Abstract
Virtually all plants and animals, including humans, are home to symbiotic microorganisms. Symbiotic interactions can be neutral, harmful or have beneficial effects on the host organism. However, growing evidence suggests that microbial symbionts can evolve rapidly, resulting in drastic transitions along the parasite–mutualist continuum. In this Review, we integrate theoretical and empirical findings to discuss the mechanisms underpinning these evolutionary shifts, as well as the ecological drivers and why some host–microorganism interactions may be stuck at the end of the continuum. In addition to having biomedical consequences, understanding the dynamic life of microorganisms reveals how symbioses can shape an organism’s biology and the entire community, particularly in a changing world.
Subject terms: Coevolution, Microbial ecology, Symbiosis, Parasite evolution
Symbiotic interactions can be neutral, harmful or have beneficial effects for host organisms. In this Review, Drew, Stevens and King discuss the evolutionary transitions of host–microorganism symbioses along the parasite–mutualist continuum, the mechanisms underlying evolutionary changes, the selective pressures involved and common empirical approaches for studying them.
Introduction
Parasitic and mutualistic microbial symbioses exist widely in nature. These interactions occur when microorganisms (that is, bacteria, fungi and viruses) take up residence in or on animals or plants, and cause damage or confer benefits to the host. Parasitic microorganisms (including pathogens) can exploit the host, and in doing so, cause harm. The term mutualist classically refers to any organism in a mutually beneficial relationship with another. However, the assumed benefits are rarely empirically tested for the symbiont1. There is thus an emerging awareness that many putative mutualisms may even be hosts exploiting symbionts2–4, in an interaction referred to as inverted parasitism5.
The continuum
The designation of entities as ‘parasite’ or ‘mutualist’ implies a simple binary system whereby species incur positive or negative impacts on fitness during interactions. However, these terms represent ends of a continuum along which an interaction between a host and symbiont can shift. These transitions occur as the relative benefits and costs to each species in the relationship strengthen or weaken (Fig. 1) across ecological or evolutionary time. Transitions can be driven by changes in the environment and ecology of the interacting species or communities. At the centre of the continuum sit commensals, which benefit from the interaction with hosts, but do not cause a detectable cost6.
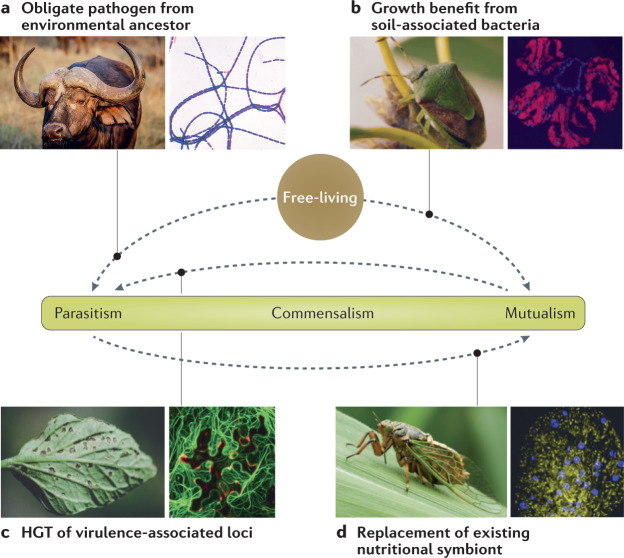
Fig. 1. Evolutionary transitions onto and along the parasite–mutualist continuum.
Examples from nature of microorganisms transitioning from free-living to host-associated lifestyles include the evolution of parasitic species in the Bacillus cereus group (for example, the causative agent of anthrax) from soil-dwelling ancestors237 (part a), and environmental Pantoea bacteria evolving obligate mutualistic roles in stink bug growth and development16 (part b). Examples involving transitions along the continuum are the widespread plant parasite Pseudomonas syringae likely evolving from mutualistic ancestors, driven by horizontal gene transfer (HGT) of type III secretion systems29,79 (part c), and entomopathogens taking over the metabolic role of an ancient and degraded endosymbiont in cicadas165 (part d). Image credits: part a (right) Getty images Smith Collection/Gado.Contributor; part b is adapted from ref.238, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/); part c (left), image courtesey of Gerald Holmes; part c (right) is adapted from ref.239, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/); part d (left), image courtesey of Yu Matsuura; part d (right) adapted with permission from ref.165, PNAS.
The concept of the parasite–mutualist continuum dates back several decades. An early discussion by Ewald7 focused on the fundamental role of transmission route in driving evolutionary transitions between parasitism and mutualism in symbiotic associations. The conditionality of symbiotic interactions was later highlighted by Bronstein8. She reviewed evidence that the costs and benefits of interspecific interactions vary greatly with ecological context, and thus the outcome of a symbiosis can change throughout an organism’s lifetime.
Evolution of microorganisms into parasites or mutualists
Microorganisms can rapidly adapt to new environments. Short generation times, large population sizes and high mutation rates combined with genome flexibility all facilitate accelerated microbial evolution9. Furthermore, their capacity for plastic responses10–12 and the dynamic nature of the communities that microorganisms are nested and interact within13,14 provide further routes for changing costs and benefits of association with hosts.
Free-living environmental microorganisms, which do not associate with hosts, were the progenitors for all symbiont diversity observed today15. Free-living microorganisms can evolve to be parasites or mutualists16–21. A new host-associated lifestyle often remains facultative for the microorganism22,23, but in some cases the microorganism evolves an obligate dependency on the host24,25. Transitions from free-living to host association are sometimes facilitated by horizontal transfer of genes, often encoding traits that facilitate immediate exploitation of, or benefit to, hosts (for example, immune evasion, toxin production, nitrogen fixation and bioluminescence)15,26. Once associated with a host, symbiotic interactions can shift along the continuum (Fig. 1). For instance, parasites can evolve to be less antagonistic to hosts. Reduced antagonism is thought to be favoured if alternative hosts are rarely available or if transmission of the parasite is enhanced by increases in host fitness27,28. Molecular phylogenetics corroborates this trajectory, showing that parasites have frequently served as progenitors for the independent descent of symbionts that now exhibit mutualistic traits15,29. In this context, microorganisms shifting into novel host taxa is an important process, often forging novel associations on the continuum30,31. Transitions can also occur if a parasite’s own selfish traits benefit a host as a by-product27, or by hosts rewarding32 or capturing33 symbiont genotypes that confer benefits. Conversely, mutualisms can break down into parasitisms. This breakdown can occur owing to the spread of cheater symbionts, which exploit the benefits of host association without paying the cost of returning a benefit27,34. However, shifts from mutualism to parasitism appear rare in nature15,29,35. More frequently, symbionts leave the host-association continuum by reverting to free-living environmental lifestyles, as demonstrated by Actinobacteria abandoning ant hosts15,36.
In this Review, we discuss the evolutionary transitions of host–microorganism symbioses along the parasite–mutualist continuum, the mechanisms underlying evolutionary changes, the selective pressures involved and common empirical approaches for studying them (Box 1). We also briefly discuss context-dependent transitions and the consequences faced by microorganisms when their symbioses are constrained to the extreme ends of the continuum. Moreover, we focus the Review on eukaryotic host–microorganism symbioses; however, we note that microbial interactions with mobile genetic elements (MGEs) can be analogous to symbioses (Box 2) given the ability of these elements to confer beneficial traits and cause harm to bacterial hosts37,38.
Box 1 Two approaches to evaluating evolution along the parasite–mutualist continuum.
Phylogenetic inference
There are challenges to judging transitions in symbiosis because ancestral partnerships no longer exist for direct comparison. Interactions that now appear mutualistic may actually reflect the result of a long period of conflict resolution or the evolution of tolerance by the host. Phylogenetic inference can shed light on the evolutionary history of transitions on the parasite–mutualist continuum. Techniques such as ancestral state reconstruction and its extensions infer characteristics of ancestral taxa based on traits exhibited by extant descendants240. In this way, symbiotic phenotypes of ancestors (for example, parasite, mutualist, commensal or free-living) can be recovered and used to infer the origins and breakdowns of associations on the continuum, in addition to the rate of such transitions29. Such approaches are heavily contingent on the quality of the underlying phylogenetic tree, and reconstruction accuracy declines with increasing evolutionary time240. However, for many lineages of bacterial symbionts this approach has been used powerfully to demonstrate the marked rarity of reversions from mutualism to parasitism over evolutionary timescales15,29.
Experimental evolution
Experimental evolution permits the direct testing of hypotheses related to the tempo and pattern of the evolution of species interactions. This approach allows for evolution to be observed in real time. An added advantage in some systems is an ability to cryopreserve the eukaryotic host (for example, Caenorhabditis elegans241 and Paramecium bursaria159) and associated microbial lineages for subsequent analysis. This characteristic allows the fitness benefit or harm for both species to be compared with past and future archived generations, for example, via time shift assays242.
In an evolution experiment, the source of selection can be hypothesized and manipulated. For example, this approach could be used to determine whether the presence or absence of an enemy could affect the position of a defensive symbiosis along the continuum40, as well as whether the evolution of the eukaryotic host or the microbial symbiont, or their coevolution was responsible for the shift218. Subsets of the population can be used to establish the next generation. One focal species can be evolved and others kept in evolutionary stasis by adding from an ancestral population each generation. Alternatively, additional community members can be reciprocally evolved, opening the arena for coevolutionary dynamics between two or more species243. The process continues for generations. At the end, phenotypic and genomic comparisons can be made between ancestral and evolved populations, and also across replicates, to assess convergence or divergence in transition outcome and the genetic basis.
Candidate molecular targets in evolved lineages can be identified for manipulation and further experimentation. Moreover, follow-on genomic analysis can be powerfully combined with phenotypic assays across evolutionary time to identify the mechanism of relative benefit or cost for each species, as well as to confirm phenotypic traits under selection. One caveat is that experimental evolution might be less likely to yield increases in parasite virulence given the potential for breaking apart of the virulence–transmission trade-off at passage points244.
Box 2 Mobile genetic elements as symbionts.
Mobile genetic elements (MGEs) can cause genomic change in their microbial hosts. These changes can affect the position of the microorganism–eukaryotic host relationship on the parasite–mutualist continuum by coding for traits that harm or benefit microbial hosts. On a smaller scale, MGEs are analogous to symbionts37,38 as they are entities with their own evolutionary interests that can parasitize hosts or confer beneficial traits that promote innovation. The effects they have on microbial host fitness can change.
Many nosocomial pathogens have acquired antibiotic resistance genes through horizontal gene transfer245, gaining a survival advantage in the presence of certain antibiotics. In the absence of the corresponding antibiotic, however, a resistance-conferring MGE can become costly to its host. For example, when large low-copy-number plasmids are cumbersome to their host, these plasmids force their maintenance through the action of resolution systems, partitioning systems and post-segregational killing systems. The latter of these includes toxin–antitoxin systems, encoding both a stable protein toxin and a less stable, but more abundant antitoxin. If a plasmid fails to be inherited by a daughter cell, the antitoxin will rapidly degrade in the host, leaving it susceptible to being killed by the toxin246. The transition of MGEs from beneficial elements conferring a survival advantage to parasites can take place over very short evolutionary timescales. In turn, in the face of antibiotic treatment and other clinical interventions, MGEs can drive the evolution of their bacterial hosts towards higher virulence over an equally short period of time58,247.
MGEs are not always maintained through natural selection. The genome of Wolbachia pipientis wMel, an obligate intracellular symbiont of the fruitfly Drosophila melanogaster, is highly streamlined from extensive gene loss during adaptation to its host; however, it is also overrun with MGEs248. Repeated population bottlenecks resulting in genetic drift and inefficient natural selection248 likely contributes to the extensive maintenance of MGEs in this genome and those of other heritable symbionts249. These elements may have contributed to the substantial phenotypic diversity among Wolbachia strains, fundamentally shaping Wolbachia evolution248. In this instance, MGEs are parasitic elements maintained within the population effectively by accident via transmission of Wolbachia from one host to the next. Ultimately, it is unclear whether these elements will cross the parasite–mutualist continuum and become permanent components of Wolbachia genomes.
For some microbial hosts, the acquisition of deleterious MGEs can be partially rescued via compensatory evolution, leading to a type of host tolerance. In such cases, the association is maintained but the host ameliorates the cost, as shown for Pseudomonas fluorescens and a mega-plasmid conferring mercury resistance. In low-mercury environments, the plasmid is costly, yet experimental evolution across a mercury gradient showed P. fluorescens consistently compensated via mutation in the gacA–gacS two-component system, downregulating chromosomal and plasmid gene expression and relieving translational cost44. Such compensatory evolution may also explain the persistence of context-dependent mutualisms in environments where they do not benefit hosts.
MGEs can also become ‘immortalized’ in host lineages. Once genomic parasites, they can become indispensable components of the host genome that are ultimately passed on to daughter cells. Vestigial MGEs in the form of cryptic phages, ancient regions of viral DNA and disrupted transposon sequences or pseudogenes can be found immortalized in the genomes of organisms throughout the tree of life250. Bacterial chromosomes, for example, can contain as much as 20% phage DNA251,252. Once parasites to their hosts, these MGEs have infected the genomes of host organisms, maintained their stability as they coevolve with their host (forcibly in some cases, for example, toxin–antitoxin systems) and finally been irreversibly integrated into the genome. Integration can occur by accident during genome rearrangements, recombination, population bottlenecks and speciation events248, or by natural selection because of a fitness benefit on which the host has become dependent68. The ubiquitous presence of vestigial viral DNA in the cells of all organisms250 is a prime example, demonstrating how MGEs have been formative in the evolution of organisms, just like many eukaryotic host–microbial symbioses. MGEs leave behind remnants of DNA in host genomes like partial segments of an ancient diary.
MGEs therefore possess the capability themselves to go from genomic parasites to mutualistic or commensal components of the genome. In many situations, this process can also drive the evolution of their bacterial hosts along the continuum. MGEs have forcibly maintained their interaction with bacteria in some cases, while in others, their maintenance has been a by-product of environmental conditions or population bottlenecks. They represent fascinating examples of entities that can be both effectors and subjects of evolutionary transitions along the parasite–mutualist continuum.
Mechanisms of evolution along the continuum
The gradual emergence of microbial mutualists from parasitic ancestors15,29,31,39 and the rapid leaps in symbiont phenotypes observed in real time40–44 provide fascinating insights into the proliferation of microbial symbiotic diversity. The genetic changes involved in microbial evolution are key contributors to the formation of mutualisms and parasitisms and their transitions along the symbiotic continuum. Mechanisms that result in these changes include, for example, selection on existing genetic variation45,46, de novo mutations40,43,47–49 and genome rearrangements50–52. Genome rearrangements include inversions, duplications, translocations and gene loss50,53,54 (for further discussion of gene loss, see later section, Stuck at the end of the line). Horizontal gene transfer (HGT) events, whereby genetic material moves between organisms in a manner other than vertically, are also important factors in microbial evolutionary transitions42,55–58. These events often involve MGEs — such as plasmids, transposons, insertion elements and phages — coding for traits that are beneficial or harmful to hosts during their interaction.
Shifts between microbial parasitism and mutualism can involve selection on existing variation. Through experimental evolution of the bacterial symbiont Parachlamydia acanthamoebae and its protist host Acanthamoeba sp., one study46 observed an evolutionary shift of the microbial symbiont towards parasitism under horizontal transmission conditions. The molecular basis of this transition was a pronounced increase in the frequency of specific genetic variants within the original symbiont population, alongside marked changes in the expression of machinery necessary for manipulating host cells, such as the type III secretion system (T3SS).
Selection on de novo mutations in bacterial populations has also been detected in evolution experiments, resulting in movement along the continuum. In these cases, experiments are started by propagating a single clone in hosts. In one study40, a clonal population of Enterococcus faecalis was introduced into nematode host populations, and mutations that arose favoured enhanced production of reactive oxygen species. This phenotype allowed E. faecalis to become highly beneficial to hosts, as production of these antimicrobials suppressed infection by Staphylococcus aureus. A similar direction of travel, but from parasite to commensal, has been observed in nematode host populations by evolving Pseudomonas aeruginosa from a single clone49. Conversely, within the guts of old mice, mutations arising in clones of commensal Escherichia coli may have resulted in evolution towards pathogenicity59. In comparison with evolution within young mice, mutational targets linked to stress-related functions and associated with virulence were under strong selection in the inflamed guts of older mice. Mutation might have a prominent role in transitions when symbionts have a low initial diversity upon colonization. This situation could occur naturally when symbionts have a low infectious dose or when transmission causes population bottlenecks (see section on Transmission below).
Wide-ranging genetic changes — HGT, gene loss and genome rearrangements — have had a profound role in Yersinia pestis becoming more virulent and adapting to new host species50,60,61. Y. pestis is the causative agent of plague in mammalian and arthropod hosts. It is thought to have diverged from its less harmful ancestor Yersinia pseudotuberculosis 1,500–55,000 years ago62,63. Sequencing of isolates of the two species revealed that both HGT and insertion sequence-mediated genome rearrangements and deletions facilitated Y. pestis evolution50,60,61. The bacterium acquired two plasmids, namely pMT1 and pPCP1, making it more virulent compared with its Y. pseudotuberculosis ancestor. The former plasmid carries the ymt gene encoding Yersinia murine toxin, required for the colonization of the flea host64,65, and the capsular antigen fraction 1, which inhibits phagocytosis65,66. These acquisitions contributed to the evolution of Y. pestis towards greater virulence. Adaptation of the parasite to new hosts was mediated by genome rearrangements, particularly via insertion sequences and gene loss. Gene loss was crucial in reducing the toxicity of Y. pestis to the flea vector, allowing biofilm to develop in the flea foregut67. Gene disruption by insertion sequences, in combination with deletion events, point mutations and frameshifts, further created an extensive number of pseudogenes within the Y. pestis genome50,60,61. Altogether, these genetic changes facilitated a shift in lifestyle, from a less harmful mammalian enteropathogen to systemic pathogen of both mammalian and arthropod hosts.
Infection by various phages (mostly lytic, λ-like phages) along with other MGEs facilitated the divergence of the highly pathogenic enterohaemorrhagic E. coli strain O157 Sakai from its ancestor. The commensal E. coli strain K12 is also descended from this common ancestor68. In strain O157 Sakai, prophages and prophage-like elements encode a variety of virulence-related genes — adhesins, tellurite resistance genes and urease — contributing to the acquisition of virulence factors that have determined this bacterium’s trajectory towards increased virulence in humans. One of these elements also encodes the major virulence factor, the locus of enterocyte effacement (LEE), which is responsible for bacterial attachment followed by development of the disease-causing effacing lesions in the intestine69. Lambda-like phages on the Sakai chromosome also encode the destructive Shiga toxin, as well as proteins involved in serum resistance and cell adhesion. Having become integral to the organism’s virulence in this way, the prophages themselves have transitioned from parasitic to mutualistic elements within the O157 Sakai genome (for further discussion of MGEs as symbionts, see Box 2).
How commonly do shifts across the continuum occur owing to de novo mutation or machinery acquired by HGT? Host environments with complex, often open, microbial communities, such as the mammalian gut, might generate more extensive opportunities for HGT70–72. For example, phage-driven HGT from the resident community can dictate the evolution of invading strains73 and instigate change more rapidly than is achievable by mutation accumulation74. HGT has had a considerable role in major evolutionary transitions of living organisms; it is increasingly confirmed as a dominant force in the evolution of host–symbiont associations20,29,54,58,65,75–80. Yet, for symbionts nested within simple microbial communities (for example, intracellular environments), scarce opportunities for HGT may mean de novo mutation is more likely to underpin shifts along the continuum. Studies reporting selection on de novo mutation during transitions40,49,59 highlight the power of this genetic means to generate remarkable change on the continuum. These experiments typically involve a small number of microbial species and/or low levels of initial genetic diversity upon colonization. When incorporating a host background with an ecologically relevant microbiota, HGT might be more dominant.
Drivers of evolution along the continuum
Ecological sources of selection can drive microbial symbiont evolution towards increasing host benefits (Table 1) or harm (Table 2). Shifts occur across generations as microbial symbionts adapt to life in a new host species, encounter different transmission opportunities and face hosts that reciprocally evolve in response. The presence or absence of additional interacting species in the community can also drive evolutionary change in a host–symbiont relationship owing to changing distribution of net benefits and costs across the community. Essentially, given a strong source of selection, genetic change can occur within just a handful of microbial generations. These transitions are often investigated using experimental evolution or over macro-evolutionary timescales via phylogenetic comparisons (Box 1).
Table 1.
Studies reporting evolution of symbioses towards the mutualism end of the continuum
| Transitiona | Host | Symbiont | Association | Condition | Mechanism and evidenceb | Approach | Refs |
|---|---|---|---|---|---|---|---|
| P → P (−) | Ciliate (Paramecium caudatum) | Holospora undulata | Mixed mode transmitted parasite | Low host density | Lower virulence and increased VT frequency | Experimental | 216 |
| P → P (−) | Actinomyces odontolyticus | Nanosynbacter lyticus (TM7x) | Epibont parasite | Naive host and co-culture passage | Host susceptibility rapidly reduced | Experimental | 151 |
| P → P (−) | Escherichia coli | F1 phage | Parasitic phage | VT only | Less virulent variants favoured | Experimental | 217 |
| P → P (−) | Nematode (Caenorhabditis elegans) | Serratia marcescens | Parasite | Coevolution over 20 generations | Increased host fecundity | Experimental | 218 |
| P → P (−) | European rabbit (Oryctolagus cuniculus) | Myxoma virus | Parasite | Novel host | Increased interferon antiviral activity (host); greater transmission traded off with virulence (virus) | Field sampling | 84,85 |
| P → P (−) | Nematode (C. elegans) | Staphylococcus aureus | Parasite | Pathogen coevolved with defensive microorganism | Siderophore production reduced | Experimental | 144 |
| P → P (−) | Mouse (Mus musculus) | Friend virus | Parasite | Heterogeneity in host resistance | Resistant hosts drove parasite specialization, reduced mean virulence across host population | Experimental | 219 |
| P → P (−) | Diamond-back moth (Plutella xylostella) | Enterobacter cloacae | Gut symbiont | Pathogen exposure | Reduced virulence in some lineages | Experimental | 220 |
| P → P (−) | Barley (Hordeum vulgare) | Barley stripe mosaic virus | Plant parasite | VT only | Substantial reduction in virulence | Experimental | 111 |
| P → C | Nematode (C. elegans) | Pseudomonas aeruginosa | Gut parasite | Serial passage | Mutation in global regulator lasR and polymerase gene rpoB | Experimental | 49 |
| P → C | Legume (Mimosa pudica) | Ralstonia solanacearum and rhizobial plasmid | Root nodulation | HGT and selection from emergent nodules | T3SS (hrcV) and master virulence regulator (hrpG) inactivated | Experimental | 221 |
| P → M | Squid (Euprymna scolopes) | Vibrio fischeri | Bioluminescence | NA | Inferred evolution from parasitic ancestors | Phylogenetic | 15,222 |
| P → M | Nematode (C. elegans) | Enterococcus faecalis | Defensive microorganism | Pathogen exposure | Increased antimicrobial superoxide production | Experimental | 40 |
| P → M | Mouse (M. musculus) | Candida albicans | Gut symbiont | Gut microbiota absent | Filamentation loss, increased cytokine response, host protection against infection | Experimental | 43 |
| P → M | Fruitfly (Drosophila simulans) | Wolbachia (wRi) | Reproductive parasite | VT and reproductive manipulation | Fecundity benefit over uninfected hosts | Experimental, field sampling | 123 |
| P → M | Cicadas (Cicadoidea spp.) | Ophiocordyceps fungi | Nutrient provisioning | Genomic decay of existing symbiont | Evolution from pathogens inferred; took over amino acid synthesis | Phylogenetic, field sampling | 165 |
| P → M | Pea aphid (Acyrthosiphon pisum) | Hamiltonella defensa | Defensive microorganism | NA | Putative parasite loci remain (T3SS and toxin homologues) | Comparative genomic, phylogenetic | 121 |
| P → M | Insect spp. | Sodalis-allied | Insect endosymbionts | NA | Mutualistic lineages inferred to stem from putative parasitic ancestor | Phylogenetic | 31 |
| P → M | Lagriinae beetles | Burkholderia gladioli | Antimicrobial producer | Host shift | Metabolite repurposed for insect defence | Phylogenetic, experimental | 89 |
| P → M | Arabidopsis thaliana | Pseudomonas protegens | Rhizophere associated | Low carbon forces dependence on host | Mutation in gacS–gacA TCS; heightened competitiveness for host exudates | Experimental | 47 |
| P → M | Amoeba proteus | Legionella-like X-bacteria | Growth benefit | Coevolution over 200 host generations | Evolved mutual dependence, altered host gene expression | Experimental | 223 |
| P → M | E. coli | Cryptic prophage | Permanent host genome integration | Long-term coevolution | Increased host resistance to environmental stress | Experimental | 224 |
| P → M | E. coli | F1 phage | Parasitic phage | Serial passage | Enhanced growth rate and resistance to superinfection | Experimental | 225 |
| P → M | E. coli | M13 phage | Growth inhibition | HT restricted | Host growth benefit | Experimental | 41 |
| P → M | Pseudomonas fluorescens | Mega-plasmid pQBR103 | Mercury resistance | Mercury gradient | Host compensated by gacA–gacS TCS disruption, alleviated translational cost | Experimental | 44 |
| FL or P → M | Stink bugs (Pentatomidae spp.) | Burkholderia spp. | Gut symbiont | Unknown | Inferred evolution from parasites, colonization of specialized gut crypts | Phylogenetic, field sampling | 15,226,227 |
| P or C → M | Bed bug (Cimex lectularius) | Wolbachia (wCle) | Nutrient provisioning | Co-infection hypothesized | HGT of biotin operon | Experimental, genomic | 80 |
| C → M | Squid (E. scolopes) | Vibrio fischeri | Bioluminescence | Host choice | Mutation in signalling protein gene (binK), protected against host immune cells and chemicals | Experimental | 48 |
| M → M (+) | Jelly fish (Cassiopea xamachana) | Alga (Symbiodinium microadriaticum) | Photosynthate provisioning | VT only | Host growth benefit | Experimental | 109 |
| M → M (+) | E. coli | M13 phage | Growth benefit | Transmission opportunity varied | Greatest benefit when VT and HT allowed | Experimental | 228 |
(−), reduced; (+), elevated (for example, M → M (+) indicates transition towards increased benefit to host); C, commensalism; FL, free-living; HGT, horizontal gene transfer; HT, horizontal transmission; M, mutualism; NA, specific drivers of transition unaccounted for owing to timescale; P, parasitism; T3SS, type III secretion system; TCS, two-component regulatory system; VT, vertical transmission. aTransitions involve reduction in virulence or increased benefit of the relationship to hosts over time. bGeneral evidence to support the inferred transition, including the molecular mechanism if known.
Table 2.
Studies reporting evolution of symbioses towards the parasitism end of the continuum
| Transitiona | Host | Symbiont | Association | Condition | Mechanism and evidenceb | Approach | Refs |
|---|---|---|---|---|---|---|---|
| M → M (−) | Legume (Ensifer medicae) | Rhizobia | Nitrogen-fixing | Host choice blocked | Cheater strains favoured | Experimental | 229 |
| M → M (−) | Legume (Trifolium spp.) | Rhizobia | Nitrogen-fixing | Elevated nitrogen | Reduced cooperation under high nitrogen | Experimental | 230 |
| M → P | Vertebrate spp. | Coxiella burnetii | Intracellular parasite | Host shift | HGT of virulence-associated genes suggested | Phylogenetic | 30 |
| M → P | Jelly fish (Cassiopea xamachana) | Alga (Symbiodinium microadriaticum) | Photosynthate provisioning | HT only | Greater proliferation in host and dispersal rates | Experimental | 109 |
| M → P | Plant spp. | Agrobacterium spp. | Plant parasite | NA | HGT of virulence loci | Phylogenetic | 29,231 |
| M → P | Plant spp. | Pseudomonas syringae | Plant parasite | NA | HGT of hopZ T3SS effectors | Phylogenetic | 29,79 |
| M → P | Escherichia coli | M13 phage | Growth benefit | Host background | Parasitic when shifted to host ancestor | Experimental | 228 |
| M → P | E. coli | F1 phage | Parasitic phage | HT allowed | Antagonistic variants favoured | Experimental | 217 |
| C → P | Pill bug (Armadillidium vulgare) | Wolbachia (wVulC) | VT endosymbiont | HT only | Titre increased in non-germline-associated tissue | Experimental | 110 |
| C → P | In vitro immune envrionment | E. coli | Commensal strain | Macrophage pressure | Heightened macrophage evasion and delayed phagosome maturation, via TE insertion | Experimental | 232 |
| C → P | Arabidopsis thaliana | Pseudomonas fluorescens species complex | Rhizophere associated | NA | Gain of putative pathogenicity island | Comparative genomics, phylogenetic | 39 |
| C → P | Plant spp. | Rhodococcus spp. | Plant associated | NA | Gain of virulence plasmid (pFID188), host growth inhibition | Experimental, comparative genomics, phylogenetic | 58 |
| P → P (+) | Plant spp. | Xanthomonadaceae spp. | Phytopathogen | NA | Gain of hydrolase gene (cbsA); localized parasite become systemic | Comparative genomics, phylogenetic | 54 |
| P → P (+) | Barley (Hordeum vulgare) | Barley stripe mosaic virus | Plant parasite | HT only | Increased virulence, independent of titre | Experimental | 111 |
| P → P (+) | House finch (Haemorhous mexicanus) | Mycoplasma gallisepticum | Emerging parasite | Adaptation to novel host | Linear increase in virulence since shift | Natural sampling | 83 |
| P → P (+) | Mouse (Mus musculus) | Cryptococcus neoformans | Opportunistic parasite | Serial passage | Increased expression of iron reductase and host mortality | Experimental | 233 |
| P → P (+) | Amoebae (Acanthamoeba sp.) | Parachlamydia acanthamoebae | Obligate intracellular symbiont | HT only | Enhanced infectivity and virulence, T3SS upregulated | Experimental | 46 |
| P → P (+) | Mammal spp. | Yersinia pestis | Enteric parasite | NA | HGT of plasmids (pMT1 and pPCP1), increased transmissibility by fleas and virulence to mammals | Genomic | 65 |
(−), reduced; (+), elevated (for example, P → P (+) indicates transition towards increased parasitism); C, commensalism; HGT, horizontal gene transfer; HT, horizontal transmission; M, mutualism; NA, specific drivers of transition unaccounted for owing to timescale; P, parasitism; T3SS, type III secretion system; TE, transposable element; VT, vertical transmission. aTransitions involve increased virulence or reduced benefit of the symbiotic relationship to hosts over time. bGeneral evidence to support the inferred transition, including the molecular mechanism if known.
Novel hosts
Microorganisms frequently encounter novel host environments. They can jump across species boundaries or colonize hosts from pools of free-living environmental microorganisms. Novel infections can generate new diversity on the symbiosis continuum through divergence and speciation81. High-profile cases of host shifts, such as the recent SARS-CoV-2 pandemic82, highlight the potential for investigating evolutionary changes in virulence upon emergence83–85. New associations are often maladaptive for both host and parasite86, and associations can move unpredictably on the continuum or burn out. This trajectory has been observed in emergences of avian influenza virus, where case fatality rates can be high but human-to-human transmission is low87.
Shifts between host species, possibly driven by HGT of virulence-associated genes, appear to have been important in the emergence of the Q fever parasite, Coxiella burnetii30,88. This proposed mutualist-to-parasite transition is a complex case for which the full evolutionary story remains unknown. However, phylogenetic analysis suggests that this highly infectious bacterium recently emerged from a clade of vertically transmitted mutualistic endosymbionts of ticks30. C. burnetii may have evolved mechanisms to infect vertebrate cells, persist in the environment and be airborne-transmitted. These traits are unlikely to be found in the arthropod-restricted ancestors30. Ticks feeding on vertebrates likely provided the ecological bridge. Similar transitions occurred within Sodalis-allied symbionts, a group of host-restricted bacteria common to insects including the tsetse fly vector. A free-living Sodalis sp. was isolated after a person suffered a wound from a tree branch, and this serendipitous finding provided evidence that symbiont lineages emerged from environmental ancestors31. Early vectoring of these environmental strains by insects was likely pivotal in the evolution of the beneficial, heritable Sodalis endosymbionts observed today.
Novel species interactions can drive rapid innovation. This might particularly be the case if a microorganism bears characteristics that can provide instant benefits. Microorganisms encoding functions of light generation, photosynthesis, nitrogen fixation or antimicrobials may provide such rapid benefits15. These characteristics may be remodelled (or act as pre-adaptations) for transitions in symbiosis15. Such repurposing may have occurred in the antifungal-producing Burkholderia symbionts associated with Lagriinae beetles. Burkholderia symbionts appear to have transitioned from a plant parasite to insect mutualist. In this context, secondary metabolites previously used as virulence factors against plants may have been repurposed for antifungal defence on beetle eggs89. Additional evidence comes from marine hosts, including within the bulbs of anglerfish and the Vibrio fischeri-filled light organs of bobtail squid. These hosts benefit from these bioluminescent bacteria to lure prey and avoid predation, respectively, and the symbionts often retain the capacity to live freely, or persist in the environment22,51.
Transmission opportunities
Transmission mode has been considered to predict the direction of a symbiont’s evolution on the continuum. When horizontally transmitted symbionts can move between unrelated host individuals, the fitness interests between species are uncoupled, a scenario thought to favour parasitism7. The degree of harm caused to hosts from infection is often framed by the virulence–transmission trade-off90,91. The relationship assumes that virulence — the reduction in host fitness caused by parasite infection — is costly to the parasite as host resources are needed for replication92. The cost of harming the host too much or too soon from replication might result in less transmission. Thus, it is predicted that transmission should be highest at intermediate virulence, which balances the costs of within‐host replication and infectious period length90. This model is particularly pertinent for symbionts that rely on a mobile host for transmission (for example, socially transmitted microorganisms). Those that do not (for example, vector- and water-borne microorganisms) can bypass trade-offs between virulence and transmission91. This conventional model goes some way to hypothesizing on general patterns of virulence, yet several extensions and alternatives have been suggested93–95.
It has been suggested that mutualists may evolve from parasitic ancestors when the frequency of horizontal transmission routes is reduced or lost7. If vertical transmission is the remaining dominant mode of transmission then the fitness of host and symbiont can become tightly coupled, reducing the arena for evolutionary conflict and thereby favouring selection for mutual benefit7,90,96. Mutualisms involving symbiont inheritance are predicted to be stable on the continuum and unlikely to revert to parasitism15,97. But exclusively vertical transmission can endanger associations via genetic bottlenecking (see section on Stuck at the end of the line). Clearly, becoming inherited is not the sole route by which bacterial mutualists evolve. Comparative analysis has found no evidence for vertical transmission preceding the origin of mutualism15. Many mutualisms involve horizontal transmission such as conjugative plasmids in bacterial populations98 and the vast networks of mycorrhizae that improve plant productivity99,100. In particular, evolution of defensive traits in symbionts are proposed to be facilitated by the genetic diversity and selection for innovation promoted by horizontal transmission101. Many horizontally transmitted microbial symbionts are obligate for host fitness16,22,102, but many can be facultative24 and confer costs in different environments.
Conversely, not all inherited microorganisms become mutualists103. Wolbachia, Spiroplasma and Arsenophonus species are common inherited parasites that manipulate host reproduction, maximizing resource allocation to the transmitting host sex (females) by feminizing hosts or killing their sons104. However, theory suggests that the spread of such reproductive parasites will be enhanced by the evolution of traits that benefit hosts105. A beneficial trait (that is, defence) may even interact with a parasitic trait (that is, reproductive manipulation) to completely exclude a natural enemy105. Indeed, cryptic benefits are now found in several systems106,107, and there is evidence that some reproductive parasites may need to also transmit horizontally just to persist108.
Transmission as a determinant of the location of a symbiosis along the continuum is complex. There are numerous exceptions to classical theory. Nonetheless, experimental manipulation of transmission modes finds general support for the theory that horizontal transmission can select for parasitism and vertical transmission for reduced antagonism (Table 1; Table 2). In a symbiosis between a jellyfish and the alga Symbiodinium microadriaticum, cooperative traits, including growth enhancement, were selected when transmission was restricted to heritable routes109. Such cooperative traits are fundamental for stable mutualisms, protecting against transitions to parasitism or abandonment events. In the reverse experiment, restriction of the alga to horizontal transmission selected for faster proliferation and dispersal (traits associated with parasitism), and declines in host fitness were detected109. Such findings are mirrored across terrestrial systems46,110,111. The common pill bug hosts a Wolbachia strain (wVulC) that feminizes genetic males112. Blocking the typical vertical route, and mimicking horizontal transmission, saw systemic increases in Wolbachia (wVulC) density and a drastic transition from a benign partner to a highly virulent one110.
The community
The drivers of transitions along the parasite–mutualist continuum can be complex and stem from the ecological and evolutionary movements of many different players. Defensive symbiosis113,114, whereby there are at least three interacting species (host, defensive symbiont and an attacking enemy) is particularly dynamic along the continuum in response to community composition changes. The absence of the symbiont or enemy can have evolutionary consequences for other species in the community, even without direct interactions115,116. Co-infections in hosts can also influence transitions in the symbiosis by providing new phenotypes via HGT of genetic material (for example, symbiosis islands, plasmids and phages)78,80,114,117.
The impact of community complexity is demonstrated by the bacterium Hamiltonella defensa and its lysogenic phage, APSE. This association protects host aphids against parasitoid wasps118,119 (Fig. 2). In this context, the fitness benefit afforded to the aphid host is contingent on parasitoid presence — in its absence, H. defensa with APSE phage is costly to the aphid120. The mechanism of protection (toxin production) hinges on the initial lateral transfer of phage from a co-infecting symbiont117,121. Subsequent loss of the phage can move the interaction between H. defensa and aphids back towards parasitism122. Theory105,116, experimental evolution40 and field studies123 have captured how microorganisms, even parasitic ones, can evolve rapidly to protect their hosts when collectively threatened, often crossing the parasite–mutualist continuum in the process. In Caenorhabditis elegans nematodes, a mildly parasitic gut bacterium was shown to evolve an enhanced ability to protect against infection by a more virulent parasite40. In the parasite’s absence, the gut bacterium did not emerge as a microbial line of host defence.
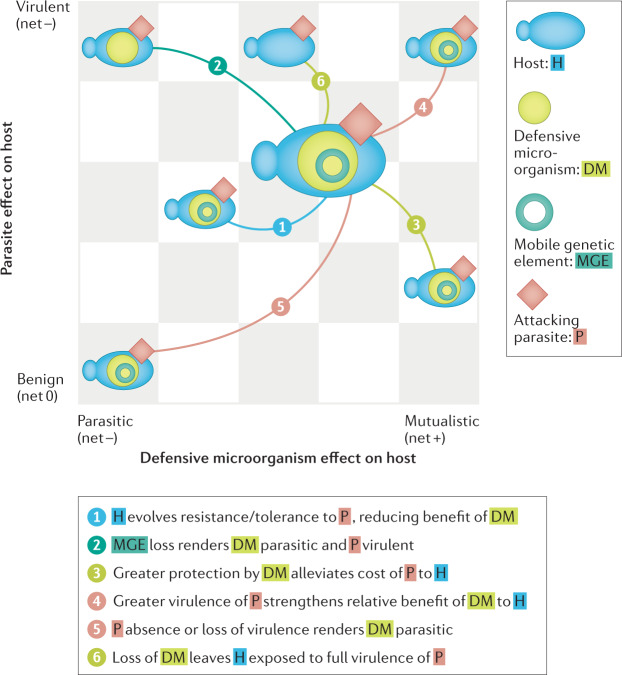
Fig. 2. Transitions in a community context.
Defensive symbioses involve multiple species, including a host (H) and defensive microorganism (DM) that protects against an attacking parasite (P)113. Often, hidden players exist within a DM, such as mobile genetic elements (MGEs; for example phages, plasmids and transposable elements) that encode factors involved in the protective function of the DM. In this community, the evolutionary and ecological moves (examples denoted by arrows) of each player can affect the relative position of another on the parasite–mutualist continuum. Players may move, resulting in an overall beneficial (net+), detrimental (net–) or negligible (net 0) effect on host fitness. For example, if a MGE encodes key protective functions, then its loss (move 2) will shift the DM’s position towards parasitism (all cost and no benefit to host). Meanwhile, the costs of P to H will increase now that H is no longer protected by the DM and its MGE. Transitions here can also alter the coevolutionary patterns and processes between players and species.
Additional symbionts, with previously unknown effects, are increasingly being identified even in iconic ‘two-player’ symbioses, such as corals124 and lichens125,126. It is thus not surprising that the complexity of a host’s whole microbiota (which often includes a diverse repertoire of bacteria, fungi and viruses) can interact to produce new outcomes for individual strains, species and the community as a whole. Members of the microbiota compete and cooperate in a myriad of ways127, influencing the virulence of one another via processes such as the suppression of public goods128 or the facilitation of biofilm formation129 and epithelial translocation130. The passage of Candida albicans in mice lacking gut microbiota has highlighted the role of communities in determining fate on the parasite–mutualist continuum. In the absence of a gut microbiota, C. albicans mutants emerge that are defective in hyphal formation, no longer requiring it for competition against other microbiota members. When compared with the wild-type ancestor that coexists with a microbiota, these C. albicans mutants are less virulent and protect their hosts against Aspergillus fumigatus infection in a manner independent of host adaptive immunity43. This transition from pathobiont to conditional mutualist in this context appears to hinge on the absence of competing microorganisms. However, given a gradient of increasing microbiome diversity, it would be valuable to understand when the selective advantage of the transition disappears. Other recent work, in microbiota-free mice, noted that when E. coli is a lone colonizer of the gut, it is consistently selected to increase metabolism of amino acids serine and threonine. A small increase in microbiome diversity (the addition of a single competing species) alters the evolutionary trajectory of E. coli substantially, instead favouring mutations associated with anaerobic metabolism131. This outcome suggests that bacteria may have low fidelity in metabolic function even within a single host generation132. Such a finding suggests host–microbial symbioses may not adhere to the idea of the ‘holobiont’ being a cohesive unit of selection133. This idea relies on high fidelity between partners134, which may easily be disrupted by changes to the surrounding microbial community.
If we can selectively drive the evolution of microorganisms and their communities, applications may improve on the already promising use of faecal microbiota transplants in medicine135, symbiont-mediated vector control136,137 and the manipulation of crop parasites42. There is, however, a pressing need to understand the long-term response of microbial communities to the engineering of symbionts. Recently, theoretical models have treated virulence as a cost shared by all symbionts coexisting in a host138,139. These models find that defence by a symbiont often drives reduced virulence across the microbial community (including in attacking parasites), an outcome dependent on the cost of defence being low and the shared cost of virulence also being low139. However, defensive microorganisms may also select for resistance mechanisms (for example, toxin production and inflammatory stimulation) in the parasites they protect against, causing collateral damage to hosts and driving increased parasite virulence140. This is akin to established predictions for co-infecting parasite species, whereby competition selects for increased virulence141–143. Promisingly though, and in line with some theory138,139, selection for reduced parasite virulence has been revealed in response to microorganism-mediated protection144. Others also report long-term efficacy of protection mechanisms despite an evolving pathosphere145.
Host control
Beyond microbial symbiont evolution, hosts can affect the position of the symbiosis on the continuum146. Hosts can be resistant (that is, reducing symbiont colonization) and tolerant (that is, coping with symbiont-associated damage without limiting colonization)147, which reduces any negative impacts of the host–symbiont interaction. Evolving control mechanisms (for example, sanctions and rewards, and microbiome modulators)146,148, or acquiring symbiotic function from an alternative source (for example, symbiont switching and HGT)100 can also limit or cause a change in the position of the interaction along the continuum.
Resistance to symbiont infection is observed ubiquitously across evolving host–parasite associations149,150. Mutations associated with membrane transporters in the bacterium Actinomyces odontolyticus coincided with a reduction in the negative effects of its ectoparasite (Nanosynbacter lyticus)151, perhaps indicating an adaptive host response to block resources to the ectoparasite or prevent its attachment151. As host resistance and tolerance strategies can affect parasitic symbiont fitness, they can counter-adapt152,153. This process may lead to a repeated back and forth along the continuum.
Hosts can also have key roles in restraining symbiont-driven shifts along the continuum. They may act to prevent the emergence of cheating symbionts, which exploit the benefits of host association without paying the cost of returning a benefit27,34. Alternatively, hosts may maintain the association at a position optimal for their own fitness. Sanction and reward strategies, spatial segregation of symbionts and partner choice mechanisms have evolved to promote and maintain cooperation27,154,155. For instance, legumes may sanction defective nitrogen-fixing bacteria by blocking resources to the respective root nodule32,154, and plants reward helpful mycorrhizal fungi with extra carbohydrate156. These mechanisms protect the host from investing in symbionts with net costs and avoid trajectories towards antagonism.
There is mounting theoretical and empirical evidence that many putative mutualisms may actually be a product of hosts exploiting symbionts2–4,33. Interactions can benefit the host, but with no reciprocity to the symbiont whose fitness is markedly reduced within the walls of host confinement1. These may be viewed as cases of inverted parasitism5. The host is the parasite of its smaller guest. This phenomenon is exemplified by zooxanthellae in which replication rates are severely compromised by host association4, rising from 3 days outside of coral hosts157 to around 70 days within158. Another example comes from Paramecium bursaria and photosynthetic Chlorella symbionts. Chlorella species provide fixed carbon in return for organic nitrogen, but the host tightly controls symbiont density in response to light conditions, ensuring the best nutrient trade for itself159. Control of the symbiont potentially occurs via digestion of Chlorella cells160. The host may win twofold, paying the workforce only when required and acquiring nutrition via digestion of surplus symbionts. The growth rate for Chlorella remains consistently better outside the host159, but inside, this symbiont avoids algal competitors161 and may be protected against its own parasites162. Research on exploitation by hosts is in its infancy, with the greatest evidence coming from interactions with photosynthetic symbionts4,159,163. Many questions remain, including the ubiquity of the phenomenon and whether some classes of symbiont are more vulnerable to exploitation than others.
Although considered relatively rare over evolutionary time, hosts may also eschew parasitic164 and mutualistic associations100. Fleeing the infectious environment is one strategy. Spatiotemporal escape by asexual rotifers prevents them interacting with fungal parasites consistently over evolutionary time. By drying up and blowing away in the wind, these animals are protected from infection, which allows them to maintain their asexual reproductive strategy164. Mutualistic associations can be abandoned via the recruitment of new symbionts100. As the Hodgkinia endosymbionts of cicadas teetered on the edge of genomic collapse, Ophiocordyceps fungi (commonly parasites) began to take over the essential roles in amino acid synthesis for the host165. Abandonment can also occur via exploitation of an alternative resource100. For example, the evolution of carnivory in plants led to several plant species deserting arbuscular mycorrhizal fungal symbionts, as the plant now gains nutrients directly from prey100. These cases chime with a growing debate over whether hosts can have the upper hand in symbioses, despite generally being the species that evolves more slowly (known as the Red King effect166,167), exploiting and imprisoning their microorganisms to gain disproportionate control and benefit2–4,33,159,168.
Context-dependent shifts
The outcome of many microbial interactions with hosts are context dependent14. Both facultative and obligate symbioses can make shifts along the parasite–mutualist continuum that do not involve evolution, often occurring within a generation and driven by ecological change or opportunity (Table 3). Abiotic factors such as temperature169, resource availability170, environmental toxicity171 and the biotic composition of the surrounding community119 or host ontogeny172,173 can all affect the distribution of costs and benefits incurred by the host and microbial symbiont. The position on the continuum can also change if the microbial symbiont becomes infected with its own symbionts (for example, phages and mycoviruses)42,122. Here, we focus on short-term disruptions to host–symbiont associations, but note that sustained alterations to context will feed back to evolutionary change for the interacting species.
Table 3.
Examples of context-dependent transitions of symbioses along the mutualist–parasite continuum
| Contexta | Species examples | Transition |
|---|---|---|
| Ontogeny | Queen conch–Symbiodinium spp.172 | Growth and survival benefit at larval stage, but photosynthetic activity of Symbiodinium spp. at adult stage potentially limited owing to shell cover (M → P) |
| Host genotype | Aphid spp.–Hamiltonella defensab189 | The longevity cost of hosting defensive symbiont differs across aphid genotypes |
| Temperature | Scleractinian coral–Symbiodinium spp.169 | Elevated temperature reduces net primary productivity of coral, but no cost to Symbiodinium spp. detected (M → P) |
| Metabolic | Chlamydomonas reinhardtii– Saccharomyces cerevisiae170 | Mutualism between microorganisms occurs only in CO2-restricted environment (FL → M) |
| Co-infecting microorganisms or microbiome | Aphids–Hamiltonella defensa– Serratia symbiotica119 | Co-infection provides additive benefit, enhancing host resistance to parasitoid wasps (M → M) (+) |
| Symbiont passengers (for example, phages and mycoviruses) | Brassica crop–Sclerotinia sclerotiorum–mycovirus42 | Mycovirus infection converts a fungal parasite (S. sclerotiorum) into a crop enhancer (P → M) |
| Enemy presence | Drosophila melanogaster–Wolbachia (wMel)234 | Wolbachia wMel variants protect against viruses. In the absence of viral threat, host pays the cost of significantly curtailed lifespan (M → P) |
| Environmental toxicity | Pseudomonas fluorescens–mercury resistance plasmid171 | Fitness effects of plasmid carriage vary with environmental mercury levels |
| Host switch | Nematodes–Xenorhabdus spp.235 | Mutualistic strains are harmful in non-native host (M → P) |
| Light | Hydra–Chlorella algae236 | Under dark conditions Chlorella is costly, indicated by a growth disadvantage over uninfected Hydra (M → P) |
(+), elevated (for example, M → M (+) indicates transition towards increased benefit for host); FL, free-living; M, mutualism; P, parasitism. aContextual variables can affect both host and symbiont processes independently, which may affect transitions. bInteractions between host genotype, symbiont genotype and the environment also operate here.
Generally, theory predicts that nutrient-limited environments, or other harsh environments, can foster beneficial interactions between compatible players27,174 via mechanisms such as cross-protection and cross-feeding. This outcome has been substantiated by empirical work175–177. For symbionts that have nutritional roles (for example, vitamin synthesis and nitrogen fixation), abundant resources can substantially undermine the net benefit gained by the host. The provisioning of mineral nitrogen from fertilizer erases the benefit Bradyrhizobium symbionts provide to legume hosts (Lotus strigosusas) as this acquisition route is less energetically costly for the legume than its symbiont-fixed equivalent178. Some hosts evade context-dependent costs by divesting themselves of associations when ecological conditions change, such as the phytoplankton that abandon their nitrogen-fixing cyanobacteria when environmental nitrogen is abundant179. For host–parasite systems, there is no evidence for a one-way effect of nutrient availability to hosts on the harm caused by infection180. One study180 suggested that the level of parasite virulence in a given environment is likely the result of a balance between the effect of host nutrition on the immune system and on parasite resources.
Temperature can affect symbiont phenotypes181,182, which directly impact symbiont virulence or benefit, such as the regulation of toxin production183 or molecules required for nutrient scavenging184. Some obligate mutualists can constitute thermally ‘weak links’ for hosts, becoming non-functional or even lost from hosts outside adapted temperature ranges, which can have catastrophic consequences for host fitness185,186. Interactions can occur between abiotic and biotic factors. For instance, a 5 °C increase in temperature diminishes H. defensa-mediated defence against parasitoids187,188. This temperature-dependent reduction in defence may be ameliorated if co-infection with an additional bacterium, known as pea aphid X-type symbiont, occurs187.
In other cases, community composition alone can temporarily cause transitions. Defensive symbioses present a clear demonstration of community context-dependent shifts, whereby benefits to the host are contingent on the presence of an enemy species113,114. In the absence of the enemy, the host pays the cost with no detectable benefit, and the association moves towards one that is parasitic114,189. Infection of a symbiont with its own symbionts (that is, hyperparasitism190) can also generate transitions. Recent work found that the devastating effects of a fungal parasite on rapeseed crop are significantly reduced if the fungus becomes infected with mycovirus SsHADV-1 (ref.42). The presence of the mycovirus appeared to affect the expression of a suite of both fungal and crop genes, including those encoding plant cell-wall-degrading enzymes and crop signalling pathways42.
Pathobionts provide an excellent example of context-dependent transitions from neutral to harmful agents191. In a host with a functional immune system and healthy microbiota, pathobionts can exist as commensals191–193. Pathobionts are well adapted to proliferate beyond their normal niche. During dysbiosis (for example, compromised immunity, disruption of the microbiota or introduction of medical devices such as catheters or surgical implants) pathobionts can cause disease in a wide variety of forms, from minor infections to more serious chronic or invasive disease194. This ability to transition from harmless to harmful in different contexts makes pathobionts hard to place on the continuum. They are neither consistent parasites nor consistent commensals, with the state of the host generally determining their transition from one to the other.
Stuck at the end of the line
At either end of the continuum lie the extremes of host-killing (or castration) and mutual dependence. What maintains an association here, and what is its future?
The ability to shift along the continuum for some parasitic microorganisms could depend on transmission route. Some infectious agents may stay hypervirulent owing to a high degree of environmental transmission or a lack of reliance on hosts to transmit and propagate. The ‘curse of the Pharoah’ hypothesis195 posits that microorganisms able to ‘sit and wait’ in the environment can be perpetual killers, whereas others suggest that traits that enable persistence in the environment will be traded off with virulence196. There may also be constraints of the parasitic life cycle that prevent a transition. Microbial parasites that must lyse host cells to transmit (for example, lytic phages and Plasmodium species in mammals) or steal resources in a way that castrates the host (Pasteuria bacterial parasite infecting Daphnia magna197) are systems in which transitions away from antagonism are unlikely.
At the opposing end of the continuum lie inherited, obligate endosymbionts, which often have nutritional roles. Although many of these associations are ancient and bestow mutual benefits, they can be risky, particularly for the endosymbiont3,53,198. The genomes of these symbionts can gradually decay as transmission bottlenecks allow deleterious mutations to become fixed by genetic drift, and mutational bias towards deletions removes genes199–202. Genomic decay can lead to extinction, unless heightened genetic and cellular support is provided by the host203 or other symbionts78,204,205. For example, leafhoppers show gene expression patterns that appear tailored to the deficiencies of each of their endosymbionts’ highly degraded genomes203. In rare cases, symbionts may transition to organelle status206, notoriously achieved by mitochondria and plastids, but this does not guarantee shelter against further gene loss or extinction207,208. Hosts may also avoid extinction alongside an endosymbiont by exploiting alternative nutritional resources or gaining new symbionts158,159.
Conclusions and future perspectives
Plants and animals, including humans, are colonized by innumerable microorganisms. This observation has sparked a revolution in studying the impacts of those microorganisms on host biology and health. Many more examples of microbial evolution causing transitions across the parasite–mutualist continuum will emerge through further research using experimental evolution and investigating the microbiome in an evolutionary context. The potential evolution of species in the human microbiome from good to bad209,210, and the degree to which beneficial interactions could be upset by microbiome perturbation211, are of clinical relevance for individuals vulnerable to infectious disease. In the future, such individuals may benefit from engineering of the microbiome or symbiont communities, via either direct genetic modifications to key transition loci in microbiome members, or exposure to selection sources with known outcomes. This approach has recently been achieved for honeybees, with the genetic modification of a core gut bacterium improving resistance to viral infection212. These are exciting applications, but we must strive to understand the evolutionary consequences for the parasites targeted too.
More fundamentally, understanding causes of transitions will provide insight into the dynamics of how an organism’s biology and its community are shaped by microbial inhabitants. The ecological and evolutionary transitions of other species, as well as environmental change, can alter the scope for conflict in symbioses involving microorganisms. Interest has grown in thinking of host–microorganism symbioses as holobionts with highly aligned selective interests134. Many associations may be also viewed in an ecological community context13,146 in which constant shifts occur back and forth on the parasite–mutualist continuum. The degree to which the host and symbiont, or both, have control over those shifts remains relatively unexplored. Research in the field has focused on the propensity of symbionts to invade unwilling hosts or cheat reciprocal arrangements. Yet an exciting new avenue is emerging, one that is exposing hosts as exploiters and imprisoners of microorganisms33,198. The extent to which microorganisms are able to evolve to counter or take advantage of that exploitation is also unclear.
Moreover, environmental changes have the potential to substantially alter selection in symbiotic interactions213. In addition to altering established symbioses, marked changes to abiotic variables can also move the boundaries of environmental constraint, fostering the evolution of new interactions on the continuum that were previously impossible or profitless. How will the collectively growing impact of humans affect the stability of beneficial associations and the emergence of parasites globally (for example, see refs214,215)? This question is particularly timely given the COVID-19 pandemic. Undoubtedly, as environmental perturbations increase in magnitude and frequency, and as the use of antimicrobials grows, understanding the effects on the real-time evolution of host–symbiont interactions will become more and more valuable.
Acknowledgements
This work was funded by a European Starter Grant (COEVOPRO 802242) to K.C.K. The authors are grateful to four reviewers for their comments.
Glossary
- Inverted parasitism
An interaction whereby the classically viewed host exploits its smaller symbiont, implementing a fitness cost to the symbiont.
- Parasitism
An antagonistic symbiotic relationship in which one species is harmed, while the other benefits.
- Mutualism
A symbiotic relationship in which both interacting species benefit, or are perceived to benefit. Benefit is often only confirmed empirically for the host.
- Symbiosis
An association between two dissimilar organisms that have some degree of physical association, which is potentially long lasting, regardless of the implications for the fitness of either organism.
- Free-living
A microbial lifestyle not dependent on association with a host for long-term survival and replication; this is the ancestral state of all symbionts.
- Mobile genetic elements
(MGEs). Sequences of genetic material that can be exchanged between chromosomes or organisms via either their own mobilizing machinery or that of their host. Examples include transposable elements, plasmids and phages.
- Horizontal gene transfer
(HGT). The movement of genetic material between organisms that does not flow from parent to offspring.
- Virulence
The damage caused to the host due to infection by a parasite, often measured as a reduction in host fitness.
- Defensive symbiosis
An interaction in which the symbiont protects the host (via direct or indirect mechanisms) against natural enemies, such as microbial parasites and eukaryotic parasitoids.
- Pathobiont
Any organism that can cause harm to its host, but normally lives as a harmless symbiont.
Author contributions
All authors researched data for the article, contributed substantially to discussion of the content, wrote the article and/or edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Georgia C. Drew, Emily J. Stevens
References
- 1.Garcia JR, Gerardo NM. The symbiont side of symbiosis: do microbes really benefit? Front. Microbiol. 2014;5:510. doi: 10.3389/fmicb.2014.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law R, Dieckmann U. Symbiosis through exploitation and the merger of lineages in evolution. Proc. Biol. Sci. 1998;265:1245–1253. [Google Scholar]
- 3.Keeling PJ, McCutcheon JP. Endosymbiosis: the feeling is not mutual. J. Theor. Biol. 2017;434:75–79. doi: 10.1016/j.jtbi.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wooldridge SA. Is the coral-algae symbiosis really ‘mutually beneficial’ for the partners? BioEssays. 2010;32:615–625. doi: 10.1002/bies.200900182. [DOI] [PubMed] [Google Scholar]
- 5.Mushegian AA, Ebert D. Rethinking ‘mutualism’ in diverse host-symbiont communities. BioEssays. 2016;38:100–108. doi: 10.1002/bies.201500074. [DOI] [PubMed] [Google Scholar]
- 6.Mathis KA, Bronstein JL. Our current understanding of commensalism. Ann. Rev. Ecol. Evol. Syst. 2020;51:167–189. [Google Scholar]
- 7.Ewald PW. Transmission modes and evolution of the parasitism-mutualism continuum. Ann. N. Y. Acad. Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- 8.Bronstein JL. Conditional outcomes in mutualistic interactions. Trends Ecol. Evol. 1994;9:214–217. doi: 10.1016/0169-5347(94)90246-1. [DOI] [PubMed] [Google Scholar]
- 9.Schu MG, Schrallhammer M. Cultivation conditions can cause a shift from mutualistic to parasitic behavior in the symbiosis between Paramecium and its bacterial symbiont Caedibacter taeniospiralis. Curr. Microbiol. 2018;75:1099–1102. doi: 10.1007/s00284-018-1493-1. [DOI] [PubMed] [Google Scholar]
- 10.Osman EO, et al. Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome. 2020;8:8. doi: 10.1186/s40168-019-0776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kümmerli R, Jiricny N, Clarke LS, West SA, Griffin AS. Phenotypic plasticity of a cooperative behaviour in bacteria. J. Evol. Biol. 2009;22:589–598. doi: 10.1111/j.1420-9101.2008.01666.x. [DOI] [PubMed] [Google Scholar]
- 12.Kumamoto CA. Niche-specific gene expression during C. albicans infection. Curr. Opin. Microbiol. 2008;11:325–330. doi: 10.1016/j.mib.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thrall PH, Hochberg ME, Burdon JJ, Bever JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol. Evol. 2007;22:120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Chamberlain SA, Bronstein JL, Rudgers JA. How context dependent are species interactions? Ecol. Lett. 2014;17:881–890. doi: 10.1111/ele.12279. [DOI] [PubMed] [Google Scholar]
- 15.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc. Natl Acad. Sci. USA. 2011;108(Suppl. 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosokawa T, et al. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol. 2016;1:1–7. doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- 17.Gupta A, Nair S. Dynamics of insect–microbiome interaction influence host and microbial symbiont. Front. Microbiol. 2020;11:1357. doi: 10.3389/fmicb.2020.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutzoni F, Pagel M. Accelerated evolution as a consequence of transitions to mutualism. Proc. Natl Acad. Sci. USA. 1997;94:11422–11427. doi: 10.1073/pnas.94.21.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaltenpoth M, et al. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc. Natl Acad. Sci. USA. 2014;111:6359–6364. doi: 10.1073/pnas.1400457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manzano-Marín A, et al. Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids’ di-symbiotic systems. ISME J. 2020;14:259–273. doi: 10.1038/s41396-019-0533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyauchi S, et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat. Commun. 2020;11:5125. doi: 10.1038/s41467-020-18795-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McFall-Ngai MJ. The importance of microbes in animal development: lessons from the squid-Vibrio symbiosis. Annu. Rev. Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown SP, Cornforth DM, Mideo N. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 2012;20:336–342. doi: 10.1016/j.tim.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher RM, Henry LM, Cornwallis CK, Kiers ET, West SA. The evolution of host-symbiont dependence. Nat. Commun. 2017;8:15973. doi: 10.1038/ncomms15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell JM. Genomes of obligate plant pathogens reveal adaptations for obligate parasitism. Proc. Natl Acad. Sci. USA. 2011;108:8921–8922. doi: 10.1073/pnas.1105802108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson BA, Salyers AA. Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 2003;11:347–350. doi: 10.1016/s0966-842x(03)00179-3. [DOI] [PubMed] [Google Scholar]
- 27.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolution of cooperation. Q. Rev. Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- 28.Bull JJ, Rice WR. Distinguishing mechanisms for the evolution of co-operation. J. Theor. Biol. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- 29.Sachs JL, Skophammer RG, Bansal N, Stajich JE. Evolutionary origins and diversification of proteobacterial mutualists. Proc. Biol. Sci. 2014;281:20132146. doi: 10.1098/rspb.2013.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duron O, et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 2015;11:e1004892. doi: 10.1371/journal.ppat.1004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clayton AL, et al. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genet. 2012;8:e1002990. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West SA, Kiers ET, Simms EL, Denison RF. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. Biol. Sci. 2002;269:685–694. doi: 10.1098/rspb.2001.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sørensen MES, et al. The role of exploitation in the establishment of mutualistic microbial symbioses. FEMS Microbiol. Lett. 2019;366:fnz148. doi: 10.1093/femsle/fnz148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trivers RL. The evolution of reciprocal altruism. Q. Rev. Biol. 1971;46:35–57. [Google Scholar]
- 35.Frederickson ME. Mutualisms are not on the verge of breakdown. Trends Ecol. Evol. 2017;32:727–734. doi: 10.1016/j.tree.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Mueller UG, Ishak H, Lee JC, Sen R, Gutell RR. Placement of attine ant-associated Pseudonocardia in a global Pseudonocardia phylogeny (Pseudonocardiaceae, Actinomycetales): a test of two symbiont-association models. Antonie Van Leeuwenhoek. 2010;98:195–212. doi: 10.1007/s10482-010-9427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dietel A-K, Kaltenpoth M, Kost C. Convergent evolution in intracellular elements: plasmids as model endosymbionts. Trends Microbiol. 2018;26:755–768. doi: 10.1016/j.tim.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Hurst GDD. Extended genomes: symbiosis and evolution. Interface Focus. 2017;7:20170001. doi: 10.1098/rsfs.2017.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melnyk RA, Hossain SS, Haney CH. Convergent gain and loss of genomic islands drive lifestyle changes in plant-associated Pseudomonas. ISME J. 2019;13:1575–1588. doi: 10.1038/s41396-019-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King KC, et al. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 2016;10:1915–1924. doi: 10.1038/ismej.2015.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro JW, Turner PE. Evolution of mutualism from parasitism in experimental virus populations. Evolution. 2018;72:707–712. doi: 10.1111/evo.13440. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, et al. A 2-kb mycovirus converts a pathogenic fungus into a beneficial endophyte for brassica protection and yield enhancement. Mol. Plant. 2020;13:1420–1433. doi: 10.1016/j.molp.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Tso GHW, et al. Experimental evolution of a fungal pathogen into a gut symbiont. Science. 2018;362:589–595. doi: 10.1126/science.aat0537. [DOI] [PubMed] [Google Scholar]
- 44.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015;25:2034–2039. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Porter SS, Faber-Hammond J, Montoya AP, Friesen ML, Sackos C. Dynamic genomic architecture of mutualistic cooperation in a wild population of Mesorhizobium. ISME J. 2019;13:301–315. doi: 10.1038/s41396-018-0266-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrera P, et al. Molecular causes of an evolutionary shift along the parasitism–mutualism continuum in a bacterial symbiont. Proc. Natl Acad. Sci. USA. 2020;117:21658–21666. doi: 10.1073/pnas.2005536117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li E, et al. Rapid evolution of bacterial mutualism in the plant rhizosphere. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.12.07.414607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pankey MS, et al. Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria. eLife. 2017;6:e24414. doi: 10.7554/eLife.24414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansen G, et al. Evolutionary transition from pathogenicity to commensalism: global regulator mutations mediate fitness gains through virulence attenuation. Mol. Biol. Evol. 2015;32:2883–2896. doi: 10.1093/molbev/msv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chain PSG, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendry TA, et al. Ongoing transposon-mediated genome reduction in the luminous bacterial symbionts of deep-sea ceratioid anglerfishes. mBio. 2018;9:e01033–18. doi: 10.1128/mBio.01033-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nygaard S, et al. Reciprocal genomic evolution in the ant–fungus agricultural symbiosis. Nat. Commun. 2016;7:12233. doi: 10.1038/ncomms12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett GM, Moran NA. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc. Natl Acad. Sci. USA. 2015;112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gluck-Thaler E, et al. Repeated gain and loss of a single gene modulates the evolution of vascular pathogen lifestyles. bioRxiv. 2020 doi: 10.1101/2020.04.24.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arredondo-Alonso S, et al. Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. mBio. 2020;11:e03284-19. doi: 10.1128/mBio.03284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Driscoll TP, et al. Evolution of Wolbachia mutualism and reproductive parasitism: insight from two novel strains that co-infect cat fleas. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.06.01.128066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frantzeskakis L, et al. Signatures of host specialization and a recent transposable element burst in the dynamic one-speed genome of the fungal barley powdery mildew pathogen. BMC Genomics. 2018;19:381. doi: 10.1186/s12864-018-4750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Savory EA, et al. Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management. eLife. 2017;6:e30925. doi: 10.7554/eLife.30925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barreto HC, Sousa A, Gordo I. The landscape of adaptive evolution of a gut commensal bacteria in aging mice. Curr. Biol. 2020;30:1102–1109.e5. doi: 10.1016/j.cub.2020.01.037. [DOI] [PubMed] [Google Scholar]
- 60.Parkhill J, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 61.Deng W, et al. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Achtman M, et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rasmussen S, et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell. 2015;163:571–582. doi: 10.1016/j.cell.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hinnebusch BJ, et al. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 65.Lindler LE, Plano GV, Burland V, Mayhew GF, Blattner FR. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du Y, Rosqvist R, Forsberg Å. Role of fraction 1 antigen of Yersinia pestis in inhibition of phagocytosis. Infect. Immun. 2002;70:1453–1460. doi: 10.1128/IAI.70.3.1453-1460.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Y-C, Jarrett CO, Bosio CF, Hinnebusch BJ. Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe. 2014;15:578–586. doi: 10.1016/j.chom.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohnishi M, Kurokawa K, Hayashi T. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 2001;9:481–485. doi: 10.1016/s0966-842x(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 69.Franzin FM, Sircili MP. Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed. Res. Int. 2015;2015:534738. doi: 10.1155/2015/534738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brito IL, et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535:435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Broaders E, O’Brien C, Gahan CGM, Marchesi JR. Evidence for plasmid-mediated salt tolerance in the human gut microbiome and potential mechanisms. FEMS Microbiol. Ecol. 2016;92:fiw019. doi: 10.1093/femsec/fiw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarthy AJ, et al. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 2014;6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frazão N, Sousa A, Lässig M, Gordo I. Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proc. Natl Acad. Sci. USA. 2019;116:17906–17915. doi: 10.1073/pnas.1906958116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niehus R, Mitri S, Fletcher AG, Foster KR. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat. Commun. 2015;6:8924. doi: 10.1038/ncomms9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koonin EV. Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000Res. 2016 doi: 10.12688/f1000research.8737.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nowack ECM, et al. Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc. Natl Acad. Sci. USA. 2016;113:12214–12219. doi: 10.1073/pnas.1608016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bordenstein SR, Bordenstein SR. Eukaryotic association module in phage WO genomes from Wolbachia. Nat. Commun. 2016;7:13155. doi: 10.1038/ncomms13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waterworth SC, et al. Horizontal gene transfer to a defensive symbiont with a reduced genome in a multipartite beetle microbiome. mBio. 2020;11:e02430-19. doi: 10.1128/mBio.02430-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma W, Dong FFT, Stavrinides J, Guttman DS. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2006;2:e209. doi: 10.1371/journal.pgen.0020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nikoh N, et al. Evolutionary origin of insect–Wolbachia nutritional mutualism. Proc. Natl Acad. Sci. USA. 2014;111:10257–10262. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheppard SK, Guttman DS, Fitzgerald JR. Population genomics of bacterial host adaptation. Nat. Rev. Genet. 2018;19:549–565. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]
- 82.Day T, Gandon S, Lion S, Otto SP. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020;30:R849–R857. doi: 10.1016/j.cub.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tardy L, Giraudeau M, Hill GE, McGraw KJ, Bonneaud C. Contrasting evolution of virulence and replication rate in an emerging bacterial pathogen. Proc. Natl Acad. Sci. USA. 2019;116:16927–16932. doi: 10.1073/pnas.1901556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alves JM, et al. Parallel adaptation of rabbit populations to myxoma virus. Science. 2019;363:1319–1326. doi: 10.1126/science.aau7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kerr PJ. Myxomatosis in Australia and Europe: a model for emerging infectious diseases. Antivir. Res. 2012;93:387–415. doi: 10.1016/j.antiviral.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 86.Longdon B, et al. The causes and consequences of changes in virulence following pathogen host shifts. PLoS Pathog. 2015;11:e1004728. doi: 10.1371/journal.ppat.1004728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Boven M, et al. Detecting emerging transmissibility of avian influenza virus in human households. PLoS Comput. Biol. 2007;3:e145. doi: 10.1371/journal.pcbi.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moses AS, Millar JA, Bonazzi M, Beare PA, Raghavan R. Horizontally acquired biosynthesis genes boost Coxiella burnetii’s physiology. Front. Cell Infect. Microbiol. 2017;7:174. doi: 10.3389/fcimb.2017.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flórez LV, et al. Antibiotic-producing symbionts dynamically transition between plant pathogenicity and insect-defensive mutualism. Nat. Commun. 2017;8:1–9. doi: 10.1038/ncomms15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- 91.Ewald PW. Host-parasite relations, vectors, and the evolution of disease severity. Annu. Rev. Ecol. Syst. 1983;14:465–485. [Google Scholar]
- 92.Bull JJ. Perspective: Virulence. Evolution. 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 93.Rafaluk C, Jansen G, Schulenburg H, Joop G. When experimental selection for virulence leads to loss of virulence. Trends Parasitol. 2015;31:426–434. doi: 10.1016/j.pt.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 94.Alizon S, Van Baalen M. Transmission-virulence trade-offs in vector-borne diseases. Theor. Popul. Biol. 2008;74:6–15. doi: 10.1016/j.tpb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 95.Cressler CE, McLeod DV, Rozins C, Hoogen JVD, Day T. The adaptive evolution of virulence: a review of theoretical predictions and empirical tests. Parasitology. 2016;143:915–930. doi: 10.1017/S003118201500092X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- 97.Yamamura N. Vertical transmission and evolution of mutualism from parasitism. Theor. Popul. Biol. 1993;44:95–109. [Google Scholar]
- 98.Hall JPJ, Brockhurst MA, Dytham C, Harrison E. The evolution of plasmid stability: are infectious transmission and compensatory evolution competing evolutionary trajectories? Plasmid. 2017;91:90–95. doi: 10.1016/j.plasmid.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 99.Kiers ET, Denison RF. Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Annu. Rev. Ecol. Evol. Syst. 2008;39:215–236. [Google Scholar]
- 100.Werner GDA, et al. Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proc. Natl Acad. Sci. USA. 2018;115:5229–5234. doi: 10.1073/pnas.1721629115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Herre EA, et al. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 102.Nussbaumer AD, Fisher CR, Bright M. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature. 2006;441:345–348. doi: 10.1038/nature04793. [DOI] [PubMed] [Google Scholar]
- 103.Dusi E, Krenek S, Petzoldt T, Kaltz O, Berendonk TU. When enemies do not become friends: experimental evolution of heat-stress adaptation in a vertically transmitted parasite. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.01.23.917773. [DOI] [Google Scholar]
- 104.Engelstädter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 2009;40:127–149. [Google Scholar]
- 105.Fenton A, Johnson KN, Brownlie JC, Hurst GDD. Solving the Wolbachia paradox: modeling the tripartite interaction between host, Wolbachia, and a natural enemy. Am. Nat. 2011;178:333–342. doi: 10.1086/661247. [DOI] [PubMed] [Google Scholar]
- 106.Zug R, Hammerstein P. Evolution of reproductive parasites with direct fitness benefits. Heredity. 2018;120:266–281. doi: 10.1038/s41437-017-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Drew, G. C., Frost, C. L. & Hurst, G. D. Reproductive parasitism and positive fitness effects of heritable microbes. in eLShttps://onlinelibrary.wiley.com/doi/abs/10.1002/9780470015902.a0028327 (2019).
- 108.Parratt SR, et al. Superparasitism drives heritable symbiont epidemiology and host sex ratio in a wasp. PLoS Pathog. 2016;12:e1005629. doi: 10.1371/journal.ppat.1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sachs JL, Wilcox TP. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proc. Biol. Sci. 2006;273:425–429. doi: 10.1098/rspb.2005.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Le Clec’h W, Dittmer J, Raimond M, Bouchon D, Sicard M. Phenotypic shift in Wolbachia virulence towards its native host across serial horizontal passages. Proc. Biol. Sci. 2017;284:20171076. doi: 10.1098/rspb.2017.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stewart AD, Logsdon JM, Kelley SE. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution. 2005;59:730–739. [PubMed] [Google Scholar]
- 112.Rigaud T, Souty-Grosset C, Raimond R, Mocquard J-P, Juchault P. Feminizing endocytobiosis in the terrestrial crustacean Armadilidium vulgare Latr. (isopoda) - recent acquisitions. Cell Res. 1991;15:259–273. [Google Scholar]
- 113.King KC. Defensive symbionts. Curr. Biol. 2019;29:R78–R80. doi: 10.1016/j.cub.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 114.Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat. Prod. Rep. 2015;32:904–936. doi: 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 115.Couret J, Huynh‐Griffin L, Antolic‐Soban I, Acevedo‐Gonzalez TS, Gerardo NM. Even obligate symbioses show signs of ecological contingency: impacts of symbiosis for an invasive stinkbug are mediated by host plant context. Ecol. Evol. 2019;9:9087–9099. doi: 10.1002/ece3.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ashby B, King K. Friendly foes: the evolution of host protection by a parasite. Evol. Lett. 2017;1:211–221. doi: 10.1002/evl3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Duron O. Arsenophonus insect symbionts are commonly infected with APSE, a bacteriophage involved in protective symbiosis. FEMS Microbiol. Ecol. 2014;90:184–194. doi: 10.1111/1574-6941.12381. [DOI] [PubMed] [Google Scholar]
- 118.Ferrari J, Darby AC, Daniell TJ, Godfray HCJ, Douglas AE. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 2004;29:60–65. [Google Scholar]
- 119.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl Acad. Sci. USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Polin S, Simon J-C, Outreman Y. An ecological cost associated with protective symbionts of aphids. Ecol. Evol. 2014;4:826–830. doi: 10.1002/ece3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl Acad. Sci. USA. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Weldon SR, Strand MR, Oliver KM. Phage loss and the breakdown of a defensive symbiosis in aphids. Proc. Biol. Sci. 2013;280:20122103. doi: 10.1098/rspb.2012.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 2007;5:e114. doi: 10.1371/journal.pbio.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kwong WK, del Campo J, Mathur V, Vermeij MJA, Keeling PJ. A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature. 2019;568:103–107. doi: 10.1038/s41586-019-1072-z. [DOI] [PubMed] [Google Scholar]
- 125.Tuovinen V, et al. Two basidiomycete fungi in the cortex of wolf lichens. Curr. Biol. 2019;29:476–483.e5. doi: 10.1016/j.cub.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 126.Spribille T, et al. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science. 2016;353:488–492. doi: 10.1126/science.aaf8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Coyte KZ, Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr. Biol. 2019;29:R538–R544. doi: 10.1016/j.cub.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lopez-Medina E, et al. Candida albicans inhibits Pseudomonas aeruginosa virulence through suppression of pyochelin and pyoverdine biosynthesis. PLoS Pathog. 2015;11:e1005129. doi: 10.1371/journal.ppat.1005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Diebel LN, Liberati DM, Diglio CA, Dulchavsky SA, Brown WJ. Synergistic effects of Candida and Escherichia coli on gut barrier function. J. Trauma. Acute Care Surg. 1999;47:1045. doi: 10.1097/00005373-199912000-00009. [DOI] [PubMed] [Google Scholar]
- 131.Barroso-Batista J, et al. Specific eco-evolutionary contexts in the mouse gut reveal Escherichia coli metabolic versatility. Curr. Biol. 2020;30:1049–1062.e7. doi: 10.1016/j.cub.2020.01.050. [DOI] [PubMed] [Google Scholar]
- 132.King KC, Stevens E, Drew GC. Microbiome: evolution in a world of interaction. Curr. Biol. 2020;30:R265–R267. doi: 10.1016/j.cub.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 133.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 134.Douglas AE, Werren JH. Holes in the hologenome: why host-microbe symbioses are not holobionts. mBio. 2016;7:e02099. doi: 10.1128/mBio.02099-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bakken JS, et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011;9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bourtzis K, et al. Harnessing mosquito–Wolbachia symbiosis for vector and disease control. Acta Tropica. 2014;132:S150–S163. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 137.O’Neill, S. L. in Dengue and Zika: Control and Antiviral Treatment Strategies (eds Hilgenfeld, R. & Vasudevan, S. G.) 355–360 (Springer, 2018).
- 138.Nelson PG, May G. Coevolution between mutualists and parasites in symbiotic communities may lead to the evolution of lower virulence. Am. Nat. 2017;190:803–817. doi: 10.1086/694334. [DOI] [PubMed] [Google Scholar]
- 139.Nelson P, May G. Defensive symbiosis and the evolution of virulence. Am. Nat. 2020;196:333–343. doi: 10.1086/709962. [DOI] [PubMed] [Google Scholar]
- 140.Ford SA, King KC. Harnessing the power of defensive microbes: evolutionary implications in nature and disease control. PLoS Pathog. 2016;12:e1005465. doi: 10.1371/journal.ppat.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc. Biol. Sci. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- 142.Alizon S, de Roode JC, Michalakis Y. Multiple infections and the evolution of virulence. Ecol. Lett. 2013;16:556–567. doi: 10.1111/ele.12076. [DOI] [PubMed] [Google Scholar]
- 143.Frank SA. Host–symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 1996;263:339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- 144.Ford SA, Kao D, Williams D, King KC. Microbe-mediated host defence drives the evolution of reduced pathogen virulence. Nat. Commun. 2016;7:1–9. doi: 10.1038/ncomms13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Engl T, et al. Evolutionary stability of antibiotic protection in a defensive symbiosis. Proc. Natl Acad. Sci. USA. 2018;115:E2020–E2029. doi: 10.1073/pnas.1719797115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 2008;8:889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Voges MJEEE, Bai Y, Schulze-Lefert P, Sattely ES. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl Acad. Sci. USA. 2019;116:12558–12565. doi: 10.1073/pnas.1820691116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gandon S, Michalakis Y. Evolution of parasite virulence against qualitative or quantitative host resistance. Proc. Biol. Sci. 2000;267:985–990. doi: 10.1098/rspb.2000.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Best A, White A, Boots M. The coevolutionary implications of host tolerance. Evolution. 2014;68:1426–1435. doi: 10.1111/evo.12368. [DOI] [PubMed] [Google Scholar]
- 151.Bor B, et al. Rapid evolution of decreased host susceptibility drives a stable relationship between ultrasmall parasite TM7x and its bacterial host. Proc. Natl Acad. Sci. USA. 2018;115:12277–12282. doi: 10.1073/pnas.1810625115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc. Natl Acad. Sci. USA. 2010;107:7359–7364. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kerr PJ, et al. Next step in the ongoing arms race between myxoma virus and wild rabbits in Australia is a novel disease phenotype. Proc. Natl Acad. Sci. USA. 2017;114:9397–9402. doi: 10.1073/pnas.1710336114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume–rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 155.Frederickson ME. Rethinking mutualism stability: cheaters and the evolution of sanctions. Q. Rev. Biol. 2013;88:269–295. doi: 10.1086/673757. [DOI] [PubMed] [Google Scholar]
- 156.Kiers ET, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333:880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 157.Fitt WK, Trench RK. The relation of diel patterns of cell division to diel patterns of motility in the symbiotic dinoflagellate Symbiodinium microadriaticum Freudenthal in culture. N. Phytol. 1983;94:421–432. [Google Scholar]
- 158.Wilkerson FP, Kobayashi D, Muscatine L. Mitotic index and size of symbiotic algae in Caribbean reef corals. Coral Reefs. 1988;7:29–36. [Google Scholar]
- 159.Lowe CD, Minter EJ, Cameron DD, Brockhurst MA. Shining a light on exploitative host control in a photosynthetic endosymbiosis. Curr. Biol. 2016;26:207–211. doi: 10.1016/j.cub.2015.11.052. [DOI] [PubMed] [Google Scholar]
- 160.Kodama Y, Fujishima M. Symbiotic Chlorella variabilis incubated under constant dark conditions for 24 hours loses the ability to avoid digestion by host lysosomal enzymes in digestive vacuoles of host ciliate Paramecium bursaria. FEMS Microbiol. Ecol. 2014;90:946–955. doi: 10.1111/1574-6941.12448. [DOI] [PubMed] [Google Scholar]
- 161.Iwai S, Fujita K, Takanishi Y, Fukushi K. Photosynthetic endosymbionts benefit from host’s phagotrophy, including predation on potential competitors. Curr. Biol. 2019;29:3114–3119.e3. doi: 10.1016/j.cub.2019.07.074. [DOI] [PubMed] [Google Scholar]
- 162.Reisser W, et al. Viruses distinguish symbiotic Chlorella spp. of Paramecium bursaria. Endocytobiosis Cell Res. 1991;7:245–251. [Google Scholar]
- 163.Ahmadjian V. The lichen symbiosis. Ann. Botany. 1993;75:101–102. [Google Scholar]
- 164.Wilson CG, Sherman PW. Anciently asexual bdelloid rotifers escape lethal fungal parasites by drying up and blowing away. Science. 2010;327:574–576. doi: 10.1126/science.1179252. [DOI] [PubMed] [Google Scholar]
- 165.Matsuura Y, et al. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc. Natl Acad. Sci. USA. 2018;115:E5970–E5979. doi: 10.1073/pnas.1803245115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bergstrom CT, Lachmann M. The Red King effect: when the slowest runner wins the coevolutionary race. Proc. Natl Acad. Sci. USA. 2003;100:593–598. doi: 10.1073/pnas.0134966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Veller C, Hayward LK, Hilbe C, Nowak MA. The Red Queen and King in finite populations. Proc. Natl Acad. Sci. USA. 2017;114:E5396–E5405. doi: 10.1073/pnas.1702020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vigneron A, et al. Insects recycle endosymbionts when the benefit is over. Curr. Biol. 2014;24:2267–2273. doi: 10.1016/j.cub.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 169.Baker DM, Freeman CJ, Wong JCY, Fogel ML, Knowlton N. Climate change promotes parasitism in a coral symbiosis. ISME J. 2018;12:921–930. doi: 10.1038/s41396-018-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hom EFY, Murray AW. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science. 2014;345:94–98. doi: 10.1126/science.1253320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hall JPJ, et al. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Env. Microbiol. 2015;17:5008–5022. doi: 10.1111/1462-2920.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Banaszak AT, García Ramos M, Goulet TL. The symbiosis between the gastropod Strombus gigas and the dinoflagellate Symbiodinium: an ontogenic journey from mutualism to parasitism. J. Exp. Mar. Biol. Ecol. 2013;449:358–365. [Google Scholar]
- 173.Nakazawa T, Katayama N. Stage-specific parasitism by a mutualistic partner can increase the host abundance. Front. Ecol. Evol. 2020 doi: 10.3389/fevo.2020.602675. [DOI] [Google Scholar]
- 174.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol. Syst. Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yurtsev EA, Conwill A, Gore J. Oscillatory dynamics in a bacterial cross-protection mutualism. Proc. Natl Acad. Sci. USA. 2016;113:6236–6241. doi: 10.1073/pnas.1523317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hoek TA, et al. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 2016;14:e1002540. doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc. Natl Acad. Sci. USA. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Regus JU, Gano KA, Hollowell AC, Sofish V, Sachs JL. Lotus hosts delimit the mutualism–parasitism continuum of Bradyrhizobium. J. Evol. Biol. 2015;28:447–456. doi: 10.1111/jeb.12579. [DOI] [PubMed] [Google Scholar]
- 179.Hay ME, et al. Mutualisms and aquatic community structure: the enemy of my enemy is my friend. Annu. Rev. Ecol. Evol. Syst. 2004;35:175–197. [Google Scholar]
- 180.Pike VL, Lythgoe KA, King KC. On the diverse and opposing effects of nutrition on pathogen virulence. Proc. Biol. Sci. 2019;286:20191220. doi: 10.1098/rspb.2019.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Corbin C, Heyworth ER, Ferrari J, Hurst GDD. Heritable symbionts in a world of varying temperature. Heredity. 2017;118:10–20. doi: 10.1038/hdy.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Thomas MB, Blanford S. Thermal biology in insect-parasite interactions. Trends Ecol. Evol. 2003;18:344–350. [Google Scholar]
- 183.Delor I, Cornelis GR. Role of Yersinia enterocolitica Yst toxin in experimental infection of young rabbits. Infect. Immun. 1992;60:4269–4277. doi: 10.1128/iai.60.10.4269-4277.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kouse AB, Righetti F, Kortmann J, Narberhaus F, Murphy ER. RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS ONE. 2013;8:e63781. doi: 10.1371/journal.pone.0063781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kishimoto M, Baird AH, Maruyama S, Minagawa J, Takahashi S. Loss of symbiont infectivity following thermal stress can be a factor limiting recovery from bleaching in cnidarians. ISME J. 2020;14:3149–3152. doi: 10.1038/s41396-020-00742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhang B, Leonard SP, Li Y, Moran NA. Obligate bacterial endosymbionts limit thermal tolerance of insect host species. Proc. Natl Acad. Sci. USA. 2019;116:24712–24718. doi: 10.1073/pnas.1915307116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guay J-F, Boudreault S, Michaud D, Cloutier C. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect Physiol. 2009;55:919–926. doi: 10.1016/j.jinsphys.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 188.Bensadia F, Boudreault S, Guay J-F, Michaud D, Cloutier C. Aphid clonal resistance to a parasitoid fails under heat stress. J. Insect Physiol. 2006;52:146–157. doi: 10.1016/j.jinsphys.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 189.Vorburger C, Gouskov A. Only helpful when required: a longevity cost of harbouring defensive symbionts. J. Evol. Biol. 2011;24:1611–1617. doi: 10.1111/j.1420-9101.2011.02292.x. [DOI] [PubMed] [Google Scholar]
- 190.Parratt SR, Laine A-L. The role of hyperparasitism in microbial pathogen ecology and evolution. ISME J. 2016;10:1815–1822. doi: 10.1038/ismej.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hajishengallis G, Lamont RJ. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016;24:477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Neville BA, d’Enfert C, Bougnoux M-E. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 2015;15:fov081. doi: 10.1093/femsyr/fov081. [DOI] [PubMed] [Google Scholar]
- 194.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bonhoeffer S, Lenski RE, Ebert D. The curse of the pharaoh: the evolution of virulence in pathogens with long living propagules. Proc. Biol. Sci. 1996;263:715–721. doi: 10.1098/rspb.1996.0107. [DOI] [PubMed] [Google Scholar]
- 196.Rafaluk-Mohr C. The relationship between parasite virulence and environmental persistence: a meta-analysis. Parasitology. 2019;146:897–902. doi: 10.1017/S0031182019000015. [DOI] [PubMed] [Google Scholar]
- 197.Ebert D, Joachim Carius H, Little T, Decaestecker E. The evolution of virulence when parasites cause host castration and gigantism. Am. Nat. 2004;164:S19–S32. doi: 10.1086/424606. [DOI] [PubMed] [Google Scholar]
- 198.McCutcheon JP, Boyd BM, Dale C. The life of an insect endosymbiont from the cradle to the grave. Curr. Biol. 2019;29:R485–R495. doi: 10.1016/j.cub.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 199.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl Acad. Sci. USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 201.Wernegreen JJ. Reduced selective constraint in endosymbionts: elevation in radical amino acid replacements occurs genome-wide. PLoS ONE. 2011;6:e28905. doi: 10.1371/journal.pone.0028905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 203.Mao M, Yang X, Bennett GM. Evolution of host support for two ancient bacterial symbionts with differentially degraded genomes in a leafhopper host. Proc. Natl Acad. Sci. USA. 2018;115:E11691–E11700. doi: 10.1073/pnas.1811932115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Husnik F, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 205.Łukasik P, et al. Multiple origins of interdependent endosymbiotic complexes in a genus of cicadas. Proc. Natl Acad. Sci. USA. 2018;115:E226–E235. doi: 10.1073/pnas.1712321115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Keeling PJ, McCutcheon JP, Doolittle WF. Symbiosis becoming permanent: survival of the luckiest. Proc. Natl Acad. Sci. USA. 2015;112:10101–10103. doi: 10.1073/pnas.1513346112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Karnkowska A, et al. A eukaryote without a mitochondrial organelle. Curr. Biol. 2016;26:1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 208.John U, et al. An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci. Adv. 2019;5:eaav1110. doi: 10.1126/sciadv.aav1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Venkova T, Yeo CC, Espinosa M. Editorial: The good, the bad, and the ugly: multiple roles of bacteria in human life. Front. Microbiol. 2018;9:1702. doi: 10.3389/fmicb.2018.01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cirstea M, Radisavljevic N, Finlay BB. Good bug, bad bug: breaking through microbial stereotypes. Cell Host Microbe. 2018;23:10–13. doi: 10.1016/j.chom.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 211.Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 2019;216:20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Leonard SP, et al. Engineered symbionts activate honey bee immunity and limit pathogens. Science. 2020;367:573–576. doi: 10.1126/science.aax9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wolinska J, King KC. Environment can alter selection in host–parasite interactions. Trends Parasitol. 2009;25:236–244. doi: 10.1016/j.pt.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 214.Kiers ET, Palmer TM, Ives AR, Bruno JF, Bronstein JL. Mutualisms in a changing world: an evolutionary perspective. Ecol. Lett. 2010;13:1459–1474. doi: 10.1111/j.1461-0248.2010.01538.x. [DOI] [PubMed] [Google Scholar]
- 215.Lafferty KD. The ecology of climate change and infectious diseases. Ecology. 2009;90:888–900. doi: 10.1890/08-0079.1. [DOI] [PubMed] [Google Scholar]
- 216.Magalon H, Nidelet T, Martin G, Kaltz O. Host growth conditions influence experimental evolution of life history and virulence of a parasite with vertical and horizontal transmission. Evolution. 2010;64:2126–2138. doi: 10.1111/j.1558-5646.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 217.Bull JJ, Molineux IJ, Rice WR. Selection of benevolence in a host-parasite system. Evolution. 1991;45:875–882. doi: 10.1111/j.1558-5646.1991.tb04356.x. [DOI] [PubMed] [Google Scholar]
- 218.Gibson AK, et al. The evolution of reduced antagonism—a role for host–parasite coevolution. Evolution. 2015;69:2820–2830. doi: 10.1111/evo.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kubinak JL, Potts WK. Host resistance influences patterns of experimental viral adaptation and virulence evolution. Virulence. 2013;4:410–418. doi: 10.4161/viru.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Matthews AC, Mikonranta L, Raymond B. Shifts along the parasite–mutualist continuum are opposed by fundamental trade-offs. Proc. Biol. Sci. 2019;286:20190236. doi: 10.1098/rspb.2019.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Marchetti M, et al. Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol. 2010;8:e1000280. doi: 10.1371/journal.pbio.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ruby EG, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Biol. Sci. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jeon KW. Genetic and physiological interactions in the amoeba-bacteria symbiosis. J. Eukaryot. Microbiol. 2004;51:502–508. doi: 10.1111/j.1550-7408.2004.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 224.Wang X, et al. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010;1:1–9. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bull JJ, Molineux IJ. Molecular genetics of adaptation in an experimental model of cooperation. Evolution. 1992;46:882–895. doi: 10.1111/j.1558-5646.1992.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 226.Kikuchi Y, Hosokawa T, Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011;5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kikuchi Y, Hosokawa T, Fukatsu T. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Env. Microbiol. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shapiro JW, Williams ESCP, Turner PE. Evolution of parasitism and mutualism between filamentous phage M13 and Escherichia coli. PeerJ. 2016;4:e2060. doi: 10.7717/peerj.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Porter SS, Simms EL. Selection for cheating across disparate environments in the legume-rhizobium mutualism. Ecol. Lett. 2014;17:1121–1129. doi: 10.1111/ele.12318. [DOI] [PubMed] [Google Scholar]
- 230.Weese DJ, Heath KD, Dentinger BTM, Lau JA. Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution. 2015;69:631–642. doi: 10.1111/evo.12594. [DOI] [PubMed] [Google Scholar]
- 231.Slater SC, et al. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J. Bacteriol. 2009;191:2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Proença JT, Barral DC, Gordo I. Commensal-to-pathogen transition: one-single transposon insertion results in two pathoadaptive traits in Escherichia coli–macrophage interaction. Sci. Rep. 2017;7:4504. doi: 10.1038/s41598-017-04081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hu G, et al. Microevolution during serial mouse passage demonstrates FRE3 as a virulence adaptation gene in Cryptococcus neoformans. mBio. 2014;5:e00941-14. doi: 10.1128/mBio.00941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chrostek E, et al. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet. 2013;9:e1003896. doi: 10.1371/journal.pgen.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sicard M, et al. When mutualists are pathogens: an experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria) J. Evol. Biol. 2004;17:985–993. doi: 10.1111/j.1420-9101.2004.00748.x. [DOI] [PubMed] [Google Scholar]
- 236.Margulis L. Words as battle cries: symbiogenesis and the new field of endocytobiology. BioScience. 1990;40:673–677. [PubMed] [Google Scholar]
- 237.Didelot X, Barker M, Falush D, Priest FG. Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 2009;32:81–90. doi: 10.1016/j.syapm.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 238.Oishi S, Moriyama M, Koga R, Fukatsu T. Morphogenesis and development of midgut symbiotic organ of the stinkbug Plautia stali (Hemiptera: Pentatomidae) Zool. Lett. 2019;5:16. doi: 10.1186/s40851-019-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kang Y, et al. HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog. 2014;10:e1004232. doi: 10.1371/journal.ppat.1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Joy JB, Liang RH, McCloskey RM, Nguyen T, Poon AFY. Ancestral reconstruction. PLoS Comput. Biol. 2016;12:e1004763. doi: 10.1371/journal.pcbi.1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rafaluk‐Mohr C, Ashby B, Dahan DA, King KC. Mutual fitness benefits arise during coevolution in a nematode-defensive microbe model. Evol. Lett. 2018;2:246–256. doi: 10.1002/evl3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ford SA, Williams D, Paterson S, King KC. Co-evolutionary dynamics between a defensive microbe and a pathogen driven by fluctuating selection. Mol. Ecol. 2017;26:1778–1789. doi: 10.1111/mec.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hall AR, Ashby B, Bascompte J, King KC. Measuring coevolutionary dynamics in species-rich communities. Trends Ecol. Evol. 2020;35:539–550. doi: 10.1016/j.tree.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 244.Betts A, Rafaluk C, King KC. Host and parasite evolution in a tangled bank. Trends Parasitol. 2016;32:863–873. doi: 10.1016/j.pt.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 245.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:e00088-17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Unterholzner SJ, Poppenberger B, Rozhon W. Toxin-antitoxin systems: biology, identification, and application. Mob. Genet. Elem. 2013;3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wu M, et al. Phylogenomics of the reproductive parasite wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Frost CL, et al. The hypercomplex genome of an insect reproductive parasite highlights the importance of lateral gene transfer in symbiont biology. mBio. 2020;11:e02590-19. doi: 10.1128/mBio.02590-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bamford DH. Do viruses form lineages across different domains of life? Res. Microbiol. 2003;154:231–236. doi: 10.1016/S0923-2508(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 251.Casjens S, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 252.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]