ABSTRACT
Background: Rapid Lateral Flow Test (LFT) has been broadly utilized in detection or diagnosis of numerous disease-related antigens and antibodies. It is the most popular format of point-of-care test (POCT) and quickest and easiest way to detect a targeted molecule. In the combat against COVID-19 pandemic, hundreds of POCTs have been developed and are commercially available now. They are designed to detect either a SARS-CoV-2 viral antigen or IgG and IgM antibodies binding to it. Among the binding antibodies, a special type of functional antibodies that block the interaction between SARS-CoV-2 virus and its human receptor, neutralizing antibodies (NAbs), are of particular interest to public as well as in vaccination management. However as of today, POCTs for the detection of SARS-CoV-2 NAbs remain under late stage of development.
Scope and method:In this review, we first summarize the importance of awareness and monitoring of SARS-CoV-2 NAbs in the combat against COVID-19 pandemic. Secondly, we compare the available methods for the detection of SARS-CoV-2 NAbs. Next, we describe challenges in the development of a rapid lateral flow test for the detection of SARS-CoV-2 NAbs. Finally, we outline its product formats and applications in research and in disease management. Conclusion:Vaccine effectiveness is unknown for an individual unless measured. NAb level is the most viable measurement for vaccine effectiveness or immunity. A broadly accessible NAb POCT is urgently needed.
KEYWORDS: Lateral flow test, point-of-care, diagnostics, SARS-CoV-2, neutralizing antibodies
1. INTRODUCTION
1.1. What is SARS-CoV-2 neutralizing antibody (NAb)?
Antibodies against SARS-CoV-2 can be generally divided into two main categories, neutralizing antibodies (NAbs) and non-neutralizing virus binding antibodies (BAbs). BAbs can be induced to all protein components of the SARS-CoV-2 virus including spike (S) protein, nucleocapsid (N) protein, envelope (E) protein, and membrane (M) protein. The most abundant N protein and antibodies against it are often the detection target in commercial tests to identify SARS-CoV-2 infected individuals, such as, Abbott’s BinaxNOW™ COVID-19 Ag Card[1] and Abbott’s SARS-CoV-2 IgG Architech[2]. In contrast, SARS-CoV-2 NAbs can be raised only against S protein. This is because SARS-CoV-2 virus invades its host via interaction of its S protein with ACE-2 protein on the surface of host cells [3,4]. All of the vaccines in the market or under development contain SARS-CoV-2 S protein or S protein-encoding gene. The capability of inducing long-lasting and high titer NAbs is one of the most important criteria in predicting the success of a SARS-CoV-2 vaccine.
To understand SARS-CoV-2 NAb, we first need to understand structure and immunogenicity of its S protein. There are a total of five functional domains on S protein which are involved in the virus invasion process to its host cells, N-terminal domain (NTD, 14–305 residues) and receptor-binding domain (RBD, 319–541 residues) on S1 subunit and the fusion peptide (FP, 788–806 residues), heptapeptide repeat sequence 1 (HR1, 912–984 residues), HR2 (1163–1213 residues) on S2 subunit. NTD and RBD are responsible for receptor binding and the three domains on S2 responsible for membrane fusion [5] (Figure 1). Theoretically any antibodies binding to these five domains may possibly interrupt the interaction between the virus and host cell. In reality, only a subset of binding antibodies shows neutralization function. McCallum, M. et al. found that among 41 monoclonal BAbs to NTD, only 15 showed neutralization function [6]. Ju, B. et al. reported 7 out of 16 RBD binding antibodies neutralized a live SARS-CoV-2 isolate [7]. Liu, L. et al. isolated 121 monoclonal antibodies binding to S trimer from 5 severe COVID-19 patients and found 61 capable of neutralization[8]. Among all domains of S protein, RBD seems to be most immunogenic and induces majority of specific antibodies in patients. For instance, Piccoli, L. et al. tested antibodies against different SARS-CoV-2 proteins and different domains of S protein from 647 SARS-CoV-2-infected subjects and found that SARS-CoV-2 RBD-specific Abs dominated IgG responses whereas much lower titers were observed to the S2 subunit and the majority of the neutralizing activity (90%) against SARS-CoV-2 is mediated by RBD-specific Abs interfering with binding to ACE2[9]. In an analysis of 278 monoclonal antibodies isolated from three COVID-19 patients, 64.6%-76.5% of them are RBD specifc[6]. The data from the above literatures are summarized in the embedded table of Figure 1.
Figure 1.
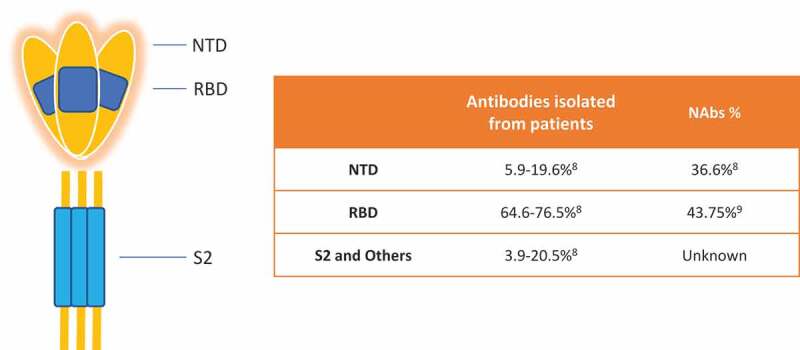
An illustrative SARS-CoV-2 S protein structure and a summary about binding and neutralizing antibodies against different domains. RBD is dominant in inducing both binding and neutralizing antibodies
SARS-CoV-2 NAbs binding to RBD isolated from convalescent patients have been grouped into four classes based on the binding site and conformation of RBD [10,11]. A supersite within NTD recognized by multiple potent NAbs has also been discovered[6]. One of these NTD NAb provided prophylactic protection against lethal SARS-CoV-2 challenge of Syrian hamsters[6]. Recently, Liu, L. et al. reported that they did not isolate any RBD binding antibodies from 1 of the 5 COVID-19 patients with high NAb titer[8]. They isolated 13 S trimer binding antibodies from this patients, but none of the antibodies bind to RBD. Seven of the 13 non-RBD binders are neutralizing antibodies and 2 of them are potent NAb targeting NTD. The authors did not disclose where the other five NABs bind to. This is the first indication that RBD-based NAb test may generate false-negative result for this type of individuals. Three domains in S2 involve in cell fusion, an important role in viral invasion and therefore it is likely that the cell fusion be interrupted by certain antibodies. However, antibody response to S2 subunit has been so far rarely studied possibly because it is less immunogenic compared to RBD and NTD and much less S2 antibodies have been isolated.
1.2. Why is important to measure SARS-CoV-2 NAbs?
We all know that the most important task in combating COVID-19 pandemic is to produce enough vaccine and vaccinate as many people as possible within a time frame. Nevertheless, the goal of this worldwide effort should be aligned to raise protective level of neutralizing antibodies in vaccine recipients. It is clear that the neutralizing antibodies can block a viral invasion at the initial access to human receptors. The cellular immunity, another important part of immunity, plays an important role after an individual is infected, but not at the first encounter to the receptors. In a foreseeable future, a measurable protective immunity for COVID-19 will be established, by a like hood of a NAb test quantifying the NAb levels. However, this process is likely to be quite challenging. First, a correlation between a protection against infection and NAb level quantified by an acceptable standard method needs to be established through large-scale clinical trials. Second, the practical and comparable NAb measurement methods need to be developed to meet the global demands. Obviously, large-scale collaborations are needed to achieve this goal.
Protection rate, efficacy, or effectiveness varies from vaccine to vaccine and also depends on the age of the recipients and even where they live. The protection rate ranges from Pfizer’s 95% to AstraZeneca’s 70%. The clinical trial data of Johnson & Johnson’s vaccine revealed that it is 72% effective in the US, 66% in Latin America and 57% in South Africa, 28 days after vaccination, 85% effective overall in preventing severe disease[12]. A key question for the vaccine recipients is which group you belong to: the 95% or the 5% if you received Pfizer’s vaccine, or the 72% or 28% if you receive a J&J vaccine. Additional unanswered key question for everyone is how long such protection will last. A continuous monitoring of NAb level is needed to address this question. You can only answer these questions by testing your NAb level. In fact, a more important question is whether it is necessary to know the answer. We will address this question in the last section of this review.
1.3. What methods are available to measure NAbs?
In order to measure NAb against a viral-to-receptor binding, a binding pair materials or molecules need to be provided, either in the forms of live virus plus its targeted host cells or a virus surface protein plus its receptor on a host cell in the case the viral infection mechanism at molecular level is well known. In this section, we will review the various methods utilized in detection of SARS-CoV-2 NAbs and provide a comparison in Table 1.
Table 1.
A comparison of different SARS-CoV–2 neutralizing antibody tests
| NAb Test | Assay time | Biosafety Level | S domain targeted | Pros | Cons |
|---|---|---|---|---|---|
| Plaque reduction neutralization test (PRNT) | Several days | 3 | Binding and fusion | Gold standard, assay condition is close to real situation, | Very slow, complicated assay leading to high variation |
| Pseudovirus neutralization test | Several days | 2 | Binding and fusion | More accessible and higher sensitivity than PRNT | Still very slow and complicated, but better than PRNT |
| ELISA | Several hours | 1 | NTD and RBD | Simple system, high throughput, high sensitivity | Unable to measure fusion blocking antibody |
| Lateral Flow | 15 min | 0-1 | NTD and RBD | Most accessible, fastest, simple, can be used outside a lab | Same as ELISA |
The most classic method is ‘plaque reduction neutralization test’ which is considered gold standard for neutralizing antibody measurement [13,14]. This test was developed from Viral Plaque Assay [15], an assay for quantitation of infectious viral particles. The assay result is the number of plaque-forming units (pfu) in a virus sample, and a pfu or a viral plaque is an infected area including multiple lysed cells in a monolayer of host cells covered with a semi-solid medium in petri dishes or multi-well plates and can be examined with an optical microscope. The viral plaques usually take 3–14 days to form and are generally counted manually. Engineered SARS-CoV-2 virus has been used to avoid manual counting and to improve assay throughput[16]. The concentration of a serum sample to reduce the number of plaques by 50% is used to determine the neutralizing capability of a serum or plasma and denoted as the PRNT50 value which is equivalent to the common in use 50% of Inhibition Concentration or IC5013. Clearly, this assay is quite cumbersome and labor-intensive and requires Biosafety Level 3 working condition. The most troublesome issue is that the live and infectious SARS-CoV–2 virus has to be included in the test to measure neutralizing antibodies against it[17].
Several virus neutralization tests using pseudovirus [18–23] have been developed to avoid the use of highly infectious SARS-CoV-2 virus. A pseudovirus is a nonpathogenic virus engineered to express spike protein of SARS-CoV-2. Certain molecules that help in signal readout, such as GFP, Luciferase, are often co-engineered into pseudovirus [24]. The signals of these pseudovirus-based neutralization tests can be measured by an instrument in a higher throughput manner. Meanwhile on the other side, engineered host cells with high level of ACE2 expression are commonly seen in literature and commercial products [21]. These tests only need a biosafety level-1 or −2 facility [19] which are more accessible to biomedical researchers.
There are several common issues with virus neutralization tests using live virus regardless of using a pseudovirus or SARS-CoV-2 virus. In these tests, the neutralization ability of the antibodies is highly dependent on the maturation state or titer of virus and the cell type and cell condition used in the assay [25]. Poor reproducibility or even false results can be generated if the virus and host cells are not at optimal assay conditions [25]. Poor correlation between them is not uncommon[8].
Because the recombinant SARS-CoV-2 S protein (and RBD) and recombinant ACE-2 protein became vastly available just a few months after the pandemic started, and because of the quick discovery of the high binding affinity (Kd (M): 1.52x10–[7] to 7.7 × 10−9 dependent on the methods used [3,26–28]) in vitro between the two molecules, a simpler and faster enzyme-linked immunosorbent assay (ELISA) that detects SARS-CoV-2 NAbs based on ACE2-RBD interaction can be readily developed. Multiple ELISA tests for SARS-CoV–2 NAbs have been developed and reported [29–35]. Most of these tests use a recombinant RBD protein or ACE2 as coating antigen and horseradish peroxidase (HRP) labeled ACE2 or HRP labeled RBD to generate detection signal [31–35]. Among them, only one has obtained the status of In Vitro Diagnostic Use Under FDA Emergency Use Authorization as of March 2021[31]. These ELISA tests can be completed within a few hours in a Biosafety Level 1 or 2 environment.
Utilizing a similar principle and the same set of recombinant proteins as used in ELISA, lateral flow tests have been developed to detect SARS-CoV-2 NAb [30,36]. Currently, they are still in late development stage and are not commercially available.
Different set of SARS-CoV-2 NAbs will be measured by different neutralization tests depending upon the number of domains on S protein involved.
2. Rapid lateral flow test for detection of SARS-CoV-2 NAb
The lateral flow test (LFT) is the fastest and most convenient test among the popular immunoassays, which typically takes only 15 minutes to complete. It can be performed either in a professional laboratory or by an individual at home, thus if available should be a necessary complement to the existing NAb tests and meanwhile play an unreplaceable role in the management of personal life.
2.1. A special type of inhibition test
A lateral flow test typically uses antibody–antigen interaction as test principle like other immunoassays. Usually, higher antibody affinity results in higher assay sensitivity. High-affinity antibody to an antigen can be obtained through modern antibody development technologies nowadays. Antibody production cost can be well managed with available mature manufacturing processes. Protein–protein interaction without antibody can also be used in lateral flow test[37]. However, it is quite rare, simply because the binding affinity between proteins is usually much lower than that between an antibody and an antigen and is impossible to increase. Lower binding affinity or lower assay sensitivity often leads to much longer assay time and use of high sensitivity detection system, such as fluorescent labeling and fluorescence detection instrument[37]. Manufacturing cost of a recombinant protein is usually much higher than that of an antibody.
The interaction between RBD and ACE2 or between S protein and ACE2 must be included to develop a lateral flow inhibition test. One of these recombinant proteins needs to be conjugated onto gold nanoparticle (GNP) or nanoshell and the other needs to be embedded into nitrocellulose membrane. This is a rare protein–protein interaction LFT. Luckily, SARS-CoV-2 RBD has a very high binding affinity to ACE2 and therefore traditional format of LFT using GNP or gold nanoshell works well [30,36]. NAb in a sample will neutralize or block the interaction between, for example, RBD-GNP and ACE2 in such an assay, diminishing or disappearing the visible GNP signal (Figure 2)[30]. Our internal comparison showed that only recombinant proteins produced from mammalian cells provided reasonable signal intensity and product stability.
Figure 2.
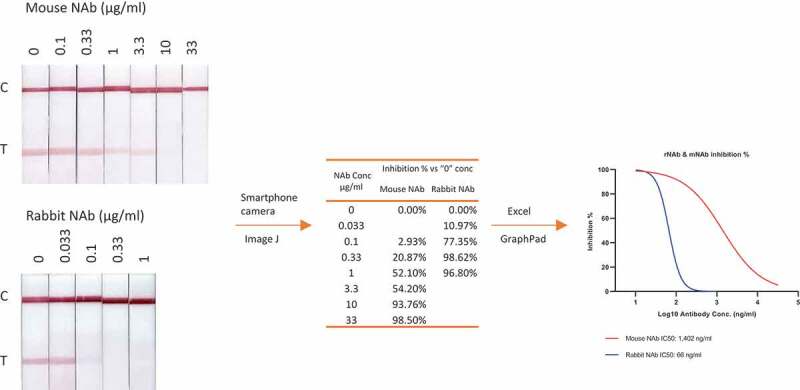
Measurement of inhibitory concentration of two monoclonal neutralizing antibodies using 60 nm GNP labeled RBD with a rabbit Fc tag. A workflow from visible LFT results to IC50 calculation is illustrated, including capturing the visible bands with a smartphone, converting the image to digital signal with ImageJ software, calculating IC50 and generating inhibition curves with GraphPad
Although an LFT using the protein pair RBD and ACE2 works well in certain applications, such as evaluation of RBD binding NAbs (Figure 2), it is not ideal to examine specimens from COVID-19 patients and vaccine recipients. As mentioned earlier in this review, there are COVID-19 patients who only developed NAbs binding to non-RBD domains[8]. To avoid potential false-negative result, S or S1 protein should be used to replace RBD. Fortunately, LFT using S1-ACE2 proteins worked as good as RBD-ACE2 as shown in Figure 3, which shows comparable results among LFT, a NAb ELISA, and a pseudovirus-based neutralization assay.
Figure 3.
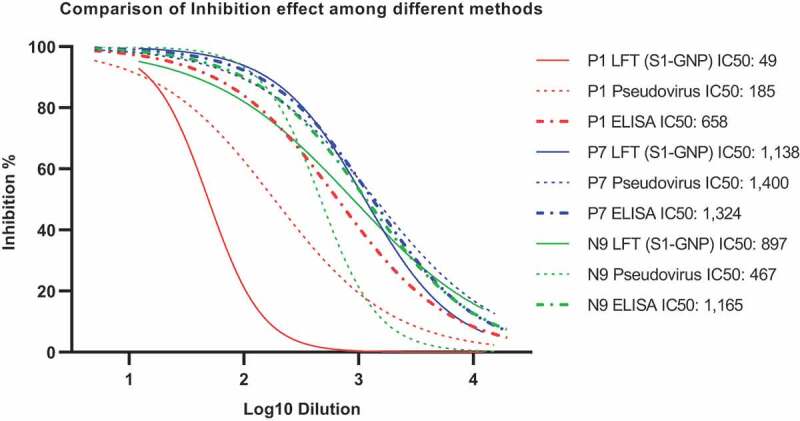
A comparison of three different inhibition tests, LFT, an in-house NAb ELISA and a pseudovirus-based neutralization test. Three plasma from COVID-19 patients were used for the comparison, one with moderate level of NAb and two with higher level of NAb. The same workflow as shown in Figure 2 legend was used to generate IC50 values and inhibition curves. Results are generally comparable
2.2. Product format
A neutralization assay in either ELISA format or PRNT format usually measure multiple dilutions of test samples in triplicate in order to calculate IC50, EC50, or PRNT50. Although cumbersome and costly, it can be easily done in a professional laboratory. A LFT can be used in the same way (Figures 2 and 3), but for an LFT aiming to serve individual users at home, it is impossible to accurately prepare different dilutions of a blood sample outside a professional lab. A single dilution has to be the choice and this dilution must cover the protection titer of NAb. However, a consensus for the protection titer is still lacking.
The clues for the protection titer can be found from some FDA guidance and the data that vaccine developers published or submitted to FDA.
In the Investigational COVID-19 Convalescent Plasma Guidance for Industry, FDA recommended ‘neutralizing antibody titers of at least 1:160. A titer of 1:80 may be considered acceptable if an alternative matched unit is not available’[38]. During such therapy, 200 ml of convalescent plasma is transfused to a severe COVID–19 patient within 1-2 hours[39]. The actual titer of NAbs will be obviously reduced more than 10 times by dilution. Pfizer and BioNTech reported that their 10 or 30 μg BNT162b led to mean neutralizing antibody titers of 168 and 267, respectively, seven days after the boost. The levels are 1.8 and 2.8 times higher than the mean neutralizing titer of 94 for convalescent sera from 38 donors, and higher than the 1:160 recommended by FDA for convalescent plasma transfusion therapy [40,41]. Therefore, a NAb LFT using a single dilution of test sample can be designed to cover a color change range equivalent to 1:20 to 1:100 in PRNT, most likely the final consensus would be in this range. Ideally, the consent protection titer is set to be the IC50 of the chosen single dilution.
Two available references about other infectious diseases have provided support for this hypothesis. Protective level of NAb has been established through large-scale clinical trials, exemplified as IC50 of 1:10 diluted serum for Japanese Encephalitis [42] and 1:22 for Mumps [43].
2.3. Readout methods
The results from a lateral flow assay can be either qualitative (‘yes/no’, if an analyte is or isn’t present within the limits of detection), semi-quantitative (analyte present at low, medium, or high levels), or quantitative. The pregnancy test is an example of a qualitative yes/no assay, by displayed visible or non-visible test line signal intensity. Semi-quantitative assay can be achieved by comparing test line intensity visually with a series of preset color scales to obtain an approximal signal level. For quantitative diagnostics, the test line intensities are compared to a calibration standard and converted to an analyte concentration value. To accurately measure the test line intensity, the LFA result signal must be first acquired by a device, and then converted to a defined quantity that is directly proportion to the resulting signal. This quantity is further computed with that of calibrated standard to determine the precise analyte concentration results.
The quantitative lateral flow assay reading can be obtained with a standalone benchtop strip reader. A tested lateral flow cartridge is inserted into such an instrument, the test and the standard line intensities are imaged with a digital camera, and the results are sent to a computer for computation, display, and interpretation [44,45]. The calibration curve for any given assay can be encoded into the software, such that the test line signal intensity can be automatically converted to analyte concentration and presented to the user. The stand-alone bench top readers are best served for laboratories and locations where high throughput readings of tested strips are needed.
For home use of point-of-care diagnostics, the most convenient and practical quantitative readout for lateral flow test is through modern mobile phones [46–48]. The digital cameras on most smartphones, nowadays, have equipped with autofocusing and automatic exposure level control. So that proper instructed photographing of tested strip with the smartphone camera should be able to replace a separated image acquisition device as an accessory. In addition, the computing power of a modern smart phone with proper app is sufficient to perform relatively simple computations and curve fittings to resolve the analyte concentrations. Therefore, smart cell phones as an integrated single device for convenient POCT readout by end users is the direction for the foreseeable future.
Most of the lateral flow test assays to SARS-CoV-2 neutralizing antibodies adopted a competitive inhibition scheme [30,36]. Quantifying the level of inhibitions requires two or dual strips running side-by-site with one serving as a no inhibition standard with signal intensity, S0, and the next strip with a test line shows inhibited signal, ST, by neutralizing antibody inhibition. The percentage of the inhibition can be expressed as:
5.0.3. I% = (1– ST/S0)*100%
That is, if ST = 0, then I% = 100%, meaning 100% inhibition; if ST = S0, then I% = 0, refers to no inhibition. If test line is half of zero inhibition standard intensity, that is ST/S0 = 50%, resulting in 50% of inhibition. For the most application needs, this level of quantification, percentile inhibition by neutralizing antibody would be sufficient. If additional calibration standards with known neutralizing antibody concentrations are provided, by plotting titration curve and curve-fitting computation, the absolution levels of neutralizing antibodies from test sample can be determined. These two types of data acquisition and computation tasks can be realized conveniently by a mobile smartphone.
Semi-quantitative evaluation of NAb lateral flow POCT in terms of percentile inhibition needs neither any strip reader nor a smartphone. An end-user may estimate percentage of inhibition by simply matching provided color palettes with different inhibition ratios by eyes.
3. Future perspectives and expert opinion
During the COVID-19 pandemic, we hear a lot of medical terms, herd immunity, vaccine efficacy, NAb protective geometric mean titer (GMT) and basic reproduction number (Ro), etc. All of them are meaningful only for large population and are often misleading for the general public. When we hear that a vaccine is 95% protective, we assume we belong to that 95%, not the 5%. In fact, unless we test ourself, we do not know. For many existing infectious diseases, we usually don’t need to worry about which percentage we belong to, because we will be protected by herd immunity even though we are part of the 5% without protective level of NAb.
Herd immunity, also known as population immunity, is the indirect protection from an infectious disease that happens when a population is immune either through vaccination or immunity developed through previous infection[49]. The percentage of people who need to be immune in order to achieve herd immunity varies with each disease. For example, herd immunity against measles requires about 95% of a population to be vaccinated. The remaining 5% will be protected by the fact that measles will not spread among those who are vaccinated[49]. The formula for calculating the herd-immunity threshold is 1–1/Ro – meaning that the more people a patient or an asymptomatic carrier can infect, the higher the proportion of the population that needs to be immune to reach herd immunity[50]. The Ro and herd immunity threshold change from country to country and from time to time. It was estimated in August 2020 that Ro for COVID-19 is about 3 [51], the herd immunity threshold for SARS-CoV-2 was expected to be about 66% in the absence of any interventions[50]. The new UK B.1.1.7 variant is 50% more transmissible [52] that means each case would on average lead to 4.5 additional people infected. Then, 78% of the population would need to be immune to SARS-CoV-2[53]. To reach this goal using a vaccine with 95% protection rate, 82% of the population would need to be vaccinated which is extremely difficult to achieve in consideration that a vaccine for those aged under 16 years old is still not available and this age group accounts for 20% of the total population in US and >25% worldwide. In addition to higher transmission rate, some SARS-CoV-2 variants, such as the E484K carrying strains, South Africa B.1.351 variant [54] and Brazil P1 variant [55], the L452R carrying strains originated from California, B.1.427 and B.1.429 [56] showed moderate to high resistance to current COVID-19 vaccines. Using LFT, reduced neutralization to South Africa variant can be visually demonstrated using blood sample from COVID–19 vaccine recipients (Figure 4).
Figure 4.
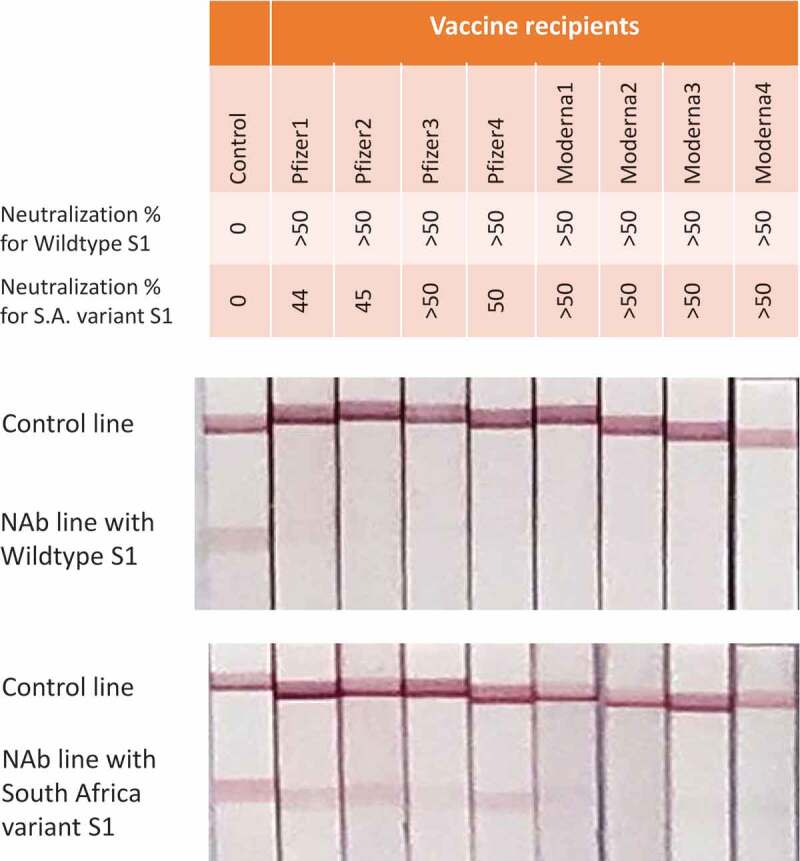
NAb measurement by LFT against wildtype and South Africa variant. Plasmas from 4 BioNTech/Pfizer vaccine recipients and 4 Moderna vaccine recipients were collected 10–20 days after second dose. These plasmas were 1:20 diluted and tested against gold nanoparticles conjugated with same amount of S1 proteins from wildtype and South Africa variant, respectively. Neutralization percentage was obtained through the same procedure as described in Figure 2 legend
Under this situation, we should not assume we would be immune after being vaccinated. We need to get this answer through a scientific way by testing whether we have developed enough NAb. It is NAb test result, not vaccination, that will provide confidence in arranging our daily work and life. For instance, the individuals who developed enough NAb against wild type SARS-CoV-2 after vaccination but not enough for South Africa variant (Figure 4), extreme caution should be taken when planning a trip to the countries where this variant prevails. We suggest that governments and healthcare systems closely monitor the effective duration of each COVID-19 vaccine and advise public in time to receive additional vaccination or other prevention when protection is approaching disappear. We also suggest an effort at government level to identify the 5% unprotected vaccine recipients and provide them with special instruction and care. Five percent in US alone means 16 million.
Funding Statement
This work was supported by R&D budget of Novodiax Inc.
Declaration of interest
J Wang, N Zhang and J Wu are co-inventors on patents or patent applications related to methods cited in this review (US 63/029,243; US 63/124,579 and CN 2,020,116,359,289), which are assigned to their current employer. J Wang, N Zhang, SA Richardson and J Wu are employees of Novodiax Inc. All authors declare no other relevant affiliations or financial conflict with the subject matter or materials discussed in the manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.7.
- 1.Abbott . Abbott’s BinaxNOW™ COVID-19 Ag Card. (2020).
- 2.Abbott . SARS-CoV-2 IgG Architech - Instructions for Use. (2020).
- 3.Walls AC, Park Y-J, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV–2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Yang C, Xu X-F, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCallum M, Marco AD, Lempp F, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv. 2021. 2021.2001.2014.426475. DOI: 10.1101/2021.01.14.426475. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This is the first publication reporting a supersite in N-terminal domain of spike protein and indicating its importance in protective immunity. This supersite can be recognized by a group of neutralizing antibodies.
- 7.Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584(7819):115–119. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584(7821):450–456. [DOI] [PubMed] [Google Scholar]
- 9.Piccoli L, Park Y-J, Tortorici MA, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high–resolution serology. Cell. 2020;183(4):1024–1042.e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Although 77% hospitalized COVID-19 patients developed NAb, only 18% and 11% of non-hospitalized symptomatic and asymptomatic individuals had detectable NAb.
- 10.Barnes CO, Jette CA, Abernathy ME, et al. Structural classification of neutralizing antibodies against the SARS-CoV-2 spike receptor-binding domain suggests vaccine and therapeutic strategies. bioRxiv:Preprint Serv Biol. 2020. 2020.2008.2030.273920. DOI: 10.1101/2020.08.30.273920. [DOI] [Google Scholar]
- 11.Barnes CO, Jette CA, Abernathy ME, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This article analyzed and defined antigenic structure of spike protein at molecular level and meanwhile provided a common communication language to scientific community.
- 12.Johnson J. Johnson & Johnson Announces Single-Shot Janssen COVID-19 Vaccine Candidate. (2021).
- 13.Thomas SJ, Endy TP, Kalayanarooj S, et al. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: how alterations in assay conditions impact performance. Am J Trop Med Hyg. 2009;81(825–833):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratnam S, Gadag V, West R, et al. Comparison of commercial enzyme immunoassay kits with plaque reduction neutralization test for detection of measles virus antibody. J Clin Microbiol. 1995;33(811–815):811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baer A, Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J Vis Exp. 2014;93:e52065–e52065. DOI: 10.3791/52065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muruato AE, Fontes-Garfias CR, Ren P, et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. bioRxiv:Preprint Serv Biol. 2020. 2020.2005.2021.109546. DOI: 10.1101/2020.05.21.109546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suthar MS, Zimmerman MG, Kauffman RC, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med. 2020;1100040. 10.1016/j.xcrm.2020.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong H, Wu Y, Cao J, et al. Robust neutralization assay based on SARS-CoV-2 S-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressed BHK21 cells. (2020). [DOI] [PMC free article] [PubMed]
- 19.Zettl F, Meister TL, Vollmer T, et al. Rapid quantification of SARS-CoV-2-neutralizing antibodies using propagation-defective vesicular stomatitis virus pseudotypes. Vaccines (Basel). 2020;8(3). DOI: 10.3390/vaccines8030386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt F, Weisblum Y, Muecksch F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217(11). DOI: 10.1084/jem.20201181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford KHD, Eguia R, Dingens AS, et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X, Muruato AE, Zhang X, et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. bioRxiv. 2020. DOI: 10.1101/2020.06.22.165712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sprangers MC, Lakhai W, Koudstaal W, et al. Quantifying adenovirus-neutralizing antibodies by luciferase transgene detection: addressing preexisting immunity to vaccine and gene therapy vectors. J Clin Microbiol. 2003;41(11):5046–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee S, Dowd KA, Manhart CJ, et al. Mechanism and significance of cell type-dependent neutralization of flaviviruses. J Virol. 2014;88(13):7210–7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun C, Tilston-Lunel NL, Nambulli S, et al. SARS-CoV-2 and SARS-CoV spike-RBD structure and receptor binding comparison and potential implications on neutralizing antibody and vaccine development. bioRxiv. 2020. 2020.2002.2016.951723. DOI: 10.1101/2020.02.16.951723. [DOI] [Google Scholar]
- 28.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan CW, Chia WN, Qin X, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. [DOI] [PubMed] [Google Scholar]
- 30.Zhang N, Chen S, Wu JV, et al. A lateral flow test detecting SARS-CoV-2 neutralizing antibodies. medRxiv. 2020. 2020.2011.2005.20222596. DOI: 10.1101/2020.11.05.20222596 [DOI] [Google Scholar]; •• First publication regarding a rapid lateral flow test detecting SARS-CoV-2 neutralizing antibody.
- 31.GenScript . cPass™ SARS-CoV-2 Neutralization Antibody Detection Kit. 2020. Available from: https://www.fda.gov/media/143583/download
- 32.CusaBio . SARS-CoV-2 Neutralizing Antibody ELISA Kit 2021. Available from: https://www.cusabio.com/ELISA-Kit/Human-Novel-Coronavirus–SARS-CoV-2—Neutralizing-Antibody-ELISA-Kit-12928647.html
- 33.Abeomics . SARS-CoV-2 neutralizing antibodies detection kit 2021. Available from: https://www.abeomics.com/sars-cov-2-neutralizing-antibodies-detection-kit
- 34.ACRO . Anti-SARS-CoV-2 neutralizing antibody titer serologic assay kit. 2021. Available from: https://www.acrobiosystems.com/P3249-Anti-SARS-CoV-2-neutralizing-antibody-titer-serologic-assay-kit.html
- 35.Cayman . SARS-CoV-2 Neutralizing Antibody Detection ELISA Kit. 2021.
- 36.Lake DF, Roeder AJ, Kaleta E, et al. Development of a rapid point-of-care test that measures neutralizing antibodies to SARS-CoV-2. medRxiv. 2020. 2020.2012.2015.20248264. DOI: 10.1101/2020.12.15.20248264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rey EG, Finkelstein JL, Erickson D. Fluorescence lateral flow competitive protein binding assay for the assessment of serum folate concentrations. PLoS One. 2019;14(6):e0217403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.FDA . Investigational COVID-19 convalescent plasma guidance for industry. 2020
- 39.FDA . EMERGENCY USE AUTHORIZATION (EUA) OF COVID-19 CONVALESCENT PLASMA FOR TREATMENT OF COVID-19 IN HOSPITALIZED PATIENTS. 2020.
- 40.BioCentury . BioNTech, Pfizer vaccine yields highest titers to dateBioNTech, Pfizer vaccine yields highest titers to date. (2020).
- 41.Sahin U, Frenck R, Falsey AR, et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv. 2020. 2020.2012.2009.20245175. DOI: 10.1101/2020.12.09.20245175. [DOI] [Google Scholar]
- 42.Kurane I. In: Barrett ADT, Stanberry LR, eds. Vaccines for biodefense and emerging and neglected diseases. Academic Press, London; 2009. p. 527–535. [Google Scholar]
- 43.Rubin SA, Plotkin SA. Vaccines (Sixth Edition). eds, Plotkin SA, Orenstein WA, Offit PA. 419–446. W.B. Saunders, London; 2013 [Google Scholar]
- 44.He Y, Zhang X, Zhang S, et al. Visual detection of single-base mismatches in DNA using hairpin oligonucleotide with double-target DNA binding sequences and gold nanoparticles. Biosens Bioelectron. 2012;34(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Mao X, Zeng Q, et al. Aptamer-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for protein analysis. Anal Chem. 2009;81(2):669–675. [DOI] [PubMed] [Google Scholar]
- 46.Hosseini S, Vázquez-Villegas P, Martínez-Chapa SO. Paper and fiber-based bio-diagnostic platforms: current challenges and future needs. Appl Sci. 2017;7(8):863. [Google Scholar]
- 47.Ahmed S, Bui M-PN, Abbas A. Paper-based chemical and biological sensors: engineering aspects. Biosens Bioelectron. 2016;77(249):249–263. [DOI] [PubMed] [Google Scholar]
- 48.Hayes B, Murphy C, Crawley A, et al. Developments in point-of-care diagnostic technology for cancer detection. Diagnostics (Basel). 2018;8(2). DOI: 10.3390/diagnostics8020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.WHO . Coronavirus disease (COVID-19). In: Herd immunity, lockdowns and COVID-19. 2020. Available from: https://www.who.int/news-room/q-a-detail/herd-immunity-lockdowns-and-covid-19 [Google Scholar]
- 50.Omer SB, Yildirim I, Forman HP, et al. Implications for SARS-CoV-2 control. JAMA. 2020;324(2095–2096):2095. [DOI] [PubMed] [Google Scholar]; • A good article about COVID-19 herd immunity.
- 51.Royal Society . Reproduction number (R) and growth rate (r) of the COVID-19 epidemic in the UK. 2020. Available from: https://royalsociety.org/-/media/policy/projects/set-c/set-covid-19-R-estimates.pdf .
- 52.Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. medRxiv. 2021. 2020.2012.2024.20248822. DOI: 10.1101/2020.12.24.20248822. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• An important publication about SARS-CoV-2 UK variant B.1.1.7.
- 53.Coronavirus Mutations SHAW,J Threaten to Worsen Pandemic. Harvard Magazine (2021).
- 54.Wang P, Liu L, Iketani S, et al. Increased resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021. 2021.2001.2025.428137. DOI: 10.1101/2021.01.25.428137. [DOI] [PubMed] [Google Scholar]; •• An important publication about SARS-CoV-2 South Africa variant B.1.135.
- 55.Wang P, Wang M, Yu J, et al. Increased resistance of SARS-CoV-2 Variant P.1 to antibody neutralization. bioRxiv. 2021. 2021.2003.2001.433466. DOI: 10.1101/2021.03.01.433466. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• An important publication about SARS-CoV-2 Brazil variant P.1.
- 56.Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021. 2021.2003.2007.21252647. DOI: 10.1101/2021.03.07.21252647. [DOI] [Google Scholar]; •• An important publication about SARS-CoV-2 California variants carrying L452R mutation.


