Abstract
The AAA+ Lon protease is conserved from bacteria to humans, performs crucial roles in protein homeostasis, and is implicated in bacterial pathogenesis and human disease. We investigated how Lon selectively degrades specific substrates among a diverse array of potential targets. We report the discovery of HspQ as a new Lon substrate, unique specificity-enhancing factor, and potent allosteric activator. Lon recognizes HspQ via a C-terminal degron, whose precise presentation, in synergy with multipartite contacts with the native core of HspQ, is required for allosteric Lon activation. Productive HspQ-Lon engagement enhances degradation of multiple new and known Lon substrates. Our studies reveal the existence and simultaneous utilization of two distinct substrate recognition sites on Lon, an HspQ binding site and an HspQ-modulated allosteric site. Our investigations unveil an unprecedented regulatory use of an evolutionarily conserved heat shock protein and present a distinctive mechanism for how Lon protease achieves temporally enhanced substrate selectivity.
INTRODUCTION
Proteolysis plays a central role in maintaining ideal concentrations of key regulatory proteins and in the disposal of misfolded, damaged, or disused proteins. The capacity of a cell to remodel its proteome thus relies on the exquisite substrate specificity of its proteolytic complement. In bacteria, ATP-driven proteases, that include Lon, ClpXP, ClpAP, HslUV, and FtsH (Gottesman, 2003), carryout the bulk of cytoplasmic proteolysis, both for regulating cellular responses and ridding the cell of unwanted proteins. The Lon protease serves as a principal proteolytic component in many bacterial species, and as such plays a pivotal role in general cellular physiology, protein quality assurance, stress response, and bacterial pathogenesis (Tsilibaris et al., 2006). Although first discovered in Escherichia coli, the Lon protease belongs to the AAA+ (ATPases associated with a variety of cellular activities) superfamily of proteins, which are widely distributed in all kingdoms of life, and contain highly conserved domain architectures. The bacterial Lon protease forms a spherical chamber composed of six Lon monomers. Each Lon monomer contains an N-terminal substrate recognition domain, a central ATPase domain involved in substrate unfolding and translocation, and a C-terminal peptidase domain that harbors the active site serine-lysine catalytic dyad. The AAA+ Lon protease is thus endowed with all the necessary attributes to form a self-compartmentalized hexameric protease capable of recognizing, unfolding, and degrading a broad range of proteins.
Spatio-temporally controlled alterations in degradation preferences of ATP-fueled proteases ensure that they act on substrates in a regulated manner to maintain their desired cellular concentration. Proteases often rely on adaptor proteins for selective substrate recognition. Adaptors act by increasing protease specificity towards certain substrates. The Clp proteases, ClpXP and ClpAP, have known adaptors. ClpS, an adaptor for N-end rule substrates of the ClpAP protease (Dougan et al., 2002), binds and delivers N-degron containing substrates to ClpA for selective degradation (Erbse et al., 2006). Similarly, SspB and RssB are adaptors for the ClpXP protease. SspB enhances the degradation of ssrA-tagged substrates (Levchenko et al., 2000), whereas RssB promotes the recognition of Sigma-S by ClpXP (Zhou et al., 2001). These adaptors function by forming a high-affinity ternary complex between the protease and its substrates, thus facilitating efficient degradation. Curiously, conserved broad-spectrum Lon protease adaptors have not been reported.
Indirect functional overlaps between Lon protease and other cytosolic chaperones have been observed in disposal of some Lon substrates (Huang et al., 2001; Jubete et al., 1996; Wu et al., 1999). Recent data suggest that unfolded proteins and some Lon recognition degrons can allosterically modulate Lon activity. For instance, comparison of the degradation properties of model substrates containing well-characterized Lon recognition tags, suggest that they modulate the ATPase and protease functions of E. coli Lon protease (Gur et al., 2009). Similarly, unfolded proteins containing a model degron allosterically activate Caulobacter cresentus Lon protease in vitro (Jonas et al., 2013). Recently, a modulator of Lon activity, SmiA, was shown to facilitate degradation of SwrA in Bacillus subtilis, in a pathway specific manner without affecting degradation of unfolded or tag-containing proteins (Mukherjee et al., 2015). Given that Lon plays a central regulatory role in the cell, it is highly likely that additional regulators and adaptors, with novel mechanistic and regulatory features, will be identified.
We report the discovery of the small heat-shock protein Q (HspQ) as a unique, long sought-after, specificity-enhancing factor for the AAA+ Lon protease. We demonstrate that HspQ allosterically regulates the substrate preference and catalytic efficiency of Lon protease. Characterization of HspQ-Lon interactions suggests that multipartite HspQ-Lon interactions, including specific contacts with the native core and the C-terminal degron of HspQ, are required for the productive engagement and allosteric activation of Lon protease.
RESULTS
HspQ is a heat-inducible protein and a Lon substrate
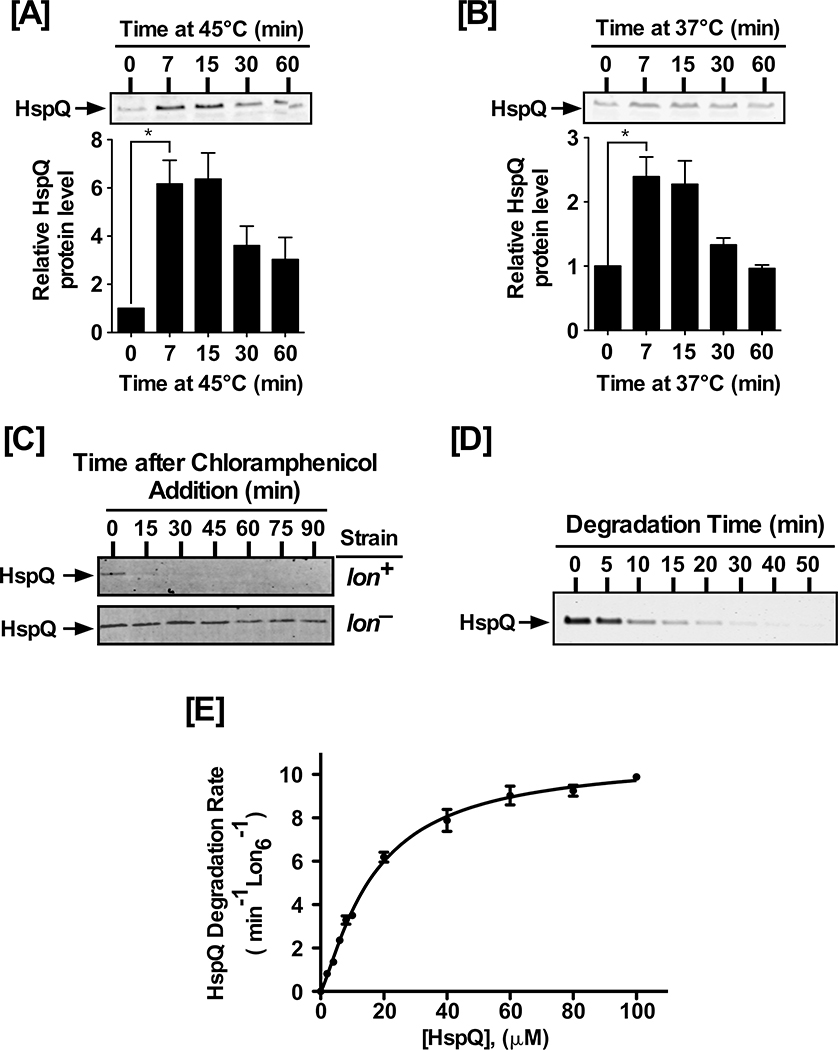
The regulatory mechanisms that modulate Lon substrate preferences remain poorly understood. We used the LonTRAP approach, where the catalytic Ser679 in the peptidase domain of Lon was changed to alanine (Van Melderen et al., 1999), for capturing Lon substrates in Yersinia pestis and identified heat-shock protein Q (HspQ) among a set of potential Lon substrates (Z. Ge and W. Karzai, unpublished data). HspQ is a highly conserved member of the YccV protein family (Fig S1A–B), with members in all kingdoms of life. Heat-stress induces HspQ expression in E. coli (Shimuta et al., 2004); therefore, we examined HspQ expression in Y. pestis under normal and heat-stress conditions (Fig 1, and Fig S1C). We observed a rapid rise in HspQ protein level upon transition from 25°C to 45°C, with a significant increase within 7 min, reaching maximal increase (~6-fold) within 15 min of heat-shock induction (Fig 1A). This rapid increase in HspQ level was transient, reminiscent of the prototypical pattern of heat-shock gene expression (Gamer et al., 1992; Herman et al., 1995; Straus et al., 1990; Straus et al., 1987; Tilly et al., 1989), which decreases to a new equilibrium within 30 min (Fig 1A). Intriguingly, we also observed a 2-fold increase in HspQ level upon transition from 25°C to 37°C, under conditions conducive to induction of the Y. pestis type-three secretion system (T3SS), with an analogous transient expression pattern (Fig 1B).
Figure 1: HspQ is a stress-induced Lon substrate.
(A) Yersinia pestis HspQ protein is induced by heat-shock at 45°C, and (B) induction of the T3SS at 37°C. Total cellular proteins, from equal number of cells at indicated time point, were examined by Western blot analysis to detect changes in HspQ protein levels, using polyclonal anti-HspQ antibodies (Key Resources Tables). p-values were calculated by performing one-tailed t-tests between HspQ protein levels at 0 min and 7 min under heat-shock (A) * p= 0.017, and T3SS induction (B) * p= 0.023. (C) In vivo degradation of HspQ requires Lon protease. lon+ and lon− Yersinia strains were cultured and chloramphenicol was added to inhibit protein synthesis. HspQ protein levels were determined by Western blot analysis. (D) In vitro proteolysis of HspQ by Lon, using 10 μM HspQ and 200 nM Lon6, was carried out in 1X Lon activity buffer in the presence of an ATP regeneration system at 37°C. Aliquots were mixed with 2X-SDS sample buffer at specified time points, resolved by electrophoresis on 15% Tris-Tricine gels, stained with Coomassie Brilliant Blue, scanned using a Li-COR Odyssey scanner, and quantified using the Image Studio software. (E) Kinetics of HspQ degradation by Lon at a range of HspQ concentrations. Degradation assays, with specified HspQ concentrations, were carried out as described in (D). Degradation rates for HspQ were fitted to a modified Hill equation. Representative Coomassie Brilliant Blue stained gels and Western blots are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).
To assess whether HspQ served as a Lon substrate in vivo, we examined the stability of HspQ in a Lon deficient (lon−) strain and an isogenic parental (lon+, or WT) strain. This examination revealed that while HspQ is rapidly degraded in the WT strain, it is substantially stabilized in the lon− strain (Fig 1C), indicating that HspQ could be a Lon substrate. To determine whether HspQ served as a direct Lon substrate, we cloned, expressed, and purified the Y. pestis HspQ and Lon proteins. We assessed the ability of Lon protease to degrade HspQ in an in vitro proteolysis assay, composed of purified components, and observed efficient HspQ recognition and degradation by Lon protease (Fig 1D). We determined the steady-state kinetic parameters of HspQ degradation by Lon, at a range of HspQ concentrations (Fig 1E, and Table S1). The HspQ degradation data fit equally well to the Michaelis-Menten and Hill equations, with a kdeg (Vmax Lon6−1) of 10.8, a K0.5 (substrate concentration at half-saturation) of 16.6 μM, and a Hill cooperativity constant of 1.3 (Table S1). These results demonstrate that the highly conserved HspQ protein is recognized directly and degraded efficiently by Lon protease.
HspQ enhances YmoA degradation by Lon protease
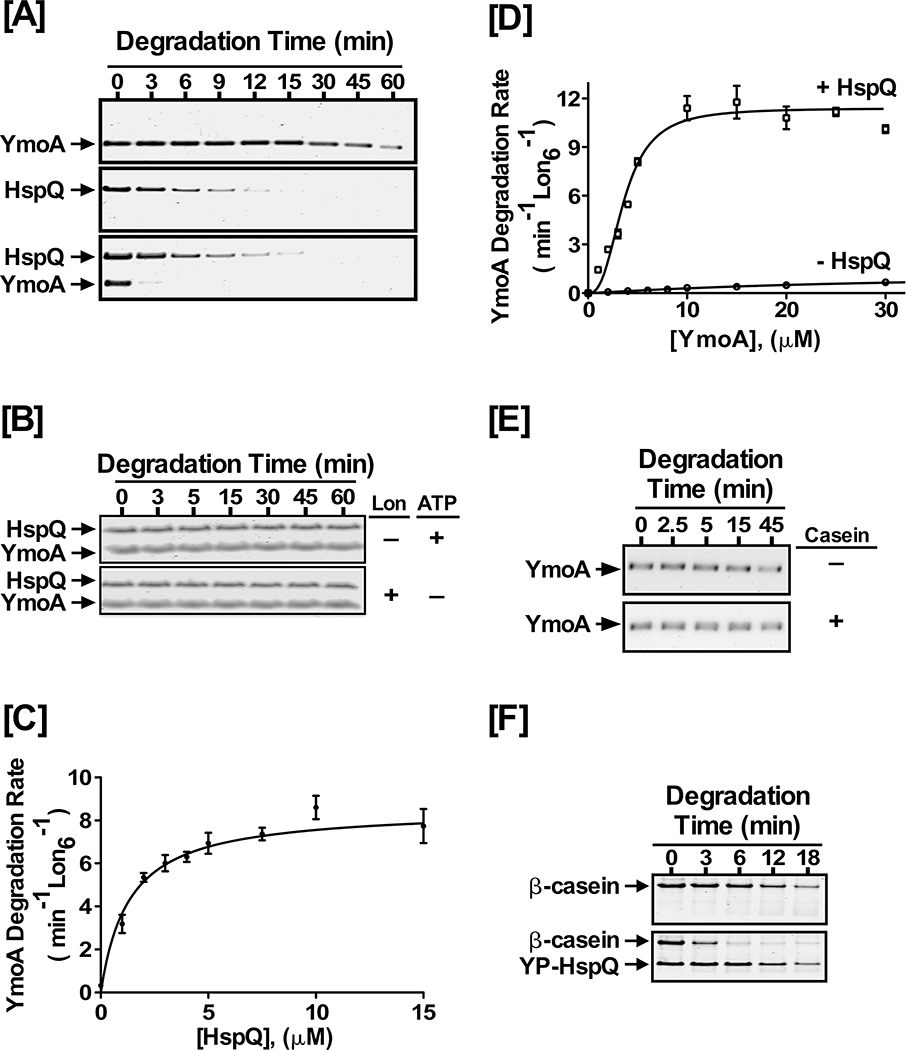
YmoA, or Yersinia modulator A, is a small histone-like protein whose efficient removal by the Lon and ClpXP proteases is required for Yersinia T3SS induction (Jackson et al., 2004). We cloned and purified YmoA and performed in vitro Lon proteolysis assays. YmoA is a poor Lon substrate in vitro, with a modest degradation rate of ~1 min−1 Lon6−1 (Fig 2A). Since YmoA is a known Lon substrate for T3SS induction and HspQ levels are elevated under these conditions, we postulated that HspQ might act as a Lon specificity-enhancing factor and increase Lon-mediated degradation of YmoA. To test this hypothesis, we included HspQ in the YmoA proteolysis assay. The presence of HspQ had a profound impact on the ability of Lon to recognize and degrade YmoA (Fig 2A). Interestingly, the presence of YmoA did not alter HspQ degradation by Lon (Fig 2A), indicating YmoA and HspQ are processed via independent substrate binding sites on Lon. The HspQ-enhanced degradation of YmoA required the presence of both Lon and ATP (Fig 2B), signifying that HspQ is not a protease and the observed enhancement in YmoA degradation is a Lon-dependent property of HspQ.
Figure 2: HspQ enhances degradation of YmoA by Lon.
Purified Yersinia YmoA protein was examined in Lon degradation assay without and with HspQ. Reactions containing equimolar concentrations of 10 μM each HspQ and YmoA were subjected to proteolysis by 200 nM Lon6 in 1X Lon activity buffer at 37°C as described in Fig 1D. (B) In vitro proteolysis assays performed in absence of Lon or ATP regeneration system are shown. Both Lon and ATP are required for HspQ-enhanced YmoA degradation. (C) The effect of HspQ on YmoA degradation by Lon was measured at a range of HspQ concentrations. Degradation rates at each HspQ concentration were measured and fitted to a Michaelis-Menton equation. (D) Kinetics of YmoA degradation by Lon. Degradation assays with a range of YmoA concentrations were carried out in absence and presence of 10 μM HspQ. Creatine Kinase (CK) is a component of the ATP regeneration system. Degradation rates at each YmoA concentration were measured and fitted to a modified Hill equation. (E) YmoA degradation by Lon was monitored in absence and presence of 100 μg/mL casein, showing that casein has no effect on YmoA degradation. (F) In vitro proteolysis of 200 μg/mL β-casein by 200 nM Lon6 was carried out in absence and presence of 10 μM HspQ. Representative Coomassie Brilliant Blue stained gels are shown.
To gain further mechanistic insights into the effect of HspQ on YmoA proteolysis by Lon, we performed detailed steady-state kinetic analyses, in the presence and absence of HspQ (Fig 2C–D, and Fig S2A). First, we examined the effect of varying HspQ concentration on enhancing Lon’s ability to degrade YmoA (Fig 2C), and selected an HspQ concentration (10 μM) that yielded maximal activation of Lon protease for further kinetic assessments. Next, we determined YmoA degradation rates, at a range of YmoA concentrations, in the absence or presence of the optimal concentration of HspQ (Fig 2D). YmoA degradation by Lon exhibited sigmoidal kinetics, indicative of cooperative allosteric behavior, and fit best to the Hill equation (Fig 2D, and Table S1). The Hill constant of 2.6 for YmoA degradation in presence of HspQ suggested that HspQ allosterically modifies Lon protease for enhanced YmoA recognition and degradation (Fig 2D, and Table S1). Indeed, the profound effect of HspQ on the ability of Lon to recognize and degrade YmoA was reflected in the 8-fold improvement in K0.5 and the 9-fold enhancement in kdeg (Fig 2D, and Table S1). These kinetic data also enabled calculation of the specific enhancement in the catalytic efficiency of Lon (kdeg/K0.5) for YmoA degradation. This analysis showed that the presence of HspQ imparts a dramatic 80-fold enhancement in Lon protease preference for YmoA (Table S1). Consistent with an allosteric activation of Lon by HspQ, we found that HspQ binds directly to Lon and forms a stable complex in vitro (Fig S2B). Furthermore, we found that HspQ does not form a stable complex with YmoA in vitro (Fig S2C), suggesting that HspQ does not operate via a substrate delivery mechanism.
Previous reports suggest that unfolded proteins activate Lon to enhance degradation of some substrates (Jonas et al., 2013). We examined whether the presence of the same unstructured protein, casein, could analogously enhance YmoA degradation by Lon. Our analysis showed that the presence of casein did not stimulate YmoA degradation by Lon (Fig 2E), suggesting that the observed allosteric activation of Lon for enhanced YmoA degradation is a unique functional attribute of native HspQ. Most intriguingly, Lon degraded casein at a rate of ~ 1 min−1 Lon6−1 in the absence of HspQ and 3.6 min−1 Lon6−1 in the presence of HspQ (Fig 2F), suggesting that HspQ also enhances degradation of unstructured proteins. Furthermore, E. coli HspQ similarly enhanced degradation of β-casein by E. coli Lon (Fig S2D), implying that the HspQ-dependent activation of Lon is widely conserved. Relatedly, E. coli HspQ was implicated in the in vivo degradation of DnaA508, a temperature-sensitive variant of the DNA replication protein DnaA (Shimuta et al., 2004). However, a link between HspQ and Lon or a role for Lon protease in DnaA508 degradation was not explored. We examined the role of HspQ and Lon in the in vivo stability of DnaA508. The dna508 strain grows well at 30°C but is progressively temperature-sensitive for growth at 39°C and 41°C (Fig S3A). In accord with the findings of Shimuta et al. (Shimuta et al., 2004), Loss of HspQ function enables the dna508/hspQ mutant strain to grow at the nonpermissive temperatures (Fig S3A). We interpreted this finding to suggest that HspQ is required for enhanced degradation of DnaA508 by Lon protease. Consistent with this inference, lon gene disruption in the dna508 strain rescued the temperature-sensitive growth defect of the dna508 mutant at 39°C but not to the same extent as the hspQ gene disruption (Fig S3B), implying that the absence of Lon protease stabilizes the less stable DnaA508 protein at the restrictive temperatures but suffers additional defects at 41°C, perhaps due to a requirement for the proteolytic or chaperone functions of Lon at the higher temperatures. To directly assess the stabilization of DnaA508 protein in the absence of Lon protease, we examined the in vivo stability of the DnaA508 protein in the parental dnaA508 and the dnaA508/lon− strains by Western blot analysis. This assessment showed that the DnaA508 protein was substantially stabilized in the dnaA508/lon− strain at 41°C as compared to the parental dnaA508 strain at 41°C (Fig S3C), supporting the conclusion that HspQ enhances the degradation of the DnaA508 mutant by Lon protease in vivo.
HspQ enhances degradation of multiple Lon substrates
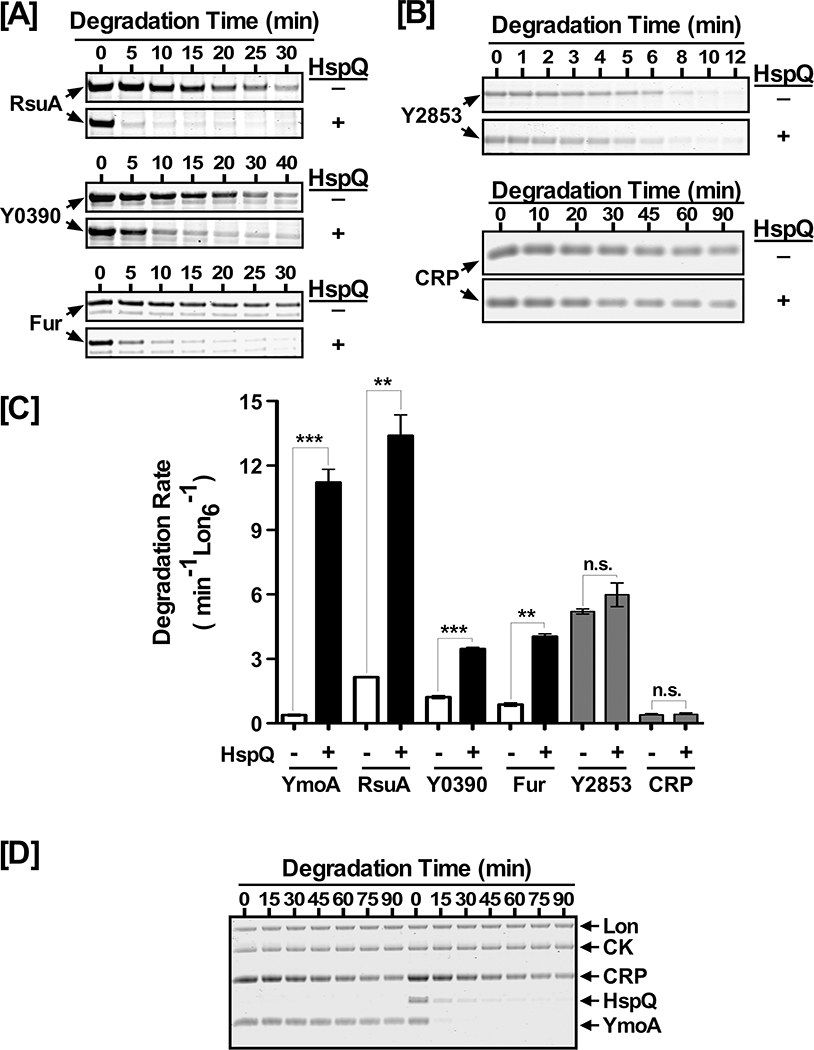
Next, we investigated whether the HspQ-mediated allosteric activation of Lon was restricted to specific proteins or influenced a broad-range of Lon substrates. To explore this possibility, we examined the effect of HspQ on the degradation of five putative Lon substrates, RsuA, Y0390, Fur, Y2853, and CRP, which were also identified by our LonTRAP approach. We cloned, expressed, and purified these proteins and evaluated their proteolytic susceptibility in the presence and absence of HspQ (Fig 3). Our analysis showed that all five proteins serve as Lon substrates in the absence of HspQ (Fig 3A–B), with individual degradation rates ranging from ~0.3 min−1 Lon6−1 to 5 min−1 Lon6−1 (Table S2). In the presence of HspQ, we observed significant enhancement in the degradation rates of RsuA, Y0390, and Fur by Lon (Fig 3C and Table S2). In sharp contrast, the presence of HspQ imparted little or no significant enhancement in degradation of Y2853 and CRP (Fig 3B–C). These findings suggest that although the HspQ-mediated allosteric activation of Lon results in enhanced degradation of a broad range of native and unstructured substrates, it does not result in universal activation of Lon protease. Consistent with this conclusion, we found that HspQ specifically enhanced degradation of YmoA by Lon even when an alternate substrate (CRP) –whose an unaided degradation rate is virtually identical to YmoA (Table S2)–was concomitantly present in the degradation reaction (Fig 3D). This result supports the notion that HspQ specifically enhances Lon substrate preferences for a select set of substrates. Among Lon substrates examined, RsuA exhibited the highest HspQ-dependent degradation rate (13 min−1 Lon6−1), whereas Y2853 exhibited the highest HspQ-independent degradation rate (5 min−1 Lon6−1) (Fig 3A–C, and Table S2).
Figure 3: HspQ allosterically activates Lon for enhanced degradation of multiple Lon substrates.
In vitro Lon proteolysis assays were performed for (A) RsuA, Y0390, and Fur and (B) Y2853 and CRP. Each protein was examined at 10 μM concentration in the presence or absence of 10 μM HspQ. Reactions were carried as described in Fig 1D. (C) YmoA, RsuA, Y0390, Fur, Y2853, and CRP degradation rates were calculated in absence and presence of HspQ and plotted to illustrate the HspQ effect on their degradation by Lon. p-values from one-tailed t-test between absence and presence of HspQ for each substrate were calculated as *** p<0.0001 (YmoA), ** p=0.0038 (RsuA), *** p=0.0008 (Y0390), ** p= 0.0041 (Fur), n.s. p= 0.1389 (Y2853), n.s. p= 0.1124 (CRP). (D) HspQ specifically enhances YmoA degradation by Lon in the presence of an additional substrate, CRP. In vitro proteolysis reactions for YmoA and CRP in absence and presence of HspQ were carried out under conditions specified in (A). Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).
HspQ and Y2853 have partially overlapping Lon-binding sites
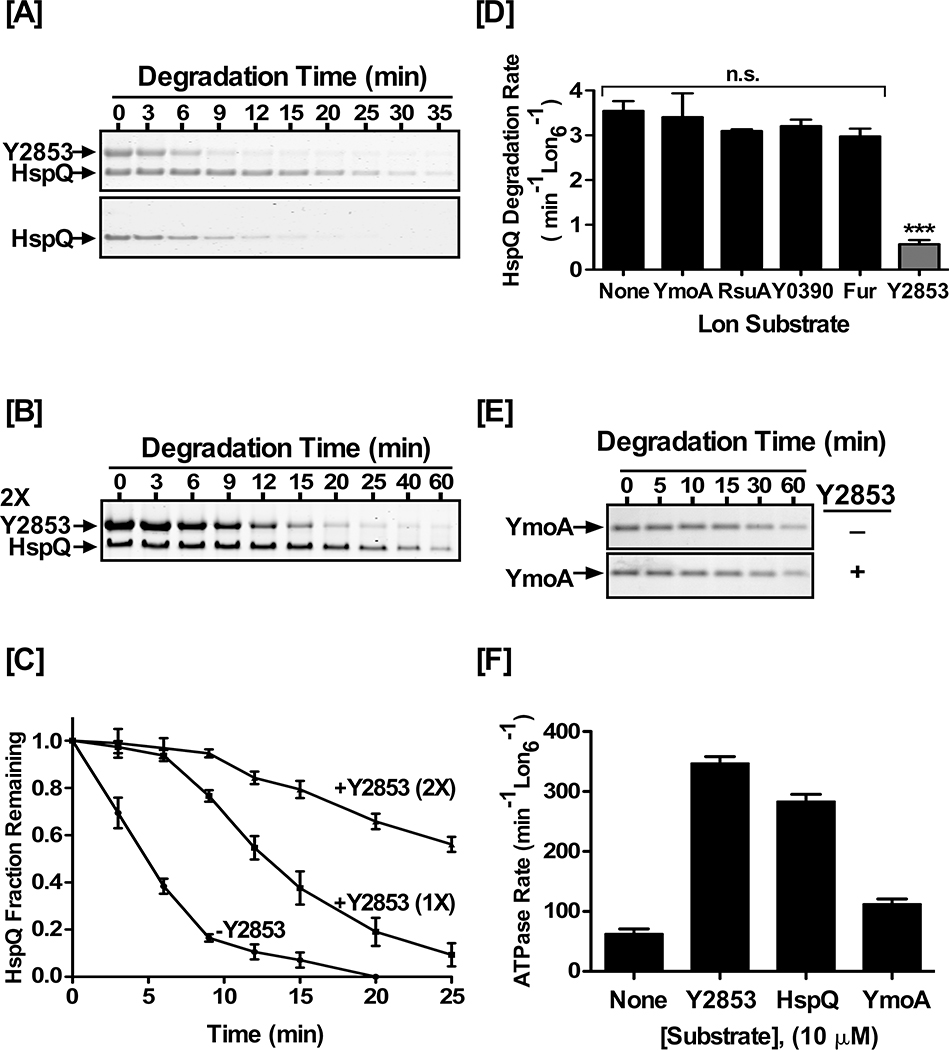
The effect of HspQ on substrate selection by Lon and the interplay between HspQ and Lon substrates (Fig 2 and Fig 3) shed new light on the existence of more than one distinct substrate-binding site on Lon. We observed that while HspQ had no significant effect on degradation of Y2853, there was a clear and significant reduction in HspQ degradation in the presence of Y2853 (Fig 4). To gain deeper insights, we focused on the effect of Y2853 on HspQ degradation, with a fixed concentration of HspQ and a range of Y2853 concentrations. In the presence of equimolar concentrations of both proteins, HspQ degradation rate was significantly reduced, as compared to HspQ alone or HspQ in the presence of YmoA (Fig 4A). In fact, HspQ proteolysis by Lon was drastically reduced (0.6 min−1 Lon−1) until most of Y2853 was degraded (Fig 4A, and Table S3). In the apparent second phase of degradation (Fig 4C, and Table S3), once most of Y2853 was depleted, HspQ degradation rate recovered to 3.2 min−1 Lon−1, approaching its degradation rate in the absence of Y2853 (3.9 min−1 Lon−1). Doubling Y2853 concentration to two molar-equivalents (2X) further exacerbated the delay in HspQ degradation (Fig 4B–C), indicating that HspQ and Y2853 have competing, overlapping or partially overlapping, binding-sites on Lon. Most interestingly, all of the HspQ-dependent Lon substrates examined had no influence on degradation of HspQ by Lon (Fig 4D, and Table S3), consistent with the conclusion that these substrates bind to a discrete allosterically activated binding-site on Lon that is separate from Y2853 and HspQ binding sites. Together, these data strongly suggest that Lon has at least two distinct substrate binding-sites and that HspQ and Y2853 have overlapping binding-sites on Lon protease.
Figure 4: Y2853 and HspQ have partially overlapping binding sites.
(A) Y2853 delays HspQ degradation by Lon. In vitro Lon proteolysis of 10 μM HspQ was carried out in absence or presence of (A) equimolar and (B) two molar equivalent (2X) concentration of Y2853. Reactions were carried out in Lon activity buffer and 200 nM Lon6 at 37°C. (C) HspQ fraction remaining from proteolysis reactions in (A) and (B) are plotted over time. Data are normalized to protein amount at the zero time point for each degradation assay. (D) Degradation rate of HspQ in absence and presence of 10 μM concentration of the indicated protein substrates. One-tailed t-test between no substrate addition and specified substrate was performed to calculate the p-values as n.s. p= 0.3851 (YmoA), n.s. p= 0.1158 (RsuA), n.s. p= 0.1328 (Y0390), n.s. p=0.0551 (Fur), *** p<0.0001 (Y2853). (E) In vitro Lon proteolysis of YmoA in absence and presence of 10 μM Y2853. Reactions were carried out as described above. (F) Substrates stimulated ATP hydrolysis by Lon. ATP hydrolysis rates were determined in the presence of 10 μM concentration of each substrate. Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).
Allosteric activation of Lon protease is a unique functional attribute of HspQ
Given that Y2853 is an efficient Lon substrate and shares an overlapping binding-site with HspQ, we inquired whether Y2853 could analogously enhance YmoA degradation by Lon. However, examination of the effect of Y2853 on YmoA degradation revealed that, unlike HspQ, the presence of Y2853 had no significant impact on YmoA degradation (Fig 4E). This result suggested that although HspQ and Y2853 have partially overlapping binding sites, HspQ must make additional unique and consequential contacts with Lon.
It was conceivable that the HspQ-mediated allosteric activation of Lon was due to its ability to vigorously stimulate Lon ATPase activity, thus transforming it to a more robust unfoldase. To assess this possibility, we examined the capacity of HspQ, Y2853, RsuA, Y0390, CRP, Fur, and YmoA to stimulate the ATPase activity of Lon. This analysis showed that Y2853 educed the highest level of Lon ATPase activation (5.5-fold), followed by HspQ (4-fold) and CRP (2.7-fold) (Fig 4F, and Table S4), whereas YmoA elicited the lowest ATPase activation at ~2.4-fold. The observed stimulation of Lon ATPase activity correlates well with the degradation rates of each individual substrate. The ATPase activation elicited by Y2853 was higher than the simultaneous presence of both HspQ and YmoA (Table S4). However, despite vigorously stimulating Lon ATPase activity, Y2853 was unable to enhance degradation of YmoA, or any other Lon substrates we have examined. Together, these data suggest that HspQ activates Lon via a unique mechanism that alters its substrate preferences, and which does not solely depend on stimulation of Lon ATPase activity.
Lon recognizes a C-terminal HspQ degron
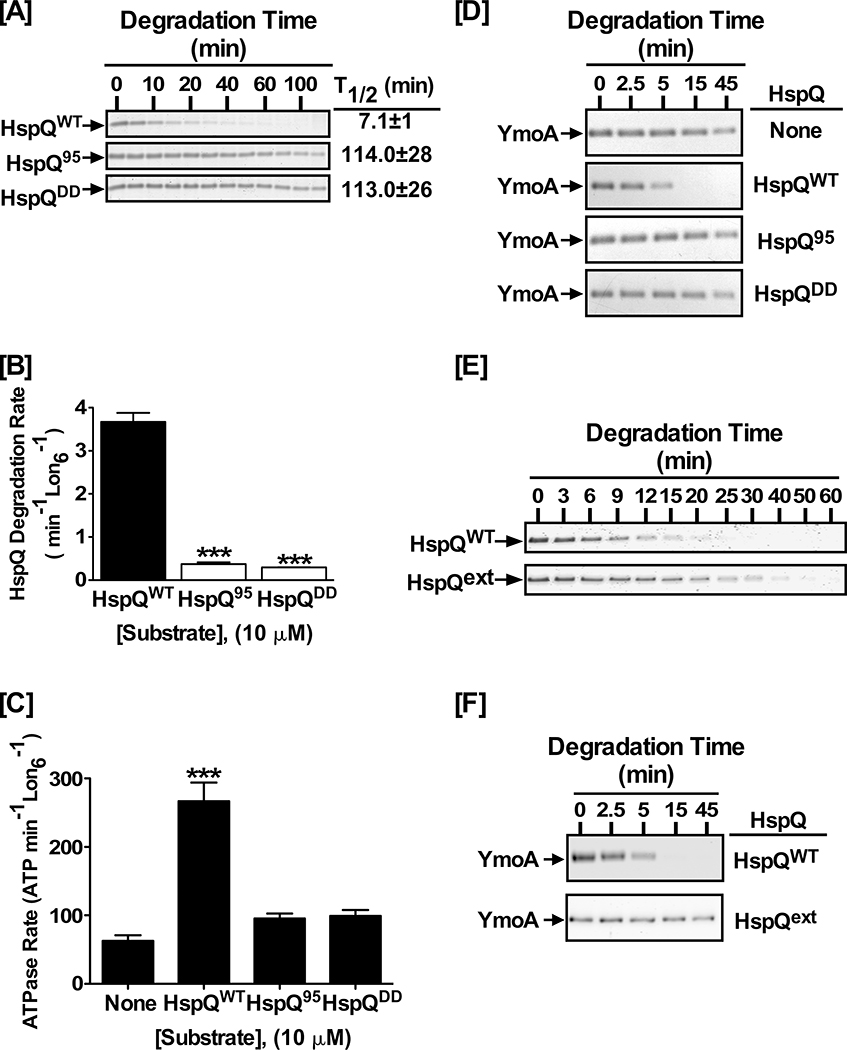
To gain deeper mechanistic insights into how HspQ functions as a Lon substrate and allosteric activator, we analyzed HspQ sequence signals required for Lon recognition. Since native untagged HspQ and the N-terminally His6-tagged HspQ serves as equally efficient Lon substrates and activators (Fig S4), we reasoned that the Lon recognition element (degron) of HspQ might reside at an internal segment or the C-terminus of the protein. The C-terminal eight amino acid residues of E. coli HspQ are not discernable in the available structural model of the protein (PDB: 1VBV, and Fig S1B). We explored the possibility that the C-terminal unstructured tail of HspQ could serve as the Lon recognition degron, and accordingly generated an HspQ truncation variant (HspQ95) that lacks the C-terminal ten residues of the protein. Additionally, to avoid complications with removing potential structural elements, and to preserve the length of the C-terminus, we designed the HspQDD variant where the last two residues, Arginine (R) 104 and Asparagine (N) 105, were replaced with Aspartic acid (D) residues, which Lon disfavors in its recognition sequences (Ge et al., 2009; Gur et al., 2008). We examined these HspQ C-terminal variants for their propensity to serve as Lon substrates. Our analysis showed that Lon did not efficiently degrade either HspQ95 or HspQDD (Fig 5A), with ~ 10-fold reduction in their individual degradation rates (Fig 5B, and Table S5). The half-life (T1/2) of HspQ95 was 114 ± 28 min and that of HspQDD 113 ± 26 min, a >15-fold increase compared to that of HspQWT (T1/2 = 7 ± 1 min). Consistent with their deficiency as Lon substrates, the HspQ95 and HspQDD variants exhibited similar defects in stimulating Lon ATPase activity (Fig 5C, and Table S5). HspQWT stimulated the ATPase rate of Lon by ~4-fold, from ~60 to ~250 min−1 Lon6−1, (Table S5). In contrast, the C-terminal degron variants HspQ95 and HspQDD elicited only a modest (~1.5-fold) increase in Lon ATPase rate. These results suggest that the C-terminal ten residues of HspQ are required for Lon recognition and likely constitute part of the HspQ degron. We further assessed the ability of the HspQ95 and HspQDD variants to specifically enhance YmoA degradation by Lon. This analysis showed that HspQ95 and HspQDD were both defective in the specific enhancement of YmoA proteolysis by Lon (Fig 5D, and Table S5), suggesting that productive Lon-HspQ engagement, via its C-terminal degron, is required not only for productive HspQ-Lon engagement but also for the ensuing allosteric activation of Lon protease.
Figure 5: Precise positioning of the HspQ degron is required for Lon activation.
(A) In vitro proteolysis of 10 μM each HspQWT, HspQ95, and HspQDD by 200 nM Lon6. Reactions were carried out in 1X Lon activity buffer at 37°C. (B) Degradation rates of reactions in panel A were calculated and plotted. One-tailed t-tests were performed to calculate p-values (*** = p<0.0001). (C) HspQ degron mutants do not stimulate ATP-hydrolysis by Lon. ATP-hydrolysis assay for Lon protease in absence and presence of 10 μM HspQ C-terminal variants, as compared to wild-type HspQ, are shown with p-values (*** = p<0.0001). (D) YmoA proteolysis assays by Lon were carried out with 10 μM each HspQWT, HspQ95, and HspQDD to assess the ability of the HspQ degron mutants to allosterically activate Lon for enhanced YmoA degradation. HspQ95 and HspQDD are defective in allosterically activating Lon protease. (E) Proteolysis assays were performed with 10 μM each HspQWT and HspQext, showing that increasing the distance between the C-terminal HspQ degron and the HspQ core (HspQext) impacts its degradation. (F) In vitro Lon proteolysis of YmoA with equimolar HspQext was carried out under conditions described in (A), showing the HspQext variant does not activate Lon protease for enhanced YmoA degradation. Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).
Allosteric activation of Lon requires precise positioning of the HspQ degron
To further elucidate the mechanism of HspQ-mediated allosteric activation of Lon, and to decipher whether the HspQ degron is necessary and sufficient for Lon recognition and allosteric activation, we attached the last 20 amino acid residues of HspQ (Q20) to the C-terminus of the model λ-cI protein to create the λ-Q20 tagged reporter. Analysis of the proteolytic susceptibility of λ-Q20 and the control untagged λ reporter showed that the C-terminal 20 residues of HspQ do indeed contain the Lon recognition degron and facilitate enhanced degradation of the λ-Q20 protein (Fig S5A). However, the λ-Q20 tagged protein was unable to enhance YmoA degradation by Lon protease (Fig S5B). Furthermore, the presence of YmoA had no significant impact on λ-Q20 degradation (Fig S5C), suggesting that the λ-Q20 substrate is recognized and processed by Lon through the same pathway as wild type HspQ. These results, while highlighting the autonomous nature and importance of the HspQ degron for recognition and degradation by Lon, underscore the need for existence of additional HspQ sequence signals required for the allosteric activation of Lon.
The insufficiency of HspQ-degron to allosterically activate Lon suggested that the native core of HspQ must make additional contacts with Lon protease. To examine the nature and spatial requirements for these additional HspQ contacts, we constructed and purified HspQext, which has an eight-residue (GSTGSTGS) flexible linker inserted between the unstructured C-terminal tail and the preceding structural helix (Fig S5D), and assessed its propensity to serve as Lon substrate and promote YmoA degradation. Lon recognized and degraded the HspQext variant, albeit with a slight reduction in its degradation rate (Fig 5E). Most interestingly, however, HspQext failed to allosterically activate Lon for enhanced YmoA degradation (Fig 5F). This result supports the conclusion that the native core of HspQ makes additional contacts with Lon, and that these contacts must be established within precise space and time to achieve allosteric Lon activation.
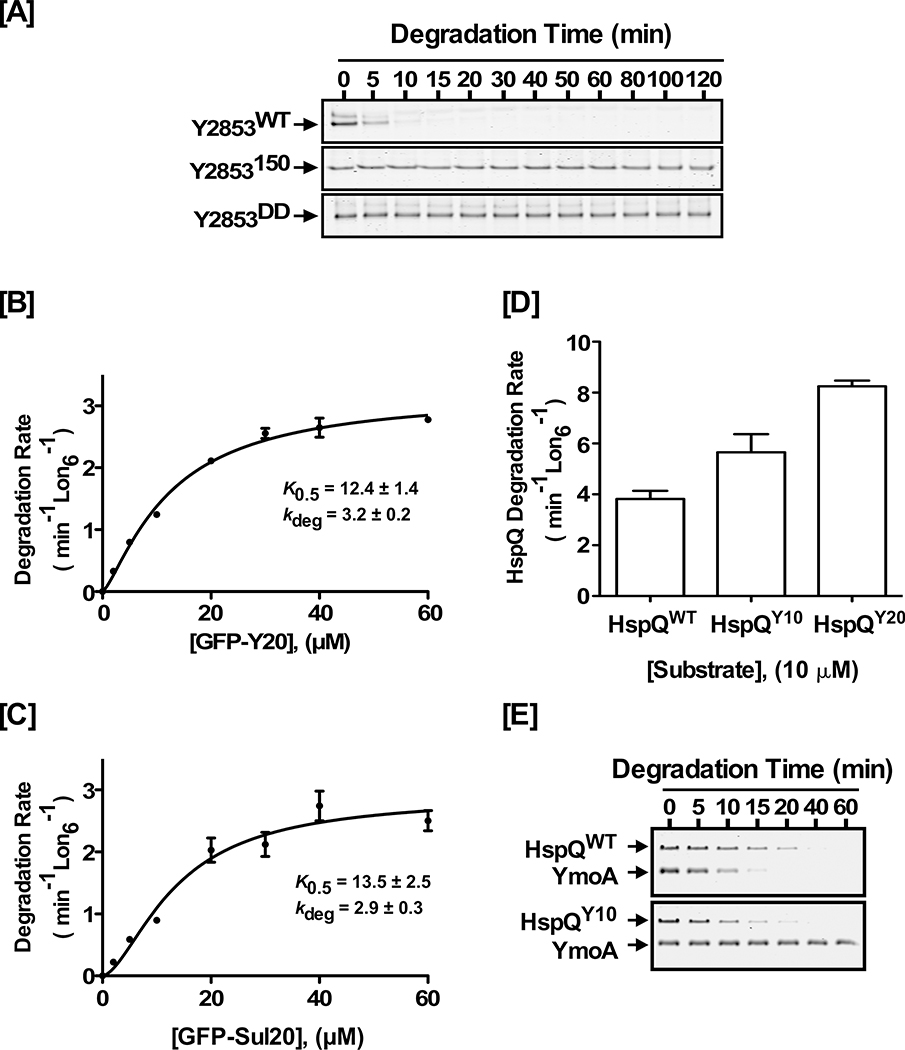
To gain further mechanistic insights, we explored whether the precise spatial orientation of the HspQ degron, during its initial engagement with Lon, had an effect on Lon activation. We also wished to ascertain whether an increase in HspQ degradation rate had an impact on Lon activation. Our studies of Y2853 showed that it forms a stable complex with Lon (Fig S6A) and has the fastest unassisted degradation rate (5.2 ± 0.1 min−1 Lon6−1) of all Lon substrates (Table S2), implying it possess an ideal Lon degron. Indeed, the C-terminal sequence of Y2853 (-SYPIIH) resembles the C-terminal sequence of SulA (-SYLYH), a well-known Lon degron (Ishii et al., 2001; Ishii et al., 2000). Since HspQ has a C-terminal degron, and Y2853 and HspQ have partially overlapping binding sites on Lon, we reasoned that Y2853 might analogously possess a C-terminal degron. Therefore, we constructed and purified Y2853150, a variant lacking the C-terminal 11 residues, and Y2853DD, a variant where the ultimate and penultimate residues were replaced with two Aspartates. Consistent with our prediction, examination of the proteolytic propensities of the Y2853150 and Y2853DD variants revealed them both to be poor Lon substrates (Fig 6A), signifying that Y2853 possesses a C-terminal Lon recognition degron. Consistent with the conclusion that HspQ and Y2853 have partially overlapping binding site on Lon, analysis of the Y2853DD variant showed that it did not have a negative effect on HspQ degradation by Lon (Fig S6B), suggesting that Y2853 must bind Lon to interfere with HspQ-Lon interactions.
Figure 6: Lon activation requires unique signals in the HspQ degron.
(A) Alterations of the Y2853 C-terminal degron impact its recognition and degradation by Lon. In vitro proteolysis of 10 μM each Y2853WT, Y2853150, or Y2853DD by 200 nM Lon6 are shown. (B) Kinetics of GFP-Y20 and (C) GFP-Sul20 degradation by Lon, at a range of GFP concentrations, show that Y2853 has an ideal Lon recognition degron. Degradation rates for GFP-Y20 and GFP-sul20 were fitted to a modified Hill equation. (D) Hybrid HspQ variants HspQY10 and HspQY20, which carry the C-terminal 10 or 20 residues of the Y2853 degron, are recognized and degraded faster by Lon. In vitro proteolysis of 10 μM each HspQWT, HspQY10, and HspQ20 are shown. (E) In vitro proteolysis demonstrating the effect of HspQWT and the HspQY10 hybrid on YmoA degradation by Lon. Lon degrades the HspQY10 hybrid faster, but it is unable to allosterically activate Lon for enhanced YmoA degradation. Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).
The C-terminal 20 residues of Y2853, when appended to GFP (GFPY20), served as a highly efficient and autonomous Lon recognition degron (Fig 6B), better than, or on par with, the well-characterized SulA degron appended to GFP (GFPSul20) (Fig 6C, and Table S6). To further validate the identity of the Y2853 degron, we constructed and purified the GFPY20(H161A) variant, where a key Lon recognition determinant, the ultimate histidine (H161), is changed to alanine. Unlike GFPY20, the GFPY20(H161A) variant is not degraded by Lon protease (Fig S6C). Having established the identity of the highly efficient Y2853 degron, we attempted to generate a faster degrading HspQ hybrid, HspQY10, by replacing the C-terminal 10 residues of HspQ with the corresponding 10 residues of Y2853. The rationale for constructing the HspQY10 hybrid was to generate a more rapidly-proteolyzed HspQ variant that retained the putative HspQ core signals for Lon activation, engaged Lon via the alternate but partially overlapping Y2853 interaction pathway, while retaining the same total length as wild type HspQ. We also constructed and purified the extended HspQY20 chimera, where the last ten residues of HspQ were replaced with the C-terminal twenty residues of Y2853. Although Lon recognized the HspQY10 and HspQY20 hybrid proteins and degraded them faster than HspQWT (Fig 6D), both variants failed to allosterically activate Lon for enhanced YmoA degradation (Fig 6E, and Table S5). These findings are consistent with the conclusion that allosteric activation of Lon requires precise engagement of the HspQ C-terminal degron, with optimal orientation and ideal spacing with respect to the core of the HspQ protein. Although Lon recognizes the Y2853 degron and enhances degradation of the HspQ hybrid, this alternate degron likely places HspQ in a non-optimal orientation, such that supplemental HspQ-core interactions required for allosteric Lon activation cannot be established.
Conserved HspQ residues contribute to Lon activation
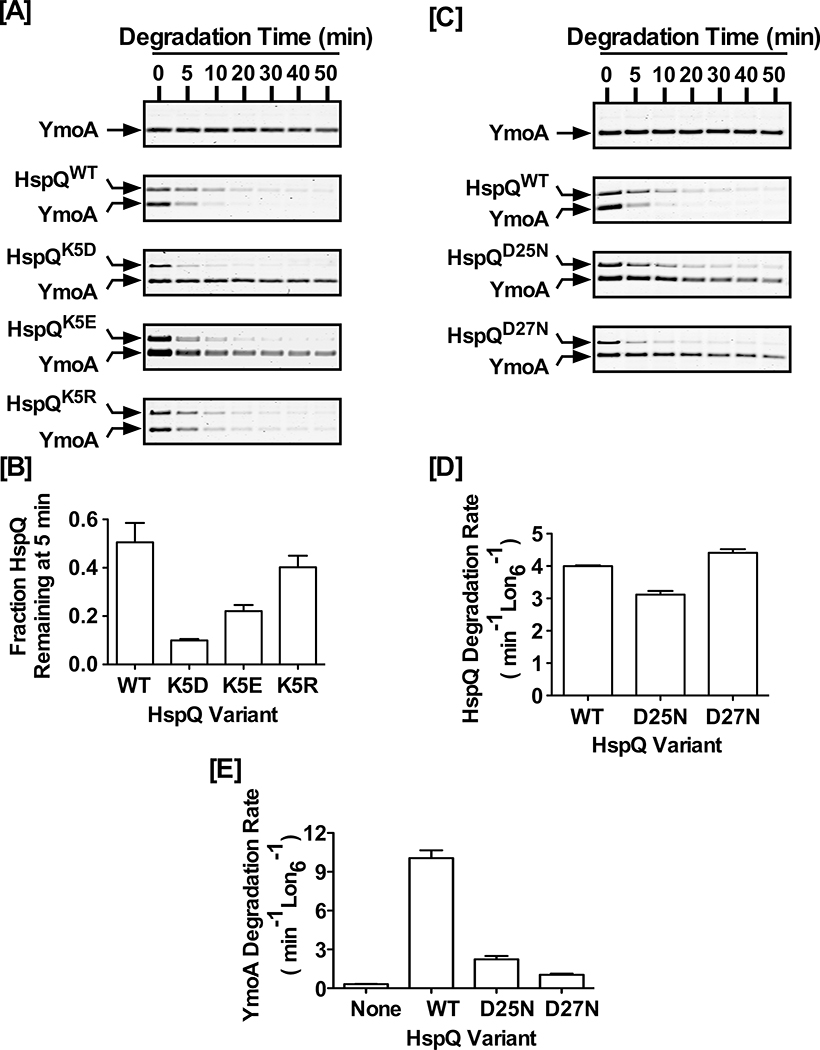
In an attempt to tease-apart HspQ-sequence signals required for allosteric activation of Lon, we explored the potential contributions of highly conserved surface-exposed residues of HspQ (Fig S1B). To assess the contribution of the conserved Lysine five (K5), we constructed and purified HspQK5D, a variant where K5 was replaced with Asp (D), and evaluated its propensity to serve as a Lon substrate and allosteric activator. We discovered that Lon degraded HspQK5D faster than HspQWT (Fig 7A), implying that the side chain of K5 contributes to HspQ stability. To ameliorate the potential deficiencies associated with the shorter side chain and head-group of the Asp substitution, we generated the HspQK5E and HspQK5R variants. The longer side chain HspQK5E variant and the longer side chain and positively charged head-group HspQK5R variant progressively improved their proteolytic sensitivity (Fig 7A–B, and Fig S7A), implying that the aliphatic side chain and head-group of K5 contribute to HspQ protein stability. The faster degrading HspQK5D variant was most defective in allosterically activating Lon for enhanced YmoA degradation (Fig 7A, and Fig S7B). This activation deficiency was in accord with a subtle structural distortion of HspQK5D (Fig S7C) that was amended with the K5E substitution and further improved with the K5R substitution (Fig 7A–B, and Fig S7B). Given the HspQK5E and HspQK5R variants promote rapid initial degradation of YmoA (Fig S7B), while sufficient concentration of HspQ variants is still available (Fig 7B), indicates that K5 is not involved in allosteric contacts but contributes to HspQ protein stability. The fact that the HspQK5D variant is recognized and rapidly degraded by Lon supports the conclusion that allosteric Lon activation requires coordinated presentation of two sets of signals, one originating from the C-terminal HspQ degron and the other from the folded native core of the protein, which constitute the second-site allosteric contacts.
Figure 7: Conserved HspQ residues contribute to allosteric activation of Lon.
(A) The proteolytic stability of HspQWT, HspQK5D, HspQK5E, and HspQK5R and their effect on Lon activation for enhanced YmoA degradation were examined at 10 μM concentration of each HspQ variant and YmoA. Reactions were carried out in Lon activity buffer at 37°C. (B) HspQ fraction remaining at 5 min was calculated for K5 variants and plotted to illustrate the effect of side-chain length and charge on HspQ stability. (C) The proteolytic stability of HspQWT, HspQD25N, and HspQD27N and their effect on Lon activation for enhanced YmoA degradation were examined at 10 μM concentration of each HspQ variant and YmoA. (D) Degradation rates for HspQWT and its second contact site variants (HspQD25N and HspQD27N) were calculated and plotted to demonstrate that Lon degrades them at similar rates. (E) YmoA degradation rates were measured in the presence of HspQWT, HspQD25N, and HspQD27N and plotted to show that these second contact site variants are defective in allosterically activating Lon. Representative Coomassie Brilliant Blue stained gels are shown, and the data presented in graphs are from three independent experiments (mean ± SEM).
Next, we assessed the potential contributions to Lon activation of conserved amino acid residues in the C-terminal helix of HspQ, located in close proximity to the HspQ degron. We constructed HspQS93A, where Ser93 was replaced with Ala, and HspQRHQ/AAA, where Arg95, His96, Gln97 were all replaced with Ala. We expressed and purified both HspQ variants and tested their propensity for degradation and allosteric activation of Lon. Both C-terminal helix variants, HspQS93A and HspQRHQ/AAA, behaved like HspQWT and specifically enhanced Lon degradation of YmoA (Fig S7D), suggesting that these conserved HspQ residues do not contribute to HspQ structural stability, HspQ-Lon interactions, or allosteric activation of Lon.
Finally, we investigated the role of the conserved Asp25 and Asp27 by constructing and purifying the HspQD25N and HspQD27N variants, where Asp25 and Asp27 were individually replaced with Asn (N). Analysis of the proteolytic propensity of the HspQD25N and HspQD27N variants showed that both proteins were efficiently recognized and degraded by Lon, with rates comparable to HspQWT (Fig 7C–D, and Table S5). Similarly, the HspQD25N and HspQD27N variants were equally capable of stimulating Lon ATPase activity (Table S5). However, both variants exhibited significant defects in allosterically activating Lon to specifically enhance YmoA degradation (Fig 7E), suggesting that the highly conserved Asp25 and Asp27 contribute to Lon activation and may constitute part of its allosteric second site contact with Lon protease.
DISCUSSION
We have uncovered HspQ as a new substrate and specificity-enhancing factor for the AAA+ Lon protease. HspQ is the founding member of the highly conserved YccV protein family, with members in all kingdoms of life. In E. coli, the hspQ and lon genes are upregulated under stress conditions (Lesley et al., 2002; Richmond et al., 1999). Consistent with these observations, our investigations show the Yersinia HspQ protein levels increase rapidly in response to heat stress at 45°C and during induction of the T3SS at 37°C. These intriguing initial findings guided us to postulate that HspQ serves as a unique regulatory factor of Lon protease that facilitates enhanced degradation of regulatory proteins and surplus unwanted or destabilized substrates. However, unlike the ClpX and ClpA adaptor proteins, SspB and ClpS respectively, which are not degraded by their cognate proteases during substrate delivery, HspQ serves as a Lon substrate, thus enabling cells to establish a new proteostastic-equilibrium. It should be noted that the C. crescentus SspB adaptor is degraded by ClpXP (Chien et al., 2007), perhaps for a similar regulatory purpose. Direct examination of the hypothesis that HspQ serves as a potent Lon regulatory factor showed that HspQ dramatically enhances the specific degradation of YmoA by Lon. Further exploration of the effect of HspQ on substrate selection by Lon protease revealed that productive HspQ-Lon engagement promotes robust allosteric activation of Lon that results in enhanced recognition and degradation of not only YmoA but also several additional native Lon substrates, including RsuA, Fur, and Y0390. Notably, HspQ enhances degradation of casein, a known unstructured Lon substrate (Fig 2F), suggesting that HspQ could similarly enhance degradation of other stress destabilized proteins, thus greatly expanding the spectrum of proteins influenced by HspQ. In accord with these findings, in vivo studies of the dnaA508 variant in E. coli showed that this temperature-sensitive protein is stabilized in the hspQ− strain and a to much greater extent in the lon− strain, supporting the conclusion that HspQ enhances the recognition and degradation of the DnaA508 protein at the restrictive temperatures. Our inquiries also revealed that HspQ does not induce an all-encompassing universal stimulation of Lon protease, as we did not detect any significant HspQ effect on degradation of Y2853 and CRP, rather HspQ elicits an allosteric and substrate-specific activation of Lon protease.
In eukaryotes, YccV-like domains are found in various mosaic combinations with other functional domains (http://supfam.org/SUPERFAMILY/). A recent report suggests that ClpF, a uvrB/C and YccV containing multidomain protein, acts in concert with the plant plastid ClpS1 and ClpC protease system (Nishimura et al., 2015). The YccV domain of ClpF appears to be dispensable for ClpS and ClpC interactions and has a much extended and divergent C-terminal sequence. These changes might represent evolutionary adaptations by the eukaryotic YccV family members. Most curiously, however, ClpS1 protein is stabilized by >3-fold in the clpf-1 mutant, without any alterations in CLPS1 gene expression at the mRNA level. Furthermore, the observed ClpS1 protein stabilization in the clpf-1 mutant is similar in magnitude to its stabilization in a clpc1 null mutant (Nishimura et al., 2013), raising the possibility that ClpF might be involved in ClpS protein homeostasis. The E. coli HspQ protein was originally reported to have a negative impact on dnaA gene expression (d’Alencon et al., 2003). Subsequently, HspQ was shown to play an important role in the in vivo degradation of DnaA508, a temperature-sensitive DnaA protein variant in vivo (Shimuta et al., 2004). Although HspQ function was not linked to Lon protease activity, and HspQ on its own did not degrade DnaA, it was nevertheless proposed to be an oligomeric protease akin to the Clp family proteases (Shimuta et al., 2004). Our studies suggest that DnaA508 is a Lon substrate whose degradation by Lon is enhanced by HspQ in vivo. The fact that HspQ enhances degradation of casein in vitro combined with the inferred HspQ function in removal of destabilized DnaA variants supports the conclusion that HspQ specifically enhances degradation of destabilized and unfolded proteins by Lon protease.
Our investigation provide mechanistic insights into how HspQ promotes Lon activation, illuminating HspQ sequence signals required for (a) recognition by Lon and (b) allosteric activation of Lon. Biochemical dissection of HspQ functions demonstrates that its C-terminus acts as a Lon degron. Deletion of the C-terminal 10 residues (HspQ95) or substitution of the ultimate and penultimate residues with aspartates (HspQDD) result in severe defects in Lon’s ability to recognize and degrade HspQ. Curiously, these C-terminal alterations also dramatically curtail the ability of HspQ to allosterically activate Lon protease. We interpret these results to mean that a functional HspQ degron is required for productive engagement and subsequent allosteric activation of Lon protease. Consistent with this conclusion, we found that although the C-terminal HspQ degron is capable of converting an unrelated reporter protein (λ-Q20) to a Lon substrate, it is unable to allosterically activate Lon for enhanced YmoA degradation. Furthermore, increasing the distance between the core of HspQ protein and its C-terminal degron, via a flexible 8-residue linker in HspQext (Fig S5D), results in a modest degradation defect but a complete loss of capacity to allosterically activate Lon. Further exploration of the mechanism of HspQ-Lon interactions, using the HspQY10 and HspQY20 chimeras (Fig 6D–E), revealed that allosteric activation of Lon requires productive engagement of the HspQ degron for precise positioning of its native core, such that additional consequential allosteric contacts can be made in the precise time and space.
We reasoned that highly conserved amino acid residues in the native core of HspQ must mediate these second-site allosteric contacts. Indeed, mutational analysis of several conserved residues strongly supports this conclusion. For instance, substitutions of the conserved Lysine 5 (K5) suggest that even subtle structural distortions in HspQ protein impact its precise presentation and capacity to allosterically engage Lon (Fig 7A), thus providing an important mechanistic clue for the temporal requirements for HspQ-Lon interactions. Most significantly, substitution analysis of D25 and D27 (HspQD25N and HspQD27N) revealed that these residues could constitute part of the allosteric contacts with Lon, as the HspQD25N and HspQD27N variants have a minimal effect on HspQ stability and proteolytic sensitivity but exhibit a dramatic effect on YmoA degradation by Lon.
Our studies of an intriguing new Lon substrate, Y2853, demonstrate that it has an autonomous C-terminal degron that resembles the well-known SulA degron. Curiously, although Y2853 is an outstanding Lon substrate, robustly stimulates Lon ATPase activity, and shares a binding site with HspQ on Lon, it is unable to allosterically activate Lon for enhanced YmoA degradation, thus supporting the conclusion that HspQ makes additional contacts to allosterically activate Lon. Recent studies show that the E. coli SulA degron and casein have independent binding sites on Lon protease (Ishii et al., 2000). A prediction of this analysis is that casein is recognized and processed via the HspQ-regulated allosteric binding site on Lon. Consistent with this conclusion, our studies demonstrate that HspQ substantially enhances degradation of casein by Lon. Our data define HspQ as the founding member of a distinct class of Lon specificity-enhancing factors. We propose that the HspQ-dependent allosteric stimulation of Lon represents a unique activation mechanism that is fundamentally different from the reported unfolded-protein activation of Lon ATPase (Jonas et al., 2013) and the SspB and ClpS mediated activation of ClpXP and ClpAP, respectively.
Our studies provide unique insights into how HspQ allosterically primes Lon protease to achieve temporally enhanced substrate selectivity by heightening Lon’s ability to differentiate target proteins among a diverse pool of potential substrates. The transient expression and rapid degradation of HspQ protein provide the first indication of how the adaptor itself is regulated, thus unveiling an unprecedented use of a highly conserved heat shock protein, with family members in all kingdoms of life. These studies thus illuminate a distinctive mechanism for protease control that might be more widely utilized not only by ATP-fueled proteases but also by the related AAA+ enzymes, which participate in a diverse array of critical biological functions in all organisms.
STAR METHODS
Strains and Culture Conditions
E. coli strain BL21 star (DE3) was used to express recombinant proteins. Unless otherwise stated, LB broth (5 g yeast extract, 10 g tryptone, and 5 g NaCl per liter) was supplemented with 100 μg/ml ampicillin, 100 μg/ml carbenicillin, 50 μg/ml kanamycin, or 25 μg/ml chloramphenicol. Cells were cultured at 37°C with shaking at 250 rpm. The Yersinia KIM5− strain was cultured in heart infusion broth (10 g beef hart infusion, 10 g tryptose, and 5 g NaCl per liter) containing the desired antibiotics.
Cloning, Protein Expression and Purification
Y. pestis genes encoding lon, hspQ, ymoA, y2853, fur, rsu, and y0390 were either sub-cloned, from existing vectors, or PCR amplified using primer sets and expression vectors listed in Key Resource Tables. The pET28b-lon plasmid was used to overexpress Yersinia Lon in E. coli strain BL21 star (DE3). Cells were cultured in LB containing 50 μg/ml kanamycin at 37°C to optical density at 600 nm (OD600) of 0.5. Protein expression was induced by 1 mM IPTG. Protein overexpression was carried out for 16 hours at 16°C. Cells were harvested by centrifugation at 3,700 xg and resuspended in buffer A (50 mM KHPO4 pH 7, 1mM EDTA, 1mM DTT, and 10% glycerol). After sonication, cleared cell lysate was prepared by centrifugation at 30,000 xg. Activated and buffer A equilibrated P11-cellulose resin was added to the cleared cell lysate to allow Lon binding. The column was washed with buffer A to remove unbound proteins, and bound Lon protein was eluted in 10 ml of elution buffer B (400 mM KHPO4 pH 7, 1mM EDTA, 1mM DTT, and 10% glycerol). Lon protein was further purified on a Source 15Q ion-exchange column using buffer C (50 mM Tris pH 7.5, 50 mM KCl, and 1 mM DTT). Bound Lon was eluted using a 20 column-volume linear-gradient (0–100%) of buffer D (50 mM Tris pH 7.5, 1 M KCl, and 1 mM DTT). Lon eluted at 200 mM KCl. Fractions containing Lon protease were pooled, concentrated, and loaded on S300 gel filtration column in buffer E (50 mM Tris pH 7.5, 100 mM KCl, 10 mM MgCl2, 1mM DTT, 20% glycerol). Aliquots of purified Lon were flash-frozen and stored at −80°C.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal anti-HspQ | This study | N/A |
| Polyclonal anti-DnaA | (Sutton and Kaguni, 1995) | PMID: 7592447 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa 680 | Invitrogen | A-21076 |
| Bacterial Strains | ||
| Y. pestis KIM5–3001 (parent) | (Jackson et al., 2004) | PMID: 15554975 |
| Y. pestis KIM5–3001 Δlon | (Jackson et al., 2004) | PMID: 15554975 |
| dnaA508 tnaA::Tn10 | (Hinds and Sandler, 2004) | PMID: 15588282 |
| dnaA508 tnaA::Tn10 lon::kan | This study | N/A |
| BL21(DE3) Star™ | Invitrogen | C601003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| NADH, disodium salt | Roche | 10107735001 |
| Pyruvate Kinase from rabbit muscle | Sigma | P9136 |
| Lactate Dehygrogenase, from rabbit muscle | Calbiochem | 427217 |
| Creatine Kinase from rabbit muscle | Roche | 10736988001 |
| Adenosine-5′triphosphate (ATP) | Roche | 10127531001 |
| Creatine Phosphate | Roche | 10621722001 |
| Phosphoenolpyruvate potassium salt | Bachem | 4014027.0005 |
| Oligonucleotides | ||
| hspQYP105 (Forward): 5′-gagcatatgatcgccagcaaattcggtatag-3′ hspQYP105 (Reverse): 5′-caggagctcttagttacgtaggtgcggagcc-3′ |
This study | N/A |
| ymoA (Forward): 5′-tgtacatatgacaaaaactgactacc-3′ ymoA (Reverse): 5′-agaggatccttatttcacatgttgccatac-3′ |
This study | N/A |
| y2853 (Forward): 5′-tacttccaatccaatgcaatggattcactcatcgtccctg-3′ y2853 (Reverse): 5′ttatccacttccaatgttattaatgaatgatggggtaagaggtgg-3′ |
This study | N/A |
| hspQEC (Forward): 5′-gtgcatatgattgccagcaaattcggtatc-3′ hspQEC (Reverse): 5′-gttgagctcttagttacgcagacgcgggg-3′ |
This study | N/A |
| hspQ95 (Top): 5′-gatgaactggcggcctcgattcgctaacagttacaggctccgcacctacg-3′ hspQ95 (Bottom): 5′-cgtaggtgcggagcctgtaactgttagcgaatcgaggccgccagttcatc-3’ |
This study | N/A |
| hspQDD (Top): 5′-ccagttacaggctccgcacctagatgactaagagctccgtcgacaagc-3′ hspQDD (Bottom): 5′-gcttgtcgacggagctcttagtcatctaggtgcggagcctgtaactgg-3 |
This study | N/A |
| hspQK5D (Top): 5′-ggcagccatatgatcgccagcgatttcggtataggccagcagg-3′ hspQK5D (Bottom): 5′-cctgctggcctataccgaaatcgctggcgatcatatggctgcc-3′ |
This study | N/A |
| hspQK5E (Top): 5′-ggcagccatatgatcgccagcgaattcggtataggccagcagg-3′ hspQK5E (Bottom): 5′-cctgctggcctataccgaattcgctggcgatcatatggctgcc-3′ |
This study | N/A |
| hspQK5R (Top): 5′-ggcagccatatgatcgccagccgtttcggtataggccagcagg-3′ hspQK5R (Bottom): 5′-cctgctggcctataccgaaacggctggcgatcatatggctgcc-3′ |
This study | N/A |
| hspQD25N (Top): 5′-cgggtatttgggagtggtgattaatatcgatcctgaatattctcttgc-3′ hspQD25N (Bottom): 5′-gcaagagaatattcaggatcgatattaatcaccactcccaaatacccg-3′ |
This study | N/A |
| hspQD27N (Top): 5′-gggagtggtgattgatatcaatcctgaatattctcttgcacc-3′ hspQD27N (Bottom): 5′-ggtgcaagagaatattcaggattgatatcaatcaccactccc-3′ |
This study | N/A |
| hspQS93A (Top): 5′-ccttcattagatgaactggcggccgcgattcgccaccagttacaggctcc-3′ hspQS93A(Bottom): 5′-ggagcctgtaactggtggcgaatcgcggccgccagttcatctaatgaagg-3′ |
This study | N/A |
| hspQRHQ/AAA (Top): 5′-gatgaactggcggcctcgattgccgccgcgttac-3′ hspQRHQ/AAA (Bottom): 3′-gttacgtaggtgcggagcctgtaacgcggcggcaatc-5′ |
This study | N/A |
| hspQext (Top): 5′-caaccttcattagatgaactggcggcctcgattcgcggcagcaccggcagcaccggcagc-3′ hspQext (Bottom): 3′-ctcttagttacgtaggtgcggagcctgtaactggtggctgccggtgctgccggtgctgcc-5′ |
This study | N/A |
| Recombinant DNA | ||
| pET28b-hspQEC | This study | N/A |
| pET28b-hspQYP | This study | N/A |
| pET28b-hspQ95 | This study | N/A |
| pET28b-hspQDD | This study | N/A |
| pET28b- hspQK5D | This study | N/A |
| pET28b- hspQK5E | This study | N/A |
| pET28b- hspQK5R | This study | N/A |
| pET28b- hspQD25N | This study | N/A |
| pET28b- hspQD27N | This study | N/A |
| pET28b- hspQS93A | This study | N/A |
| pET28b- hspQRHQ/AAA | This study | N/A |
| pET28b- hspQext | This study | N/A |
| pET28b-ymoA | This study | N/A |
| pET28b-lonYP | This study | N/A |
| pET28b-lonEC | This study | N/A |
| 2BT- hspQY10 | This study | N/A |
| 2BT- hspQY20 | This study | N/A |
| 2BT-y2853 | This study | N/A |
| 2BT-y2853150 | This study | N/A |
| 2BT-y2853DD | This study | N/A |
| pDEST17- rsuA | This study | N/A |
| pDEST17- crp | This study | N/A |
| pDEST17- y0390 | This study | N/A |
| pDEST17- fur | This study | N/A |
| 2BT-cp60Y20 | This study | N/A |
| 2BT-cp6-Sul20 | This study | N/A |
| Software and Algorithms | ||
| GraphPad Prism for data organization, analysis, and presentation | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Imagestudio™ for densitometry | LI-COR Biosciences | https://www.licor.com/bio/products/software/image_studio/ |
| Canvas™ Draw 3 for Mac for creation of technical illustrations | ACD Systems | http://www.canvasgfx.com/en/products/canvasdraw |
HspQ and its variants were purified using a combination of Ni-NTA affinity, ion exchange, and size exclusion chromatography steps. BL21 star (DE3) harboring pET28b-hspQ plasmid was grown in LB containing 50 μg/ml kanamycin, and protein expression was induced with 1 mM IPTG at OD600 of 0.5. Cultures were allowed to grow for 3 h while shaking. Harvested cells were resuspended in lysis buffer (50 mM Tris pH 8, 1M NH4Cl, 2 mM beta-mercaptoethanol (β-ME), and 10 mM imidazole). After sonication, buffer-equilibrated Ni-NTA beads were added to cleared cell lysates. After 1 h end-to-end rocking at 4°C, unbound proteins were removed and the beads were washed extensively, and the bound proteins were eluted using a step elution with lysis buffer containing 250 mM imidazole. HspQ protein containing fractions were combined, buffer exchanged to buffer F (50 mM Tris pH 8, 50 mM KCl, 2 mM β-ME), loaded on a Source15Q column, and eluted using a 20 column-volume linear-gradient (0–100%) of buffer G (50 mM Tris pH 8, 1M KCl, 2 mM β-ME). HspQ eluted at 250 mM KCl. Fractions containing HspQ were pooled, concentrated and loaded on a Superdex 75 column equilibrated in buffer H (50 mM Tris pH 8, 50 mM KCl, 2 mM β-ME). Protein aliquots containing 10% glycerol were flash-frozen and stored at −80°C.
In vitro Proteolysis Assay
Each in vitro proteolysis assay reaction was carried out in Lon activity buffer (50 mM Tris-HCl pH 8, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, and 10% Glycerol), ATP regeneration system (16 mM creatine phosphate, 0.32 mg/ml creatine kinase, and 4 mM ATP). Substrate and Lon hexamer (Lon6) concentrations were as specified in figure legends. All reaction components except ATP regeneration system were mixed and incubated at 37°C. Warm ATP regeneration system was added to initiate the reaction. Aliquots at specific time were mixed with 2X-SDS sample buffer to terminate the reaction. Reaction products were resolved by electrophoresis on 15% Tris-Tricine gels and scanned using a Li-COR Odyssey scanner and quantified using the Image Studio software. Fraction of substrate remaining was estimated and the data was normalized to creatine kinase as loading control. These data were fit to a straight line and the slope was extracted to calculate the rate of degradation. For GFP assays, the circularly permuted GFP-CP6 variant (Wohlever et al., 2013) (herein referred to as GFP) was used to construct the GFPY2853 reporter, carrying the Y2853 C-terminal 20 residues, and the GFPsul20 reporter, carrying the SulA C-terminal 20 residues. These GFP variants were subjected to proteolysis by Lon, and the GFP protein levels were evaluated by fluorescence measurements obtained using a SpectraMax M5e microplate reader at excitation and emission wavelengths of 480 nm and 510 nm, respectively. All reaction components were pre-warmed to 37°C. Degradation rates for a range of substrate concentrations were fit to a modified Hill Equation Y=Vmax*XH/(K0.5H + XH) where X is the substrate concentration, Y is degradation rate, and H is the Hill constant.
In vitro ATP-Hydrolysis Assay
Coupled ATP hydrolysis assay was carried out in Lon activity buffer (50 mM Tris-HCl pH 8, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, and 10% Glycerol). Reactions contained 100 nM Lon6, 1 mM NADH, 10 U/ml lactate dehydrogenase, 20 mM phosphoenol pyruvate, 10 U/ml pyruvate kinase, and 2 mM ATP (Key Resources Tables). Lon and other reaction components were warmed separately at 30°C. Reactions were initiated by adding Lon, and NADH disappearance was monitored at 340 nm.
In vivo HspQ protein detection
To monitor HspQ protein level in Yersinia pestis under heat-shock (45°C), KIM5− cells were grown overnight at 25°C in heart infusion (HI) broth containing 50 μg/ml ampicillin. Cells were sub-cultured in HI to a starting OD600 of 0.1, and grown to OD600 of 0.6 at 25°C. At this point, the zero-minute sample was collected–by adding 1X SDS sample buffer to cell pellets–and the remaining culture was placed in a shaking water bath at 45°C. Aliquots containing equal number of cell were taken at indicated time points. To monitor HspQ level under T3SS inducing condition, overnight KIM5− cells were sub-cultured in T3SS permissive (HI with 20 mM MgCl2, 20 mM NaOxalate) and T3SS non-permissive (HI with 2.5 mM CaCl2) media. Both cultures were grown to OD600 of 0.6 at 28°C. T3SS permissive cultures were switched to 37°C at the zero-minute time. To monitor HspQ protein levels, equal number of cells was removed and prepared at specified time-points. All samples were resolved by electrophoresis on 15% Tris-Tricine gels, proteins were transferred to PVDF membrane, and Western hybridization was performed using anti-HspQ serum and an Alexa-680 conjugated goat anti-Rabbit IgG secondary antibody (Key Resources Tables). Blots were scanned using a Li-COR Odyssey scanner and quantified using the Image Studio software. Data were normalized to Coomassie stained gels as loading controls. Fold change in HspQ protein level was calculated by normalizing data to the zero time-point.
In vivo HspQ stability
To monitor HspQ degradation in vivo, KIM5− 3001 lon+ and lon− strains (Jackson et al., 2004) were grown overnight at 25°C in HI media. Cells were sub-cultured in HI/amp to a starting OD600 of 0.1, and grown to OD600 of 0.6. At this point, cultures were shifted to 37°C, allowed to grow for an additional ten minutes, and 200 μg/ml chloramphenicol was added to inhibit protein synthesis. To monitor HspQ, equal number of cells was removed and samples were prepared at specified time-points. All samples were resolved on 15% Tris-Tricine gels and subjected to Western blot analysis using anti-HspQ serum. Blots were scanned using a Li-COR Odyssey scanner.
In vivo DnaA stability
To monitor DnaA508 degradation in vivo, E. coli dnaA508 (Hinds et al., 2004), its hspQ− (dnaA508/hspQ−), and lon− (dnaA508/lon−) derivatives (Key Resources Tables) were grown overnight at 30°C on LB/Tet plates. Cells were sub-cultured in LB/Tet and grown to OD600 of 0.5. Chloramphenicol at 200 μg/mL was added to stop new rounds of protein synthesis. The cultures were immediately transferred to 41°C and equal number of cells were harvested at the indicated time points, resolved by electrophoresis on 10% Tris-Tricine gels, and subjected to Western blot analysis using DnaA antiserum (Sutton et al., 1995). DnaA protein bands were quantified and plotted. Blots were scanned using a Li-COR Odyssey scanner.
CD Measurements
Applied Photophysics Chirascan circular dichroism spectrometer was used to perform CD wavelength scans. Measurements were performed in a 1 mm path length quarts cuvette from 190 to 260 nm in buffer containing 10 mM potassium phosphate pH 8.0, and 50 mM NaF. Three scans were averaged to record the final scans. Molar ellipticity for each protein against wavelength is presented.
Plots and statistical evaluation
GraphPad Prism software was used for data analysis. Mean and standard error of mean (SEM) was calculated by performing column statistics. Paired or unpaired one-tailed t-test analysis was performed and p-values were calculated from relevant data sets (n≥3). Statistical significance is reported as n.s. = not statistically significant, * significant at p < 0.05, ** significant at p < 0.005, *** significant at p < 0.001.
KEY RESOURCES TABLE
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arnav Choksi, Arthur Korman, and Hui Shi for helpful discussions, and Zhiyun Ge for generating protein expression constructs and performing preliminary experiments. We thank David Thanassi and Steven Glynn for critical reading of the manuscript, Steven Sandler, Jon Kaguni, and Gregory Plano for bacterial strains and antisera, and Jorge Benach for his continued support.
REFERENCES
- Chien P, et al. (2007). Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci U S A 104, 6590–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Alencon E, et al. (2003). Isolation of a new hemimethylated DNA binding protein which regulates dnaA gene expression. J Bacteriol 185, 2967–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, et al. (2002). ClpS, a substrate modulator of the ClpAP machine. Molecular cell 9, 673–683. [DOI] [PubMed] [Google Scholar]
- Erbse A, et al. (2006). ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439, 753–756. [DOI] [PubMed] [Google Scholar]
- Gamer J, et al. (1992). Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69, 833–842. [DOI] [PubMed] [Google Scholar]
- Ge Z, and Karzai AW (2009). Co-evolution of multipartite interactions between an extended tmRNA tag and a robust Lon protease in Mycoplasma. Mol Microbiol 74, 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S (2003). Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 19, 565–587. [DOI] [PubMed] [Google Scholar]
- Gur E, and Sauer RT (2008). Evolution of the ssrA degradation tag in Mycoplasma: specificity switch to a different protease. Proc Natl Acad Sci U S A 105, 16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, and Sauer RT (2009). Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proceedings of the National Academy of Sciences of the United States of America 106, 18503–18508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C, et al. (1995). Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci U S A 92, 3516–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds T, and Sandler SJ (2004). Allele specific synthetic lethality between priC and dnaAts alleles at the permissive temperature of 30 degrees C in E. coli K-12. BMC Microbiol 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, et al. (2001). The molecular chaperone DnaJ is required for the degradation of a soluble abnormal protein in Escherichia coli. J Biol Chem 276, 3920–3928. [DOI] [PubMed] [Google Scholar]
- Ishii Y, and Amano F (2001). Regulation of SulA cleavage by Lon protease by the C-terminal amino acid of SulA, histidine. Biochem J 358, 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, et al. (2000). Regulatory role of C-terminal residues of SulA in its degradation by Lon protease in Escherichia coli. Journal of biochemistry 127, 837–844. [DOI] [PubMed] [Google Scholar]
- Jackson MW, et al. (2004). The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol Microbiol 54, 1364–1378. [DOI] [PubMed] [Google Scholar]
- Jonas K, et al. (2013). Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154, 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubete Y, et al. (1996). Role of the heat shock protein DnaJ in the lon-dependent degradation of naturally unstable proteins. J Biol Chem 271, 30798–30803. [DOI] [PubMed] [Google Scholar]
- Lesley SA, et al. (2002). Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng 15, 153–160. [DOI] [PubMed] [Google Scholar]
- Levchenko I, et al. (2000). A specificity-enhancing factor for the ClpXP degradation machine. Science 289, 2354–2356. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, et al. (2015). Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci U S A 112, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, et al. (2015). Discovery of a Unique Clp Component, ClpF, in Chloroplasts: A Proposed Binary ClpF-ClpS1 Adaptor Complex Functions in Substrate Recognition and Delivery. Plant Cell 27, 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, et al. (2013). ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25, 2276–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond CS, et al. (1999). Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimuta TR, et al. (2004). Novel heat shock protein HspQ stimulates the degradation of mutant DnaA protein in Escherichia coli. Genes Cells 9, 1151–1166. [DOI] [PubMed] [Google Scholar]
- Straus D, et al. (1990). DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev 4, 2202–2209. [DOI] [PubMed] [Google Scholar]
- Straus DB, et al. (1987). The heat shock response of E. coli is regulated by changes in the concentration of sigma 32. Nature 329, 348–351. [DOI] [PubMed] [Google Scholar]
- Sutton MD, and Kaguni JM (1995). Novel alleles of the Escherichia coli dnaA gene are defective in replication of pSC101 but not of oriC. J Bacteriol 177, 6657–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, et al. (1989). Modulation of stability of the Escherichia coli heat shock regulatory factor sigma. J Bacteriol 171, 1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibaris V, et al. (2006). Biological roles of the Lon ATP-dependent protease. Res Microbiol 157, 701–713. [DOI] [PubMed] [Google Scholar]
- Van Melderen L, and Gottesman S (1999). Substrate sequestration by a proteolytically inactive Lon mutant. Proc Natl Acad Sci U S A 96, 6064–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlever ML, et al. (2013). Engineering fluorescent protein substrates for the AAA+ Lon protease. Protein Eng. Des. Sel. 26, 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WF, et al. (1999). Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J Bacteriol 181, 3681–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, et al. (2001). The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev 15, 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.