Abstract
Root-associated bacteria are able to influence plant fitness and vigor. A key step in understanding the belowground plant-bacteria interactions is to quantify root colonization by the bacteria of interest. Probably, genetic engineering with fluorescence markers is the most powerful way to monitor bacterial strains in plant. However, this could have some collateral problems and some strains can be challenging to label. In this sense, bacterial inoculation under properly controlled conditions can enable reliable and reproducible quantification of natural bacterial strains. In this protocol, we describe a detailed procedure for quantification of root-associated bacteria. This method applies non-aggressive samples processed with morphological identification and PCR-based genetic fingerprinting. This easy-to-follow protocol is suitable for studying bacterial colonization of plants grown either in artificial medium or in soil.
Keywords: Plant-bacteria interaction, Rhizobacteria, PGPR, Pathogen, Colonization, Root, Soil
Background
Plants naturally live with various soil bacteria in the rhizosphere, which refers to a thin layer of soil adhering to the roots. While some rhizobacteria have no observable effects on plants, others are either pathogens that cause detrimental effects or growth-promoting rhizobacteria (PGPR) that promote plant vigor ( Mendes et al., 2013 ; Olanrewaju et al., 2019 ). The capacity of bacterial pathogens or PGPR to impact plant growth is tightly correlated with their level of bacterial root colonization. Therefore, the investigation of bacterial root colonization is an important stepping stone to understanding the belowground plant-bacteria interactions.
The abundance of bacteria strain can be assessed by visualization of fluorescence signals by modifying them to express a transgenic marker gene encoding the fluorescent protein such as GFP ( Rochat et al., 2010 ; Krzyzanowska et al., 2012 ; Saad et al., 2018 ). The abundance of bacteria strain can also be measured by PCR-based amplification of the bacterial genomic DNA (Maciá- Vicente et al., 2009 ; Mendis et al., 2018 ). These two methods detect the bacterial strain of interest regardless of the presence or absence of other bacterial species; nevertheless, both methods have their potential limitations. The fluorescence-assisted detection may be limited by technical difficulties during genetic transformation, by the possibility that the introduction of the transgene may interfere with wild-type bacterial behavior, and by the risks posed to environmental safety. On the other side, PCR-based bacterial detection requires that levels of the target DNA templates be above the PCR detection limits, and that the target sequence be specific enough to represent the bacterial strain of interest only.
In addition to the fluorescence-based and the PCR-based methods, root-associated bacteria can also be quantified by in vitro culturing. This increases the detection limits, followed by counting the colony-forming units (CFUs), if the bacterial strain of interest is culturable with an artificial growth medium. With proper experimental setups, the method of counting CFUs is efficient, affordable, reliable, and reproducible for quantification of root-associated bacteria. By using Arabidopsis thaliana and B. megaterium YC4-R4 as a model system for studying plant-bacteria interactions, herein we describe a protocol for bacteria quantification based on counting CFUs from in vitro bacterial cultures. This protocol, which includes an optional genetic fingerprinting step by Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR, has been applied to different bacterial species in studies where plants were either grown in artificial medium or in soil, and under control or stress conditions ( Vílchez et al., 2020 ). In addition to studying root-associated bacteria, this methodology can be applied to studies involving other types of plant organs or other types of bacteria-host systems under optimized conditions.
Materials and Reagents
1.5 ml Eppendorf tubes (ShangYu Yite Plastic Co., Ltd, catalog number: MCTB015, or similar)
15 ml tubes
Surgical blade and scalpellum (Qingdao Sinoland International Trade Co., Ltd, catalog number: SS-002021, or similar)
Circular-shape (9 × 1.5 cm) and square-shape (56 × 35 × 30 cm) Petri dishes (ShangYu Yite Plastic Co., Ltd, catalog number: PD0009, or similar; Haimen Laiboreike Experiment Instrument Manufacturing Co., Ltd, catalog number: LB077, or similar)
Disposable tissue grinder plastic pestle for 1.5 ml tubes (Corning, Axygen®, catalog number: PES-15-B-SI, or similar)
Inoculation Loops (Renon Laboratory Experiment Co., Ltd, catalog number; 52150000 or similar)
Tape (3M MicroporeTM, 1530C-0, or similar)
Seeds of Arabidopsis thaliana
Murashige and Skoog Basal Salt Mixture (MS) (Sigma-Aldrich, catalog number: M5524)
Tryptone (Sigma, catalog number: T7293)
Yeast extract (Sigma, catalog number: Y1625)
NaCl (Sangon Biotech, catalog number: A501218)
NaOH or KOH (Sangon Biotech, catalog numbers: A100583 and A610441)
Agar (Sigma, catalog number: L2897)
Agarose (Sigma, catalog number: A9539)
RedSafeTM Nucleic Acid Staining Solution (Invitrogen, catalog number: S33102)
2K Plus Ladder (Transgenbiotech, Trans2K® Plus DNA Marker, catalog number: BM121)
Double-distilled sterilized H2O
100% Ethanol (Sangon Biotech, catalog number: A500737)
Bleach (commercial 5% NaClO diluted 1:5 with water)
DNA extraction kit (Qiagen DNA Blood&Tissue, Qiagen, catalog number: 69504 or similar)
Master Mix solution for PCR (Transgenbiotech, 2× EasyTaq® PCR SuperMix, AS111-01)
-
Enterobacterial Repetitive Intergenic Consensus fingerprinting and 16S rRNA PCR amplification primers:
ERIC (5’-ATGTAAGCTCCTGGGGATTCAC-3’)
27F (5’-AGAGTTTGATCMTGGCTCAG-3’)
1492R (5’-TACGGYTACCTTGTTACGACTT-3’)
Equipment
Stainless steel tweezer with long fine point (Labdirect, catalog number: SA-Q2090 or similar)
Analytical balance (Sartorius, Practum224-1S or similar)
Mechanical pipettes (Eppendorf Research Plus, P1000, P100 and P10, or similar)
Benchtop vortex mixer (DragonLab MX-S, or similar)
Benchtop centrifuge (Eppendorf, Centrifuge 5810R with A-4-62 Model Rotor, or similar; up to 4,000 rpm/3,220 × g)
Clean bench (BCM-1000A Biological Clean Bench, Airtech, or similar)
Shaker incubator (Eppendorf, New BrunswickTM I26 Stackable Incubator Shakers, or similar)
Rolling incubator (Kylin-Bell Lab Instruments Co., Ltd., Qilinbeier QB-128, or similar)
Autoclave
Pasteur oven
Plant growth chamber (Percival CU36L5 or similar) or growth room.
Culture oven (Shanghai JingHong DHG-9038A, or similar)
Electrophoresis devices (Bio-Rad, PowerPacTM Basic Power Supply + Wide Mini-Sub Cell GT Cell, or similar)
A gel imaging system (Bio-Rad, ChemiDoc XRS+TM Gel Imaging System, or similar)
Thermocycler (Bio-Rad, T100 Thermal Cycler, or similar)
Procedure
-
Bacterial culture using B. megaterium YC4-R4 as an example
Refresh YC4-R4 from glycerol stock on LB-Agar plate and incubate at 37 °C for 24 h*/**.
Take a single colony to inoculate 5 ml LB medium (in vitro) (recommended 15 ml tubes). Incubate overnight in a shaker (220 rpm, 37 °C). For soil inoculation, scale up by using 1 ml of pre-culture per liter of fresh culture (final volume depends on soil initial moisture and pot volume)*.
Prepare 0.45% NaCl solution by mixing 4.5 g per liter of ddH2O. Autoclave for 20 min at 121 °C. Cool to room temperature before use.
Centrifuge the culture at 3,220 × g for 10 min. Resuspend the pellet in the same volume of 0.45% NaCl. OD600 ~0.8-1.1 (log growth phase; generally, equals to 106-108 CFU/ml) is recommended.
Notes:- *This step shall be done under sterile conditions.
- **For tests using antibiotic-resistant strains, LB plates may be prepared with the appropriate antibiotics.
-
Prepare bacteria growth medium
Mix 10 g of tryptone, 5 g of yeast extract, and 10 g NaCl per liter of ddH2O.
For making LB agar plates, add 15 g agar per liter.
Autoclave for 20 min at 121 °C. For LB agar plates, pour the liquid medium into Petri dishes and allow it to cool down and solidify.
-
Prepare plant growth medium
Mix MS powder in half strength (2.21 g) and agar (0.7% and 1% for circular and square plates, respectively, depending on test requirements) per liter of ddH2O.
Adjust pH to 5.7 with NaOH or KOH.
Autoclave for 20 min at 121 °C. Pour the medium into the Petri dishes and allow it to cool down and solidify.
-
Seed sterilization and plant growth conditions
Mix seeds with absolute ethanol in 1.5 ml tubes and keep in agitation for 1 min. For 100 seeds, 1 ml of ethanol is recommended.
Discard ethanol and add 20% bleach solution (from commercial bleach stock). For 100 seeds, 1 ml of 20% bleach solution is recommended. Agitate in a spinner wheel for 10-15 min.
Discard bleach and wash three times with sterile ddH2O in a clean bench*.
Plant the seeds on MS medium one by one with the aid of pipet tips*.
-
Stratify the seeds at 4 °C for 48 h. After stratification, place the plates vertically in the growth chamber for 14 days under the following conditions: 22 °C; 12 h light/12 h dark cycle; 40% relative humidity; and up to 155 µmol·m−2s−1 light intensity.
Note: *This step shall be done under sterile conditions.
-
In vitro colonization test
Under sterile conditions, place 15-20 14-days-after-germination (DAG) seedlings grown in MS 1% agar (in squared Petri plates) per replica in a 1.5 ml sterile tube (volume of sterile tube depends on plant size). Add 1 ml of bacteria in 0.45% NaCl solution.
Incubate the tube overnight on a shaker (220 rpm, 26 °C*).
Discard the bacterial solution. Place the seedlings on a sterile surface, such as sterilized aluminum foil or paper, or a Petri dish. Cut roots with sterile razor blades and keep them in 1.5 ml tubes.
Sterilize the root surface by adding 1 ml of 75% ethanol, treating for 7-10 min with continuous agitation (spin wheel or equivalent).
Discard the ethanol and wash the roots for three times with sterile ddH2O.
Discard the ddH2O and grind roots with sterile pistils (for 1.5 ml tubes).
Add 1 ml of 0.45% NaCl to the tube, homogenize by vortexing, and prepare serial dilutions (usually 5 times 10-fold dilution should be enough).
Plate drop by drop (10 drops of 10 µl approx. up to a total volume of 100 µl) each dilution in LB-agar plates, let them dry for 2 min and incubate at 30 °C** for 24 h (Figure 1).
Keep and centrifuge the original solution at the maximal speed. Once root debris is pelleted, discard the supernatant and fully dry at 60 °C in an oven for 48 h and record this weight as the dry weight (DW), by using Pasteur oven.
Count bacteria colony numbers and calculate the colony forming units (CFUs) by taking into account the dilution and plated volume. Normalize the CFU values with root DW to obtain the colonization rates. Colonization rate = (CFUs × dilution factor)/mg of root DW.
Notes:- All work shall be done under sterile conditions.
- *This temperature has been tested to ensure non-stressing conditions for plants; at the same time, bacteria growth under the same temperature should also be considered to avoid imposing stress to the bacteria.
- **The temperature used herein is optimal for colony-counting of the example bacteria. It is highly recommended to optimize the temperature for different strains in order to avoid overgrowth/overlapped colonies.
-
Determination of colonization ratio in soil-grown plants
Grow Arabidopsis seedlings as described above.
Prepare soil based on the conditions required. In our tests, soil is prepared by meshing a mix 1:3 (v:v) of vermiculite and soil substrate. To minimize unwanted microbes in the soil, the soil needs to be tyndalized (75 °C for 3 h + cooling interval for 10 h + 75 °C for another 3 h). Transfer 5 DAG seedlings to the soil.
Inoculate with bacteria in 0.45% NaCl solution. As reference, in a regular 0.4 L pot, inoculate 50 ml of bacteria solution around planted seedlings.
Allow the plants to grow under conditions as required by the desired tests (e.g., with or without certain abiotic stress).
Harvest the roots (usually 20-40 mg of roots) at the time when colonization quantification is desired. For root harvesting, manually detach the soil around each root (with the help of tweezers) and then use a soft tissue to detach the smallest and most adhered pieces of soil. Fine soil particles must be removed by 3 × ddH2O washing*. Once roots are clean, follow indications for sterilizing, grinding, preparing dilutions, and counting CFUs in Part E.
Note: *This step shall be done under sterile conditions.
-
Strain isolation and identification
Although the soil was tyndalized, typically Part F will yield a small portion of bacteria other than the inoculated strain. To confirm the identity of the inoculated strain and to calculate its proportion in the total bacterial population, first classify bacterial colonies by morphology (use https://microbeonline.com/colony-morphology-bacteria-describe-bacterial-colonies/, as reference), then use sterile toothpicks to pick single colonies with similar morphology and transfer them to liquid LB medium for culturing. Usually, 20-30 colonies should be randomly sampled to avoid bias (Figure 2).
Culture the individual colonies in 5 ml of liquid LB on a shaker (220 rpm, 30 °C) for 24 h.
Centrifuge the culture (suggestion in bench centrifuge: 3,220 × g, 10 min), discard supernatant, and extract DNA from pellets by using a high-performance method. Here, a DNA extraction kit is suggested (see methods).
Amplify and sequence 16S rRNA genes with the 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’) (or equivalent), and BLAST the sequence to identify isolated species.
To further distinguish the inoculated strain from other strains of the same species, perform PCR with ERIC primer (5’-ATGTAAGCTCCTGGGGATTCAC-3’) to generate fingerprinting phenotype patterns for each strain. Use the stock strains employed in the test as the reference. Run the samples in a 2% agarose gel to maximize band separation. Use GelJ to compare the patterns with the reference and obtain similarity indexes. Figure 3 includes a representative gel image output.
Figure 1. Example of drop-by-drop seeding of a serial dilution in a quarter-divided growth plate.
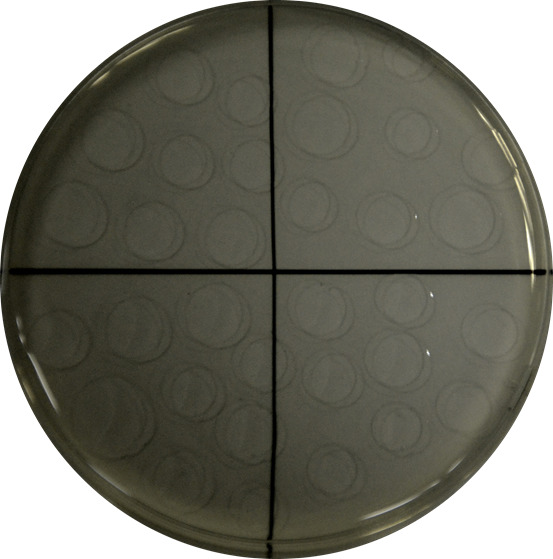
Drops are carefully placed on the dry growth medium plate to avoid interferences.
Figure 2. An example of colonies with different morphologies in a regular root colonization quantification process.
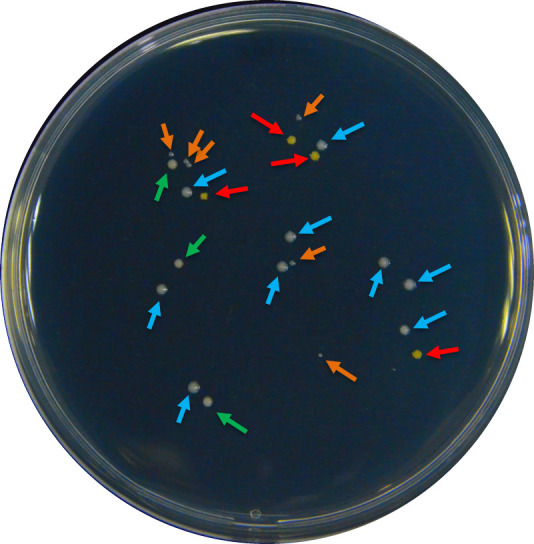
Different morphologies were selected by color, size, or brightness of the colonies. The colour of the arrows indicates a pre-group of strains: according to the following criteria: red – intense yellow color; blue – white color, big size; orange – white color, small size; and green – beige color, matte surface.
Figure 3. Example of a representative ERIC-PCR gel image output(modified from Vílchez et al., 2020 ).
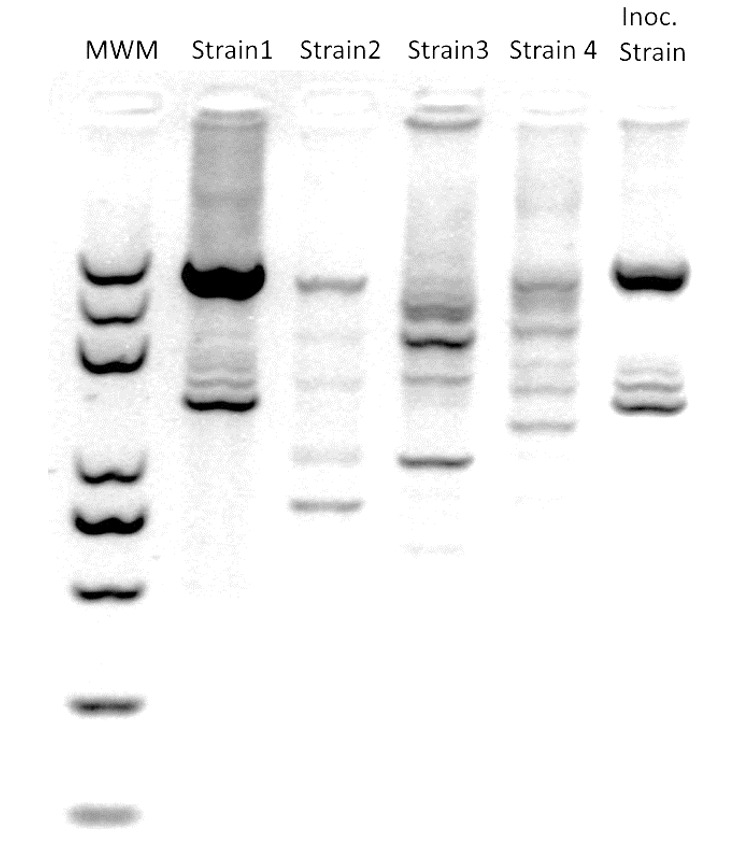
MWM: molecular weight marker; Strains 1 to 4 were recovered from roots in inoculated soil; Inoc. Strain: originally soil-inoculated strain.
Data analysis
Each test should be carried out with at least three replicates per condition prepared as indicated above (in vitro or in soil). Results will be considered statistically different when P-value 0.05 according to Student’s t-test. The relative colonization rates are calculated as: RC = (colony numbers × dilution factor)/mg of root DW.
Notes
If applying this method to other plant species, scale the recipe to fit the plant size. For grinding samples, it is recommended to use 1.5 ml tubes and the matching pistils. Automatic grinding may be performed at low vibration frequencies. In the case of using mechanical disruption devices such as TissueLyser, use low frequencies of disruption in order to avoid overheating and mechanical stress that may cause disruption of bacterial membranes. Mechanical disruption at high frequencies is used to extract total DNA from soil, sludge, or other complex samples. Aggressive grinding/disruption methods increase the likelihood of introducing biases in the final counting and identification of bacterial strains.
The ethanol-based sterilization process has been tested in roots of Arabidopsis and tomato as the reference plants. In the case of Arabidopsis (Figure 4), 7-10 min (about 5 min for longer and thicker roots, as in tomato) was assessed as optimal root surface sterilization time (these tests were performed with Gram+ bacteria; consider shorter time for Gram-). Longer incubation periods could interfere with the quantification of the colonizing bacteria.
Nevertheless, it should be noted that this set up is for Arabidopsis plants. Sterilization time depends on the type of plant and bacterial population tested, so it is highly recommended to optimize sterilization time depending on the actual experimental set up.
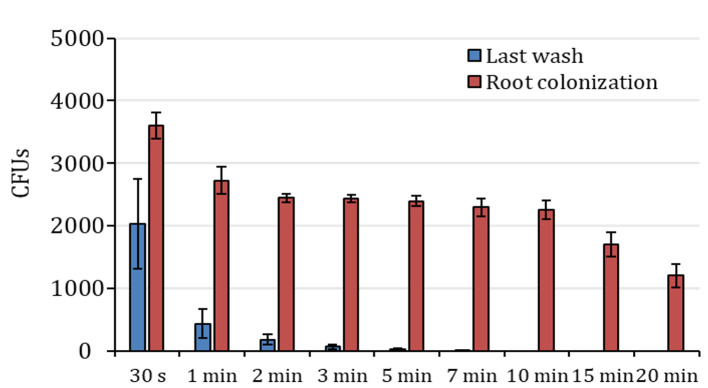
Figure 4. Colony counts after ethanol treatment in Arabidopsis roots.
Colony forming units (CFUs) counted in the last wash solution after treatment with ethanol (blue columns) and respective root colonization counting (red columns). The X-axis indicates ethanol treatment time. n=3, data are represented as mean ± SE.
Acknowledgments
Research in the laboratory of H.Z. has been supported by the Chinese Academy of Sciences (CAS). J.I.V. was supported by the CAS President’s International Fellowship Initiative fellowship. We thank Prof. Rosa Lozano-Duran at PSC and the anonymous reviewers for critical reading of the protocol.
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Krzyzanowska D., Obuchowski M., Bikowski M., Rychlowski M., and Jafra S.(2012). Colonization of potato rhizosphere by GFP-tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 shown on large sections of roots using enrichment sample preparation and confocal laser scanning microscopy . Sensors 12(12): 17608-17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maciá-Vicente J. G., Jansson H. B., Talbot N. J., and Lopez-Llorca L. V.(2009). Real-time PCR quantification and live-cell imaging of endophytic colonization of barley(Hordeum vulgare) roots by Fusarium equiseti and Pochonia chlamydosporia . New Phytol 182(1): 213-228. [DOI] [PubMed] [Google Scholar]
- 3. Mendes R., Garbeva P., and Raaijmakers J. M.(2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37(5): 634-663. [DOI] [PubMed] [Google Scholar]
- 4. Mendis H. C., Thomas V. P., Schwientek P., Salamzade R., Chien J. T., Waidyarathne P., Kloepper J. and De La Fuente L.(2018). Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I-1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions . Plos One 13(2): e0193119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olanrewaju O. S., Ayangbenro A. S., Glick B. R. and Babalola O. O.(2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl Microbiol Biot 103(1): 1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rochat L., Péchy-Tarr M., Baehler E., Maurhofer M. and Keel C.(2010). Combination of fluorescent reporters for simultaneous monitoring of root colonization and antifungal gene expression by a biocontrol pseudomonad on cereals with flow cytometry. Mol Plant Microbe Interact 23(7): 949-961. [DOI] [PubMed] [Google Scholar]
- 7. Saad M. M., De Zelicourt A., Rolli E., Synek L. and Hirt H.(2018). Quantification of Root Colonizing Bacteria. Bio-protocol 8: e2927. [Google Scholar]
- 8. Vílchez J. I., Yang Y., He D., Zi H., Peng L., Lv S., Kaushal R., Wang W., Huang W., Liu R., Lang Z., Miki D., Tang K., Paré P. W., Song C. P., Zhu J. K. and Zhang H.(2020). DNA demethylases are required for myo-inositol-mediated mutualism between plants and beneficial rhizobacteria. Nature Plants 6: 983-995. [DOI] [PubMed] [Google Scholar]