Abstract
While much of this volume focuses on mammalian DNA repair systems that are directly involved in genome stability and cancer, it is important to still be mindful of model systems from prokaryotes. Herein we review the Red recombination system of bacteriophage λ, which consists of an exonuclease for resecting dsDNA ends, and a single-strand annealing protein (SSAP) for binding the resulting 3′-overhang and annealing it to a complementary strand. The genetics and biochemistry of Red have been studied for over 50 years, in work that has laid much of the foundation for understanding DNA recombination in higher eukaryotes. In fact, the Red exonuclease (λ exo) is homologous to Dna2, a nuclease involved in DNA end-resection in eukaryotes, and the Red annealing protein (Redβ) is homologous to Rad52, the primary SSAP in eukaryotes. While eukaryotic recombination involves an elaborate network of proteins that is still being unraveled, the phage systems are comparatively simple and streamlined, yet still encompass the fundamental features of recombination, namely DNA end-resection, homologous pairing (annealing), and a coupling between them. Moreover, the Red system has been exploited in powerful methods for bacterial genome engineering that are important for functional genomics and systems biology. However, several mechanistic aspects of Red, particularly the action of the annealing protein, remain poorly understood. This review will focus on the proteins of the Red recombination system, with particular attention to structural and mechanistic aspects, and how the lessons learned can be applied to eukaryotic systems.
Keywords: DNA recombination, Exonuclease, Single-strand annealing, Homologous recombination, Recombineering, Single-stranded DNA binding protein
1. Overview of red recombination
The recombination system of phage λ, called Red for recombination defective, was originally discovered from mutants of phage λ that could not recombine in a recA− E. coli host (Echols and Gingery, 1968; Signer and Weil, 1968). It consists of two proteins (Fig. 1): a 5′-3′ exonuclease that binds to dsDNA ends and processively digests the 5′-strand into mononucleotides (Little, 1967; Carter and Radding, 1971), and a single-strand annealing protein (SSAP) that binds the resulting 3′-overhang and anneals it to a complementary strand from another DNA molecule (Kmiec and Holloman, 1981; Muniyappa and Radding, 1986; Stahl et al., 1997). The exonuclease is encoded by the redα (or exo) gene, and has been called λ exonuclease or λ exo (we will use the latter). The SSAP is encoded by the redβ (or bet) gene, and has been called Redβ or β protein (we will use Redβ). Intriguingly, the two proteins form a complex with one another that somehow couples the two steps of the reaction, end-resection and annealing, by an unknown mechanism (Radding et al., 1971; Muyrers et al., 2000). A third protein called Gam, encoded by the redγ (or gam) gene, does not participate directly in recombination, but binds and inactivates the host RecBCD enzyme, to prevent it from digesting the phage chromosome (Karu et al., 1975; Wilkinson et al., 2016). The Red system is not required for viability of phage λ, but it boosts the levels of genome concatemers, which are the substrates for genome packaging (Enquist and Skalka, 1973), and promotes genetic diversity by recombining homologous regions of infecting phage and resident prophage (De Paepe et al., 2014).
Fig. 1.
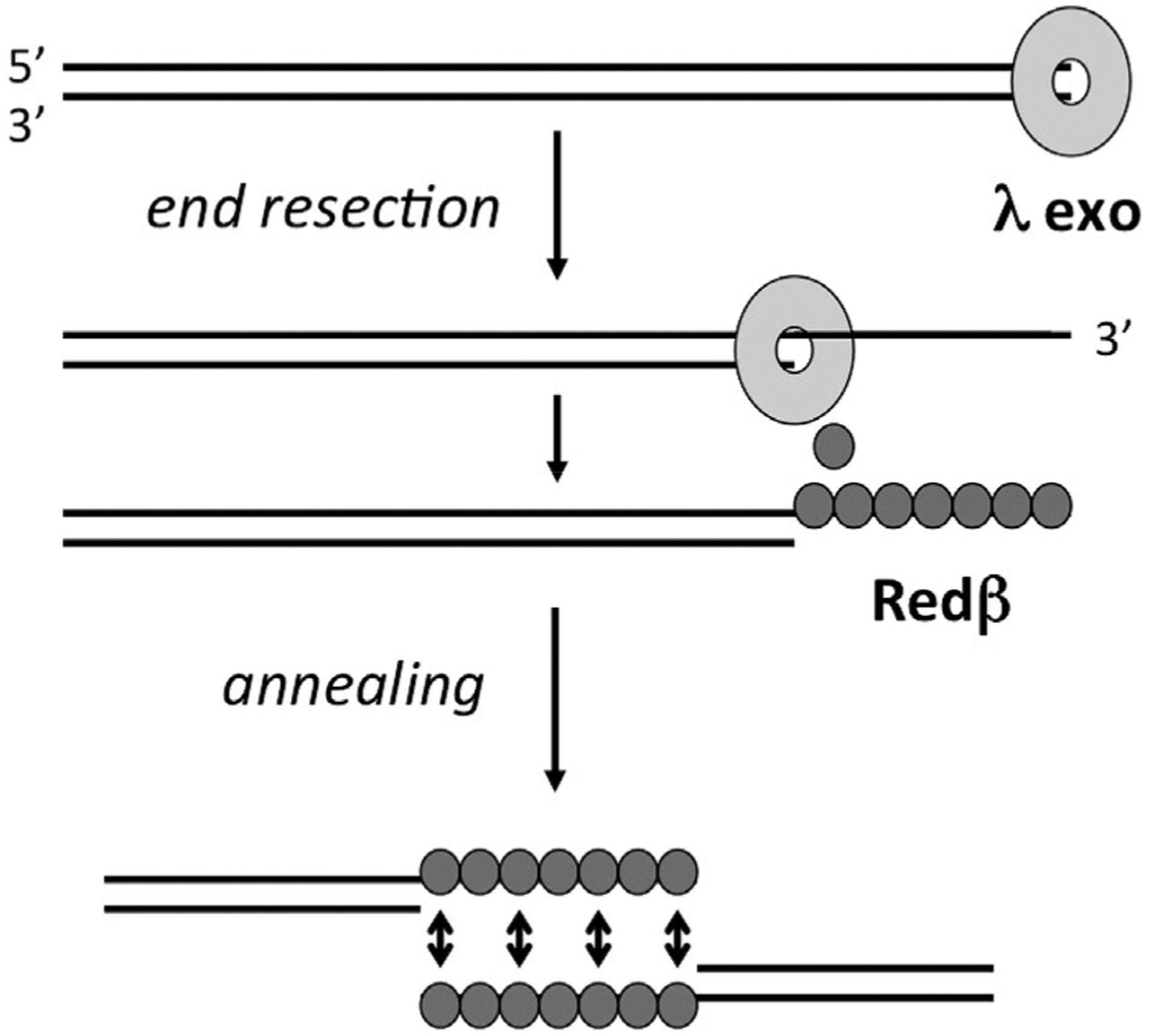
Overview of the Red recombination system from bacteriophage λ. λ exo forms a ring-shaped homotrimer that binds to dsDNA ends and processively digests the 5′-strand into mononucleotides. Through a specific protein-protein interaction, λ exo is thought to load the Redβ SSAP directly onto the 3′-overhang as it is formed. The Redβ SSAP then assembles on the 3′-overhang to promote its annealing with a complementary strand from another DNA molecule, such as another copy of the genome.
Homologs of the Red proteins, often encoded as “SynExo” pairs (for synaptase-exonuclease; Vellani and Myers, 2003), are found predominantly within phage genomes, or in bacterial genomes as active or defective prophage (Datta et al., 2008). They are also found in certain types of mobile genetic elements, such as integrating conjugative elements, where they contribute to the spread of antibiotic resistance (Garriss et al., 2013). The Pfam database (version 32.0, as of September 2018; Eberhardt et al., 2016) lists 2237 sequence homologs of λ exo, in the “YqaJ-like viral recombinase domain” family PF09588, and 1181 sequence homologs of Redβ, in the “RecT” family PF03837. Several of these have been purified and characterized, most notably the RecET proteins from the Rac prophage of E. coli, where RecE is the exonuclease and RecT is the annealing protein (Kolodner et al., 1994). Proteins from several other Red-related systems that have been characterized, either in vitro or in vivo, are summarized in Table 1. Homologs of λ exo are also found in oncogenic dsDNA viruses that infect mammalian cells, such as Herpes simplex virus 1 (HSV1), Kaposis sarcoma-associated herpesvirus (KSHV), and Epstein-Barr virus (Reuven et al., 2003). These enzymes also partner with a SSAP, but one that is much larger and not homologous to the phage annealing proteins (Tolun et al., 2013).
Table 1.
Red-related SynExo proteins that have been characterized biochemically and/or tested for recombineering activity in either E. coli or their native or closely related bacterial host.a
| Bacterial host | Phage (or prophage) | Exo/SSAP | Accessory | Reference |
|---|---|---|---|---|
| S. enterica | phage P22 | Abc2b-RecBCD/Erfb | Abc2, Arf | Poteete & Fenton (1993) |
| E. coli | Rac prophage | RecEb/RecTb | Kolodner et al. (1994) | |
| E. coli | phage λ | λ exob/Redβb | gam, rap, orf | Yu et al. (2000) |
| B. subtilisd | phage SPP1 | G34.1Pb/G35Pb | G36P | Martínez-Jiménez et al. (2005); Datta et al. (2008) |
| M. smegmatis | phage Che9c | gp61b/g60b | van Kessel and Hatfull (2007); Datta et al. (2008) | |
| E. faecalisc | prophage | EF2131/EF2132 | Datta et al. (2008) | |
| L. pneumophilad | prophage | orfB/orfC | Datta et al. (2008) | |
| V. choleraec | SXT (ICE)e | s066b/s065 | Datta et al. (2008); Chen et al. (2011) | |
| P. luminescensd | prophage | Plu2936/Plu2935 | Plu2934 | Datta et al. (2008); Yin et al. (2015) |
| L. monocytogenesd | phage A118 | orf47/orf48 | Datta et al. (2008) | |
| L. lactisc | phage ul36.2 | ———/Orf245 | Datta et al. (2008) | |
| S. aureusc | phage phiNM3 | ——/GP20 | Datta et al. (2008) | |
| P. syringae | prophage | RecEPsy/RecTPsy | Swingle et al. (2010) | |
| L. honkogenesis | prophage | LHK-Exob/LHK-Bet | Yang et al. (2011) | |
| L. reuterif,g | prophage | —/RecT1 | van Pijkeren & Britton (2012) | |
| C. perfringensg,h | prophage | —/CPF0939 | Dong et al. (2014) | |
| P. aeruginosa | D3 phage | exo/erf | De Paepe et al. (2014) | |
| L. planatarum | P1 prophage | lp_0642/lp_0641 | lp_0640 | Yang et al. (2015) |
| P. putidae | prophage | T1E1404/T1E1405 | Aparicio et al. (2016) |
Similar tables have been provided in Datta et al. (2008), and Yin et al. (2015).
These proteins have been purified and characterized for DNA binding and nuclease activities in vitro.
These systems were tested in E. coli for ssDNA recombineering.
These systems were tested in E. coli for ssDNA and dsDNA recombineering.
ICE: integrating conjugative element (a mobile genetic element).
The RecT1 protein from L. reuiteri was also tested in L. lactis, L. gasseri, and L. plantarum.
For these SSAPs only ssDNA recombineering was tested.
The recT homolog from C. perfringens was tested for ssDNA recombineering in C. acetobutylicum.
The λ Red system has already been reviewed extensively. Examples include “one geneticisťs historical perspective” (Stahl, 1998), a section within an extensive review on bacterial recombination (Kuzminov, 1999), a description of the features of λ Red that make it useful for genetic engineering (Poteete, 2001), comprehensive reviews of the biochemistry of the Red proteins, mechanisms of in vivo recombination and iťs role in propagation of phage λ (Murphy, 2012, 2016; Szczepanska, 2009), and reviews and protocols on recombineering (Sharan et al., 2009; Fu et al., 2010; Thomason et al., 2014; Pines et al., 2015; Wang et al., 2016). This review will focus on structural and mechanistic aspects of λ exo and Redβ, with particular attention to how the information gained can help in understanding eukaryotic systems.
2. Use of red in powerful methods for bacterial genome engineering
A key aspect of Red (and RecET) recombination is that it can occur with high efficiencies between regions of homology as short as 35–50 bp, which means that the necessary homology can be encoded on synthetic PCR primers (Zhang et al., 1998; Yu et al., 2000). Thus, for “insertional” recombination, a linear dsDNA encoding a gene of interest (such as for antibiotic resistance) can be amplified with primers containing terminal ~50 bp homologies to a target site on a chromosome or plasmid, and then electroporated into cells expressing λ exo and Redβ (or RecE and RecT), which recombine it into the target site (Zhang et al., 1998; Yu et al., 2000; Datsenko and Wanner, 2000; Murphy et al., 2000). The method can also be used in a “gap repair” reaction to copy and subclone a chromosomal gene into a linearized electroporated plasmid with terminal targeting homologies, without the need for PCR amplification of the target gene (Zhang et al., 2000; Lee et al., 2001). These technologies, which became known as “recombineering”, do not require unique restriction sites, and are thus particularly powerful for cloning involving large DNA constructs, such as the bacterial artificial chromosomes used for mouse functional genomics (Muyrers et al., 2001; Copeland et al., 2001; Skarnes et al., 2011). The remarkable fidelity of recombineering has made it useful for several high-throughput genomics applications, such as the Keio collection of E. coli single gene knockouts (Baba et al., 2006), and the C. elegans “TransgeneOme” resource in which 73% of the proteome was fused to a fluorescence-affinity tag for functional analysis (Sarov et al., 2012). Although most recombineering applications have used the Red proteins from phage λ, the E. coli RecE and RecT proteins have proven to be particularly powerful in a “linear plus linear” method of recombineering for cloning large biosynthetic gene clusters for bioprospecting (Fu et al., 2012). Curiously, this latter method requires the large N-terminal portion of full-length RecE protein (866 amino acids total, with the nuclease domain starting at residue ~600), which has unknown function and is not present in λ exo (266 amino acids total). The linear plus linear RecET method has been further improved by pre-resection and in vitro annealing of the two linear DNAs prior to their co-electroporation, which has allowed for cloning of even larger genome segments (Wang et al., 2018).
Soon after development of the original recombineering methods, which used PCR-generated dsDNA for targeting, it was realized that Red (or RecET) mediated recombination could also occur with electroporation of single-stranded oligonucleotides, which could encode single base changes or larger insertions and deletions (Ellis et al., 2001; Zhang et al., 2003). This technology, known as oligo-mediated recombineering, requires the annealing protein, but not the exonuclease, as the oligos are already single-stranded. Observation of a significant strand bias for the electroporated oligonucleotide led to the realization that it was annealing to a target site that was exposed as ssDNA at the lagging strand of a replication fork. It has since been realized that recombination of electroporated dsDNA cassettes occurs by a similar mechanism, in which one strand of the duplex is completely digested by the exonuclease, while the opposing strand is annealed to the replication fork, presumably in the same manner as oligonucleotides (Maresca et al., 2010; Mosberg et al., 2010).
With the knowledge that the targeting DNA is annealed at the replication fork, recombination levels were significantly increased by strategies to evade the host mismatch repair system, either by mutation of the relevant repair genes, or by appropriate design of the targeting oligonucleotide (Costantino and Court, 2003). Recombineering has also been optimized by deletion of host nucleases to protect the DNA substrate that is electroporated (Sawitzke et al., 2011; Mosberg et al., 2012), and by manipulation of the host replisome components, to increase the amount of ssDNA that is exposed at the replication fork (Lajoie et al., 2012). The Red system has also been combined with CRISPR-Cas9, to cut and eliminate unaltered (non-recombinant) chromosomes, to further increase the percentage of recombinants (Jiang et al., 2013). While Red has primarily been limited to engineering of bacterial genomes, recent efforts have begun to develop related technologies in cell lines from yeast (Barbieri et al., 2017) and humans (Valledor et al., 2018), the latter using the ICP8 SSAP from human herpes virus 1.
A particularly powerful extension of oligonucleotide recombineering has been developed, called MAGE for multiplex automated genome engineering, in which degenerate libraries of oligos directed towards multiple target sites are used in an iterative, selective manner to manipulate and evolve entire biosynthetic pathways (Wang et al., 2009). For example, MAGE was used to optimize the expression levels of 24 genes to boost the production of lycopene by 5-fold, and to replace all known TAG stop codons in E. coli with TAA, to enable its reassignment for incorporation of non-standard amino acids (Lajoie et al., 2013). With the development of MAGE, the Red system can now be used not just to make cloning applications more convenient, but rather for full-scale genome engineering, for applications in synthetic biology and metabolic engineering. A future challenge will be to develop efficient recombineering systems for a wider range of host species, particularly those that are well suited for particular biosynthetic purposes. A significant obstacle to this is the observation that SynExo proteins from one species often work in closely related species of bacteria, but not in more distant organisms, possibly due to interactions with as yet un-identified host proteins (Datta et al., 2008). To address this gap, increased understanding of the mechanisms of the proteins behind Red recombination will be imperative, as we will now expand on.
3. Structure and mechanism of λ exo, a processive 5′-3′ exonuclease
λ exo (226 aa; Mr 24.9 kDa) is a 5′-3′ exonuclease that binds with ~20 nM affinity to dsDNA ends. Once bound, it processively digests the 5′-strand into mononucleotides to form a long 3′-overhang. Notable features are a rate of 10–30 nt/s, maximal activity under alkaline conditions (optimal pH of 9.4), requirement of a 5′-phosphate on the DNA to initiate digestion, and sensitivity to high salt (Little, 1967; Mitsis and Kwagh, 1999). In a single molecule study, λ exo was seen to pause at a specific GGCGATTCT sequence on the DNA (Perkins et al., 2003), similar to pausing of RecBCD at χ sites (GCTGGTGG), but without any of the enzymatic changes in exonuclease function that occur for RecBCD at χ (Anderson and Kowalczykowski, 1997). However, this sequence is not present in the λ chromosome, and the physiological relevance of pausing is unknown. The crystal structure of λ exo revealed a ring-shaped homotrimer with a central channel for tracking along the DNA substrate (Kovall and Matthews, 1997). The channel is funnel-shaped, such that dsDNA can enter on one side, but only ssDNA can exit at the other. The structure thus provided a compelling explanation for the high processivity of the enzyme, as the trimer could remain topologically tethered to the 3′-strand as it digests the 5′-strand. Structures of exonuclease enzymes from related SynExo systems have also revealed toroidal oligomers, suggesting that encircling of the DNA is of fundamental importance (Yang et al., 2011). Interestingly, the nuclease domain of RecE exonuclease forms a toroidal tetramer instead of a trimer, and has different inter-subunit packing interactions, suggesting that the oligomeric ring structures have evolved independently, despite having monomers that share a common core fold (Zhang et al., 2009).
Our group determined the crystal structure of λ exo in complex with a 12 bp DNA duplex containing a 2-nucleotide overhang with a 5′-phosphate (Fig. 2; Zhang et al., 2011). This confirmed the general binding mode proposed above, but the DNA was significantly tilted, to project the end with the 5′-overhang into the active site of one subunit of the trimer. The other two subunits also form contacts with the DNA, primarily with the sugar-phosphate backbone. A notable interaction is the insertion of Arg45 into the minor groove of the downstream portion of the DNA. Arg45 may act as a rudder to help keep the enzyme on track as it spirals around the DNA. The structure also revealed that the enzyme unwinds exactly two base pairs from the end of the DNA, to feed the 5′-strand into the active site for cleavage, and the 3′-strand through the central channel and out the back of the trimer. This observation is consistent with a single molecule study, which concluded that unwinding of the terminal base pair is the rate-limiting step of the reaction cycle (van Oijen et al., 2003). Unwinding of the DNA is facilitated by a loop crossing over the active site cleft that splits into the base pairs to form hydrophobic interactions (see Leu78 in Fig. 2b).
Fig. 2.
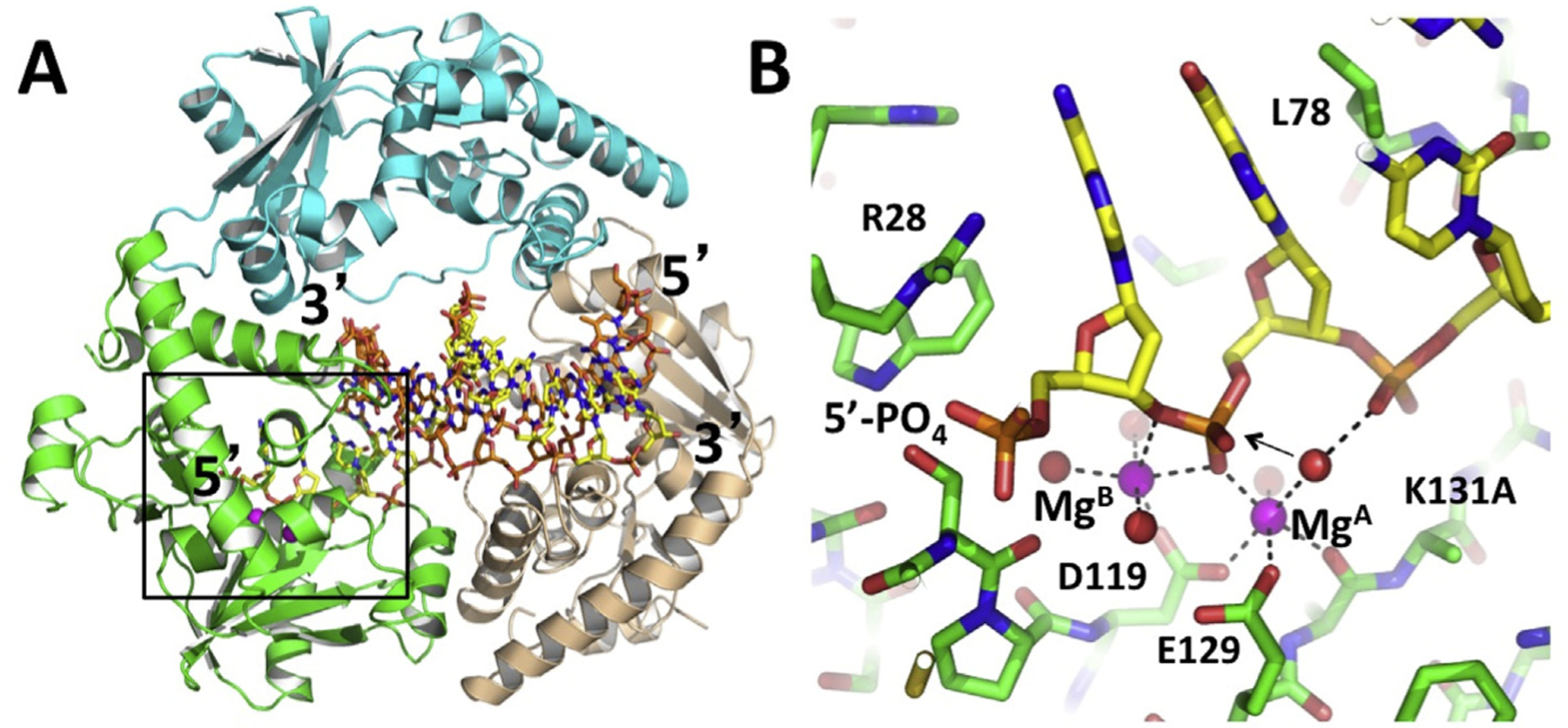
Structure of the λ exo trimer in complex with DNA. (A) Each subunit of the trimer is colored differently. The 5′-phosphorylated overhang of the DNA inserts into the active site of the green subunit (boxed region), while the 3′-end of the DNA pokes through the back of the central channel. (B) Close-up view of the active site interactions. The two Mg2+ ions are shown as magenta spheres, with coordinating interactions shown in dashed lines. The arrow indicates the presumed in-line attack of the water molecule bound to MgA. The reaction is prevented from occurring in the crystal by mutation of the active site lysine (K131A), which would deprotonate the attacking water. The 5′-phosphate of the DNA is bound to a positively charged pocket formed by Arg28, three –OH groups, and a helix N-dipole. Leu78 wedges into the base pairs to help unwind the DNA. The coordinates are taken from PDB code 3SM4 (Zhang et al., 2011). Structural figures were generated using PyMOL (PyMol Molecular Graphics System, Version 2.0 Schrödinger, LLC.).
The scissile phosphate on the DNA, between the first and second nucleotides of the 5′-strand, binds in the active site with two Mg2+ ions (MgA and MgB), in a configuration that is poised for catalysis, complete with a water molecule bound to MgA that is in suitable position for in-line attack. Turnover of the complex was prevented by a K131A mutation to remove the active site lysine, which is presumably required to deprotonate the hydrolytic water molecule. Trapping the complex in this way allowed for minimal disruption of the network of other active-site interactions, particularly those involving the metals. We subsequently determined the structure of WT λ exo with the same DNA, but with Ca2+ in place of Mg2+ to prevent cleavage (Zhang et al., 2014). This structure did not have correct metal binding geometry (only one Ca2+ ion was bound instead of two), but did show the interactions of the active site lysine. Together, the two structures were used to generate a composite model showing the likely interactions in the ground state. This model is consistent with the classic two-metal mechanism (Yang et al., 2006), in which MgA and Lys-131 bind and activate the hydrolytic water molecule for in-line attack, MgA and MgB bind the scissile phosphate to stabilize the negative charge of the transition state, and MgB, which coordinates the scissile phosphate twice, stabilizes the trigonal bipyramidal geometry of the transition state, and positions a water to facilitate deprotonation of the 3′–OH leaving group of the cleaved mononucleotide. A recent single molecule study provides evidence that MgB dissociates from the active site after each round of cleavage, while MgA remains bound continuously (Hwang et al., 2018). In the absence of DNA substrate, and in the two unoccupied active sites in the complex, neither Mg2+ is bound, although Ca2+ or Mn2+ are often bound to site A in crystal structures.
A set of interactions that is critical for processivity involves the 5′-phosphate on the DNA, which binds to a positively charged pocket at the end of the active site cleft formed by Arg-28, three OH groups, and a helix N-dipole (Subramanian et al., 2003). In all structures determined in the absence of DNA, this site on the enzyme is occupied by a free phosphate or sulfate ion, even when such ions are absent from the buffers used for protein purification and crystallization. This suggests an unusually strong interaction, and accordingly the R28A variant of λ exo is inactive (Pan et al., 2015). As noted above, a 5′-phosphate on the DNA is absolutely required for activity, which is perplexing because after cleavage of the first nucleotide, the next nucleotide would have a 5′-phosphate, and the reaction could proceed as normal. It is likely that the interaction of the 5′-phosphate with the positively charged pocket is needed to overcome the energetic penalty of unwinding the base pairs, to feed the 5′-strand into the active site. Consistent with this hypothesis, a structure of λ exo bound to a non-phosphorylated 12-mer duplex showed that the DNA binds in the central channel, but is not unwound and inserted into the active site (Zhang et al., 2011). Based on this, we proposed that during a processive digestion, electrostatic attraction of the 5′-phosphate exposed on the next nucleotide to be cleaved to the positively charged pocket provides a force to generate forward movement of the enzyme along the DNA at each step of the reaction. Similar interactions with the 5′-phosphate are critical for the activity of XrnI, a processive ribonuclease that digests and unwinds partially structured RNA substrates for mRNA decay (Jinek et al., 2011).
λ exo is a member of the type 2 restriction endonuclease (T2RE) family of nuclease enzymes, which have a conserved PD-(D/E)XK motif that contains the active-site lysine and the residues for binding the Mg2+ ions (Pingoud et al., 2005; Steczkiewicz et al., 2012). This large family includes the majority of restriction enzymes, and several other types of enzymes involved in DNA repair and RNA processing. Structures of several T2RE enzymes have been determined, many in complex with nucleic acid substrates, but usually not with appropriately bound metals. From the few structures in which the metals are appropriately bound, such as MutH (Lee et al., 2005) and BglI (Newman et al., 1998), a precise metal-binding geometry emerges, which is common to other nuclease folds such as RNaseH (Nowotny et al., 2005), indicating that it is fundamentally conserved (Yang et al., 2006). Table 2 lists some notable structures of T2RE enzymes related to λ exo, in order of their Dali rmsd alignment score (Holm and Laakso, 2016). These enzymes act on a wide variety of substrates, including RNA or DNA, and either exonucleolytically or endonucleolytically, but contain the same αβββαβ core fold and a conserved active site. The structure of the λ exo-DNA complex could thus be used as a template to model the active site interactions of these other enzymes, and to design strategies to trap more native-like complexes.
Table 2.
Proteins of the PD-(D/E)-XK family that are structurally homologous to λ exo.a
| Protein Name (organism) | Oligomeric state | PDB code | #residues | %ident. | rmsd (Å) | Ligands | Reference |
|---|---|---|---|---|---|---|---|
| LHK-exo (L. Hongkongensis) | 3-mer ring | 3sz5 | 186 | 27 | 1.3 | dT4-Mg2+ | Yang et al. (2011) |
| Phage exo (H. somnus) | 3-mer ring | 3k93 | 184 | 18 | 2.4 | ||
| Cas4 (Mimivirus Virophage) | dimer | 5yeu | 186 | 20 | 2.8 | Mg2+ | Dou et al. (2018) |
| Epstein-Barr virus nuclease | monomer | 2w4b | 188 | 12 | 2.9 | Buisson et al. (2009) | |
| KSHV-SOX | monomer | 3pov | 179 | 16 | 3.1 | dsDNA | Bagnéris et al. (2011) |
| KSHV-SOX | monomer | 5hsw | 179 | 14 | 2.9 | 31-mer RNA | Lee et al. (2017) |
| Cas4 (S. solfataricus) | 2×5-ring | 4ic1 | 152 | 10 | 3.3 | Mn2+ | Lemak et al. (2013) |
| Exonuclease (E. rectale) | 3-mer ring | 3loa | 167 | 9 | 3.4 | Zn2+ | |
| RecE exo (E. coli rac) | 4-mer ring | 3h4r | 153 | 10 | 3.4 | Zhang et al. (2009) | |
| HsMGME1 (mitochondrial) | monomer | 5zyu | 150 | 9 | 3.6 | ssDNA-Mn2+ | Yang et al. (2018) |
| Dna2 nuclease (Mouse) | monomer | 5eax | 157 | 8 | 3.9 | ssDNA | Zhou et al. (2015) |
| Cas4 nuc (P. calidifontis) | monomer | 4r5q | 138 | 12 | 3.5 | Mg2+ | |
| RecB (E. coli) | heterotrim. | 1w36 | 132 | 9 | 3.7 | Ca2+ | Singleton et al. (2004) |
| AddA (B. subtilis) | heterodim | 4ceh | 113 | 15 | 3.1 | Krajewski et al. (2014) | |
| eIF3d (N. vitripennis) | monomer | 5k4d | 159 | 8 | 3.6 | Lee et al. (2016) | |
| Rai1 nuc (A. gossypii) | monomer | 5bud | 143 | 8 | 4.1 | pU5-Mn2+ | Wang et al. (2015) |
| HJC resolvase (P. furiosis) | dimer | 1gef | 90 | 14 | 3.2 | Nishino et al. (2001) | |
| Dom3Z (DXO, human) | monomer | 4j7l | 144 | 9 | 4.0 | RNA-Mg2+ | Jiao et al. (2013) |
| tRNA splicing (M. jannaschii) | dimer | 1a79 | 93 | 11 | 3.6 | Li et al. (1998) | |
| NucS/EndoMS (E. kodakaren) | dimer | 5gke | 80 | 16 | 3.8 | dsDNA | Nakae et al. (2016) |
| FAN1 nuclease (mouse) | monomer | 4rib | 93 | 9 | 2.4 | 5′P-FlapDNA | Wang et al. (2014) |
The listed protein structures are selected from a Dali Search with the structure of λ exo (Chain B of 3SM4) as the query. They are listed in order of decreasing Z-score from the Dali search. In cases where there are multiple PDB entries for the same protein, the structure with the most functionally comparable ligand is listed. Only some of the protein hits from the search are listed, to minimize redundancy of similar proteins. The following structures form ring-shaped oligomers for processing dsDNA breaks (like λ exo): LHK-exo, H. somnus exo, E. rectale exo, and RecE. EBV nuclease and KSHV-SOX perform the same function, but do so as monomers. RecB and AddA have nuclease domains that are homologous to λ exo, and function as larger helicase/nuclease complexes for processing dsDNA breaks. eIF3d, Rai1, and Dom3Z all act on 5′-caps of mRNA substrates.
A notable example is the group of exonucleases from oncogenic dsDNA viruses, such as Herpes simplex virus 1 (HSV-1), Epstein Barr virus (EBV), and Kaposis sarcoma-related herpes virus (KSHV), which are of interest for anti-viral drug discovery. Like λ exo, these enzymes digest the 5′-strand of dsDNA, and require a 5′-phosphate to get started, although they are monomeric and less processive. They also have an RNase activity for degradation of host mRNAs during “shutoff”, and are thus called “SOX” enzymes, for shutoff and exonuclease. While crystal structures are available for the SOX enzymes from EBV (Buisson et al., 2009) and KSHV (Dahlroth et al., 2009), none of the structures contain a substrate bound in a catalytic configuration. Structures of KSHV-SOX bound to DNA and RNA have been determined (Bagnéris et al., 2011; Lee et al., 2017), but neither has the scissile phosphate bound with correctly positioned metal ions. The complex of KSHV-SOX with DNA was trapped by omitting the 5′-phosphate from the DNA, but this resulted in the 5′-strand not being inserted fully into the active site (Fig. 3). Modeling of the 5′-phosphorylated DNA from the λ exo structure into these structures shows that essentially all of the residues mentioned above are conserved, including the active site lysine and metal binding residues, the positively charged pocket for binding the 5′-phosphate, and residues of hydrophobic wedge on the crossover loop for unwinding the base pairs (although this loop is disordered for KSHV-SOX). Remarkably, the dsDNA substrates of KSHV-SOX and λ exo follow the same trajectories along surfaces of the protein structures, despite their different oligomeric states. Moreover, both enzymes insert an arginine into the minor grove at the same position on the downstream portion of the DNA, even though the arginine comes from different regions of the respective protein folds (from another subunit of the λ exo trimer and from an insertion within the KSHV-SOX monomer).
Fig. 3.
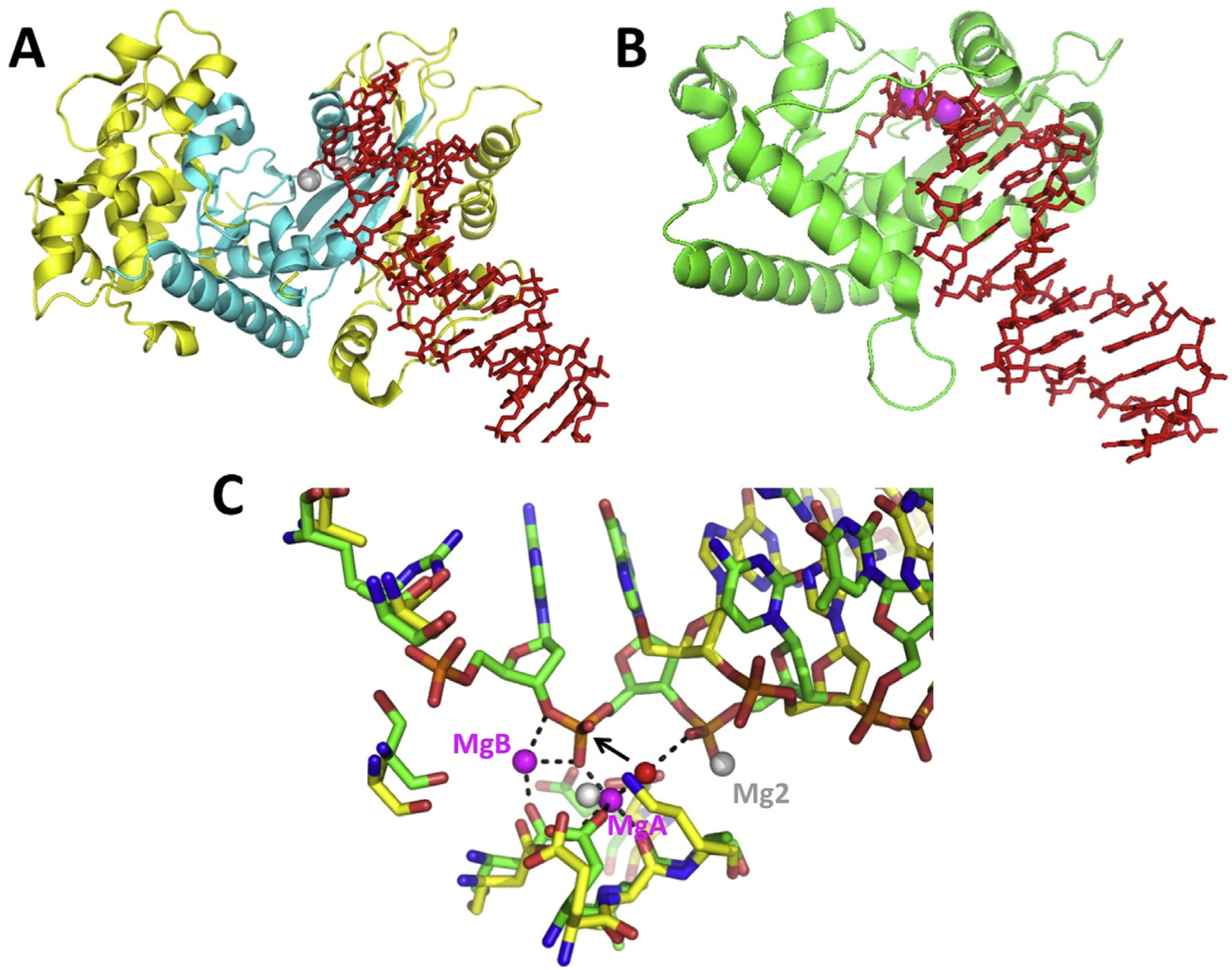
Structure of KSHV-SOX bound to DNA and comparison with λ exo. (A) The KSHV-SOX monomer is shown in yellow, with the core of the fold common to λ exo in cyan. The dsDNA is in red, with a 5′-end projecting into the active site, which is marked by two Mg2+ ions (grey spheres). The coordinates are taken from PDB code 3POV (Bagnéris et al., 2011). (B) Monomer of λ exo from the structure of the trimer bound to DNA, in a similar orientation as panel A. The two Mg2+ ions are shown as magenta spheres. (C) Overlay of the active sites. The 50-phosphorylated DNA from the λ exo structure (green bonds) inserts fully into the actives site to interact with two Mg2+ ions (magenta spheres; interactions shown with dashed lines). The 5′-nucleotide of the DNA in the KSHV-SOX structure (yellow bonds), which has a 5′–OH instead of a phosphate, does not insert fully into the active site. This is actually the second nucleotide of the crystallized DNA, as the first was disordered. Mg1 of the KSHV-SOX structure overlaps with MgA of λ exo, but Mg2 binds to a different, likely non-catalytic position.
Other notable examples are the human Dna2 nuclease (Zhou et al., 2015), which is involved in degradation of Okazaki fragments and DNA end-resection for homology-dependent repair of dsDNA breaks, and the FAN1 nuclease of the Fanconi aneamia pathway for repair of inter-strand crosslinks (Wang et al., 2014). Dna2 differs from λ exo in that it is not highly processive (at least on its own), and generates short fragments by endonuclease activity from either end of a ssDNA. In the structure of Dna2 bound to a 15-mer single-stranded oligonucleotide, one Ca2+ is bound to the scissile phosphate, to a position near MgA of λ exo, but the other Ca2+ is bound to a likely non-catalytic position, and the scissile phosphate is not fully engaged with the active site. Alignment of Dna2 with λ exo provides a model for the likely binding of the metals in Dna2 (Fig. 4). Structures of FAN1 nuclease have been determined in complex with a nicked and severely bent DNA duplex with various 5′-flap structures. Interestingly, interactions of FAN1 with the 5′-phosphate of the flap are important for binding, but in this case it is the 3rd phosphate from the 5′-end of the DNA that is cleaved, instead of the adjacent one. This allows FAN1 to cleave off trinucleotides instead of mononucleotides, which is consistent with its role in unhooking inter-strand crosslinks. The FAN1 structure contained a single Ca2+ ion bound to the scissile phosphate, in a position corresponding to MgA in λ exo. Alignment with λ exo shows the interactions that are likely to occur with two Mg2+ ions in the ground state of FAN1 (Fig. 5).
Fig. 4.
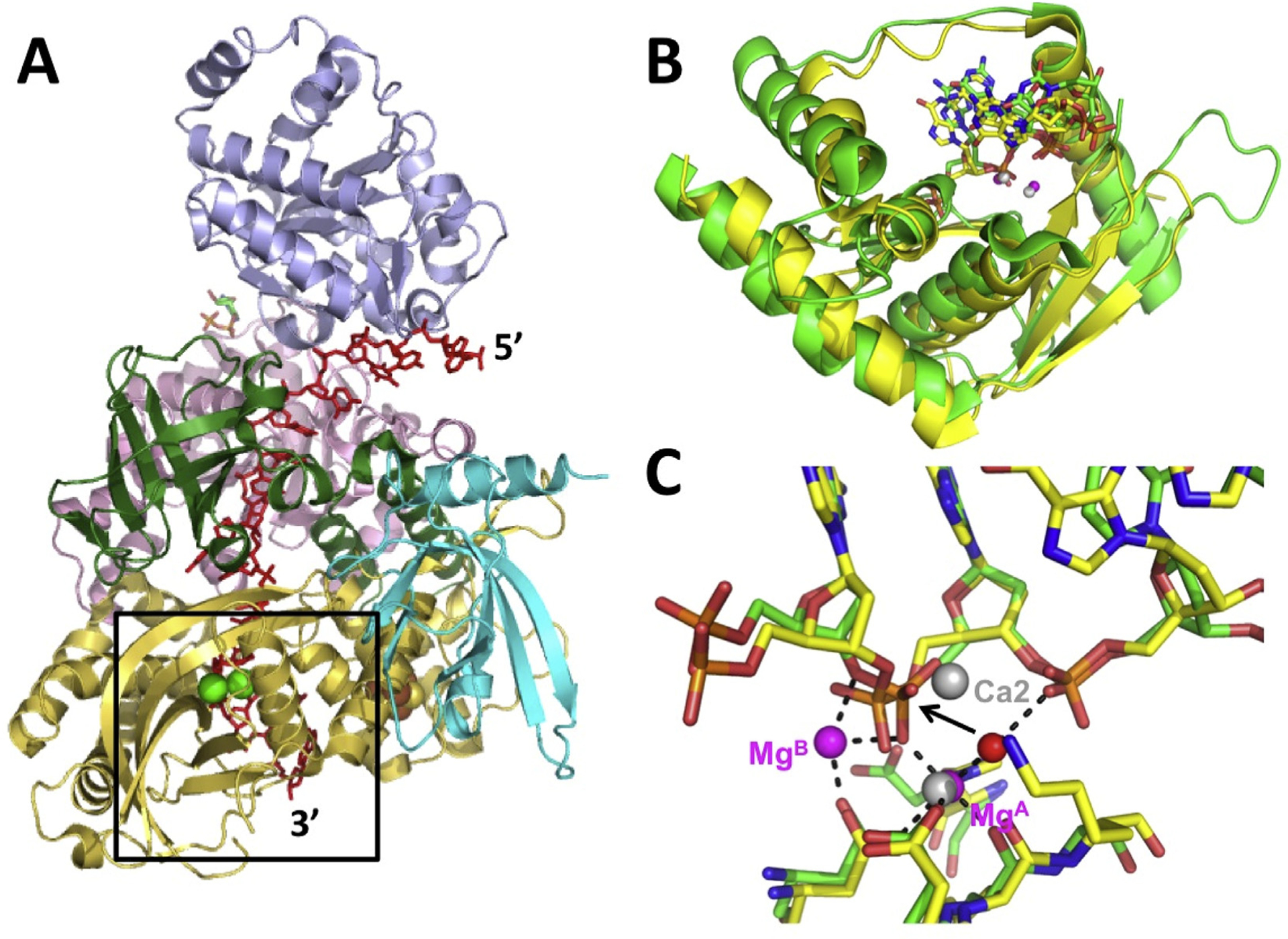
Structure of Dna2 nuclease and comparison with λ exo. (A) Structure of Dna2 nuclease in complex with a 15-mer ssDNA (red) and two Ca2+ ions (green spheres) (PDB code 5EAX; Zhou et al., 2015). The structure is colored by domain, with the nuclease domain in yellow. The active site is poised to cleave between the 11th and 12th nucleotides of the ssDNA, 4 nucleotides from the 3′-end. (B) Rmsd alignment of the core portions of the nuclease domains of Dna2 (yellow) and λ exo (green). The core folds share 5 α-helices and 6 β-strands. The 4 nucleotides from the 3′-end of the ssDNA in the Dna2 structure align (in the same direction) with four nucleotides from the 5′-end of the DNA in the λ exo structure (PDB code 3SM4; Zhang et al., 2011). (C) Overlay of the active sites. The scissile phosphates and metal binding residues of the two structures overlap closely. Ca1 of the Dna2 structure overlaps with MgA of the λ exo structure, but Ca2 is bound in a different, likely non-catalytic position.
Fig. 5.
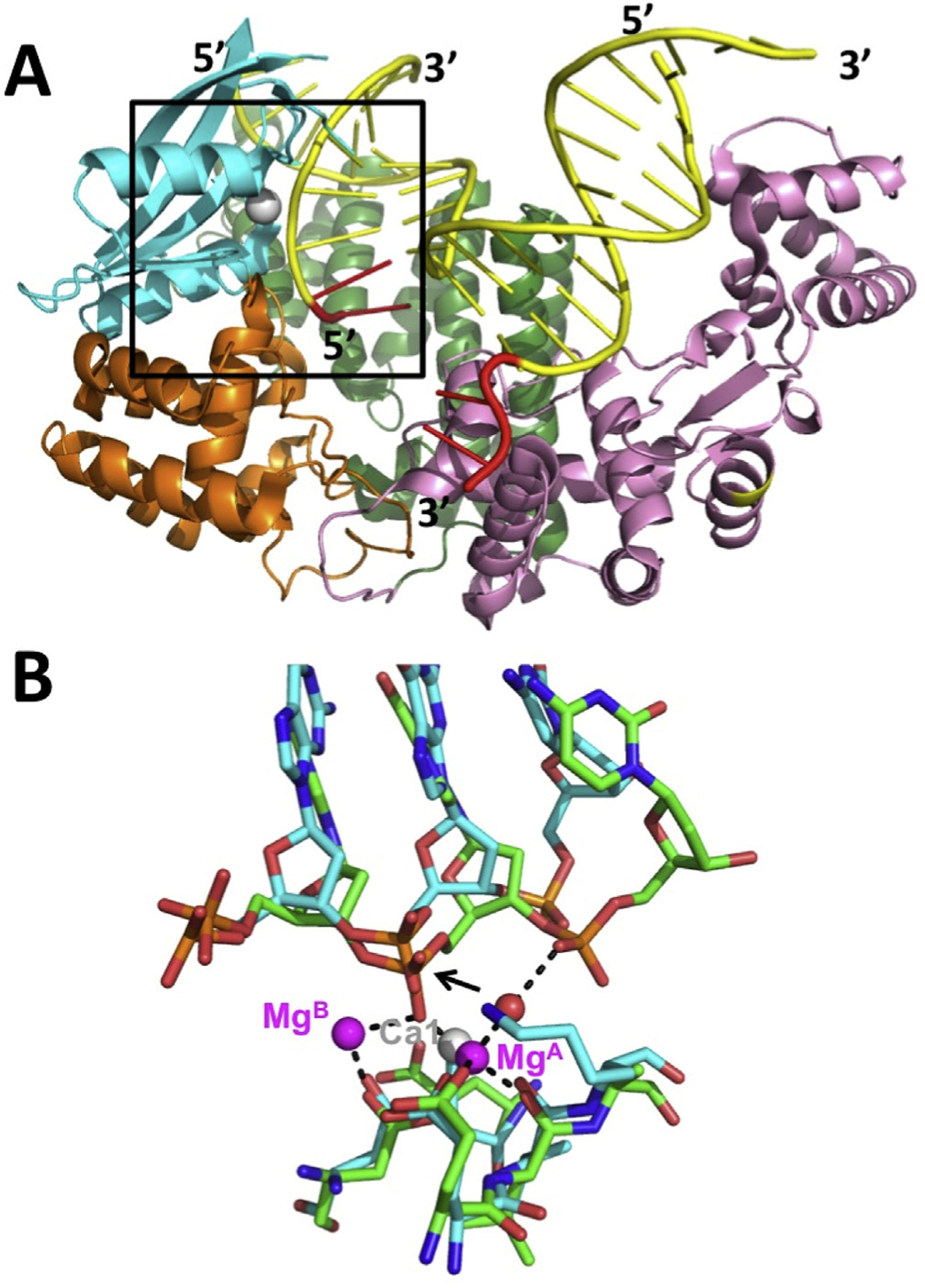
Structure of FAN1 nuclease and comparison to λ exo. (A) FAN1 structure bound to a nicked DNA (PDB code 4RI8; Wang et al., 2014). The DNA is shown in yellow, with 50- and 30-flaps in red. The T2RE domain is in cyan, with a single Ca2+ ion in grey, poised to cleave between the 3rd and 4th nucleotides from end of the 5′-flap. (B) Alignment of the active site of FAN1 (cyan bonds) with λ exo (green bonds). The 5′-3′ direction of both DNAs goes from left to right. The black dashed lines show the interactions of the metals in the λ exo structure. The single Ca2+ ion in the FAN1 structure (grey sphere) overlaps with MgA from the λ exo structure. The red sphere shows the position of the attacking water molecule, as indicated by the black arrow.
4. Structure and mechanism of the Redβ single strand annealing protein
Redβ is a 261 amino acid protein (Mr 29.7 kDa) that binds to ssDNA and promotes the annealing of complementary strands. It is distinct from RecA in that it is ATP-independent, and under normal conditions does not promote strand invasion reactions to form D-loops. The protein exhibits an unusual and intriguing DNA binding behavior: it binds weakly to ssDNA, not at all to pre-formed dsDNA, but tightly to the product (or intermediate) of annealing formed when two complementary oligos are added to the protein sequentially (i.e. when the protein is first incubated with one oligo, and then the complementary oligo is added subsequently; Karakousis et al., 1998). Structural analysis of Redβ by negative stain transmission electron microscopy (TEM) revealed distinct oligomeric forms that closely parallel this DNA binding behavior (Passy et al., 1999): the protein forms rings of 10–12 subunits on its own, slightly larger rings in the presence of ssDNA, and left-handed helical filaments in the presence of heat-denatured dsDNA, which presumably underwent annealing. For the latter complex, the filaments were seen to extend from rings, suggesting that the annealing reaction started on a ring, and then continued on a filament.
Based on these data, a compelling model for the annealing reaction was proposed, in which the ring form of the protein binds to ssDNA along its sugar phosphate backbone, exposing the bases for homology recognition (Fig. 6A; Passy et al., 1999). Once a complementary strand of ssDNA is found, and an annealing reaction initiated, the nascent duplex peels off of the ring as it is formed, and is then bound by free monomers of Redβ, which assemble on the nascent duplex to form a stable helical nucleoprotein filament. In this way, tighter binding of Redβ to the duplex product of annealing than to the ssDNA substrate drives the annealing reaction forward. In agreement with this model, the crystal structure of the N-terminal DNA-binding domain of human Rad52 revealed an 11-mer ring with a narrow positively charged groove (Kagawa et al., 2002; Singleton et al., 2002). The groove can accommodate ssDNA, which was modeled into the groove, but is not wide enough to bind dsDNA. Mutagenesis studies identified a secondary “outer” ssDNA-binding site on the Rad52 ring (Kagawa et al., 2008), and recent crystal structures show that ssDNA binds to both of these sites (Saotome et al., 2018). The “inner” primary site around the ring has the ssDNA bound in a repeating conformation, with four-nucleotide B-form segments separated by more extended spacers (Fig. 7). Such a pattern is highly reminiscent of the trinucleotiderepeating conformation of ssDNA bound to RecA (Chen et al., 2008), although the latter forms a helical filament instead of a planar ring. Earlier hydroxyl radical footprinting of the Rad52-ssDNA complex had revealed a precise binding stoichiometry of four nucleotides per monomer (Parsons et al., 2000), which is in close agreement with the structures. The stoichiometry of Redβ on ssDNA has not been established by this method, but in titration experiments it appears to be very similar (Mythili et al., 1996; Smith & Bell, unpublished results). Interestingly however, the complex of Redβ with annealed duplex appears to have a different stoichiometry of 10–11 bp per monomer, reflective of the structural transition observed by TEM and AFM (Erler et al., 2009; Ander et al., 2015).
Fig. 6.
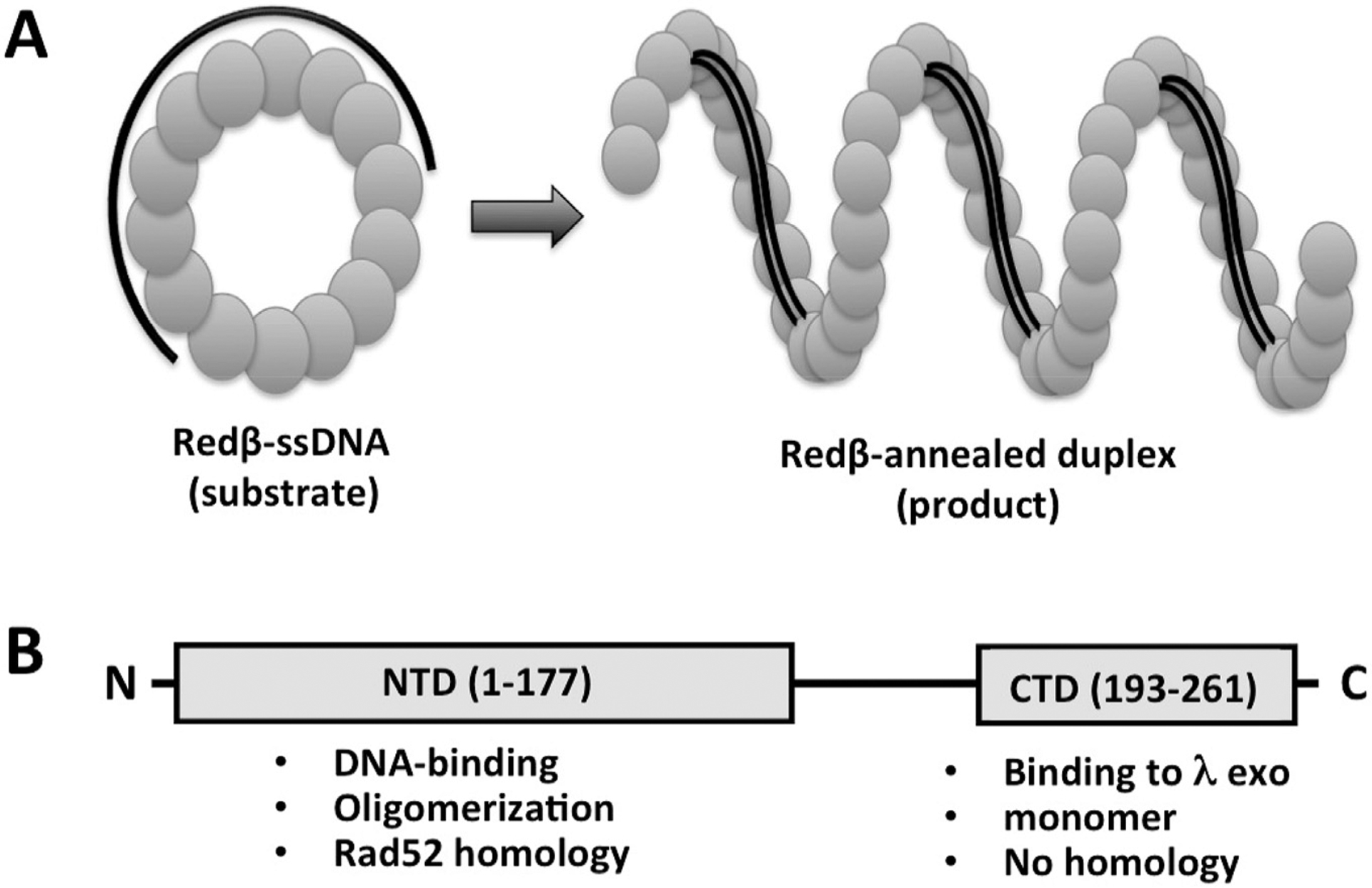
Domain structure and model for the mechanism of the Redβ single strand annealing protein. (A) Model for the oligomeric ring and filament forms of Redβ, based on TEM images (Passy et al., 1999). In the model, the ring form of the protein binds to ssDNA and presents it in a conformation that is poised for homology recognition, while the helical filament form of the protein assembles on the duplex product of annealing as it is being formed, to help drive the annealing reaction forward. (B) Domain structure of Redβ, based on limited proteolysis and functional properties (Wu et al., 2006; Smith and Bell, 2016).
Fig. 7.
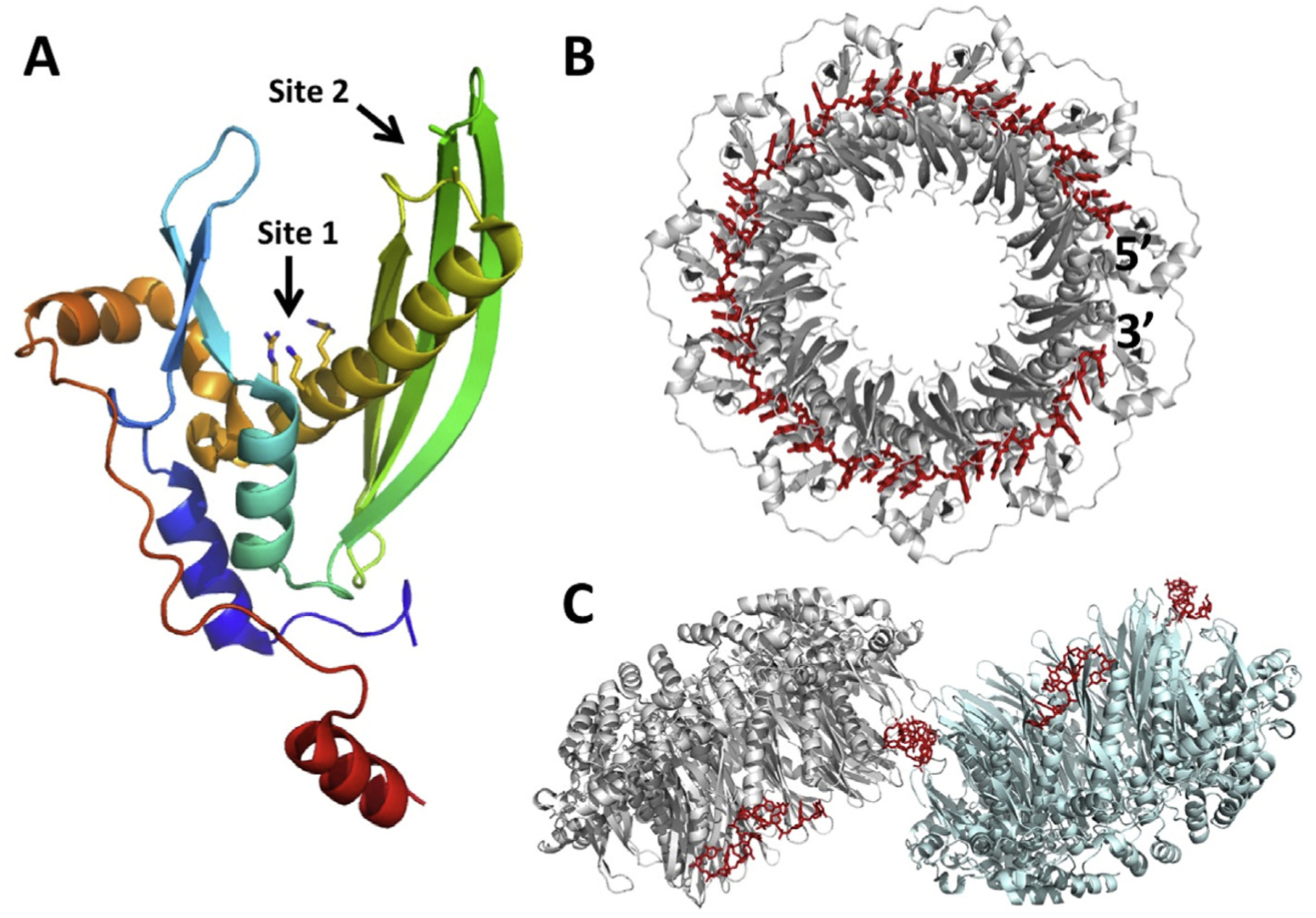
Crystal structure of the Rad52 DNA-binding domain in complex with ssDNA. (A) Monomer of Rad52 colored in rainbow from blue (N-terminus) to red (C-terminus). The primary ssDNA-binding site (Site 1) is at the base of the deep cleft, lined by positively charged residues (Lys152, Arg153, Arg156). The secondary site (Site 2) is at the tip of a β-hairpin. (B) 11-mer ring of Rad52 bound to a 40-mer ssDNA (PDB code 5XRZ). The ssDNA is bound in the “inner” primary site, deep in the groove, primarily by its sugar-phosphate backbone, with the bases exposed for homology recognition. (C) Two 11-mer rings bound to segments of four different 40-mer oligonucleotides (PDB code 5XS0). Each ssDNA is bound to the “outer” secondary site, which bridges neighboring rings. One bridge is shown in the center, and the others would form with neighboring rings in the crystal lattice. Coordinates used to make the figure are from Saotome et al. (2018).
While the model for annealing involving rings and filaments of Redβ is compelling, not all of the data are in agreement. Structural studies of Redβ by atomic force microscopy showed that it forms a split lock washer (or a gapped ellipse) of 11 subunits in the absence of DNA, disperse monomers (instead of oligomeric rings) on ssDNA, and helical filaments on short annealed duplex (Erler et al., 2009). The authors concluded that the oligomeric ring is not the species that binds to ssDNA. An oligomeric ring of approximately 11 subunits does seem to be a strikingly ubiquitous structure among SSAPs, having also been seen for phage P22 Erf (Poteete et al., 1983), E. coli RecT (Thresher et al., 1995), Sak protein of lactoccocal phage ul36 (Ploquin et al., 2008), and the mitochondrial Mgm101 protein (Mbantenkhu et al., 2011). However, as seen for Redβ in the AFM study, the ring is not always the species that binds to ssDNA: RecT and Mgm101, for example, form oligomeric rings on their own, and helical filaments on ssDNA. In addition, while many SSAPs form helical filaments, their features vary widely, and the remarkably stable filament of Redβ on annealed duplex appears to be unique. The ICP8 annealing protein of HSV-1, which is much larger and not homologous to Redβ, forms bipolar helical filaments without DNA (Makhov et al., 2009), suggesting a side-by-side anti-parallel mechanism of annealing, but a double 9-mer ring with ssDNA (Tolun et al., 2013), suggesting a completely different mechanism. Thus, if the model for annealing proposed above is correct for Redβ, it is not at all clear that it would be generally applicable to all SSAPs, or even to other members of the RecT/Redβ superfamily. A summary of the oligomers seen for different types of SSAPs is given in Table 3.
Table 3.
Oligomeric structures formed by single-strand annealing proteins in their different DNA-bound states.a
| Protein name (organism) | Oligomer without DNA | Oligomer on ssDNA | Oligomer on dsDNA (annealed duplex) | Method and References |
|---|---|---|---|---|
| Erf (P22 phage) | 10–14-mer rings | large aggregates | large aggregates | nsTEM; Poteete et al. (1983) |
| RecT (Rac prophage) | 7–8-mer rings | twisted filaments | inter-twined filaments | nsTEM; Thresher et al. (1995) |
| Redβ (phage λ) | 12-mer rings | 15–18-mer rings | left-handed helical filaments | nsTEM; Passy et al. (1999) |
| Redβ (phage λ) | 11-mer split lock washer | disperse monomers | left-handed helical filaments | AFM; Erler et al. (2009) |
| G35P (B. subtilis phage SPP1) | 7–8-mer rings | left-handed helical filaments | nsTEM; Ayora et al. (2002) | |
| hRad52 (human) | 7-mer ring | nsTEM/STEM; Stasiak et al. (2000) | ||
| hRad52 DNA-binding domain | 11-mer rings | XRAY; Singleton et al. (2002); Kagawa et al. (2002) | ||
| hRad52 DNA-binding domain | 11-mer ring, networks of rings | XRAY; Saotome et al. (2018) | ||
| SAK (L. lactis phage ul36) | 11-mer rings, stacks of rings | nsTEM; Ploquin et al. (2008) | ||
| Mgm101 (mitochondrial) | 14-mer rings, compressed filaments | distinct nucleoprotein filaments | nsTEM; Mbantenkhu et al. (2011) | |
| ICP8 (HSV-1) | Bipolar filaments | nsTEM; Makhov et al. (2009) | ||
| ICP8 (HSV-1) | 2 stacked 9-mer rings | nsTEM; Tolun et al. (2013) | ||
| DdrB (D. radiodurans) | 5-mer ring, higher-ordered assemblies | 5-mer ring (with partially annealed intermediate) | XRAY; Sugiman-Marangos et al. (2013), 2016 | |
| RecO (D. radiodurans) | monomer | XRAY; Makharashvili et al. (2004); Leiros et al. (2005) |
The table lists several SSAPs and the oligomeric states that they are seen to occur in the different DNA-bound states, by different experimental methods. The table includes four distinct families of SSAPs, which all have distinct folds. The Rad52 family includes Rad52, the phage annealing proteins (RecT, Redβ, G35P, SAK), and the mitochondrial annealing protein Mgm101. The ICP8 family includes the annealing proteins from oncogenic dsDNA virsus (HSV1, KSHV, EBV, etc.). DdrB and RecO each have distinct folds of their own.
On top of this significant variation in the types of oligomers seen for different SSAPs, it has been questioned whether or not the oligomers of Redβ seen by TEM are even present at the lower concentrations of the protein that are likely to be present in the cell, (Ander et al., 2015). The authors used Western blotting to determine that Redβ is present at concentrations of less than 150 nM in cells active for recombineering (where Red proteins were expressed from a pBAD promoter), far below the concentrations at which oligomers are seen to form in vitro by gel filtration (30 μM or higher), and TEM (0.8–3.6 μM) (Subramaniam et al., 2016; Matsubara et al., 2013; Smith and Bell, 2016; Passy et al., 1999). The authors also used a sensitive method, fluorescence correlation spectroscopy of labeled Redβ and ssDNA, to examine oligomerization of Redβ at very low concentrations, and found that Redβ is predominantly monomeric at concentrations below 1 μM (50-fold excess of monomer to oligomer at 1 μM). Finally, the authors used an optical tweezers instrument to observe the binding of single monomers of Redβ to short DNA hairpins as they underwent cycles of unzipping and annealing. A key observation from this work was that the remarkably stable complex presumed to be responsible for annealing could be formed by only two monomers of Redβ (on a 25 bp hairpin), as opposed to the eleven subunits that are seen in the oligomeric rings. Similar results were seen in gel-based experiments (Ander et al., 2015; Erler et al., 2009). Based on these experiments, and on earlier work showing that Redβ binds to ssDNA as disperse monomers, a model for annealing involving clamping of two adjacent monomers at an initial ~20 bp region of annealed duplex was proposed (see Fig. 6 of Erler et al., 2009). A key aspect of the model is the structural change that takes place in going from the initial Redβ-ssDNA complex, which is relatively weak (~12 pN), to the clamped dimer complex with ~20 bp of annealed duplex, which is remarkably stable (~200 pN). A high energy barrier for the formation of the stably clamped dimer complex is thought to ensure the fidelity of annealing, as a single mismatch would reduce the stabilization energy needed to complete the transition. According to this model, the oligomeric rings of Redβ seen at higher concentrations in vitro are not relevant in vivo, although the interactions that form them are likely to reflect the lateral interactions between adjacent monomers in the stably clamped dimer.
A caveat to the experiments of Ander et al. was the low-salt buffer that was used to monitor oligomerization (20 mM Tris, 10 mM NaCl, 10 mM MgCl2, pH 7.5), which could significantly weaken the effect of hydrophobic interactions, should they be important for oliogomer formation. Similarly, in Erler et al. the free Redβ protein, for which fairly uniform 11-mer ring-like complexes were seen, was prepared in high salt buffer (20 mM KH2PO4, pH 6.0, 10 mM MgCl2, 200 mM KCl), whereas the ssDNA complex, for which predominantly monomers were seen, was prepared in low salt buffer (20 mM KH2PO4, pH 6.0, 10 mM MgCl2). If anything, one would expect the presence of ssDNA to strengthen the lateral association of Redβ monomers, through cooperativity, so it is curious that stable oligomers were seen without ssDNA, but not with ssDNA.
It is also difficult to account for the effects of molecular crowding that would occur in the cell. Many recombineering systems in use express the Red proteins from the native pL promoter in the context of a defective λ prophage (Yu et al., 2000), which has been shown to give higher levels of recombination than paraBAD, in a way that correlates with higher expression of Red proteins (Datta et al., 2006). Although the stable clamped dimer complex can form on annealed duplex as short as 20 bp (Erler et al., 2009; Ander et al., 2015), the efficiency of recombination increases by approximately 100-fold as the length of the homology arms is increased from 20 to 50 bp (Yu et al., 2000; Erler et al., 2009). This coincides with the minimal length of 36 nucleotides required for binding of Redβ to ssDNA in vitro (Mythili et al., 1996), and is also the approximate length of ssDNA that would fit around an 11-mer ring at a stoichiometry of 4 nucleotides per monomer. Binding of Redβ to ssDNA has been observed to be cooperative (Smith and Bell, 2016), which would tend to favor oligomer formation, at any protein concentration. Nonetheless, the observation that Redβ is present at such low concentrations (<150 nM) in cells that are active for recombineering does call into question the relevance of in vitro experiments performed at higher protein concentrations.
Despite much effort, there is still no crystal structure available for Redβ, or any member of the RecT/Redβ superfamily. While initial sequence comparisons suggested that RecT and Redβ are not related to Rad52 (Iyer et al., 2002), three groups have since used threading approaches to conclude that the N-terminal domain of Redβ (residues 1–177) does have the same core fold as Rad52 (Erler et al., 2009; Lopes et al., 2010; Matsubara et al., 2013). However, the resulting sequence alignments differ considerably, making it difficult to map specific residues onto the structure. Domain analysis by limited proteolysis, chemical modification, and mass spectrometry identified six lysine residues of Redβ, all within the N-terminal domain, as being implicated in DNA binding (Wu et al., 2006). The C-terminal region of Redβ (residues 183–261) has recently been purified, and shown to form a folded, α-helical, monomeric domain (Smith and Bell, 2016). This C-terminal domain (CTD) shows no interaction with ssDNA or dsDNA, but forms the complex with λ exo. It has no detectable sequence homology with any other protein in the database, including other members of the RecT/Redβ superfamily. The linker connecting the two domains (residues 178–193) is likely to be flexible, such that the domains move independently of one another. When expressed separately, the two domains show no interaction, as judged by Ni-spin pull down. This type of domain structure (Fig. 6B), with a conserved N-terminal domain for oligomerization and DNA-binding, and a more variable C-terminal region, is common to many SSAPs, including Rad52 (Singleton et al., 2002), Erf (Poteete et al., 1983), Sak (Scaltriti et al., 2010), and many other RecT/Redβ homologs that we have purified (Smith & Bell, unpublished results).
In summary, our understanding of the mechanism of Redβ, and of SSAPs in general, is currently limited by a lack of structural information for the relevant DNA complexes. The range and inconsistency of the different types of oligomers that are formed is particularly bewildering, and whether or not the oligomers are functionally relevant, and if there is a conserved, unifying mechanism for SSAPs, are key outstanding questions.
5. Structure and mechanism of the Redβ – λ exo complex
While the individual functions of Redβ and λ exo have been well characterized, the role of the complex formed by the two proteins is not well established. The complex was first discovered by Radding and colleagues, who observed co-purification of Redβ and λ exo as a 1:1 complex from cell lysates (Radding et al., 1971). SynExo proteins with homology to Redβ and λ exo are often encoded as pairs in bacteriophage genomes, suggesting that their functions are coordinated. An interaction between RecE and RecT, the other well-studied SynExo system, has been demonstrated (Muyrers et al., 2000). The importance of the interaction was inferred from experiments showing that while λ exo and Redβ, or RecE and RecT could function similarly in recombination, the pairs of proteins were not interchangeable: Redβ could not function with RecE, and λ exo could not function with RecT.
The complex may serve to directly load Redβ onto the 3′-over-hang as it is being formed by λ exo. This would help to ensure that Redβ gets onto the ssDNA, before it is coated by E. coli single stranded DNA binding protein (SSB), and would be closely analogous to the loading of RecA onto the 3′ overhang generated by RecBCD (Anderson and Kowalczykowski, 1997). Based on this function, the 1:1 stoichiometry, and the fact that the interaction involves the Redβ CTD, we have developed an α3β3 hexamer model for the complex, as shown in Fig. 8. This model predicts a relatively transient interaction, such that as each monomer of Redβ is released onto the ssDNA, another monomer would take its place on the λ exo trimer, and so on. Consistent with the transient interaction posed by this model, a relatively weak interaction between λ exo and Redβ (Kd of 8 μM) has been measured by isothermal titration calorimetry (Subramaniam et al., 2016).
Fig. 8.
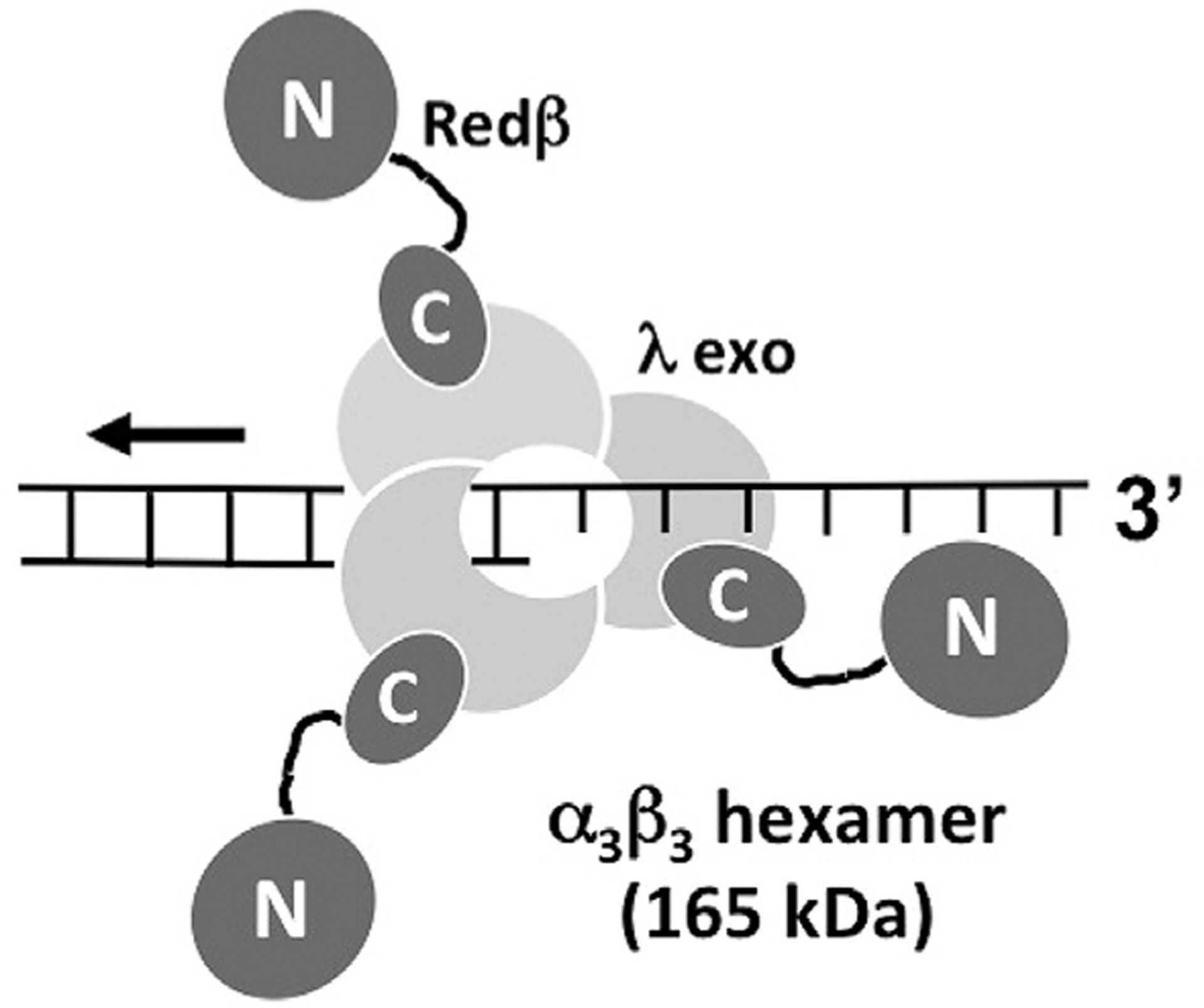
Model for the complex formed between λ exo and Redβ. According to the model, the λ exo trimer digests the 5′-strand of the duplex, moving from right to left, loading monomers of Redβ on to the 3′-overhang, one at a time. Redβ is depicted as a dumbbell structure, with an N-terminal domain for binding DNA, and a C-terminal domain for binding to λ exo.
However, the size of the complex is not yet clear. The α3β3 hexamer proposed above would have a molecular weight of 165 kDa. As measured by dynamic light scattering, however, the complex may be as large as 800 kDa (Tolun, 2007). Based on this measurement, the observed 1:1 stoichiometry, and the fact that the individual proteins form trimers (λ exo) and rings of 10–13 subunits (Redβ), a model was proposed in which a 12-mer ring of Redβ binds to four trimeric rings of λ exo (Tolun, 2007). This model tends to suggest a more stable “synaptasome” complex that could simultaneously resect two dsDNA ends and promote their annealing directly on the complex as the complementary regions between the strands became exposed. Whether or not such a complex exists at the physiological concentrations of the two proteins in the cell is unclear. Moreover, overexpression of the annealing protein relative to the exonuclease has been shown to favor recombination, suggesting that the 1:1 complex is not obligatory (Muyrers et al., 2000).
Recently, our group determined the crystal structure of a minimal λ exo–Redβ complex, consisting of the λ exo trimer bound to three copies of the Redβ CTD (Caldwell et al., 2019, Fig. 9). Since the N-terminal DNA-binding domain of Redβ shows no interaction with λ exo on its own, this minimal complex likely captures the essential features of the complex formed by the full-length proteins. The structure represents the first architectural information for a complex from a SynExo recombination system, and thus provides the first clues for how the interaction coordinates the annealing reaction. Notably, the structure shows that the Redβ CTD binds to the side of the λ exo trimer from which the 3′-overhang of a dsDNA substrate would emerge (Fig. 9B). This places the N-terminal DNA binding domains, connected by their flexible linkers, in perfect position to load onto the nascent 3′-overhang. Thus, the overall architecture of the complex is in perfect agreement with the loading mechanism that has been proposed.
Fig. 9.
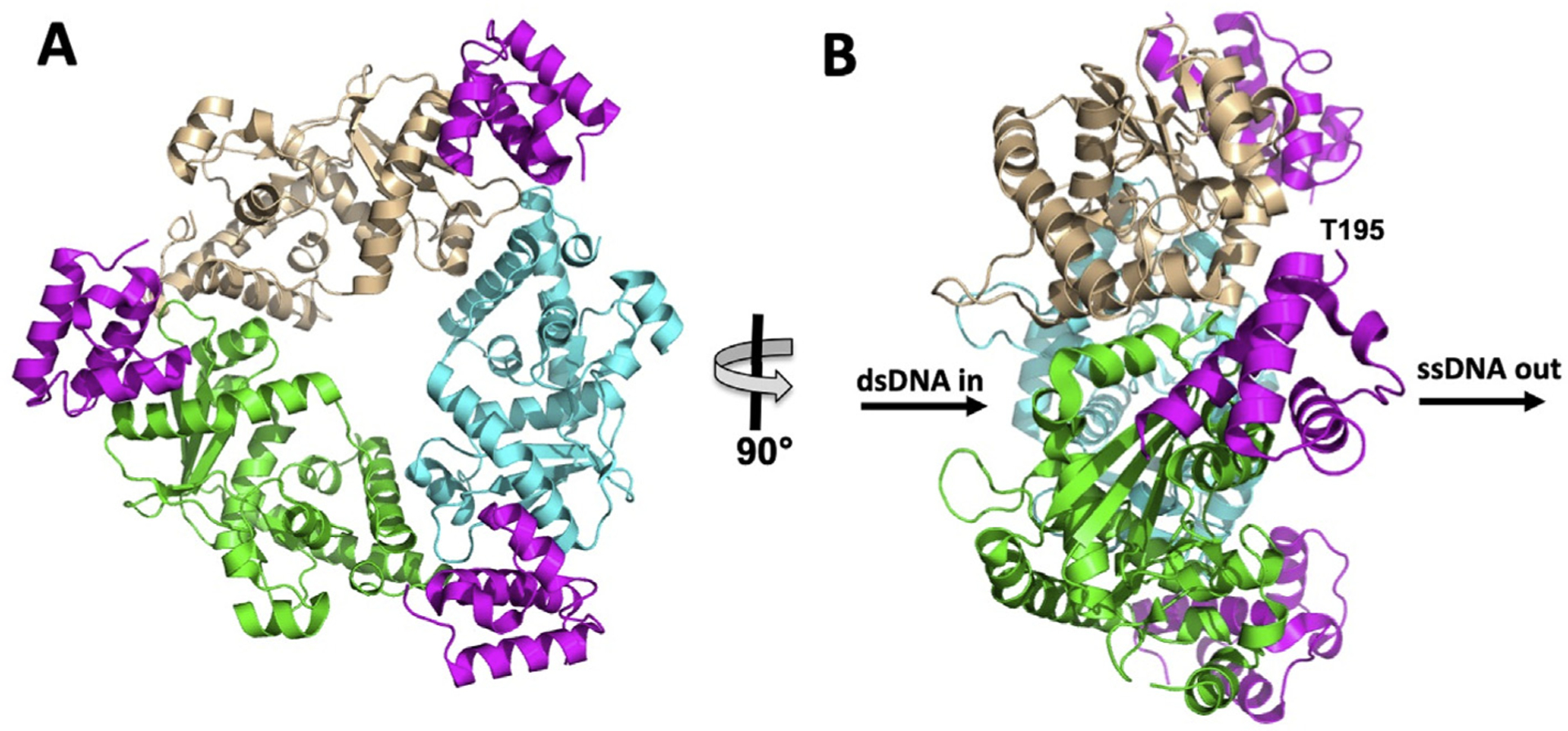
Structure of the minimal λ exo–Redβ complex (PDB code 6M9K; Caldwell et al., 2019). (A) View facing the back of the λ exo trimer, the side from which the 3′-overhang of the DNA would emerge. The three monomers of λ exo are colored wheat, cyan, and green, while the three Redβ CTDs (residues 195–260) are colored in magenta. (B) Side view showing that the three Redβ CTDs, and their DNA-binding domains, are positioned on the side of the trimer from which the 3′-overhang would emerge (indicated by “ssDNA out”). Residue Thr-195, to which the N-terminal DNA binding domain would connect, is labeled for the Redβ CTD in front, but similarly positioned for all three CTDs.
The structure also revealed the detailed atomic interactions at the interface of the two proteins, and thus provided a framework for mutational analysis, to test the role of the interaction in promoting recombination in vivo. Briefly, the interaction is formed by a patch of apolar residues exposed on the surface of each λ exo monomer, which binds to a hydrophobic pocket formed on the surface of each Redβ CTD. This central core of hydrophobic interactions is surrounded by a network of electrostatic interactions, including several hydrogen bonds and ion pairs. Mutational analysis revealed that the apolar residues at the hydrophobic core of the interaction are particularly important for stabilizing the complex, and further that the complex is indeed required for dsDNA recombination in vivo (Caldwell et al., 2019).
Curiously, the mutations that blocked formation of the complex were found to disrupt recombination involving both dsDNA and ssDNA targeting substrates. The latter result was unexpected, because ssDNA recombination doesn’t even require the presence of λ exo, let alone its interaction with Redβ, and the NTD of Redβ seems to posses all of the residues needed for DNA binding, oligomerization, and annealing in vitro (Wu et al., 2006). We hypothesized, therefore, that the site on the Redβ CTD that is used for binding to λ exo may overlap with another function of the protein, such as binding to an unknown host protein. But what could this unknown protein be?
The structure of the CTD itself provided an important clue. The CTD forms a small three-helix bundle, a seemingly simple and common fold. However a DALI search for structural homologs revealed a striking and unexpected homology with another protein from phage λ called Orf. Orf binds to DNA and to E. coli SSB to function as a recombination mediator, to allow RecA to gain access to SSB-coated ssDNA in the absence of RecFOR (Maxwell et al., 2005). By analogy with Orf, could Redβ use its Orf-like CTD to bind to SSB and mediate the annealing activity of the NTD? We tested for such an interaction, and found that indeed, the site on the CTD for binding to λ exo is also used for binding to SSB, specifically to its C-terminal tail, including its terminal Phe-177 residue (Caldwell et al., 2019). Moreover, mutational analysis indicated that binding to SSB is required for ssDNA (and dsDNA) recombination, as the mutations noted above that were defective for recombination also blocked binding to SSB. Altogether, these results suggest a dual role of the Redβ CTD, first in binding to λ exo to facilitate loading of Redβ directly onto the 3′-overhang as it is formed on the first DNA molecule, and second in binding to SSB to facilitate access of the 3′-overhang to SSB-coated ssDNA, likely at the lagging strand of the replication fork. This is a striking example of how discovering the fold of a protein (or protein domain) can provide critical new insights into its function.
6. Future directions
From the wealth of structural and biochemical data, we have a good understanding of how λ exo operates, but moving forward, the high-resolution crystal structure of the λ exo-DNA complex in a near catalytic configuration with two Mg2+ ions could be leveraged to gain deeper insights, such as with molecular dynamics simulations to analyze the energetics of the chemical mechanism, or with infrared spectroscopy to probe the role of electrostatics. By comparison, the Redβ SSAP still holds many secrets. Does it indeed share a common fold, and by extension a common mechanism with Rad52? Are both the ring and filament forms of Redβ involved in the mechanism of annealing, and if so, are these structures also used by other SSAPs? What are the residues of Redβ that contact the DNA in the different states that are relevant to the reaction (oligomeric rings and helical filaments) and to what extent are these different binding sites overlapping? Are these oligomeric structures even relevant to the mechanism of annealing, or do they simply reflect the interactions that occur between two adjacent monomers in the dimer clamping model? What is the architecture of the complex formed between λ exo and Redβ, and how exactly does it couple the two fundamental steps of the reaction, end-resection and annealing? The answers to these questions will certainly require high-resolution structural information, which has been difficult to achieve with x-ray crystallography. However, with the dramatic improvements in cryo-electron microscopy, the relevant structures will presumably be forthcoming. A better mechanistic understanding of the proteins behind the magic of Red recombineering will no doubt help to advance the utility of this system for applications in genome engineering, in particular with regard to expanding the methods to a wider variety of organisms for applications in systems biology and metabolic engineering.
Acknowledgements
This work was supported by the National Science Foundation [MCB-1616105 to C.E.B.] and the National Institutes of Health [T32GM118291 to B.J.C]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation or the National Institutes of Health.
References
- Ander M, Subramaniam S, Fahmy K, Stewart AF, Schöffer E, 2015. A single-strand annealing protein clamps DNA to detect and secure homology. PLoS Biol. 13 e1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DG, Kowalczykowski SC, 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90, 77–86. [DOI] [PubMed] [Google Scholar]
- Aparicio T, Jensen SI, Nielsen AT, de Lorenzo V, Martinez-Garcia E, 2016. The Ssr protein (T1E_1405) from Pseudomonas putida DOT-T1E enables oligonucleotide-based recombineering in platform strain P. putida EM42. Biotechnol. J 11, 1309–1319. 10.1002/biot.201600317. [DOI] [PubMed] [Google Scholar]
- Ayora S, Missich R, Mesa P, Lurz R, Yang S, Egelman EH, Alonso JC, 2002. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem 277, 35969–35979. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H, 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol 2 10.1038/msb4100050, 2006.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnéris C, Briggs LC, Savva R, Ebrahimi B, Barrett TE, 2011. Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff. Nucleic Acids Res. 39, 5744–5756. 10.1093/nar/gkr11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri EM, Muir P, Akhuetie-Oni BO, Yellman CM, Isaacs FJ., 2017. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell 171,1453–1467. 10.1016/j.cell.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson M, Géoui T, Flot D, Tarbouriech N, Ressing ME, Wiertz EJ, Burmeister WP, 2009. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J. Mol. Biol 391, 717–728. [DOI] [PubMed] [Google Scholar]
- Caldwell BJ, Zakharova E, Filsinger GT, Wannier TM, Hempfling JP, Chun-Der L, Pei D, Church GM, Bell CE, 2019. Crystal structure of the Redβ C-terminal domain in complex with λ exonuclease reveals and unexpected homology with λ Orf and an interaction with Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 10.1093/nar/gky1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DM, Radding CM, 1971. The role of exonuclease and β protein of phage λ in genetic recombination. II. Substrate specificity and the mode of action of λ exonuclease. J. Biol. Chem 246, 2502–2512. [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP, 2008. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494. 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Chen WY, Ho JW, Huang JD, Watt RM, 2011. Functional characterization of an alkaline exonuclease and single strand annealing protein from the SXT genetic element of Vibrio cholera. BMC Mol. Biol 12, 16. 10.1186/1471-2199-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL, 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet 2, 769–779. [DOI] [PubMed] [Google Scholar]
- Costantino N, Court DL, 2003. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants. Proc. Natl. Acad. Sci. U.S.A 100, 15748–15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlroth S-L, Gurmu D, Haas J, Erlandsen H, Nordlund P, 2009. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 276, 6636–6645. 10.1111/j.1742-4658.2009.07374.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL, 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL, 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379, 109–115. [DOI] [PubMed] [Google Scholar]
- Datta S, Costantino N, Zhou X, Court DL, 2008. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc. Natl. Acad. Sci. U.S.A 105, 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe M, Hutinet G, Son O, Amarir-Bouhram J, Schbath S, Petit M-A, 2014. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinases. PLoS Genet. 10 e100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Tao W, Gong F, Li Y, Zhang Y, 2014. A functional recT gene for recombineering of Clostridium. J. Biotechnol 173, 65–67. 10.1016/j.jbiotec.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Dou C, Yu M, Gu Y, Wang J, Yin K, Nie C, Zhu X, Qi S, Wei Y, Cheng W, 2018. Structural and mechanistic analyses reveal a unique Cas4-like protein in the mimivirus virophage resistance element system. iScience 3, 1–10. 10.1016/j.isci.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A, 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44. D289–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H, Gingery R, 1968. Mutants of bacteriophage λ defective for vegetative genetic recombination. J. Mol. Biol 34, 239–249. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Yu D, DiTizio T, Court DL, 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. U.S.A 98, 6742–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist LW, Skalka A, 1973. Replication of bacteriophage λ DNA dependent on the function of host and viral genes. J. Mol. Biol 75, 185–212. [DOI] [PubMed] [Google Scholar]
- Erler A, Wegmann S, Ellie-Caille C, Bradshaw CR, Maresca M, Seidel R, Habermann B, Muller DJ, Stewart AF, 2009. Conformational adaptability of Redβ during DNA annealing and implications for its structural relationship with Rad52. J. Mol. Biol 391, 586–598. [DOI] [PubMed] [Google Scholar]
- Fu J, Teucher M, Anastassiadis K, Skarnes W, Stewart AF, 2010. A recombineering pipeline to make conditional targeting constructs. Methods Enzymol. 477, 125–144. [DOI] [PubMed] [Google Scholar]
- Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, et al. , 2012. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol 30, 440–446. [DOI] [PubMed] [Google Scholar]
- Garriss G, Poulin-Laprade D, Burrus V, 2013. DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R291 family. J. Bacteriol 195, 1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Laakso LM, 2016. Dali server update. Nucleic Acids Res. 44, W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W, Yoo J, Lee Y, Park S, Hoang PL, Cho H, Yu J, Hoa VT, Shin M, Jin MS, Park D, Hyeon C, Lee G, 2018. Dynamic coordination of two-metalions orchestrates λ-exonuclease catalysis. Nat. Commun 9, 4404. 10.1038/s41467-018-06750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L, 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF, and RAD52. BMC Genomics 3 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA, 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol 31, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chang JH, Kilic T, Tong L, Kiledgian M, 2013. A mammalian pre-mRNA 5’-end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell 50, 104–115. 10.1016/j.molcel.2013.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Coyle SM, Doudna JA, 2011. Coupled 5’ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600. 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S, 2002. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol. Cell 10, 359–371. [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kagawa A, Saito K, Ikawa S, Shibata T, Kurumizaka H, Yokoyama S, 2008. Identification of a second DNA binding site in the human Rad52 protein. J. Biol. Chem 283, 24264–24273. 10.1074/jbc.M802204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakousis G, Ye N, Li Z, Chiu SK, Reddy G, Radding CM, 1998. The beta protein of phage λ binds preferentially to an intermediate in DNA renaturation. J. Mol. Biol 276, 721–731. 10.1006/jmbi.1997.1573. [DOI] [PubMed] [Google Scholar]
- Karu AE, Sakaki Y, Echols H, Linn S, 1975. The γ protein specified by bacteriophage λ. Structure and inhibitory activity for the recBC enzyme of Escherichia coli. J. Biol. Chem 250, 7377–7387. [PubMed] [Google Scholar]
- Kmiec E, Holloman WK, 1981. β protein ofbacteriophage λ promotes renaturation of DNA. J. Biol. Chem 256, 12636–12639. [PubMed] [Google Scholar]
- Kolodner R, Hall SD, Luisi-DeLuca C, 1994. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol. Microbiol 11, 23–30. [DOI] [PubMed] [Google Scholar]
- Kovall R, Matthews BW, 1997. Toroidal structure of λ-exonuclease. Science 277, 1824–1827. [DOI] [PubMed] [Google Scholar]
- Krajewski WW, Fu X, Wilkinson M, Cronin NB, Dillingham MS, Wigley DB, 2014. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature 508, 416–419. 10.1038/nature13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A, 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Gregg CJ, Mosberg JA, Washington GC, Church GM, 2012. Manipulating replisome dynamics to enhance lambda Red-mediated multiplex genome engineering. Nucleic Acids Res. 40 10.1093/nar/gks751 e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ, 2013. Genomically recoded organisms expand biological functions. Science 342, 357–360. 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG, 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAD DNA. Genomics 73, 56–85. [DOI] [PubMed] [Google Scholar]
- Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W, 2005. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell 20, 155–166. 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JH, 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536,96–99. 10.1038/nature18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Patschull AOM, Bagnéeris C, Ryan H, Sanderson CM, Ebrahimi B, Nobeli I, Barret TE, 2017. KSHV SOX mediated host shutoff: the molecular mechanism underlying mRNA transcript processing. Nucleic Acids Res. 45, 4756–4767. 10.1093/nar/gkw1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros I, Timmins J, Hall DR, McSweeney S, 2005. Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 24, 906–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak S, Beloglazova N, Nocek B, Skarina T, Flick R, Brown G, Popovic A, Joachimiak A, Savchenko A, Yakunin AF, 2013. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc 135, 17476–17487. 10.1021/ja408729b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Trotta CR, Abelson J, 1998. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280, 279–284. [DOI] [PubMed] [Google Scholar]
- Little JW, 1967. An exonuclease induced by bacteriophage λ II. Nature of the enzymatic reaction. J. Biol. Chem 242, 679–686. [PubMed] [Google Scholar]
- Lopes A, Amarir-Bouhram J, Faure G, Petit M-A, Guerois R, 2010. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51, and Gp2.5 remote homologs. Nucleic Acids Res. 38, 3952–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Koroleva O, Bera S, Grandgenett DP, Korolev S, 2004. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure 12, 1881–1889. [DOI] [PubMed] [Google Scholar]
- Makhov AM, Sen A, Yu X, Simon MN, Griffith JD, Egelman EH, 2009. The bipolar filaments formed by herpes simplex virus type SSB/recombination protein (ICP8) suggest a mechanism for DNA annealing. J. Mol. Biol 386, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF, 2010. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol. Biol 11, 54. 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Jiménez MI, Alonso JC, Ayora S, 2005. Bacillus subtilis bacteriophage SPP1-encoded gene 341 product is a recombination-dependent DNA replication protein. J. Mol. Bol 351, 1007–1019. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Malay AD, Curtis FA, Sharples GJ, Heddle JG, 2013. Structural and functional characterization of the Redβ recombinase from bacteriophage λ. PLoS One 8, e78869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL, Reed P, Zhang RG, Beasley S, Walmsley AR, Curtis FA, Joachimiak A, Edwards AM, Sharples GJ, 2005. Functional similarities between phage λ Orf and Escherichia coli RecFOR in initiation of genetic exchange. Proc. Natl. Acad. Sci. U.S.A 102, 11260–11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbantenkhu M, Wang X, Nardozzi JD, Wilkens S, Hoffman E, Patel A, Cosgrove MS, Chen XJ, 2011. Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J. Biol. Chem 286, 42360–42370. 10.1074/jbc.M111.307512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsis PG, Kwagh JG, 1999. Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Res. 27, 3057–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosberg JA, Lajoie MM, Church GM, 2010. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics 186, 791–799. 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM, 2012. Improving lambda red genome engineering in Escherichia coli via rational removal of endogenous nucleases. PLoS One 7, 344638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa K, Radding CM, 1986. The homologous recombination system of phage λ. Pairing activities of β protein. J. Biol. Chem 261, 7472–7478. [PubMed] [Google Scholar]
- Murphy KC, 2012. Phage recombinases and their applications. Adv. Virus Res 83, 367–414. [DOI] [PubMed] [Google Scholar]
- Murphy KC, 2016. λ recombination and recombineering. EccoSal Plus 7. ESP-0011–2015. [DOI] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG, Poteete AR, 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246, 321–330. [DOI] [PubMed] [Google Scholar]
- Muyrers JPP, Zhang Y, Buchholz F, Stewart AF, 2000. RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 14, 1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Stewart AF, 2001. Techniques: recombinogenic engineering-new options for cloning and manipulating DNA. Trends Biochem. Sci 26, 325–331. [DOI] [PubMed] [Google Scholar]
- Mythili E, Kumar KA, Muniyappa K, 1996. Characterization of the DNA-binding domain of β protein, a component of phage λ Red-pathway, by UV catalyzed cross-linking. Gene 182, 81–87. [DOI] [PubMed] [Google Scholar]
- Nakae S, Hijikata A, Tsuji T, Yonezawa K, Kouyama KI, Mayanagi K, Ishino S, Ishino Y, Shirai T, 2016. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure 24, 1960–1971. 10.1016/j.str.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Newman M, Lunnen K, Wilson G, Greci J, Schildkraut I, Phillips SEV, 1998. Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J. 17, 5466–5476. 10.1093/emboj/17.18.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K, 2001. Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition. Structure 9, 197–204. [DOI] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SS, Crouch RJ, Yang W, 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Pan X, Smith CE, Zhang J, McCabe KA, Fu J, Bell CE, 2015. A structure-activity analysis for probing the mechanism of processive double-stranded DNA digestion by λ exonuclease trimers. Biochemistry 54, 6139–6148. [DOI] [PubMed] [Google Scholar]
- Parsons CA, Baumann P, Van Dyck E, West SC, 2000. Precise binding of single-stranded DNA termini by human RAD52 protein. EMBO J. 19, 4175–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Egelman EH, 1999. Rings and filaments of β protein from bacteriophage λ suggest a superfamily of recombination proteins. Proc. Natl. Acad. Sci. U.S.A 96, 4279–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins TT, Dalal RV, Mitsis PG, Block SM, 2003. Sequence-dependent pausing of single lambda exonuclease molecules. Science 301, 1914–1918. 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines G, Freed EF, Winkler JD, Gill RT, 2015. Bacterial recombineering: genome engineering via phage-based homologous recombination. ACS Synth. Biol 4, 1176–1185. [DOI] [PubMed] [Google Scholar]
- Pingoud A, Fuxreiter M, Pingoud V, Wende W, 2005. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci 62, 685–707. 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson Y-Y, 2008. Functional and structural basis for a bacteriophage homolog ofhuman Rad52. Curr. Biol 18,1142–1146. 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, 2001. What makes the bacteriophage λ Red system useful for genetic engineering: molecular mechanism and biological function. FEMS Microbiol. Lett 201, 9–14. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC, 1993. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages λ and P22. Genetics 134,1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR, Sauer RT, Hendrix RW, 1983. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J. Mol. Biol 171, 401–418. [DOI] [PubMed] [Google Scholar]
- Radding CM, Rosenweig J, Richards F, Cassuto E, 1971. Separation and characterization of exonuclease, β protein, and a complex of both. J. Biol. Chem 246, 2510–2512. [Google Scholar]
- Reuven NB, Staire AE, Myers RS, Weller SK, 2003. The herpes simplex virus type λ alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol 77, 7425–7433. 10.1128/JVI.77.13.7425-7433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome M, Saito K, Yasuda T, Ohtomo H, Sugiyama S, Nishimura Y, Kurumizaka H, Kagawa W, 2018. Structural basis of homology-directed DNA repair mediated by RAD52. iScience 3, 50–62. 10.1016/j.isci.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Costantino N, Li XT, Thomason LC, Bubunenko M, Court C, Court DL, 2011. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J. Mol. Biol 407, 45–59. 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaltriti E, Moineau S, Launay H, Masson JY, Rivetti C, Ramoni R, Campanacci V, Tegoni M, Cambillau C, 2010. Deciphering the function of lactococcal phage ul36 Sak domains. J. Struct. Biol 170, 462–469. 10.1016/j.jsb.2009.12.02. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Thomason LC, Kuznetsov SG, Court DL, 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc 4, 206–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer ER, Weil J, 1968. Recombination in bacteriophage λ I. Mutants deficient in general recombination. J. Mol. Biol 34, 261–271. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB, 2002. Structure of the single-strand annealing domain of human RAD52 protein. Proc. Natl. Acad. Sci. U.S.A 99, 13492–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wiley DB, 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. , 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 471, 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Bell CE, 2016. Domain structure of the Redβ single-strand annealing protein: the C-terminal domain is required for fine-tuning DNA-binding properties, interaction with the exonuclease partner, and recombination in vivo. J. Mol. Biol 428, 561–578. [DOI] [PubMed] [Google Scholar]
- Stahl FW, 1998. Recombination in phage λ: one geneticisťs historical perspective. Gene 223, 95–102. [DOI] [PubMed] [Google Scholar]
- Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW, 1997. Annealing vs. invasion in phage λ recombination. Genetics 147, 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, Stasiak A, Müller S, Engel A, Van Dyck E, West SC, Egelman EH, 2000. The human Rad52 protein exists as a heptameric ring. Curr. Biol 10, 337–340. [DOI] [PubMed] [Google Scholar]
- Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K, 2012. Sequence, structure, and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 40, 7016–7045. 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Erler A, Fu J, Kranz A, Tang J, Gopalswamy M, Ramakrishnan S, Keller A, Grundmeier G, Müller D, Sattler M, Stewart AF, 2016. DNA annealing by Redβ is insufficient for homologous recombination and the additional requirements involve intra- and inter-molecular interactions. Sci. Rep 6, 34525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K, Rutvissuttinunt W, Scott W, Myers RS, 2003. The enzymatic basis of processivity in λ exonuclease. Nucleic Acids Res. 15,1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiman-Marangos SN, Peel JK, Weiss YM, Ghirlando R, Junop MS, 2013. Crystal structure of the DdrB/ssDNA complex from Deinococcus radiodurans reveals a DNA binding surface involving higher-order oligomeric states. Nucleic Acids Res. 41, 9934–9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiman-Marangos SN, Weiss YM, Junop MS, 2016. Mechanism for accurate, protein-assisted DNA annealing by Deinococcus radiodurans DdrB. Proc. Natl. Acad. Sci. U.S.A 113, 4308–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle B, Bao Z, Markel E, Chambers A, Cartinhour S, 2010. Recombineering using RecTE from Psudomonas syringae. Appl. Environ. Microbiol 76, 4960–4968. 10.1128/AEM.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanska AK, 2009. Bacteriophage-encoded functions engaged in initiation of homologous recombination events. Crit. Rev. Microbiol 35, 197–220. 10.1080/10408410902983129. [DOI] [PubMed] [Google Scholar]
- Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL, 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol 106, 1–30. 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- Thresher RJ, Makhov AM, Hall SD, Kolodner R, Griffith JD, 1995. Electron microscopic visualization of RecT protein and its complexes with DNA. J. Mol. Biol 254, 364–371. 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- Tolun G, 2007. More than the Sum of its Parts: Physical and Mechanistic Coupling in the Phage Lambda Red Recombinase. Ph.D. thesis. University of Miami, FL. [Google Scholar]
- Tolun G, Makhov AM, Ludtke SJ, Griffith JD, 2013. Details of ssDNA annealing revealed by an HSV-1 ICP8-ssDNA binary complex. Nucleic Acids Res. 41, 5927–5937. 10.1093/nar/gkt266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valledor M, Myers RS, Schiller PC, 2018. Herpes ICP8 protein stimulates homologous recombination in human cells. PLoS One 13. 10.1371/journal.pone.0200955 e0200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Hatfull GF, 2007. Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152. 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS, 2003. Single-moledule kinetics of λ exonuclease reveal base dependence and dynamics of disorder. Science 301, 1235–1238. 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- van Pijkeren J-P, Britton RA, 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 40, e76. 10.1093/nar/gks14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellani TS, Myers RS, 2003. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J. Bacteriol 185, 2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM, 2009. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Persky NS, Yoo B, Ouerfelli O, Smogorzewska A, Elledge SJ, Pavletich NP, 2014. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science 346, 1127–1130. 10.1126/science.1258973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Jiao X, Kiledjian M, Tong L, 2015. Structural and biochemical studies of distinct activity profiles of Rai1 enzymes. Nucleic Acids Res. 43, 6596–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Z, Jia R, Hou Y, Yin J, Bian X, Li A, Müller R, Stewart AF, Fu J, Zhang Y, 2016. RecET direct cloning and Redαβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nat. Protoc 11, 1175–1190. [DOI] [PubMed] [Google Scholar]
- Wang H, Li Z, Jia R, Yin J, Li A, Xia L, Yin Y, Müller R, Fu J, Stewart AF, Zhang Y, 2018. ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct cloning from complex genomes. Nucleic Acids Res. 46, e28. 10.1093/nar/gkx1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M, Troman L, Wan Nur Ismah WA, Chaban Y, Avison MB, Dillingham MS, Wigley DB, 2016. Structural basis for the inhibition of RecBCD by Gam and its synergistic antibacterial effect with quinolones. Elife 5, e22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xing X, Wisler JW, Dalton JT, Bell CE, 2006. Domain structure and DNA binding regions of β protein from bacteriophage λ. J. Biol. Chem 281, 25205–25214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lee JY, Nowotny M, 2006. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13. 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Yang W, Chen WY, Wang H, Ho JW, Huang JD, Woo PC, Lau SK, Yuen KY, Zhang Q, Zhou W, Bartlam M, Watt RM, Rao Z, 2011. Structural and functional insight into the mechanism of an alkaline exonuclease from Lar-ibacter hongkongensis. Nucleic Acids Res. 39, 9803–9819. 10.1093/nar/gkr660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang J, Qi Q, 2015. Prophage recombinases-mediated genome engineering in Lactobacillus planatarum. Mocriob. Cell Fact 14, 154. 10.1186/s12934-015-0344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wu R, Liu H, Chen Y, Gao Y, Chen X, Li Y, Ma J, Li J, Gan J, 2018. Structural insights into DNA degradation by human mitochondrial nuclease MGME1. Nucleic Acids Res. 46, 11075–11088. 10.1093/nar/gky855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Zhu H, Xia L, Ding X, Hoffmann T, Hoffmann M, Bian X, Müller R, Fu J, Stewart AF, Zhang Y, 2015. A new recombineering system for Photorhabdus and Xenorhabdus. Nucleic Acids Res. 43, e36. 10.1093/nar/gku1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL, 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Buckholz F, Muyrers JPP, Stewart AF, 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet 20, 123–128. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JPP, Testa G, Stewart AF, 2000. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol 18, 1314–1317. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JP, Rientjes J, Stewart AF, 2003. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol. Biol 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xing X, Herr AB, Bell CE, 2009. Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks. Structure 17, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McCabe KA, Bell CE, 2011. Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U.S.A 108, 11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Pan X, Bell CE, 2014. Crystal structure of λ exonuclease in complex with DNA and Ca(2+). Biochemistry 53, 7415–7425. [DOI] [PubMed] [Google Scholar]
- Zhou C, Pourmal S, Pavletich NP, 2015. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife 4, e09832. 10.7554/eLife.09832. [DOI] [PMC free article] [PubMed] [Google Scholar]


