Ranst M., Bossuyt N., Siira L., Sane J., Tegmark-Wisell K., Palmérus M., Broberg E.K., Beauté J., Jorgensen P., Bundle N., Pereyaslov D., Adlhoch C., Pukkila J., Pebody R., Olsen S., Ciancio B.C. First cases of coronavirus disease 2019 (COVID-19) in the WHO European Region, 24 January to 21 February 2020. Eurosurveillance. 2020;25(9):2000178. doi: 10.2807/1560-7917.ES.2020.25.9.2000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson H. COVID-19 case clusters offer lessons and warnings for reopening. Sci. News. 2020;198(3) [Google Scholar]
- U.S. Census Bureau . 2020. Small Area Health Insurance Estimates.https://www.census.gov/content/dam/Census/library/publications/2019/demo/p30-05.pdf [Google Scholar]
- U.S. Census Bureau . 2020. Small Area Income and Poverty Estimates.https://www.census.gov/programs-surveys/saipe/about.html [Google Scholar]
- van Dorn A., Cooney R.E., Sabin M.L. COVID-19 exacerbating inequalities in the US. Lancet. 2020;395(10232):1243–1244. doi: 10.1016/S0140-6736(20)30893-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Inst . Vera Institute of Justice; 2020. Incarceration Trends Dataset.https://github.com/vera-institute/incarceration_trends [Google Scholar]
- Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect. Dis. 2020;20(6):669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N., Kvalsvig A., Barnard L.T., Baker M.G. Case-fatality risk estimates for COVID-19 calculated by using a lag time for fatality. Emerg. Infect. Dis. 2020;26(6):1339–1441. doi: 10.3201/eid2606.200320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.N. arXiv:2005.02090; 2020. Did COVID-19 Infections Decline before UK Lockdown? [Google Scholar]
- World Health Organization . 2020. Novel Coronavirus (2019-nCoV) Situation Report – 11. [Google Scholar]
- Wu M.S., Li B., Zhu L., Zhou C. Culture change and affectionate communication in China and the United States: evidence from Google digitized books 1960–2008. Front. Psychol. 2019;10:1110. doi: 10.3389/fpsyg.2019.01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Nethery R.C., Sabath B.M., Braun D., Dominici F. 2020. Exposure to Air Pollution and COVID-19 Mortality in the United States: A Nationwide Cross-Sectional Study. medRxiv:2020.04.05.20054502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusef D., et al. Large outbreak of coronavirus disease among wedding attendees, Jordan. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2609.201469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Qian B. Making decisions to mitigate COVID-19 with limited knowledge. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and relevant code for this research work are stored in GitHub repository: https://github.com/damianruck/EWS-pandemic-socioeconomic-variables.